Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Controlling the structure of supramolecular fibre formation for benzothiazole based hydrogels with antimicrobial activity against methicillin resistant Staphylococcus aureus†‡
Kira L. F.
Hilton
a,
Antonis A.
Karamalegkos
b,
Nyasha
Allen
b,
Lauren
Gwynne
c,
Bree
Streather
b,
Lisa J.
White
a,
Karen B.
Baker
b,
Samantha A.
Henry
b,
George T.
Williams
 d,
Helena J.
Shepherd
d,
Helena J.
Shepherd
 a,
Mark
Shepherd
b,
Charlotte K.
Hind
e,
Mark J.
Sutton
e,
Toby A.
Jenkins
*c,
Daniel P.
Mulvihill
*b,
Jennifer M. A.
Tullet
*b,
Marina
Ezcurra
*b and
Jennifer R.
Hiscock
a,
Mark
Shepherd
b,
Charlotte K.
Hind
e,
Mark J.
Sutton
e,
Toby A.
Jenkins
*c,
Daniel P.
Mulvihill
*b,
Jennifer M. A.
Tullet
*b,
Marina
Ezcurra
*b and
Jennifer R.
Hiscock
 *a
*a
aSchool of Chemistry and Forensics, University of Kent, Canterbury, CT2 7NH, UK. E-mail: J.R.Hiscock@Kent.ac.uk
bSchool of Biosciences, University of Kent, Canterbury, CT2 7NJ, UK. E-mail: d.p.mulvihill@kent.ac.uk; j.m.a.tullet@kent.ac.uk; m.ezcurra@kent.ac.uk
cDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: chsataj@bath.ac.uk
dSchool of Chemistry, University of Southampton, Highfield, Southampton, SO17 1BJ, UK
eResearch and Evaluation, Porton Down, UKHSA, Porton Down, Salisbury SP4 0JG, UK
First published on 12th April 2023
Abstract
Antimicrobial resistance is one of the greatest threats to human health. Gram-positive methicillin resistant Staphylococcus aureus (MRSA), in both its planktonic and biofilm form, is of particular concern. Herein we identify the hydrogelation properties for a series of intrinsically fluorescent, structurally related supramolecular self-associating amphiphiles and determine their efficacy against both planktonic and biofilm forms of MRSA. To further explore the potential translation of this hydrogel technology for real-world applications, the toxicity of the amphiphiles was determined against the eukaryotic multicellular model organism, Caenorhabditis elegans. Due to the intrinsic fluorescent nature of these supramolecular amphiphiles, material characterisation of their molecular self-associating properties included; comparative optical density plate reader assays, rheometry and widefield fluorescence microscopy. This enabled determination of amphiphile structure and hydrogel sol dependence on resultant fibre formation.
Introduction
Antimicrobial resistance (AMR) is one of the world's most underestimated global health threats and as such is increasingly referred to as the ‘silent pandemic’.1 Data collected as part of a 2022 study by Naghavi and co-workers, over 204 countries, confirmed ∼5 million people died as a result of AMR in 2019 alone, a greater number than those who died from malaria or HIV/AIDS over that same time period.2 In addition, there has been a global increase in the use of antimicrobials as a result of the COVID-19 pandemic, due to factors which include misdiagnosis and co-infection treatment, further exacerbating the prevalence of AMR.3,4As part of the same 2022 study, methicillin resistant Staphylococcus aureus (MRSA) infections were found to be directly responsible for over 3.5 million disability adjusted life years and > 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2019 within high-income countries.2 One of the main reasons for this is that MRSA can remain viable on dry and inanimate objects/surfaces for months.5,6
000 deaths in 2019 within high-income countries.2 One of the main reasons for this is that MRSA can remain viable on dry and inanimate objects/surfaces for months.5,6
With AMR strains now presenting resistance to all antimicrobials currently marketed,7 including colistin, the antibiotic of last resort,8 the innovation of new antimicrobial agents displaying efficacy towards these strains is vital. Here the field of supramolecular chemistry has provided a number of unique systems, demonstrating the potential for development as antimicrobial agents into the clinic.9–11
This includes the production of novel compounds that can selectively disrupt the bacterial membrane due to inherent differences between bacterial and eukaryotic cell structures.12 This mode of antimicrobial action has been successfully targeted by researchers such as Vale and co-workers, who have developed a series of gemini serine-based surfactants that have demonstrated antimicrobial activity against several different MRSA strains.13 Bala and co-workers also targeted membrane disruption as a mode of antimicrobial action when developing a series of imino-functionalised 1,3-diazolium salts, demonstrating similar levels of antimicrobial activity to that of antibiotics gentamicin and kanamycin against both methicillin sensitive Staphylococcus aureus (S. aureus) and MRSA.14–16 Naturally derived antimicrobial peptides (AMPs) self-assemble to form larger structures that have intrinsic antimicrobial properties.17–19 The AMPs overall positive charge created due to the presence of cationic amino acid residues, causes these compounds to interact with negatively charged bacterial membranes.12,20 For example, melittin is an AMP found in bee venom and has been shown to act as a broad spectrum antimicrobial,21 firstly accumulating on the membrane, before forming pores or fissures.18 Additionally, Koksch and co-workers produced synthetic AMPs based on a 3,5-diaminobenzoic acid scaffold that achieved antimicrobial efficacy against S. aureus and other Gram-positive bacteria including Micrococcus luteus.22
Supramolecular self-associating amphiphiles (SSAs), such as those shown in Fig. 1, represent a novel class of antimicrobial agent with activity against MRSA. The hypothesised mode of action for these SSAs include selective membrane coordination and disruption.23
Furthermore, SSAs have been shown to: (i) act as (potential) drug delivery systems;24,25 (ii) enhance the activity of commonly used antimicrobials including; novobiocin and rifampicin against a variety of bacteria such as the ESKAPE pathogen, Pseudomonas aeruginosa;26,27 (iii) enhance the activity of cisplatin against ovarian cancer cells;28 and (iv) exhibit a druggable profile when administered intravenously to mice.23
In general, the anionic portion of an SSA is known to self-associate, through the formation of (thio)urea-anion intermolecular hydrogen bonds, to form anionic dimers in the solid state (Fig. 2), gas phase and in polar organic solutions, such as DMSO.29,30 In aqueous ethanol solutions these same SSAs self-associate to form spherical aggregates with hydrodynamic diameters (dH) ∼100–550 nm. However, in previous work, we also showed SSA 2 (Fig. 1) to form a hydrogel upon the addition of aqueous salts such as NaCl, followed by an annealing process in which the aqueous sample was heated to ∼313 K before being allowed to cool to room temperature. As both the spherical aggregate and the hydrogel, this SSA was shown to retain antimicrobial activity against planktonic MRSA.25 Herein, we investigate how structural alterations of 2 (1, 3–7) affect SSA self-association events and resultant material formation. In addition, we quantify the antimicrobial efficacy of those hydrogels formed against MRSA in both a planktonic form and when contained within a homogenous biofilm for the first time. Finally, the toxicity of selected SSAs was established against nematode worms (Caenorhabditis elegans) as model multicellular eukaryotic organisms.31
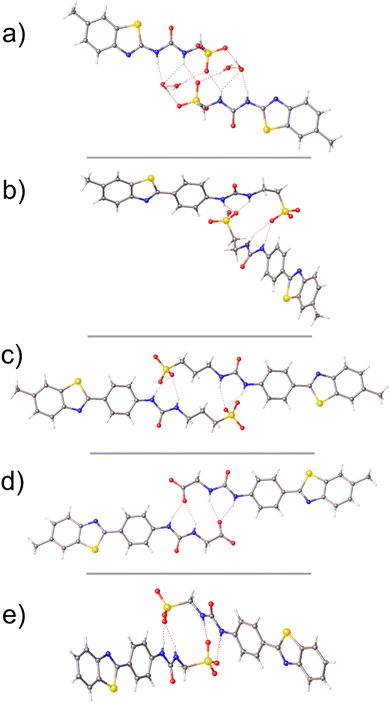 | ||
| Fig. 2 Single crystal X-ray structure of SSAs 1 (a), 3 (b), 4 (c), 6 (d) and 7. The TBA counter cations and any molecular disorder have been omitted for clarity. Grey = carbon, blue = nitrogen, red = oxygen, yellow = sulfur, white = hydrogen, red dashed lines = hydrogen bonds.† | ||
Results and discussion
Synthesis
SSAs 1–7 were prepared in line with previously published syntheses.23,29,32Solid state single crystal X-ray diffraction studies
Single crystals of 1 and 3–7 were obtained through slow evaporation of an EtOH:H2O 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 solution containing the relevant SSA. The resultant crystal structures obtained (Fig. 2) all show the anionic component of these SSAs to form urea-anion dimers, in line with previous results obtained for 2.29 However, unlike the majority of SSAs,30 including those within Fig. 2, the urea-anion dimer produced with 1 is stabilised through the formation of two urea-anion intermolecular hydrogen bonds only, one from each of the two urea NH's. The remaining urea NH's form intermolecular hydrogen bonds with additional water molecules (Fig. 2a). This produces a planar dimer with a 180.00° internal angle of dimerisation.
19 solution containing the relevant SSA. The resultant crystal structures obtained (Fig. 2) all show the anionic component of these SSAs to form urea-anion dimers, in line with previous results obtained for 2.29 However, unlike the majority of SSAs,30 including those within Fig. 2, the urea-anion dimer produced with 1 is stabilised through the formation of two urea-anion intermolecular hydrogen bonds only, one from each of the two urea NH's. The remaining urea NH's form intermolecular hydrogen bonds with additional water molecules (Fig. 2a). This produces a planar dimer with a 180.00° internal angle of dimerisation.
The addition of a phenyl ring and elongation of the urea-anion alkyl linking group from methyl (2) to ethyl (3), resulted in the formation of an SSA dimer exhibiting an internal angle of dimerisation 135.40(40)°, stabilised through the formation of four hydrogen bonds, one from each of urea NH's to the sulfonate ion. Further elongation of the urea-anion alkyl linker from ethyl (3) to propyl (4) produced an anion dimer exhibiting the same hydrogen bonding mode as 3 however, this dimer is planar due to crystal packing forces.
Replacing the sulfonate ion (2) with a carboxylate functional group (6) produced a hydrogen bonded dimer very similar to that produced with 4. However, removal of the methyl group appended from the benzothiazole functionality (7) resulted in the formation of an SSA hydrogen bonded dimer with an internal angle of dimerisation 95.12(13)°, making this structure comparable to that previously obtained for 2.29 Although not directly comparable to the solution state, understanding the self-associative properties determined via single crystal X-ray diffraction methods enables insight into the self-associative properties of this class of compounds that we hypothesise to drive nanostructure and material formation events within the solution state.
Solution state self-association studies
In line with our previous work and standard characterisation procedures for this class of compound,24 we initially studied the self-association properties of 1 and 3–7 in polar organic solvents (DMSO/DMSO-d6) to establish the low level self-association properties of these SSAs. Analogous studies for 2 have been previously published.29 These data are summarised within the ESI,‡ Sections 7 and 8. Quantitative 1H NMR studies were used to confirm the presence of larger self-associated species in solution through comparative integration with an internal standard, in this instance DCM. Here any apparent ‘loss’ of SSA 1H NMR signal is attributed to the formation of higher order self-associated aggregates which exhibit solid-like properties and thus become invisible in the solution state NMR experiment. As communicated within our previous work,24 we first use quantitative 1H NMR spectroscopy to ascertain whether the self-associated species present can be observed using traditional solution state NMR techniques. Should all of the species present in solution be visible via NMR spectroscopy then we perform 1H NMR DOSY experiments, if this is not the case then we perform DLS experiments to estimate the size of those species present. Here, only 7 showed any evidence of higher order aggregation events (Fig. S14, ESI‡), dynamic light scattering (DLS) estimated a dH for those species present to be 196–>1000 nm (obtained from peak maxima of an intensity weighted distribution). However, these data should be treated with caution due to high levels of polydispersity and the amorphous nature of those aggregated species produced, as indicated by the quality of the data collected (Fig. S58 and S59, ESI‡). The DLS experiment also confirmed the presence of lower order species in solution, comparable in size to those previously obtained for the formation of the SSA anion hydrogen bonded dimer (dH = 1.15 nm).For 1 and 3–6, where no apparent ‘loss’ in 1H NMR SSA signal could be identified through comparative quantitative 1H NMR integration, the size of the SSA species present in solution was determined through 1H NMR DOSY experiments. The data obtained confirmed that the SSA anion and cation were not strongly coordinated, while the dH of the SSA anion (Table S9, ESI‡) suggested the formation of hydrogen bonded anionic dimers in solution. The strength of SSA anion dimerisation for 1 and 3–6 (Table S1, ESI‡), under these same DMSO-d6 (0.5% H2O) solvent conditions, was calculated using data obtained from 1H NMR dilution studies and subsequent fitting to the equal K model using Bindfit v0.5.33
SSAs 1 and 3 exhibit dimerisation constants <0.01 M−1 whereas 2 and 4 exhibit dimerisation constants of 2.7 M−1 and 6.26 M−1 respectively. SSA 6 was shown to form the strongest hydrogen bonded dimers with a dimerisation constant of 92.72 M−1. We believe that the lack of hydrogen bonded self-association events observed for 1 and 3 is due to the decreased acidity of the hydrogen bond donating groups (Section 19, ESI‡34,35) and intramolecular hydrogen bond formation between the sulfonate and urea NH as hypothesised previously for SSAs containing a urea-anion ethyl linking group respectively.30 The increase in alkyl chain length (2 and 4) was also shown to increase the dimerisation constant which we hypothesise may be due to the preferential packing of the longer alkyl chains (see Fig. 2c). The substitution of the sulfonate for the carboxylate ion (6) resulted in an order of magnitude increase in dimerisation constant (Section 7, ESI‡) due to enhanced anion basicity and preferential spatial arrangement (Section 19, ESI‡).
Moving into biologically relevant aqueous conditions, the self-association properties of the 1–7 were established through a combination of quantitative 1H NMR, DLS, zeta potential and tensiometry (to derive critical aggregation concentration – CAC), the results of which are summarised in Table 1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 or D2O/EtOH 95
5 or D2O/EtOH 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (NMR only) solution. Aggregate stability and dH were obtained via zeta potential and DLS measurements respectively, at a concentration of 5.56 mM and a temperature of 298 K, following an annealing process unless otherwise stated. The dH of the aggregates listed were obtained from intensity distribution peak maxima. CAC was derived at approximately 291 K from surface tension measurements. All quantitative 1H NMR experiments were conducted with a delay time (d1) of 60 s at 298 K and a concentration of 5.56 mM unless stated. The values given in % represent the observed proportion of compound to become NMR silent
5 (NMR only) solution. Aggregate stability and dH were obtained via zeta potential and DLS measurements respectively, at a concentration of 5.56 mM and a temperature of 298 K, following an annealing process unless otherwise stated. The dH of the aggregates listed were obtained from intensity distribution peak maxima. CAC was derived at approximately 291 K from surface tension measurements. All quantitative 1H NMR experiments were conducted with a delay time (d1) of 60 s at 298 K and a concentration of 5.56 mM unless stated. The values given in % represent the observed proportion of compound to become NMR silent
| SSA | Quantitative 1H NMR (%) | d H (nm) | CAC (mM) | Surface tension at CAC (mN m−1) | Zeta potential (mV) | |
|---|---|---|---|---|---|---|
| SSA anion | SSA cation | |||||
| a Study conducted at 0.56 mM (1) or 2.79 mM (6) due to compound solubility. b Could not be calculated due to signal overlap. c Solubility prevented data acquisition. | ||||||
| 1 | 62a | 181a | >0.56a | N/A | −42.71a | |
| 229 | 10 | 8 | 300 | 0.50 | 46.50 | −101.00 |
| 3 | 48 | 45 | 851 | 1.9223 | 56.2423 | −88.29 |
| 4 | 19 | 115 | 14.7723 | 45.1623 | −74.80 | |
| 5 | 4.5623 | 44.9223 | ||||
| 6 | 53a | 47a | 127 | 3.0323 | 49.9023 | −84.22a |
| 7 | 55 | 51 | 255 | 5.1623 | 66.9723 | −67.13 |
Quantitative 1H NMR experiments, using 5% EtOH as the internal standard in D2O, confirmed the presence of higher-order aggregated species for all SSAs at the concentrations tested. The presence of the carboxylate functionality was found to increase the proportion of SSA to become incorporated into the higher-order self-associated species, as does removal of the methyl group from the benzothiazole functionality and the presence of the ethyl sulfonate-urea linker.
Zeta potential measurements of 1–7 confirm the presence of stable self-associated aggregates (where ±30 mV is considered stable),36,37 which have a dH = 94–300 nm, except 3 which exhibits a dH > 800 nm. SSA 2 demonstrated the lowest CAC, with increasing urea-anion alkyl linker length shown to produce the most detrimental effects towards decreasing CAC.
SSA hydrogel characterisation
The addition of an aqueous salt solution to 2, followed by an annealing process, has been shown to induce hydrogel formation.25,38 Following these same gel formation processes (Section 2, ESI‡), 1 and 3–7 were tested for their propensity to act as hydrogelators. The aqueous salts solutions (0.505 M) used within the scope of this study include NaCl – the lead hydrogelator solution as identified from previous studies,24 KCl – to investigate change in cation and NaNO3, NaH2PO4, NaOBz, Na2SO4 – to investigate change in anion basicity/geometry. In initial screening experiments, each SSA (5 mg mL−1) was added to the relevant aqueous salt solution (0.505 M, 1 mL), heated to ∼323 K and allowed to cool to room temperature. The sample was then inverted, and should the mixtures flow be impeded, the sample was taken forward for further analysis.Following this, the minimum gelation concentration (MGC) was determined. The results of these studies are summarised in Table 2. In comparison to lead hydrogelator 2, removal of the benzothiazole methyl group (7) was found to have the least detrimental effect on hydrogelator efficacy, followed by elongation of the urea-anion alkyl linking group (3 and 4). These data also show that the benzene ring between the benzothiazole and urea functionality (1) is essential for hydrogelation as it is responsible for increasing hydrophobicity, activating the hydrogen bond donating NH's, enabling intermolecular hydrogen bond formation, and increasing propensity for preferential intermolecular π–π stacking interactions. The replacement of the urea (2) for the thiourea (5) was also found to prevent gelation. Although the thiourea is known to increase hydrogen bond donor acidity, this functionality also decreases molecular planarity which we hypothesise is a driving factor for material formation in this instance. Changing the SSA anion from a sulfonate (2) to a carboxylate (6), also has a detrimental effect on hydrogelation, which we hypothesise is due to anion geometry/resultant hydrogen bonding mode and increased anion hydrophilicity.
Although removal of the benzothiazole methyl group (7) had the least effect on hydrogelation, the change in hydrogel properties indicates that the methyl functionality is important for material formation. We hypothesise this is due to a reduction in hydrophobicity and steric effects, phenomena often exhibited in proteins.39–41
Following this, the formation of a hydrogel and the physical properties of that material were confirmed through completion of a series of rheology experiments. Here, the formation of a solid material is confirmed where; the storage modulus (G′) the elastic component of the material, is greater than the loss modulus (G′′), the liquid component of the material (Section 13, ESI‡). In these amplitude experiments, a linear viscoelastic region is identified where G′ and G′′ are parallel to one another (G′ > G′′). Where an increase in shear strain (γ) results in G′ becoming less than G′′, this cross-over point is identified as the gel/sol phase transition.42 These data have been summarise in Table 3.
| Salt | Gel/sol transition point (%) | |
|---|---|---|
| 2 | 7 | |
| NaCl | 46.6 | 31.6 |
| NaNO3 | 21.5 | 31.6 |
| NaOBz | 21.5 | 6.8 |
Next, the material melting temperature (Tm) of the SSA hydrogels were determined. The results of these studies are summarised in Table S4 (ESI‡). Hydrogels of 2 exhibited a Tm of 324–327 K, whereas hydrogels of 7 exhibited Tm of 308–310 K. Again, this shows that the methyl functionality appended to the benzothiazole group increases hydrogel stability with respect to increasing temperature. Hydrogels containing 7 and NaOBz were found to produce the least stable materials with a Tm = 308 K. The formation of these materials was further characterised in line with our novel plate reader technique.38 Here, hydrogel samples are formed in a 96-well plate through heating a solution past the Tm and allowing the solution to cool back to 298 K within the plate reader. Optical density measurements at 450 nm (OD450) were taken for 177 individual sections of each well at set temperatures. As the gel fibres were formed the optical density of these sections increased, thus enabling the mapping of fibre formation. The results of these studies are detailed within Section 18 of the ESI.‡
SSA hydrogel formation
The presence of the intrinsically fluorescent benzothiazole unit within the structure of 2–7 enables the direct observation of hydrogel fibre formation using widefield fluorescent microscopy. SSAs 2–7 were visualised using a 457/50 nm (blue channel) emission filter, the results of these studies are exemplified in Fig. 3 and can be found in full in Section 14 (ESI‡). Here, a chamber was created on a microscope slide using a coverslip which was secured on three of its four sides (Section 2, ESI‡). The relevant SSA (5 mg mL−1) in the appropriate aqueous salt solution (0.505 M) was heated to ∼323 K, pipetted into the microscopy slide chamber and allowed to cool on the microscope stage to enable the observation of fibre formation in real time. No fibre formation was observed for aqueous salt solutions containing 5, showing the urea moiety to drive fibre formation.To enable the comparison of those data contained within the microscopy images (Section 14, ESI‡) the gel fibres were sorted by fibre characteristic. A minimum of 4 microscopy images for each SSA sample were analysed, and when characterising the individual hydrogel fibre, a minimum of 15 fibres were analysed for each image. These fibre characteristics included whether the structures were curled (Fig. 3d) or straight (Fig. 3c), linked (Fig. 3c) or independent (Fig. 3a and f), short (<20 nm, Fig. 3c) or long (>20 nm, Fig. 3d and f), packing density (densely packed – minimum solvent visible, Fig. 3d and e, medium packing – solvent visible and crosslinking clearly discernible, Fig. 3a and c, low packing density – mostly solvent visible and little evidence of crosslinking, Fig. 3b), uniform (Fig. 3c) or stochastic (Fig. 3d–f), unidirectional (Fig. 3a) or multidirectional (Fig. 3e).
The results of the gel fibre characterisation are summarised in Table 4. The majority of fibres observed were curled rather than straight, however, some samples showed a combination of both curled and straight fibres, for example, 3 in aqueous NaOBz and Na2SO4 and 7 in NaH2PO4. In addition, most fibres were stochastic rather than uniform, with fibre uniformity increasing when the SSA alkyl linker length is increased from methyl (2) to ethyl (3) and to a lesser extent when extended from methyl (2) to propyl (4). This increase in alkyl linker length also saw a switch from mostly unidirectional fibres to multidirectional fibres when the alkyl linker was extended from methyl (2) to propyl (4). This switch from unidirectional to multidirectional fibre characteristic was also observed upon the removal of the methyl group from the benzothiazole functionality (7).
The gel fibres of 2, 3 and 7 were relatively unaffected by the change of added aqueous salt cation from NaCl to KCl, with the characteristics of the fibres exhibiting similar properties. However, the fibres of 4 and 6 were affected by the change of added salt cation to a greater extent. The fibres that were formed in aqueous KCl tended to be linked rather than independent. In changing the inorganic anion from NaCl to NaH2PO4, only samples containing 2 and 7 were found to exhibit fibre formation. Hydrogel fibres of 2 in NaH2PO4 were curled, independent and uniform, whereas fibres of 7 are almost the exact opposite; straight and curled, linked, and multidirectional. Extending the SSA alkyl linker from methyl (2) to ethyl (3) or propyl (4) and swapping the sulfonate SSA anion to a carboxylate (6) saw no fibre formation in aqueous solutions of NaH2PO4, supporting the results of the original MGC experiments (Table 2). Gel fibres formed in aqueous NaOBz produced stochastic fibres, which we hypothesise may be due to the increased hydrophobicity of the added salt anionic component or the enhanced basicity of the carboxylate anion, affecting how the SSAs self-associate. Increasing the anionic strength of the salt solution (Na2SO4) caused the gel fibres produced to be curled and crosslinked in all instances where an SSA hydrogel was formed. These studies show that fibre formation can be controlled through step-wise alteration of the SSA anion structure and the presence of additional salts.
When comparing the results obtained from these microscopy studies with the results from rheological measurements obtained from analogous systems, the following structure–material property relationships were determined, confirming that the characteristic of an SSA hydrogel fibre or resulting network to influence resultant material properties. For example, the gel/sol transition point is highest for the NaCl hydrogels of 2 (46.6%). We believe this is due to the following hydrogel fibre characteristics – fibres formed from 2 in NaCl are curled, short and multidirectional, compared to fibres formed from 2 in NaNO3 and NaOBz which are straight, long and unidirectional.
When comparing hydrogels formed from 7, all gel fibre characteristics are the same with the exception of fibre packing density. The hydrogels formed from NaCl and NaNO3 exhibit medium packed and densely packed fibres respectively however, hydrogels formed in the presence of NaOBz exhibit a low packing density. This transition from medium to low packing density was found to correspond to decreasing gel/sol transition points from 31.6% to 6.8%.
SSA hydrogel antimicrobial efficacy
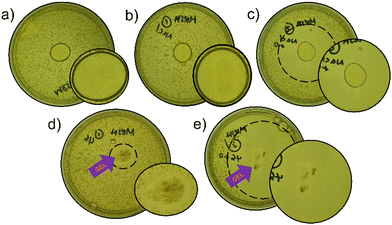
To determine the antimicrobial efficacy of the materials within this study, a series of gel plate assays, as described by White et al., were performed.25 Here, the NaCl hydrogel of 7 was chosen as the lead material from those novel materials identified based on the following: (i) biological compatibility of the additional salt solution; (ii) presence of a hydrogel material. In addition, the potential for this material to act as a delivery vehicle for the antibiotic oxacillin was also explored.Within the scope of these initial studies, a ∼50 mg aliquot of the appropriate SSA hydrogel mixture was transferred to the surface of an agar plate inoculated with MRSA USA300, Fig. 4. In addition, control experiments were performed, where the SSA material was replaced with a 1% agarose gel containing either H2O (Fig. 4a), NaCl (0.505 M, Fig. 4b) or oxacillin (8 mM) and NaCl (0.505 M – Fig. 4c). These control experiments showed MRSA growth in the presence of the agarose gel containing H2O or NaCl (0.505 M) only. However, the oxacillin impregnated agarose gel did exhibit a zone of inhibition (ZOI), demonstrating antimicrobial activity, as the oxacillin diffused out of the material. The hydrogel of 7 in NaCl (0.505 M) also exhibited antimicrobial properties (Fig. 4d). The resultant hydrogel produced through co-formulation of 7 in NaCl (0.505 M) and oxacillin (8 mM, Fig. 4e) resulted in a ZOI comparable to the 1% agarose gel impregnated with oxacillin (Fig. 4c). Due to the difference in material characteristics between the agarose and the SSA material, direct comparison of additive antimicrobial activity cannot be inferred from this experiment.
Biofilms are aggregations of bacterial cells that are surrounded by an extracellular polymeric substance (EPS).43 These aggregations of bacteria can form on almost any surface and can be classed as pseudo-multicellular organisms, which can offer bacteria increased fitness and resistance to antimicrobials and antibiotics.44–46 The EPS, which is secreted by the bacterial cells, acts as a barrier to the environment, including a hosts immune system,47 making penetrating the EPS one of the biggest challenges for antimicrobials and antibiofilm agents.48,49 For example, vancomycin, a common antibiotic, has an MIC that is 10 times greater against MRSA biofilms, compared to the planktonic bacteria.50
To test the antibiofilm efficacy of the SSA hydrogels/heterogenous gel–sol mixtures for which efficacy had previously been determined against planktonic MRSA USA300,23 a porcine skin biofilm model was adapted, see Fig. 5.51–54 Here, blank agar, absent of nutrients, was poured into a Petri dish, onto which the porcine skin was placed. The skin was then inoculated with MRSA USA300 and the relevant NaCl material was placed on top of the skin. After 18 hours incubation at 305 K, the skin was placed in phosphate-buffered saline (PBS) and sonicated. The PBS was then diluted and plated to determine the colony forming units (CFU) per mL (Fig. 5). A lower CFU observed is indicative of increased anti-biofilm efficacy. The hydrogels of 2 and 7 showed potential antibiofilm activity, as did the heterogeneous gel/sol mixture of 3 when compared to the control sample. However, the heterogenous gel/sol mixture of 4 showed no antibiofilm activity compared to that control sample.
When comparing the antibiofilm properties of these hydrogels and heterogeneous gel/sol mixtures (Fig. 5) with the structure of the fibres formed within these materials (Table 4) we find that the heterogenous material formed from 4 was the only sample to contain fibres that were both ‘long’ and ‘densely packed’ in addition to being ‘curled’, ‘stochastic’ and ‘multidirectional’ in nature. The mode of action for antimicrobial hydrogels such as those detailed herein, normal includes a mechanical action by which the material fibres are found to mechanically disrupt the microbial membrane.9 The combination of fibre characteristics details for the NaCl heterogenous hydrogel of 4, leads us to hypothesise that in comparison with those other SSA materials tested, these fibres would lack the physical properties necessary permeate the cell, hence leading to decreased antimicrobial activity.
SSA toxicity
Finally, the model organism Caenorhabditis elegans (C. elegans) was used to assess the toxicity of 1–7. Fluorescent microscopy was used in combination with developmental and brood size assays to establish the deleterious effects of 1–7 on this model eukaryotic multicellular organism.31 The adult hermaphrodite genome of C. elegans has 60–80% conservation to humans, and ∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 protein coding genes.55–58 Recent reviews on C. elegans have shown that this model organism is becoming an increasingly useful tool in toxicology.31,56,59,60 This is due to a relatively short life cycle (Fig. 6) of 3 days, their hermaphroditic nature and their ability to produce large numbers of offspring, making this organism easy to work with in the lab. C. elegans are also transparent, enabling internal visualisation through accessible microscopy techniques however, the wild type (WT) N2 C. elegans are intrinsically auto-fluorescent, exhibiting an excitation wavelength of 340 nm, and an emission range of 360–600 nm.61 In this instance, 2–7 exhibit overlapping fluorescence properties with the WT N2 C. elegans, thus, the fluorescence of the SSAs could not be distinguished from the natural fluorescence of the WT N2 C. elegans. Therefore, the GLO-1 mutant, which exhibits a reduced degree of autofluorescence, was employed in these studies.62
000 protein coding genes.55–58 Recent reviews on C. elegans have shown that this model organism is becoming an increasingly useful tool in toxicology.31,56,59,60 This is due to a relatively short life cycle (Fig. 6) of 3 days, their hermaphroditic nature and their ability to produce large numbers of offspring, making this organism easy to work with in the lab. C. elegans are also transparent, enabling internal visualisation through accessible microscopy techniques however, the wild type (WT) N2 C. elegans are intrinsically auto-fluorescent, exhibiting an excitation wavelength of 340 nm, and an emission range of 360–600 nm.61 In this instance, 2–7 exhibit overlapping fluorescence properties with the WT N2 C. elegans, thus, the fluorescence of the SSAs could not be distinguished from the natural fluorescence of the WT N2 C. elegans. Therefore, the GLO-1 mutant, which exhibits a reduced degree of autofluorescence, was employed in these studies.62
To introduce the SSAs into the C. elegans, the SSAs were added to the organism's food source, which in this instance is Escherichia coli (E. coli) OP50 (Section 17, ESI‡). GLO-1 mutant C. elegans worms were incubated on solid agar plates seeded with E. coli OP50 supplemented with the relevant SSA (0.10 mM in 5% EtOH, which was found to have a minimal effect on E. coli OP50 S123–129, ESI‡) for 24 hours at 293 K. At this point, the C. elegans were placed onto a microscopy slide to enable analysis. The microscopy slide was prepared by pipetting molten 2% agarose onto a slide and flattening to form a mounting pad, on to which ∼10 individual worms were placed. Levamisole (10 μL, 100 mM) was used to immobilise the worms for viewing, and a glass cover slide was placed on top of the sample. We can confirm that 2–7 were ingested by the worms as fluorescence can be seen inside the gut lumen as exemplified in Fig. 7 and further detailed within Section 17 (ESI‡). No observable effect on worm health was identified for 1–7, however, two further assays were performed with 4 and 6 to substantiate this initial observation. Here, the effect of 4 and 6 on the development of C. elegans was evaluated through a development and a brood size assay.
The developmental assay was carried out by growing GLO-1 mutant C. elegans worms on E. coli OP50 seeded solid agar plates supplemented with the relevant SSA (0.10 M). The results of these experiments were compared to suitable controls, where the development of these worms was observed in the absence of SSA. A comparative measure of development was achieved by ratioing the number of worms in L3 to the L4 developmental stage after 52 hours incubation (Fig. 6). The results of these experiments are shown in Fig. 8a. Here, there is no significant difference between the results of the developmental assays conducted in the presence or absence of SSA. This shows the C. elegans to develop normally with no evidence of these organisms showing signs of stress and thus entering the dauer developmental stage of growth.
 | ||
| Fig. 8 Effect of SSAs 4 and 6 on C. elegans GLO-1 mutant. (a) Development stage after 52 hours. Purple = L3, orange = L4. (b) Brood size. Error = standard deviation. Control = 0.05% EtOH. | ||
The impact of SSAs 4 and 6 on the brood size of C. elegans was then evaluated (Fig. 8b). In brief, ten worms in the L4 developmental stage (Fig. 6) were plated on individual E. coli OP50 seeded solid agar plates in the presence or absence of the relevant SSA (0.10 mM), allowed to produce eggs and moved to a new plate each day. The number of eggs produced by each worm was then counted after 3 days (Section 17, ESI‡). No effect on brood size was observed upon the addition of 4 and 6, further demonstrating that at the concentrations tested, these SSAs show limited toxicity against C. elegans.
Conclusions
Herein, we successfully determined several structure activity relationships between fibre formation, hydrogelation, antimicrobial activity, and anti-biofilm activity. The removal of the benzene ring in the centre of the SSA structure (1) is detrimental to: (i) SSA self-association; (ii) hydrogelation; (iii) antimicrobial activity against planktonic MRSA USA300; and (iv) antibiofilm activity against MRSA USA300 biofilms.Increasing the alkyl linker length between the SSA anion and urea groups from methyl (2) to ethyl (3) to propyl (4) was found to dramatically alter the structure of fibres formed in aqueous salt solutions, with the aqueous salt solutions themselves also shown to impact the morphology of any resultant structures.
Substituting the urea (2) for the thiourea (5) functionality was found to be detrimental to hydrogel formation, which we attribute to the decrease in planarity of the thiourea when compared to the urea functionality. Changing the SSA anion from a sulfonate (2) to a carboxylate (6) was also found to be detrimental to hydrogel formation. These observations combined with others from this family of compounds show the structure of fibres formed within supramolecular self-association processes can be controlled and tailored towards specific applications. In addition, we show that fibre/network structure confirmed through fluorescence microscopy techniques, does impact on the physical characteristics of the result hydrogel, specifically the gel/sol transition point determined via rheological amplitude sweep measurements.
Finally, initial toxicology studies were undertaken against the model eukaryotic multicellular organism, C. elegans. Here, the results of microscopy studies conducted with 1–7 showed no evidence of toxicity at those concentrations tested, with 4 and 6 also demonstrating no impact on C. elegans development or brood size. Given these promising results, we are now focusing our efforts on establishing mode of action to enable the production of ever more effective antimicrobial SSA hydrogels.
Author contributions
KLFH: investigation; validation; writing – original draft, review & editing. AAK, BS, NA, LG, LJW, KKB, SAH, GTW, HJS, CKH, JMS: investigation; validation; writing – review & editing. ATAJ, DPM, JMAT, MS, ME: supervision; validation; writing – review & editing; funding acquisition. JRH: conceptualization; funding acquisition; project administration; supervision; Writing – original draft, review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
KLFH, LJW and JRH would like to thank the University of Kent for funding. AAK would like to thank the GCDC for funding. KLFH and JRH would like to thank Dr John Stolz for his help reviewing this document. LJW and JRH would also like to thank the UKRI (MR/T020415/1) for funding.Notes and references
- E. Charani, M. McKee, R. Ahmad, M. Balasegaram, C. Bonaconsa, G. B. Merrett, R. Busse, V. Carter, E. Castro-Sanchez, B. D. Franklin, P. Georgiou, K. Hill-Cawthorne, W. Hope, Y. Imanaka, A. Kambugu, A. J. Leather, O. Mbamalu, M. McLeod, M. Mendelson, M. Mpundu, T. M. Rawson, W. Ricciardi, J. Rodriguez-Manzano, S. Singh, C. Tsioutis, C. Uchea, N. Zhu and A. H. Holmes, Lancet Reg. Health: Eur., 2021, 7, 100161 Search PubMed.
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, N. Mturi, T. Munera-Huertas, P. Musicha, M. M. Mussi-Pinhata, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, A. Y. Peleg, C. Perrone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, K. E. Rudd, N. Russell, J. Schnall, J. A. G. Scott, M. Shivamallappa, J. Sifuentes-Osornio, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, H. Vu, T. Walsh, S. Waner, T. Wangrangsimakul, T. Wozniak, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- C.-C. Lai, S.-Y. Chen, W.-C. Ko and P.-R. Hsueh, Int. J. Antimicrob. Agents, 2021, 57, 106324 CrossRef CAS PubMed.
- G. M. Knight, R. E. Glover, C. F. McQuaid, I. D. Olaru, K. Gallandat, Q. J. Leclerc, N. M. Fuller, S. J. Willcocks, R. Hasan, E. van Kleef and C. I. Chandler, eLife, 2021, 10, e64139 CrossRef CAS PubMed.
- D. R. Bhatta, S. Koirala, A. Baral, N. M. Amatya, S. Parajuli, R. Shrestha, D. Hamal, N. Nayak and S. Gokhale, Can. J. Infect. Dis. Med. Microbiol., 2022, 2022, e1023241 Search PubMed.
- A. Kramer, I. Schwebke and G. Kampf, BMC Infect. Dis., 2006, 6, 130 CrossRef PubMed.
- E. D. Brown and G. D. Wright, Nature, 2016, 529, 336–343 CrossRef CAS PubMed.
- K. Talkington, C. Shore and P. Kothari, The Pew Charitable Trust: Philadelphia, PA, USA, 2016, 1, 33–39.
- J. A. Doolan, G. T. Williams, K. L. F. Hilton, R. Chaudhari, J. S. Fossey, B. T. Goult and J. R. Hiscock, Chem. Soc. Rev., 2022, 51, 8696–8755 RSC.
- G. T. Williams, C. J. E. Haynes, M. Fares, C. Caltagirone, J. R. Hiscock and P. A. Gale, Chem. Soc. Rev., 2021, 50, 2737–2763 RSC.
- T. Ohashi, M. Nagashima, N. Kawai, N. Ohmagari and K. Tateda, Expert Rev. Anti-Infect. Ther., 2022, 20, 1603–1614 CrossRef CAS PubMed.
- K. L. F. Hilton, C. Manwani, J. E. Boles, L. J. White, S. Ozturk, M. D. Garrett and J. R. Hiscock, Chem. Sci. J., 2021, 12, 13273–13282 CAS.
- S. G. Silva, M. Pinheiro, R. Pereira, A. R. Dias, R. Ferraz, C. Prudêncio, P. J. Eaton, S. Reis and M. L. C. do Vale, Biochim. Biophys. Acta, Biomembr., 1864, 2022, 183969 Search PubMed.
- A. N. W. Kadafour, H. Ibrahim and M. D. Bala, J. Mol. Struct., 2022, 1262, 132997 CrossRef CAS.
- X.-C. Yang, C.-F. Hu, P.-L. Zhang, S. Li, C.-S. Hu, R.-X. Geng and C.-H. Zhou, Bioorg. Chem., 2022, 124, 105855 CrossRef CAS PubMed.
- Z. Deng, H. Sun, R. R. Y. Bheemanaboina, Y. Luo and C.-H. Zhou, Bioorg. Med. Chem. Lett., 2022, 64, 128695 CrossRef CAS PubMed.
- R. J. Dubos, J. Exp. Med., 1939, 70, 1–10 CrossRef CAS PubMed.
- S. Pandidan and A. Mechler, Sci. Rep., 2019, 9, 10841 CrossRef PubMed.
- J. Zhang, Y. Zhao, S. Han, C. Chen and H. Xu, Sci. China: Chem., 2014, 57, 1634–1645 CrossRef CAS.
- E. Glukhov, M. Stark, L. L. Burrows and C. M. Deber, J. Biol. Chem., 2005, 280, 33960–33967 CrossRef CAS PubMed.
- A. Tossi, L. Sandri and A. Giangaspero, J. Pept. Sci., 2000, 55, 4–30 CrossRef CAS.
- C. K. Thota, A. A. Berger, B. Harms, M. Seidel, C. Böttcher, H. von Berlepsch, C. Xie, R. Süssmuth, C. Roth and B. Koksch, J. Pept. Sci., 2020, 112, e24130 CAS.
- J. E. Boles, C. Bennett, J. Baker, K. L. F. Hilton, H. A. Kotak, E. R. Clark, Y. Long, L. J. White, H. Yuk Lai, C. K. Hind, J. Mark Sutton, M. D. Garrett, A. Cheasty, J. L. Ortega-Roldan, M. Charles, C. J. E. Haynes and J. R. Hiscock, J. Chem. Sci., 2022, 13, 9761–9773 CAS.
- L. J. White, J. E. Boles, K. L. F. Hilton, R. J. Ellaby and J. R. Hiscock, Molecule, 2020, 25, 4126 CrossRef CAS PubMed.
- L. J. White, J. E. Boles, N. Allen, L. S. Alesbrook, M. J. Sutton, C. K. Hind, K. L. F. Hilton, L. R. Blackholly, R. J. Ellaby, G. T. Williams, D. P. Mulvihill and J. R. Hiscock, J. Mat. Chem. B, 2020, 8, 4694–4700 RSC.
- J. E. Boles, R. J. Ellaby, H. J. Shepherd and J. R. Hiscock, RSC Adv., 2021, 11, 9550–9556 RSC.
- J. E. Boles, G. T. Williams, N. Allen, L. J. White, K. L. F. Hilton, P. I. A. Popoola, D. P. Mulvihill and J. R. Hiscock, Adv. Ther., 2022, 5, 2200024 CrossRef CAS.
- N. O. Dora, E. Blackburn, J. E. Boles, G. T. Williams, L. J. White, S. E. G. Turner, J. D. Hothersall, T. Askwith, J. A. Doolan, D. P. Mulvihill, M. D. Garrett and J. R. Hiscock, RSC Adv., 2021, 11, 14213–14217 RSC.
- L. J. White, N. J. Wells, L. R. Blackholly, H. J. Shepherd, B. Wilson, G. P. Bustone, T. J. Runacres and J. R. Hiscock, J. Chem. Sci., 2017, 8, 7620–7630 CAS.
- L. J. White, S. N. Tyuleva, B. Wilson, H. J. Shepherd, K. K. L. Ng, S. J. Holder, E. R. Clark and J. R. Hiscock, Eur. J. Chem., 2018, 24, 7761–7773 CrossRef CAS PubMed.
- W. A. Boyd, M. V. Smith and J. H. Freedman, Methods Mol. Biol., 2012, 889, 15–24 CrossRef CAS PubMed.
- A. Rutkauskaite, L. J. White, K. L. F. Hilton, G. Picci, L. Croucher, C. Caltagirone and J. R. Hiscock, Org. Biomol. Chem., 2022, 20, 5999–6006 RSC.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- C. A. Hunter, Angew. Chem., Int. Ed., 2004, 43, 5310–5324 CrossRef CAS PubMed.
- J. J. P. Stewart, J. Mol. Model., 2007, 13, 1173–1213 CrossRef CAS PubMed.
- J. D. Clogston and A. K. Patri, in Characterization of Nanoparticles Intended for Drug Delivery, ed. S. E. McNeil, Humana Press, Totowa, NJ, 2011, pp. 63–70 Search PubMed.
- R. Vogel, A. K. Pal, S. Jambhrunkar, P. Patel, S. S. Thakur, E. Reátegui, H. S. Parekh, P. Saá, A. Stassinopoulos and M. F. Broom, Sci. Rep., 2017, 7, 17479 CrossRef PubMed.
- L. J. White, C. Wark, L. Croucher, E. R. Draper and J. R. Hiscock, Chem. Commun., 2020, 56, 9557–9560 RSC.
- C. S. Leung, S. S. F. Leung, J. Tirado-Rives and W. L. Jorgensen, J. Med. Chem., 2012, 55, 4489–4500 CrossRef CAS PubMed.
- K. Feng, R. E. Quevedo, J. T. Kohrt, M. S. Oderinde, U. Reilly and M. C. White, Nature, 2020, 580, 621–627 CrossRef CAS PubMed.
- H. Schönherr and T. Cernak, Angew. Chem., Int. Ed., 2013, 52, 12256–12267 CrossRef PubMed.
- F. Zad Bagher Seighalani, D. J. McMahon and P. Sharma, Food Hydrocolloids, 2021, 120, 106886 CrossRef CAS.
- M. Vert, Pure Appl. Chem., 2012, 84, 377–410 CrossRef CAS.
- C. de la Fuente-Núñez, F. Reffuveille, L. Fernández and R. E. Hancock, Curr. Opin. Microbiol., 2013, 16, 580–589 CrossRef PubMed.
- P. Bowler, C. Murphy and R. Wolcott, Antimicrob. Resist. Infect. Control, 2020, 9, 162 CrossRef PubMed.
- P. S. Stewart and J. William Costerton, Lancet, 2001, 358, 135–138 CrossRef CAS PubMed.
- H. J. Busscher, H. C. van der Mei, G. Subbiahdoss, P. C. Jutte, J. J. A. M. van den Dungen, S. A. J. Zaat, M. J. Schultz and D. W. Grainger, Sci. Transl. Med., 2012, 4, 153rv10 Search PubMed.
- M. Kostakioti, M. Hadjifrangiskou and S. J. Hultgren, Cold Spring Harbor Perspect. Med., 2013, 3, a010306 Search PubMed.
- J. G. Leid, C. J. Willson, M. E. Shirtliff, D. J. Hassett, M. R. Parsek and A. K. Jeffers, J. Immunol., 2005, 175, 7512–7518 CrossRef CAS PubMed.
- A. H. Salem, W. F. Elkhatib and A. M. Noreddin, J. Pharm. Pharmacol., 2011, 63, 73–79 CrossRef CAS PubMed.
- L. Gwynne, G. T. Williams, K. C. Yan, J. E. Gardiner, K. L. F. Hilton, B. L. Patenall, J. R. Hiscock, J. Y. Maillard, X. P. He, T. D. James, A. C. Sedgwick and A. T. A. Jenkins, Isr. J. Chem., 2021, 61, 234–238 CrossRef CAS.
- Q. Yang, P. L. Phillips, E. M. Sampson, A. Progulske-Fox, S. Jin, P. Antonelli and G. S. Schultz, Wound Repair Regen., 2013, 21, 704–714 CrossRef PubMed.
- T. P. Sullivan, W. H. Eaglstein, S. C. Davis and P. Mertz, Wound Repair Regen., 2001, 9, 66–76 CrossRef CAS PubMed.
- D. R. Alves, S. P. Booth, P. Scavone, P. Schellenberger, J. Salvage, C. Dedi, N.-T. Thet, A. T. A. Jenkins, R. Waters and K. W. Ng, Front. Cell Infect., 2018, 8, 196 CrossRef PubMed.
- P. M. Meneely, C. L. Dahlberg and J. K. Rose, Curr. Protoc., 2019, 19, e35 Search PubMed.
- M. C. K. Leung, P. L. Williams, A. Benedetto, C. Au, K. J. Helmcke, M. Aschner and J. N. Meyer, Toxicol. Sci, 2008, 106, 5–28 CrossRef CAS PubMed.
- P. Shen, Y. Yue, J. Zheng and Y. Park, Annu. Rev. Food Sci. Technol., 2018, 9, 1–22 CrossRef PubMed.
- T. Kaletta and M. O. Hengartner, Nat. Rev. Drug Discovery, 2006, 5, 387–399 CrossRef CAS PubMed.
- K. J. Helmcke, D. S. Avila and M. Aschner, Neurotoxicol. Teratol., 2010, 32, 62–67 CrossRef CAS PubMed.
- P. H. Harlow, S. J. Perry, S. Widdison, S. Daniels, E. Bondo, C. Lamberth, R. A. Currie and A. J. Flemming, Sci. Rep., 2016, 6, 22965 CrossRef CAS PubMed.
- T. Komura, M. Yamanaka, K. Nishimura, K. Hara and Y. Nishikawa, npj Aging Mech. Dis., 2021, 7, 1–11 CrossRef PubMed.
- C. Morris, O. K. Foster, S. Handa, K. Peloza, L. Voss, H. Somhegyi, Y. Jian, M. V. Vo, M. Harp, F. M. Rambo, C. Yang and G. J. Hermann, PLoS Genet., 2018, 14, e1007772 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- O. Dolomanov, L. Bourhis, R. Gildea, J. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
Footnotes |
| † A suitable crystal was selected and mounted on a Rigaku Oxford Diffraction Supernova diffractometer. Data were collected using Cu Kα radiation at 100 K. The structure was solved with the ShelXS63via Direct Methods and refined with ShelXL64 on least squares minimisation. Olex265 was used as an interface to all ShelX programs. |
| ‡ Electronic supplementary information (ESI) available: This includes experimental details and DLS, zeta potential, tensiometry, rheometry, microscopy, mass spectrometry, NMR spectroscopy, crystallography, molecular characterisation and biological data. CCDC 1997675, 2022536, 2023004, 2033371 and 2106928. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3tb00461a |
| This journal is © The Royal Society of Chemistry 2023 |