Open Access Article
Open Access ArticleHeteroatom-directed supramolecular helical-rich architectures in N-terminal protected pyridyl aromatic amino acids†
Thangavel
Vijayakanth
 a,
Bin
Xue
a,
Bin
Xue
 c,
Sarah
Guerin
c,
Sarah
Guerin
 d,
Sigal
Rencus-Lazar
d,
Sigal
Rencus-Lazar
 a,
Natalia
Fridman
e,
Damien
Thompson
a,
Natalia
Fridman
e,
Damien
Thompson
 d,
Yi
Cao
c and
Ehud
Gazit
d,
Yi
Cao
c and
Ehud
Gazit
 *ab
*ab
aShmunis School of Biomedicine and Cancer Research, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv – 6997801, Israel. E-mail: ehudg@post.tau.ac.il
bDepartment of Materials Science and Engineering, Iby and Aladar Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv – 6997801, Israel
cNational Laboratory of Solid-State Microstructure, Department of Physics, Nanjing University, Nanjing 210000, China
dDepartment of Physics, Bernal Institute, University of Limerick, Limerick V94 T9PX, Ireland
eSchulich Faculty of Chemistry, Technion-Israel Institute of Technology, Haifa – 32000, Israel
First published on 8th March 2023
Abstract
Supramolecular helical structures formed by the assembly of biological and bio-inspired building blocks (typically amino acids, peptides and proteins) are an intriguing class of materials with prospective applications in sustainable biomedical technologies and electronics. Specifically, short peptide or single amino acid building blocks can give rise to ideal materials candidates in terms of low cost, adjustability, and compatibility. Yet, to date, reliable helical topologies with specific handedness have been highly challenging to obtain. Herein, we present simple N-terminal protected aromatic pyridyl amino acids that display helicity at the molecular level confirmed by single-crystal X-ray diffraction analysis. The helical structure is stabilized by strong intermolecular hydrogen bonding between the pyridyl nitrogen and carboxylic acid groups. By comparing the specific L and D isomers with the DL racemic mixture, we explicitly demonstrate the influence of amino acid chirality on supramolecular crystal packing, self-assembly, and electromechanical properties. Atomic force microscopy (AFM) nanoindentation analysis confirms the strong rigidity of the DL assembly with very high Young's modulus (31.8 ± 11.9 GPa) attributed to the stacked face-to-face dimers with macrocyclic architectures. The present study provides an effective strategy for precisely formulating supramolecular helical structures, which could pave the way for the development of new bio-electronic applications of smart chiroptical materials from functionalised amino acids.
Introduction
Chirality is a well-known natural materials property with significant implications from evolutionary biology to sensing technologies.1–4 A chiral molecule exists in two stereoisomers, termed enantiomers, that are non-superimposable and comprise mirror images of each other, which are often denoted as “D” or “L” forms. Chiral molecules such as L-amino acids and D-sugars are utilized in many biological and bio-inspired systems to execute essential biological functions and control of chirality is essential for crystal engineering, chiroptical, pharmaceutical and biomedical applications.5–12 Beyond single-molecule chirality based on differences in the single-molecular structure, supramolecular chirality is also observed in materials composed of periodic asymmetric assemblies of molecules (typically, helical and spiral geometry) packed via non-covalent interactions. These include electrostatic binding forces, principally hydrogen bonds (H-bonds), and weaker van der Waals forces which are mainly London dispersion forces between transient dipoles.13–15 Supramolecular secondary structures consist primarily of α-helix and β-sheet motifs. In contrast to the β-sheet, the α-helix conformation is more challenging to obtain, requiring longer amino acid sequences comprising specific residues.16–20 While proteins typically show right-handed P-helices with rare occurrence of left-handed M-helices, designed supramolecular helical structures are becoming more extensively explored for materials science and technological applications.21,22 Recently, we have shown that the supramolecular helical structure formed by a minimalistic peptide (Pro-Phe-Phe) confers interesting optical, mechanical and electrical properties.23,24 With directed H-bond interactions between donor and acceptor atoms, other materials have been similarly used to engineer smart crystalline materials.25–31 However, to date, supramolecular helical structures comprising of ultra-short single amino acids or short (typically, <4) residue peptide sequences have been rarely reported.32–34Here, we addressed the unmet demand for minimalistic supramolecular helical assemblies by developing a strategy based on simple heteroatom-derived aromatic amino acid derivatives. We designed a 9-fluorenylmethoxycarbonyl (Fmoc) protected aromatic pyridyl alanine (PyA) amino acid that self-assembles into a supramolecular helical structure as evidenced by single-crystal X-ray diffraction study. Further, by exploring specific isomers, namely L, D, and DL, we can directly track the hierarchical effect of amino acid molecular chirality on supramolecular crystal packing, self-assembly, and macroscopic mechanical and piezoelectric properties. Moreover, the non-covalent interactions that direct and stabilize the assembly were mapped using digital Hirshfeld 3D-color mapping and 2D-finger print analysis. The DL isomer shows a very high Young's modulus (31.8 ± 11.9 GPa), similar to some metals as well as cortical bones,35 arising from the constructed strong dimeric macrocyclic H-bonded structures. To the best of our knowledge, this is the first report of supramolecular minimalistic helical structures containing pyridyl nitrogen formed by the use of simple Fmoc-protected aromatic amino acids.
Results and discussions
Structural analysis of Fmoc-PyA
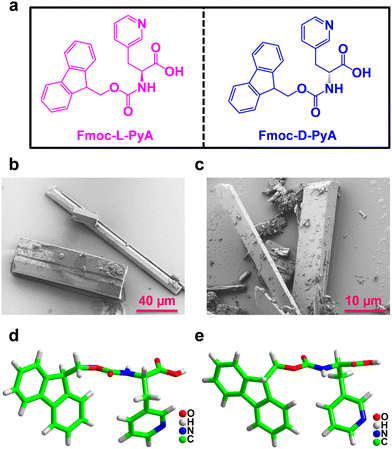
The pure enantiomers Fmoc-L-PyA and Fmoc-D-PyA were crystallized in methanol at room temperature using a slow evaporation technique. For the crystallization of the Fmoc-DL-PyA racemic mixture, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 equivalent of Fmoc-L-PyA and Fmoc-D-PyA isomers was combined in a methanolic solution. Within three days, colourless crystals were formed for all combinations (Fig. 1b and c and Fig. S1–S3, ESI†). Single-crystal X-ray diffraction analysis revealed that Fmoc-L-PyA and Fmoc-D-PyA crystallized in the isostructural non-polar acentric orthorhombic space group P212121 (Table S1, ESI†). Moreover, Fmoc-DL-PyA crystallized in the centrosymmetric monoclinic space group P21/c confirming the racemic mixture of Fmoc-L-PyA and Fmoc-D-PyA isomers. Interestingly, it also produced a variety of polymorphic forms with notable differences in the unit cell parameters. Detailed single crystal data collection and structural refinement parameters obtained for the enantiomers and their conjugate assemblies are summarized in Tables S1–S8 (ESI†).
1 equivalent of Fmoc-L-PyA and Fmoc-D-PyA isomers was combined in a methanolic solution. Within three days, colourless crystals were formed for all combinations (Fig. 1b and c and Fig. S1–S3, ESI†). Single-crystal X-ray diffraction analysis revealed that Fmoc-L-PyA and Fmoc-D-PyA crystallized in the isostructural non-polar acentric orthorhombic space group P212121 (Table S1, ESI†). Moreover, Fmoc-DL-PyA crystallized in the centrosymmetric monoclinic space group P21/c confirming the racemic mixture of Fmoc-L-PyA and Fmoc-D-PyA isomers. Interestingly, it also produced a variety of polymorphic forms with notable differences in the unit cell parameters. Detailed single crystal data collection and structural refinement parameters obtained for the enantiomers and their conjugate assemblies are summarized in Tables S1–S8 (ESI†).
The asymmetric unit of Fmoc-L-PyA and Fmoc-D-PyA contained one molecule without any co-crystallised solvent (Fig. 1d and e and Fig. S4–S6, ESI†). ORTEP (Oak Ridge Thermal Ellipsoid Plot) analysis showed the ellipsoid thermal behaviour of the enantiomers with similar sizes, shapes and coherent orientations (Fig. S7 and S8, ESI†).
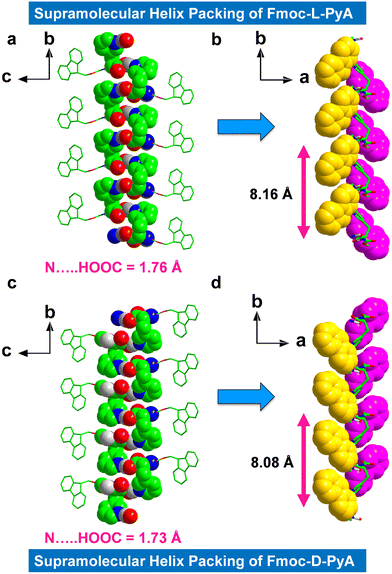
The higher-order packing arrangement revealed intermolecular H-bonded assemblies, including inter-amide and pyridine-carboxylic acid contacts (Fig. 2 and 3a, b). The first formed with a short bond distance of 2.35 Å (N–H⋯O, bond angle = 144.19°), and the second was mediated by very strong H-bonds in length 1.76 Å (O–H⋯N, helical bond angle = 167.51°) between the acceptor (pyridyl atom) and the donor (carboxylic acid) motifs (in this case of the L isomer) (Fig. 2a and Fig. S9a and b, ESI†). The observed bond distance is comparable to typical donor-acceptor distances.36 Moreover, no significant H-bonded networks were observed between the N-terminal amide NH-group and the C-terminal COOH group.
The asymmetric orientation and directional H-bonded molecular organization of these isomers formed the prominent supramolecular helical chiral structures. The L-isomer displayed right-handedness (P-helix) with a helical pitch of ∼8.16 Å (Fig. 2b) which is ∼1.66 times higher than the known Fmoc-protected phenylalanine (Phe) derivative that shows a typical helical pitch of 4.91 Å (Fig. S10, ESI†) (A helical pitch value of 8.08 Å was observed for the D-isomer, Fig. 2d). Due to the strong interactions of the pyridyl nitrogen with the adjacent carboxylic acid, Fmoc-L-PyA showed a higher helix angle and pitch compared to most known Fmoc-protected amino acids (Fig. 2b and Fig. S10, ESI†).7,32–34
It should be highlighted that the fluorene segment displays inverse helicity of M-handedness to that of pyridine-carboxylic acid H-bonded structures for Fmoc-L-PyA in the higher-order packing topologies (Fig. 2b). As expected, Fmoc-D-PyA showed mirror handedness to Fmoc-L-PyA, assembling in a left-handed M-helix (both pyridine-carboxylic acid and fluorene segment), indicating that amino acid chirality modulates the supramolecular handedness via asymmetrical H-bonds (Fig. 2c, d and Fig. S9c, d, ESI†). Such features demonstrate the significance of heteroatom, such as nitrogen, in the emergence of molecular packing, symmetry, and higher-order structures. The aryl groups in both structures showed M-handedness with tilted chirality (Fig. 2b and d). This unexpected lack of mirror-handedness between the fluorene segments of Fmoc-L-PyA and Fmoc-D-PyA suggests that these pyridyl amino acids are distinct from previously-reported Fmoc-protected amino acids.32–34
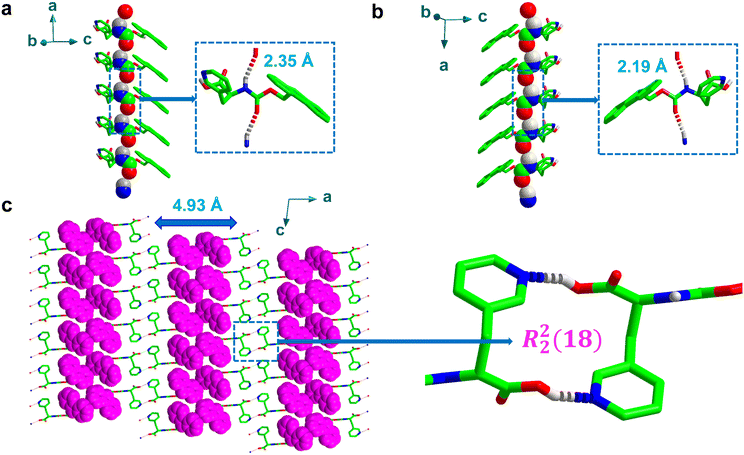
Furthermore, the asymmetric H-bonds between adjacent inter amide sites (typically, N–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group) were weaker than the rigid helical structures generated between pyridyl-nitrogen and carboxylic acid O–H, resulting in longer H-bond distances of 2.35 Å and 2.19 Å for Fmoc-L-PyA and Fmoc-D-PyA, respectively (O2⋯H1N1, Fig. 3a and b), with O2⋯N1 distances of 3.11 Å and 3.06 Å for Fmoc-L-PyA and Fmoc-D-PyA, respectively, comparable to previously-reported distances.32–34 Moreover, the backbone torsion angles for Fmoc-L-PyA and Fmoc-D-PyA were found to be −157 (Φ), −172 (ψ) and 156 (Φ), 170 (ψ), respectively. Fig. 3a and b displays the spatially induced higher-order structure directed by the asymmetric inter amide H-bonded network highlighting the formation of right and left-handed helix topologies.
O group) were weaker than the rigid helical structures generated between pyridyl-nitrogen and carboxylic acid O–H, resulting in longer H-bond distances of 2.35 Å and 2.19 Å for Fmoc-L-PyA and Fmoc-D-PyA, respectively (O2⋯H1N1, Fig. 3a and b), with O2⋯N1 distances of 3.11 Å and 3.06 Å for Fmoc-L-PyA and Fmoc-D-PyA, respectively, comparable to previously-reported distances.32–34 Moreover, the backbone torsion angles for Fmoc-L-PyA and Fmoc-D-PyA were found to be −157 (Φ), −172 (ψ) and 156 (Φ), 170 (ψ), respectively. Fig. 3a and b displays the spatially induced higher-order structure directed by the asymmetric inter amide H-bonded network highlighting the formation of right and left-handed helix topologies.
To gain further insight into the molecular arrangement of the conjugate assemblies of Fmoc-DL-PyA, the single-crystal structures were analysed in detail (Table S1, ESI†).37 Unlike the chiral single component crystals, the racemic mixtures were found to crystallize in monoclinic space group P21/c validating the formation of a racemic mixture. Interestingly, these racemic mixtures displayed two polymorphic forms [P1: a = 25.92 Å, b = 4.93 Å, c = 15.67 Å, volume = 1985 Å3; P2: a = 12.51 Å, b = 30.00 Å, c = 5.78 Å, volume = 2151 Å3], with iso-structural characteristics of an identical crystal system with the same space group in the crystal lattice (Table S1, ESI†). Fig. 3c shows the P1 polymorph consisting of a series of discrete strong face-to-face dimers containing macrocycles as indicated by the formation of R22(18)-type rings. The notation ring (R), which is used to denote the macrocycles formed by intermolecular H-bonds, is frequently used to represent the number of donors (here, 2) and acceptors (2) as well as the overall number of atoms (18) involved in the formation of macrocycles.38,39 This structure was not apparent in either of the individual chiral crystals. In the higher-order crystal packing diagram of Fmoc-DL-PyA (P1), consecutive fluorene segments were separated by a distance of 4.93 Å (Fig. 3c). It should be highlighted that while the P1 polymorph has been reproduced, multiple attempts to reproduce the P2 polymorph have been unsuccessful. Although the presence of true polymorphism is thus unknot yet proven, the occurrence of P2, likely as a minor state, suggests that individual enantiomers in self-assembled structures are dynamic in nature (Fig. S11, ESI†). Moreover, future characterisation studies using techniques such as thermal analysis, microscopy, solid–solid NMR could lead to a better understanding of the polymorphism in these amino acids.
Circular dichroism (CD) analysis further confirmed the asymmetrical H-bonding packing and handedness of the Fmoc-protected pyridyl amino acids in solution (Fig. S12, ESI†). The CD signal for the Fmoc-L-PyA and Fmoc-D-PyA enantiomers displayed mirrored images of the positive and negative cotton effects, indicating their distinct chirality. The racemic mixture, on the other hand, showed a flat baseline that is optically inactive. Moreover, Fourier-transform infrared (FT-IR) and Raman spectroscopic investigations were employed to examine the structural integrity of the neat enantiomers and the racemic conjugate in solid-state samples (Fig. S13–S18, ESI†). The results showed the major vibrational peaks, notably the O–H stretching (ca. 3395 cm−1), N–H stretching (ca. 3310 cm−1), and the aromatic vibrational modes of the fluorenyl segment C–H bendings (ca. 1477 and 1324 cm−1), fingerprinting the pyridyl amino acids (Fig. S13–S15, ESI†). The solution-state absorption spectra of the individual and conjugate assemblies were analysed and the absorption region typically ranged from 250 to 300 nm. Further, the optical bandgap (Eg) was calculated using the Tauc equation (αhν)2 = A(hν − Eg), where A is a constant, hν is the photon energy, and α is the absorption coefficient. The estimated Eg values ranging from 4.40 to 5.60 eV demonstrate the semiconductive nature of the enantiomers and the conjugate mixtures (Fig. S19–S21, ESI†). Such large bandgap values suggest the potential utilization of these systems in next-generation optoelectronic device applications.40
To further investigate the presence of diverse intermolecular interactions (C⋯H, H⋯C, N⋯H, H⋯N, O⋯H, H⋯O, C⋯C, and H⋯H), the Crystal Explorer 21.5 software was used to generate three-dimensional (3D) Hirshfeld surface maps and two-dimensional (2D) fingerprint plots based on their single-crystal structures (Fig. S22–S30 and Table S9, ESI†).41 The Hirshfeld surface was used to quantify and decode the non-covalent interactions using dnorm (normalized contact distance), de (nearest exterior), and di (nearest interior). The 2D and 3D plots of the Fmoc-D-PyA and Fmoc-DL-PyA (P1) systems showed very similar intermolecular interaction characteristics (Fig. S25–S30 and Table S9, ESI†). For Fmoc-L-PyA, the strong H-bonding provided ∼¼ of the overall molecular interactions, with a typical stabilizing energy of ∼12–30 kJ mol−1 per H-bond known to drive the assembly,42,43 here comprising mainly of pyridyl nitrogen (acceptor)–carboxylic acid (donor) and amino N–H (donor)–amide O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (acceptor). The remaining interactions stem from dispersion and van der Waals's contacts, individually weak (∼0.4–4.0 kJ mol−1) but here covering a large fraction of the intermolecular contact area mainly due to the large fluorene units. Further, we employed powder X-ray diffraction (PXRD) and compared the results with simulated data from single-crystal XRD to validate the bulk phase purity of each individual and racemic mixture form. In comparison to the computer-simulated data, the finely ground powdered crystallites displayed a similar crystalline nature, peak-pattern matching and single-phase identification, confirming the individual structural purity of the samples (Fig. S31–S33, ESI†). In addition to their extensive directional H-bonding arrangement at the molecular level, thermogravimetric analysis (TGA) was used to evaluate the thermal stability of Fmoc-L-PyA, Fmoc-D-PyA, and Fmoc-DL-PyA (P1), demonstrating high thermal stability (>500 K) (Fig. S34–S36, ESI†).
C (acceptor). The remaining interactions stem from dispersion and van der Waals's contacts, individually weak (∼0.4–4.0 kJ mol−1) but here covering a large fraction of the intermolecular contact area mainly due to the large fluorene units. Further, we employed powder X-ray diffraction (PXRD) and compared the results with simulated data from single-crystal XRD to validate the bulk phase purity of each individual and racemic mixture form. In comparison to the computer-simulated data, the finely ground powdered crystallites displayed a similar crystalline nature, peak-pattern matching and single-phase identification, confirming the individual structural purity of the samples (Fig. S31–S33, ESI†). In addition to their extensive directional H-bonding arrangement at the molecular level, thermogravimetric analysis (TGA) was used to evaluate the thermal stability of Fmoc-L-PyA, Fmoc-D-PyA, and Fmoc-DL-PyA (P1), demonstrating high thermal stability (>500 K) (Fig. S34–S36, ESI†).
Self-assembly studies of Fmoc-PyA
To test for environmental factors, we investigated the self-assembly process of individual chiral and conjugate systems using three different solvent combinations: 100% methanol (MeOH), 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10% MeOH/water (H2O), and 95
10% MeOH/water (H2O), and 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5% dimethyl sulfoxide (DMSO)/H2O while maintaining a constant overall concentration of Fmoc-protected pyridyl amino acids of 1 mg ml−1 (Fig. S37–S45, ESI†). Enantiomers and their racemic mixtures completely dissolved in MeOH and in DMSO. The self-assembled aggregates were visually observed in both the MeOH/H2O and DMSO/H2O solvent systems indicating the highly hydrophobic nature of these Fmoc-protected pyridyl amino acids.
5% dimethyl sulfoxide (DMSO)/H2O while maintaining a constant overall concentration of Fmoc-protected pyridyl amino acids of 1 mg ml−1 (Fig. S37–S45, ESI†). Enantiomers and their racemic mixtures completely dissolved in MeOH and in DMSO. The self-assembled aggregates were visually observed in both the MeOH/H2O and DMSO/H2O solvent systems indicating the highly hydrophobic nature of these Fmoc-protected pyridyl amino acids.
The structures self-assembled in pure MeOH formed kinetically-controlled spherical products that varied in length from nano to micrometer scales (Fig. S37–S39, ESI†). Interestingly, the addition of 10% water to produce the MeOH/water mixture transformed the enantiomer spheres into meta-stable ribbon-like structures whereas the racemic assemblies yielded both micro half-spheres and stable self-assembled crystallites (Fig. S40–S42, ESI†). The enantiomers demonstrate the kinetically controlled pathway, but racemic assemblies exhibit both kinetically and thermodynamically controlled by-products. These results indicate the competitive relationship between the kinetic and thermodynamic states of self-assembled products. In addition, for all of L, D, and DL, the H2O ratio in DMSO determined the self-assembled structure of thermodynamically-stable crystallites (Fig. S43–S45, ESI†). Such diverse self-assembled structures reflect the synergism and cooperativity of diverse weak non-covalent interactions in the self-assembly process, and ease of directing and controlling the solid forms by varying the experimental conditions.44–48
Mechanical properties analysis of Fmoc-PyA
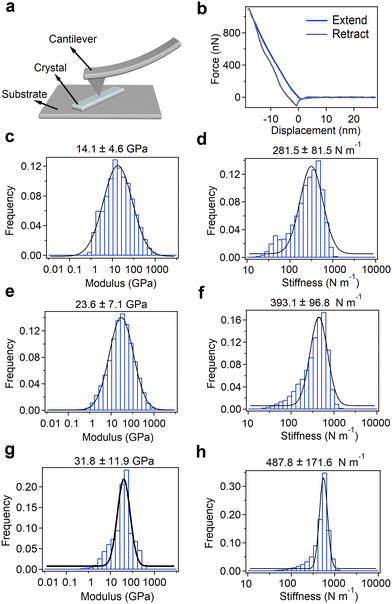
Finally, to investigate their potential for utilization as electromechanical device materials, we examined the micromechanical rigidity and stiffness of the crystals using AFM-based nanoindentation experiments (Fig. 4 and Fig. S46, ESI†). The AFM cantilever was positioned to scan the surface of microcrystalline samples spread on a pristine mica substrate and retrieved at a continuous speed of 80 m s−1 (Fig. 4a). Further, the Young's modulus was determined using the Hertz model to fit the force-displacement traces (Fig. 4b and Fig. S46, ESI†).Young's modulus and point stiffness distributions of the enantiomers and racemic mixture are shown in Fig. 4c–h. The measured elasticity of the Fmoc-L-PyA sample showed a Young's modulus of 14.1 ± 4.6 GPa along the elongated direction, indicating lower mechanical stability compared to Fmoc-Phe,32–34 and a point stiffness value of 281.5 ± 81.5 N m−1 (Fig. 4c and d). Fmoc-D-PyA revealed higher Young's modulus and point stiffness values of 23.6 ± 7.1 GPa and 393.1 ± 96.8 N m−1, respectively (Fig. 4e and f). Moreover, Fmoc-DL-PyA (P1) displayed a highly increased Young's modulus of 31.8 ± 11.9 GPa, with a point stiffness of 487.8 ± 171.6 N m−1, reflecting the more rigid supramolecular packing network in the racemic crystal (Fig. 4g and h). The order of Young's modulus and point stiffness values is DL > D > L, indicating that the molecular chirality dramatically alters the properties of the crystals. The higher Young's modulus of the DL isomer reflects the formation of the strong discrete dimeric H-bonded structures (Fig. 3c). Remarkably, the measured Young's modulus value is comparable to many well-known bio-inspired and synthetic materials such as collagen, actin, amyloids, bone, wood, silk, Phe–Phe, gold, steel, glass and metallic alloys,35,49,50 suggesting their potential use for printed circuit boards (PCBs) and biomedical engineering applications.
Density functional theory simulations of Fmoc-PyA
Bio-inspired materials have emerged as appealing piezoelectric alternatives to conventional inorganic oxides due to their sustainability, easy solution-phase synthesis, biocompatibility, biodegradability, and robust mechanical properties.51–54 As discussed above, the individual L and D isomers crystallize in non-centrosymmetric orthorhombic space group P212121. To explore the potential piezoelectric properties, we used density functional theory (DFT) calculations and the obtained magnitude of polarization values are summarized and compared with the well-known supramolecular piezoelectric materials in Tables S10–S12 (ESI†). As expected, crystals of Fmoc-DL-PyA assembled in a centrosymmetric space group, specifically monoclinic P21/c, that precludes piezoelectricity. The specific orthorhombic symmetry allows for the presence of three non-zero shear piezoelectric tensor components only: d14, d25 and d36. Both enantiopure compounds showed a similar trend in computed piezoelectric response with a maximum predicted strain constant of 11.3 pC N−1 in Fmoc-L-PyA (Fig. 5). Additionally, using the relationship between the piezoelectric coefficient (dij) and the calculated absolute permittivity (ε) of the material (gij = dij/εij), the piezoelectric voltage coefficient (gij) characteristics were also predicted. Using the computed relative permittivity of 3.36 and 3.43 for the Fmoc-L-PyA and Fmoc-D-PyA systems, respectively. The maximal g values were g14 = 444 and 349 V m N−1 for Fmoc-L-PyA and Fmoc-D-PyA, respectively (Fig. 5). Thus, the predicted piezoelectric characteristics reflect the asymmetrical crystal packing of hetero aromatic nitrogen acceptor and carboxylic acid donor atoms of the pyridyl amino acids, which can in the future be utilized to complement inorganic ceramic oxides counterparts, piezo-polymers, and organic and organic–inorganic hybrid materials.55,56 These characteristics suggest that if a device design allows for the application of shear forces, the enantiopure crystals could display technologically useful energy harvesting capabilities. The significant g14 response is oriented along the pyridyl nitrogen–carboxylic acid directed helical structure on the a axis of the crystals (Fig. 2 and Fig. S9, ESI†). Thus, the predicted piezoelectric properties reflect the asymmetrical crystal packing of hetero-aromatic nitrogen acceptor and carboxylic acid donor atoms of the pyridyl amino acids. This type of structure-based design can direct the engineering of new piezoelectric bio-organic and hybrid materials.55,56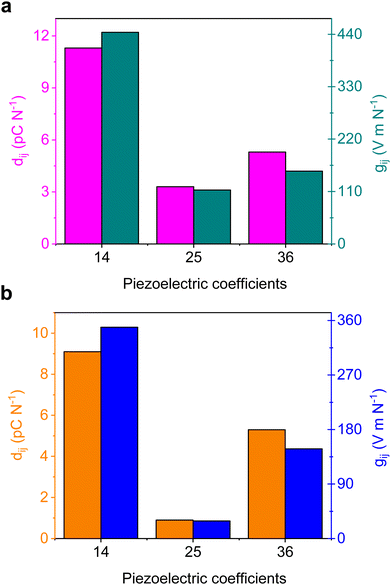 | ||
| Fig. 5 (a and b) DFT-predicted piezoelectric coefficients of the non-centrosymmetric (a) L- and (b) D-Fmoc-PyA crystal structures. | ||
Conclusions
In summary, we introduce a simple N-terminal protected aromatic pyridyl amino acid that exhibits supramolecular helicity with specific handedness, self-assembled structures, and mechanical and piezoelectric properties. The high-order organization of helicity at the molecular level is revealed by single-crystal X-ray diffraction analysis. The enantiopure form of Fmoc-L-PyA has two helical modalities: amide to amide and pyridine to carboxylic acid (P-helix), and the fluorene groups have distinctive supramolecular tilted chirality (M-helix). Interestingly, the racemic DL mixture was crystallised in two alternative polymorphic forms, demonstrating dynamic interactions between individual enantiomers. Nanoindentation measurements showed that the DL crystal is more mechanically stable than typical biological materials49,50 due to the rigid structures formed between the donor (carboxylic acid) and acceptor (pyridyl nitrogen atom) sites. Furthermore, DFT calculations predict a significant piezoelectric response in the enantiopure crystals that could be utilized for energy harvesting. This study provides the first demonstration of supramolecular chirality by introducing a heteroatom such as nitrogen into a minimalistic Fmoc-protected aromatic amino acid. The demonstrated effect of chirality in these minimalistic helical assemblies makes them attractive for emerging technologies, such as spin filters for electron transport and enantio-selectors for purification and asymmetric catalysis.57–60Conflicts of interest
There are no conflicts to declare.Acknowledgements
T. V. thanks Tel Aviv University for the post-doctoral fellowship. E. G. acknowledges the support of the European Research Council PoC project PiezoGel (966813) and Ministry of Science and Technology (MOST) Israel-China Program (3-19130). We thank the members of the Gazit laboratory for helpful discussions. D. T. and S. G. acknowledge Science Foundation Ireland for funding under award number 12/RC/2275_P2 (SSPC) and thank the Irish Center for High-End Computing (ICHEC) for supercomputing resources. S. G. would like to acknowledge funding from Science Foundation Ireland under grant number 21/PATH-S/9737. S. G. is funded by the European Union. Views and opinions expressed are however those of the author only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them.Notes and references
- W. Ma, L. Xu, A. F. de Moura, X. Wu, H. Kuang, C. Xu and N. A. Kotov, Chem. Rev., 2017, 117, 8041–8093 CrossRef CAS PubMed.
- N. A. Kotov, L. M. Liz-Marzán and P. S. Weiss, ACS Nano, 2021, 15, 12457–12460 CrossRef CAS.
- S. M. Morrow, A. J. Bissette and S. P. Fletcher, Nat. Nanotechnol., 2017, 12, 410–419 CrossRef CAS PubMed.
- S. Zhang, Nat. Biotechnol., 2003, 21, 1171–1178 CrossRef CAS PubMed.
- S. F. Mason, Nature, 1984, 311, 19–23 CrossRef CAS PubMed.
- J. Yeom, B. Yeom, H. Chan, K. W. Smith, S. Dominguez Medina, J. H. Bahng, G. Zhao, W. S. Chang, S. J. Chang, A. Chuvilin, D. Melnikau, A. L. Rogach, P. Zhang, S. Link, P. Kral and N. A. Kotov, Nat. Mater., 2015, 14, 66–72 CrossRef CAS PubMed.
- J. Liang, A. Hao, P. Xing and Y. Zhao, ACS Nano, 2021, 15, 5322–5332 CrossRef CAS PubMed.
- D. Yan, Chem. – Eur. J., 2015, 21, 4880–4896 CrossRef CAS PubMed.
- M. C. Cringoli, O. Bellotto, R. D. Zorzi, A. V. Vargiu and S. Marchesan, Synlett, 2020, 434–438 CAS.
- D. Yan and D. G. Evans, Mater. Horiz., 2014, 1, 46–57 RSC.
- B. Lu, S. Liu and D. Yan, Chin. Chem. Lett., 2019, 30, 1908–1922 CrossRef CAS.
- Y. J. Ma, G. Xiao, X. Fang, T. Chen and D. Yan, Angew. Chem., Int. Ed., 2023, e202217054 CAS.
- P. Xiang and Y. Zhao, Acc. Chem. Res., 2018, 51, 2324–2334 CrossRef PubMed.
- E. Yashima, N. Ousaka, D. Taura, K. Shimomura, T. Ikai and K. Maeda, Chem. Rev., 2016, 116, 13752–13990 CrossRef CAS PubMed.
- M. Liu, L. Zhang and T. Wang, Chem. Rev., 2015, 115, 7304–7397 CrossRef CAS PubMed.
- J. Venkatraman, S. C. Shankaramma and P. Balaram, Chem. Rev., 2001, 101, 3131–3152 CrossRef CAS PubMed.
- M. Zelzer and R. V. Ulijn, Chem. Soc. Rev., 2010, 39, 3351–3357 RSC.
- T. E. Creighton, Nature, 1987, 326, 547–548 CrossRef CAS PubMed.
- X. Yan, P. Zhu and J. Li, Chem. Soc. Rev., 2010, 39, 1877–1890 RSC.
- Z. Luo and S. Zhang, Chem. Soc. Rev., 2012, 41, 4736–4754 RSC.
- S. Mondal and E. Gazit, ChemNanoMat, 2016, 2, 323–332 CrossRef CAS.
- W. Ji, B. Xue, S. Bera, S. Guerin, L. J. W. Shimon, Q. Ma, S. A. M. Tofail, D. Thompson, Y. Cao, W. Wang and E. Gazit, Mater. Today, 2021, 42, 29–40 CrossRef CAS.
- S. Bera, S. Mondal, B. Xue, L. J. W. Shimon, Y. Cao and E. Gazit, Nat. Mater., 2019, 18, 503–509 CrossRef CAS PubMed.
- S. Bera, S. Guerin, H. Yuan, J. O’Donnell, N. P. Reynolds, O. Maraba, W. Ji, L. J. W. Shimon, P. A. Cazade, S. A. M. Tofail, D. Thompson, R. Yang and E. Gazit, Nat. Commun., 2021, 12, 2634 CrossRef CAS PubMed.
- O. Dumele, J. Chen, J. V. Passarelli and S. I. Stupp, Adv. Mater., 2020, 32, 1907247 CrossRef CAS PubMed.
- T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813–817 CrossRef CAS PubMed.
- P. Rozhin, C. Charitidis and S. Marchesan, Molecules, 2021, 26, 4084 CrossRef CAS PubMed.
- K. Ariga, J. Li, J. Fei, Q. Ji and J. P. Hill, Adv. Mater., 2016, 28, 1251–1286 CrossRef CAS PubMed.
- M. P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50, 2440–2448 CrossRef CAS PubMed.
- B. Lu, X. Fang and D. Yan, ACS Appl. Mater. Interfaces, 2020, 12, 31940–31951 CrossRef CAS PubMed.
- S. Li, Y. Lin and D. Yan, J. Mater. Chem. C, 2016, 4, 2527–2534 RSC.
- Z. Zong, A. Hao and P. Xing, Nanoscale, 2020, 12, 20610–20620 RSC.
- Z. Wang, A. Hao and P. Xing, Chin. Chem. Lett., 2021, 32, 1390–1396 CrossRef CAS.
- E. R. Draper, K. L. Morris, M. A. Little, J. Raeburn, C. Colquhoun, E. R. Cross, T. O. McDonald, L. C. Serpell and D. J. Adams, CrystEngComm, 2015, 17, 8047–8057 RSC.
- V. Basavalingappa, S. Bera, B. Xue, J. O’Donnell, S. Guerin, P. A. Cazade, H. Yuan, E. U. Haq, C. Silien, K. Tao, L. J. W. Shimon, S. A. M. Tofail, D. Thompson, S. Kolusheva, R. Yang, Y. Cao and E. Gazit, ACS Nano, 2020, 14, 7025–7037 CrossRef CAS PubMed.
- G. J. Jeffrey, An Introduction to Hydrogen Bonding, Oxford University Press, New York, 1997 Search PubMed.
- G. M. Sheldrick, SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- M. C. Etter, Acc. Chem. Res., 1990, 23, 120–126 CrossRef CAS.
- G. Bolla, B. Sarma and A. K. Nangia, Chem. Rev., 2022, 122, 11514–11603 CrossRef CAS PubMed.
- A. Yoshikawa, H. Matsunami and Y. Nanishi, Development and applications of wide bandgap semiconductors, Wide Bandgap Semiconductors, Springer, New York, 2007, pp. 1–24 Search PubMed.
- M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, P. R. Spackman, D. Jayatilaka and M. A. Spackman, Crystal Explorer 3.1, The University of Western Australia, 2012 Search PubMed.
- P. Hobza and J. Řezáč, Chem. Rev., 2016, 116, 4911–4912 CrossRef CAS PubMed.
- K. Müller-Dethlefs and P. Hobza, Chem. Rev., 2000, 100, 143–167 CrossRef PubMed.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- P. Ke, M. A. Sani, F. Ding, A. Kakinen, I. Javed, F. Separovic, T. P. Davis and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 6492–6531 RSC.
- J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC.
- S. Gilead and E. Gazit, Supramol. Chem., 2005, 17, 87–92 CrossRef CAS.
- G. Wei, W. Xi, R. Nussinov and B. Ma, Chem. Rev., 2016, 116, 6516–6551 CrossRef CAS PubMed.
- I. Azuri, E. Meirzadeh, D. Ehre, S. R. Cohen, A. M. Rappe, M. Lahav, I. Lubomirsky and L. Kronik, Angew. Chem., Int. Ed., 2015, 54, 13566–13570 CrossRef CAS PubMed.
- T. P. J. Knowles and M. J. Buehler, Nat. Nanotechnol., 2011, 6, 469–479 CrossRef CAS PubMed.
- H. Y. Zhang, Z. X. Zhang, X. G. Chen, X. J. Song, Y. Zhang and R. G. Xiong, J. Am. Chem. Soc., 2021, 143, 1664–1672 CrossRef CAS PubMed.
- T. Vijayakanth, A. K. Srivastava, F. Ram, P. Kulkarni, K. Shanmuganathan, B. Praveenkumar and R. Boomishankar, Angew. Chem., Int. Ed., 2018, 57, 9054–9058 CrossRef CAS PubMed.
- T. Vijayakanth, F. Ram, B. Praveenkumar, K. Shanmuganathan and R. Boomishankar, Angew. Chem., Int. Ed., 2020, 59, 10368–10373 CrossRef CAS PubMed.
- Y. M. Wang, Q. Zeng, L. He, P. Yin, Y. Sun, W. Hu and R. Yang, iScience, 2021, 24, 102274 CrossRef CAS PubMed.
- T. Vijayakanth, D. J. Liptrot, E. Gazit, R. Boomishankar and C. R. Bowen, Adv. Funct. Mater., 2022, 32, 2109492 CrossRef CAS.
- S. Guerin, A. Stapleton, D. Chovan, R. Mouras, M. Gleeson, C. McKeown, M. R. Noor, C. Silien, F. M. F. Rhen, A. L. Kholkin, N. Liu, T. Soulimane, S. A. M. Tofail and D. Thompson, Nat. Mater., 2017, 17, 180–186 CrossRef PubMed.
- A. K. Mondal, N. Brown, S. Mishra, P. Makam, D. Wing, S. Gilead, Y. Wiesenfeld, G. Leitus, L. J. W. Shimon, R. Carmieli, D. Ehre, G. Kamieniarz, J. Fransson, O. Hod, L. Kronik, E. Gazit and R. Naaman, ACS Nano, 2020, 14, 16624–16633 CrossRef CAS PubMed.
- W. Mtangi, F. Tassinari, K. Vankayala, A. V. Jentzsch, B. Adelizzi, A. R. A. Palmans, C. Fontanesi, E. W. Meijer and R. Naaman, J. Am. Chem. Soc., 2017, 139, 2794–2798 CrossRef CAS PubMed.
- L. D. Pachón, I. Yosef, T. Z. Markus, R. Naaman, D. Avnir and G. Rothenberg, Nat. Chem., 2009, 1, 160–164 CrossRef PubMed.
- K. B. Ghosh, O. B. Dor, F. Tassinari, E. Capua, S. Yochelis, A. Capua, S. H. Yang, S. S. P. Parkin, S. Sarkar, L. Kronik, L. T. Baczewski, R. Naaman and Y. Paltiel, Science, 2018, 360, 1331–1334 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2211500–2211503. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2tc05320a |
| This journal is © The Royal Society of Chemistry 2023 |