Enhanced immunogenicity induced by mRNA vaccines with various lipid nanoparticles as carriers for SARS-CoV-2 infection†
Yanhao
Zhang
,
Ji
Wang
,
Hanlei
Xing
,
Chao
Liu
,
Wenhui
Zha
,
Shuo
Dong
,
Yuhao
Jiang
and
Xinsong
Li
 *
*
School of Chemistry and Chemical Engineering, Southeast University, Nanjing 211189, P. R. China. E-mail: lixs@seu.edu.com
First published on 5th July 2023
Abstract
mRNA vaccines have emerged as a highly promising approach for preventing cancer and infectious diseases, attributed to their superior immunogenicity, rapid development speed, and quality-controlled scale production. While homologous mRNA vaccine administration is currently the most prevalent method employed in clinical settings, heterologous administration is a promising avenue worth exploring. In this report, two types of mRNA vaccine formulations for SARS-CoV-2 infection were developed based on different lipid nanoparticle (LNP) delivery systems, and heterologous and homologous mRNA vaccinations were administered to explore the levels of immune responses comparatively. First, five novel H-series ionizable lipids were synthesized and confirmed by NMR and MS. Subsequently, six SARS-CoV-2 receptor-binding domain (RBD) mRNA-encapsulated LNP formulations were prepared using a microfluidic mixer based on H-series and MC3 lipids. These formulations exhibited spherical structures with an average diameter ranging from 90–140 nm, as characterized by dynamic light scattering (DLS) and transmission electron microscopy (TEM). The safety of these formulations was confirmed in vitro by the cytotoxicity assay. Moreover, transfection assay, lysosomal escape test, and western blot, and in vivo biodistribution analyses collectively demonstrated that lipids H03 and MC3 exhibited superior in vitro and in vivo delivery efficacy in comparison to other H-series lipids. Notably, H03-Fluc mRNA exhibited an approximately 2.2-fold higher in vivo bioluminescence signal intensity than MC3-Fluc mRNA. Additionally, evaluation of humoral immunity demonstrated that homologous H03-mRNA vaccination elicited an immune response that was approximately 3-fold higher than that of homologous MC3-mRNA vaccination. More significantly, the heterologous H03-mRNA/MC3-mRNA vaccination elicited an immune response that was approximately 2-3-fold higher than that of homologous H03-mRNA vaccination and 6–9-fold higher than that of homologous MC3-mRNA vaccination, without any observable adverse effects. These results suggest that heterologous mRNA vaccination is superior to homologous mRNA vaccination and may be attributed to differences in LNP carriers. Therefore, our research may inspire further exploration of different delivery systems to enhance mRNA-based therapeutics.
1. Introduction
Since coronavirus disease 2019 (COVID-19) swept across the globe, mRNA-based vaccines such as Pfizer-BioNTech BNT162b2 and Moderna COVID-19 mRNA-1273 have gained widespread usage in clinical settings due to their rapid manufacturing speed and high protection efficiency.1–5 The mechanism underlying mRNA vaccines entails the translation of mRNA into antigens by the cytoplasmic ribosomal translation machinery. This, in turn, elicits adaptive immunity against the virus.6 However, the primary challenges to effective systemic in vivo delivery of mRNA include instability, immune activation, and limited cell uptake.7–10Over the past few decades, various delivery systems have been developed to facilitate the delivery of mRNA into cells including protamine,11,12 liposomes,13,14 polymer micelles15,16 and lipid nanoparticles (LNPs).17,18 Among them, LNPs exhibit tremendous potential for clinical applications due to the simple molecular structure of lipids,18 high mRNA encapsulation efficiency19–21 and low toxicity in vivo.6 LNPs are typically composed of cholesterol, phospholipids, and lipid modified polyethyleneglycol and an ionizable lipid, the latter playing a key role in complexation of mRNA and subsequent release into the cytosol. At physiological pH, ionizable lipids with neutral charge reduce non-specific lipid–protein interactions, while in the acidic environment of endosomes following cellular uptake, protonated lipids promote endosomal destabilization and mRNA release into the cytosol.22–24 Dlin-MC3-DMA (MC3) was the first ionizable lipid applied in a marketed siRNA drug (Onpattro®).17 Notably, the recent approval of mRNA-1273 and BNT162b2 vaccines, and other mRNA vaccines against SARS-CoV-2 infections in clinical settings all employed ionizable lipids to prepare LNPs as mRNA delivery systems.25–27 The chemical structures of ionizable lipids in various mRNA vaccines are different, which can affect the immunogenicity in vivo and storage methods in vitro.28–32
Usually, two doses of homologous vaccines are recommended to evaluate the levels of immune response in animal or human vaccination practice. However, recent reports have suggested that two doses of heterologous SARS-CoV-2 vaccines may elicit a stronger immune response compared to that induced by two doses of homologous vaccines. For instance, heterologous mRNA-1273/BNT162b2 vaccinations are superior to the homologous BNT162b2/BNT162b2 administration and heterologous vaccination with ChAdOx1-S vaccine (Vaxzevria, AstraZeneca) and BNT162b2 vaccine was superior to BNT162b2/BNT162b2 vaccines.33,34 It is noteworthy that the three types of vaccines employ distinct delivery systems, with mRNA-1273 and BNT162b2 vaccines utilizing various ionizable lipids to prepare LNPs as delivery systems, which may account for the superior efficacy of heterologous vaccination compared to homologous use. However, it is regrettable that the aforementioned studies did not unequivocally establish whether the enhanced effect was solely attributable to the difference in ionizable lipids, as the amount of mRNA encapsulated in LNPs varied between the studies.
In this study, two types of mRNA vaccine formulations for SARS-CoV-2 infection were developed based on different lipid nanoparticle (LNP) delivery systems, and heterologous and homologous mRNA vaccinations were administered to explore the levels of immune responses comparatively. Importantly, the only difference in the vaccines is the lipid components in LNP systems. First, six LNP-mRNA formulations were prepared by mixing ethanol solution containing ionizable lipid, DSPC, cholesterol and DMG-PEG2000 with citrate buffer containing mRNA. After that, the physicochemical properties of LNP-mRNA formulations were thoroughly studied. Next, a transfection assay, lysosomal escape assay, and western blot analysis were performed to investigate the in vitro delivery effect of LNP-mRNA formulations. The intramuscular injection of LNP-Fluc mRNA was carried out to evaluate the in vivo delivery effect. Finally, following in vitro and in vivo screening, H03-mRNA and MC3-mRNA formulations were selected as candidate vaccines to assess their immunogenicity in mice administered with heterologous or homologous mRNA vaccines. The toxicity of these vaccines was also evaluated in a mouse model. The storage stability was assessed under different physical conditions for various periods.
2. Materials and methods
2.1. Materials
All reagents and solvents used for compound synthesis were provided by Macklin Biochemical Co., Ltd and Aladdin Biochemical Technology (Shanghai, China) without further purification. 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethyleneglycol-2000 (DMG-PEG2000), cholesterol, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and Dlin-MC3-DMA were obtained from A.V.T.Pharmaceutical Tech Co., Ltd (Shanghai, China). In addition, HEK293T and DC2.4 cells were supplied by American Type Culture Collection (ATCC). Gel DNA Extraction Mini Kit (Cat: DC301-01) and FastPure EndoFree Plasmid Maxi Kit (Cat: DC202-01) were bought from Vazyme Biotech Co. Ltd (Nanjing, China). Six- to eight-week-old female BALB/c mice (15–20 g) were supplied by Qinglongshan Experimental Animal Center (Nanjing, China). All animal studies were approved by the Experiment Animal Ethic Committee of Southeast University and were performed according to institutional ethics standards and regulations.2.2. Synthesis of H-series ionizable lipids
The synthesis of H-series ionizable lipids is shown in the ESI.†2.3. Synthesis of mRNA
The pVAX1 (GENEWIZ) plasmid containing a 5′ UTR sequence, signal peptide sequence, SARS-CoV-2 (B.1.617.2) receptor-binding domain (RBD) mRNA sequence, 3′ UTR sequence, and poly-A tail sequence was prepared according to the literature.35–38 The linearized plasmid DNA as templates for in vitro transcription was obtained by treating pVAX1 plasmid with Xho I (Absin, Shanghai) at 37 °C for 30 min. The linearized plasmid DNA templates were used to synthesize linear mRNA precursors using a T7 High Yield RNA Synthesis Kit (Novoprotein, Cat No. E131). The modified linear RNA was prepared by replacing all uridine with N1-methylpseudouridine, and was capped and purified using the Vaccinia Capping System (Novoprotein, Cat No. M082). The enhanced green fluorescent protein-encoding mRNA (eGFP mRNA) and the firefly luciferase-encoding mRNA (Fluc mRNA) were prepared using the same procedure as above.2.4. Preparation and characterization of lipid nanoparticles (LNP-mRNA)
Six formulations were prepared according to the literature.35 In brief, a volume of 200 μL SARS-CoV-2 (B.1.617.2) RBD-mRNA solution (concentration: 0.5 μg μL−1) was dissolved in 700 μL of 20 mM citrate buffer with pH = 4. MC3 (1.194 mg), DSPC (0.294 mg), cholesterol (0.553 mg) and DMG-PEG2000 (0.141 mg) at molar ratios of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 were dissolved in 300 μL of ethanol. The LNP-mRNA formulation was prepared by mixing the above aqueous and organic phases through a microfluidic mixer (N/P = 6
1.5 were dissolved in 300 μL of ethanol. The LNP-mRNA formulation was prepared by mixing the above aqueous and organic phases through a microfluidic mixer (N/P = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Nano S, Unigen Biotech Co. Ltd, Suzhou, China). The prepared LNP-mRNA solution was quickly added into the ultrafiltration tube with 10 times volume of PBS (pH = 7.4), and centrifuged at 5000 rpm for 30 min to remove the ethanol and citrate buffer. The desired concentration was obtained using Amicon Ultra centrifugal filters. The other five LNP-mRNA formulations were prepared in the same way as above, with the exception that MC3 was replaced by H01, H02, H03, H04 and H05. Six LNP-RBD mRNA formulations were named MC3M, H01M, H02M, H03M, H04M and H05M, respectively. Moreover, RBD-mRNA was used instead of eGFP mRNA and Fluc mRNA to prepare LNP-eGFP mRNA and LNP-Fluc mRNA formulations, which were used to evaluate the in vitro and in vivo delivery efficacy. Finally, all formulations were passed on a 220 nm filter and stored at −20 °C in PBS containing 5% sucrose.
1, Nano S, Unigen Biotech Co. Ltd, Suzhou, China). The prepared LNP-mRNA solution was quickly added into the ultrafiltration tube with 10 times volume of PBS (pH = 7.4), and centrifuged at 5000 rpm for 30 min to remove the ethanol and citrate buffer. The desired concentration was obtained using Amicon Ultra centrifugal filters. The other five LNP-mRNA formulations were prepared in the same way as above, with the exception that MC3 was replaced by H01, H02, H03, H04 and H05. Six LNP-RBD mRNA formulations were named MC3M, H01M, H02M, H03M, H04M and H05M, respectively. Moreover, RBD-mRNA was used instead of eGFP mRNA and Fluc mRNA to prepare LNP-eGFP mRNA and LNP-Fluc mRNA formulations, which were used to evaluate the in vitro and in vivo delivery efficacy. Finally, all formulations were passed on a 220 nm filter and stored at −20 °C in PBS containing 5% sucrose.
The size and zeta-potential of nanoparticles were analyzed by dynamic light scattering (DLS) using a NanoBrook Omni Particle Size Analyzer (Brookhaven, Holtsville, NY). The morphology of nanoparticles was observed by transmission electron microscopy (TEM) (JEOL Inc., Tokyo, Japan). The mRNA encapsulation efficiency (EE) was detected with a RiboGreen reagent (Thermo Fisher Scientific, Cat No. R11491). First, the TE buffer and 2% Te-Triton buffer solution were prepared. To determine free mRNA in the LNP-mRNA formulations, the LNP-mRNA formulation solution was diluted with 1 × TE buffer and 100 μL of the diluted solution was added into the 96-well plate. For the total mRNA assay, the LNP-mRNA formulation solution was diluted with 2% TE-Triton buffer and 100 μL of diluted solution was added to the 96-well plate. Next, 100 μL of RiboGreen dye was added into the plate well and incubated for 5 min. The optical density (OD) of each well was measured using a spectrophotometer (Thermo Fisher Scientific, USA) with an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The corresponding mRNA concentrations were calculated using the standard curve method. The values of EE (%) were calculated based on the following formula: EE (%) = (total mRNA concentration − free mRNA concentration)/the total mRNA concentration × 100%.
2.5. pKa of LNP-mRNA measurement
Apparent pKa was measured using the 2-(ptoluidino)-6-naphthalene sulfonic acid (TNS) according to the literature.25,39 Briefly, to a 96-well plate was added 4 mM LNP-mRNA formulation, 2 μM TNS (Sigma) and buffers containing 20 mM HEPES, 20 mM 4-morpholineethanesulfonic acid, 20 mM ammonium acetate, and 260 mM NaCl at pH values ranging from 3 to 11 and each pH experiment was carried out in triplicate. The fluorescence intensity was recorded using a spectrophotometer (RF-6000, SHIMADZU) with an excitation wavelength of 321 nm and an emission wavelength of 445 nm. The relative fluorescence intensity was plotted as a function of Log pH. When the relative fluorescence intensity reaches 0.5, the corresponding pH value is the apparent pKa.2.6. Cytotoxicity of LNP-mRNA formulations
The cytotoxicity of LNP-mRNA formulations against cell lines including HEK293T and DC 2.4 cells was evaluated by the Cell Counting Kit-8 (CCK-8, APExBIO, USA). Cells were seeded into 96-well plates at a density of 2 × 103 cells per well and were exposed to medium containing free mRNA and LNP-mRNA (1 μg mRNA/well) at 37 °C for 24 h. Cells incubated with medium culture group was used as the control. Next, the CCK-8 reagent was added into the medium for 2 h. The ultraviolet absorbance of each well was measured using a spectrophotometer (Thermo Fisher Scientific, USA) with a test wavelength of 450 nm.2.7. In vitro transfection assay
The in vitro delivery effect of LNP-mRNA formulations was evaluated by transfection assay of eGFP mRNA against HEK293T and DC 2.4 cells. Cells (1 × 104 cells/dish) were seeded into a confocal dish and incubated with Opti-MEM I Reduced Serum Medium containing naked eGFP mRNA and LNP-eGFP mRNA formulations, respectively. Cells incubated with medium culture and medium culture containing naked eGFP mRNA groups were used as controls. The eGFP expression was detected using an inverted fluorescence microscope (Olympus, Japan). Furthermore, the eGFP positive cells were measured by flow cytometry (FACS Aria II, BD Biosciences). Briefly, HEK293T cells were seeded into 6-well plates. After attachment to the wall, the cells were incubated in culture medium containing naked eGFP mRNA and LNPs-eGFP mRNA formulations for 24 h. After that, cells of each well were collected and analyzed via flow cytometry.2.8. Lysosome escape assay
To verify that LNP-mRNA formulations can successfully deliver mRNA into cells and release mRNA into the cytoplasm, a lysosome escape assay was performed using DC2.4 cells. Cells were seeded into confocal dishes at a density of 1 × 104 cells per well and incubated with culture medium containing formulations (mRNA labeled by FITC) for 4 h. After the mixed medium culture was discarded, cells were washed by cold PBS 2 times and treated by Lyso-Tracker Red (KeyGEN BioTECH, China) for 15 min. Finally, the cells were washed with PBS 3 times and observed by confocal laser scanning microscopy (CLSM) (Olympus,Tokyo, Japan). In the meantime, naked mRNA was used as the control.2.9. Western blot analysis
The expression levels of SARS-Cov-2 (B.1.617.2) RBD in cells treated by H03M and MC3M formulations were evaluated by Western Blot Assay against cell lines (HEK293T, DC 2.4 cells). The in vitro transfection assays were carried out as in section 2.7. The eGFP mRNA was replaced by RBD mRNA. After 24 h of transfection, cells were collected and lysed with RIPA lysis buffer. The total protein concentration was measured using a BCA kit. The concentration of SDS-PAGE gel should be determined according to the size of the protein band, and the preparation method followed the instructions of the kit. After the SDS-PAGE gel was prepared, the same amount of protein was added into each well of SDS-PAGE gel, running at 90 V for 40 min, and then 120 V for another 1 h. After that, the gel was transferred to a PVDF membrane, cleaned with TBST buffer 3 times, and sealed with skim milk for 1 h. Following cleaning with TBST buffer 3 times again, the primary antibody of the SARS-CoV-2 (B.1.617.2) was added, and incubated at 4 °C overnight. On the second day, the PVDF membrane was added to the secondary antibody and incubated at room temperature for 1 h. Finally, the ECL substrate was added and the protein blots were visualized on the BioSpectrum Gel Imaging System (Tanon-5200, China).2.10. In vivo delivery profiles of LNP-Fluc mRNA
To investigate the in vivo mRNA delivery profiles of these LNPs upon intramuscular injection, firefly luciferase mRNA (Fluc mRNA) encoding firefly luciferase was encapsulated in LNP systems and determined in female BALB/c mice (n = 3). The mice were injected intramuscularly with 100 μL of buffer solution containing LNP-Fluc mRNA with 20 μg of Fluc mRNA encapsulated. At 12 h post injection, mice were injected intraperitoneally with luciferase substrate and reacted for 5 min. Their bioluminescence signal images were obtained using an IVIS Lumina Series III(PerkinElmer).2.11. In vivo humoral immunity response
The female BALB/c mice were randomly divided into six groups (n = 6/group) that were intramuscularly vaccinated with two doses of saline/saline, empty LNPs/empty LNPs (placebo, H03/MC3), H03M/H03M, H03M/MC3M (a second dose of MC3M formulations after a first dose of H03M formulations), MC3M/MC3M and MC3M/H03M (a second dose of H03M formulations after a first dose of MC3M formulations) (SARS-CoV-2 (B.1.617) RBD mRNA, 10 μg per dose). At day 14 after the first injection, the booster injection was finished. At days 14, 21, and 28 after vaccination, sera samples were collected from the orbital vein of mice to detect IgG antibody and neutralizing antibody titers. The specific IgG antibody was evaluated using an ELISA kit (Sino Biological Inc, Beijing, China). Briefly, SARS-CoV-2 RBD antigen and diluted serum were added in 96-well microtiter plates and co-incubated for 2 h at 37 °C. Next, microtiter plates were washed with PBS twice and were added with HRP-conjugated goat anti-mouse IgG for 1 h co-incubation. The plates were washed again, and TMB solution was added and co-incubated at 37 °C for 20 min. Finally, the absorbance was measured using a spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts) with a test wavelength of 450 nm. The specific neutralizing antibody of serum samples was measured according to the literature.35 In brief, HEK293T cells were seeded in 96-well plates at a density of 2 × 104 cells per well. The SARSCoV-2 (B.1.617.2) pseudovirus was diluted to 500–1000 TCID50 with medium and incubated with the serum samples in 96-well plates at 37 °C for 1 h. Finally, after removing the supernatant, the luciferase substrate was added to each well and the luciferase intensity was analyzed using a Microplate Luminometer (Promega, USA).2.12. In vivo effector memory T cell immune response
Fluorochrome-conjugated antibodies in mice: for effector memory T cell detection, CD3 (PE/Cyanine7), CD4 (FITC), CD8 (FITC), CD44 (PE) and CD62L (APC) were bought from BioLegend. Firstly, the splenocytes of the vaccinated mouse were incubated with a protein transport inhibitor (Brefeldin A, MCE) at 37 °C for 4 h. Next, the splenocytes were washed with PBS twice and stained with fluorescent antibodies. Finally, the effector memory T cells’ proliferation was analyzed by flow cytometry (FACS Aria II, BD Biosciences).2.13. Safety analysis
To evaluate the in vivo safety of vaccines, the weights, hepatorenal function, pathology and immune activated cytokines of mice administered with heterologous or homologous vaccines were tested. The weights of mice were recorded every four days in the above section of the in vivo humoral immunity response study. Another batch of female BALB/c mice (n = 6/group) was treated with heterologous or homologous vaccines (SARS-CoV-2 (B.1.617) RBD mRNA, 10 μg per dose). After booster vaccination for 24 h, serum was collected for measuring blood biochemical parameters and immune activated cytokines. The immune activated cytokines were checked using an IL-6 ELISA kit and TNF-α ELISA kit (Thermo Fisher Scientific). Then, all the mice were sacrificed following institutional guidelines and the hearts, livers, spleens, lungs and kidneys were separated and stained with hematoxylin and eosin (H&E) to assess the systemic toxicity.2.14. Storage stability of H03-mRNA and MC3-mRNA formulations
H03-Fluc mRNA and MC3-Fluc mRNA formulation solutions containing 5% sucrose were stored at 25 °C, 4 °C and −20 °C under nitrogen in triplicate. In vivo bioluminescence signals were measured at specific intervals. The experiment procedures were the same as in the Section 2.10. Moreover, to further verify the storage stability of H03-mRNA and MC3-mRNA formulations, the stability of the H03-RBD mRNA and MC3-RBD mRNA formulations containing 5% sucrose stored at −20 °C for 90 days was assessed by humoral immune response. The experiment procedures were the same as in Section 2.11.2.15. Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.01 software. The statistical analysis was performed by using one-way ANOVA. P < 0.05 was considered significant.3. Results and discussion
3.1. Design and synthesis of ionizable lipids
The chemical structure of lipid components directly affects the delivery effect of LNPs in vitro and in vivo. The ideal ionizable lipids usually contain three parts: (i) tertiary amine headgroups for condensing mRNA to form complexes, (ii) hydrophobic hydrocarbon chains generally between 8 to 18 carbon units in length with various branching degrees capable of promoting self-assembly, adjusting lipophilicity and delivering mRNA into cytoplasm, and (iii) linker groups including esters, ethers, amides etc. with properties of circulation stability within a biological environment and safe clearance after mRNA delivery. Additionally, the incorporation of a hydroxyl group in the neighboring headgroup is beneficial for the encapsulation of nucleic acids via hydrogen bonding interactions with lipids.18 The cone-shaped structure of the lipid contributes to the destruction of the lysosomal membrane and improves the ability of LNPs to escape from the lysosome, in accordance with the molecular shape hypothesis.40,41Based on the understanding of the chemical structure of ionizable lipids on the influence of mRNA delivery effect,18,42–45 we designed and synthesized a series of novel ionizable lipids, named H01, H02, H03, H04 and H05. As shown in Fig. 1b, all lipids had a cone-shaped structure, with differences in the aspect of tail length, degree of branching, linker group and the number of hydroxyl groups. Compared with H01, H02 increased a branching chain using an ether bond as the linker. H03, H04 and H05 had a more symmetrical cone-shaped structure than H01 and H02. One branching chain of H03 used an ester bond as the linker and another one was linked via an ether bond. H04 increased a butyl hydroxy group based on the H03 structure and H05 further modified H03 by using two ester bonds as linkers.
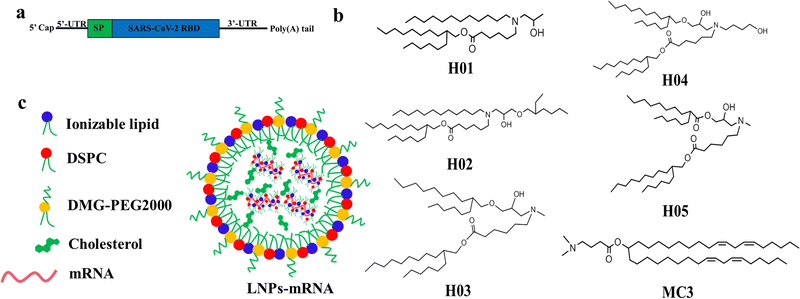 | ||
| Fig. 1 The construction diagrams: (a) RBD-mRNA construction, (b) ionizable lipids structures, and (c) LNP-mRNA construction and components. | ||
The H01 and H02 were synthesized using a two-step process as follows: dodecan-1-amine reacted with 2-methyloxirane or 2-(((2-ethylhexyl)oxy)methyl)oxirane by ring opening reaction of epoxy; the above products were reacted with 2-hexylundecyl 6-bromohexanoate by bromine substitution reaction. The H03 was obtained by a three-step process as follows: 2-hexyldecan-1-ol was reacted with epichlorohydrin through an epoxy open-loop and closed-loop process; the product of the first step was reacted with ethanamine by ring opening reaction of epoxy; the product of the second step was reacted with 2-hexylundecyl 6-bromohexanoate by bromine substitution reaction. The H04 was obtained by a two-step process as follows: 3-(methylamino)propane-1,2-diol was reacted with 2-hexylundecyl 6-bromohexanoate by bromine substitution reaction; the above product was reacted with 2-hexyl-1-decanol by esterification reaction. The H05 was obtained by a two-step process as follows: 2-(((2-hexyldecyl)oxy)methyl)oxirane was reacted with 4-aminobutan-1-ol by ring opening reaction of epoxy; the above product was reacted with 2-hexylundecyl 6-bromohexanoate by bromine substitution reaction. The detailed procedures are given in the ESI.† The resultant products were characterized by 1H NMR and MS (Fig. S1 and S2, ESI†). In order to evaluate the immunogenicity in mice administered with heterologous mRNA vaccines, the commercial MC3 was used as lipid components of another type of LNP system due to the large structural difference between MC3 and H-series lipids.
3.2. Preparation and characterization of mRNA-encapsulated lipid nanoparticle (LNP-mRNA) formulations
The SARS-CoV-2 (B.1.617.2) receptor-binding domain (RBD) mRNA sequence is shown in Fig. 1a and Fig. S6 (ESI†). Six LNP-RBD mRNA formulations (Fig. 1c) were prepared based on H-series lipids and MC3. In brief, MC3, DSPC, cholesterol and DMG-PEG2000 at molar ratios of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 were dissolved in ethanol. The LNP-RBD mRNA formulations were prepared by mixing the above lipid mixture with 20 mM citrate buffer (pH 4.0) containing mRNA at a ratio of 1
1.5 were dissolved in ethanol. The LNP-RBD mRNA formulations were prepared by mixing the above lipid mixture with 20 mM citrate buffer (pH 4.0) containing mRNA at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (ethanol
3 (ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) aqueous) using a microfluidic mixer. The ethanol and citrate buffer were removed by dialysis method in PBS (pH 7.4). Another five LNP-RBD mRNA formulations were prepared using the same procedures, with the exception that MC3 was replaced by H-series lipids. Six LNP-RBD mRNA formulations were named H01M, H02M, H03M, H04M, H05M, and MC3M, respectively. The size, distribution and zeta-potential of six formulations were analyzed by DLS. The morphology of H03M formulation was observed by TEM. The pKa values of six formulations were measured using TNS and the mRNA encapsulation efficiency (EE) was detected with RiboGreen reagent. As shown in Fig. 2a and Table 1, the DLS results exhibited that H01M, H02M, H03M, H04M, H05M, and MC3M formulations have average particle sizes of 97.13 ± 2.78, 122.74 ± 3.26, 103.44 ± 3.95, 138.52 ± 4.65, and 107.85 ± 3.53 nm, respectively, with narrow distributions and negative surface charge within a physiological environment. It was found that the diameter of formulations increased with the increment of tail length and branching degree of lipid components. Notably, the diameter of H04M reached a maximum value of 138.52 nm, which might be attributed to the increased hydrophilicity caused by its double hydroxyl chemical structure of lipid H04. However, the large particle size of formulations is not conducive to LNP delivery.46 The TEM image confirmed the spherical structure of H03M with a diameter of approximately 100 nm (Fig. 2b).
aqueous) using a microfluidic mixer. The ethanol and citrate buffer were removed by dialysis method in PBS (pH 7.4). Another five LNP-RBD mRNA formulations were prepared using the same procedures, with the exception that MC3 was replaced by H-series lipids. Six LNP-RBD mRNA formulations were named H01M, H02M, H03M, H04M, H05M, and MC3M, respectively. The size, distribution and zeta-potential of six formulations were analyzed by DLS. The morphology of H03M formulation was observed by TEM. The pKa values of six formulations were measured using TNS and the mRNA encapsulation efficiency (EE) was detected with RiboGreen reagent. As shown in Fig. 2a and Table 1, the DLS results exhibited that H01M, H02M, H03M, H04M, H05M, and MC3M formulations have average particle sizes of 97.13 ± 2.78, 122.74 ± 3.26, 103.44 ± 3.95, 138.52 ± 4.65, and 107.85 ± 3.53 nm, respectively, with narrow distributions and negative surface charge within a physiological environment. It was found that the diameter of formulations increased with the increment of tail length and branching degree of lipid components. Notably, the diameter of H04M reached a maximum value of 138.52 nm, which might be attributed to the increased hydrophilicity caused by its double hydroxyl chemical structure of lipid H04. However, the large particle size of formulations is not conducive to LNP delivery.46 The TEM image confirmed the spherical structure of H03M with a diameter of approximately 100 nm (Fig. 2b).
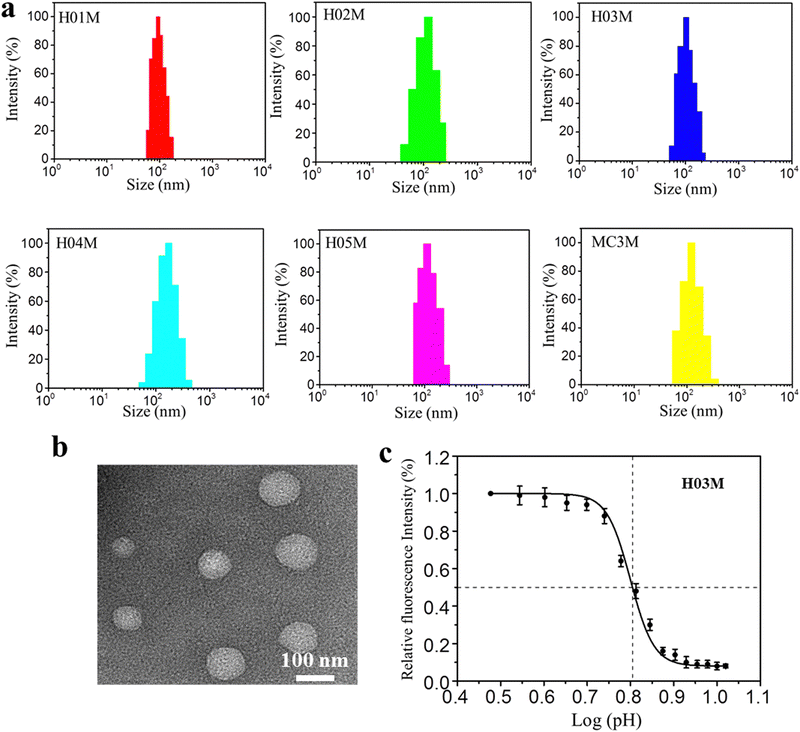 | ||
| Fig. 2 Physicochemical characteristics of LNP-mRNA formulations: (a) particle size of formulations by DLS. (b) Morphology of H03M by TEM. (c) pKa value of H03M by TNS assay. | ||
| LNP-mRNA formulations | Size/nm | PDI | Zeta potential/mV | EE/% | pKa |
|---|---|---|---|---|---|
| H01M | 97.13 ± 2.78 | 0.165 ± 0.021 | −4.43 ± 0.34 | 76 ± 2 | 5.9 |
| H02M | 122.74 ± 3.26 | 0.154 ± 0.028 | −5.23 ± 0.26 | 88 ± 2 | 5.3 |
| H03M | 103.44 ± 3.95 | 0.167 ± 0.032 | −3.58 ± 0.31 | 94 ± 3 | 6.3 |
| H04M | 138.52 ± 4.65 | 0.186 ± 0.026 | −4.40 ± 0.29 | 97 ± 2 | 5.7 |
| H05M | 107.85 ± 3.53 | 0.183 ± 0.031 | −4.14 ± 0.25 | 91 ± 2 | 6.5 |
| MC3M | 115.15 ± 2.84 | 0.175 ± 0.029 | −3.23 ± 0.27 | 96 ± 3 | 6.4 |
The mRNA encapsulation efficiency (EE) under physiological conditions showed a remarkable difference among six formulations. H04M showed the maximum EE value of 97% and H01M showed the minimum EE value of 76%. We concluded that the EE value increases with increasing tail length, branching degree and the amounts of hydroxyl group. The pKa value has a drastic influence on the potency of LNP-mRNA formulations in vivo and the most appropriate pKa value of LNP-mRNA formulations is between 6.0 and 6.5.47,48Table 1 and Fig. 2c showed the pKa values of the H-series formulations and MC3M are 5.9, 5.3, 6.3, 5.7, 6.5 and 6.4. Among them, the pKa values of H03M, H05M and MC3M are between 6.2 and 6.5, which should contribute to the protein expression in vitro and in vivo. Taken together, these data suggested that the monodisperse and stable LNP-mRNA formulations can be formed based on H-series lipids and MC3.
3.3. Cytotoxicity, transfection assay, western blot analysis and lysosome escape assay
The cytotoxicity, transfection assay, western blot analysis and lysosome escape assay of LNP-mRNA formulations were evaluated against HEK-293T and DC 2.4 cells. The cytotoxicity was checked by CCK-8 assay using the naked mRNA as a control. As shown in Fig. 3a and b, cells treated with naked mRNA and LNP-mRNA formulations exhibited minimal decrease in viability and remained comparable to non-treated cells, indicating the safety of the six formulations in vitro. Next, the in vitro delivery effect of LNP-mRNA was evaluated by transfection assay of LNP-eGFP mRNA formulations using naked eGFP mRNA as the control. As shown in Fig. 3c, cells treated by LNP-eGFP mRNA formulations displayed varying degrees of green fluorescence signal intensity while the non-treated and naked mRNA-eGFP treated cells showed no green fluorescence signal whatsoever. Importantly, the H03-eGFP mRNA and MC3-eGFP mRNA group exhibited a stronger signal intensity than other groups. To quantify the transfection efficiency, the eGFP positive cells were measured by flow cytometry. As shown in Fig. 3d, e and Fig. S3 (ESI†), the percentage of eGFP positive cells in the H01-eGFP mRNA, H02-eGFP mRNA, H03-eGFP mRNA, H04-eGFP mRNA, H05-eGFP mRNA and MC3-eGFP mRNA groups were 21.3%, 64.3%, 98.1%, 43.2%, 74.6% and 84.6%, respectively, further demonstrating the H03-mRNA and MC3-mRNA formulations could efficiently deliver mRNA into cells and achieve protein expression.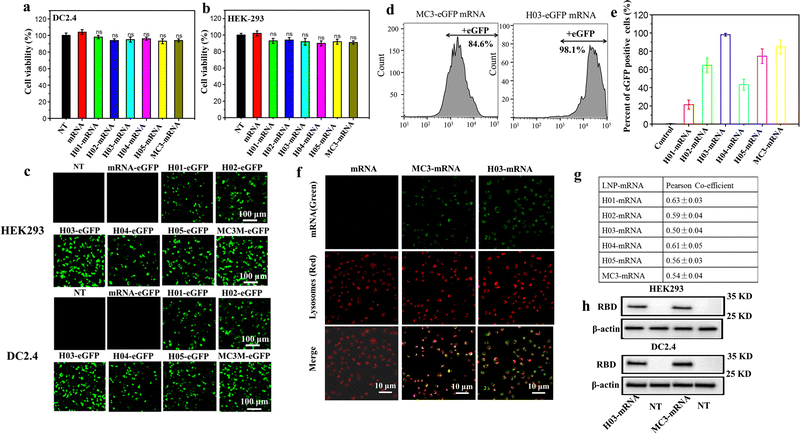 | ||
| Fig. 3 Cytotoxicity and protein expression of LNP-mRNA: (a) and (b) viabilities of HEK293T and DC 2.4 cells inoculated with LNP-mRNA formulations after 24 h. (c) Confocal laser scanning microscopy (CLSM) images of the eGFP protein expression after transfection. Scale bar = 100 μm. (d) Percentages of eGFP positive cells after transfection analyzed by flow cytometry. (e) Percentages of eGFP positive cells of six groups according to Fig. S3 (ESI†). (f) Representative images of lysosomal escape analysis. (g) Pearson's correlation coefficient was calculated using ImageJ software according to (f) and Fig. S4 (ESI†). (h) Western blotting analysis of the SARS-CoV-2 RBD protein expression. | ||
It was reported that LNP-mRNA formulations must escape from lysosomes and release mRNA into the cytoplasm to translate antigen protein. Therefore, the delivery efficacy of LNPs has a strong correlation with lysosome escape capability.49 To further explore the reason for the difference in transfection effect of six formulations, a lysosome escape assay was executed by observing the distribution of FITC-labeled mRNA in cells incubated with LNP-FITC mRNA formulations for 4 h. Cells treated with naked FITC-mRNA was used as the control. As shown in Fig. 3f and Fig. S4 (ESI†), all formulation treated groups appeared with a green fluorescence signal (FITC mRNA) in the cytoplasm. In contrast, the naked mRNA group showed no green signal intensity in cells. In order to further analyze the endosomal release capability of LNP-mRNA formulations, Pearson's correlation coefficient was calculated using ImageJ software to study the correlation between FITC-mRNA (Green) and Lysotracker (Red). The results showed all formulations have a moderate correlation at 4 h (Fig. 3g). Importantly, the H03-FITC mRNA and MC3-FITC mRNA groups had a lower Pearson's coefficient than other groups, suggesting their superior endosomal escape capability. This result confirmed that the transfection efficiency of LNP-mRNA formulations is dependent on their lysosome escape capability.
Obviously, H03-mRNA, H05-mRNA and MC3-mRNA formulations with appropriate pKa values and particle size exhibited better escape capability than other formulations. When the particle size and pKa value are similar, H03-mRNA formulation showed a better escape ability than H05-mRNA and MC3-mRNA formulations. The superiority of H03 over H05 implied that the introduction of an ether bond in one branching chain can enhance the mRNA delivery effect. We speculated that in the low pH environment of lysosomes, the hydrolysis rate of the ester bond was accelerated, resulting in H05 being less stable than H03 in lysosomes. Consequently, H05-based formulations were less effective in protecting mRNA from degradation by lysosomes and lysosomal membrane destruction than H03-based formulations. Compared to MC3, H03 possesses a better cone-shaped structure due to the two branching chains, resulting in more lysosomal membrane disruption. Furthermore, the hydroxyl group incorporated in the neighboring headgroup of H03 facilitates the encapsulation of mRNA through hydrogen bonding interactions with lipids. Based on the aforementioned research findings, H03-RBD mRNA and MC3-RBD mRNA formulations were selected for the detection of the RBD expression levels in vitro, and both formulations successfully expressed the RBD proteins (Fig. 3h).
3.4. In vivo delivery profiles of LNP-Fluc mRNA
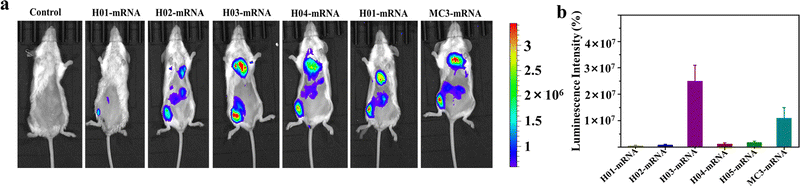
Vaccines are typically administered via intramuscular injection. To investigate the in vivo delivery profiles of these LNPs upon intramuscular injection, Fluc mRNA was encapsulated in LNPs systems. BALB/c mice (n = 3) were intramuscularly injected with 100 μL of buffer solution containing LNP-Fluc mRNA with encapsulated 20 μg of Fluc-mRNA. Normal saline was used as a control. The results in Fig. 4a indicated that H03-mRNA and MC3-mRNA formulations exhibited stronger bioluminescence signals than other lipids. The quantification analysis of signal intensity confirmed the above conclusion (Fig. 4b). Significantly, the H03-Fluc mRNA formulation exhibited approximately 2.2-fold higher in vivo signal intensity than the MC3-Fluc mRNA formulation. Therefore, H03-mRNA and MC3-mRNA formulations were selected as candidate vaccines to evaluate immunogenicity in mice administered with heterologous or homologous mRNA vaccines.3.5. Humoral immune response of heterologous or homologous vaccinations
To evaluate immunogenicity in mice induced by heterologous or homologous H03M and MC3M vaccines, BALB/c mice of four groups (n = 6/group) were intramuscularly vaccinated with H03M/H03M, H03M/MC3M (a second dose of MC3M formulation after a first dose of H03M formulation), MC3M/MC3M and MC3M/H03M (a second dose of H03M formulation after a first dose of MC3M formulation). The BALB/c mice of two groups administered with two doses of saline/saline and empty LNPs/empty LNPs (placebo) were used as controls. At day 14 after the first injection, the booster injection was provided (Fig. 5a). No inflammatory response or other adverse effects appeared during all injections. At 14, 21, and 28 days after vaccination, sera samples were collected from the orbital vein of mice to detect IgG antibody and neutralizing antibody titers. The specific IgG antibody was evaluated using an ELISA kit (Sino Biological Inc, Beijing) and neutralizing antibody titers (NT50) were tested according to luciferase intensity analyzed using a Microplate Luminometer (Promega, USA).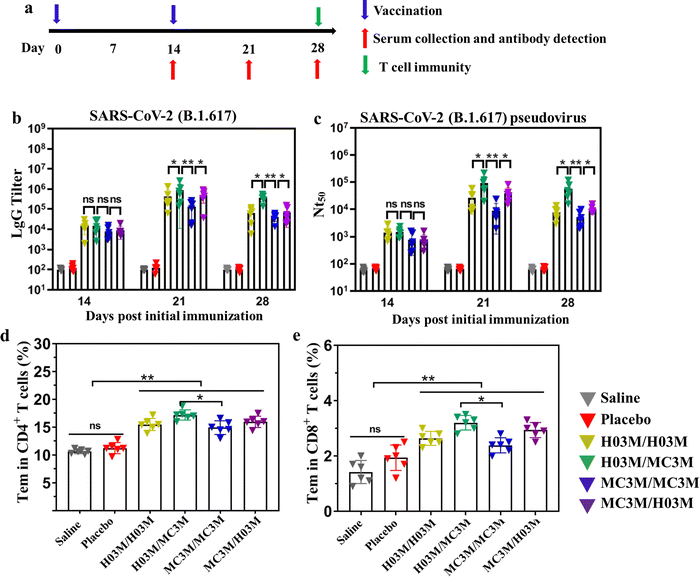 | ||
| Fig. 5 Humoral immune response and T-cell immune response in mice inoculated with heterologous or homologous H03M and MC3M vaccines. (a) Schematic diagram of the immunization strategy, including vaccination, serum collection and antibody detection, and immunogenicity detection. (b) The SARS-CoV-2 (B.1.617.2) RBD-specific IgG antibody titer was determined by ELISA. (c) The serum neutralization antibody titer (NT50) was tested against SARS-CoV-2 (B.1.617.2) pseudovirus. SARS-CoV-2 (B.1.617.2) RBD-specific CD4+ (d) and CD8+ (e) Tem cells in splenocytes of mice inoculated with heterologous or homologous H03M and MC3M vaccines were detected by flow cytometry (Fig. S5, ESI†) (** p < 0.01, * p < 0.05; ns: p > 0.05). | ||
As shown in Fig. 5b and c, IgG and NT50 antibody levels obviously increased in vaccination groups at day 14 post vaccination, and reached the maximum values at day 21. However, the control groups consistently showed no increase in specific IgG tilter and NT50 antibody levels. Importantly, we found that IgG and NT50 antibody levels showed a significant difference in vaccination groups. The maximum values of IgG antibody titer and NT50 in the H03M/H03M group approached approximately 1/397![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 1/26
000 and 1/26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 respectively, which were higher than the values in the MC3M/MC3M group (1/125
800 respectively, which were higher than the values in the MC3M/MC3M group (1/125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 1/9100). The result indicated that lipid H03 had a better in vivo delivery effect than MC3. More importantly, the maximum values of IgG antibody titer and NT50 in the H03M/MC3M group approached approximately 1/90
000 and 1/9100). The result indicated that lipid H03 had a better in vivo delivery effect than MC3. More importantly, the maximum values of IgG antibody titer and NT50 in the H03M/MC3M group approached approximately 1/90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3200 and 1/81
3200 and 1/81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600, which were approximately 2-3-fold higher than the values in the H03M/H03M group and 6-9-fold higher than the values in the MC3M/MC3M group. Meanwhile, the maximum values of IgG antibody titer and NT50 in the H03M/MC3M group were also higher than the values in MC3M/MC3M and H03M/H03M groups. These data confirmed that heterologous H03M and MC3M vaccination can induce a higher level of immune response than homologous H03M and MC3M vaccination. Therefore, heterologous vaccination could stimulate a stronger immunogenicity with the right combination of different lipid nanoparticles. mRNA vaccination can lead to transfection of various cells including muscle cells, epidermal cells, tissue-resident immune cells at the injection sites and immune cells in the secondary lymphoid tissues.6,50,51 H03M and MC3M formulations based on different LNP carriers have various physicochemical properties such as particle size, zeta potential and pKa value, which can affect the efficiency and cell selectivity of the delivery vector.6 Therefore, we speculate that the two type of mRNA vaccination could deliver the antigen into more different antigen-presenting cells, thereby leading to enhanced immune responses.
600, which were approximately 2-3-fold higher than the values in the H03M/H03M group and 6-9-fold higher than the values in the MC3M/MC3M group. Meanwhile, the maximum values of IgG antibody titer and NT50 in the H03M/MC3M group were also higher than the values in MC3M/MC3M and H03M/H03M groups. These data confirmed that heterologous H03M and MC3M vaccination can induce a higher level of immune response than homologous H03M and MC3M vaccination. Therefore, heterologous vaccination could stimulate a stronger immunogenicity with the right combination of different lipid nanoparticles. mRNA vaccination can lead to transfection of various cells including muscle cells, epidermal cells, tissue-resident immune cells at the injection sites and immune cells in the secondary lymphoid tissues.6,50,51 H03M and MC3M formulations based on different LNP carriers have various physicochemical properties such as particle size, zeta potential and pKa value, which can affect the efficiency and cell selectivity of the delivery vector.6 Therefore, we speculate that the two type of mRNA vaccination could deliver the antigen into more different antigen-presenting cells, thereby leading to enhanced immune responses.
3.6. T-cell immune response of heterologous or homologous vaccinations
T-cell immunity induced by heterologous or homologous H03M and MC3M vaccines was evaluated using a flow cytometer. To explore T-cell immunity, the SARS-CoV-2 RBD-specific CD4+ and CD8+ effector memory T cells (Tem) in splenocytes were checked. As shown in Fig. 5d and e, upon stimulation with peptide pools (Fig. S8, ESI†), CD4+ and CD8+ Tem cells in vaccination groups displayed a noteworthy increase compared with control groups and especially, effector memory T cells induced by heterologous H03M and MC3M vaccination were more abundant than those induced by homologous H03M and MC3M vaccination. These results confirmed that T-cell immunity was successfully induced by heterologous and homologous vaccination, and heterologous vaccines can stimulate higher levels of T-cell immunity.3.7. Safety of heterologous or homologous vaccination
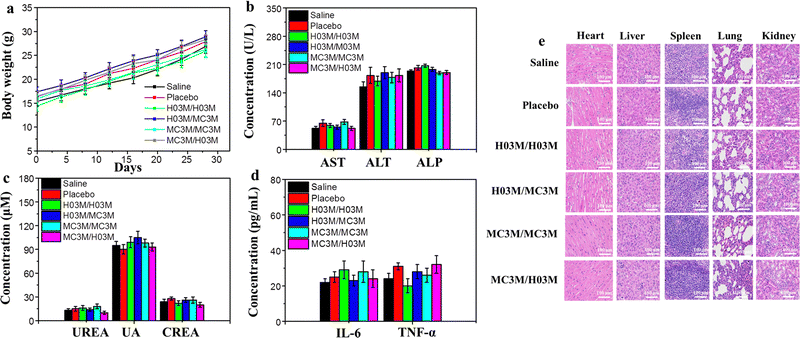
To evaluate the safety of heterologous or homologous vaccination in vivo, the changes in body weight of the mice in the above section of the in vivo humoral immune response study were recorded every four days. Moreover, another batch of female BALB/c mice (n = 6/group) treated with heterologous or homologous vaccines (SARS-CoV-2 (B.1.617) RBD mRNA, 10 μg per dose) were used to analyze blood biochemical parameters, immune activated cytokines and the pathology of mice. After booster vaccination for 24 h, the serum of the mice was collected to measure blood biochemical parameters and immune activated cytokines. The pathology of the mice was analyzed by observing the organ tissue sections stained with H&E. As shown in Fig. 6b–d, the blood biochemical parameters and the immune activated cytokines in mice treated with heterologous or homologous vaccines had no remarkable change compared with the saline group. Moreover, the H&E staining results of organ tissues from the six groups of mice after vaccination indicated no apparent injury, and the weights of the mice continued to slightly increase, as shown in Fig. 6a and e. These outcomes demonstrate that both heterologous and homologous vaccinations are safe in vivo.3.8. Storage stability of H03-mRNA and MC3-mRNA vaccines
The storage stability of H03-mRNA and MC3-mRNA formulations was measured at 25 °C, 4 °C and −20 °C using Fluc mRNA as a reporter gene. In Table 2 and Fig. S6 (ESI†), H03-Fluc mRNA and MC3-Fluc mRNA formulations remained stable for over 90 days at −20 °C and for 20 –30 days at 4 °C. However, the formulations displayed limited short-term stability at 25 °C. Additionally, the stability of H03-RBD mRNA and MC3-RBD mRNA formulations stored at −20 °C for 90 days was assessed by analyzing the humoral immune response. As shown in Fig. 7a and b, IgG antibody in the storage groups had no significant difference with the fresh groups, indicating that the H03-RBD mRNA and MC3-RBD mRNA formulations were stable for over 90 days at −20 °C.| Stability at −20 °C | Stability at 4 °C | Stability at 25 °C | |
|---|---|---|---|
| H03-Fluc mRNA | over 90 days | 30 days | 18 h |
| MC3-Fluc mRNA | over 90 days | 20 days | 12 h |
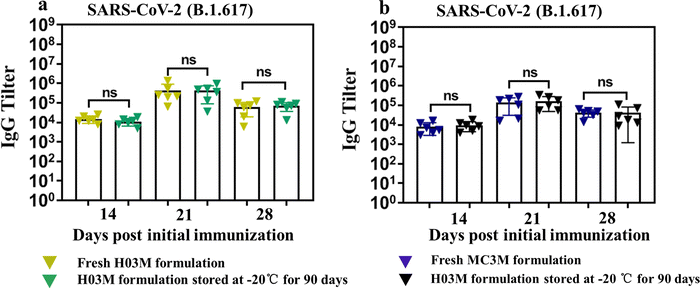 | ||
| Fig. 7 Storage stability of H03-mRNA and MC3-mRNA vaccines. (a) and (b) The SARS-CoV-2 (B.1.617.2) RBD-specific IgG antibody titer was determined by ELISA. | ||
4. Conclusions
In summary, we successfully developed six LNP-mRNA formulations based on synthetic lipids and MC3. Among them, H03M and MC3M formulations had more appropriate particle size and pKa values and better in vitro or vivo delivery effects than other formulations. Notably, H03 displayed approximately 2.2-fold higher in vivo delivery efficacy than MC3. Therefore, H03M and MC3M formulations were used as candidate vaccines for heterologous and homologous administration in mice. Finally, the humoral and T-cell immunity results demonstrated that homologous H03M vaccines induced a higher level of immune response than homologous MC3M vaccines. Importantly, the heterologous H03M and MC3M vaccinations were significantly superior to their homologous counterparts. Furthermore, the safety evaluation indicated that both homologous and heterologous vaccinations were safe in vivo. Together, our research might inspire more combinations of different delivery systems to augment mRNA-based therapeutics.Author contributions
Xinsong Li: conceptualization, methodology, formal analysis, data curation, resource, writing – review and editing, supervision, project administration, and funding acquisition. Yanhao Zhang: investigation, validation, formal analysis, data curation, and writing – original draft. Ji Wang, Chao Liu, Hanlei, and Xing: formal analysis, investigation, and data curation. Wenhui Zha, Shuo Dong, Yuhao, and Jiang: investigation and data curation.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Key Research and Development Program of Jiangsu Province of China (Grant No. BE2022826).References
- P. L. Truong, Y. Yin, D. Lee and S. H. Ko, Exploration, 2023, 3, 20210232 CrossRef PubMed
.
- L. J. Kubiatowicz, A. Mohapatra, N. Krishnan, R. H. Fang and L. Zhang, Exploration, 2022, 2, 20210217 CrossRef PubMed
.
- A. E. Nel and J. F. Miller, ACS Nano, 2021, 15, 5793–5818 CrossRef CAS
.
- M. Papi, D. Pozzi, V. Palmieri and G. Caracciolo, Nano Today, 2022, 43, 101403 CrossRef CAS PubMed
.
- Z. Wang, K. Cui, U. Costabel and X. Zhang, Exploration, 2022, 2, 20210082 CrossRef
.
- J. Kim, Y. Eygeris, M. Gupta and G. Sahay, Adv. Drug Delivery Rev., 2021, 170, 83–112 CrossRef CAS PubMed
.
- N. Chen, P. Xia, S. Li, T. Zhang, T. T. Wang and J. Zhu, IUBMB Life, 2017, 69, 297–304 CrossRef CAS
.
- M. Tatematsu, K. Funami, T. Seya and M. Matsumoto, J. Innate Immun., 2018, 10, 398–406 CrossRef CAS PubMed
.
- M. A. Islam, Y. Xu, W. Tao, J. M. Ubellacker, M. Lim, D. Aum, G. Y. Lee, K. Zhou, H. Zope and M. Yu, Nat. Biomed. Eng., 2018, 2, 850–864 CrossRef CAS PubMed
.
- C. Lorenz, M. Fotin-Mleczek, G. Roth, C. Becker, T. C. Dam, W. P. Verdurmen, R. Brock, J. Probst and T. Schlake, RNA Biol., 2011, 8, 627–636 CrossRef CAS PubMed
.
- I. Hoerr, R. Obst, H. G. Rammensee and G. Jung, Eur. J. Immunol., 2000, 30, 1–7 CrossRef CAS PubMed
.
- M. Fotin-Mleczek, K. M. Duchardt, C. Lorenz, R. Pfeiffer, S. Ojkic-Zrna, J. Probst and K.-J. Kallen, J. Immunother., 2011, 34, 1–15 CrossRef CAS PubMed
.
- G. J. Dimitriadis, Nature, 1978, 274, 923–924 CrossRef CAS
.
- R. W. Malone, P. L. Felgner and I. M. Verma, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 6077–6081 CrossRef CAS
.
- A. K. Patel, J. C. Kaczmarek, S. Bose, K. J. Kauffman, F. Mir, M. W. Heartlein, F. DeRosa, R. Langer and D. G. Anderson, Adv. Mater., 2019, 31, 1805116 CrossRef PubMed
.
- A. K. Blakney, Y. Zhu, P. F. McKay, C. R. Bouton, J. Yeow, J. Tang, K. Hu, K. Samnuan, C. L. Grigsby and R. J. Shattock, ACS Nano, 2020, 14, 5711–5727 CrossRef CAS PubMed
.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope and T. D. Madden, Nat. Nanotechnol., 2019, 14, 1084–1087 CrossRef CAS
.
- Y. Zhang, C. Sun, C. Wang, K. E. Jankovic and Y. Dong, Chem. Rev., 2021, 121, 12181–12277 CrossRef CAS
.
- A. K. Leung, Y. Y. C. Tam, S. Chen, I. M. Hafez and P. R. Cullis, J. Phys. Chem. B, 2015, 119, 8698–8706 CrossRef CAS PubMed
.
- E. Samaridou, J. Heyes and P. Lutwyche, Adv. Drug Delivery Rev., 2020, 154, 37–63 CrossRef
.
- M. Ripoll, M.-C. Bernard, C. Vaure, E. Bazin, S. Commandeur, V. Perkov, K. Lemdani, M.-C. Nicolaï, P. Bonifassi and A. Kichler, Biomaterials, 2022, 286, 121570 CrossRef CAS
.
- X. Cheng and R. J. Lee, Adv. Drug Delivery Rev., 2016, 99, 129–137 CrossRef CAS PubMed
.
- J. A. Kulkarni, D. Witzigmann, J. Leung, Y. Y. C. Tam and P. R. Cullis, Nanoscale, 2019, 11, 21733–21739 RSC
.
- Y. Sato, H. Hatakeyama, Y. Sakurai, M. Hyodo, H. Akita and H. Harashima, J. Controlled Release, 2012, 163, 267–276 CrossRef CAS PubMed
.
- FDA. Fact Sheet for Healthcare Providers Administering Vaccine (Vaccination Providers) Emergency Use Authorization (EUA) of the Moderna COVID-19 Vaccine to Prevent Coronavirus Disease 2019 (COVID-19). https://www.fda.gov/media/144637/download (accessed 2021-05-19).
- EMA. EMA Recommends COVID-19 Vaccine Moderna for Authorisation in the EU. https://www.ema.europa.eu/en/news/emarecommends-covid-19-vaccine-moderna-authorisation-eu (accessed on 2021-05-19).
- FDA. FDA Takes Key Action in Fight Against COVID-19 by Issuing Emergency Use Authorization for First COVID-19 Vaccine. https://www.fda.gov/news-events/press-announcements/fda-takeskey-action-fight-against-covid-19-issuing-emergency-useauthorization-first-covid-19 (accessed 2021-05-19).
- K. J. Hassett, K. E. Benenato, E. Jacquinet, A. Lee, A. Woods, O. Yuzhakov, S. Himansu, J. Deterling, B. M. Geilich and T. Ketova, Mol. Ther.--Nucleic Acids, 2019, 15, 1–11 CrossRef CAS
.
- E. J. Anderson, N. G. Rouphael, A. T. Widge, L. A. Jackson, P. C. Roberts, M. Makhene, J. D. Chappell, M. R. Denison, L. J. Stevens and A. J. Pruijssers, N. Engl. J. Med., 2020, 383, 2427–2438 CrossRef CAS PubMed
.
- Novel Lipids and Lipid Nanoparticle Formulations for Delivery of Nucleic Acids. Available online: https://patents.google.com/patent/WO2017075531A1/en (accessed on 18 December 2020).
- U. Sahin, A. Muik, E. Derhovanessian, I. Vogler, L. M. Kranz, M. Vormehr, A. Baum, K. Pascal, J. Quandt and D. Maurus, Nature, 2020, 586, 594–599 CrossRef CAS
.
- P. F. McKay, K. Hu, A. K. Blakney, K. Samnuan, J. C. Brown, R. Penn, J. Zhou, C. R. Bouton, P. Rogers and K. Polra, Nat. Commun., 2020, 11, 3523 CrossRef CAS
.
- C. Janssen, M. Cachanado, L. Ninove, M. Lachatre, J. Michon, O. Epaulard, Z. Maakaroun-Vermesse, C. Chidiac, B. Laviolle and H. Aumaitre, EClinicalMedicine, 2022, 48, 10144 CrossRef
.
- A. M. Borobia, A. J. Carcas, M. Pérez-Olmeda, L. Castaño, M. J. Bertran, J. García-Pérez, M. Campins, A. Portolés, M. González-Pérez and M. T. G. Morales, Lancet, 2021, 398, 121–130 CrossRef CAS PubMed
.
- N.-N. Zhang, X.-F. Li, Y.-Q. Deng, H. Zhao, Y.-J. Huang, G. Yang, W.-J. Huang, P. Gao, C. Zhou and R.-R. Zhang, Cell, 2020, 182, 1271–1283 CrossRef CAS PubMed
.
- T. Strong, T. Hampton, I. Louro, G. Bilbao, R. Conry and D. Curiel, Gene Ther., 1997, 4, 624–627 CrossRef CAS PubMed
.
- J. Jemielity, J. Kowalska, A. M. Rydzik and E. Darzynkiewicz, New J. Chem., 2010, 34, 829–844 RSC
.
- T. Schlake, A. Thess, M. Fotin-Mleczek and K.-J. Kallen, RNA Biol., 2012, 9, 1319–1330 CrossRef CAS
.
- S. C. Semple, A. Akinc, J. Chen, A. P. Sandhu, B. L. Mui, C. K. Cho, D. W. Sah, D. Stebbing, E. J. Crosley and E. Yaworski, Nat. Biotechnol., 2010, 28, 172–176 CrossRef CAS PubMed
.
- D. Witzigmann, J. A. Kulkarni, J. Leung, S. Chen, P. R. Cullis and R. van der Meel, Adv. Drug Delivery Rev., 2020, 159, 344–363 CrossRef CAS
.
- C. Tilcock, Chem. Annu. Rev. Biophys. Biophys. Chem., 1985, 14, 211–238 CrossRef
.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. Mcfadyen and J. Chan, Mol. Ther., 2018, 26, 1509–1519 CrossRef CAS
.
- K. A. Hajj, R. L. Ball, S. B. Deluty, S. R. Singh, D. Strelkova, C. M. Knapp and K. A. Whitehead, Small, 2019, 15, e1805097 CrossRef
.
- H. Ni, M. Z. Hatit, K. Zhao, D. Loughrey, M. P. Lokugamage, H. E. Peck, A. D. Cid, A. Muralidharan, Y. Kim and P. J. Santangelo, Nat. Commun., 2022, 13, 4766 CrossRef CAS
.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye, C. Huang, J. Evans and E. P. Henske, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(8), e2116271119 CrossRef CAS PubMed
.
- A. Akinc, M. Goldberg, J. Qin, J. R. Dorkin, V. C. Gamba, M. Maier, K. N. Jayaprakash, M. Jayaraman, K. G. Rajeev and M. Manoharan, Mol. Ther.: J. Am. Soc. Gene Ther., 2009, 17, 872–879 CrossRef CAS
.
- K. A. Whitehead, J. R. Dorkin, A. J. Vegas, P. H. Chang, O. Veiseh, J. Matthews, O. S. Fenton, Y. Zhang, K. T. Olejnik, V. Yesilyurt and D. Chen, Nat. Commun., 2014, 5, 4277 CrossRef CAS PubMed
.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 1–8 CrossRef
.
- P. R. Cullis and M. J. Hope, Mol. Ther., 2017, S1525001617301119 Search PubMed
.
- N. Pardi, S. Tuyishime, H. Muramatsu, K. Kariko, B. L. Mui, Y. K. Tam, T. D. Madden, M. J. Hope and D. Weissman, J. Controlled Release, 2015, 217, 345–351 CrossRef CAS PubMed
.
- J. Probst, B. Weide, B. Scheel, B. Pichler, I. Hoerr, H. Rammensee and S. Pascolo, Gene Ther., 2007, 14, 1175–1180 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S8 include the 1H NMR spectra, MS spectra, flow cytometry spectra and images of lysosomal escape assay and storage stability. See DOI: https://doi.org/10.1039/d3tb00303e |
| This journal is © The Royal Society of Chemistry 2023 |