Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Carbohydrate–protein interaction-based detection of pathogenic bacteria using a biodegradable self-powered biosensor†
Swati
Panda
a,
Sugato
Hajra
a,
Hang Gyeom
Kim
a,
Haejin
Jeong
b,
P. G. R.
Achary
c,
Seonki
Hong
 b,
Bhaskar
Dudem
d,
S. Ravi P.
Silva
b,
Bhaskar
Dudem
d,
S. Ravi P.
Silva
 d,
Venkateswaran
Vivekananthan
d,
Venkateswaran
Vivekananthan
 *de and
Hoe Joon
Kim
*de and
Hoe Joon
Kim
 *af
*af
aDepartment of Robotics and Mechatronics Engineering, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu-42988, Republic of Korea. E-mail: joonkim@dgist.ac.kr
bDepartment of Physics and Chemistry, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu-42988, Republic of Korea
cDepartment of Chemistry, Siksha O Anusandhan University, Bhubaneswar-751030, India
dAdvanced Technology Institute, Department of Electrical and Electronic Engineering, University of Surrey, Guildford, Surrey GU2 7XH, England, UK. E-mail: v.venkateswaran@surrey.ac.uk; vivek@kluniversity.in
eCenter for Flexible Electronics, Department of Electronics and Communication Engineering, Koneru Lakshmaiah Education Foundation, Andhra Pradesh-522302, India
fRobotics and Mechatronics Research Center, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu-42988, South Korea
First published on 11th October 2023
Abstract
Battery-free and biodegradable sensors can detect biological elements in remote areas. The triboelectric nanogenerator (TENG) can potentially eliminate the need for a battery by simply converting the abundant vibrations from nature or human motion into electricity. A biodegradable sensor system integrated with TENG to detect commonly found disease-causing bacteria (E. coli) in the environment is showcased herein. In this system, D-mannose functionalized 3D printed polylactic acid (PLA) with the brush-painted silver electrode was used to detect E. coli by a simple carbohydrate–protein interaction mechanism. The adsorption capacity of D-mannose is generally altered by varying the concentration of E. coli resulting in changes in resistance. Thus, the presented biosensor can detect bacterial concentrations by monitoring the output current. The PLA TENG generates an output of 70 V, 800 nA, and 22 nC, respectively. In addition, tap water and unpasteurized milk samples are tested for detecting bacteria, and the output is measured at 6 μA and 5 μA, respectively. Further, the biosensor was tested for biodegradability in soil compost by maintaining constant temperature and humidity. This study not only proposes an efficient and fast method for screening E. coli but also gives important insights into the ability to degrade and long-term reliability of TENG-based sensor platforms.
1. Introduction
Escherichia coli (E. coli) is a bacteria, frequently found in human and animal intestinal tracts.1,2 While most E. coli strains are not harmful, a few of them can result in serious sickness, such as diarrhea and kidney failure.3–5 The bacteria that cause E. coli infections produce a Shiga toxin as part of their infection process. This toxin can harm the lining of the intestines, causing symptoms like vomiting, diarrhea, and stomach pain.6,7 The toxin can also enter the bloodstream and harm the kidneys, resulting in hemolytic uremic syndrome (HUS), a potentially fatal illness.8,9 Consumption of contaminated food or water, particularly undercooked or raw meat, unpasteurized dairy products, or fruits and vegetables infected with animal feces, is the most typical way to contract the disease.10,11 Fecal-oral transfer from one person to another is also possible, particularly in unhygienic environments.12 Proper food handling and hygiene procedures are necessary to prevent E. coli infection. These procedures include washing hands before handling or eating food, properly cooking meat, avoiding unpasteurized dairy products, cleaning fruits and vegetables before consumption, and detecting bacteria.Early detection and treatment of symptoms are essential to preventing infection and subsequent complications. Quickly identifying potentially hazardous strains in food, water, and other surroundings makes rapid detection of E. coli crucial,13,14 providing quicker reaction times to limit the spread of the bacteria and aid in preventing disease outbreaks. Several studies have been performed to eradicate the disease. Liu et al. have proposed a highly sensitive immunosensor for quick IgE detection, with potential applications in clinical diagnostics, allergy monitoring, and possibly early cancer detection, providing varied healthcare solutions.15 Wang et al. treat drug-resistant bacterial infections using multifunctional nano-assembly consisting of a conjugated oligomer with ferrous ions. The nano-assembly efficiently converts light to heat, catalyzes H2O2 into hydroxyl radicals, and uses low H2O2 dosages to remove Staphylococcus aureus in vitro and in vivo.16
In this context triboelectric nanogenerators (TENGs) are emerging technologies for detecting pathogenic bacteria due to their high sensitivity and rapid response time without additional power sources.17,18 TENGs are nanogenerators that convert mechanical energy into electrical energy and power low-power electronics.19–21 In the case of pathogenic bacteria detection, a TENG-based sensor is designed to generate a voltage signal when bacteria come into contact with the surface of the sensor.22,23 This voltage signal can then be measured and used to detect the presence of pathogenic bacteria. One of the advantages of TENG-based sensors is their high sensitivity.24 They can detect even small amounts of bacteria, making them useful for early detection of infections.25 Additionally, TENG-based sensors are inexpensive and easy to fabricate, making them accessible for widespread use.26–28 There are also challenges in using TENGs for pathogenic bacteria detection, such as the need to optimize the design of the sensor and the need to ensure specificity toward pathogenic bacteria.29,30 However, ongoing research is focused on addressing these challenges and improving the performance of TENG-based sensors for pathogenic bacteria detection.31,32 Overall, using TENGs for pathogenic bacteria detection has significant potential for advancing healthcare by providing a rapid, sensitive, and affordable method for detecting bacterial infections.
This work presents a sensor system that uses D-mannose functionalized 3D printed polylactic acid (PLA) and brush-painted silver electrodes platform to capture E. coli bacteria through a simple carbohydrate–protein interaction mechanism. Changes in E. coli concentration alter the adsorption capacity of D-mannose, leading to variations in resistances and allowing for effective detection of bacterial concentrations through monitoring of output current. The biosensor was also validated for its performance in detecting S. aureus, a Gram-positive bacterium without pili. Real-time tap water samples and unpasteurized milk samples were also tested for bacterial detection. The linear relation between peak current and bacteria concentration in the detection range 2 × 10−8 to 2 × 10−2 ml−1 was established. Furthermore, the biosensor's biodegradability was tested in soil compost under constant temperature and humidity, demonstrating potential environmental sustainability. This research offers a promising strategy for the rapid and effective detection of E. coli and sheds light on the progress of biodegradable and self-powered sensor systems.
2. Materials and method
2.1. Chemicals and materials
Polylactic acid (PLA), polydimethylsiloxane (PDMS), silver (Ag) paint, aluminum (Al), hydrogen peroxide (H2O2), distilled water (DI), 0.1 mM 3-mercaptonic acid (MPA), D-mannose, concanavalin-A (Con-A), phosphate buffer saline (PBS), 2% v/v glutaraldehyde, and gelatin were obtained from Sigma Aldrich, Korea. The pure culture of E. coli strain W1485 (ATCC 12435) in broth media and Luria Bertani (LB) broth was purchased from Biozoa, Korea.2.2. Bacteria strain, sample preparation
Desired concentrations of bacteria were made in PBS buffer using the stock solution by conventional serial dilution method. The various concentration of the bacterial solutions was incubated for further use. Con A lectin was added to the bacterial solution and incubated for 30 min at 37 °C.2.3. Biosensor preparation
PLA substrate and IDT patterned masking of PLA were made by 3D printing. Ag paint was used as the electrode by the brush painting method and stored for 1 hour at room temperature for drying. The conductivity of the silver electrode was tested after drying. The silver electrode surface was thoroughly cleaned to remove contaminants and dirt and dried using a stream of N2 gas. D-Mannose was dissolved in PBS buffer at 1 mg ml−1, creating a solution of D-mannose molecules in the buffer, ready for immobilization. A suitable linker molecule, 0.1 mM 3-mercaptonic acid solution with PBS buffer layer, was applied to facilitate D-mannose binding to the silver electrode surface, where the thiol group (-SH) of 3-MPA plays a key role. The sensor was incubated for a few hours to allow the D-mannose to bind to the linker molecules on the surface. 2% V/V glutaraldehyde solution was also applied to the sensor and incubated for 1 hour at room temperature. The glutaraldehyde helps to activate the D-mannose and provides reactive functional groups for subsequent attachment to the linker molecules on the Ag electrode surface. The electrode was rinsed with DI water to remove excessive D-mannose and dried using a stream of N2 gas. Gelatin solution was used to block unreacted sites on the silver electrode surface to prevent the non-specific binding of bacteria or other unwanted molecules to the immobilized D-mannose surface. The surface with the blocking agent was incubated for 1–2 hours, and the electrode was dried and stored for further use.2.4. Fabrication of TENG device
The operating mode of TENG was selected as vertical contact separation mode. The active area of the TENG was 2 cm × 2 cm, and the structure was 3D printed like an eye shape. The substrate layer of the TENG is PLA, which is prepared using the 3D printing technique. The opposite triboelectric layer comprises PET, PDMS, or porous PDMS. Adhesive aluminum (Al) tape was used as the electrode pasted upon the inner side of the eye shaped TENG. Kapton tape was used to paste the copper wires drawn from both sides of the electrode. The PLA and opposite layer were uniformly fixed onto the Al tape. Finally, the vertical contact separate mode based TENG with PLA and the opposite triboelectric layer was designed. The plain PDMS was processed using the monomer to hardener at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, while for porous PDMS (pPDMS abbreviation further), an additional 5 ml of DI water was mixed with the solution of clear PDMS and blended properly. The plain and pPDMS underwent curing for 4 hours at 70 °C. In the case of the pPDMS, the water molecules evaporated, leaving non-uniform pores in the PDMS matrix.
1 ratio, while for porous PDMS (pPDMS abbreviation further), an additional 5 ml of DI water was mixed with the solution of clear PDMS and blended properly. The plain and pPDMS underwent curing for 4 hours at 70 °C. In the case of the pPDMS, the water molecules evaporated, leaving non-uniform pores in the PDMS matrix.
2.5 Experimental techniques
An electrometer (M/S Keithley 6514) and a homemade LabVIEW program help trace TENG's output. The contact and separation motion of the TENG was done with the aid of a linear motor (M/S LINMOT, USA). The surface morphology of the sensors employed was taken employing a scanning electron microscope (M/S Hitachi, SU-8020) and elemental mapping using OXFORD EDS. The Raman band was traced using a Raman imaging system bearing a laser module of 532 nm (M/S Thermo Scientific, Nicolet Almega XR). The Fast Fourier Infrared spectra (FT-IR) were captured using Thermo Scientific Nicolet Continuum from a wavelength 500 cm−1 to 3000 cm−1. The UV-vis spectrum was measured with a UV-Vis spectrophotometer (M/S Edinburgh Instruments, UK). The X-ray spectra were traced using an X-ray diffractometer (M/S Bruker, Germany) with Cu-Kα radiation. The substrate was 3D printed using an FFT-based 3D printer (M/S Ultimaker, Netherlands).3. Results and discussions
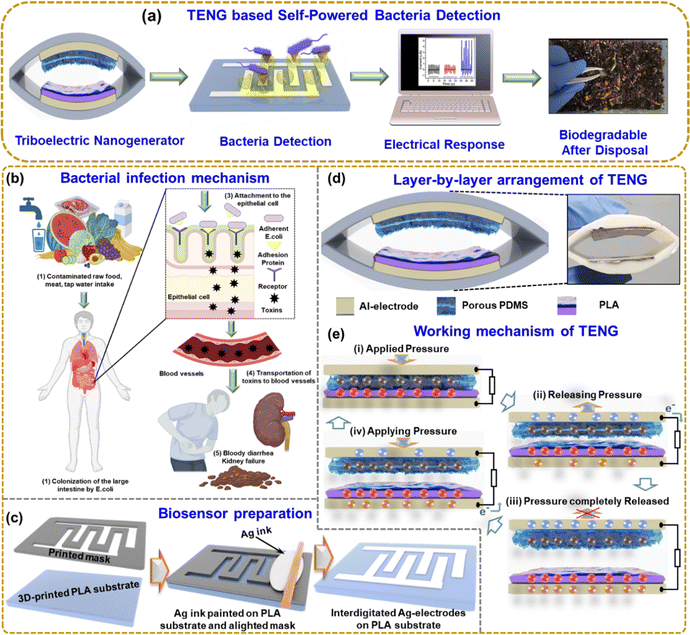
Fig. 1(a) shows the overall schematic of self-powered bacteria detection using TENG. The system implements the carbohydrate–protein interaction mechanism for bacteria detection. The TENG electrical output depends on the bacteria concentration, so the output can be monitored to identify the bacteria growth, which could be a very effective approach to eradicate diseases or infections. Fig. 1(b) shows the E. coli infection mechanism step by step. When contaminated food, such as undercooked meat, raw vegetables, or unpasteurized milk, is consumed, E. coli can enter the body Fig. 1(b1). E. coli can colonize the large intestine by utilizing adhesion, allowing the bacteria to attach to the epithelial cells lining the intestinal wall, as shown in Fig. 1(b2). Adhesion is facilitated by specialized structures called pili or fimbriae present on the surface of E. coli. Pili are filamentous appendages composed of proteins that extend from the bacterial surface. They had an important role in the initial attachment of E. coli to the epithelial cells. Pili have specific adhesin proteins at their tips that recognize and bind to receptors on the surface of the intestinal epithelial cells. This interaction promotes the tight attachment of E. coli to the cell surface Fig. 1(b3). Once attached, E. coli can release toxins, such as Shiga toxins, responsible for the severe symptoms observed in certain E. coli strains. These toxins damage the cells lining the intestines, leading to inflammation and the breakdown of blood vessels, resulting in bloody diarrhea. The damaged intestinal cells release inflammatory chemicals that attract immune cells, exacerbating the inflammation. In some cases, these toxins can also enter the bloodstream, causing systemic effects Fig. 1(b4). The toxins primarily target the kidneys, leading to a condition known as hemolytic uremic syndrome (HUS), which can result in kidney failure Fig. 1(b5). So, it is crucial to detect E. coli by a rapid and cost-effective method.Fig. 1(c) shows the 3D-printed biosensor substrate and mask, and the brush-painted method was used to prepare the Ag-electrode. Fig. 1(d) shows the layer-by-layer arrangement of the TENG with its photo. Fig. 1(e) illustrates the working mechanism of the TENG device. At the initial state, the two layers of the TENG are fully separated, and there is no generation of charges. When the two layers are fully in contact, positive and negative charges are generated on the triboelectric layer as per the polarity (i). While releasing (ii), the electrical charges flow from the top electrode to the bottom electrode. When the two layers of TENG are fully released (iii), the device is at an equilibrium state, and no electrical charges flow. Again, when pressing the two layers (iv), the electrons flow from the bottom to the top electrode. This periodic contact separation leads to the generation of the TENG output.
Fig. S1(a) (ESI†) shows the room-temperature XRD analysis of PLA, pPDMS, and aluminum. XRD pattern of PLA shows a wide diffraction peak centered at 2θ = 16°. The absence of a distinctive peak in the PLA shows the structure is amorphous.33 In the case of PDMS, a smaller and broader at 2θ = 22.6° is depicted. These findings show that the material is amorphous.34 The peak at (2θ = 38.54, 44.78, 65.12°)corresponds to the aluminum peaks.35 The morphological properties of pPDMS, aluminum, and 3D printed PLA substrate are shown in Fig. S1(b)–(d) (ESI†). The EDS spectra and at% of Si, C, and O in pPDMS reveal the homogeneous distribution of elements on the PDMS surface, as shown in Fig. S1(e) (ESI†). The EDS spectra of Al is shown in Fig. S1(f) (ESI†).
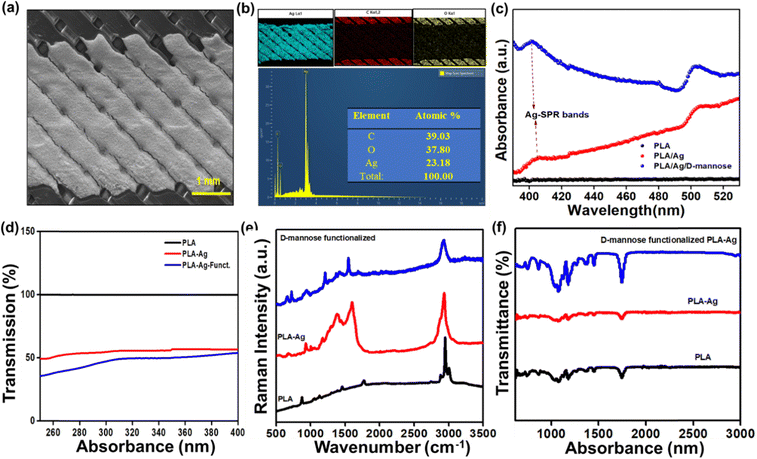
As previously discussed, the biosensor was fabricated by a cost-effective method and characterized for bacteria sensing. Fig. 2(a) shows the SEM image of a brush-painted Ag electrode upon a PLA substrate. The elemental color mapping and EDS spectra in Fig. 2(b) reveal the distribution of Ag, C, and O in the system, and no evidence of additional impurities can be seen. Fig. S2(a) (ESI†) shows the digital image of the biosensor. The conductivity of the IDT patterned electrodes is shown in Fig. S2(b) and (c) (ESI†).
Fig. 2(c) shows the UV-visible absorption spectra of the PLA, PLA/Ag, and PLA/Ag/D-mannose between 390 nM to 530 nM. The 3D-printed PLA polymer matrix did not show any noticeable absorption bands. However, after connecting the electrode to the neat PLA with conducting Ag-paint, for the PLA/Ag system, the absorption increases above 400 nm. The well-known surface plasmon resonance (SPR) band due to silver nanoparticles was detected around 400 nm for both PLA/Ag and the D-mannose modified PLA/Ag samples.36 However, the observed Ag-SPR band around 400 nm is weak for both PLA/Ag and the modified PLA/Ag/D-mannose. The intensity of such an Ag-SPR band depends on the size of Ag-nanoparticles. The present study uses silver paste instead of synthesizing the Ag-nanoparticles on the PLA polymer.36 The intensity of the Ag-SPR band is highest for the PLA/Ag/D-mannose, it is because, during the fabrication of the sensor with MPA solution and D-mannose probable, the Ag-paste degrades to release greater Ag-particles as compared to plain PLA/Ag sample. Also, between 400 and 490 nm, the absorbance for the PLA/Ag increases with wavelength, whereas for the D-mannose modified PLA/Ag, it decreases with the increase in wavelength. This behavior can also be attributed to the interaction of the thiol group (S–H) of Ag particles and –COOH of MPA with D-mannose.
Fig. 2(d) shows the transmission % of PLA, PLA-Ag, and PLA-Ag-functionalized, where the transmission % of neat PLA is 100%, and the % decreases with electrode painting and functionalizing. Raman spectroscopy was performed to study the PLA, PLA/Ag, and PLA/Ag/D-mannose systems, focusing on specific intensity shifts in different spectrum regions. Fig. 2(e) shows the Raman spectra for neat PLA. PLA spectrum shows the characteristic Raman-bands at 3001 cm−1, 2946 cm−1, and 2880 cm−1, related to asymmetric and symmetrical stretching vibrations of the C–H (νas/sCH3) bond of the PLA chain; 1769 cm−1, corresponds to the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (νC
O (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bond; 1453 cm−1, corresponds to the asymmetric deformation vibration of the CH3 bound (δasCH3); 872 cm−1, corresponding to the stretching vibration of the C–COO (νC–COO) bond.37 Raman spectra of PLA/Ag do not contain any specific peaks corresponding to Ag-NPs due to the formation of some s Ag–O bonds38 arising from the interface between AgNPs with the carboxylate groups (n Ag–OCO–) of PLA.38 Also, the spectra show the characteristic signature Raman peaks of PLA at 549 cm−1 (C–O–C vibration), 674 cm−1 (C
O) bond; 1453 cm−1, corresponds to the asymmetric deformation vibration of the CH3 bound (δasCH3); 872 cm−1, corresponding to the stretching vibration of the C–COO (νC–COO) bond.37 Raman spectra of PLA/Ag do not contain any specific peaks corresponding to Ag-NPs due to the formation of some s Ag–O bonds38 arising from the interface between AgNPs with the carboxylate groups (n Ag–OCO–) of PLA.38 Also, the spectra show the characteristic signature Raman peaks of PLA at 549 cm−1 (C–O–C vibration), 674 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration), 935 cm−1 (C–COO vibration), 1277 and 1382 cm−1 (CH deformation vibration), 1598 cm−1 (asymmetric C
O stretching vibration), 935 cm−1 (C–COO vibration), 1277 and 1382 cm−1 (CH deformation vibration), 1598 cm−1 (asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of carboxylate groups of PLA), and 2923 cm−1 (CH3 symmetric and asymmetric stretching vibration).38
O stretching vibrations of carboxylate groups of PLA), and 2923 cm−1 (CH3 symmetric and asymmetric stretching vibration).38
The stretching vibrations (ν) of the CH2 groups of the monosaccharides were analyzed in the 2800–3000 cm−1 region. Additionally, the bending vibrations (δ) of the CH2 and C–OH groups were observed in the 1200 to 1500 cm−1 region.39 This study found that D-mannose exhibited similar vibrational behavior to the monosaccharide in the aforementioned regions, indicating the presence of these functional groups in the PLA/Ag/D-mannose system. The fingerprint region, spanning approximately 950–1200 cm−1, provided characteristic bands that represented the conformation of the rings and the relative orientation of several substituents.40 The vibrations observed in this region were associated with the deformation vibration (δ) of C–OH, C–CH, and O–CH groups. The interval between 600 and 950 cm−1 allowed for specifying the β and α anomers for D-mannose and the characterization of pyranose and furanose rings. Specifically, the intensities of the peaks at ∼960 and ∼974 cm−1 were analyzed to determine the anomeric proportions of α and β forms.40 In the PLA/Ag/D-mannose system studied, the intensity of the peak at ∼974 cm−1 was higher compared to the ∼960 cm−1 peak, which exhibited a weaker signal. Based on this observation, it can be concluded that the PLA/Ag/D-mannose electrode contained a higher proportion of the β anomeric form. This Raman study provides valuable information about the vibrational behavior and composition of the PLA/Ag/D-mannose system, shedding light on its molecular structure and characteristics.
FT-IR spectroscopy was employed to analyze the Ag-NP interaction with PLA. Fig. 2(f) displays the FT-IR peaks corresponding to PLA, PLA/Ag, and PLA/Ag/D-mannose. Specifically, the peaks observed at 1179 (1074), 2994 (2945), and 3522 cm−1 were attributed to the stretching of C–O, C–H (double), and O–H groups, respectively, present in the –CH (CH3)–OH end group of PLA. The band at 1744 cm−1, representing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O carbonyl stretching, likely indicated the presence of the –CH–CO–O– group. Furthermore, the 1445 and 1350 cm−1 peaks were associated with –CH3 and –CH– bending, encompassing both symmetric and asymmetric bending.36 The interaction between PLA chain molecules and Ag-NPs was linked to the peak at 3489 cm−1.36 The slight broadening of this peak can be attributed to van der Waals interactions between the –O–H group of PLA and the partially positive charge on the surface of the Ag-NPs.36 Additionally, the interaction of Ag-NPs with the carbonyl group in PLA is evident from the shift of the carbonyl peak from 1744 cm−1 in PLA to 1748 cm−1 in the PLA/Ag system.
O carbonyl stretching, likely indicated the presence of the –CH–CO–O– group. Furthermore, the 1445 and 1350 cm−1 peaks were associated with –CH3 and –CH– bending, encompassing both symmetric and asymmetric bending.36 The interaction between PLA chain molecules and Ag-NPs was linked to the peak at 3489 cm−1.36 The slight broadening of this peak can be attributed to van der Waals interactions between the –O–H group of PLA and the partially positive charge on the surface of the Ag-NPs.36 Additionally, the interaction of Ag-NPs with the carbonyl group in PLA is evident from the shift of the carbonyl peak from 1744 cm−1 in PLA to 1748 cm−1 in the PLA/Ag system.
The FTIR spectra of PLA/Ag/D-mannose provide insight into the modified material's molecular composition and structural characteristics. One notable feature observed is the presence of free hydroxyl groups associated with the terminal ends of the structure, which exhibit a peak at 3343 cm−1. These hydroxyl groups are likely derived from D-mannose, indicating the successful incorporation of the sugar moiety into the PLA/Ag composite. The spectra also reveal distinct stretching vibrations related to different functional groups in the modified material. The C–H stretching groups originating from the aromatic protective group within the sugar skeleton are observed in the 3000 to 3100 cm−1 range. This region signifies the presence of these aromatic C–H bonds within the PLA/Ag/D-mannose structure. Additionally, signals corresponding to the CH2 groups within the overall structure can be observed at approximately 2900 to 3000 cm−1. Furthermore, the intense vibration associated with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in the ester bond is evident at around 1753 cm−1. This peak confirms the presence of ester linkages within the PLA/Ag/D-mannose material. The frequencies related to the C–O bonds fall within the range of 1363 to 1076 cm−1, indicating the characteristic stretching vibrations of these bonds in the composite.41
O group in the ester bond is evident at around 1753 cm−1. This peak confirms the presence of ester linkages within the PLA/Ag/D-mannose material. The frequencies related to the C–O bonds fall within the range of 1363 to 1076 cm−1, indicating the characteristic stretching vibrations of these bonds in the composite.41
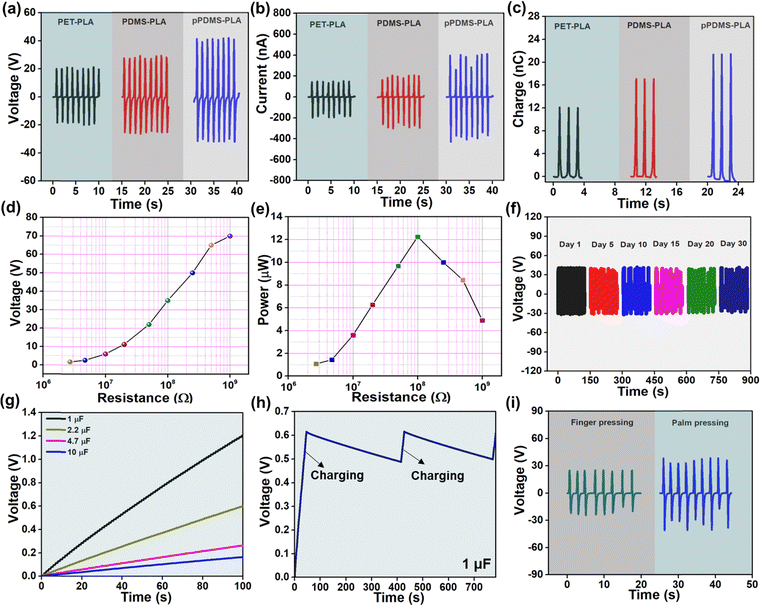
The electrical output (voltage, current, and charge) of a PLA-based TENG with several opposite layers is shown in Fig. 3(a–c). As PLA shows positive triboelectric polarity, the negative triboelectric layers were considered PET, plain PDMS, and pPDMS. The results vary depending on the position in the triboelectric series and the roughness; the pPDMS-PLA-based TENG produces the best results. The pPDMS-PLA TENG's output voltage, current, and charge are 70 V, 800 nA, and 22 nC, respectively.
The porosity in the PDMS can produce a high surface area, which enhances charge transfer and promotes efficient energy conversion. Porosity improves air circulation and contact between surfaces, enhancing TENG performance. For further experiments, PLA-pPDMS-based TENG is selected for further studies. Fig. 3(d) shows the voltage output of PLA-pPDMS TENG at different load resistance. It follows the trend of Ohm's law: the resistance value increases, and the voltage rises. By varying the load resistances, the corresponding power output of the TENG was presented in Fig. 3(e). The highest power of 12 μW across a resistance of 108 Ω. The long-term stability of the device was tested for over 30 days, and the constant output was obtained for a constant frequency of 1 Hz and 5 N, presented in Fig. 3(f). Fig. 3(g) shows the charging of different capacitors (1, 2.2, 4.7, 10 μF) for 100 s using the TENG and bridge rectifier circuit. The stable electrical output can also be confirmed in Fig. 3(h), elucidating the charge and discharge plot of a 1 μF capacitor. TENG device was used to harvest energy from body motions such as finger and palm pressing, shown in Fig. 3(i). The performance of the TENG demonstrates its usability and stability for effective energy harvesting.
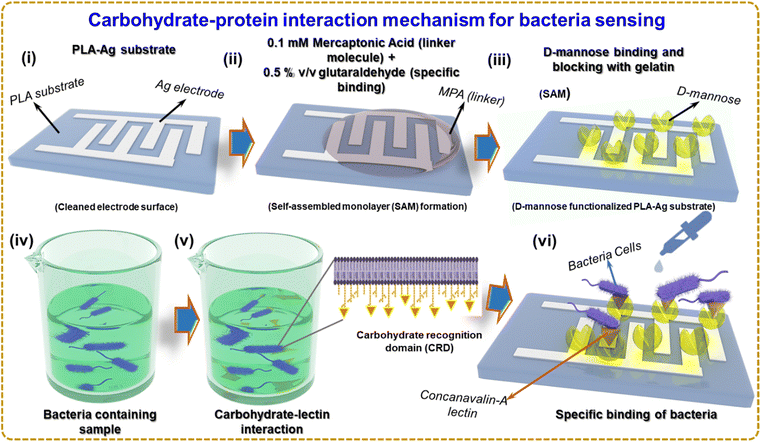
Fig. 4 illustrates the sensing mechanism of bacteria based on a carbohydrate–protein bond. Self-assembled monolayer (SAM) formation is a technique that can be used to immobilize D-mannose on the surface of a silver electrode, as shown in Fig. 4(i) and (ii). One of the most widely investigated types of SAMs is based on the attachment of alkanethiols (in the present case 3-MPA) to gold, silver, copper, palladium, platinum, and mercury surfaces. The strong attraction of thiols to these metals allows the creation of well-defined organic surfaces with versatile and easily modifiable chemical functions displayed at the exposed interface.42 Using SAMs to functionalize noble metal surfaces provides a simple route to functionalize electrode surfaces by organic molecules (both aliphatic and aromatic) containing free anchor groups such as thiols, disulfides, amines, acids or silanes for improved stability of biomolecules.43 Due to the high affinity of SH groups to metals, thiol-terminated SAMs have attracted tremendous attention for construction of biomolecular electronic devices.44 The thiol group (-SH) of 3-MPA has a lone pair of electrons on sulfur. When the 3-MPA solution is applied to the silver electrode surface, the thiol groups bind covalently to the silver surface, forming a covalent bond between the sulfur atom of 3-MPA and the silver atoms on the surface. This step is called chemisorption. The sensor is incubated for a few hours to allow D-mannose to bind to the linker molecules (3-MPA) on the surface. A 2% v/v glutaraldehyde solution is applied to the sensor and incubated for 1 hour at room temperature. Glutaraldehyde serves as a cross-linker and activates D-mannose on the surface. Gelatin solution blocks unreacted sites on the silver electrode surface, preventing the non-specific binding of bacteria or other unwanted molecules to the immobilized D-mannose surface. The surface with the blocking agent is incubated for 1–2 hours. The immobilized D-mannose on the surface of the Ag electrode selectively binds to lectins that recognize mannose on the surface of bacteria and recognize the bacterium. The sample containing the bacteria was supplemented with Concanavalin-A (Con A), a lectin, a class of proteins that recognizes certain carbohydrates through a process known as carbohydrate–protein binding.45 Con A is an effective material for sensing bacteria because it has a strong affinity for mannose and glucose residues found on the surface of several microorganisms, including bacteria such as E. coli.46 Con A could be involved in identifying and reacting with carbohydrates found on bacterial surfaces.47 Like several other organisms, bacteria have carbohydrates on the surface of their cells. These carbohydrates can perform various tasks, such as signaling, cell recognition, and adhesion.48
Fig. 4(iv) and (v) depicts several stages during Con A interactions with bacteria. First, Con A recognizes the carbohydrate recognition domain (CRD) and binds to mannose residues on the surface of E. coli pili. van der Waals forces, electrostatic interactions, hydrogen bonds, and other non-covalent interactions play a role in this bonding. The lectin then undergoes a conformational change in response to Con A binding to E. coli pili, forming a hydrophobic pocket that contacts hydrophobic molecules on the bacterial surface. The relationship between Con A and E. coli pili is further stabilized by this interaction. Now that D-mannose has been immobilized on the Ag electrode surface, it can be used to identify bacteria that selectively bind to D-mannose. As shown in Fig. 4(vi), the surface of the biosensor was exposed to the bacterial suspension containing Con A in the PBS buffer solution, and the impedance change of the Ag electrode surface can be seen. Bacteria binding D-mannose can affect the electrical properties of the surface, which can be monitored and used for bacterial detection. Fig. S3(a) and (b) (ESI†) shows the molecular diagram of the reaction which occurs at (ii) and (iii) of Fig. 4.
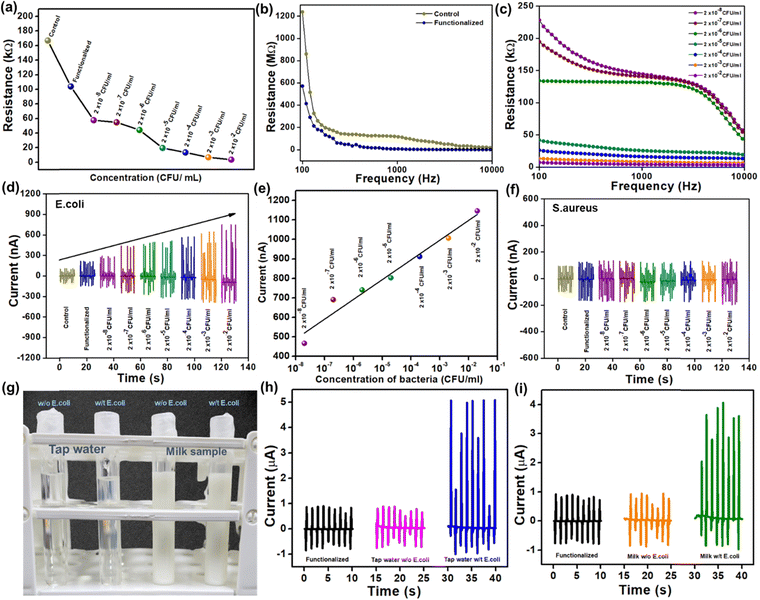
Fig. 5(a) shows the shift in resistance of the sensor at control, functionalized, and different bacterial concentrations. The biosensor was further connected with the impedance analyzer, and 100 μl of bacterial suspension with PBS buffer was added drop by drop with different concentrations ranging from 2 × 10−8 to 2 × 10−2 ml−1. Fig. 5(b) and (c) shows an instant resistance drop, where the control and functionalized samples have higher resistances at lower frequency regions. The resistance decreases to the concentrations as per the frequency-dependent impedance measurement. The biosensor is now connected with PLA-pPDMS-based TENG in parallel connection to detect the output changes to the concentration of bacteria.
As shown in Fig. 5(d), the current output of the PLA-TENG increases when the bacteria suspension of 2 × 10−8 to 2 × 10−2 ml−1 is added. The biosensor response could be directly related to the current output of TENG. In simple terms, as the concentration of bacteria on the biosensor increases, the resistance of the biosensor reduces, causing the current of PLA TENG to grow following Ohm's law (V = IR). The self-powered biosensing device offered an outstanding linear response to E. coli concentration, as shown in Fig. 5(e). The corresponding regression coefficient (R2) is R2 =![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.962. As can be observed in Fig. 5(f), Staphylococcus aureus (S. aureus) detection was used as a negative control to test the selectivity of the self-powered biosensing system. Changes in S. aureus concentrations do not result in any detectable changes in the current output. Only bacteria with pili containing the lectin protein, which is lacking in the case of S. aureus, can be detected by the synthesized biosensor with the mechanism of carbohydrate–protein interaction. This outcome indicates the specific sensing mechanism of the designed biosensor.
0.962. As can be observed in Fig. 5(f), Staphylococcus aureus (S. aureus) detection was used as a negative control to test the selectivity of the self-powered biosensing system. Changes in S. aureus concentrations do not result in any detectable changes in the current output. Only bacteria with pili containing the lectin protein, which is lacking in the case of S. aureus, can be detected by the synthesized biosensor with the mechanism of carbohydrate–protein interaction. This outcome indicates the specific sensing mechanism of the designed biosensor.
Finally, Fig. 5(g) shows a digital photo of real-time bacteria sensing using materials including tap water and unpasteurized milk. The current suddenly increases with E. coli in both the sample and the output current, reaching up to 6 μA and 5 μA, respectively. However, the output current from the control sample and the tap water and milk samples without E. coli are practically identical. In conclusion, the self-powered biosensing system can quickly and efficiently detect specific Gram-negative bacteria (E. coli), aiding in timely medical interventions and disease control.
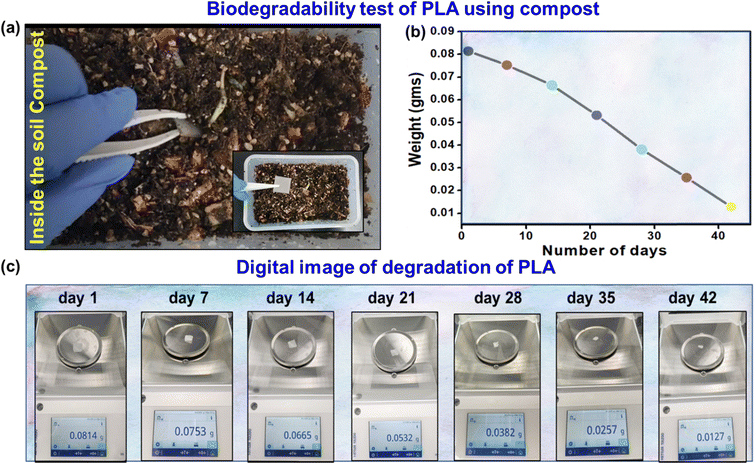
Biodegradable sensors are essential to solve environmental problems and advance sustainable technologies.49–51 The spontaneous biodegradability of these sensors after use minimizes the accumulation of waste and has no adverse effects on the environment.52,53 Due to their bio-friendly nature, they can also be used for biomedical applications as implanted devices. Biodegradable sensors offer an attractive option for eco-friendly and healthcare purposes.54,55 Therefore, the biodegradability test of the PLA-based biosensor was demonstrated using soil compost. The 3D-printed PLA was placed in the soil compost, as shown in Fig. 6(a), and the temperature and moisture content were maintained at 40 °C and 50%, respectively, for 42 days. Fig. 6(b) illustrates the estimated degradation percentage and a graph showing the decreasing weight of PLA with increasing time. Fig. 6(c) shows a digital image of PLA degradation with a 7 day interval. The result indicates that the fabricated sensor is naturally biodegradable and supports sustainable device engineering options.
4. Conclusions
In conclusion, a self-powered TENG-based biosensor has been developed for the rapid detection of bacteria, implementing D-mannose as a sensing layer and a simple carbohydrate–protein binding mechanism. E. coli, a Gram-negative bacterium with pill-like structural characteristics, was used as a prototype to demonstrate the viability of self-powered bio-sensing. The biosensor can accurately detect E. coli concentrations by monitoring the output current because changes in E. coli concentration cause changes in resistance. In addition, real-time samples such as tap water and unpasteurized milk were used to detect E. coli. S. aureus, a Gram-positive bacterium without pili, was chosen as a negative control to demonstrate the selectivity of the self-powered biosensor. Further, the biosensor was tested for biodegradability in soil compost by maintaining a consistent temperature and humidity. This study presents a fast and efficient method for detecting E. coli and offers valuable information for advancing TENG-based self-powered sensor systems.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work is supported by the National Research Foundation of Korea (2021R1C1C101158813) and Ministry of Trade, Industry and Energy of Korea (RS-2023-00231350). VV thank Department of Science and Technology, India for the financial support through PURSE program (SR/PURSE/2023/196: TPN: 88213), KLEF for the support through KLEF R&D Funding (KLEF/IFP/2022-2023/ECE/007) and the Royal Society of Chemistry for the Researcher Grant R22-7381975972. Support from the EPSRC (EP/S02106X/1) UK is also acknowledged.References
- S. Ramos, V. Silva, M. D. L. E. Dapkevicius, M. Caniça, M. T. Tejedor-Junco, G. Igrejas and P. Poeta, Animals, 2020, 10, 2239 CrossRef PubMed.
- S. Ishii and M. J. Sadowsky, Microb. Environ., 2008, 23, 101–108 CrossRef PubMed.
- C. M. Thorpe, Clin. Infect. Dis., 2004, 38, 1298–1303 CrossRef PubMed.
- G. Ekici and E. Dümen, Escherichia coli and food safety, The universe of Escherichia coli, IntechOpen, 2019 Search PubMed.
- S. D. Manning, Escherichia coli infections, Infobase Publishing, 2010 Search PubMed.
- J. B. Kaper, J. P. Nataro and H. L. Mobley, Nat. Rev. Microbiol., 2004, 2, 123–140 CrossRef CAS PubMed.
- T. G. Obrig, Toxins, 2010, 2, 2769–2794 CrossRef CAS PubMed.
- J. Detzner, G. Pohlentz and J. Müthing, Toxins, 2020, 12, 373 CrossRef CAS PubMed.
- R. A. Exeni, R. J. Fernandez-Brando, A. P. Santiago, G. A. Fiorentino, A. M. Exeni, M. V. Ramos and M. S. Palermo, Pediatr. Nephrol., 2018, 33, 2057–2071 CrossRef PubMed.
- A. J. Linscott, Clin. Microbiol. Newsletter, 2011, 33, 41–45 CrossRef.
- S. Ünüvar, Microbial foodborne diseases, Foodborne Diseases, Elsevier, 2018, pp. 1–31 Search PubMed.
- M.-A. Lim, J.-Y. Kim, D. Acharya, B. B. Bajgain, J.-H. Park, S.-J. Yoo and K. Lee, Int. J. Environ. Res. Public Health, 2020, 17, 3149 CrossRef PubMed.
- T. Kim and J.-I. Han, J. Environ. Manage., 2013, 130, 267–275 CrossRef CAS.
- J.-C. Cheng, C.-L. Huang, C.-C. Lin, C.-C. Chen, Y.-C. Chang, S.-S. Chang and C.-P. Tseng, Clin. Chem., 2006, 52, 1997–2004 CrossRef CAS PubMed.
- Y. Liu, M. Cao, Z. Huang, C. Yu, N. Yang, Q. Wu, L. Shi, W. Duan, Y. Zhu, J. Wei, L. Li and W. Huang, Chin. Chem. Lett., 2022, 33, 1855–1860 CrossRef CAS.
- Y. Wang, C. Zhang, H. Zhang, L. Feng and L. Liu, Chin. Chem. Lett., 2022, 33, 4605–4609 CrossRef CAS.
- C. Wang, P. Wang, J. Chen, L. Zhu, D. Zhang, Y. Wan and S. Ai, Nano Energy, 2022, 93, 106828 CrossRef CAS.
- A. A. Mathew, A. Chandrasekhar and S. Vivekanandan, Nano Energy, 2021, 80, 105566 CrossRef CAS.
- S. Panda, S. Hajra, K. Mistewicz, P. In-na, M. Sahu, P. M. Rajaitha and H. J. Kim, Nano Energy, 2022, 100, 107514 CrossRef CAS.
- S. Hajra, M. Sahu, A. M. Padhan, I. S. Lee, D. K. Yi, P. Alagarsamy, S. S. Nanda and H. J. Kim, Adv. Funct. Mater., 2021, 31, 2101829 CrossRef CAS.
- S. Panda, H. Shin, S. Hajra, Y. Oh, W. Oh, J. Lee, P. Rajaitha, B. K. Panigrahi, J. Shukla and A. K. Sahu, J. Materiomics, 2023, 9, 609–617 CrossRef.
- G. Khandelwal, S. Deswal and R. Dahiya, ACS Omega, 2022, 7, 44573–44590 CrossRef CAS PubMed.
- S. Du, N. Zhou, G. Xie, Y. Chen, H. Suo, J. Xu, J. Tao, L. Zhang and J. Zhu, Nano Energy, 2021, 85, 106004 CrossRef CAS.
- X. Qu, P. Qi, P. Wang, J. Li, C. Wang, D. Zhang, Y. Wan, S. Ai and X. Wang, Sens. Actuators, B, 2023, 390, 133917 CrossRef CAS.
- Z. Zhou, P. Wang, J. Li, C. Wang, J. Chen, L. Zhu, H. Zhu and D. Zhang, Nano Energy, 2022, 98, 107317 CrossRef CAS.
- M. Sahu, S. Hajra, S. Panda, M. Rajaitha, B. K. Panigrahi, H.-G. Rubahn, Y. K. Mishra and H. J. Kim, Nano Energy, 2022, 97, 107208 CrossRef CAS.
- W. Oh, S. Hajra, S. Divya, S. Panda, Y. Oh, Z. Jaglic, P. Pakawanit, T. H. Oh and H. J. Kim, Mater. Sci. Eng., B, 2023, 292, 116397 CrossRef CAS.
- S. Hajra, P. In-na, C. Janpum, S. Panda and H. J. Kim, Electron. Mater. Lett., 2023, 19, 367–373 CrossRef CAS.
- X. Ma, W. Ding, C. Wang, H. Wu, X. Tian, M. Lyu and S. Wang, Sens. Actuators, B, 2021, 331, 129422 CrossRef CAS.
- M. Alafeef, P. Moitra and D. Pan, Biosens. Bioelectron., 2020, 165, 112276 CrossRef CAS PubMed.
- A. Li, H.-H. Ho, S. R. Barman, S. Lee, F. Gao and Z.-H. Lin, Nano Energy, 2022, 93, 106826 CrossRef CAS.
- L. Lan, J. Ping, J. Xiong and Y. Ying, Adv. Sci., 2022, 9, 2200560 CrossRef CAS.
- V. Giita Silverajah, N. A. Ibrahim, W. M. Z. W. Yunus, H. A. Hassan and C. B. Woei, Int. J. Mol. Sci., 2012, 13, 5878–5898 CrossRef PubMed.
- P. Ferreira, Á. Carvalho, T. R. Correia, B. P. Antunes, I. J. Correia and P. Alves, Sci. Technol. Adv. Mater., 2013, 14, 055006 CrossRef CAS PubMed.
- F. Peng, H. Wang, H. Yu and S. Chen, Mater. Res. Bull., 2006, 41, 2123–2129 CrossRef CAS.
- K. Shameli, M. B. Ahmad, W. M. Z. W. Yunus, N. A. Ibrahim, R. A. Rahman, M. Jokar and M. Darroudi, Int. J. Nanomed., 2010, 573–579 CrossRef CAS PubMed.
- A. M. Pandele, A. Constantinescu, I. C. Radu, F. Miculescu, S. Ioan Voicu and L. T. Ciocan, Materials, 2020, 13, 274 CrossRef CAS PubMed.
- N. Vidakis, M. Petousis, E. Velidakis, M. Liebscher and L. Tzounis, Biomimetics, 2020, 5, 42 CrossRef CAS.
- M. Dudek, G. Zajac, E. Szafraniec, E. Wiercigroch, S. Tott, K. Malek, A. Kaczor and M. Baranska, Spectrochim. Acta, Part A, 2019, 206, 597–612 CrossRef CAS PubMed.
- V. Dumouilla and C. G. Dussap, Bioengineered, 2021, 12, 4420–4431 CrossRef CAS PubMed.
- A.-M. Pană, L.-M. Rusnac, G. Bandur, M. Silion, C. Deleanu and M. Bălan, e-Polymers, 2011, 11, 004 Search PubMed.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103–1170 CrossRef CAS PubMed.
- Z. Shen, M. Huang, C. Xiao, Y. Zhang, X. Zeng and P. G. Wang, Anal. Chem., 2007, 79, 2312–2319 CrossRef CAS PubMed.
- S. K. Arya, P. R. Solanki, M. Datta and B. D. Malhotra, Biosens. Bioelectron., 2009, 24, 2810–2817 CrossRef CAS PubMed.
- F. A. Jaipuri, B. Y. Collet and N. L. Pohl, Angew. Chem., 2008, 120, 1731–1734 CrossRef.
- R. Bryce, I. Hillier and J. Naismith, Biophys. J., 2001, 81, 1373–1388 CrossRef CAS PubMed.
- O. A. Loaiza, P. J. Lamas-Ardisana, E. Jubete, E. Ochoteco, I. Loinaz, G. Cabanero, I. García and S. Penadés, Anal. Chem., 2011, 83, 2987–2995 CrossRef CAS.
- N. Sharon and H. Lis, Glycobiology, 2004, 14, 53R–62R CrossRef CAS PubMed.
- J. Min, Y. Jung, J. Ahn, J. G. Lee, J. Lee and S. H. Ko, Adv. Mater., 2023, 2211273 CrossRef PubMed.
- N. P. Maria Joseph Raj, N. R. Alluri, V. Vivekananthan, A. Chandrasekhar, G. Khandelwal and S.-J. Kim, Appl. Energy, 2018, 228, 1767–1776 CrossRef CAS.
- V. Vivekananthan, N. R. Alluri, Y. Purusothaman, A. Chandrasekhar, S. Selvarajan and S.-J. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 18650–18656 CrossRef CAS.
- E. S. Hosseini, S. Dervin, P. Ganguly and R. Dahiya, ACS Appl. Bio Mater., 2020, 4, 163–194 CrossRef PubMed.
- S. Panda, S. Hajra, H. Jeong, B. K. Panigrahi, P. Pakawanit, D. Dubal, S. Hong and H. J. Kim, Nano Energy, 2022, 102, 107682 CrossRef CAS.
- N. Ashammakhi, A. L. Hernandez, B. D. Unluturk, S. A. Quintero, N. R. de Barros, E. Hoque Apu, A. Bin Shams, S. Ostrovidov, J. Li, C. Contag, A. S. Gomes and M. Holgado, Adv. Funct. Mater., 2021, 31, 2104149 CrossRef CAS.
- T. A. Baldo, L. F. de Lima, L. F. Mendes, W. R. de Araujo, T. R. L. C. Paixão and W. K. T. Coltro, ACS Appl. Electron. Mater., 2021, 3, 68–100 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01820b |
| This journal is © The Royal Society of Chemistry 2023 |