Open Access Article
Open Access ArticleSpermine-starch nanoparticles with antisense vicR suppress Streptococcus mutans cariogenicity†
Lei
Lei‡
a,
Yue
Zhang‡
a,
Yichen
Xu‡
b,
Yuting
Tian
a,
Jialiang
Zhao
a,
Yong
Xiang
a,
Huiyu
Yang
a,
Yingming
Yang
*a and
Tao
Hu
 *a
*a
aState Key Laboratory of Oral Diseases, Department of Preventive Dentistry, West China Hospital of Stomatology, Sichuan University, 14# Third Section Renmin South Road, Chengdu, 610041, China. E-mail: ymyang@scu.edu.cn; hutao@scu.edu.cn
bState Key Laboratory of Oral Diseases, Department of Oral Prosthodontics, West China Hospital of Stomatology, Sichuan University, 14# Third Section Renmin South Road, Chengdu, 610041, China
First published on 23rd May 2023
Abstract
Dental caries of permanent teeth is a common public health concern and has the second-highest incidence among global diseases. The exopolysaccharides (EPS) synthesized by Streptococcus mutans (S. mutans) are the principal virulence factor for cariogenic etiology. We previously discovered that an endogenous antisense vicR RNA (ASvicR) could significantly inhibit EPS synthesis in S. mutans and reduce its cariogenicity. However, ASvicR cannot be directly applied in the oral environment. An appropriate vector is of great need to protect ASvicR from being degraded by nucleases for effective gene delivery to S. mutans. Functionally modified starches shed light on this field because of their excellent biocompatibility and biodegradability. In this study, a biocompatible and biodegradable spermine-starch nanocomposite (SSN) was constructed for ASvicR delivery. Starch was cationically functionalized by grafting endogenous spermine to closely bind with the recombinant ASvicR plasmid. The SSN not only protected the recombinant ASvicR plasmid from DNase I but also achieved highly efficient gene transformation to S. mutans via the hydrolysis of α-amylase in the saliva. In addition, SSN-ASvicR was shown to endow ASvicR with an increasing transformation efficiency approximately four times that of the naked ASvicR plasmid, as well as allowing for targeting specificity to the transcription of the vicR gene and the suppression of biofilm organization via EPS digestion. In particular, SSN-ASvicR nanoparticles exhibited excellent biological safety and maintained oral microbiota homeostasis in vivo. The SSN can be prepared in a ready-to-use formulation for targeting cariogenic bacteria, thus demonstrating important prospects in the prevention of dental caries.
1. Introduction
As one of the most prevalent oral diseases, dental caries threatens the well-being of every individual. The incidence of permanent tooth caries was the second-highest among global diseases affecting the world's population.1 Etiologically, dental caries is an infectious disease on the tooth surface. Streptococcus mutans (S. mutans) is the principal decay-causing bacterium due to its capability to synthesize exopolysaccharides (EPS) that can further form a cariogenic biofilm on the tooth surface.2 The S. mutans VicRK two component signal transduction system (TCS) regulates genes that are associated with EPS synthesis, genetic competence, oxidative stress response, and cell wall homeostasis, and they are conserved in other Gram-positive coccus species.3,4 In particular, these EPS synthesis-associated genes, including gtfB/C and gbpB, are positively regulated by VicR. Moreover, vicR deletion mutants have been unable to be constructed, thus suggesting that VicR is essential for viability and controls the expression of virulence genes.5 Notably, in our previous studies, we identified an endogenous antisense vicR RNA (ASvicR) and verified that the overexpression of ASvicR could inhibit the transcription and translation of the vicR gene.6,7Via RNA interference (RNAi), ASvicR can downregulate the expression of glucosyltransferases (GtfB, GtfC, and GtfD), thus significantly inhibiting EPS synthesis and reducing the cariogenicity of S. mutans.7 On the other hand, dextranase A (DexA) participates in the degradation of exopolysaccharides by hydrolyzing glycosidic bonds.8,9 Nevertheless, ubiquitous deoxyribonucleases (DNases) can severely limit the application of ASvicR in the oral environment. Therefore, an appropriate vector is urgently needed for efficient ASvicR delivery.Although various materials (such as viral vectors, lipid-based particles, and polymeric particles) have been explored as protective nucleic acid carriers, the biocompatibility and safety of these vectors remain controversial.10,11 For Gram-positive cocci, a cell-penetrating peptide agent was used to facilitate oligonucleotide transformation, but their inherent toxicity and poor stability have restricted their applications.12 Recent progress in materials science has indicated that naturally obtainable starch is promising for biomedical applications, such as blood-absorbing collagen sponges,13 nano-encapsulating agents,14 and drug delivery systems.15 As a gene vector applied to the oral cavity, starch may possess superior biocompatibility and biodegradability16 compared to virus-based and artificially synthesized vectors. The starch vector can also be hydrolyzed by amylase in saliva, thus achieving gene delivery in an innovative manner. In our recently published work,17 we constructed the aminated dendritic mesoporous silica nanoparticles (DMSNs-NH2) carrying plasmid-ASvicR. However, the use of DMSNs-NH2 is restricted by its non-degradable properties.
Attempts have previously been made to modify starch molecules with positively charged quaternary ammonium groups.18 However, toxicological reports of quaternary ammonium compounds have raised significant concerns regarding their harmful effects on human health, such as oxidative stress, DNA damage, mitochondrial apoptosis, and cell death.19,20 For biomedical applications, spermine is a biofriendly and endogenous tetraamine with two primary and two secondary amino groups. Madeo et al. reported that dietary supplementation with polyamines is a protective strategy to increase longevity.21 In addition, polyamines have been shown to promote mitochondrial respiration and delay brain aging.22 The positive charge of spermine under physiological conditions allows it to bind and stabilize nucleic acids, which provides favorable conditions for DNA transport.
In this study, we utilized starch and spermine to construct a gene vector for ASvicR plasmid delivery. The starch molecule was cationically functionalized by grafting the spermine molecule via 1,1′-carbonyldiimidazole (CDI) activation. The spermine-starch nanocomposite (SSN) was capable of loading ASvicR via electrostatic interactions. The SSN was effective in protecting ASvicR from nuclease degradation, thus achieving highly efficient gene transformation to S. mutans. After treatment with SSN-ASvicR by mouth rinsing, the EPS synthesis of S. mutans was inhibited. An in vivo animal study further demonstrated the impediment effect of SSN-ASvicR on dental caries. Innovatively, this biocompatible and biodegradable SSN material shows important prospects in the prevention of dental caries. More importantly, the vector construction strategy may provide a novel approach for targeted nonantibiotic treatment against bacterial infection.
2. Materials and methods
2.1 Materials
Pullulanase, anhydrous dimethylsulfoxide (DMSO), CDI, and chemically pure grade PEG20000 were purchased from Macklin Reagent Company (Shanghai, China). α-Amylase, spermine, dialysis bags (7000D), cornstarch, DNase I, and the calcein-AM/PI kit were commercially obtained from Solaibio Reagent Company (Beijing, China). Agarose, brain heart infusion, and Dulbecco's modified Eagle medium (DMEM) were bought from Thermo Fisher Scientific (MA, USA). Additionally, the EndoFree Plasmid Kit (Tiangen, Beijing, China), the MasterPure RNA purification Kit (Epicentre, MD, USA), the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific), and the Cell Counting Kit-8 (CCK-8, Dojindo, Japan) were used in the present study.2.2 Sample preparation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 for 2 h at room temperature. Then, spermine (CDI
6 for 2 h at room temperature. Then, spermine (CDI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) spermine molar ratio of 1
spermine molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added and the reactions were continued at 35 °C for 24 h. When the reactions were complete, the products were dialyzed against pure water (molecular weight cutoff of 7000) for three days and lyophilized. The obtained samples were named SSN-1, SSN-2, and SSN-3.
1) was added and the reactions were continued at 35 °C for 24 h. When the reactions were complete, the products were dialyzed against pure water (molecular weight cutoff of 7000) for three days and lyophilized. The obtained samples were named SSN-1, SSN-2, and SSN-3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 to prepare the dispersed phase. Polyethylene glycol (PEG) was dissolved in deionized water to produce a PEG solution with a final concentration of 33.1 g/100 mL. The PEG solution (continuous phase) was agitated at 500 rpm in a water bath at 70 °C for 1 h. When the temperature of this continuous phase dropped to 40 °C, the solution was transferred to centrifuge tubes, and then the dispersed phase was gently poured into the continuous phase (v/v = 1
8 to prepare the dispersed phase. Polyethylene glycol (PEG) was dissolved in deionized water to produce a PEG solution with a final concentration of 33.1 g/100 mL. The PEG solution (continuous phase) was agitated at 500 rpm in a water bath at 70 °C for 1 h. When the temperature of this continuous phase dropped to 40 °C, the solution was transferred to centrifuge tubes, and then the dispersed phase was gently poured into the continuous phase (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8).
8).
A vortex oscillator was used to vibrate the solution for 10 min. The obtained emulsion was refrigerated at 4 °C for 24 h to solidify the droplets. Finally, the prepared emulsion was centrifuged at 5000 rpm for 15 min. The sediment (SSN-ASvicR nanoparticles) was washed twice with 75% ethanol, dehydrated with absolute ethanol, and dried in an oven at 50 °C for 24 h. The dried microspheres were sealed and stored in a desiccator for further investigations.23
2.3 Materials characterization
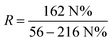 | (1) |
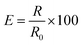 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, respectively. The weight percent (wt%) of spermine in SSN-1, SSN-2, and SSN-3 was calculated using eqn (3):
6, respectively. The weight percent (wt%) of spermine in SSN-1, SSN-2, and SSN-3 was calculated using eqn (3):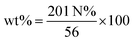 | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and 160
000× and 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× magnifications at an accelerating voltage of 20 kV.
000× magnifications at an accelerating voltage of 20 kV.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 8
1, and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 30 min to investigate the binding ability of the materials. The SSN-ASvicR nanoparticles are mixed with α-amylase or/and DNase at 60 °C in a water bath for 15 min. Next, the mixtures were analyzed by 1% agarose gel electrophoresis in tris-acetate (TAE) buffer at 100 V for 30 min. The resulting DNA products were observed under UV irradiation using a Bio-Rad electrophoresis system (Bio-Rad Laboratories, CA, USA).
1 for 30 min to investigate the binding ability of the materials. The SSN-ASvicR nanoparticles are mixed with α-amylase or/and DNase at 60 °C in a water bath for 15 min. Next, the mixtures were analyzed by 1% agarose gel electrophoresis in tris-acetate (TAE) buffer at 100 V for 30 min. The resulting DNA products were observed under UV irradiation using a Bio-Rad electrophoresis system (Bio-Rad Laboratories, CA, USA).
2.4 Bacterial biofilm experiments
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm = 0.2).25,26 For the transformation of the naked ASvicR-gfp plasmid without the competence stimulating peptide (CSP), two hundred nanograms of recombinant ASvicR-gfp were added to 500 μL of S. mutans culture (OD600
nm = 0.2).25,26 For the transformation of the naked ASvicR-gfp plasmid without the competence stimulating peptide (CSP), two hundred nanograms of recombinant ASvicR-gfp were added to 500 μL of S. mutans culture (OD600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm = 0.2) and incubated for 2 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). For the transformation of the naked ASvicR-gfp plasmid with the competence stimulating peptide (CSP), CSP was added to 500 μL of S. mutans culture (OD600
nm = 0.2) and incubated for 2 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). For the transformation of the naked ASvicR-gfp plasmid with the competence stimulating peptide (CSP), CSP was added to 500 μL of S. mutans culture (OD600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm = 0.2) to achieve a final concentration of 1 μg mL−1. Two hundred nanograms of recombinant ASvicR-gfp were added to the culture and incubated for 2 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). For the transformation of SSN-ASvicR-gfp, 1.8 micrograms of SSN-ASvicR-gfp, which contained 200 ng ASvicR-gfp, were added to 500 μL of S. mutans culture (OD600
nm = 0.2) to achieve a final concentration of 1 μg mL−1. Two hundred nanograms of recombinant ASvicR-gfp were added to the culture and incubated for 2 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). For the transformation of SSN-ASvicR-gfp, 1.8 micrograms of SSN-ASvicR-gfp, which contained 200 ng ASvicR-gfp, were added to 500 μL of S. mutans culture (OD600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm = 0.2) and incubated for 2 h. For the transformation of SSN (the nanoparticle vector minus plasmid), 1.6 micrograms of SSN and 200 ng recombinant ASvicR-gfp were added to 500 μL of S. mutans culture (OD600
nm = 0.2) and incubated for 2 h. For the transformation of SSN (the nanoparticle vector minus plasmid), 1.6 micrograms of SSN and 200 ng recombinant ASvicR-gfp were added to 500 μL of S. mutans culture (OD600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm = 0.2) and incubated for 2 h. Untreated S. mutans was set as blank control.
nm = 0.2) and incubated for 2 h. Untreated S. mutans was set as blank control.
After SSN-ASvicR-gfp, ASvicR-gfp with CSP, ASvicR-gfp without CSP or SSN plus ASvicR-gfp treatment, 50 μL bacterial suspensions (n = 5) were dropped on coverslips and dried in air at room temperature for 30 minutes. Then, the planktonic bacterial cells were stained with 5 μg mL−1 Alexa-555 WGA (Invitrogen) for 10 minutes in the dark. Confocal laser scanning microscopy (CLSM) was applied to observe the expression level of eGFP. The transformation of ASvicR-gfp was qualitatively assessed by observing green fluorescence intensities. The quantification of ASvicR-gfp transformation has been compared with the expressions of gfp genes by quantitative real-time PCR described in 4.5.
After SSN-ASvicR-gfp, ASvicR-gfp with CSP, ASvicR-gfp without CSP or SSN plus ASvicR-gfp treatment, the selective plating cell counting assay was conducted. One hundred microliters of bacterial suspensions were coated on BHI plates containing 1000 μg mL−1 spectinomycin for selection and incubated for 48 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). For the flow cytometry assays, 500 μL bacterial suspensions were collected after SSN-ASvicR-gfp, ASvicR-gfp with CSP, ASvicR-gfp without CSP or SSN plus ASvicR-gfp treatment. Untreated S. mutans was set as background control. The average fluorescence intensity of GFP was detected immediately, analyzed using a flow cytometer (Beckman Coulter, USA) and the fluorescence above the background level was calculated.27
Total RNAs of S. mutans in biofilm or planktonic cultures were extracted using a MasterPure RNA purification Kit (Epicentre, MD, USA). After biofilms were established for 24 h, the biofilm samples were harvested by scraping using a cell scraper and resuspended in PBS buffer. The bacterial cells were collected by centrifugation (4500 rpm, 4 °C) for 15 min and then resuspended in tissue and cell lysis solution (MasterPure RNA purification Kit). The planktonic cells were collected by centrifugation (4500 rpm, 4 °C) for 15 min, washed with PBS buffer and resuspended in tissue and cell lysis solution (MasterPure RNA purification Kit). The genomic DNA was removed by digestion with Turbo RNase-free DNase I (Ambion, Austin, TX, USA) according to the manufacturer's instructions. The purity (A260/A280) and concentration of RNA were determined using a NanoDrop2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA samples were reverse transcribed to complementary DNA (cDNA) using random 6 mers or gene-specific primer for ASvicR (Table S1, ESI†) with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA). Real-time PCR was carried out with a Bio-Rad Applied Biosystems ABI 7500 System (Bio-Rad Laboratories, Hercules, CA, USA). For each real-time PCR reaction, 20 μL of a mixture containing 10 μL SYBR Premix Ex TaqII, 2.0 μL template cDNA, 1 μL 10 μM PCR Forward Primer, 1 μL 10 μM PCR Reverse Primer, 0.4 μL ROX Reference Dye, and 5.6 μL deionized water was placed in each well. Primers used in real-time PCR reactions are shown in ESI,† Table S1 and were obtained commercially (Sangon Biotech, Shanghai, China). Conditions for real-time PCR were as follows: 95 °C, 3 min (initial denaturation), followed by 35 cycles at 95 °C for 30 s (denaturation), 55 °C for 30 s (primer annealing), and 72 °C for 30 s (extension). Threshold cycle values (CT) were quantified and the expression of each gene was normalized relative to the expression of the gyrA gene, which was used as an internal reference. Data were calculated according to the 2−ΔΔCT method.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 2 min, and 4 °C) and protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (BioRad, Hercules, CA, USA) according to the manufacturer's instructions.
000 rpm, 2 min, and 4 °C) and protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (BioRad, Hercules, CA, USA) according to the manufacturer's instructions.
For western blotting analysis, equal amounts of protein (50 μg) were mixed with 2× SDS-PAGE sample loading buffer (Beyotime Biotech, Shanghai, China) in boiling water for 10 min and loaded on 10% SDS-PAGE gels (Bio-Rad). Proteins were fractionated and then electrotransferred to polyvinylidene fluoride (PVDF) membranes (Biosharp Biotech, Shanghai, China). Membranes were blocked in TSBT buffer (25 mM Tris, 140 mM NaCl, 3 mM KCl, 0.1% Tween 20) containing 5% w/v nonfat dry milk at room temperature for 1 h. The membranes were incubated with purified DexA or VicR-specific polyclonal antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, AbMax Biotechnology, Beijing, China) overnight at 4 °C, washed with TSBT buffer, and incubated with horse-radish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1
1000, AbMax Biotechnology, Beijing, China) overnight at 4 °C, washed with TSBT buffer, and incubated with horse-radish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for 2 h at room temperature as previously described.6 Protein immunoreactive bands were visualized using an Immobilon Western Chemiluminescent kit (Millipore, Billerica, MA, USA). The ChemiDoc MP Imaging system (Bio-Rad) was used to detect the signal density of the protein bands.
000) for 2 h at room temperature as previously described.6 Protein immunoreactive bands were visualized using an Immobilon Western Chemiluminescent kit (Millipore, Billerica, MA, USA). The ChemiDoc MP Imaging system (Bio-Rad) was used to detect the signal density of the protein bands.
Electrophoretic mobility shift assays (EMSA) were used to determine whether promoter regions of the dexA gene were regulated by VicR as previously described.7 Amplicons of the promoter regions of the dexA gene were labeled with FAM (Qingke Biological Technology Co., Ltd, Beijing, China; the primers are listed in ESI† Table S1). Binding reactions of labeled DNA (20 fmol) with recombinant VicR protein (0, 20, 40, 60, and 80 pmol) were carried out in volumes of 20 μL containing 1× binding buffer [100 mM Tris, 500 KCl, 10 mM DTT; pH 7.5], with one-hundred-fold excess of unlabeled DNA fragments (cold DNA) as a competitor.4 After incubation for 30 min on ice, the reactants were loaded onto non-denaturing 6% acrylamide gels in 0.5× TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, 1 mM EDTA, pH 8.0). Protein–DNA complexes were separated at 4 °C (100 V, 3 h) in 0.5x TBE buffer. The signals were directly detected using the ChemiDoc MP system (Bio-Rad) according to the manufacturer's instructions.
2.5 Cytotoxicity assessment
Human gingival epithelial cells (HGECs) were obtained from clinical samples. The Ethics Committee of West China Hospital of Stomatology approved the use of HGECs in the present study. HGECs were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), penicillin (100 U mL−1), and streptomycin (100 μg mL−1) at 37 °C under 5% CO2 air conditions. For the cytotoxicity assays, HGECs in the logarithmic growth phase were seeded in a 96 well plate at a density of 5000 cells per well overnight. Next, the HGECs were treated with different concentrations (3.6, 14.4, and 28.8 μg mL−1) of SSN-ASvicR suspension. After incubation for 24 h, relative metabolic activity was assessed using the Cell Counting Kit-8 (Dojindo, Japan) as previously described.29 Live/dead staining was performed using the calcein-AM/PI kit (Solarbio, Beijing, China). Fluorescently labeled cells were observed using a fluorescence microscope (Leica, Germany), where the live and dead cells were stained with calcein-AM and PI into green and red, respectively.2.6 Animal study
To investigate the effects of SSN-ASvicR nanoparticles on the formation of dental plaque and cariogenicity in vivo, a rat model was used as previously described.24,30,31 Animal experiments were reviewed and approved by the Ethics Committee of West China Hospital of Stomatology, Chengdu, China (Permit number: WCHSIRB-D-2021-336). Twenty-four specific-pathogen-free Wistar rats (male, 3 weeks of age) were included (Experimental Animal Center of Public Health, Sichuan University, Chengdu, China). Before the S. mutans inoculation, rats were randomized into four experimental groups: UA159 as a positive control, SSN group, ASvicR plasmid group, and SSN-ASvicR group (n = 6 rats per group). SPF (specified pathogen-free) animals were housed, 6 rats per cage in a room on a 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 light/dark cycle. On days 1 to 7, each rat was infected orally with 200 μL of S. mutans mid-exponential phase (1 × 108 cells per CFU). The animals received both a cariogenic (sucrose-rich) diet and 5% sucrose water. The cariogenic diet (Trophic Animal Feed High-Tech Co. Ltd, China; NO. TP100352D20) comprises of milk powder, sucrose, starch, wheat flour and protein powder, which contains 56% sucrose. Sugar drinks were given using sterile water containing 5% sucrose. For treatment, a sterile cotton bud (Labor Import) was used to swab the rat teeth with 200 μL of SSN (1 mg mL−1), ASvicR plasmid (0.12 mg mL−1) or SSN-ASvicR (1 mg mL−1) suspensions respectively on days 1 to 14 as per our recently published study.17 Rats were weighed once a week and then their physical appearance was routinely recorded. On day 28, the animals were sacrificed by CO2 asphyxiation. The lower jaws were aseptically removed and divided into two pieces. After sonication in a PBS bath, the lower jaws were dissected and immersed in 10% buffered formalin phosphate for 48 h. The researchers were blinded to the group analyzed. The teeth were observed under a stereomicroscope and caries were scored according to a modified Keyes score.32,33 For micro-CT reconstruction, the mandibular samples were scanned by micro-CT at 7 μm voxels with a resolution of 500 projections/180°. The total scanning time of each sample was 14.3 minutes. After scanning, the images were obtained using Scco Evaluation software (Scco Medical AG).
12 light/dark cycle. On days 1 to 7, each rat was infected orally with 200 μL of S. mutans mid-exponential phase (1 × 108 cells per CFU). The animals received both a cariogenic (sucrose-rich) diet and 5% sucrose water. The cariogenic diet (Trophic Animal Feed High-Tech Co. Ltd, China; NO. TP100352D20) comprises of milk powder, sucrose, starch, wheat flour and protein powder, which contains 56% sucrose. Sugar drinks were given using sterile water containing 5% sucrose. For treatment, a sterile cotton bud (Labor Import) was used to swab the rat teeth with 200 μL of SSN (1 mg mL−1), ASvicR plasmid (0.12 mg mL−1) or SSN-ASvicR (1 mg mL−1) suspensions respectively on days 1 to 14 as per our recently published study.17 Rats were weighed once a week and then their physical appearance was routinely recorded. On day 28, the animals were sacrificed by CO2 asphyxiation. The lower jaws were aseptically removed and divided into two pieces. After sonication in a PBS bath, the lower jaws were dissected and immersed in 10% buffered formalin phosphate for 48 h. The researchers were blinded to the group analyzed. The teeth were observed under a stereomicroscope and caries were scored according to a modified Keyes score.32,33 For micro-CT reconstruction, the mandibular samples were scanned by micro-CT at 7 μm voxels with a resolution of 500 projections/180°. The total scanning time of each sample was 14.3 minutes. After scanning, the images were obtained using Scco Evaluation software (Scco Medical AG).
In addition, another group of rats (n = 4 rats) was used to verify the biological genetic safeties of SSN-ASvicR. On days 1 to 3, each rat was infected orally with 200 μL of the mid-exponential phase bacterial suspension that contained the Streptococcus mutans UA159 strain, Streptococcus gordonii, and Streptococcus sanguis. Then, a sterile cotton bud was used to swab the molars of the rats with 200 μL of SSN-ASvicR suspension. Swabs were used to collect the salivary samples from the rats’ mouth before and five hours after SSN-ASvicR treatment. The samples were soaked in DNA/RNA Shield Reagent (ZYMO Research) prior to DNA extraction. Then, we followed the manufacturer's protocol for DNA extraction. The extracted DNA was amplified with universal bacterial primers 8F (5′-AGAGTTTGATCATGGCTCAG-3′) and 1492R (5′-CGGTTACCTTGTTACGACTT-3′) to amplify the full length of the 16S rRNA gene. To avoid errors during cluster identification because of the high similarity of bases across all amplicons, we tagged a 24 bp barcode to forward primers. The 20 μL PCR volume contained 10 ng of extracted DNA and 0.3 μM of primers with Kodaq 2× PCR MasterMix (G497, Applied Biological Materials Inc.). PCR was carried out with an activation step at 95 °C for 3 min followed by 10 touchdown cycles at 95 °C for 10 s, 65 °C (decrease of 1 °C for each cycle) for 20 s, and 68 °C for 90 s, followed by 20 cycles at 95 °C for 10 s and 68 °C for 90 s and a final elongation for 7 min. Amplifications were quantified using a Qubit dsDNA HS Assay Kit (Q32854, Thermo Fisher Scientific) on a Tecan F200 luminescent plate reader, and samples were pooled to equimolar amounts of DNA. The library was prepared as recommended by Nanopore (SQK-LSK109, Oxford Nanopore Technologies, ONT) and loaded on a GridION × 5 flow cell (FLO-MIN106 R9.4.1, Oxford Nanopore Technologies) for a 72 h sequencing protocol using MinKNOW software at Rhonin BioSciences Company (Chengdu, China).
2.7 The effects of pDL278 plasmid containing ASvicR on S. gordonii and S. sanguinis
The S. gordonii and S. sanguinis strains were provided by the State Key Laboratory of Oral Diseases (Sichuan University, Chengdu, China). S. gordonii and S. sanguinis strains were routinely grown in brain heart infusion (BHI) media overnight at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2). Then, S. gordonii and S. sanguinis suspensions were diluted into fresh BHI media at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio and cultured into optical density at 600 nm [OD600] 0.2. For the transformation, the competence stimulating peptide (CSP) was added to the S. gordonii and S. sanguinis suspension cultures to achieve a final concentration of 1 μg mL−1. Recombinant pDL278 plasmid containing ASvicR were simultaneously added to the cultures and incubated for 60 min. S. gordonii-ASvicR and S. sanguinis-ASvicR were isolated using BHI plates containing 1000 μg mL−1 spectinomycin for selection and incubated for 48 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2).
20 ratio and cultured into optical density at 600 nm [OD600] 0.2. For the transformation, the competence stimulating peptide (CSP) was added to the S. gordonii and S. sanguinis suspension cultures to achieve a final concentration of 1 μg mL−1. Recombinant pDL278 plasmid containing ASvicR were simultaneously added to the cultures and incubated for 60 min. S. gordonii-ASvicR and S. sanguinis-ASvicR were isolated using BHI plates containing 1000 μg mL−1 spectinomycin for selection and incubated for 48 h at 37 °C anaerobically (90% N2, 5% CO2, and 5% H2).
The transcription analyses of S. gordonii-ASvicR and S. sanguinis-ASvicR strains respectively were prepared as described in 4.5. For planktonic cultures, S. gordonii-ASvicR and S. sanguinis-ASvicR cultures were grown overnight to the mid-exponential phase in BHI medium. The suspensions of S. gordonii-ASvicR and S. sanguinis-ASvicR were followed by diluting 100 μL culture into 2 mL of fresh BHI. Total RNAs of S. gordonii-ASvicR and S. sanguinis-ASvicR planktonic cultures were extracted using a MasterPure RNA purification Kit (Epicentre, MD, USA). The planktonic cells were collected by centrifugation (4500 rpm, 4 °C) for 15 min, washed with PBS buffer and resuspended in tissue and cell lysis solution (MasterPure RNA purification Kit). The genomic DNA was removed by digestion with Turbo RNase-free DNase I (Ambion, Austin, TX, USA) according to the manufacturer's instructions. RNA samples were reverse transcribed to complementary DNA (cDNA) using random 6 mers with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA). Real-time PCR was carried out as described in 4.5. Primers used in real-time PCR reactions are shown in ESI,† Table S1 and were obtained commercially (Sangon Biotech, Shanghai, China). Threshold cycle values (CT) were quantified and the expression of each gene was normalized relative to the expression of the 16sR genes of S. gordonii and S. sanguinis, which was used as an internal reference. Data were calculated according to the 2−ΔΔCT method.
2.8 Data analysis
Statistical analyses of the data were performed using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). Shapiro–Wilk tests were conducted to examine the normal distribution of the experimental data, and Bartlett's tests were performed to assess the homogeneity of variances. For parametric testing, one-way analysis of variance (ANOVA) was used to detect the significant effects of variables followed by the Student–Newman–Keuls test to compare the means of each group. For nonparametric testing, the Kruskal–Wallis test and least significant difference (LSD) multiple comparisons were used for comparison. A value of p < 0.05 was considered statistically significant.3. Results
3.1 Materials characterization of SSN and SSN-ASvicR
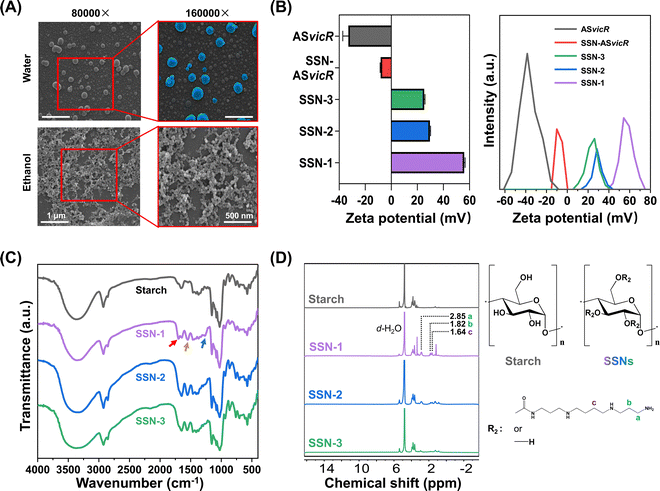
The morphology of SSN-ASvicR nanoparticles was greatly affected by the dispersion liquid (Fig. 1A). The Mw of degraded starch was 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112 g mol−1, and the molecular weight distribution is shown in ESI,† Fig. S2. Zeta potential measurements demonstrated that the mean surface charges of ASvicR, SSN-ASvicR, SSN-1, SSN-2, and SSN-3 were −32.9 mV, −8.1 mV, 55.9 mV, 29.6 mV, and 25.2 mV, respectively (Fig. 1B). The grafting of spermine onto the starch chain was verified by the results of FTIR. Additional signal peaks were detected in the spectra of SSN-1, SSN-2, and SSN-3, such as the amide I band associated with the C
112 g mol−1, and the molecular weight distribution is shown in ESI,† Fig. S2. Zeta potential measurements demonstrated that the mean surface charges of ASvicR, SSN-ASvicR, SSN-1, SSN-2, and SSN-3 were −32.9 mV, −8.1 mV, 55.9 mV, 29.6 mV, and 25.2 mV, respectively (Fig. 1B). The grafting of spermine onto the starch chain was verified by the results of FTIR. Additional signal peaks were detected in the spectra of SSN-1, SSN-2, and SSN-3, such as the amide I band associated with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1699 cm−1, the amide II band arising mainly from the in-plane N–H bending vibration at 1548 cm−1, and the amide III band located at 1264 cm−1 (Fig. 1C).34 The 1H NMR analysis results further confirmed the successful conjugation of spermine (Fig. 1D). In addition to the peaks belonging to the starch backbone, which appeared at δ = 5.38 ppm and δ = 3.5–3.9 ppm, new peaks were observed at δ = 1.1–3.3 ppm, and were assigned to the protons on the spermine chains. Table 1 shows the results of quantitative analysis of EA. Among the three SSN materials, SSN-1 displayed the highest amount of grafted spermine. As the grafting amount increased, the grafting efficiency decreased accordingly.
O stretching vibration at 1699 cm−1, the amide II band arising mainly from the in-plane N–H bending vibration at 1548 cm−1, and the amide III band located at 1264 cm−1 (Fig. 1C).34 The 1H NMR analysis results further confirmed the successful conjugation of spermine (Fig. 1D). In addition to the peaks belonging to the starch backbone, which appeared at δ = 5.38 ppm and δ = 3.5–3.9 ppm, new peaks were observed at δ = 1.1–3.3 ppm, and were assigned to the protons on the spermine chains. Table 1 shows the results of quantitative analysis of EA. Among the three SSN materials, SSN-1 displayed the highest amount of grafted spermine. As the grafting amount increased, the grafting efficiency decreased accordingly.
| Nitrogen content (%) | Molar ratio of spermine to glucose unit | Grafting efficiency of spermine (%) | Weight percent of spermine (%) | |
|---|---|---|---|---|
| SSN-1 | 4.21 ± 0.77 | 0.15 ± 0.03 | 29.23 ± 6.34 | 15.10 ± 2.77 |
| SSN-2 | 3.29 ± 0.72 | 0.11 ± 0.03 | 43.79 ± 10.97 | 11.79 ± 2.59 |
| SSN-3 | 2.56 ± 0.45 | 0.08 ± 0.02 | 49.50 ± 9.66 | 9.20 ± 1.63 |
3.2 The binding capacity, biodegradation behavior, and protective effect of the SSN
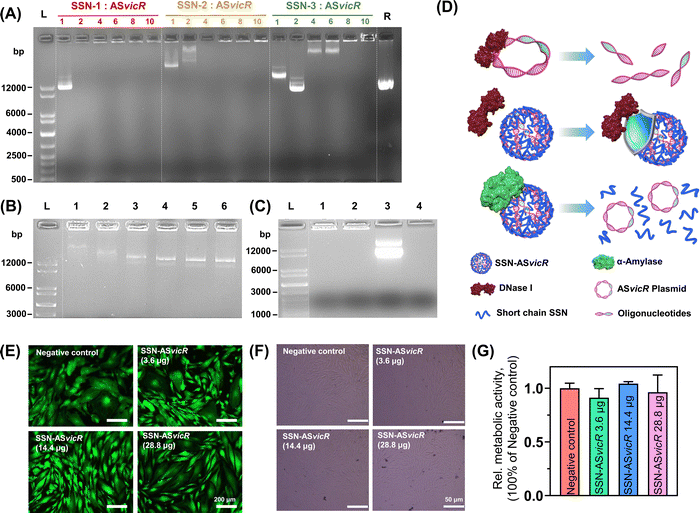
SSN-1 could reduce the migration of the ASvicR plasmid at or above a w/w ratio of 2, whereas clear ASvicR plasmid bands could be observed in the agarose gel for SSN-2 and SSN-3, thus indicating that their ASvicR plasmid binding abilities were weaker than that of SSN-1 (Fig. 2A). For this reason, SSN-1 was selected for SSN-ASvicR fabrication in this study. Subsequently, SSN-ASvicR was mixed with α-amylase in a water bath to analyze the biodegradable properties. The ASvicR plasmid bands were detected in all of the groups, and the dose of α-amylase was positively correlated with the clearness of the bands, thus indicating that the ASvicR plasmid was released due to the hydrolyzation of the starch chain in SSN-ASvicR (Fig. 2B). After DNase I treatment, the naked ASvicR plasmid was completely degraded and could not be detected in the gel (Fig. 2C, Lane 4). In contrast, the ASvicR plasmid bound with the SSN was still retained in the well (Fig. 2C, Lane 2). A schematic diagram of enzymatic digestion of SSN-ASvicR by α-amylase or DNase I is shown in Fig. 2D.3.3 Biocompatibility and transformation efficiency of SSN-ASvicR
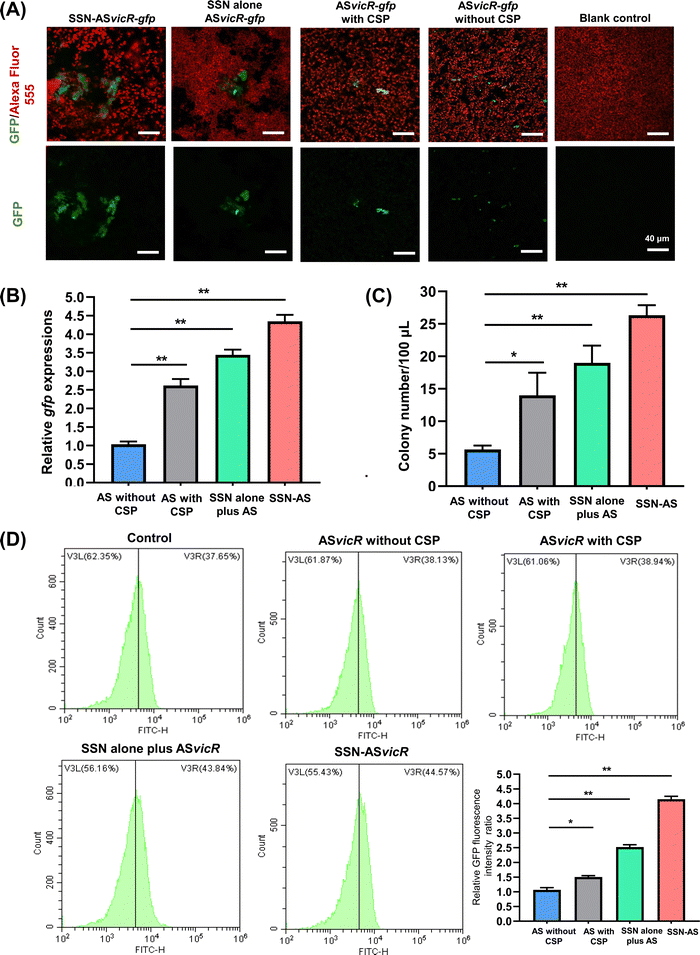
As shown in Fig. 2E and F, the cell viability and morphology of human gingival epithelial cells (HGECs) treated with the SSN-ASvicR suspension showed no significant difference from those of the negative control. The cells were stained green (live cells), and few red-stained cells (dead cells) could be observed. The CCK-8 assay showed that HGEC cells treated with the SSN-ASvicR suspension did not show a statistically significant difference from that of the negative control group (Fig. 2G), thus suggesting that SSN-ASvicR had favorable cytocompatibility. To ensure the transformation of the ASvicR plasmid into S. mutans cells, the recombinant ASvicR plasmid was labeled with a gfp tag. As shown in Fig. 3A, S. mutans cell walls were stained red with Alexa Fluor 555, and GFP was observed (green fluorescence). The quantification of the gfp gene expression via quantitative RT-PCR and the transformation assays with selective plating cell counting indicated that the transformation efficiency of SSN-ASvicR transformation was four times that of the naked ASvicR plasmid (Fig. 3B and C). Additionally, the flow cytometry assay indicated that the SSN-ASvicR transformation efficiency was the highest (Fig. 3D).3.4 Suppression effect of SSN-ASvicR on biofilm organization of S. mutans
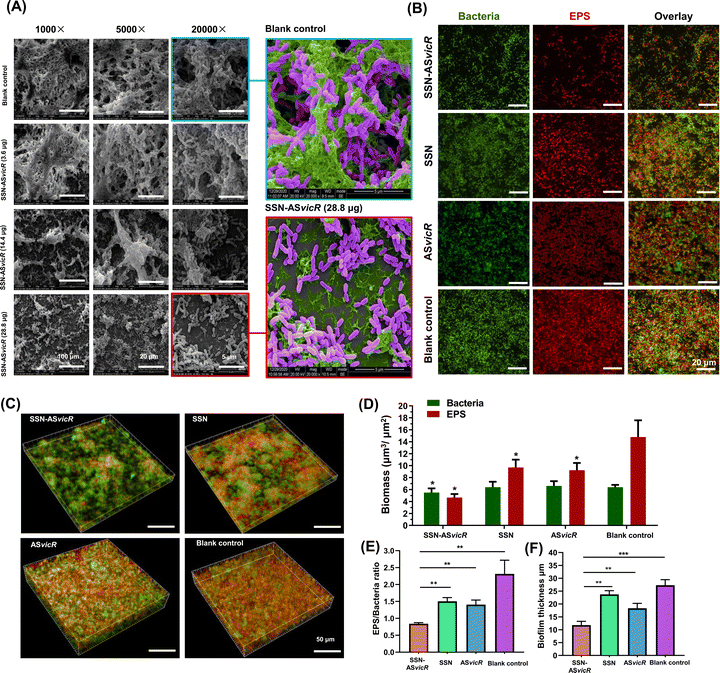
The SEM results showed that the untreated S. mutans were densely packed with an enriched extracellular matrix (ECM), whereas SSN-ASvicR-treated bacteria appeared to be scattered with reduced ECM interspersed among empty areas (Fig. 4A). Moreover, the CLSM observation showed that the EPS production of the SSN-treated, ASvicR-treated, and SSN-ASvicR-treated S. mutans decreased compared to that of the untreated S. mutans (Fig. 4B and C). Interestingly, we found that EPS biomass was variable between the treatments, and the cell biomass in the SSN-ASvicR group was changed compared with the untreated S. mutans (Fig. 4D). Further quantitative analysis showed that the EPS/bacteria ratios of the SSN-ASvicR-treated S. mutans biofilm were significantly lower than those of the ASvicR-treated biofilm (Fig. 4E). The thickness of biofilms was also variable between the treatments, indicating that the biofilm of the SSN-ASvicR group was the lowest (Fig. 4F).3.5 Mechanism analyses of the suppression effect of SSN-ASvicR on the biofilms of S. mutans
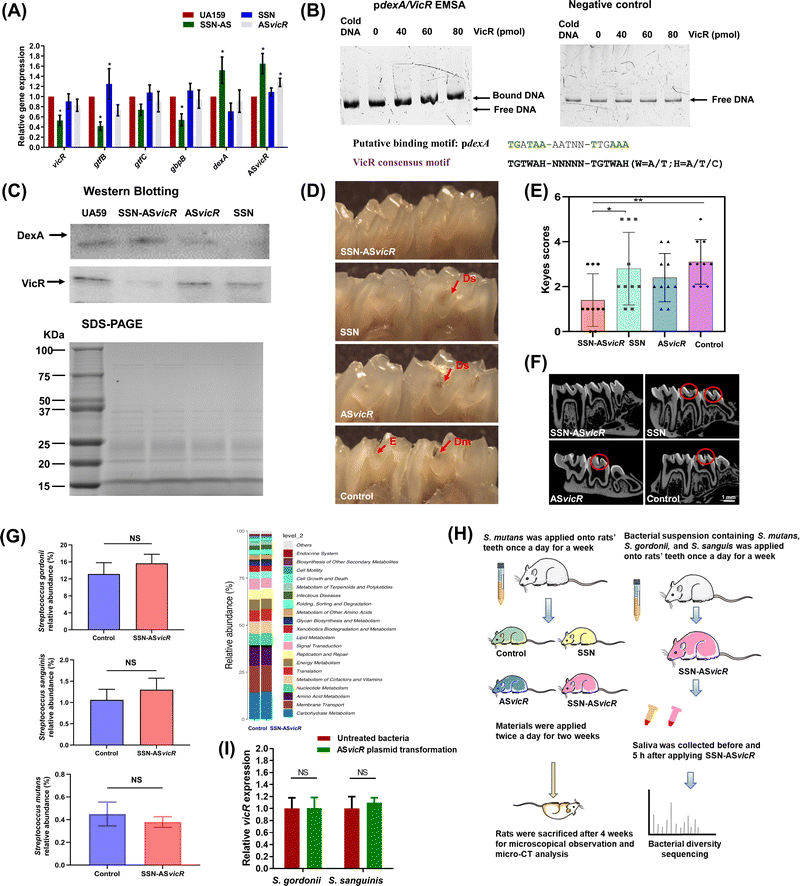
Quantitative RT-PCR assays showed that S. mutans biofilm cells treated with SSN-ASvicR were different from those observed after treatment with naked ASvicR. Interestingly, there was upregulation of gtfB in S. mutans treated with SSN alone (Fig. 5A and Fig. S3, ESI†). The transcription levels of the vicR, gtfB/C, and gbpB genes in the SSN-ASvicR-treated group exhibited the lowest values. Furthermore, EMSA was performed on the promoter regions of the dexA gene, which demonstrated that the VicR protein was directly bound to the dexA promoter regions (Fig. 5B). Western blot analysis of VicR and DexA proteins revealed that VicR expression levels were decreased in the ASvicR-treated and SSN-ASvicR-treated biofilms (Fig. 5C). The quantitative RT-PCR assays indicated that dexA transcription was significantly decreased in a vicK knockout mutant (Fig. S4, ESI†), which would strengthen the proposal that dexA is directly regulated by VicR as well as VicRK TCS.3.6 In vivo effects of SSN-ASvicR on the cariogenicity of S. mutans and oral microbiota homeostasis
The treatments were applied daily for two weeks to investigate the effects on the oral microbiota and incidence of caries after S. mutans colonization. As shown in Fig. 5D, a lower cariogenic incidence was observed in dentinal (Dm) and extensive dentinal (Ds) regions of the rat teeth treated with ASvicR and SSN-ASvicR. Particularly, the SSN-ASvicR-treated rat teeth showed the lowest cariogenic incidence, which was calculated by using the modified Keyes’ counting method (Fig. 5E and Table S2, ESI†). Micro-CT analysis showed that pit and fissure caries occurred in rat teeth without SSN-ASvicR treatment (Fig. 5F), which was consistent with the results of Keyes’ counting method. To address the potential effects of ASvicR on commensal streptococci, the abundances of S. mutans, S. gordonii and S. sanguinis were compared in the control and test groups, which indicated that there was no significant difference. In addition, KEGG pathway impact analysis demonstrated similar functional prediction results of metabolic pathways between control and SSN-ASvicR treated groups (Fig. 5G). The experimental flowchart of animal study is listed in Fig. 5H. We transformed the pDL278 plasmid containing ASvicR into S. gordonii and S. sanguinis. The quantitative RT-PCR results indicated that the expression of the homologous vicR gene in both S. gordonii and S. sanguinis was not affected by transformation of the pDL278 plasmid containing ASvicR (Fig. 5I).4. Discussion
In the present study, SSN materials were developed by grafting spermine onto the starch molecule via CDI activation. Due to its excellent binding capacity, SSN-1 was selected to effectively encapsulate the ASvicR plasmid. The morphology of SSN-ASvicR nanoparticles dispersed in deionized water was larger and more uneven in size than those dispersed in ethanol, which could be explained by the swelling effect. The presence of nitrogen in SSN materials was confirmed by EA, signifying the successful grafting of spermine onto the starch chain.35 This could be further verified by the results of FTIR and 1H NMR, demonstrated by the additional signal peaks on the spectra.16,36 With an increase in the grafting amount of spermine, we observed a decrease in grafting efficiency (Table 1). This might be explained by the steric hindrance and electrostatic repulsion phenomenon in polymer grafting.37 The zeta potential results suggest the possible binding mechanism between the SSN materials and the ASvicR plasmid. Before binding, the SSN materials exhibited a distinct positive charge (55.9 mV, 29.6 mV, and 25.2 mV), while the ASvicR plasmid carried a noticeable negative charge (−32.9 mV). The zeta potential of the resulting complex (SSN-ASvicR) lies between the two (−8.1 mV), indicating that the SSN and ASvicR may bind together through electrostatic adsorption. The favorable cytocompatibility of SSN-ASvicR may be attributed to the raw materials used for SSN fabrication; specifically, starch is a naturally available and environmentally friendly material with excellent biocompatibility, and spermidine is an endogenous polyamine that is beneficial in extending lifespans through epigenetic modifications and necrosis suppression.38To ensure the transformation of the ASvicR plasmid into S. mutans cells, we chose a green fluorescent protein (GFP) as a model protein for investigation,39 and S. mutans cell walls were marked via Alexa Fluor 555.40 Regarding the treatment with SSN-ASvicR and its effects on promoting a gene transcript profile, it has been reported that the source of carbohydrate significantly impacts the stochastic behaviors that can induce competence in S. mutans.41,42 In the present study, the SSN could be bio-suitably hydrolyzed by the endogenous α-amylase of S. mutans,43 which may stimulate the progression to the competent state. Taken together, we speculated that carbohydrate sources not only induce genetic competence but also alter the expression of EPS-related genes.
The results of the SEM observation and CLSM analysis revealed that SSN-ASvicR could significantly reduce the S. mutans biofilm due to its inhibitory effect on EPS production. Via complementary base pairing, ASvicR was able to form a double-stranded structure with vicR mRNA, thereby inhibiting the transcription and translation of the vicR gene.6,7 Because two component systems (TCSs)44 are widely present in most lower eukaryotes and prokaryotes, ASvicR can target the S. mutans VicRK TCS without affecting mammalian cells. Etiologically, EPS plays a key role in the development of dental caries by promoting bacterial adhesion and aggregation.45 As the long-term application of antibiotics may induce drug resistance and dysbacteriosis,46 the EPS reduction in cariogenic biofilms is beneficial for the prevention and treatment of dental caries. With fewer adverse complications, SSN-ASvicR nanoparticles may be a promising alternative or supplement to the currently used antibiotics.
Our previous study indicated that the expression of the ASvicR RNA transcript was elevated in planktonic cultures and repressed during biofilm growth.6 In the present study, we detected changes in AS vicR in planktonic and biofilm cultures (Fig. S3, ESI†), which indicated that the increase in ASvicR expression in S. mutans treated with SSN-ASvicR in the planktonic state was greater than that in the biofilm state. We speculated that the SSN system improves the efficiency of ASvicR transformation by inducing genetic competence, especially in the planktonic state. Regarding the SSN-ASvicR nanoparticle stability, we detected the quantity of released nucleic acid (Fig. S5, ESI†) from the SNN-ASvicR complex at RT (room temperature) and 4 °C as well as with or without FBS (10% FBS representing in vitro and in vivo conditions respectively).
Interestingly, the SSN alone inhibited EPS/bacteria ratios, and treatment with the SSN alone also reduced VicR expression. One possible explanation was consistent with the results of our previous study,6 which showed that the transcripts of ASvicR were elevated when S. mutans biofilm cultures were complemented with sucrose or glucose. As the SSN can be hydrolyzed by endogenous α-amylase of S. mutans and can serve as a carbohydrate source, S. mutans treated with the SSN alone likely reduced VicR expression and inhibited EPS production.6,7,24 Dental caries is a complex process that is also associated with other bacteria. The dynamic balance between commensals and opportunistic pathogens was disrupted, which causes microbial community shifts and disrupts tooth-enamel mineral homeostasis.47,48 The SSN nanoparticles have a slight inhibitory effect on the Enterococcus faecalis or Streptococcus sanguis (Fig. S6 and S7, ESI†). Another possible explanation was that the SSN (as a positively charged surface material) is supposed to capture or affect bacteria,49 thus warranting further investigation.
In this study, we found that the vicR, gtfB/C, and gbpB expression levels were significantly downregulated by ASvicR and SSN-ASvicR treatment. The upregulation of gtfB in S. mutans treated with the SSN alone was consistent with the results of a previous study,4 which indicated that the expression of the gtfB gene was elevated when S. mutans was in biofilms supplemented with sucrose. In contrast, the dexA gene expression was significantly elevated in the SSN-ASvicR-treated group. We previously reported that dexA-encoded dextranase (DexA) could partially inhibit the synthesis of and promote the digestion of glucans and eventually cause the breakdown of biofilm organization.50 As the dexA gene expression increased in the SSN-ASvicR-treated group, we speculated that VicR negatively regulates the dexA gene transcription and that SSN-ASvicR represses EPS biosynthesis by digesting glucans. As a VicR-binding box was found in the dexA promoter, further investigations are needed to explore the interactions of the specific VicR-binding box in the dexA promoter and the VicR protein.
Hu et al. sampled saliva at 10 to 60 min intervals to track the high-resolution microbiome dynamics during the course of human eating activities for 5 days.51 Indeed, it is uncertain whether the abundance of streptococcal species analyzed only 5 h after SSN-ASvicR treatment has biological significance, and further investigations are needed. Regarding the high degree of conservation of vicR genes, there is a 71.5% similarity of vicR genes between S. mutans and S. gordonii, as well as a 72.6% similarity of vicR genes between S. mutans and S. sanguinis. However, there is only a 23.9% similarity between S. mutans ASvicR and the S. gordonii vicR gene, as well as only a 24.9% similarity between S. mutans ASvicR and the S. sanguinis vicR genes. However, the potential limitation is that the cariogenic biofilm may be more complex, and additional species should be considered to provide cariogenicity ability. Future research is needed to investigate a prolonged observation time for rat caries with or without high sucrose conditions.
In our recently published work,17 we constructed the aminated dendritic mesoporous silica nanoparticles (DMSNs-NH2) carrying plasmid-ASvicR. The plasmid-ASvicR loaded on DMSNs-NH2 could be transformed into S. mutans, which reduced EPS synthesis and biofilm organization and showed preliminary anti-caries potential. However, the use of DMSNs-NH2 is different from that of the materials in the present study. Firstly, the properties of materials are improved and enhanced, in which DMSNs-NH2 were non-degradable while the SSN is made from natural corn starch and spermine with degradability. The SSN is more biofriendly as a gene carrier. Secondly, the potential applications of the DMSNs-NH2 and SSN nanoparticles are different. The DMSNs-NH2 has silicon dioxide as the carrier with a certain hardness and non-degradable properties, and can be used for filling resin or toothpaste filler. The main use of the SSN nanoparticles is developed for mouthwash. Regarding the chemical synthesis process, DMSNs-NH2 is based on the silicon dioxide while SSN nanoparticles are based on spermine. From this point of view, the SSN nanoparticles in the present article are more innovative and more promising for caries-prevention transformation applications.
5. Conclusions
A SSN material was developed by grafting spermine onto the starch molecule via CDI activation. The positively charged SSN exhibited an excellent ability to bind with nucleic acids and protect them from degradation by nucleases. When applied to the oral cavity, the SSN achieved high transformation efficiency in delivering the ASvicR plasmid to S. mutans. Via the targeted posttranscriptional regulation of the vicR gene, the SSN-ASvicR complex inhibited EPS synthesis and cariogenicity in S. mutans without disrupting oral microbiota homeostasis (Scheme 1). Thus, this SSN-ASvicR system could be prepared as a ready-to-use formulation targeting cariogenic bacteria, showing great potential in the prevention of dental caries. More importantly, taking into consideration the global antibiotic abuse and multiple drug resistance issues, this SSN material design strategy provides a novel approach for anti-infective treatments based on gene transformation and targeted RNAi.Data availability statement
The data that support the findings of this study are available from the corresponding author upon a reasonable request.Author contributions
Lei, L: contributed to conception and design, acquisition and analysis, and drafted and critically revised the manuscript. Zhang, Y: contributed to conception and design, acquisition and analysis, and drafted and critically revised the manuscript. Xu, Y: contributed to conception and design, acquisition and interpretation, drafted the manuscript, and critically revised the manuscript. Tian, Y: contributed to conception and analysis, and critically revised the manuscript. Zhao, J: contributed to conception and analysis, and critically revised the manuscript. Xiang, Y: contributed to conception and analysis, and critically revised the manuscript. Yang, H: contributed to conception and analysis, and critically revised the manuscript. Yang, Y: contributed to conception and design, analysis and interpretation, drafted the manuscript, and critically revised the manuscript. Hu, T: contributed to conception and design, analysis and interpretation, drafted the manuscript, and critically revised the manuscript. All authors gave their final approval and agreed to be accountable for all aspects of the work.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 82170948, 31971196 and 32201096), the Sichuan Science and Technology Program (Grant no. 2021YFS0085 and 2023NSFSC0557), the Research and Development Program of West China Hospital of Stomatology Sichuan University (Grant no. RD-03-202002, RD-03-202103 and RD-02-202001), and the Research Funding from West China School/Hospital of Stomatology Sichuan University (Grant no. RCDWJS2021-13, RCDWJS2020-24 and RD-02-202114).References
- GBD 2016 Disease and Injury Incidence and Prevalence Collaborators, Lancet, 2017, 390, 1211–1259 CrossRef PubMed.
- W. H. Bowen and H. Koo, Caries Res., 2011, 45, 69–86 CrossRef CAS PubMed.
- M. D. Senadheera, A. W. C. Lee, D. C. I. Hung, G. A. Spatafora, S. D. Goodman and D. G. Cvitkovitch, J. Bacteriol., 2007, 189, 1451–1458 CrossRef CAS PubMed.
- R. N. Stipp, H. Boisvert, D. J. Smith, J. F. Höfling, M. J. Duncan and R. O. Mattos-Graner, PLoS One, 2013, 8, e58271 CrossRef CAS PubMed.
- M. D. Senadheera, B. Guggenheim, G. A. Spatafora, Y.-C. C. Huang, J. Choi, D. C. I. Hung, J. S. Treglown, S. D. Goodman, R. P. Ellen and D. G. Cvitkovitch, J. Bacteriol., 2005, 187, 4064–4076 CrossRef CAS PubMed.
- L. Lei, R. N. Stipp, T. Chen, S. Z. Wu, T. Hu and M. J. Duncan, J. Dent. Res., 2018, 97, 1477–1484 CrossRef CAS PubMed.
- L. Lei, B. Zhang, M. Mao, H. Chen, S. Wu, Y. Deng, Y. Yang, H. Zhou and T. Hu, J. Dent. Res., 2020, 99, 204–213 CrossRef CAS PubMed.
- S. E. Cross, J. Kreth, L. Zhu, R. Sullivan, W. Shi, F. Qi and J. K. Gimzewski, Microbiology (Reading), 2007, 153, 3124–3132 CrossRef CAS PubMed.
- M. F. Hayacibara, H. Koo, A. M. Vacca-Smith, L. K. Kopec, K. Scott-Anne, J. A. Cury and W. H. Bowen, Carbohydr. Res., 2004, 339, 2127–2137 CrossRef CAS PubMed.
- H. Shen, X. Huang, J. Min, S. Le, Q. Wang, X. Wang, A. A. Dogan, X. Liu, P. Zhang, M. S. Draz and J. Xiao, Curr. Top. Med. Chem., 2019, 19, 2507–2523 CrossRef CAS PubMed.
- S. Y. Neshat, S. Y. Tzeng and J. J. Green, Curr. Opin. Biotechnol, 2020, 66, 1–10 CrossRef CAS PubMed.
- A. Ghosal, A. Vitali, J. E. M. Stach and P. E. Nielsen, ACS Chem. Biol., 2013, 8, 360–367 CrossRef CAS PubMed.
- A. B. Castro-Ceseña, T. A. Camacho-Villegas, P. H. Lugo-Fabres, E. E. Novitskaya, J. McKittrick and A. Licea-Navarro, Carbohydr. Polym., 2016, 148, 78–85 CrossRef PubMed.
- S. Y. Cheuk, F. F. Shih, E. T. Champagne, K. W. Daigle, J. A. Patindol, C. P. Mattison and S. M. Boue, Food Chem., 2015, 174, 585–590 CrossRef CAS PubMed.
- M. J. Majcher, A. Babar, A. Lofts, A. Leung, X. Li, F. Abu-Hijleh, N. M. B. Smeets, R. K. Mishra and T. Hoare, J Controlled Release, 2021, 330, 738–752 CrossRef CAS PubMed.
- X. Huang, X. Li, L. Chen and L. Li, Carbohydr. Polym., 2017, 173, 690–700 CrossRef CAS PubMed.
- Y. Tian, Y. Zhang, M. Zhang, X. Chen, L. Lei and T. Hu, Int. J. Nanomedicine, 2022, 17, 1255–1272 CrossRef CAS PubMed.
- S. A. Engelberth, N. Hempel and M. Bergkvist, Bioconjug. Chem., 2015, 26, 1766–1774 CrossRef CAS PubMed.
- Y. Jiao, S. Ma, Y. Wang, J. Li, L. Shan, J. Sun and J. Chen, Int. J. Biol. Sci., 2016, 12, 580–593 CrossRef CAS PubMed.
- Y. Jiao, L.-N. Niu, S. Ma, J. Li, F. R. Tay and J.-H. Chen, Prog. Polym. Sci., 2017, 71, 53–90 CrossRef CAS PubMed.
- F. Madeo, T. Eisenberg, F. Pietrocola and G. Kroemer, Science, 2018, 359, eaan2788 CrossRef PubMed.
- Y. Liang, C. Piao, C. B. Beuschel, D. Toppe, L. Kollipara, B. Bogdanow, M. Maglione, J. Lützkendorf, J. C. K. See, S. Huang, T. O. F. Conrad, U. Kintscher, F. Madeo, F. Liu, A. Sickmann and S. J. Sigrist, Cell Rep., 2021, 35, 108941 CrossRef CAS PubMed.
- H. Yang, Y. Yang, B.-Z. Li, B. Adhikari, Y. Wang, H.-L. Huang and D. Chen, Carbohydr. Polym., 2020, 231, 115692 CrossRef CAS PubMed.
- L. Lei, Y. Yang, M. Mao, H. Li, M. Li, Y. Yang, J. Yin and T. Hu, Front. Microbiol., 2015, 6, 1432 Search PubMed.
- F. C. Petersen and A. A. Scheie, Methods Mol. Biol., 2010, 666, 167–180 CrossRef PubMed.
- Z. Xie, T. Okinaga, F. Qi, Z. Zhang and J. Merritt, Appl. Environ. Microbiol., 2011, 77, 8025–8033 CrossRef CAS PubMed.
- F. Vromman, M. Laverrière, S. Perrinet, A. Dufour and A. Subtil, PLoS One, 2014, 9, e99197 CrossRef PubMed.
- Y. Deng, Y. Yang, B. Zhang, H. Chen, Y. Lu, S. Ren, L. Lei and T. Hu, Int. J. Oral. Sci., 2021, 13, 45 CrossRef CAS PubMed.
- Y. Xu, A. B. Xepapadeas, B. Koos, J. Geis-Gerstorfer, P. Li and S. Spintzyk, Dent. Mater., 2021, 37, e314–e327 CrossRef CAS PubMed.
- W. H. Bowen, K. M. Madison and S. K. Pearson, J. Dent. Res., 1988, 67, 1316–1318 CrossRef CAS PubMed.
- M.-Y. Mao, Y.-M. Yang, K.-Z. Li, L. Lei, M. Li, Y. Yang, X. Tao, J.-X. Yin, R. Zhang, X.-R. Ma and T. Hu, Front. Microbiol., 2016, 7, 00687 Search PubMed.
- H. Koo, P. L. Rosalen, J. A. Cury, Y. K. Park, M. Ikegaki and A. Sattler, Caries Res., 1999, 33, 393–400 CrossRef CAS PubMed.
- L. Su, F. Yu, Z. Li, C. Zeng, Q. Xu and M. Fan, Acta Pharmacol. Sin., 2014, 35, 592–598 CrossRef CAS PubMed.
- Y. Ji, X. Yang, Z. Ji, L. Zhu, N. Ma, D. Chen, X. Jia, J. Tang and Y. Cao, ACS Omega, 2020, 5, 8572–8578 CrossRef CAS PubMed.
- H. Li, B. Zhang, S. Lü, H. Ma and M. Liu, Int. J. Biol. Macromol., 2018, 120, 1225–1231 CrossRef CAS PubMed.
- Y. Huang, H. Hu, R.-Q. Li, B. Yu and F.-J. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 3919–3927 CrossRef CAS PubMed.
- Y. Chu, Y. Sun, W. Wu and H. Xiao, Carbohydr. Polym., 2020, 250, 116892 CrossRef CAS PubMed.
- T. Eisenberg, H. Knauer, A. Schauer, S. Büttner, C. Ruckenstuhl, D. Carmona-Gutierrez, J. Ring, S. Schroeder, C. Magnes, L. Antonacci, H. Fussi, L. Deszcz, R. Hartl, E. Schraml, A. Criollo, E. Megalou, D. Weiskopf, P. Laun, G. Heeren, M. Breitenbach, B. Grubeck-Loebenstein, E. Herker, B. Fahrenkrog, K.-U. Fröhlich, F. Sinner, N. Tavernarakis, N. Minois, G. Kroemer and F. Madeo, Nat. Cell Biol., 2009, 11, 1305–1314 CrossRef CAS PubMed.
- Y. Jiang, X. Pan, J. Chang, W. Niu, W. Hou, H. Kuai, Z. Zhao, J. Liu, M. Wang and W. Tan, J. Am. Chem. Soc., 2018, 140, 6780–6784 CrossRef CAS PubMed.
- H. Chan, B. Söderström and U. Skoglund, MicrobiologyOpen, 2020, 9, e999 CrossRef PubMed.
- Z. D. Moye, M. Son, A. E. Rosa-Alberty, L. Zeng, S.-J. Ahn, S. J. Hagen and R. A. Burne, Appl. Environ. Microbiol., 2016, 82, 4821–4834 CrossRef CAS PubMed.
- Y. Sato, K. Okamoto-Shibayama and T. Azuma, J. Oral. Microbiol., 2013, 5, 21285 CrossRef PubMed.
- C. L. Simpson and R. R. Russell, J. Bacteriol., 1998, 180, 4711–4717 CrossRef CAS PubMed.
- M. Kruppa and R. Calderone, FEMS Yeast Res., 2006, 6, 149–159 CrossRef CAS PubMed.
- A. Gedif Meseret, Int. J. Oral Dent. Health, 2021, 7, 127 Search PubMed.
- H. Wu, Y. Ma, X. Peng, W. Qiu, L. Kong, B. Ren, M. Li, G. Cheng, X. Zhou and L. Cheng, Arch. Oral Biol., 2020, 114, 104730 CrossRef CAS PubMed.
- F. Manji, G. Dahlen and O. Fejerskov, Caries Res., 2018, 52, 548–564 CrossRef PubMed.
- N. Philip, B. Suneja and L. J. Walsh, Caries Res., 2018, 52, 153–165 CrossRef PubMed.
- W. Li, E. S. Thian, M. Wang, Z. Wang and L. Ren, Adv. Sci., 2021, 8, e2100368 CrossRef PubMed.
- Y. Yang, M. Mao, L. Lei, M. Li, J. Yin, X. Ma, X. Tao, Y. Yang and T. Hu, Mol. Oral Microbiol., 2019, 34, 51–63 CrossRef CAS PubMed.
- Y. Hu, A. Amir, X. Huang, Y. Li, S. Huang, E. Wolfe, S. Weiss, R. Knight and Z. Z. Xu, Genome Res., 2022, 32, 1112–1123 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb02628g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |