 Open Access Article
Open Access ArticleMetal–organic framework-derived photoelectrochemical sensors: structural design and biosensing technology
Yanting
Shi†
a,
Yutao
Zou†
a,
M. Shahnawaz
Khan
 b,
Mengge
Zhang
a,
Jiang
Yan
c,
Xiaohua
Zheng
b,
Mengge
Zhang
a,
Jiang
Yan
c,
Xiaohua
Zheng
 *a,
Weiqi
Wang
*a,
Weiqi
Wang
 *a and
Zhigang
Xie
*a and
Zhigang
Xie
 *d
*d
aSchool of Pharmacy, Nantong University, Nantong 226001, Jiangsu Province, China. E-mail: wwq1990@ntu.edu.cn; xiaohuaz@ntu.edu.cn
bPillar of Engineering Product Development, Singapore University of Technology and Design, 8 Somapah Road, 487372, Singapore
cDepartment of Cardiology, Affiliated Hospital of Nantong University, Nantong, 226001, China
dState Key Laboratory of Polymer Physics and Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, China. E-mail: xiez@ciac.ac.cn
First published on 13th February 2023
Abstract
Metal–organic frameworks (MOFs) take on highly ordered self-assembled structures and have gained much attention due to their intriguing structural features and enormous applications. Typical features of MOFs include large surface areas, nanoporosity, tunable size and shape, and reliable host–guest interactions, making them demanding candidates in sensor applications. Moreover, MOFs offer an opportunity to upgrade selectivity toward certain analytes in response to various chemical and physical stimulations. In this review, we have elaborated on the structural features of MOFs for biological sensing and the MOF-analyte interaction originating from the inherent porosity, vacancy defects, or abundance functional groups. Also, we summarize the state-of-the-art progress on MOF-based sensors applied in healthcare, food safety, environmental monitoring, and pharmaceutics, category by transduction mechanisms involving optical, electrochemical, and plasmons. In addition, we have discussed in detail the possible applications of MOF-based material for identifying high performance and detecting food freshness, industrial gas leakage, and medical diagnostics. Finally, current limitations regarding sensing performance in practical applications are also overviewed. Thus, this literature review will open new doors for chemists or bioengineers to tune the structural features further to modulate the sensing ability of low-cost materials for biosensing applications in future endeavors.
1. Introduction
Metal–organic frameworks (MOFs) have intriguing properties such as large surface area, tailored pore size, exclusive topology, structural diversity, and exposed active sites.1,2 Taking the structural advantages, MOFs have been demonstrated for their feasibility in practical applications involving gas storage and separations,3,4 pervaporation,5 heterogeneous catalysis,6–8 optoelectronic sensing,9 and theranostics treatment.10–12 It is noteworthy that the inherent porosity makes MOFs a demanding candidate in reliable molecular recognition and high-performance sensor applications. In recent decades, MOF-derived sensor devices have made rapid breakthroughs due to their high selectivity and sensitivity in point-of-care clinical analysis.13–16 Additionally, MOF-derived sensors have been utilized in various emerging fields such as environmental monitoring,17–21 food safety,22–25 pharmaceutics,26–29 and medical diagnostics.30–33 In particular, MOFs with a predictable arrangement have shown outstanding performance in detecting inorganic anions, heavy metal ions, and volatile organic compounds.34–37 Based on the structure-determined property, MOFs have been proven to bring about high selectivity, sensitivity, and reproducibility in constructing sensing devices.38–42 The structure and composition of MOFs are closely correlated with the sensing performance parameters in terms of the limit of detection (LOD) and relative standard deviation (RSD) for stability and reproducibility.Of note, the porous nature of MOF structures provides an exclusive topological paradigm that lets them relate sturdily with analytes depending on the aperture size, geometry, and conformation.43–47 Such spatial confinement for selective identification of guest molecules results in easily measurable outputs, even in detecting trace amounts of analytes or enantiomer detections.48–52 In addition to the inherent porosity, post-synthetic modulations endow MOF with reliable structural identification through plentiful functional group chemistry.53,54 Besides, vacancy defects within MOF may serve as a nanoreactor that allows for site-specific catalytic transformations and result in optical/electrical readout system.55–59 In response to environmental stimulation, MOF constructed from periodic coordination offers external selectivity for target analytes in practical sensor applications with high anti-interference performance.
There have been review articles providing comprehensive explanations on the role of MOFs as potential sensory materials.65–68 Pioneering work from Zhou's group has already proposed the application of MOF-based sensing platforms with an emphasis on chemical species,69 and the applications for environmental contaminant sensing have been reported by Rasheed et al.70 Increasing attention has been put into exploring the application of MOF-based sensory materials for biomedical applications. In this review, device architectures of MOF-based nanostructures categorized by the photoelectrochemical-transduction mechanism are comprehensively summarized (Scheme 1), with an emphasis on the measurement of optical spectroscopies, plasmonic activation, Raman scattering, and electric signals. Here, we propose an outline of the structural features of MOF-derived composites associated with the sensing presentation and introduce their synthetic methodology, structure–function relationship, and practical applications for targeted analytes. We conclude by providing insights into the limitations of the current bioanalytical applications and highlighting the promising prospects of leveraging optoelectronic MOF structures.
 | ||
| Scheme 1 Signal-transduction mechanisms from the photoelectrochemical category classify MOFs-derived sensor devices. Reproduced with permission.60 Copyright 2022, ELSEVIER. Reproduced with permission.61 Copyright 2020, ACS publications. Reproduced with permission.30 Copyright 2022, ACS publications. Reproduced with permission.62 Copyright 2021, ELSEVIER. Reproduced with permission.63 Copyright 2021, Wiley. Reproduced with permission.64 Copyright 2020, ACS publications. | ||
2. Methods and mechanisms of MOF-based sensing devices
Structural features endow MOF materials with immense feasibility in the construction of sensing platforms with the advantages of facilitating adsorbing and preconcentrating the analytes, target-oriented molecular recognition, and real-time self-reference detection. Herein, MOF-based sensory materials were classified by the signal response, such as luminescence enhancement/quenching,71–73 multi-emission/ratiometric fluorescence,74–76 surface plasmon resonance (SPR),77,78 surface-enhanced Raman scattering (SERS),79 electrical impedance,80,81 and cyclic voltammetry.82–84 In the following sections, we will introduce these methods in the application fields of agricultural field operations, explosive detection, and point-of-care clinical analysis.2.1 Luminescent sensor
For sensing purposes, the luminescent material is highly desired, particularly that material with high selectivity and sensitivity even in complex biological fluids. According to the luminescent source, the signals are routinely divided into photoluminescence,85,86 electrochemiluminescence (ECL),87 and bioluminescence.88 Luminescence signals are generally branded as fluorescence and phosphorescence due to different exciton migration mechanisms.89 In this section, sensing applications of various biological analytes in terms of MOF-derived sensor devices relying on optical spectroscopies are briefly summarized (Table 1). As a rapidly evolving class of sensory materials, MOF-incorporated lanthanide metal ions or polydentate photosensitizers, as organic building units with periodically arranged chromophores may dramatically facilitate signal amplification in terms of monochromatic quenching or enhancement responses. Besides, the luminescent MOFs offer great control over the chromophore engagements, enabling label-freed luminescent centers or labeled with fluorescent tags, thus fulfilling the requirement of self-calibrating sensors through multiple emissions or colorimetric shifts. In addition, the examination results are recognizable to the naked eye, which is of great value in real-life applications.| Materials | Mechanism | LOD | Analyte | Source | Ref. |
|---|---|---|---|---|---|
| Abbreviations: ABDC – 2-amino-1,4-benzenedi-carboxylic acid; ALP – akaline phosphatase; ATP – adenosine triphosphate; BBDC – 3,5-dicarboxybenzeneboronic acid; BTB – 2,4,6-tris(4-carboxyphenyl)aniline; BTDB, 4,4′-(benzoic)i[1,2,5]thiadiazole-4,7-diyl) dibenzoic acid; CEA – carcinoembryonic antigen; CIP, ciprofloxacin; Cyt C – cytochrome c; DNBT – 3,3′-dinitro-5,5′-bis(1H-1,2,4-triazole); ENR – enrofloxacin; GSH – gutathione; H2L, 5-(pyridin-4-ylmethoxy) isophthalic acid; IM – imidazole-2-carboxaldehyde; NANA – N-acetylneuraminic acid; NOR – norfloxacin; NTO – 5-nitro-2,4-dihydro-3H-1,2,4-triazole-3-one; OVA, ovalbumin; PFOA – perfluorooctanoic acid; TCPE – trakis[4-(4-carboxyphenyl)phenyl]ethene; TCPP – trakis (4-carboxyphenyl) porphyrin; TC – tetracycline antibiotics; 1-HP – 1-hydroxybenzene. | |||||
| dsDNA/UiO-67-NH2 | Turn on | 5.03 fM | ATP | Human serum | 60 |
| 6.11 fM | Cyt C | ||||
| UiO-68-An/Ma | Turn on | 50 μM | Biothiols | Solution | 90 |
| DNA/UiO-66 | Turn on | 1.87 fM | DNA | Solution | 91 |
| 40 CFU mL−1 | Escherichia coli O157:H7 | ||||
| AR/ZIF-67 | Turn on | 31 nM | L-cysteine | Disease cell | 92 |
| ZIF-8/DNAzyme | Turn on | 47.85 pM | miRNA-21 | Living cell | 93 |
| Ce-HMMOF | Turn on | 0.04 μM | OVA | Human serum | 94 |
| Co-TCPE | Turn on/colorimetric | 0.011 mM | NTO | Mixture | 95 |
| 0.059 mM | DNBT | ||||
| In-TCPP | Turn on | 1.3 mg L−1 | F− | Drinking water | 96 |
| Turn off | 19 mg L−1 | PFOA | |||
| Eu-H2L/Fe2+ | Turn off | 3.7 μM | Bromate | Wheat flour | 97 |
| Eu-doped MOF | Turn off | 0.69 μg L−1 | 1-HP | Urine | 98 |
| Eu-BBDC | Ratiometric | 22.0 nM | Gallic acid | Fruit juice | 74 |
| NA/Zr-BTB/F/R | Ratiometric | — | H+/GSH | Living cell | 99 |
| Eu MOF | Ratiometric | 15 nM | Arginine | Mixture | 100 |
| UiO-66@COF | Ratiometric | 0.067 μM | PO43− | Mixture | 101 |
| 0.038 μM | ATP | ||||
| dZIF-8 BH | Colorimetric | — | glucose | Blood | 102 |
| UiO-66-NH2-IM | Colorimetric | 8.63 μM | S2O82− | Groundwater | 103 |
| 4.95 μM | Fe3+ | ||||
| MOF-525(Zn) ionogels | Colorimetric | — | H+/glyphosate | Human body | 104 |
| Cu-ABDC | Ratiometric/multicolor | 0.078 mU mL−1 | ALP | Human serum | 105 |
| DNA/Au/Cu-TCPP(Fe) | Colorimetric | 0.742 pg mL−1 | CEA | Human serum | 106 |
| B-EuMOF | Colorimetric | 23.3 mM | NANA | Urine | 107 |
| ZIF-8@aptamer | Colorimetric | 0.036 μM | Fipronil | Environmental pollutants | 108 |
| BUC-88 | Colorimetric | 0.12 μM | ENR | Environmental pollutants | 109 |
| 0.52 μM | NOR | ||||
| 0.75 μM | CIP | ||||
| 0.08 μM | TC | ||||
The detection based on fluorescence enhancement or quenching using optical spectroscopy methods is a commercially available technology, enabling immediate response toward biological analytes in handling complex samples. In this regard, the designed Co-based MOF (ZIF-67) served as a ‘‘turn-on’’ sensor for detecting L-cysteine (L-Cys).92 The guest recognition involves the following steps: the presence of Co-based MOF (ZIF-67) to trigger the oxidation of 3,30,5,50-tetramethylbenzidine (TMB) and amplex red (AR), relying on the enhancement of the fluorescence signal. The as-designed ZIF-67 takes on a dodecahedral structure, with a Brunauer–Emmett–Teller (BET) surface area of up to 1833.26 m2 g−1. According to the Michaelis–Menten curve, the obtained Km is about 22 μM, and Vmax is 412 nM s−1 for highly active oxidase-mimicking nanozyme ZIF-67 (the fitting equation: F/F0 = 0.56CL-Cys + 0.97, R2 = 0.97). The dose-response curve demonstrated that the detection limit was 31 nM with AR as the substrate, and two redox processes (AR and oxAR1, ox AR 1 and ox AR2) accompanied by turn-on fluorescence, performed by alternating the application of ZIF-67 and L-Cys. This provides a reasonable analysis method using AR and ZIF-67 to detect L-Cys via a fluorescence “turn-on” assay.
MOFs consisting of lanthanide ions (Ln-MOF) are promising sensory materials taking the advantage of large Stokes shifts, higher quantum yields, and high photostability.110,111 Generally, Ln-MOF was used for the ‘‘signal-off’’ type of the fluorescent sensor attributed to the energy transfer such as photoinduced electron transfer, resonance energy transfer, and inner filter effect.112,113 However, luminescence switching takes on the ‘‘turn-off’’ type are difficult to quantitative signal decrease and ensure its sensing sensitivity.114–117 To address this issue, Yan et al. prepared a luminescent Eu-doped MOF for the quantitative analysis of urinary chemical carcinogens.98 The mechanism for 1-hydroxytryptamine (1-HP) detection depends on the ligand-to-metal resonance energy transfer. Due to the large overlap within the UV-absorption spectrum of ligand and 1-HP, resulting in the block of the energy transfer from the ligand to Eu3+. According to the “turn-off” strategy, the LOD of 1-HP was calculated to be 0.69 μg L−1 (the benchmark value of 1.0 μg L−1), suggesting the high sensitivity of this portable 1-HP urine test paper for practical biological carcinogen evaluation.
To balance the cost-effectiveness, Eu-based MOF (BUC-88) was designed and developed as a fluorescent sensor for antibiotic detection.109 By using 2,6-naphthalenedicarboxylic acid (2,6-NDC), as a bridging ligand, BUC-88 exhibited a lower lowest unoccupied molecular orbital (LUMO) when in recognition to quinolone antibiotics and tetracycline antibiotics. Besides, this sensory process can be distinguished by the fluorescence discoloration process for the recognition of quinolone antibiotics, while significantly enhanced fluorescence was observed after tetracycline antibiotics recognition. The main mechanisms for this switchable fluorescence can be attributed to the light competition effect and light-induced electron transfer. The detection limits for enrofloxacin, norfloxacin, ciprofloxacin, and tetracycline are 0.12 μM, 0.52 μM, 0.75 μM, and 0.08 μM, respectively. Overall, BUC-88 with highly reproducible (RSD < 0.83%) can be used as water-stable fluorescent sensors to detect quinolone antibiotics and tetracycline hydrochloride.
For intracellular microRNA (miRNA) detection, Su et al. designed the complexes (ZIF-8@DNAzyme) by combining DNAzyme with ZIF-8 to generate fluorescence intracellularly.93 The DNAzyme, which is a complementary fragment of the target miRNA complementarily, pairs with the substrate strand modified by the fluorescent tags. Through the electrostatic interaction, ZIF-8 binds tightly to DNAzyme and forms an annular catalytic domain to protect DNAzyme from loop breaking and drive the substrate strand breakage. The fluorescence detection limit of miRNA-21 by the DNAzyme probe is 47.85 pm, which is much lower than the conventional detection method.118 Overall, the ZIF-8@DNAzyme can accurately distinguish cells with different miRNA-21 expression levels and corresponding dynamic changes.
The “turn-on” and/or “turn-off” sensing modes inevitably lead to false-positive results. To guarantee accuracy, CRISPR-Cas9 triggers the identification of ssDNA genome realized enrichment of target sequence through strand displacement amplification (SDA) and rolling circle amplification (RCA), a two-step isothermal amplification system.91 The fluorescence quenching/recovery arises from the adsorption/desorption of the fluorophore-labeled double-stranded DNA (ssDNA) from the microporous zirconium MOF. Finally, the fluorescence signal is sent out by using the adsorbed ssDNA molecular probe and quenched by UiO-66 to realize the detection of E. coli o157:h7. According to the electrophoresis results, the RCA reaction can occur only when there are many components, forming a long chain structure and leaving it in the glue hole. The linear range of the sensor for the bacteria sample detection is from 1.3 × 102 CFU mL−1 to 6.5 × 104 CFU mL−1, with detection limits of 1.87 fm and 40 CFU mL−1. Overall, the two-step isothermal amplification method has great application potential in pathogenic bacterial detection, food safety, and clinical diagnosis.
In physical health, nearly 20% of incurable diseases are directly related to heavy metal ions and organic matter pollutants. For example, the 3-nitro propionic acid (3-NPA) in mouldy sugarcane has been proven harmful. Thus, the highly sensitive detection of 3-NPA in sugarcane deterioration is important for global food security. Given that, Li et al. proposed a novel Eu-MOF with intrinsic dual-emission from 2,6-naphthalene dicarboxylate (NDC) ligands and Eu3+ ions for ratiometric detecting 3-NPA (Fig. 1(A)).119 As shown in Fig. 1(B), trinuclear europium has seven ligands, and this coordination environment makes the structure of Eu-MOF extremely stable. The emission peak at 615 nm (Eu3+) consistently drops, and the emission band for the NDC ligand (459 nm) gradually increased even at the 3-NPA concentration of as low as 0.1 mM (Fig. 1(C)). The Eu-MOF will switch from red to blue in the detection through pH-regulated ratiometric luminescence, enabling a moderate anti-interference ability for practical applications. Another example of MOFs for multichannel detection is based on aggregation-induced emission (AIE) characteristics.95 The AIE-MOF was constructed with tetrakis[4-(4-carboxyphenyl)phenyl]ethene (TCPE), and Co(NO3)2·6H2O was employed to Co-load fluorescence quencher and color-changing agent for nitrogen-rich heterocyclic compounds (NRHCs) detection. Following the competitive substitution between NRHCs and the TCPE ligand, the AIE-MOF exhibits a turn-on fluorescence emission and color changes from blue to yellow (Fig. 1(D)). According to quantification, the fluorescence intensities (I/I0) and the NRHC concentration takes on good exponential relationships ranging from 0 to 100 μM, and this process can be intuitively observed and referred to the International Commission on Illumination (Fig. 1(E)). Compared with the reported methods, Co-TCPE selectively recognized both single- and dual-ring NRHCs, while there were no signal responses when mixed with other types of explosives (Fig. 1(F)). Overall, the AIE-MOF provides insight for designing a high-performance fluorescent and colorimetric dual-responses sensing system.
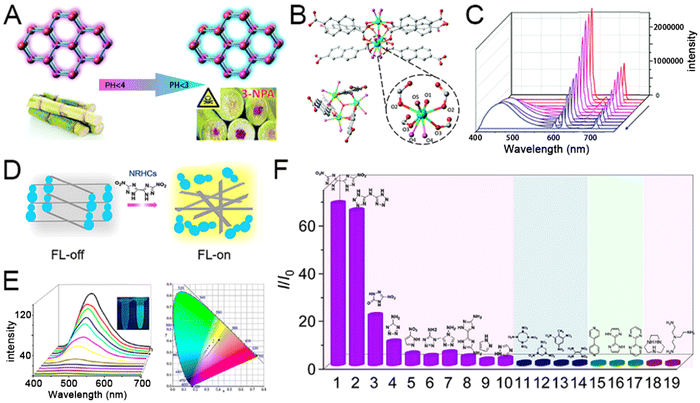 | ||
| Fig. 1 (A) Graphical representation of pH-Regulated ratiometric luminescence rapid detection of toxic mycotoxin in moldy sugarcane; (B) structure of Eu-MOF coordinated by trinuclear europium and six bis-bidentate bridgings NDC ligands. (C) Emission spectra of 3-NPA solutions of different concentrations; reprinted from ref. 119 with permission of Royal Society of Chemistry, Copyright 2020. (D) Description of the sensing mechanism based on competitive coordination substitution. (E) FL spectra of Co-TCPE with different concentrations of DNBT and coordinates derived from part d in the CIE 1931 color space. (F) I/I0 results of the sensing system toward different analytes. 1, DNBT; 2, bis(1H-tetrazol-5-yl) amine (H3bta); 3, NTO; 4, 5H-tetrazole-5-amine; 5, 3-nitro-1H-pyrazole; 6, 1-methyl-1H-tetrazol-5-amine; 7, 4H-1,2,4-triazol-4-amine; 8, 3,3′-diamino-5,5′-bis(1H-1,2,4-triazole) (DABT); 9, pyrazole; 10, 4H-1,2,4-triazole; 11, octahydro-1,3,5,7-tetrazocine (HMX); 12, hexahydro-1,3,5-trinitro triazine (RDX); 13, trinitrotoluene (TNT); 14, 1,1-diamino-2,2-dinitroethylene (FOX-7); 15, 4,4′-bipyridine; 16, 2,2′-biimidazole; 17, 2,2′-bipyrimidine; 18, 1,4,7-triazacyclononane; and 19, tris(2-aminoethyl)amine. Reprinted from ref. 95 with permission of the American Chemical Society, Copyright 2021. | ||
Portable biosensors are widely used due to their simplicity and versatility. In this regard, Chen et al. put forward a defective ZIF biohybrid hydrogel (dZIF-8 BH) for portable glucose biosensing.102 Glucose oxidase (GOx) and peroxidase (POD) were co-encapsulated into dZIF-8 BH, thus it could efficiently convert glucose into a blue-violet product through the biocatalytic cascade. Of note, this portable sensor can be combined with a smartphone for colorimetric biosensing of glucose distinguished by the naked eye with high sensitivity, selectivity, and stability. Similar to the colorimetric detection, Ce-based MOFs (Ce-MOFs) exhibited alkaline phosphatase (ALP)-like activity and have been considered candidate sensory materials. To improve the sensing of bulk phosphorylated proteins, Gao et al. prepared a hierarchically mesoporous Ce-MOF (Ce-HMMOF) with multiple active sites.94 Exploiting polymeric F127 as a removable template is practical for the synthesis of Ce-HMMOF. In parallel, the phosphorylated compound can be selectively dephosphorylated by Ce clusters, and the resulting phosphate further complexed with molybdate produces a recognizable signal (green) to the naked eye, exhibiting astonishing sensitivity.
Considering the increasing number of biological targets in complex analytes, multi-target detection is critical for disease diagnosis and treatment. Given that, Wang et al. developed a novel platform based on target-regulated competitive binding and exonuclease I (Exo I)-driven release of signaling molecules.60 The single-stranded DNA (ssDNA) Sx (X = ATP or Cyt C) with a carboxyl group can be linked to the UiO-67-NH2 surface, which was synthesized by a one-step hydrothermal method. Two dyes, Rho 6G and Cy5, were wrapped in the mesopore of the MOFs by the aptamer ssDNA Ax (X = ATP or Cyt C) and hybridized with Sx to form a dsDNA-functionalized MOF. In the absence of a detection target, the double-stranded structure prevented the release of signal molecules. On the contrary, in the presence of the detection targets, the aptamer ssDNA Ax bonded to the target to form a complex leaving the MOFs, and thus the dye molecule was released. The release of signal molecules was further facilitated when the free Sx of the MOFs were degraded by Exo I reducing the steric hindrance. Rho 6G @UiO-67 and Cy5@UiO-67 were used to detect ATP and Cyt C, respectively, and the fluorescence intensity increased with the increasing concentration of the detectors. The efficient performance and highly selective detection make the proposed method a promising platform for multiple-target diagnostics.
High-performance electrochemiluminescence (ECL) emitters are important in improving the sensitivity for immunoassay.121,122 In this aspect, Wei et al. proposed an innovative orientation to employ dumbbell plate-like MOF (Zr-TCBPE) with coordination-triggered ECL enhancement behavior as a quenching-typed neuron-specific enolase (NSE) detection immunosensor.120 For sensor construction, polyethyleneimine (PEI) is anchored to Zr-TCBPE MOF to form a self-enhanced ECL complex, and the AuPd@SiO2 composite was designed as a high-efficiency quencher. Using scanning electron microscopy, AgNPs were on the surface of the synthetic MOF, while AuPd NPs were dispersed on the surface of the SiO2 nanospheres. In comparison with other interfering substances, including carcinoembryonic antigen (CEA), brain natriuretic peptide (BNP), procalcitonin (PCT), and glucose (Glu), the Zr-TCBPE-PEI-Ag substrate showed excellent selectivity when compared with the ECL response (Fig. 2(B) and (C)). Of note, there is no difference in the ECL response even with the mixture comprising tenfold interfering substances, demonstrating the robust anti-interfering capacity. Taken together, the intramolecular self-catalytic ECL system with the LOD of 52 fg mL−1, exhibits excellent selectivity, stability, and recyclability as an immunosensor for NSE detection.
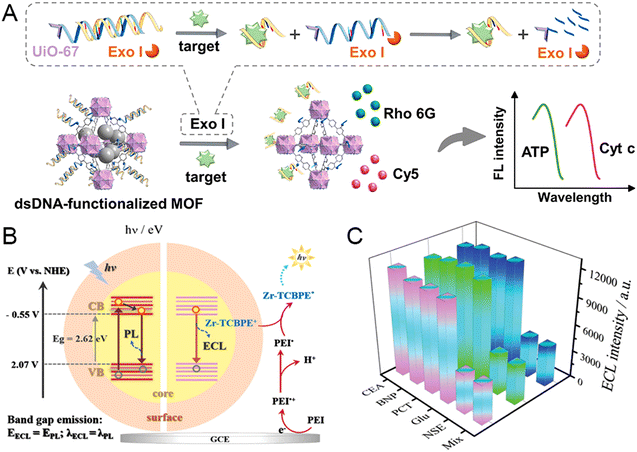 | ||
| Fig. 2 (A) Schematic illustration of the target-modulated competitive binding and exonuclease I-powered strategy for the simultaneous and rapid detection of biological targets. Reprinted from ref. 60 with permission of ELSEVIER, Copyright 2022. (B) Schematic illustration for the ECL and PL emission mechanism of Zr-TCBPE-MOF. (C) The selectivity of the developed immunosensor for NSE detection compared with the other four interfering substances or the mixture. Reprinted from ref. 120 with permission of Wiley, Copyright 2022. | ||
2.2 Plasmonic sensor
Existing methods for luminescent detection generally require molecule absorption, limiting the range of species that can be detected. In contrast, plasmonic sensors with advantageous properties such as real-time and label-free detection of various biomarkers, such as proteins, nucleic acids, exosomes, and pathogens even in complex biological media.123–125 The plasmonic-based platform is generally classified into SPR and SERS, and this sensing modality is widely applied in bioanalysis owing to the advantageous properties of molecular fingerprint information, resistance to photobleaching, low interference, and precise control over multi-component analysis.116–118 The representative biosensor based on SPR or SERS, summarized in Table 2, involves strategies for enhancing the sensitivity of plasmonic nanostructures that are highly desired to facilitate progress in point-of-care testing, as well as mimicking the physiological environment.| Materials | Structure | LOD | Analyte | Recognization | Ref. |
|---|---|---|---|---|---|
| Abbreviations: BPTCN – 4′-mercapto biphenyl carbonitrile; Cre – creatinine; DA – dopamine; HOE – hoechst H 33258; NHA – nanohole array; PCS – plastic-clad silica; PE – pseudoephedrine; THC – tetrahydrocortison; 4-ATP – 4-amino thiophenol; MB – methylene blue; R6G – rhodamine 6G; OPPs – organophosphorus pesticides; VOC – volatile organic compounds. | |||||
| Cu-TCPP/Au chip/peptide | Surface coating | 16.7 particles/mL | PD-L1 exosomes | Peptide targeting | 126 |
| Cu-BTC NHA | Surface coating | 500 ppb | VOA | Pore adsorption | 77 |
| MOF-5/Au PCS | Surface coating | ∼1 pM | OPPs | Pore adsorption | 78 |
| Au-AlaZnCl | Surface coating | 1 pM | PE | Chiral | 127 |
| 3D ZnO/Ag nanostructure | Surface coating | 0.1 pM | R6G | Pore adsorption | 136 |
| Au/ZIF-8 | Core–shell | 0.1 mM | 4-ATP | Ionic ligands | 128 |
| Au/ZIF-8/PMA | Core–shell | — | HOE | Bioimaging | 131 |
| MOF-5 coated gold gratings | Core–shell | 10 fM | OPPs | Pore adsorption | 135 |
| Ag/MPBA | Hybrids | 0.1 fM | THC/pregnane | Chemical bond | 61 |
| D-MIL-125-NH2 | Hybrids | 0.66 μM | DA | Vacancy defects | 129 |
| Au/MIL-101(Fe) | Hybrids | 0.1 μM | Cre | Electrostatic/π–π interactions | 130 |
| Au/Fe-MOF | Hybrids | 9.3 pM | MB | Electronic transfer | 132 |
| M-TCPP MOF | Sheet-like | 10 nM | R6G | Electronic transfer | 133 |
To reach high-throughput analyses, Hu et al. put forward a direct SPR assay for the programmed death ligand-1 (PD-L1) exosomes.126 The two-dimensional Cu-TCPP with improved photoelectric properties can increase the sensitivity and provide precise detection for trace biomarkers. Generally, the SPR angle increased as the refractive index value of the solution flowing through the chip surface increased. As a comparison, the SPR angle of the lamellar Cu-TCPP changed more drastically, reaching up to 137.67°/RIU at a concentration of 10 μg mL−1. The SPR angular displacement shows a good linear relationship (Δθ = 0.0324![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lgCExo − 0.11984, R2 = 0.989) with the logarithm of the concentration of PD-L1 exosomes, with a LOD of 16.7 particles per mL. Overall, the lamellar Cu-TCPP is a promising SPR sensor chip for detecting PD-L1 exosomes, demonstrating excellent specificity, accuracy, and reproducibility for actual sample detection.
lgCExo − 0.11984, R2 = 0.989) with the logarithm of the concentration of PD-L1 exosomes, with a LOD of 16.7 particles per mL. Overall, the lamellar Cu-TCPP is a promising SPR sensor chip for detecting PD-L1 exosomes, demonstrating excellent specificity, accuracy, and reproducibility for actual sample detection.
SERS platforms offer the possibility of detecting biomarker molecules at low concentrations from hot spots on plasmonic nanocrystals.132,137 For example, Yang et al. proposed a superficial wet-chemistry tactic for the site-selective deposition of zeolitic imidazolate framework 8 (ZIF-8) on anisotropic Au nanobipyramids (NBPs).128 As depicted in Fig. 3(A), ZIF-8 was placed at the ends, waist, and around the entire surface of NBPs, denoted as NBP/end-ZIF, NBP/waist-ZIF, and NBP@ZIF, respectively. The Au NBPs and three kinds of nanostructures possess uniform shapes and high purity. By using 4-amino thiophenol (4-ATP) as the probe molecule, the distinct Raman signals of the NBP/end-ZIF sample are stronger than those from the NBP/waist-ZIF and NBP@ZIF nanostructures (Fig. 3(B)). Moreover, the porous ZIF-8 shell only allows analytes smaller than the flexible aperture size of ZIF-8 (4–6 Å) to approach the Au NBPs and contribute to the SERS spectrum. Overall, the NBP/end-ZIF taken on unique nanotopography is a promising candidate for practical SERS detection. Li et al. demonstrated a core–shell Au nanorod@Zr-MOF nanostructures based on wet chemistry to improve the SERS sensitivity.79 The Zr6-octahedron clusters are linked by 1,3,6,8-tetrakis (p-benzoic acid) pyrene (TBAPy), so the concentration of TBAPy can change the thickness of the MOF coating. Raman signals were not observed for the substrate with a shell thickness larger than 45 nm for detecting 4′-mercapto biphenyl carbonitrile (BPTCN) molecules, and 3 nm thickness exhibits dramatically improved SERS spectra. The Au nanorod@Zr-MOF substrate exhibits a good linear relationship, high RSD, excellent sensitivity, and suitable reproducibility, providing a promising strategy to remarkably improve the SERS performance.
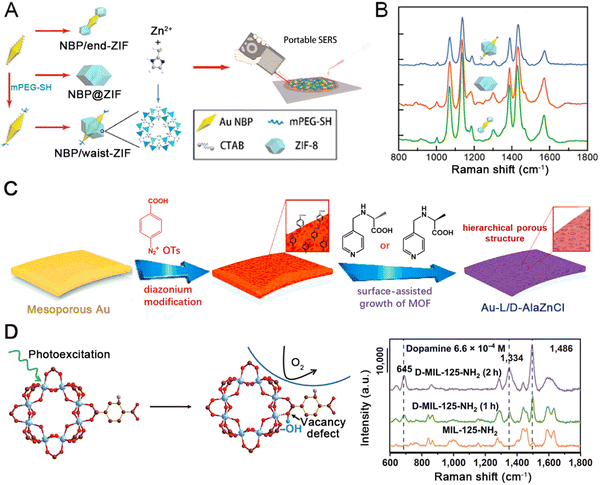 | ||
| Fig. 3 (A) Three routes to the site-selective deposition of ZIF-8 on anisotropic Au nanobipyramids (NBPs) and nanorods (NRs). (B) SERS spectrum of 4-ATP on the different kinds of nanostructures. Reprinted from ref. 128 with permission of American Chemical Society, Copyright 2021. (C) Schematic diagram showing the preparation of the Au-L/D-AlaZnCl. (B) Schematic illustrations of enantiomer detections. Reprinted from ref. 127 with permission of ELSEVIER, Copyright 2021. (D) Illustration of the stabilization of photo-induced vacancy defects in MOFs and SERS spectra of dopamine (6.6 × 10−4 M) on substrates deposited with initial MIL-125-NIL, D-MIL-125-NH2, (1 h), D-MIL-125-NH2 (2 h). Reprinted from ref. 138 with permission of Springer, Copyright 2022. | ||
Olga et al. proposed a class of hierarchically-porous Au films based on chiral recognition for enantioselective SERS sensing (Fig. 3(C)).127 The hierarchical films consist of mesoporous Au with homochiral MOF grown inside, which is extremely appealing for enantioselective detection. Surface-assisted growth of the nanoporous homochiral MOF AlaZnCl was performed via diazonium modification to enable hierarchical MOF growth leading to two types of films. The homochiral MOF has already been shown to selectively capture pseudoephedrine (PE). In contrast, (+)-PE can be selectively adsorbed by L-AlaZnCl, while (−)-D-AlaZnCl dominantly recognizes PE enantiomer. Au-L-AlaZnCl is selective towards (+)-PE sensing with ultra-high sensitivity, even in the analysis of complex biosystems. The Au-L/D-AlaZnCl is a promising plasmonic material to monitor the enantiomers of monoamine neurotransmitters.
Vacancy defects are of critical importance in improving SERS sensitivity. However, generally used semiconductor materials lack stability in vacancy defects due to their rapid self-healing upon air exposure. Sun et al. demonstrated that the photoexcitation of MOF materials could stabilize photo-induced oxygen vacancy (PIVO).138 As depicted in Fig. 3(D), the steric hindrance and vacancy defects in D-MIL-125-NH2 greatly facilitate the electronic transfer, thus contributing to the SERS performance. In contrast to numerous other substrate materials, dopamine's Raman intensity is greatly enhanced using the D-MIL-125-NH2. The lowest detectable dopamine concentration on the photo-excited MIL-125-NH2 was 6.6 × 10−7 M, showing extremely high sensitivity. D-MIL-125-NH2 exhibited stable SERS behaviors, such as stable enhancement of the intensity in Raman spectra of the characteristic peak of bisphenol A within 60 days at 100 °C. In conclusion, D-MIL-125-NH2 is an ideal material for evaluating the relationship between vacancy defects and sensing performance.
Creatinine is a credible indicator of nephritic disease speculated to be theoretically distinguishable in Raman spectra. Traditional detection methods for urinary creatinine levels have many drawbacks, such as high cost and low sensitivity. Thereby, Jiang et al. fabricated Au@MIL-101(Fe) as a SERS substrate using the solution impregnation method to monitor creatinine precisely.139 The SERS substrate was constructed by mixing pre-synthesized MIL-101(Fe) with chloroauric acid to form ultra-small nanoparticles on the surface. The framework structure could drag creatinine into the neighborhood of Au by the strong electrostatic interaction. Compared to Au NPs, the Raman intensity of R6G was greatly enhanced using Au@MIL-101(Fe). Besides, the SERS intensity correlated with the creatinine concentration ranging from 1 μM to 100 μM. High recoveries and clear Raman spectra also showed satisfactory accuracy when applied to actual urine samples. The selective SERS substrate is a promising material for further sensitive and robust bio-detection applications. For urinalysis, Kao et al. proposed a plasmonic-active super-hydrophobic SERS platform to concentrate analytes into confinement physically.61 The innovative platform successfully correlated SERS readouts with miscarriage risks among pregnant women by quantifying the relative concentrations of these two metabolites: pregnancy and tetrahydrocortisone (THC). As depicted in Fig. 4(A), the stable superhydrophobic (SPHB) plasmonic nanostructured surface uses a binary mixture of monodisperse Ag nanocubes and octahedra. The 4-mercapto phenylboronic acid (MPBA)-grafted Ag nanocubes function as probes to selectively capture pregnane and THC via a boronate ester bond under alkaline conditions (Fig. 4(B)). The partial least-squares regression (PLS) analyses of the experimental mixture and real pregnant women's urine samples display a high linear coefficient (Fig. 4(C)). The SPHB plasmonic nanostructure gives rise to good predictive accuracy for multiplex detection, which holds great promise for clinical ultrasensitive metabolite screening.
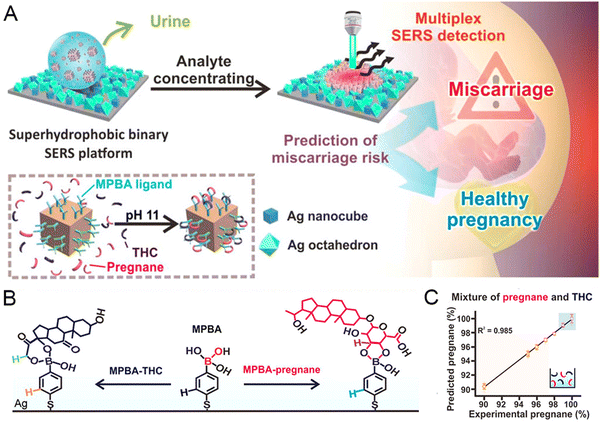 | ||
| Fig. 4 (A) Schematic illustration of the SPHB SERS platform to stratify the miscarriage risk. (B) Description of MPBA capture function. (C) PLS analysis of a mixture comprising pregnane and THC. Reprinted from ref. 61 with permission of the American Chemical Society, Copyright 2020. | ||
The combination of confocal microscopy and surface-enhanced Raman spectroscopy holds great promise in current medical applications. Given that, gold nanostar/ZIF-8 nanocomposites with plasmonic absorption were developed for the visualization of the thermoplastic-driven release of encapsulated active molecules inside the living cells.131 To further improve the stability of such materials, the amphiphilic polymer poly [isobutylene-maleic anhydride]-graft-dodecyl was selected to modify the nanocomposite comprising gold nanostars and ZIF-8. The core–shell nanocomposite exhibited high drug loading capacity and superior stability, enabling spatiotemporal controllable release of the cargo upon near-infrared (NIR) irradiation (Fig. 5(A)). According to the fluorescence analysis, much higher fluorescence signals from Hoechst (HOE) were observed after Au/ZIF-8/PMA incubation when treated with NIR illumination (Fig. 5(B)). The system is very stable and enables fast response; once the NIR irradiation is stopped, HOE is no longer released (Fig. 5(C)). The PMA samples present black shades due to the lack of HOE leaking, while cells irradiated with the NIR having the nuclei staining show that the HOE can localize subcellular organelles such as cytoplasm or nucleus (Fig. 5(D)–(F)). The Au/ZIF-8/PMA system demonstrated a promising synthetic methodology for controllable drug delivery, making MOF-based intracellular vehicles for drug vectorization.
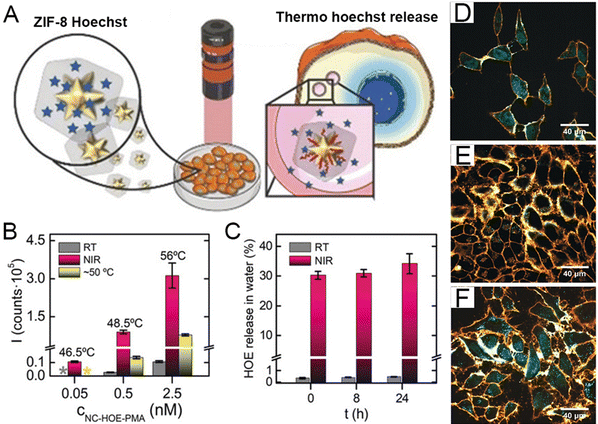 | ||
| Fig. 5 (A) Schematic diagram of PMA/ZIF-8 system. (B) Fluorescence signals from HOE's release after different treatments (thermostatic batch). (C) The release profile of HOE with/without NIR illumination. Confocal microscopy images of cells incubated with (D) NC-HOE, (E) NC-HOE-PMA, and (F) NC-HOE-PMA after NIR treatment. Reprinted from ref. 131 with permission of Wiley, Copyright 2019. | ||
Exfoliating MOFs into ultrathin nanosheets endows outperforming SERS enhancement, because of the fully exposed cluster nodes with accessible ligand interaction.140,141 The layered MOF serving as SERS substrates with inherent ligand-cluster charge transfer would directly contribute to signal amplification. To fully exploit the ligand-cluster charge transfer effect, the ultrasonic-assisted stripping strategy was employed to construct ultrathin 2D cobalt-tetrakis(4-carboxyphenyl) porphyrin (Co-TCPP) compress complete activated ligand-cluster units under incident photons.133 Taking rhodamine 6G (R6G) as the probe molecule, Co-TCPP showed a significant increase in the SERS spectrum (Fig. 6(A)). Co-TCPP MOFNs have the highest Raman-enhanced intensity of the R6G signal compared to other substrates (Co-TCPP MOF, SiO2/Si) (Fig. 6(B)). The MOFN-based SERS substrate has shown extremely high sensitivity, with detection limits of as low as 10−9 M (Fig. 6(C)). To further improve the enzyme mimics activity, Zhao et al. synthesized a hybrids sensor (Au NRs/Fe-MOF) for ultra-sensitive toxic substances analysis.132 This biosensor takes on localized surface plasmon resonance properties, thus enhancing the catalytic performance through thermal charge transfer within the MOF (Fig. 6(D)). As shown in Fig. 6(E), the electrons migrating from Au NRS to Fe-MOF are practical for H2O2 reduction. When methylene blue (MB) was chosen for the analysis, logarithmic signal intensities of Raman spectroscopy at 1628 cm−1versus logarithmic MB concentrations (9.4 × 10−11 M–9.4 × 10−6 M) exhibited a linear change. (Fig. 6(F)). In comparison to current detection methods, the detection limit reached up to 9.3 × 10−12 M. Five repeated cycles of MB molecules yielded an RSD of 2.2%, indicating good reproducibility of the substrate (Fig. 6(G)). Overall, the biosensor significantly enhances the chemical catalytic capability and has great potential for the detection of toxic substances.
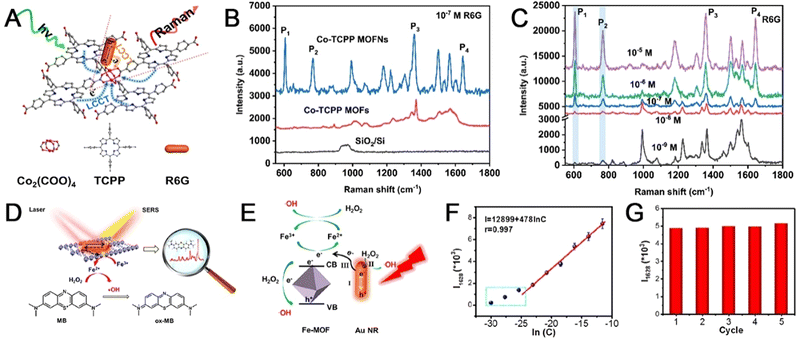 | ||
| Fig. 6 (A) Illustration of the L-type CT pathway contributing to enhanced SERS sensitivity of ultrathin two-dimensional MOF nanosheets. (B) SERS spectra of R6G (10−7 M) on different surfaces. (C) SERS spectra of R6G on the Co-TCPP MOFNs substrate at six different concentrations ranging from 10−5 to 10−9 M. Reprinted from ref. 133 with permission of the American Chemical Society, Copyright 2021. (D) Schematic diagram of the Au NRs/Fe-MOF biosensor. (E) Schematic illustration of the photo-enhanced peroxidase-like performance of Au NRs/Fe-MOF. (F) Corresponding linear relationship of logarithmic signal intensities versus logarithmic MB concentrations. (G) The intensity distribution of the band after five cycles. Reprinted from ref. 132 with permission of the American Chemical Society, Copyright 2022. | ||
Immune checkpoint blockade (ICB) therapy, which enhances the immune anti-tumor response by selectively blocking the immune checkpoint signaling pathway, has been used on a large scale but some patients may experience serious immune-related adverse effects for individual reasons.142,143 To predict the ICB response for rational selection of target patients in advance. Li et al. proposed a method to predict the therapeutic effect of checkpoint blockade treatment using the SERS technique to analyze tumor-related markers obtained from body fluids, such as circulating tumor cells (CTCs).134 Highly-sensitive SERS nanotags were prepared using metal nanoboxes conjugated to specific antibodies. The nanotags were used to identify CTCs, and checkpoint blockade responders were identified by obtaining the average Raman intensity changes of the four tumor biomarkers at the ensemble and single-cell levels (Fig. 7(A)). Interferon-γ (IFN-γ) stimulation can simulate the secretion of signaling molecules in vivo after ICB (Fig. 7(B)). Thus, the response of patient-derived CTCs to IFN-γ stimulation can fully reflect the tumor status after treatment. The use of SERS nanotags targeting four tumor biomarkers produced higher Raman intensities compared to cells labeled with immunoglobulin G (IgG)-functionalized nanotags, indicating specific binding of these four SERS nanotags to LM-MEL-33 cells. In addition, the expression levels of the four biomarkers on LM-MEL-33 cells showed different responses after the IFN-γ treatment (Fig. 7(C)). A heatmap showing the CTCs characteristics of 14 patients using the IFN-γ-stimulated Raman signals of melanoma tumor markers indicated the potential of patients to respond to ICB (Fig. 7(D)). Overall, SERS assay may have a potential role in predicting treatment response, and MOF-based substrate for SERS detection may be applied in many bio-related applications.
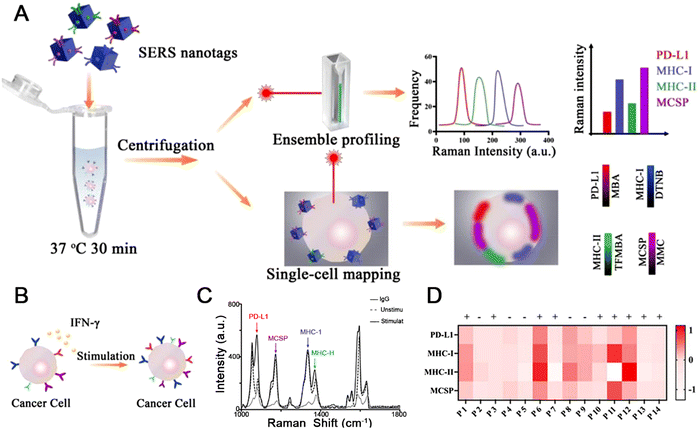 | ||
| Fig. 7 (A) Schematic illustration of SERS assay for profiling melanoma CTCs. (B) CTC phenotypic alteration in response to IFN-γ stimulation. (C) Ensemble cell measurements showing average SERS intensities. (D) Heatmap of four biomarker expression levels with IFN-γ stimulation for patients. Reprinted from ref. 134 with permission of American Chemical Society, Copyright 2022. | ||
2.3 Electronic sensor
MOFs have unsaturated metal coordination sites, uniform network structure, and unique topologies that can be engineered through the judicious combination of metal ions/clusters and organic linkers for functional conductive properties. These distinct features made MOFs capable of strengthening the electric signal response.144,145 Motivated by these fascinating properties, pristine MOFs with excellent electrocatalytic activities have been used as electrocatalysts to detect various small-molecule compounds such as H2O2, glucose, hazardous gases, and enantiomers. The analytical performances are listed in Table 3.| Material | Analyte | Response | LOD | RSD | Application | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: ATP – adenosine triphosphate; CNT – carbon nanotube; DA – dopamine; DABDT – 2,5-diamino1,4-benzene dithiol dihydrochloride; DPV – differential pulse voltammetry; GCE – glassy carbon electrode; HiTP – 2,3,6,7,10,11-hexaaminotriphenylene; HQ – hydroquinone; ITO – indium tin oxide; MIP – molecularly imprinted polymer; MSMC – microstructured mixed cellulose; NB – nitrobenzene; PMMA – poly(methyl methacrylate); PT – patulin; CE – tumor markers; CA153 – carbohydrate antigen 153; OTC – oxytetracycline; VC – vitamin C. | ||||||
| Ni-DABDT | X-ray | Current density | 7.2 μGya s−1 | — | Wearable device | 146 |
| MIP/Au@PANI/SeS2/Co-MOF electrode | PT mycotoxin | DPV current | 0.66 pM | 2.49% (reuse) | Food safety | 62 |
| 3.22% (reproduce) | ||||||
| Mn-PCN-222/ITO | NB | Cyclic voltammetry | 0.106 mM | 2.67% | Water pollutant | 63 |
| Ce-MOF/CNTs/GCE | HQ | Cyclic voltammetry | 5.3 μM | 5.76% (reuse) | Isomer distinction | 147 |
| 3.24% (reproduce) | ||||||
| CC | 3.5 μM | 4.85% (reuse) | ||||
| 4.25% (reproduce) | ||||||
| Cu2O/ZIF-67 | Glucose | Cyclic voltammetry | 6.5 μM | 4.6% | Public health | 148 |
| Ni-HiTP/MSMC | Pressure/temperature | Impedance | 1 Pa | — | Electronic skin | 149 |
| Co-MOF-74 electrodes | NO2 | Impedance | 0.5–5 ppm | — | Industry and public health | 150 |
| Zn-MOF PEC aptasensor | CEA | Current | 0.0275 U mL−1 | 1.1–3.6% | Tumor diagnosis | 151 |
| CA153 | 2.85 pg mL−1 | |||||
| MV/ZIF-90 | ATP | Photocurrent | 3.2 pM | 4.1% | Food safety | 152 |
| Ce-MOF/MCA | OTC | Impedance | 17.4 fg | 1.16% | Food safety | 153 |
| Ni3HHTP2 | VC | Current | — | 13.4% | Public health | 154 |
| Au/RP1/Ni3HHTP2 | DA | Current | 63 nM | 5.3% | Public health | 155 |
Flexible flat panel X-ray detectors are portable and shape tuneable, but active sensing materials mainly utilize toxic heavy metals.156,157 Li et al. designed a flexible X-ray detector using Ni-DABDT as the absorbing layer.146 As depicted in Fig. 8(A), the coordination reaction between the nickel(II) cations and DABDT in an alkaline solution for several hours led to the formation of Ni-DABDT MOF, and the Ni-DABDT realized a direct X-ray-to-carrier conversion. The flexible X-ray detector, a gold/Ni-DABDT/gold sandwich symmetric structure, adheres to the polyimide substrates, forming the X-ray imager. Compared with other flexible X-ray detectors, the Ni-DABDT X-ray detector is the first to use semiconductive MOFs with low Z (atomic number) for X-ray detection and shows good detection sensitivity (Fig. 8(B)). Calculated using the formula for sensitivity (S) = ∂J/∂D (S = 98.6), the Ni-DABDT detector has extremely high sensitivity. The lowest detection limit of the Ni-DABDT X-ray detector is 7.2 μGya s−1, meeting most medical diagnoses. The flexible Ni-DABDT detector exhibited great mechanical stability after 10 h of X-ray irradiation and 500 repetitions of bending. Overall, the Ni-DABDT MOF is a potential material for emerging radiation monitoring and imaging applications.
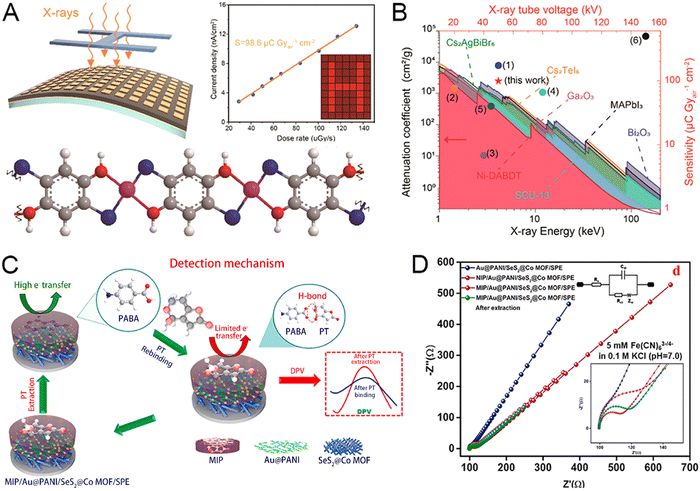 | ||
| Fig. 8 (A) Schematic diagram of the flexible Ni-DABDT X-ray detector. (B) Performance chart with the comparison of current flexible X-ray detectors. Reprinted from ref. 146 with permission of the American Chemical Society, Copyright 2021. Concept and theoretical calculation of the hydrogen-sensing materials. The prototype and concept of the Pd/MOF/PMMA nanocomposite-based hydrogen sensor. (C) Mechanism of PT detection. (D) EIS Nyquist plot of different SPE-modified layers. Reprinted from ref. 62 with permission of ELSEVIER, Copyright 2021. | ||
To achieve the reliable detection of ATP, Wang et al. developed the methylene violet (MV)-encapsulated zeolitic imidazolate framework-90 (ZIF-90) (MV@ZIF-90).152 Due to the strong coordination between ATP and Zn(II), MV@ZIF-90 could be disintegrated by ATP. The released MV with the recovery of the fluorescence emission had a sensitization effect on the ZnIn2S4 photoanode. Photoelectrochemical and fluorescence-involved dual-mode bioanalysis permits cross-checking, which was positively correlated with the ATP concentration. The proposed sensing system exhibited good performance in terms of low detection limit, wide linear range, and simple operation. Therefore, this study will inspire the construction and implementation of smart MOF materials for dual-mode bioanalysis, especially for high feasibility in real sample detection.
Traditional methods for patulin detection have many drawbacks, such as high operational costs, time-consuming procedures, and sophisticated instrumentation.158,159 To overcome these issues, Selvam et al. developed a SeS2@Co MOF and Au@PANI-containing sensor platform utilized in the molecularly-imprinted polymer (MIP)-based patulin sensor.62 As depicted in Fig. 8(C), SeS2@Co MOF and Au@PANI were synthesized by hydrothermal synthesis and interfacial polymerization strategies, combining them with MIP and SPE to form the electrochemical sensor platform. Patulin can be adsorbed on p-aminobenzoic acid (PABA) through strong H-bond interactions, thus enabling targeted detection of patulin. The high charge transfer resistance of the bare SPE is greatly reduced by the modification of Co-MOF@SeS2 and then assembled with Au@PANI. Eventually, patulin-solubilized MIP/Au@PANI/SeS2@Co-MOF/SPE displayed excellent conductivity, indicating the fabrication of MIP/Au@PANI/SeS2@Co-MOF/SPE (Fig. 8(D)). This novel sensor platform has good detection sensitivity because of the variation in the DPV current response (ΔI), and the logarithmic patulin concentration has a high linear coefficient. The sensor possessing high selectivity can specifically detect the target substance patulin, and other interfering substances have little effect on the results. Thus, the electrochemical sensor is a promising platform for the detection of patulin. To guarantee the sensitivity, Zhou et al. designed and synthesized COF-on-MOF integrated nanohybrid compound (Ce-MOF@MCA) as an electrochemical aptamer sensor for the detection of oxytetracycline (OTC).153 OTC targeting aptamer anchored to the surface of the Ce-MOF@MCA, G-quadruplex structure was formed when OTC was present, and the resulting conformational change would cause spatial site resistance. The charge-transfer resistance value increases with the increase of OTC concentration, and the OTC content was calculated from the electrochemical impedance spectroscopy calculations. The method has a detection limit of 17.4 fg for OTC, which is lower than that of any other currently reported methods, with a relative standard deviation (RSD) of around 1.16%. Overall, Ce-MOF@MCA has great promise for practical antibiotic detection.
To avoid the inactivation due to carbon monoxide (CO) adsorption, a palladium-NC film-MOF-polymer hybrid (denoted as Pd/MOF/polymer) was constructed for H2 detection (Fig. 9(A)).160 As depicted in Fig. 9(B), the dissociation energy of Pd/ZIF-67/PMMA is small, which means that when ZIF-67 covers PD-NCS, the hydrogen dissociation rate is high, indicating that the adverse effect of the PMMA interface on the hydrogen dissociation rate has been eliminated. The addition of ZIF-67 significantly improved the sensing dynamics and sensitivity, Pd/ZIF-67/PMMA and Pd/ZIF-67 showed higher response amplitude and sensitivity than Pd/PMMA and Pd, respectively (Fig. 9(C)). Overall, the experimental results and theoretical calculations revealed the strengthening mechanism of the ZIF-67 coating on the Pd–NC film in a faster and more sensitive direction.
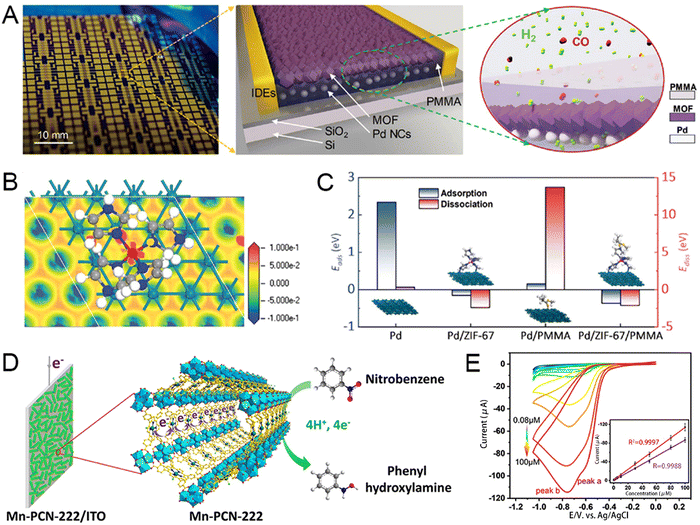 | ||
| Fig. 9 (A) Concept and theoretical calculation of the hydrogen-sensing materials. The prototype and concept of the Pd/MOF/PMMA nanocomposite-based hydrogen sensor. (B) The structure and charge distribution of Pd/ZIF-67 were calculated using the electron-density difference. (C) Calculated adsorption and dissociation energies for Pd, Pd/ZIF-67, Pd/PMMA, and Pd/ZIF-67/PMMA. Reprinted from ref. 146 with permission of American Chemical Society, Copyright 2021. (D) Description of the plausible mechanism of NB reduction on the Mn-PCN-222/ITO electrode. (E) CV curves of the Mn-PCN-222/ITO electrode towards the detection of NB and 1,3-DNB at different concentrations; the inset shows a linear trend of the peak current versus the concentration of corresponding analytes. Reprinted from ref. 63 with permission of Wiley, Copyright 2021. | ||
To enhance sensitivity, Xu et al. proposed a core–shell structure of Cu2O@ZIF-67 expected to improve the electrical conductivity and amplify the voltammetric current through their intense electron interaction.39 Cu2+ and Co2+ have a synergistic effect, and both lose electrons to higher states, and the increase of Co3+ is beneficial to glucose oxidation. The as-designed Cu2O@ZIF-67 showed a wider liner range (0.01–10 mM) and lower detection limit (6.5 μM), offering a hopeful platform for the highly-efficient determination of glucose. Given the porphyrins' selective redox catalytic properties, Zhou et al. developed an electrode of conductive indium tin oxide (ITO) surface depositing the porphyritic MOF to enable high currents and highly sensitive analysis.63 The porous coordination network-222 (PCN-222) is a prototypical porphyritic MOF constructed from a hexanuclear Zr cluster and tetrakis(4-carboxyphenyl)-porphyrin (TCPP) ligand. The Mn-PCN-222/ITO electrode was made by liquid-phase deposition of Mn-PCN-222 (manganese metalloporphyrin) films on the conductive indium tin oxide (ITO) glass using a modular assembly technique. Nitrobenzene (NB), chosen as a model toxin, can be directly reduced to benzylamine with four electron and proton transfers in a neutral medium, resulting in an electrochemical signal (Fig. 9(D)). The peak potentials are enhanced with increasing concentrations of NB, showing the feasibility of Mn-PCN-222/ITO for NB detection. The novel electrode is universal, and similar results are obtained for other nitroaromatic compounds (NACs). The novel electrode can also identify the number of nitro groups in a single NAC analyte, confirming the presence of two reduction peaks in detecting 1,3-DNB (Fig. 9(E)). Mn-PCN-222/ITO is a promising electrochemical sensor because of its simple preparation method and great voltammetric detection performance.
For simultaneous multicomponent detection, Huang et al. put forward a Ce-MOF/carbon nanotube composite for the simultaneous discrimination of hydroquinone (HQ) and catechol (CC).147 CeMOF without NaOH/H2O2 treatment shows a typical straw-sheaf structure according to scanning electron microscopy. The morphology was also maintained after adding carbon nanotubes. For the Ce-MOF/carbon nanocomposites, the presence of carbon nanotube dramatically decreases the aggregation of Ce-MOF granules, leading to ideal dispersity and stability. Besides, the post-treatment facilitated the formation of the rod-like structure into particles. The oxidation peak current increases were proportional to the increase in the concentrations of hydroquinone and catechol. The presence of two substances does not interfere with the corresponding detection. The addition of commonly present interferences does not influence the current response, exhibiting excellent selectivity in the simultaneous detection of catechol and hydroquinone. To verify the reproducibility, five Ce-MOF/carbon nanotube/glassy carbon electrodes were fabricated and used to measure catechol and hydroquinone with a concentration of 20 μM. Overall, the Ce-MOF/carbon nanotube composite is a promising electrochemical sensor for discriminating between hydroquinone and catechol.
To fulfill multi-analyte detection, Yang et al. prepared a wearable body fluid sensor for detecting vitamin C.154 The electronic sensor is composed of a nanocellulose film, two working electrode films, and an isolation film. Separately, the nanocellulose film is highly breathable and can easily bind to the active substance, while the isolation layer can avoid direct contact between the circuit and sweat. Ag/AgCl paste was chosen as the pseudo-reference electrode, the working electrode was prepared from a conductive MOF (Ni3HHTP2) doped Au counter electrode. The sensor stayed in place on the sweat-prone head, demonstrating extreme skin adhesion even during exercise (Fig. 10(A)). During detection, the analog-to-digital converter (ADC) and the digital-to-analog converter (DAC) will read and process the data obtained from the sensor. Since the output current is linearly related to the concentration of the detector, the concentration of vitamin C is easily monitored. Finally, the real-time vitamin C concentration can be displayed directly on the cell phone via Bluetooth transmission (Fig. 10(B)). The results obtained by MOF-based sensor detection of vitamin C were only 13.4% in error with those obtained by conventional HPLC, indicating high accuracy. A significant increase in vitamin concentration in body fluids can be seen after three hours of vitamin C intake (Fig. 10(C)). The device can detect vitamin levels in the body at any time, and this detection is non-invasive and highly compliant (Fig. 10(D)). Overall, this MOF-based electrochemical sensing holds great promise for practical detection in the medical field.
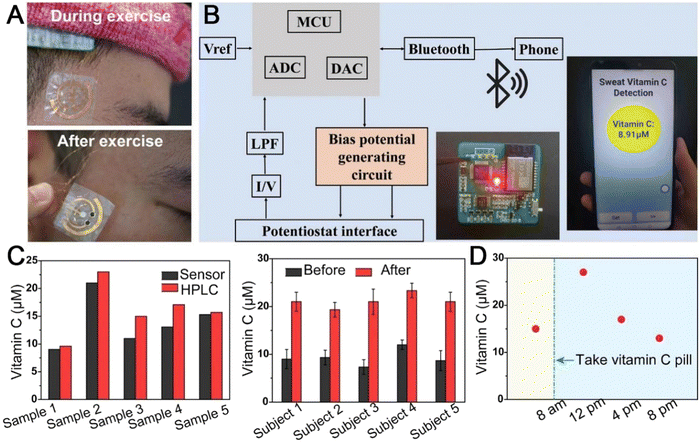 | ||
| Fig. 10 Demonstration of the Ni3HHTP2-based wearable nutrition tracker for wireless sweat analysis. (A) Photograph of the sensor during and after aerobic exercise, respectively. (B) Illustration of the wireless transmission components containing signal transduction, conditioning, processing, and wireless transmission from the sensors to a mobile phone. (C) Comparison of the vitamin C concentrations of five sweat samples detected using the MOF-based sweat sensor and HPLC. Comparison of the sweat vitamin C levels of five subjects before and 3 h after vitamin C intake (1000 mg). (D) a volunteer was measured before 8 a.m. prior to vitamin C consumption, and the vitamin C levels were then measured every four hours. Reprinted from ref. 154 with permission of Wiley, Copyright 2022. | ||
3. Conclusion
Huge pioneering works indicate that MOFs are excellent candidate materials for advanced bioanalytical applications, which have markedly exhibited great potential in various sensing scenarios, such as environmental contaminants, explosive substances, food safety, biomarkers, and corresponding medical diagnostics. Elaborate blends of MOFs with various transduction architectures and unrevealed benefits have greatly expanded the sensing performance and counteracted operational costs, time-consuming procedures, and sophisticated instrumentation. Despite the boundless potential, the sensing act of the MOF-derived composites is yet to be considered in the aspects of industrial production and artificial intelligence: (1) the role of MOFs in each sensing platform should be investigated in-depth for rational device design and performance optimization; (2) large-scale manufacture of MOF should be understood by improving integration progression and synthesis machinery to broaden their commercialization; (3) the refined structural composition, as well as spatial information, provides reliable insight for corresponding mechanistic investigation and robust models for theoretical calculations. Rigorous high-throughput theoretical modeling coupled with the analytical assessment of structure–property correlations via in situ microscopic and spectroscopic skills can be priceless. Machine learning-derived radiomics signatures could easily obtain MOF-derived sensors possessing irregularity detection, self-calibration, and lifetime expectation. This reported photoelectrochemical switching process was successfully integrated into an advanced analytical device with an accompanying logic gate for the detection of biomarkers. In the future, we firmly believe that these currently known developments will accelerate the expansion of MOF sensors, and extensive research will lead to better performance, applicability, and tangible commercialization. Furthermore, we believe that future biosensor technologies will inevitably require using artificial intelligence algorithms to process the vast amounts of information generated.Author contributions
YS: writing – original draft. YZ: draft revision and picture processing. M. S. K: draft revising. MZ: writing part of the original draft and copyright permission. LY: review and support the foundation. WW: conceptualization and draft revision. XZ: review and support the foundation. ZX: supervision and foundation.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors acknowledge grants from the Natural Science Foundation of China (Grant no. 31900990, 52103169, and 51973214), and the Natural Science Foundations of Nantong City (Grant no. MSZ20088). The Large Instruments Open Foundation of Nantong University (Grant no. KFJN2269 and KFJN2271) and the College Students’Innovation and Entrepreneurship Training Program of Nantong University (Grant no. 2022239) are gratefully acknowledged.References
- X. Qi, K. Liu and Z. Chang, Chem. Eng. J., 2022, 441, 135953 CrossRef CAS.
- R. Matheu, E. Gutierrez-Puebla, M. A. Monge, C. S. Diercks, J. Kang, M. S. Prevot, X. Pei, N. Hanikel, B. Zhang, P. Yang and O. M. Yaghi, J. Am. Chem. Soc., 2019, 141, 17081–17085 CrossRef CAS PubMed.
- H. Daglar, H. C. Gulbalkan, G. Avci, G. O. Aksu, O. F. Altundal, C. Altintas, I. Erucar and S. Keskin, Angew. Chem., Int. Ed., 2021, 60, 7828–7837 CrossRef CAS PubMed.
- Q. Qian, P. A. Asinger, M. J. Lee, G. Han, K. Mizrahi Rodriguez, S. Lin, F. M. Benedetti, A. X. Wu, W. S. Chi and Z. P. Smith, Chem. Rev., 2020, 120, 8161–8266 CrossRef CAS PubMed.
- X. Xu, D. Nikolaeva, Y. Hartanto and P. Luis, Sep. Purif. Technol., 2021, 278, 119233 CrossRef.
- Z. Li, M. Song, W. Zhu, W. Zhuang, X. Du and L. Tian, Coord. Chem. Rev., 2021, 439, 213946 CrossRef CAS.
- M. O. Cichocka, Z. Liang, D. Feng, S. Back, S. Siahrostami, X. Wang, L. Samperisi, Y. Sun, H. Xu, N. Hedin, H. Zheng, X. Zou, H. C. Zhou and Z. Huang, J. Am. Chem. Soc., 2020, 142, 15386–15395 CrossRef CAS PubMed.
- C. Zhang, D. Lei, C. Xie, X. Hang, C. He and H. L. Jiang, Adv. Mater., 2021, 33, e2106308 CrossRef PubMed.
- B. Jie, H. Lin, Y. Zhai, J. Ye, D. Zhang, Y. Xie, X. Zhang and Y. Yang, Chem. Eng. J., 2023, 454, 139931 CrossRef CAS.
- S. Mallakpour, E. Nikkhoo and C. M. Hussain, Coord. Chem. Rev., 2022, 451, 214262 CrossRef CAS.
- Y. Kang, C. Xu, L. Meng, X. Dong, M. Qi and D. Jiang, Bioact. Mater., 2022, 18, 26–41 CrossRef CAS PubMed.
- H. Peng, X. Zhang, P. Yang, J. Zhao, W. Zhang, N. Feng, W. Yang and J. Tang, Bioact. Mater., 2023, 19, 1–11 CrossRef CAS PubMed.
- J. Liu, T. Liu, P. Du, L. Zhang and J. Lei, Angew. Chem., Int. Ed., 2019, 58, 7808–7812 CrossRef CAS PubMed.
- Y. Wang, M. Chao, P. Wan and L. Zhang, Nano Energy, 2020, 70, 104560 CrossRef CAS.
- H. Guo, S. Yi, K. Feng, Y. Xia, X. Qu, F. Wan, L. Chen and C. Zhang, Chem. Eng. J., 2021, 403, 119233 Search PubMed.
- N. A. Mohamed, H. Abou-Saleh, Y. Kameno, I. Marei, G. de Nucci, B. Ahmetaj-Shala, F. Shala, N. S. Kirkby, L. Jennings, D. E. Al-Ansari, R. P. Davies, P. D. Lickiss and J. A. Mitchell, Sci. Rep., 2021, 11, 4336 CrossRef CAS PubMed.
- X. Fang, B. Zong and S. Mao, Nano-Micro Lett., 2018, 10, 64 CrossRef PubMed.
- X. Tang, C. Zhou, W. Xia, Y. Liang, Y. Zeng, X. Zhao, W. Xiong, M. Cheng and Z. Wang, Chem. Eng. J., 2022, 446, 137299 CrossRef CAS.
- L. Li, J. Zou, Y. Han, Z. Liao, P. Lu, A. Nezamzadeh-Ejhieh, J. Liu and Y. Peng, New J. Chem., 2022, 46, 19577–19592 RSC.
- X. Zhao, S. Liu, Z. Tang, H. Niu, Y. Cai, W. Meng, F. Wu and J. P. Giesy, Sci. Rep., 2015, 5, 11849 CrossRef PubMed.
- M. Liu, Y. Gao, Y. Wang, Y. Li and D. Zou, Mater. Today Chem., 2022, 26, 101209 CrossRef CAS.
- H. Yang, Z. Sun, X. Qin, H. Wu, H. Zhang and G. Liu, Food Chem., 2021, 376, 131906 CrossRef PubMed.
- C. Li, W. Yang, X. Zhang, Y. Han, W. Tang, T. Yue and Z. Li, J. Mater. Chem. C, 2020, 8, 2054–2064 RSC.
- W. Cheng, X. Tang, Y. Zhang, D. Wu and W. Yang, Trends Food Sci. Technol., 2021, 112, 268–282 CrossRef CAS.
- J. Wang, D. Li, Y. Ye, Y. Qiu, J. Liu, L. Huang, B. Liang and B. Chen, Adv. Mater., 2021, 33, e2008020 CrossRef PubMed.
- L. Li, Y. Chen, L. Yang, Z. Wang and H. Liu, Coord. Chem. Rev., 2020, 411, 213235 CrossRef CAS.
- C. Wang, Y. Luo, X. Liu, Z. Cui, Y. Zheng, Y. Liang, Z. Li, S. Zhu, J. Lei, X. Feng and S. Wu, Bioact. Mater., 2022, 13, 200–211 CrossRef CAS PubMed.
- S. Liang, X. Xiao, L. Bai, B. Liu, M. Yuan, P. Ma, M. Pang, Z. Cheng and J. Lin, Adv. Mater., 2021, 33, e2100333 CrossRef PubMed.
- Z. Fan, H. Liu, Y. Xue, J. Lin, Y. Fu, Z. Xia, D. Pan, J. Zhang, K. Qiao, Z. Zhang and Y. Liao, Bioact. Mater., 2021, 6, 312–325 CrossRef CAS PubMed.
- L. Huang, Y. Zhu, C. Xu, Y. Cai, Y. Yi, K. Li, X. Ren, D. Jiang, Y. Ge, X. Liu, W. Sun, Q. Zhang and Y. Wang, ACS Sens., 2022, 7, 1439–1450 CrossRef CAS PubMed.
- N. Singh, J. Kim, J. Kim, K. Lee, Z. Zunbul, I. Lee, E. Kim, S. G. Chi and J. S. Kim, Bioact. Mater., 2023, 21, 358–380 CrossRef PubMed.
- M. Xue, S. Ye, X. Ma, F. Ye, C. Wang, L. Zhu, Y. Yang and J. Chen, J. Am. Chem. Soc., 2022, 144, 20278–20287 CrossRef CAS PubMed.
- T. Leelasree, V. Selamneni, T. Akshaya, P. Sahatiya and H. Aggarwal, J. Mater. Chem. B, 2020, 8, 10182–10189 RSC.
- Y. Cheng, S. J. Datta, S. Zhou, J. Jia, O. Shekhah and M. Eddaoudi, Chem. Soc. Rev., 2022, 51, 8300–8350 RSC.
- A. K. Ipadeola, K. Eid, A. M. Abdullah, R. S. Al-Hajri and K. I. Ozoemena, Nanoscale Adv., 2022, 4, 5044–5055 RSC.
- Q. Liu, J. Chen, F. Yu, J. Wu, Z. Liu and B. Peng, New J. Chem., 2021, 45, 16714–16721 RSC.
- M. Chen, N. Gan, Y. Zhou, T. Li, Q. Xu, Y. Cao and Y. Chen, Sens. Actuators, B, 2017, 242, 1201–1209 CrossRef CAS.
- L. Diestel, N. Wang, B. Schwiedland, F. Steinbach, U. Giese and J. Caro, J. Membr. Sci., 2015, 492, 181–186 CrossRef CAS.
- J. Guo, Y. Qin, Y. Zhu, X. Zhang, C. Long, M. Zhao and Z. Tang, Chem. Soc. Rev., 2021, 50, 5366–5396 RSC.
- E. H. Kwon, M. Kim, C. Y. Lee, M. Kim and Y. D. Park, ACS Appl. Mater. Interfaces, 2022, 14, 10637–10647 CrossRef CAS PubMed.
- X. Zha, W. Yang, L. Shi, Y. Li, Q. Zeng, J. Xu and Y. Yang, ACS Appl. Mater. Interfaces, 2022, 14, 37843–37852 CrossRef CAS PubMed.
- P. Qin, B. A. Day, S. Okur, C. Li, A. Chandresh, C. E. Wilmer and L. Heinke, ACS Sens., 2022, 7, 1666–1675 CrossRef CAS PubMed.
- J. N. Hao, D. Niu, J. Gu, S. Lin, Y. Li and J. Shi, Adv. Mater., 2020, 32, e2000791 CrossRef PubMed.
- T. Bao, Y. Zou, C. Zhang, C. Yu and C. Liu, Angew. Chem., Int. Ed., 2022, 61, e202209433 CAS.
- Z. Gao, S. Yang, B. Xu, T. Zhang, S. Chen, W. Zhang, X. Sun, Z. Wang, X. Wang, X. Meng and Y. S. Zhao, Angew. Chem., Int. Ed., 2021, 60, 24519–24525 CrossRef CAS PubMed.
- L. Jiao, J. Y. R. Seow, W. S. Skinner, Z. U. Wang and H.-L. Jiang, Mater. Today, 2019, 27, 43–68 CrossRef CAS.
- A. Nishijima, Y. Kametani and T. Uemura, Coord. Chem. Rev., 2022, 466, 214601 CrossRef CAS.
- X. Zhang, Z. Chen, X. Liu, S. L. Hanna, X. Wang, R. Taheri-Ledari, A. Maleki, P. Li and O. K. Farha, Chem. Soc. Rev., 2020, 49, 7406–7427 RSC.
- T. He, X. J. Kong, Z. X. Bian, Y. Z. Zhang, G. R. Si, L. H. Xie, X. Q. Wu, H. Huang, Z. Chang, X. H. Bu, M. J. Zaworotko, Z. R. Nie and J. R. Li, Nat. Mater., 2022, 21, 689–695 CrossRef CAS PubMed.
- J. Shen, X. He, T. Ke, R. Krishna, J. M. van Baten, R. Chen, Z. Bao, H. Xing, M. Dinca, Z. Zhang, Q. Yang and Q. Ren, Nat. Commun., 2020, 11, 6259 CrossRef CAS PubMed.
- M. S. Zavakhina, D. G. Samsonenko, D. N. Dybtsev and V. P. Fedin, Polyhedron, 2019, 162, 311–315 CrossRef CAS.
- H. Wang, X. Dong, J. Lin, S. J. Teat, S. Jensen, J. Cure, E. V. Alexandrov, Q. Xia, K. Tan, Q. Wang, D. H. Olson, D. M. Proserpio, Y. J. Chabal, T. Thonhauser, J. Sun, Y. Han and J. Li, Nat. Commun., 2018, 9, 1745 CrossRef PubMed.
- G. Cai, X. Ma, M. Kassymova, K. Sun, M. Ding and H. L. Jiang, ACS Cent. Sci., 2021, 7, 1434–1440 CrossRef CAS PubMed.
- H. Yan, H. Ni, J. Jia, C. Shan, T. Zhang, Y. Gong, X. Li, J. Cao, W. Wu, W. Liu and Y. Tang, Anal. Chem., 2019, 91, 5225–5234 CrossRef CAS PubMed.
- S. Dissegna, K. Epp, W. R. Heinz, G. Kieslich and R. A. Fischer, Adv. Mater., 2018, 30, e1704501 CrossRef PubMed.
- J. Yang, X. Yin, L. Zhang, X. Zhang, Y. Lin, L. Zhuang, W. Liu, R. Zhang, X. Yan, L. Shi, W. Di, L. Feng, Y. Jia, J. Wang, K. Qian and X. Yao, Adv. Mater., 2022, e2201422, DOI:10.1002/adma.202201422.
- Y. Yang, W. Yan, X. Wang, L. Yu, J. Zhang, B. Bai, C. Guo and S. Fan, Biosens. Bioelectron., 2021, 177, 113000 CrossRef CAS PubMed.
- J. Ren, M. Ledwaba, N. M. Musyoka, H. W. Langmi, M. Mathe, S. Liao and W. Pang, Coord. Chem. Rev., 2017, 349, 169–197 CrossRef CAS.
- X. Wang, H. Fan, P. Shen, Y. Yao, Y. Chen, S. Lu, B. Teng and X. Liao, Microporous Mesoporous Mater., 2021, 327, 114402 Search PubMed.
- Y. Wang, D. Zhang, Y. Zeng and P. Qi, Biosens. Bioelectron., 2022, 198, 113817 CrossRef CAS PubMed.
- Y. C. Kao, X. Han, Y. H. Lee, H. K. Lee, G. C. Phan-Quang, C. L. Lay, H. Y. F. Sim, V. J. X. Phua, L. S. Ng, C. W. Ku, T. C. Tan, I. Y. Phang, N. S. Tan and X. Y. Ling, ACS Nano, 2020, 14, 2542–2552 CrossRef CAS PubMed.
- S. P. Selvam, A. N. Kadam, K. R. Maiyelvaganan, M. Prakash and S. Cho, Biosens. Bioelectron., 2021, 187, 113302 CrossRef CAS PubMed.
- Z. Zhou, S. Mukherjee, S. Hou, W. Li, M. Elsner and R. A. Fischer, Angew. Chem., Int. Ed., 2021, 60, 20551–20557 CrossRef CAS PubMed.
- Z. Wei, D. Chen, Z. Guo, P. Jia and H. Xing, Inorg. Chem., 2020, 59, 5386–5393 CrossRef CAS PubMed.
- R. Gui, H. Jin, X. Bu, Y. Fu, Z. Wang and Q. Liu, Coord. Chem. Rev., 2019, 383, 82–103 CrossRef CAS.
- Y. Liu, H. Zhang, B. Li, J. Liu, D. Jiang, B. Liu and N. Sojic, J. Am. Chem. Soc., 2021, 143, 17910–17914 CrossRef CAS PubMed.
- Y. Zhao, H. Zeng, X. W. Zhu, W. Lu and D. Li, Chem. Soc. Rev., 2021, 50, 4484–4513 RSC.
- M. Daniel, G. Mathew, M. Anpo and B. Neppolian, Coord. Chem. Rev., 2022, 468, 214627 CrossRef CAS.
- Y. Zhang, S. Yuan, G. Day, X. Wang, X. Yang and H.-C. Zhou, Coord. Chem. Rev., 2018, 354, 28–45 CrossRef CAS.
- T. Rasheed and F. Nabeel, Coord. Chem. Rev., 2019, 401, 213065 CrossRef CAS.
- L. Chen, J. W. Ye, H. P. Wang, M. Pan, S. Y. Yin, Z. W. Wei, L. Y. Zhang, K. Wu, Y. N. Fan and C. Y. Su, Nat. Commun., 2017, 8, 15985 CrossRef CAS PubMed.
- K. He, Z. Li, L. Wang, Y. Fu, H. Quan, Y. Li, X. Wang, S. Gunasekaran and X. Xu, ACS Appl. Mater. Interfaces, 2019, 11, 26250–26260 CrossRef CAS PubMed.
- Z. Sun, S. Wu, Y. Peng, M. Wang, M. Jalalah, M. S. Al-Assiri, F. A. Harraz, J. Yang and G. Li, Chem. Eng. J., 2021, 405, 126707 CrossRef CAS.
- L. Pan, X. Li, Q. Zhang, S. Xu, L. Yang, F. Yang and C. Jiang, Chem. Eng. J., 2022, 450, 138283 CrossRef CAS.
- H. Q. Yin and X. B. Yin, Acc. Chem. Res., 2020, 53, 485–495 CrossRef CAS PubMed.
- Y.-H. Wang, Z.-S. Shao, C. Cheng, J.-L. Wang, Z. Song, W.-J. Song, F. Zheng and H.-S. Wang, Chem. Eng. J., 2022, 437, 135296 CrossRef CAS.
- Y. Zhao, K. Mukherjee, K. D. Benkstein, L. Sun, K. L. Steffens, C. B. Montgomery, S. Guo, S. Semancik and M. E. Zaghloul, Nanoscale, 2019, 11, 11922–11932 RSC.
- E. Miliutina, O. Guselnikova, V. Burtsev, R. Elashnikov, P. Postnikov, V. Svorcik and O. Lyutakov, Talanta, 2020, 208, 120480 CrossRef CAS PubMed.
- J. Li, Z. Liu, D. Tian, B. Li, L. Shao and Z. Lou, Nanoscale, 2022, 14, 5561–5568 RSC.
- Z. W. Jiang, T. T. Zhao, C. M. Li, Y. F. Li and C. Z. Huang, ACS Appl. Mater. Interfaces, 2021, 13, 49754–49761 CrossRef CAS PubMed.
- Q. Wei, C. Wang, X. Zhou, T. Wu, Y. Wang, C. Li and N. Yang, Biosens. Bioelectron., 2019, 142, 111540 CrossRef CAS PubMed.
- H. Dong, S. Liu, Q. Liu, Y. Li, Y. Li and Z. Zhao, Biosens. Bioelectron., 2022, 195, 113648 CrossRef CAS PubMed.
- L. Dai, Y. Li, Y. Wang, X. Luo, D. Wei, R. Feng, T. Yan, X. Ren, B. Du and Q. Wei, Biosens. Bioelectron., 2019, 132, 97–104 CrossRef CAS PubMed.
- Y. Chen, X. Z. Meng, H. W. Gu, H. C. Yi and W. Y. Sun, Biosens. Bioelectron., 2019, 134, 117–122 CrossRef CAS PubMed.
- J. Dong, D. Zhao, Y. Lu and W.-Y. Sun, J. Mater. Chem. A, 2019, 7, 22744–22767 RSC.
- W. R. Algar, M. Massey, K. Rees, R. Higgins, K. D. Krause, G. H. Darwish, W. J. Peveler, Z. Xiao, H. Y. Tsai, R. Gupta, K. Lix, M. V. Tran and H. Kim, Chem. Rev., 2021, 121, 9243–9358 CrossRef CAS PubMed.
- Z. Liu, W. Qi and G. Xu, Chem. Soc. Rev., 2015, 44, 3117–3142 RSC.
- Y. Su, J. R. Walker, Y. Park, T. P. Smith, L. X. Liu, M. P. Hall, L. Labanieh, R. Hurst, D. C. Wang, L. P. Encell, N. Kim, F. Zhang, M. A. Kay, K. M. Casey, R. G. Majzner, J. R. Cochran, C. L. Mackall, T. A. Kirkland and M. Z. Lin, Nat. Methods, 2020, 17, 852–860 CrossRef CAS PubMed.
- C. Zhu, J. Xin, J. Li, H. Li, X. Kang, Y. Pei and M. Zhu, Angew. Chem., Int. Ed., 2022, 61, e202205947 CAS.
- B. Gui, Y. Meng, Y. Xie, J. Tian, G. Yu, W. Zeng, G. Zhang, S. Gong, C. Yang, D. Zhang and C. Wang, Adv. Mater., 2018, 30, 1802329 CrossRef PubMed.
- X. Sun, Y. Wang, L. Zhang, S. Liu, M. Zhang, J. Wang, B. Ning, Y. Peng, J. He, Y. Hu and Z. Gao, Anal. Chem., 2020, 92, 3032–3041 CrossRef CAS PubMed.
- T. Jin, Y. Li, W. Jing, Y. Li, L. Fan and X. Li, Chem. Commun., 2020, 56, 659–662 RSC.
- J. Su, J. Du, R. Ge, C. Sun, Y. Qiao, W. Wei, X. Pang, Y. Zhang, H. Lu and H. Dong, Anal. Chem., 2022, 94, 13108–13116 CrossRef CAS PubMed.
- R. Gao, N. Ye, X. Kou, Y. Shen, H. Yang, T. Wu, S. Huang, G. Chen and G. Ouyang, Chem. Commun., 2022, 58, 12720–12723 RSC.
- B. Zhu, L. Zhu, T. Hou, K. Ren, K. Kang, C. Xiao and J. Luo, Anal. Chem., 2022, 94, 3744–3748 CrossRef CAS PubMed.
- H. Q. Yin, K. Tan, S. Jensen, S. J. Teat, S. Ullah, X. Hei, E. Velasco, K. Oyekan, N. Meyer, X.-Y. Wang, T. Thonhauser, X.-B. Yin and J. Li, Chem. Sci., 2021, 12, 14189–14197 RSC.
- X. Zhang, Q. Ma, X. Liu, H. Niu, L. Luo, R. Li and X. Feng, Food Chem., 2022, 382, 132379 CrossRef CAS PubMed.
- J.-N. Hao and B. Yan, Adv. Funct. Mater., 2017, 27, 1603856 CrossRef.
- X. Ling, D. Gong, W. Shi, Z. Xu, W. Han, G. Lan, Y. Li, W. Qin and W. Lin, J. Am. Chem. Soc., 2021, 143, 1284–1289 CrossRef CAS PubMed.
- H. Q. Yin, X. Y. Wang and X. B. Yin, J. Am. Chem. Soc., 2019, 141, 15166–15173 CrossRef CAS PubMed.
- X. Y. Wang, H. Q. Yin and X. B. Yin, ACS Appl. Mater. Interfaces, 2020, 12, 20973–20981 CrossRef CAS PubMed.
- N. Zhong, R. Gao, Y. Shen, X. Kou, J. Wu, S. Huang, G. Chen and G. Ouyang, Anal. Chem., 2022, 94, 14385–14393 CrossRef CAS PubMed.
- S. Y. Zhu and B. Yan, Dalton Trans., 2018, 47, 11586–11592 RSC.
- S. Pal, Y. Z. Su, Y. W. Chen, C. H. Yu, C. W. Kung and S. S. Yu, ACS Appl. Mater. Interfaces, 2022, 14, 28247–28257 CrossRef CAS PubMed.
- L. Hou, Y. Qin, J. Li, S. Qin, Y. Huang, T. Lin, L. Guo, F. Ye and S. Zhao, Biosens. Bioelectron., 2019, 143, 111605 CrossRef CAS PubMed.
- Y. Zeng, M. Wang, Z. Sun, L. Sha, J. Yang and G. Li, J. Mater. Chem. B, 2022, 10, 450–455 RSC.
- Y. Zhang, H. Lu and B. Yan, Sens. Actuators, B, 2021, 349, 130736 CrossRef CAS.
- X. Zhang, Y. Zhou, X. Huang, X. Hu, X. Huang, L. Yin, Q. Huang, Y. Wen, B. Li, J. Shi and X. Zou, Food Chem., 2023, 407, 135115 CrossRef CAS PubMed.
- C. Y. Wang, C. C. Wang, X. W. Zhang, X. Y. Ren, B. Yu, P. Wang, Z. X. Zhao and H. Fu, Chin. Chem. Lett., 2022, 33, 1353–1357 CrossRef CAS.
- H. S. Wang, J. Li, J.-Y. Li, K. Wang, Y. Ding and X.-H. Xia, NPG Asia Mater., 2017, 9, e354–e354 CrossRef CAS.
- W. P. Lustig, S. Mukherjee, N. D. Rudd, A. V. Desai, J. Li and S. K. Ghosh, Chem. Soc. Rev., 2017, 46, 3242–3285 RSC.
- S. Wang, B. Sun, Z. Su, G. Hong, X. Li, Y. Liu, Q. Pan and J. Sun, Inorg. Chem. Front., 2022, 9, 3259–3266 RSC.
- D. Zhao, W. Li, R. Wen, N. Lei, W. Li, X. Liu, X. Zhang and L. Fan, Inorg. Chem., 2023, 62, 2715–2725 CrossRef CAS PubMed.
- J. Li, J. L. Wang, W. L. Zhang, Z. Tu, X. F. Cai, Y. W. Wang, C. Y. Gan, H. J. Deng, J. Cui, Z. C. Shu, Q. X. Long, J. Chen, N. Tang, X. Hu, A. L. Huang and J. L. Hu, Biosens. Bioelectron., 2022, 209, 114226 CrossRef CAS PubMed.
- I. Ortiz-Gomez, S. Gonzalez-Alfaro, A. Sanchez-Ruiz, I. de Orbe-Paya, L. F. Capitan-Vallvey, A. Navarro, A. Salinas-Castillo and J. C. Garcia-Martinez, ACS Sens., 2022, 7, 37–43 CrossRef CAS PubMed.
- H. Kurz, C. Hils, J. Timm, G. Horner, A. Greiner, R. Marschall, H. Schmalz and B. Weber, Angew. Chem., Int. Ed., 2022, 61, e202117570 CAS.
- S. Wang, H. Li, H. Huang, X. Cao, X. Chen and D. Cao, Chem. Soc. Rev., 2022, 51, 2031–2080 RSC.
- S. Song, Z. Liang, J. Zhang, L. Wang, G. Li and C. Fan, Angew. Chem., Int. Ed., 2009, 48, 8670–8674 CrossRef CAS PubMed.
- Y. Zhao, H. Zeng, K. Wu, D. Luo, X.-W. Zhu, W. Lu and D. Li, J. Mater. Chem. C, 2020, 8, 4385–4391 RSC.
- J. Li, H. Jia, X. Ren, Y. Li, L. Liu, R. Feng, H. Ma and Q. Wei, Small, 2022, 18, e2106567 CrossRef PubMed.
- X. Dong, Y. Du, G. Zhao, W. Cao, D. Fan, X. Kuang, Q. Wei and H. Ju, Biosens. Bioelectron., 2021, 192, 113505 CrossRef CAS PubMed.
- X. Gan, D. Han, J. Wang, P. Liu, X. Li, Q. Zheng and Y. Yan, Biosens. Bioelectron., 2021, 171, 112735 CrossRef CAS PubMed.
- Y. Wang, S. Zeng, A. Crunteanu, Z. Xie, G. Humbert, L. Ma, Y. Wei, A. Brunel, B. Bessette, J. C. Orlianges, F. Lalloue, O. G. Schmidt, N. Yu and H. P. Ho, Nano-Micro Lett., 2021, 13, 96 CrossRef CAS PubMed.
- A. Vazquez-Guardado, S. Barkam, M. Peppler, A. Biswas, W. Dennis, S. Das, S. Seal and D. Chanda, Nano Lett., 2019, 19, 449–454 CrossRef CAS PubMed.
- Z. Li, L. Leustean, F. Inci, M. Zheng, U. Demirci and S. Wang, Biotechnol. Adv., 2019, 37, 107440 CrossRef CAS PubMed.
- Y. Wang, Z. Mao, Q. Chen, K. Koh, X. Hu and H. Chen, Biosens. Bioelectron., 2022, 201, 113954 CrossRef CAS PubMed.
- O. Guselnikova, H. Lim, J. Na, M. Eguchi, H. J. Kim, R. Elashnikov, P. Postnikov, V. Svorcik, O. Semyonov, E. Miliutina, O. Lyutakov and Y. Yamauchi, Biosens. Bioelectron., 2021, 180, 113109 CrossRef CAS PubMed.
- X. Yang, Y. Liu, S. H. Lam, J. Wang, S. Wen, C. Yam, L. Shao and J. Wang, Nano Lett., 2021, 21, 8205–8212 CrossRef CAS PubMed.
- H. Sun, G. Song, W. Gong, W. Lu, S. Cong and Z. Zhao, Nano Res., 2022, 15, 5347–5354 CrossRef CAS.
- Y. Jiang, Y. Cai, S. Hu, X. Guo, Y. Ying, Y. Wen, Y. Wu and H. Yang, J. Innovative Opt. Health Sci., 2021, 14, 2141003 CrossRef CAS.
- C. Carrillo-Carrion, R. Martinez, M. F. Navarro Poupard, B. Pelaz, E. Polo, A. Arenas-Vivo, A. Olgiati, P. Taboada, M. G. Soliman, U. Catalan, S. Fernandez-Castillejo, R. Sola, W. J. Parak, P. Horcajada, R. A. Alvarez-Puebla and P. Del Pino, Angew. Chem., Int. Ed., 2019, 131, 7152–7156 CrossRef.
- X. Zhao, T. Yang, D. Wang, N. Zhang, H. Yang, X. Jing, R. Niu, Z. Yang, Y. Xie and L. Meng, Anal. Chem., 2022, 94, 4484–4494 CrossRef CAS PubMed.
- H. Sun, W. Gong, S. Cong, C. Liu, G. Song, W. Lu and Z. Zhao, ACS Appl. Mater. Interfaces, 2022, 14, 2326–2334 CrossRef CAS PubMed.
- J. Li, A. Wuethrich, Z. Zhang, J. Wang, L. L. Lin, A. Behren, Y. Wang and M. Trau, Anal. Chem., 2022, 94, 14573–14582 CrossRef CAS PubMed.
- O. Guselnikova, P. Postnikov, R. Elashnikov, E. Miliutina, V. Svorcik and O. Lyutakov, Anal. Chim. Acta, 2019, 1068, 70–79 CrossRef CAS PubMed.
- G. Su, L. Dang, G. Liu, T. Feng, W. Wang, C. Wang and H. Wei, Spectrochim. Acta, Part A, 2022, 270, 120818 CrossRef CAS PubMed.
- Z. Du, Y. Qi, J. He, D. Zhong and M. Zhou, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1672 CAS.
- H. Sun, G. Song, W. Gong, W. Lu, S. Cong and Z. Zhao, Nano Res., 2022, 15, 5347–5354 CrossRef CAS.
- Y. Jiang, Y. Cai, S. Hu, X. Guo, Y. Ying, Y. Wen, Y. Wu and H. Yang, J. Innovative Opt. Health Sci., 2021, 14, 2141003 CrossRef CAS.
- C. Tan, X. Cao, X. J. Wu, Q. He, J. Yang, X. Zhang, J. Chen, W. Zhao, S. Han, G. H. Nam, M. Sindoro and H. Zhang, Chem. Rev., 2017, 117, 6225–6331 CrossRef CAS PubMed.
- A. Dhakshinamoorthy, A. M. Asiri and H. Garcia, Angew. Chem., Int. Ed., 2016, 55, 5414–5445 CrossRef CAS PubMed.
- B. Hou, J. Ye, J. Li, Z. Xu and H. Yu, Nano Today, 2022, 47, 101661 CrossRef.
- G. Morad, B. A. Helmink, P. Sharma and J. A. Wargo, Cell, 2021, 184, 5309–5337 CrossRef CAS PubMed.
- X. Liu, T. Yue, K. Qi, Y. Qiu, B. Y. Xia and X. Guo, Chin. Chem. Lett., 2020, 31, 2189–2201 CrossRef CAS.
- L. T. Zhang, Y. Zhou and S. T. Han, Angew. Chem., Int. Ed., 2021, 60, 15192–15212 CrossRef CAS PubMed.
- Z. Li, S. Chang, H. Zhang, Y. Hu, Y. Huang, L. Au and S. Ren, Nano Lett., 2021, 21, 6983–6989 CrossRef CAS PubMed.
- H. Huang, Y. Chen, Z. Chen, J. Chen, Y. Hu and J. J. Zhu, J. Hazard. Mater., 2021, 416, 125895 CrossRef CAS PubMed.
- N. Yang, K. Guo, Y. Zhang and C. Xu, J. Mater. Chem. B, 2020, 8, 2856–2861 RSC.
- Y. Li, R. Wang, G. E. Wang, S. Feng, W. Shi, Y. Cheng, L. Shi, K. Fu and J. Sun, ACS Nano, 2022, 16, 473–484 CrossRef CAS PubMed.
- L. J. Small, S. E. Henkelis, D. X. Rademacher, M. E. Schindelholz, J. L. Krumhansl, D. J. Vogel and T. M. Nenoff, Adv. Funct. Mater., 2020, 30, 2006598 CrossRef CAS.
- Y. Zhong, X. Wang, R. Zha, C. Wang, H. Zhang, Y. Wang and C. Li, Nanoscale, 2021, 13, 19066–19075 RSC.
- L. Zhang, F. Z. Chen, H. Sun, R. Meng, Q. Zeng, X. Wang and H. Zhou, ACS Appl. Mater. Interfaces, 2022, 14, 46103–46111 CrossRef CAS PubMed.
- N. Zhou, Y. Ma, B. Hu, L. He, S. Wang, Z. Zhang and S. Lu, Biosens. Bioelectron., 2019, 127, 92–100 CrossRef CAS PubMed.
- X. Yang, J. Yi, T. Wang, Y. Feng, J. Wang, J. Yu, F. Zhang, Z. Jiang, Z. Lv, H. Li, T. Huang, D. Si, X. Wang, R. Cao and X. Chen, Adv. Mater., 2022, 34, e2201768 CrossRef PubMed.
- Y. Wang, Y. Qian, L. Zhang, Z. Zhang, S. Chen, J. Liu, X. He and Y. Tian, J. Am. Chem. Soc., 2023, 145, 2118–2126 CrossRef CAS PubMed.
- S. Demchyshyn, M. Verdi, L. Basirico, A. Ciavatti, B. Hailegnaw, D. Cavalcoli, M. C. Scharber, N. S. Sariciftci, M. Kaltenbrunner and B. Fraboni, Adv. Sci., 2020, 7, 2002586 CrossRef CAS PubMed.
- C. Liang, S. Zhang, L. Cheng, J. Xie, F. Zhai, Y. He, Y. Wang, Z. Chai and S. Wang, Angew. Chem., Int. Ed., 2020, 59, 11856–11860 CrossRef CAS PubMed.
- D. Moreno-Gonzalez, P. Jac, P. Riasova and L. Novakova, Food Chem., 2021, 334, 127607 CrossRef CAS PubMed.
- M. Zhang, Y. Wang, X. Sun, J. Bai, Y. Peng, B. Ning, Z. Gao and B. Liu, Anal. Chim. Acta, 2020, 1106, 161–167 CrossRef CAS PubMed.
- B. Xie, B. Ding, P. Mao, Y. Wang, Y. Liu, M. Chen, C. Zhou, H. M. Wen, S. Xia, M. Han, R. E. Palmer, G. Wang and J. Hu, Small, 2022, 18, e2200634 CrossRef PubMed.
Footnote |
| † YS and YZ contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |
