Open Access Article
Open Access ArticleIntracellular behavior of nanodiamonds functionalized with a zwitterionic shielding moiety†
Alina
Sigaeva‡
 a,
Viktor
Merz‡
b,
Rokshana
Sharmin
a,
Romana
Schirhagl
a,
Viktor
Merz‡
b,
Rokshana
Sharmin
a,
Romana
Schirhagl
 *a and
Anke
Krueger
*bc
*a and
Anke
Krueger
*bc
aDepartment of Biomedical Engineering, University of Groningen, Groningen, The Netherlands. E-mail: romana.schirhagl@gmail.com
bInstitute for Organic Chemistry, Julius-Maximilians University Würzburg, Würzburg, 97074, Germany. E-mail: anke.krueger@oc.uni-stuttgart.de
cInstitute of Organic Chemistry, University of Stuttgart, 70569 Stuttgart, Germany
First published on 28th April 2023
Abstract
Fluorescent nanodiamonds are versatile tools, which can be employed as perfectly photostable labels as well as for intracellular sensing of a wide array of properties and chemical products due to their fluorescent nitrogen vacancy centres. Since these nanoparticles only probe their environment within a few nanometres, it is essential to deliver them inside the cells and control their intracellular location, which is often achieved by surface functionalization. Another important aspect is the interaction of nanoparticles with the highly complex extra- and intracellular environment. In this paper, we report on the combined functionalization of fluorescent nanodiamond with moieties for the control of surface interactions, and the azide groups, allowing for subsequent covalent attachment of additional functional groups (e.g., antibodies) via click chemistry. Surface functionalized nanodiamonds (fNDs) were delivered into cells using two approaches: incubation with cells and transfection-inspired, polycationic lipid-assisted internalization. The effect of different surface chemistry and different uptake protocols on the cell viability, internalization timing and efficiency, the nanoparticle aggregation inside cells and the intracellular distribution of the fNDs was studied. We showed that both the surface functionalization and the internalization approach had dramatic effects on the fate and behaviour of fluorescent nanodiamonds inside the cells. Our results give an insight into the complex processes underlying the uptake and trafficking of nanodiamonds by the cells, as well as emphasize the need for thorough analysis of the interactions between different functional groups on the nanodiamond surface in cell-based assays. This work lays the foundation for the synthesis of fND derivatives with tailored, multifunctional surfaces.
Introduction
In the field of nanosensing, fluorescent nanodiamonds (fNDs) have attracted attention owing to their outstanding fluorescent and magnetosensitive properties.1 The internal nitrogen-vacancy (NV) centre is sensitive to changes in the physical parameters of its environment. Quantities that have been measured include strain,2 temperature,3 and magnetic or electric fields.4,5 In addition, the fluorescence of the NV centre does not bleach or blink, making it ideal for long term labelling and imaging.6 Diamond particles themselves are biocompatible and non-genotoxic.7 Ultimately these properties make fNDs well suitable for the intracellular sensing of chemical products or physical parameters,8,9 as has been demonstrated in several recent studies.10–12 Pristine fNDs typically enter cells via the endosomal pathway and in some cases it has been shown that they eventually escape the endosomes.13,14To use fNDs for the above-mentioned applications, one has to ensure that the diamond probe is close to the molecule or cellular structure of interest. For biological applications, this often requires internalization of diamond particles and their subsequent targeting to specific regions inside the cell. Over the years, different approaches have been used to achieve this goal.9,15 They can be broadly divided into techniques based on the physical properties of intracellular environment in various compartments (e.g., the negative charge of DNA16 or the combination of negative charge and high lipid content of mitochondria17), and biologically inspired approaches. The latter group includes functionalization of the diamond surface with the biological molecules that are and naturally targeted to reach specific structures and compartments of a live cell – antibodies,12,18–21 viral components,22 or intracellular localization sequences.18
Another important aspect is the stable and controlled interaction of the functionalized nanodiamond conjugates with cell fluids and culture media. Especially the formation of a protein corona is a dominating effect that directs the actual properties of the particles.17,23–28 Strategies to gain control over the surface interactions include the coating with a defined protein,9,29–31 the functionalization with e.g. branched polyglycerols32–34 or multilayer polymeric shells.35 Recently, we have reported on the grafting of short, molecular structures with terminal zwitterions that prevent the non-specific adsorption of serum proteins on the surface of functionalized nanodiamonds and ensured the colloidal stability of these particles in buffer solutions containing serum proteins.36
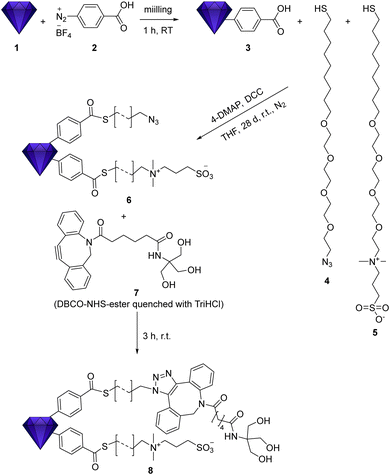
Here, we report on the combined functionalization of fluorescent nanodiamond with moieties for the control of surface interactions, and the azide groups, allowing for subsequent covalent attachment of additional functional groups (e.g., antibodies, other proteins/peptides, drugs, labels etc.) via click chemistry. The zwitterionic moieties in combination with tetraethylene glycol (TEG) improve the colloidal dispersion in physiological media with strong ion background. As a consequence, they prevent non-specific interactions with proteins, as some of us have recently reported for a different type of nanodiamond.36 The terminal azide group on the linker architecture of the pre-functionalized nanodiamond (Scheme 1) can be used for covalent attachment of functional moieties by the click chemistry with DBCO. To ensure the accessibility of the clickable groups, the azide unit is presented on a linker architecture with the same length and polarity as the zwitterions.
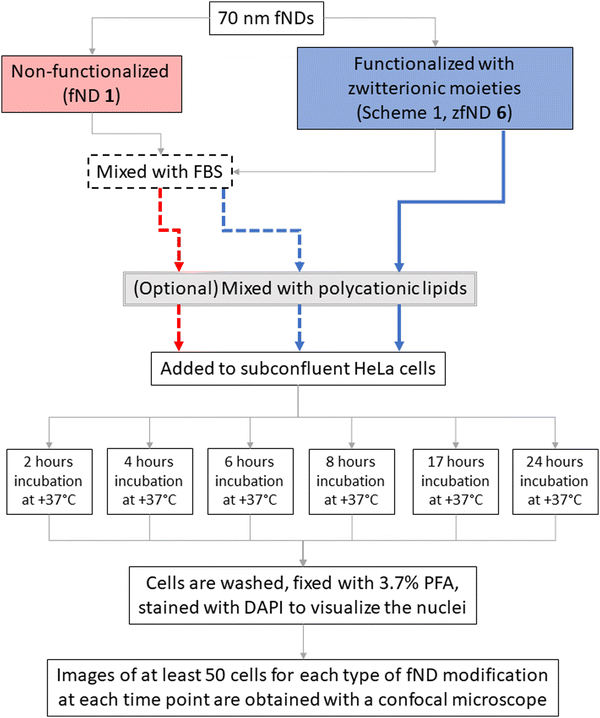
We investigated the uptake of zwitterion-functionalized fNDs in HeLa cells, as compared with non-functionalized fNDs, using two approaches. In the first case, fNDs were added to the cell culture medium and incubated with the cells to allow for endocytosis of the nanoparticles. In the second case, we used the protocol for chemical transfection, in which fNDs are pre-mixed with polycationic lipids and then incubated with the cells afterwards. For both protocols, we examined efficiency and timing of fND internalization, distribution of internalized fNDs in relation to the cell nucleus, the effects on metabolic activity and levels of reactive oxidative species in the cells. A summary of the experiments with cells is shown in Scheme 2.
Results and discussion
fND functionalization and characterization
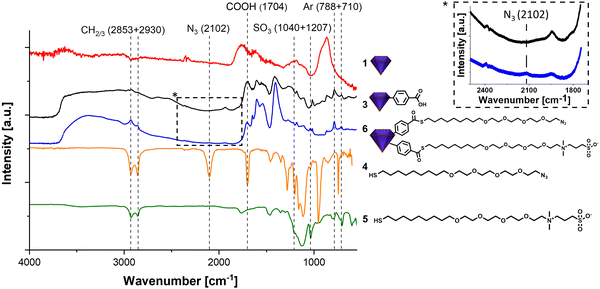
fNDs were functionalized using an adapted procedure reported in our previous work.36 However, in that work the functionalization was carried out on detonation nanodiamond (DND). These contain far less NV centers and are thus not suitable for imaging applications. As HPHT fND are less reactive, the reaction procedures had to be adapted. Harsher conditions were required leading to the replacement of ultrasonic treatment by mechanochemical reaction37 of the HPHT nanodiamond 1 and diazonium compound 2 using a mini mill at 50 Hz with steel beads. The successful functionalization is shown in the FT-IR spectra in Fig. 1 by strong signals of the surface groups, e.g. the aromatic bands at 788 and 710 cm−1. The subsequent functionalization with the two chains 4 and 5 is simultaneously performed leading to a stoichiometric distribution of the two moieties when assuming comparable reactivity. The IR spectrum shows changes at 2102 cm−1 due to the azide band and the sulfonic acid at 1040 and 1207 cm−1. The surface loading for product fND 6 obtained from the TGA data (see ESI†), shows that about 50% of the acid moieties were converted to thioesters, which also explains the remaining signal for the carboxylic groups.The functionalization of the fND with the chains containing TEG not only increases the colloidal stability but also the dispersibility. Drying the functionalized samples for the recording of IR spectra and subsequent redispersion was possible and led to again fully deagglomerated nanoparticles. This is of great importance for the generation of a homogeneous material with a fully controlled surface with as many primary particles as possible for subsequent reactions. Functionalization with long chains, as already shown in our recently published work,36 lead to a significant increase of the hydrodynamic diameter of the nanoparticles measured by DLS. In contrast, the particles show their actual, only slightly increased diameter in AFM measurements.36 Here, the successful functionalization did not lead to increased particle sizes and no agglomeration was observed. For fNDs 1 to 6 the hydrodynamic diameter (D(50) = 287(1) → 266(6) nm) remained largely the same. In addition, the mechanochemical functionalization to ND 3 using a mill (see ESI† ND 3) not only made the reaction possible, but also achieved a small particle size (D(50) = 287(1) →184(3) nm). The typically observed increasing particle size in the DLS (but not in the AFM) during the functionalization of detonation NDs was not observed here when using the same functionalization of fNDs. The reason is that the aggregate size was continuously controlled by in situ milling, which enables the continuous generation of accessible surface and overcome the agglomeration by passivating the surface groups by attachment of the linker molecules. To achieve stabilization in physiological media, the NDs were functionalized with a zwitterion chain. The zwitterionic betaine derivative provides effective protection against ions on the surface thanks to a neutrally charged surface, which not only stabilizes the ND physiologically but also leads to protein repulsion. The entire zwitterionic chain system follows the so called “Whitesides rules” and is therefore suitable for forming a protective layer against a protein corona.38 Without this protective layer, a protein corona or other molecules would attach onto the surface of the particle and disrupt the functionality of additional targeting moieties (e.g., antibodies). Thus, the additional zwitterion chains, in addition to the azide chains, are important for targeting.36
fND uptake and biocompatibility after incubation
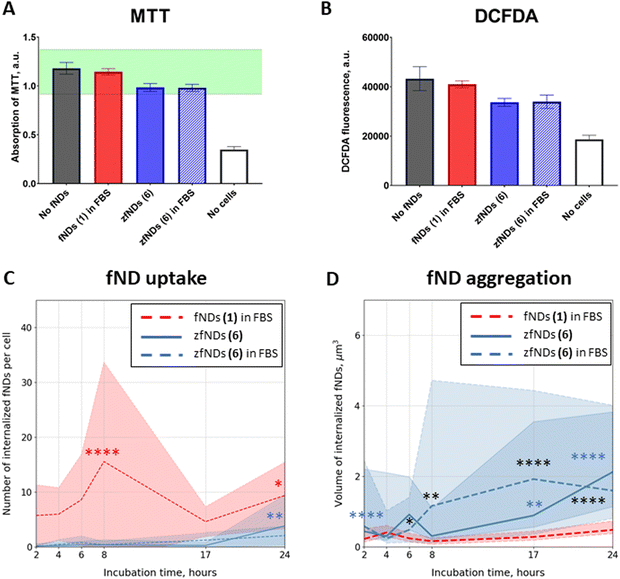
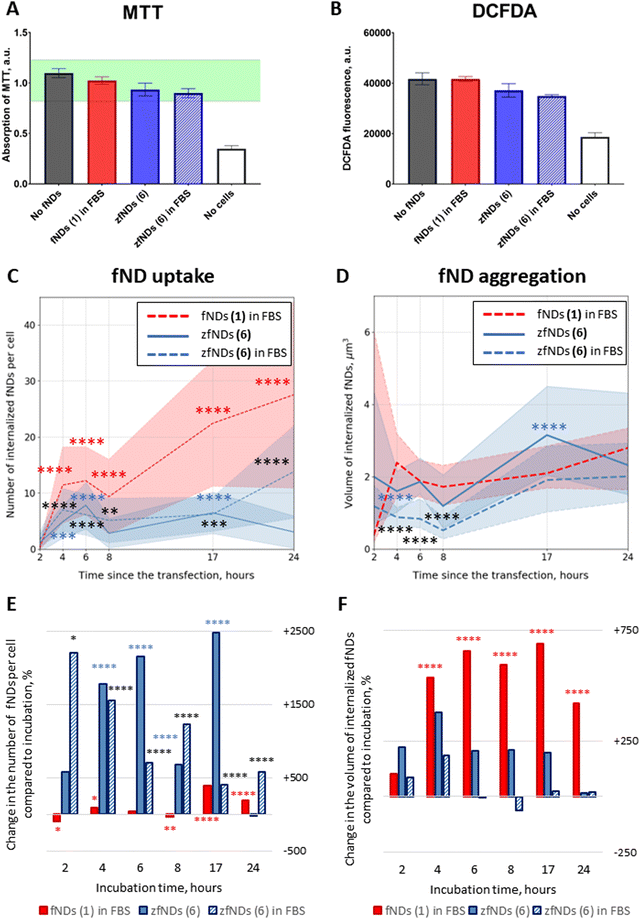
fNDs added to the cell culture medium were successfully internalized by HeLa cells. The particles could be observed in the cytoplasm already after 2 hours of incubation (Fig. S15–S17, ESI†). We did not observe significant morphological changes or pronounced cell death in any of the groups even after 24 hours of incubation. The metabolic activity of the cells was slightly decreased after the 24 hour exposure to zfNDs (6), as compared to non-functionalized fNDs (1) (Fig. 2A). Nevertheless, the metabolic activity remains within the normal physiological range (80–120%). Moreover, the DCFDA assay (Fig. 2B) does not show an increase in the intracellular production of reactive oxygen species in presence of zfNDs (6). These results indicate that zfNDs (6) are generally not toxic to HeLa cells.The uptake rates were significantly lower for 6 (zfNDs) than for non-functionalized fNDs (1) at all timepoints (Fig. 2C), even though the density of additional surface moieties is likely not high enough to completely cover the fND surface. The zwitterionic moieties might impede the initial electrostatic interaction of the particles with the cell surface, which is necessary for the uptake.
As expected, the number of internalized fNDs generally increased with longer incubation. For non-functionalized fNDs (1), the number of particles per cell significantly increased from 2 to 8 hours of incubation. For 6 (zfNDs), significant increase in the particle counts could only be seen after 24 hours of incubation. We observed a substantial difference in the size of internalized fNDs (Fig. 2D). Generally the volume of zfNDs (6) was higher than that of non-functionalized fNDs (1) (Fig. 4A). Therefore, the total amount of fND material introduced into the cell might not differ in case of non-functionalized fNDs (1) and zFNDs (6).
fND uptake and biocompatibility after transfection
The low internalization efficiency of 6 (zfNDs) could be mitigated by using a modified experimental procedure, inspired by the cell transfection protocols (Fig. S18–S20, ESI†). In this case, the material that needs to be delivered inside the cell is pre-mixed with polycationic lipids, which form a complex around the particles. Such lipoplexes are easier internalized by the cells, leading to improved uptake rates. They destabilize the endosomal membranes after the uptake, with the cargo being delivered to the cytoplasm.39 Just like incubation with fNDs, transfection does not affect cell viability or metabolic activity (Fig. 3A and B). As expected, using the transfection protocol significantly increased the number of internalized 6 (zfNDs) (Fig. 3C). FBS-free 6 (zfNDs) showed higher internalization rates at 4–17 hours (p < 0.0001). The cellular uptake of 6 (zfNDs) premixed with FBS was higher at all timepoints. At the same time, the uptake efficiency of non-functionalized fNDs (1) in FBS increased less dramatically (Fig. 3E and Fig. S18, ESI†). At shorter incubation times (2–8 hours), the number of internalized fNDs was similar to that for the non-transfected group. Pronounced increase in uptake could be seen only at longer incubation times (17–24 hours, p < 0.0001).Effects of protein corona on fND uptake and aggregation
As previously described in the literature,27 pre-mixing non-functionalized fNDs (1) with FBS generally prevents their aggregation in cell culture medium. This happens due to the serum proteins, such as bovine serum albumin, forming a corona on the fND surface. We investigated whether FBS has any effects in case of zfNDs (6).Pre-coating zfNDs (6) with FBS had no effect on HeLa cell viability or oxidative stress levels, both in case of incubation (Fig. 2A and B) and transfection (Fig. 3A and B). There was also no significant difference in the cellular uptake between zfNDs (6) added directly to the medium and pre-dissolved in FBS (Fig. 2C and 3C), regardless of the protocol used for fND internalization. Additionally, we observed certain aggregation of zfNDs (Fig. 2D and 3D), whether zfNDs were pre-dissolved in FBS or not, under both incubation and transfection.
This is in stark contrast with non-functionalized fNDs in FBS (1) that did not aggregate, if they were added directly to the cell culture medium for incubation (Fig. 2D), but formed large aggregates upon interaction with polycationic lipids necessary for transfection (Fig. 3D and F). During the transfection protocol, the volume of non-functionalized fND (1) aggregates was similar or higher than that of zfNDs (6), except for the 17-hour timepoint. The aggregation of non-functionalized fNDs (1) has increased approximately 6-fold within the first 4 hours, whereas zfNDs (6) did not exhibit further aggregation at a statistically significant level (Fig. 3F). Additional aggregation might result from the polycationic lipids displacing the protein corona from the surface of non-functionalized fNDs (1). The zwitterionic moieties of zfNDs might prevent the formation of the protein corona on the fND surface in the first place, even in such a protein-rich environment like pure serum.
Intracellular transport of fNDs and zfNDs
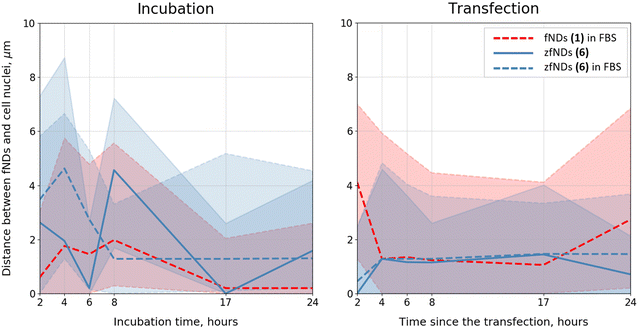
Both non-functionalized fNDs (1) and zfNDs (6) tended to be in the close vicinity (<5 μm) of the cell nucleus (Fig. 4). zfNDs (6) were located a bit further away than non-functionalized particles 1. In case of transfection, zfNDs (6) approach the nucleus very quickly, within the first 2 hours, and stay in the perinuclear region throughout the experiment. fNDs (1) in FBS exhibit similar behaviour, but the range of the distances between the particles and the nuclei is wider and varies more over time (in particular, at the very early and the very late timepoints). This inherent ability for nuclear targeting might prove useful, when designing zfND-based nuclear probes. It is worth noting that the transport of fNDs inside the cell (and towards the nucleus) occurs via endosomal pathway. The differences in the subcellular localization of non-functionalized fNDs (1) and zfNDs (6) might, therefore, stem from the differences in the particles’ ability for endosomal escape. Our data would suggest that zfNDs (6) are retained in the endosomes for longer periods of time, being eventually transported towards the nucleus. If this is the case, it might be necessary to employ additional strategies to enable the endosomal escape of zfNDs (6) at appropriate cell locations. However, additional studies on the intracellular transport and distribution of modified fNDs would be necessary to answer these questions.Overall, the functionalized fNDs 6 exhibit excellent behaviour during the interactions with cell culture medium and live cells. The above-mentioned properties of zwitterions explain why the NDs functionalized with these moieties do not agglomerate even in the presence of a strong ionic background and do not form strong protein coronas. The zwitterionic coating is beneficial during the internalization of fNDs mediated by polycationic lipids: it both improves the uptake rates and prevents additional aggregation of the nanoparticles in the cell culture medium and inside the cytoplasm of the live cells. zfNDs (6) do not exhibit substantial cell toxicity and do not induce an oxidative stress response. These properties, together with an easy and straightforward way to introduce additional functional groups on the zfND (6) surface (via click chemistry), might be useful for the design of functionalized fNDs for in vivo applications.
Experimental section
Methods
fNDs (ND-NV-70) were purchased from Adámas Nanotechnologies (North Carolina, the United States of America) as aqueous suspension with a concentration of 1 mg mL−1. Chemicals were purchased from Thermo Fisher Scientifics (Acros Organics, Fluka), Merck (Sigma Aldrich) and Grüssing and used without further purification unless noted. The DBCO-NHS-ester was purchased from Lumiprobe (batch info: 7D58H(1)). All solvents were freshly distilled and dried using standard procedures. For the purification of the fND, bidistilled water (DD-water, 18.2 MΩ cm) at a pH of 5.5 and T = 20 °C) was used. Nuclear magnetic resonance spectra were measured with a Bruker AVANCE 400 FT-NMR spectrometer at 27 °C. The chemical shift of the deuterated solvents was used as internal reference for the calibration of spectra. Electrospray ionization (ESI) mass spectrometry was performed using a Bruker Daltonics micOTOF focus. FT-IR spectroscopy was carried out in a Jasco FT/IR-430 using ∼1 mg ND. ATR spectra were recorded using the same spectrometer equipped with an ATR unit MIRacleTM from PIKE Technologies. Diffuse reflectance IR spectra (DRIFT) of ND samples were additionally measured using a DRIFT spectrometer Nicolet iS5. Thermogravimetric analysis (TGA) was performed on a PerkinElmer STA 6000 with a nitrogen flow rate of 60 mL min−1 and a heating rate of 10 K min−1 from 50 °C to 130 °C. We used a plateau time of 60 min at 130 °C and a heating phase from 130 °C to 700 °C at 5 K min−1. The particle size and zeta potential were measured in bidistilled water at the intrinsic pH of the sample colloid using a Malvern Zetasizer Nanoseries Nano-ZS (dynamic light scattering, backscattering mode). The size distribution is given as volume distribution (Dv(10), Dv(50) and Dv(90)) and was obtained using the Marquardt method. The reaction with HPHT ND and diazonium compounds was carried out in a mini mill Pulverisette 23 (Fritsch) for 1 h at 50 Hz vibration mode with 10 pieces of 2 mm stainless steel beads. The sonication for washing of the samples was carried out in a Bandelin Sonorex Digitec Typ DT52 (max. 80 W, 35 kHz). Centrifugation was performed in a Hettich Mikro 220 R at 15 °C and a maximum of 31 514 RCF (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM) using 2 mL Eppendorf safelock-cap tubes PP.
000 RPM) using 2 mL Eppendorf safelock-cap tubes PP.
Synthesis of organic linkers and the prefunctionalized nanodiamond are shown together with the respective spectroscopic data in the ESI.†
HeLa cells were cultured under standard culturing conditions until the moment of the experiment in DMEM complete medium. This is Dulbecco's Modified Eagle Medium with 4500 mg L−1 glucose supplemented with 10% Fetal Bovine Serum (FBS), 1% penicillin/streptomycin and 1% Glutamax (Gibco, ThermoFisher Scientific, Etten–Leur, The Netherlands) at 37 °C, 5% CO2. Cells were grown in glass bottom CELLview™ cell culture dishes (Greiner Bio-One B. V., Alphen aan den Rijn, the Netherlands) until approximately 60–90% confluency on the day of the experiment.
On the day of the experiment, the cells were exposed to various fND solutions in the cell culture medium, as summarized in Scheme 2. The final concentration of fND in all cases was 1 μg mL−1.
In case of non-functionalized fNDs (1), the stock fND solution was mixed with FBS, and, after 2 minutes, serum-free DMEM-HG was added to reach the desired volume and concentration of fNDs (1 μg mL−1) and FBS (10% of the total volume of the solution). In case of zfNDs (6), the stock solution was either mixed with FBS or diluted directly in complete DMEM-HG.
For the transfection, the particles (non-functionalized fNDs (1) in FBS, zfNDs (6), or zfNDs in FBS) were mixed with the polycationic lipids (transfection reagent sc-29528, Santa Cruz), according to the manufacturer's protocol. Briefly, the stock solution of polycationic lipids was diluted in transfection medium (sc-36868, Santa Cruz). The particles were also diluted in transfection medium in a separate tube. The concentration of polycationic lipids was 3 times higher than that of the nanoparticles. The two solutions were then gently mixed together and left for 15 minutes at room temperature to allow for the formation of fND-lipid complexes. Afterwards, the transfection medium was added to reach the desired concentration of fNDs, and the cell culture medium was replaced with the fND solution. As the transfection medium had a reduced concentration of serum, the cells were supplemented with DMEM-HG with double serum concentration after 4 hours of incubation. This prevents cell cycle arrest and cell death, if longer incubation is required.
After the respective incubation time (2, 4, 6, 8, 17 or 24 hours at +37 °C, 5% CO2), the cells were rinsed with phosphate buffered saline (PBS, pH = 7.4) and fixed in 3.7% paraformaldehyde (PFA) for 15 minutes. Fixed cells were washed with PBS and their nuclei were stained with DAPI. The samples were stored in 1% PFA until the microscopic imaging and analysis.
All samples were imaged with LSM780 laser scanning confocal microscope (Zeiss, Sliedrecht, the Netherlands), using a 405 nm laser for a DIC image and DAPI and a 561 nm laser to excite the fluorescence of the fNDs, which was detected in the red and far-red region (653–758 nm). As compared to the more commonly used 600 nm long-pass filters, this detection window removes some of the fND fluorescence and most of the noise – e.g., the autofluorescence of intracellular structures, especially the endosomes filled with the fluorescent components coming from the standard cell culture medium. For each sample, we have collected three-dimensional z-stacks of images of at least 50 cells.
All confocal images were processed in FIJI 2.0.0 (https://fiji.sc), using “Diffraction PSF 3D” and “Iterative Deconvolve 3D” plugins.40 fNDs were then detected with the “3D Objects Counter” plugin, which allowed us to obtain information on the number and the detected volumes of the internalized particles. All fNDs that were not internalized were excluded from the analysis. Finally, we examined the distribution of fNDs inside each individual cell in relation to the nucleus. The distances between each individual particle and the closest cell nucleus were measured using the 3D Euclidean Distance Maps (EDM) method.41 The 3D EDM approach allows assessing the degree of colocalization of different objects regardless of their relative brightness. This technique works on a single-object basis, with each particle being measured individually. It thus provides more detailed information on the spatial distribution of particles inside the cell, and can be used even in case of low particle counts.
GraphPad Prism 6 was used for statistical analysis of the results. The plots were produced with Python 3.8.
Author contributions
V. M. conducted the chemical experiments under the supervision of A. K.; A. S. conducted the biological experiments with the help of R. Sh under the supervision of R. Sc. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
R.Sc. is thankful for support via the ERC starting grant ERC-2016-StG-714289. A. K. and V. M. thank the Volkswagen Foundation for financial support under grant 88393.References
- A. Mzyk, A. Sigaeva and R. Schirhagl, Acc. Chem. Res., 2022, 1818–1825 Search PubMed.
- T. Müller, I. Aharonovich, L. Lombez, Y. Alaverdyan, A. N. Vamivakas, S. Castelletto, F. Jelezko, J. Wrachtrup, S. Prawer and M. Atatüre, New J. Phys., 2011, 13, 075001 CrossRef.
- M. W. Doherty, V. M. Acosta, A. Jarmola, M. S. J. Barson, N. B. Manson, D. Budker and L. C. L. Hollenberg, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 90, 041201 CrossRef CAS.
- S. Hong, M. S. Grinolds, L. M. Pham, D. le Sage, L. Luan, R. L. Walsworth and A. Yacoby, MRS Bull., 2013, 38, 155–161 CrossRef CAS.
- J. R. Maze, P. L. Stanwix, J. S. Hodges, S. Hong, J. M. Taylor, P. Cappellaro, L. Jiang, M. V. G. Dutt, E. Togan, A. S. Zibrov, A. Yacoby, R. L. Walsworth and M. D. Lukin, Nature, 2008, 455, 644–647 CrossRef CAS PubMed.
- S. Haziza, N. Mohan, Y. Loe-Mie, A. M. Lepagnol-Bestel, S. Massou, M. P. Adam, X. L. Le, J. Viard, C. Plancon, R. Daudin, P. Koebel, E. Dorard, C. Rose, F. J. Hsieh, C. C. Wu, B. Potier, Y. Herault, C. Sala, A. Corvin, B. Allinquant, H. C. Chang, F. Treussart and M. Simonneau, Nat. Nanotechnol., 2017, 12, 322–328 CrossRef CAS.
- K. van der Laan, M. Hasani, T. Zheng and R. Schirhagl, Small, 2018, 14, 1703838 CrossRef.
- T. Zhang, G. Pramanik, K. Zhang, M. Gulka, L. Wang, J. Jing, F. Xu, Z. Li, Q. Wei, P. Cigler and Z. Chu, ACS Sens., 2021, 6, 2077–2107 CrossRef CAS PubMed.
- Y. Wu and T. Weil, Adv. Sci., 2022, 9 Search PubMed.
- A. Morita, A. C. Nusantara, F. P. P. Martinez, T. Hamoh, V. G. Damle, K. J. van der Laan, A. Sigaeva, T. Vedelaar, M. Chang, M. Chipaux and R. Schirhagl, Nano Today, 2023, 48, 101704 CrossRef CAS.
- A. Sigaeva, H. Shirzad, F. P. Martinez, A. C. Nusantara, N. Mougios, M. Chipaux and R. Schirhagl, Small, 2022, 18, 2105750 CrossRef CAS PubMed.
- L. Nie, A. C. Nusantara, V. G. Damle, R. Sharmin, E. P. P. Evans, S. R. Hemelaar, K. J. van der Laan, R. Li, F. P. Perona Martinez, T. Vedelaar, M. Chipaux and R. Schirhagl, Sci. Adv., 2021, 7, 573 Search PubMed.
- Z. Chu, S. Zhang, B. Zhang, C. Zhang, C. Y. Fang, I. Rehor, P. Cigler, H. C. Chang, G. Lin, R. Liu and Q. Li, Sci. Rep., 2014, 4, 1–9 Search PubMed.
- Y. Zhang, R. Sharmin, A. Sigaeva, C. W. M. Klijn, A. Mzyk and R. Schirhagl, Nanoscale, 2021, 13, 13294–13300 RSC.
- D. Terada, T. Genjo, T. F. Segawa, R. Igarashi and M. Shirakawa, Biochim. Biophys. Acta, Gen. Subj., 2020, 1864, 129354 CrossRef CAS.
- K. Bray, L. Cheung, K. R. Hossain, I. Aharonovich, S. M. Valenzuela and O. Shimoni, J. Mater. Chem. B, 2018, 6, 3078–3084 RSC.
- Y. Zou, M. Nishikawa, H. G. Kang, G. Cheng, W. Wang, Y. Wang and N. Komatsu, Mol. Pharmacol., 2021, 18, 2823–2832 CrossRef CAS.
- R. Sharmin, A. C. Nusantara, L. Nie, K. Wu, A. Elias Llumbet, W. Woudstra, A. Mzyk and R. Schirhagl, ACS Sens., 2022, 7, 3326 CrossRef CAS.
- M. Mkandawire, A. Pohl, T. Gubarevich, V. Lapina, D. Appelhans, G. Rödel, W. Pompe, J. Schreiber and J. Opitz, J. Biophotonics, 2009, 2, 596–606 CrossRef CAS PubMed.
- A. Morita, T. Hamoh, A. Sigaeva, N. Norouzi, A. Nagl, K. J. van der Laan, E. P. P. Evans and R. Schirhagl, Nanomaterials, 2020, 10, 1–10 Search PubMed.
- M. P. Lake and L. S. Bouchard, PLoS One, 2017, 12, e0179295 CrossRef PubMed.
- K. Wu, T. A. Vedelaar, V. G. Damle, A. Morita, J. Mougnaud, C. Reyes San Martin, Y. Zhang, D. P. I. van der Pol, H. Ende-Metselaar, I. Rodenhuis-Zybert and R. Schirhagl, Redox Biol., 2022, 52, 102279 CrossRef CAS PubMed.
- D. Khanal, Q. Lei, G. Pinget, D. A. Cheong, A. Gautam, R. Yusoff, B. Su, S. Yamaguchi, A. Kondyurin, J. C. Knowles, G. Georgiou, L. Macia, J. H. Jang, I. Ramzan, K. W. Ng and W. Chrzanowski, Nanoscale Adv., 2020, 2, 4798–4812 RSC.
- I. Machova, M. Hubalek, T. Belinova, A. Fucikova, S. Stehlik, B. Rezek and M. H. Kalbacova, Carbon, 2020, 162, 650–661 CrossRef CAS.
- A. E. Garcia-Bennett, A. Everest-Dass, I. Moroni, I. das Rastogi, L. M. Parker, N. H. Packer and L. J. Brown, J. Mater. Chem. B, 2019, 7, 3383–3389 RSC.
- F. Perona Martínez, A. C. Nusantara, M. Chipaux, S. K. Padamati and R. Schirhagl, ACS Sens., 2020, 5, 3862–3869 CrossRef.
- S. R. Hemelaar, A. Nagl, F. Bigot, M. M. Rodríguez-García, M. P. de Vries, M. Chipaux and R. Schirhagl, Microchim. Acta, 2017, 184, 1001–1009 CrossRef CAS.
- E. Mayerhoefer and A. Krueger, Acc. Chem. Res., 2022, 55, 3594–3604 CrossRef CAS PubMed.
- Y. Wu, A. Ermakova, W. Liu, G. Pramanik, T. M. Vu, A. Kurz, L. McGuinness, B. Naydenov, S. Hafner, R. Reuter, J. Wrachtrup, J. Isoya, C. Förtsch, H. Barth, T. Simmet, F. Jelezko and T. Weil, Adv. Funct. Mater., 2015, 25, 6576–6585 CrossRef CAS.
- B. M. Chang, H. H. Lin, L. J. Su, W. der Lin, R. J. Lin, Y. K. Tzeng, R. T. Lee, Y. C. Lee, A. L. Yu and H. C. Chang, Adv. Funct. Mater., 2013, 23, 5737–5745 CrossRef CAS.
- R. A. Shimkunas, E. Robinson, R. Lam, S. Lu, X. Xu, X. Q. Zhang, H. Huang, E. Osawa and D. Ho, Biomaterials, 2009, 30, 5720–5728 CrossRef CAS.
- Y. Zou, S. Ito, F. Yoshino, Y. Suzuki, L. Zhao and N. Komatsu, ACS Nano, 2020, 14, 7216–7226 CrossRef CAS.
- D. Terada, S. Sotoma, Y. Harada, R. Igarashi and M. Shirakawa, Bioconjugate Chem., 2018, 29, 2786–2792 CrossRef CAS.
- M. C. Lukowiak, B. Ziem, K. Achazi, G. Gunkel-Grabole, C. S. Popeney, B. N. S. Thota, C. Böttcher, A. Krueger, Z. Guan and R. Haag, J. Mater. Chem. B, 2015, 3, 719–722 RSC.
- I. Rehor, H. Mackova, S. K. Filippov, J. Kucka, V. Proks, J. Slegerova, S. Turner, G. van Tendeloo, M. Ledvina, M. Hruby and P. Cigler, ChemPlusChem, 2014, 79, 21–24 CrossRef CAS PubMed.
- V. Merz, J. Lenhart, Y. Vonhausen, M. E. Ortiz-Soto, J. Seibel and A. Krueger, Small, 2019, 1901551 CrossRef CAS.
- Y. Liang, M. Ozawa and A. Krueger, ACS Nano, 2009, 3, 2288–2296 CrossRef CAS.
- Q. Wei, T. Becherer, S. Angioletti-Uberti, J. Dzubiella, C. Wischke, A. T. Neffe, A. Lendlein, M. Ballauff and R. Haag, Angew. Chem., Int. Ed., 2014, 53, 8004–8031 CrossRef CAS.
- I. M. Hafez, N. Maurer and P. R. Cullis, Gene Ther., 2001, 8, 1188–1196 CrossRef CAS PubMed.
- R. P. Dougherty, in Collection of Technical Papers - 11th AIAA/CEAS Aeroacoustics Conference, 2005, vol. 3, pp. 2036–2048.
- A. Sigaeva, A. Morita, S. R. Hemelaar and R. Schirhagl, Nanoscale, 2019, 11, 17357–17367 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tc00590a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |