3D printed reduced graphene oxide-GelMA hybrid hydrogel scaffolds for potential neuralized bone regeneration†
Xinwei
Zhang
 ab,
Hao
Zhang
ab,
Yi
Zhang
ab,
Huimin
Huangfu
ab,
Yixin
Yang
ab,
Qiuyue
Qin
ab,
Yidi
Zhang
ab,
Hao
Zhang
ab,
Yi
Zhang
ab,
Huimin
Huangfu
ab,
Yixin
Yang
ab,
Qiuyue
Qin
ab,
Yidi
Zhang
 *ab and
Yanmin
Zhou
*ab
*ab and
Yanmin
Zhou
*ab
aHospital of Stomatology, Jilin University, Changchun 130000, China. E-mail: zhangyidi@jlu.edu.cn; zhouym@jlu.edu.cn
bJilin Provincial Key Laboratory of Tooth Development and Bone Remodeling, Jilin University, Changchun 130000, China
First published on 9th January 2023
Abstract
Peripheral nerves participate in bone growth and repair by secreting neurotransmitters, and enable new bone to possess physiological bone-sensing capability. However, it is difficult to achieve synchronized nerve regeneration during the healing process of large bone defects at present. As a bioactive nanomaterial, reduced graphene oxide (rGO) can promote neuronal differentiation and myelination of Schwann cells (SCs), while enhancing the adhesion and osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) through its strong non-covalent binding ability. In this study, 3D printing-based rGO/GelMA hydrogels with enhanced osteogenic and neurogenic dual differentiation were used to simultaneously load SCs and BMSCs. By changing the concentration of rGO(0.03%/0.05%/0.1%), the compressive strength, rheological properties and aperture of the hydrogel can be improved. In vitro, cell live/death staining, phalloidin staining and SEM showed that cells loaded on the hydrogel had a high survival rate (85%) and good adhesion ability. In vivo, we found that the rGO/GelMA hydrogel exhibited the same low inflammatory response compared to the pure-GelMA group and the cell-only group, but surrounded by collagen fibers. Meanwhile, the osteogenic and neural proteins in the rGO/GelMA group were found to be highly expressed in immunohistochemistry and immunofluorescence. In this study, a scaffold material containing double cells was used to promote synergistic regeneration of nerves and bone, providing a promising strategy for the preparation of personalized and functionalized biomimetic bone material.
1. Introduction
Bone tissue is a structurally complex part of the human body, and the nerves and blood vessels in the bone are widely distributed in the Haversian canal and the Volkmann canal of the cortical bone.1–3 On the one hand, sensory and sympathetic nerves participate in the growth and repair of bones by secreting substances such as neurotransmitters and neuropeptides.4,5 On the other hand, intraosseous nerves sense changes in bone by conducting mechanical or electrical signals to maintain their normal physiological activities.6 Sensory denervation may lead to loss of trabecular bone, resulting in decreased bone strength and impairment of new bone formatio.7,8 In addition, for the bone lacking bone sensing ability, due to the lack of accurate and timely conduction to external stimuli, pathological responses may occur (for example, it is difficult to perceive whether the tooth is subjected to traumatic stress after the loss of nerves in the alveolar bone).9 Therefore, simultaneous repair of intraosseous nerves is critical for functional bone regeneration. However, there are few reports on the synergistic regeneration of bone and nerve with biomimetic bone scaffolds.10,11Cell therapy is considered a strategy to replace, repair or enhance the biological function of damaged tissues or systems by autologous or allogeneic cells.12–14 Among them, bone marrow mesenchymal stem cells (BMSCs) are stem cells with the self-proliferation ability and multi-directional differentiation potential extracted from bone marrow. They are often loaded by scaffold materials for bone tissue regeneration for the study of osteogenesis.15,16 Schwann cells (SCs) decompose and phagocytose myelin debris from distal nerve segments during functional recovery of injured peripheral nerves and proliferate to form Büngner's ribbons, which in turn guide axon regeneration in proximal segments.17,18 BMSCs and SCs can promote the differentiation of each other.19 Previous studies have shown that SCs co-cultured with BMSCs on osteogenic scaffolds exhibited better proliferative capacity and nerve growth factor (NGF), s100, TrkA and brain-derived neurotrophic factor(BDNF) expression.1
Graphene oxide (GO) is a nanomaterial with a large number of hydrophilic functional groups and a large surface area. Due to its excellent mechanical properties and topographic features, it has attracted extensive attention in cell engineering applications.20,21 However, many studies have shown that GO may have cytotoxicity and low electrical conductivity to a certain extent, and toxicity is related to exposure time, dose, and surface chemistry.22 Reduced graphene oxide (rGO) is obtained by reducing GO to remove part of the oxygen functional groups. Reduction of GO to rGO reduces water dispersibility, making rGO more safe, stable, and conductive than GO in vivo.23 rGO is rich in π–π stacking, hydrogen bonding, and electrostatic binding. It enables cell adhesion by promoting fibronectin extracellular protein adsorption, while also regulating stem cell growth and migration.24 Studies have also shown that the high conductivity of reduced graphene oxide can modulate neural cell mobility and differentiation under electrical stimulation (ES), which will be beneficial to support the growth of neuronal cells in neural tissue engineering.25
Here, we combined 3D printing technology to prepare an rGO composite hydrogel scaffold for loading SCs and BMSCs simultaneously for synergistic regeneration of bone and nerve. By changing the concentration of rGO, the compressive strength, rheological properties and porosity of the hydrogel can be well improved. The adhesion of two different stem cells on the 3D-printed hydrogel scaffolds was improved, and they were induced to differentiate into osteogenic and neurogenic cells. In vivo, the rGO hydrogel scaffolds achieved multicellular delivery and differentiation (Scheme 1). Scaffolding materials based on 3D printing technology can achieve rapid, precise and personalized applications. At the same time, the loading of various stem cells makes functionalized biomimetic bone materials provide a promising strategy.
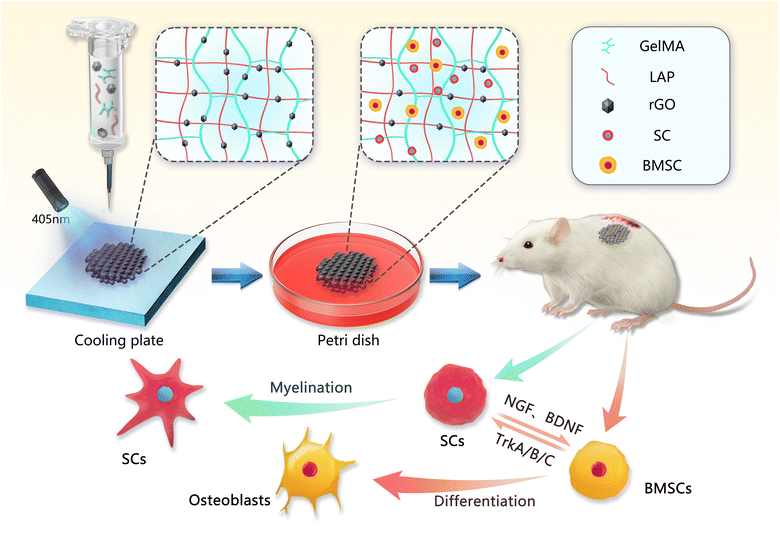 | ||
| Scheme 1 Schematic diagram of the 3D bioprinting process of the rGO composite hydrogel scaffold, and the subcutaneous ectopic osteogenesis model in the rat in vivo experiment. | ||
2. Materials and methods
2.1. Preparation of materials
Methacrylate anhydride gelatin (GelMA, 10w/v, Sunp Biotech, China) and photoinitiator (LAP, 0.25%w/v, Sunp Biotech, China) were dissolved in PBS and fully dissolved at 50 °C. Then rGO (0.03 mg ml−1, 0.05 mg ml−1, 0.1 mg ml−1, XF Nano, China) was added to the GelMA precursor solution. Cyclic magnetic stirring (30 min) and ultrasonic vibration (30 min) 3 times, and finally completely dissolved. Finally, four different concentrations of bioinks (10% GelMA, 0.03% rGO/GelMA, 0.05% rGO/GelMA and 0.1% rGO/GelMA) were prepared as described above.2.2. Scaffold fabrication
Before 3D printing, different concentrations of bioinks were transferred into cartridges. A bioprinter was used to make the scaffolds. The size of the scaffold is 15 × 15 × 1.6 mm. The temperature is 25 °C; the layer height is 0.2 mm; the line spacing is 1 mm; the printing speed is 3 mm3 s−1. The scaffolds were permanently cross-linked after 60 s cross-linking with 405 nm blue light on a cooling plate.2.3. Material characterization
| SR = (Wt − W0)/W0 × 100% |
W 0 and Wt are the initial weight and the swollen weight of the hydrogel, respectively. (n = 3)
| Degradation rate (%) = Wt/W0 × 100% |
2.4. In vitro cytocompatibility
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and the stents of different concentrations in each group were immersed in 10 ml of physiological saline for 24 h. Then add 0.2 ml of diluted blood to each group of normal saline, shake gently, and let stand at 37 °C for 60 min. After centrifugation at 1000 rpm for 5 min, the absorbance of the supernatant was measured at 545 nm. The hemolysis rate is calculated as follows:
5, and the stents of different concentrations in each group were immersed in 10 ml of physiological saline for 24 h. Then add 0.2 ml of diluted blood to each group of normal saline, shake gently, and let stand at 37 °C for 60 min. After centrifugation at 1000 rpm for 5 min, the absorbance of the supernatant was measured at 545 nm. The hemolysis rate is calculated as follows:| Hemolysis rate (%) = (Dt − Dnc)/(Dpc − Dnc) × 100% |
Scanning electron microscopy was also used to directly observe the morphology of cells on the scaffold surface. The cells seeded for 48 h were soaked in electron microscope fixative solution at 4 °C overnight. Then, the samples were dehydrated step by step with different concentrations of ethanol (30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, 100% for 10 minutes each) and then sprayed with gold for SEM detection.
2.5. In vitro osteogenic and neurogenic differentiation
2.6. In vivo experiments – subcutaneous heterotopic transplantation in rats
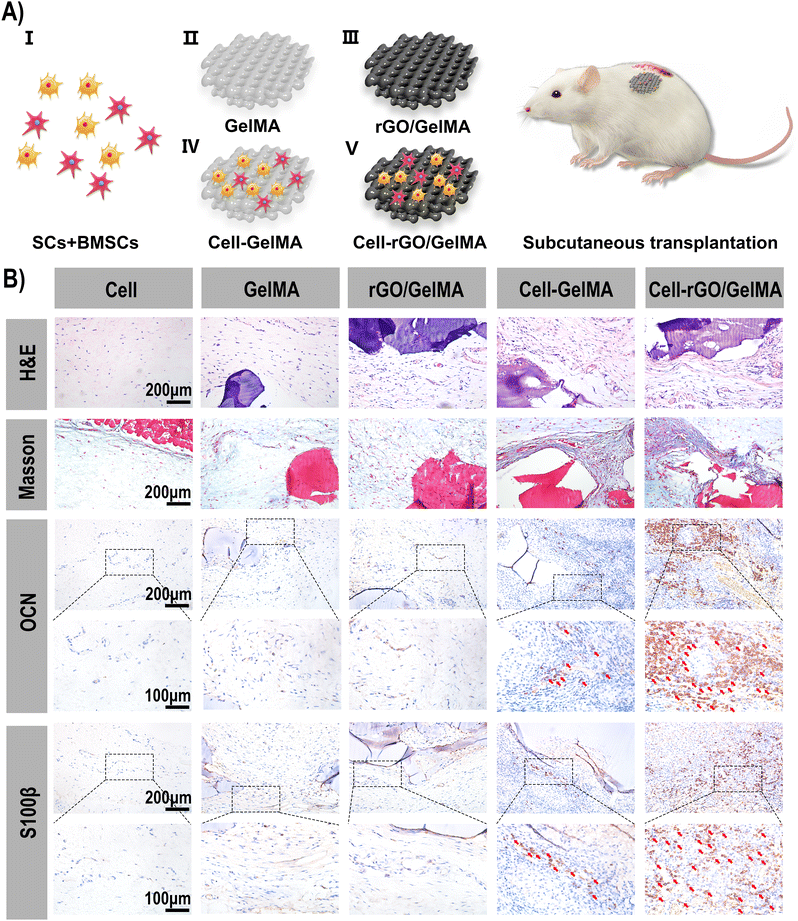
To evaluate the viability and function of the transplanted cells and the biological properties of the biomaterials, ectopic subcutaneous implantation was performed using a hydrogel scaffold loaded with two types of cells (rBMSCs and SCs). All experimental animal procedures were approved by Jilin University according to international standards for animal welfare (authorization number: KT202003217). Twenty SD rats (male, 12 weeks old, with an average weight about of 250 g) were randomly divided into 5 groups, including group I (BMSCs/SCs), group II (GelMA), group III (rGO/GelMA), group IV (Cell-loaded GelMA) and group V (Cell-loaded rGO/GelMA) (4 each). Cells were seeded on scaffolds and cultured for 7 days before surgery. Surgery is performed under anesthesia in a sterile environment. A 1.5 cm incision was made in the dorsal skin with a scalpel, the subcutaneous tissue was separated, and the hydrogel scaffold was transplanted under the skin and sutured tightly. At week 8 post-transplantation, whole regenerated tissue specimens were removed and fixed in 10% formalin. Specimens were dehydrated in graded ethanol and embedded in paraffin. Excised from the center of the regeneration site and stained with hematoxylin and eosin (H&E), Masson staining, immunohistochemical staining (s100β and OCN) and immunofluorescence (CD31, NGF, Runx2 and Col-1) according to standard protocols. All tissue sections were observed and analyzed under a light microscope/confocal microscope. The degree of inflammatory/positive cells in implant tissue sections in vivo were evaluated and observed under 5 areas with 1000 μm2 were selected from 3 replicate slides randomly, examined, and photographed through a light microscope/confocal microscope at 200 × magnification for the inflammatory/positive areas, and 400 × magnification to count the immune/positive cells.The degree of inflammatory cells in implant tissue sections and animal internal organs (heart, liver, spleen, lung and kidney) and the metabolism of graphene in vivo were evaluated and observed by H&E staining.
Masson staining. Cell nuclei stained with hematoxylin (3 min), ponceau red stained (5 min), phosphomolybdic acid treated (3 min), aniline blue stained (1 min), differentiated (1% glacial acetic acid for 1 min), dehydrated and mounted, and examined for new collagen fibers under a microscope.
Immunofluorescence (CD31, NGF, Runx2 and Col-1): Antigen retrieval (adding to 0.01 M citrate buffer and repairing by heating), permeabilization with 0.5% Triton X-100 at room temperature for 20 min), blocking (blocking with 2% BSA for 30 min), incubation with primary antibody (overnight at 4 °C)), incubated with secondary antibody (2 h at room temperature), stained cell nuclei (30 s at room temperature with DAPI), and mounted (anti-fade glycerol). The distribution and number of positive cells (red fluorescence/green fluorescence) were observed under confocal laser microscopy.
Immunohistochemistry (s100β and OCN). Antigen retrieval, blocking (incubate with 3% hydrogen peroxide at room temperature for 15 minutes), localization (determination of staining range with immunohistochemical pen), incubate primary antibody (overnight at 4 °C), incubate secondary antibody (incubate for 1 hour after rewarming at room temperature), color development (DAB chromogenic solution, stop staining after observing under the microscope until the color is suitable), hematoxylin counterstaining (3 min), dehydration and sealing. The distribution and number of positive cells (dark brown) were observed under a microscope.
2.7. Statistical analysis
Statistical analysis was performed using Prism 8.0. Data are presented as mean ± standard deviation. Each group of samples was repeated three times. A t-test or one-/two-way ANOVA was performed to determine statistical significance. A p-value less than 0.05 was considered statistically significant.3. Results and discussion
3.1. Physical properties of bioinks and 3D printed scaffolds
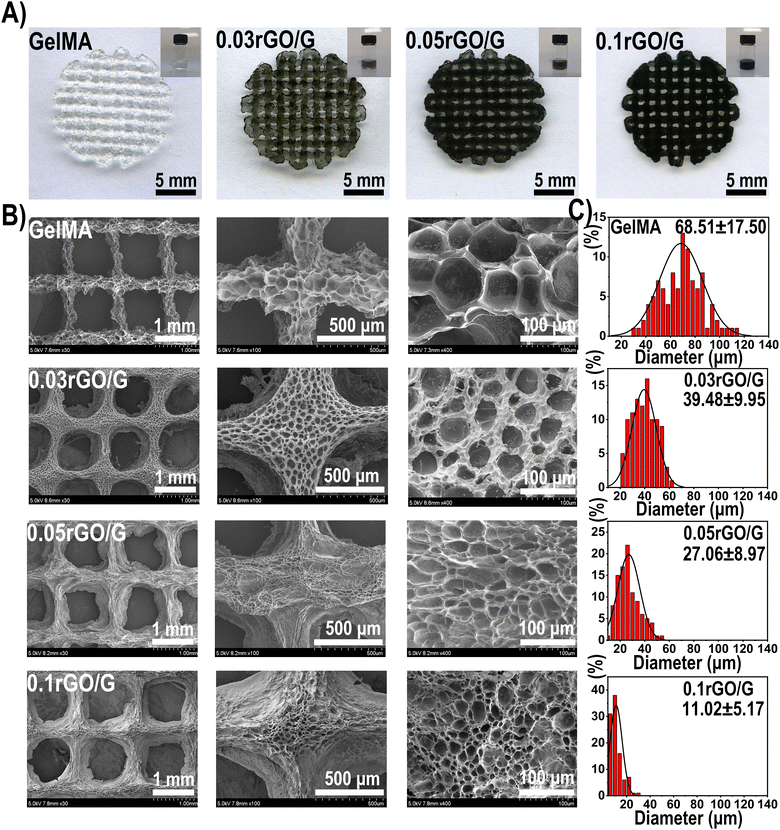
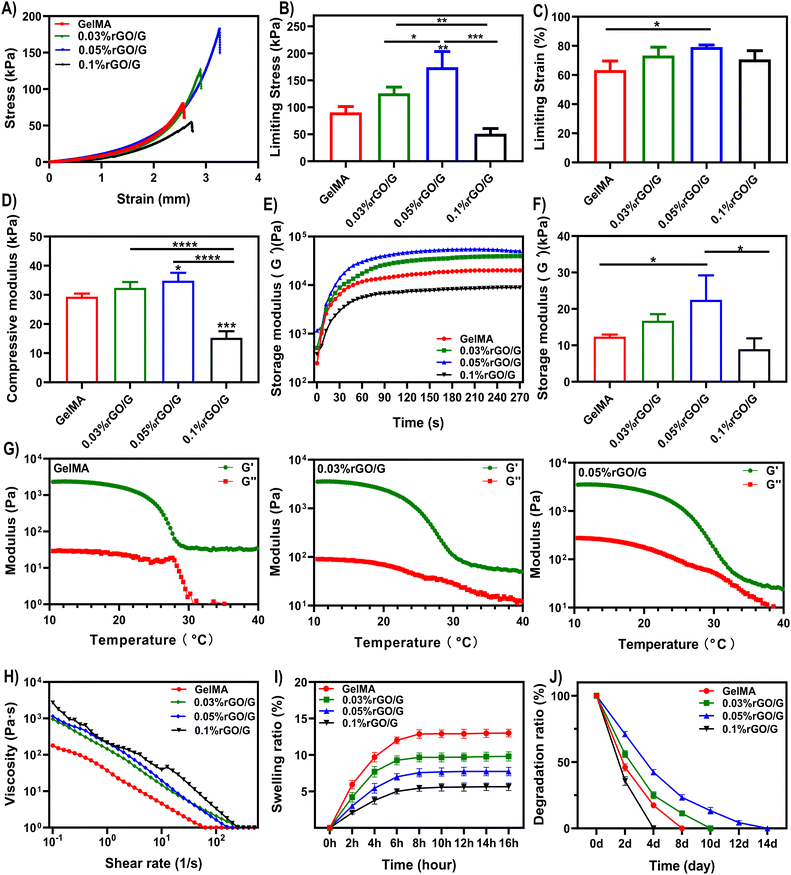
According to previous reports, reduced graphene oxide with hydrophobic properties is difficult to dissolve in an aqueous phase. However, rGO in GelMA precursor solution can be completely dissolved under ultrasonic vibration and magnetic stirring.26 With increasing rGO concentration in the hydrogel system (0.03%, 0.05% and 0.1%), the color of the composite hydrogel gradually deepened (Fig. 1A). The morphology of the lyophilized scaffolds were observed by SEM (Fig. 1B). All scaffolds have a regular grid structure, and the grid space is about 1 mm, which corresponds to the parameters set by 3D printing. Interestingly, we found that with the addition of rGO, the pore size of the scaffolds decreased significantly (GelMA 68.51 ± 17.50 μm; 0.03%rGO/GelMA 39.48 ± 9.95 μm; 0.05%rGO/GelMA 27.06 ± 8.97 μm; 0.1%rGO/GelMA 11.02 ± 5.17 μm) (Fig. 1C). The reason for the smaller pore size of the scaffold may be that the graphene nanosheets increase the density of crosslinks inside the gel.In the testing of mechanical properties, we found that the compressive strength of 0.03%rGO/GelMA group and 0.05%rGO/GelMA group (GelMA 90.52 kPa; 0.03%rGO/GelMA 125.90 kPa; 0.05%rGO/GelMA 174.26 kPa; 0.1%rGO/GelMA 51.01 kPa) and ultimate strain (GelMA 63.33%; 0.03%rGO/GelMA 73.33%; 0.05%rGO/GelMA 79.17%; 0.1%rGO/GelMA 70.67%) were significantly better (Fig. 2A–D) (Table S2, ESI†). Interestingly, we observed lower mechanical properties in the 0.1%rGO/GelMA group instead. As the concentration of rGO increases, due to its strong light-shielding ability, it is difficult to achieve complete photo-crosslinking in the interior of the hydrogel, resulting in insufficient mechanical properties.26 The effect of photocuring time on the storage modulus of the hydrogel was further explored. We found that the storage modulus of the hydrogels in the rGO/GelMA group (0.03%rGO 39.69 kPa; 0.05%rGO 54.84 kPa) was significantly higher than that of the pure GelMA group (20.11 kPa) after 405 nm light curing for 60 s (Fig. 2E and F) (Table S3, ESI†). Notably, a high concentration of rGO resulted in a decrease in the storage modulus (0.05%rGO: 88.88 kPa) of the hydrogel, which was even much lower than that of the pure GelMA group. This is consistent with the previous compression test results.
Rheology describes the deformation and flow properties of materials under the influence of external forces. And it is the physicochemical parameter that has the greatest impact on the printability of hydrogels.27 In extrusion printing, the shear-thinning properties simplify printing and affect the initial shape of the bioscaffold, and the viscosity decreases during the extrusion phase, where the shear force increases significantly. After extrusion, the shear rate decreases and the viscosity increases accordingly, helping to maintain the original shape of the print.28 The rGO/GelMA composite bioink showed higher viscosity than other curves (5974.09 Pa·s) at the low shear rate, while at a high shear rate, the viscosity rapidly decreased to almost the same level as the other curves (0.12 Pa·s) (Fig. 2H). All groups of bioinks have excellent shear thinning properties. Due to the shear-thinning behavior, all concentrations of rGO hydrogels can pass through the 0.2 mm printer needle and 3D print smoothly. In addition to good shear thinning properties and suitable viscosity, an ideal printable bioink should also have excellent temperature sensitivity.29,30 In our results, the storage moduli (G′) of all inks at low temperature were much larger than the loss moduli (G′′) (Fig. 2G). For freshly printed scaffolds, temporary physical cross-linking at low temperature is crucial to maintain the accuracy of the printed morphology.31 The swelling properties of hydrogels have a significant impact on maintaining the initial shape and mechanical stability of 3D printed structures. Excessive swelling ratios may cause the structure to swell and become brittle.32 The results show that with the increase of the concentration, the swelling ratio is greatly reduced, and the rGO-added scaffold retains the original morphology to the greatest extent (Fig. 2I). The rapid degradation of hydrogels used in bone tissue engineering has always been a difficulty worth improving.33,34 Relevant literature indicates that the uncontrolled release of loaded bioactive growth factors and stem cells is directly related to the rapid degradation of hydrogels. This is difficult to match with a bone repair time of more than 3 months.35 Collagenase type II [matrix metalloproteinase 8 (MMP-8)] is the major proteolytic enzyme involved in bone wound healing.36 GelMA is enzymatically degraded, as gelatin contains sequences that are recognized by collagenases.37 To assess degradation, the hydrogels were immersed in collagenase type II solution (1.25 U ml−1). Except for 0.1%rGO/GelMA, which could not be fully photocrosslinked, the degradation rate of other groups was significantly lower than that of the GelMA group due to changes in pore structure and scaffold crosslinking density (Fig. 2J). In summary, during the preparation process of 3D printing, we found that the hydrogel precursor was easier to pass through the 0.2 mm nozzle smoothly, as the concentration of rGO increased. In rheological experiments, hydrogels containing 0.05% and 0.1% rGO/GelMA performed better than the control group. Considering the physical properties and the differentiation potential of cells, we chose a concentration of 0.05% rGO/GelMA as the experimental group for our subsequent in vivo and in vitro experiments.
3.2. In vitro cytocompatibility
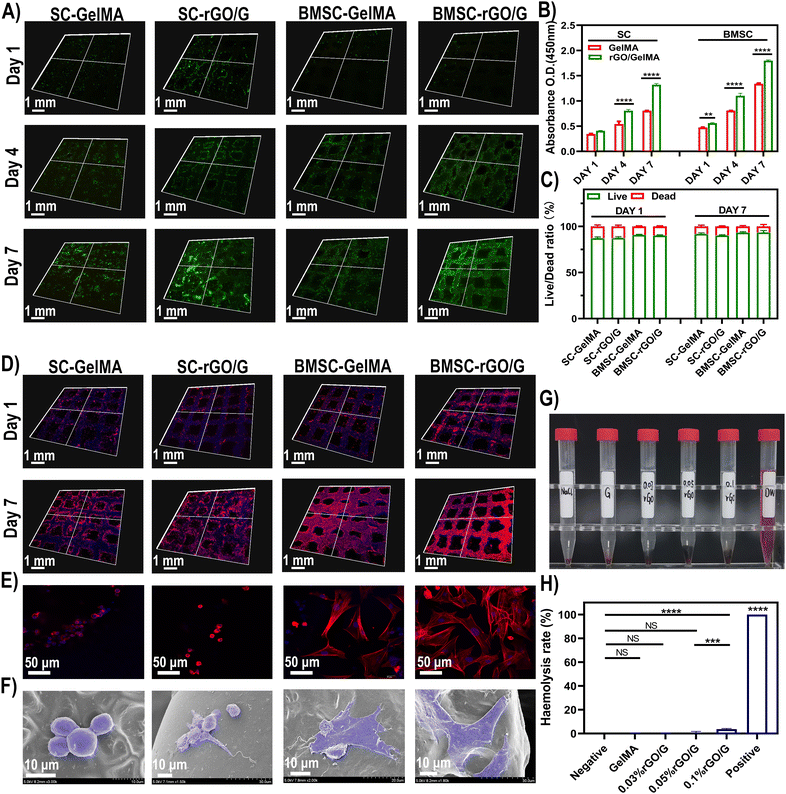
Stem cells are expected to be effectively utilized in damaged tissues because of their dual potentials of proliferation and differentiation. But how to achieve long-term survival and retention of transplanted stem cells is a big problem.38,39 To study the cell viability of rBMSCs in the 3D printed scaffolds, live (green)/dead (red) staining was performed in culture medium for 1 day, 4 days and 7 days, respectively. On the 7th day, the proportion of living cells in each group was more than 85%. At the same time, the fluorescence intensity of SCs and BMSCs loaded in rGO/GelMA is higher than that of the GelMA group. It is worth noting that the live/dead staining already showed a similar trend on day 1 (Fig. 3A, C). The resulting analysis of CCK-8 detection showed a similar tendency to that of live/dead staining (Fig. 3B), suggesting that the rGO in the scaffolds promotes cell proliferation. These results indicate that the addition of rGO improves the cell adhesion ability of the hydrogel.40 Further the morphology of BMSCs and SCs on the scaffold was detected by cytoskeletal protein F-actin (red)/DAPI (blue) staining at 1 and 7 days. To our surprise, BMSCs on the rGO/GelMA exhibited a more stretched shape compared with the G group in the early stage (day 1), which indicates the excellent cytocompatibility of rGO in the scaffold. A more prominent tendency with the larger spreading area (Fig. 3D and E) and faster proliferation rate (Fig. 3B) was shown in the rGO/GelMA group for both BMSCs and SCs. We further observed the morphology of the cells by SEM after cell seeding for 48 hours, both SC and BMSCs exhibit a smooth cell membrane, and the cell on the rGO/GelMA spread better compared with the GelMA group (Fig. 3F). According to previous reports, rGO hydrogels with electrical conductivity can support cell viscosity and migration by mediating cell bioelectricity signals.41 At the same time, our results show that after the addition of rGO, hydrogels have improved cell proliferation and cell adhesion. The reason is that rGO retains some oxygen-containing groups that can adhere to serum proteins and endow the scaffold with better cell attachment ability.42 Promoting the adhesion of stem cells by changing the surface morphology or functional groups of biomaterials is a common method for stem cell transplantation. For example the RGD sequence consisting of arginine, glycine and aspartic acid mediates cell attachment via fibronectin.43 Although RGD can improve the adhesion, it is easy to disappear in the physiological environment. The controlled release design can alleviate its rapid dissolution in the surrounding environment.44 Polydopamine (PDA) can also improve cell adhesion in tissue engineering. PDA contains many hydrophilic functional groups, such as hydrophilic amino groups and hydroxyl groups, which can provide hydrophilic groups on the hydrophobic surface, thereby improving the hydrophilicity of the surface of biomaterials, thereby enhancing the ability of cell adhesion.45 Natural polymers such as silk fibroin with better biocompatibility are also commonly used to improve cell migration and adhesion.46 As a carbon allotrope, reduced graphene oxide has a two-dimensional and honeycomb structure with strong π bonds and large surface area. Based on the above structure, rGO can improve the cell adhesion and conductivity of the material surface. Among them, the conductivity is closely related to the number of layers of graphene. In fact, conductive materials usually rely on external stimuli from the environment to improve the growth, attachment, and differentiation of nerve cells. And the three-dimensional preparation of graphene nanocomposite channels can also significantly up-regulate the biological activity of neural stem cells.47Additionally, hemocompatibility testing was performed to test the effects of different scaffolds on red blood cells (RBCs), platelets, thrombosis, and coagulation, to determine whether the biomaterial is suitable for animal studies or clinical trials.48 Through the hemolysis experiment, the hemolysis rate slightly increased with the increase of rGO concentration (Fig. 3G and H). However, according to the standard hemolytic index [ASTM F756-00(2000)], all groups of materials can be defined as non-hemolytic (<2%).
3.3. In vitro osteogenic and neurogenic differentiation
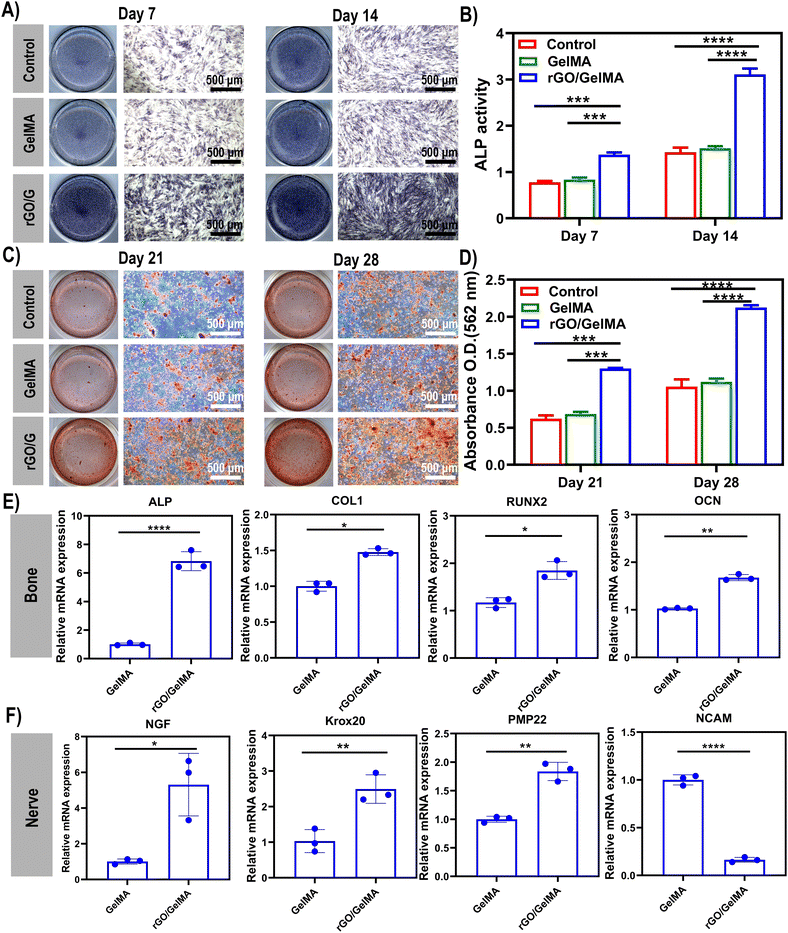
Bone scaffolds contact directly with the cells and provide the extracellular matrix (ECM) for cell adhesion, proliferation, and differentiation.49–51 rGO has been reported to increase the stability of adsorbed proteins, such as bone morphogenetic protein 2 (BMP-2), and eventually improve cell adhesion, proliferation, and osteogenic differentiation.52 The osteoblast phenotype is specifically obtained in two stages. In the first stage, the cells proliferate and the matrix matures. During 10–15 days of cell proliferation and matrix maturation, specific proteins associated with the osteocyte phenotype, such as ALP, can be detected. In the second stage of osteoblastic phenotype acquisition, matrix mineralization, the production of late bone markers and the production of calcium nodules after up to 25–30 days.53 To investigate the osteogenic differentiation ability of rGO decorated GelMA scaffolds, the ALP activity and calcium deposition were tested. In our results, ALP staining was darker on days 7 and 14 in the rGO/GelMA group compared to the control and GelMA groups, representing high expression in the osteogenic direction. Notably, there was no difference in ALP staining between the GelMA group and the control group (Fig. 4A). In the activity experiment of ALP, the absorbance of the rGO group was also significantly higher than that of the other two groups on day 7 and 14 (Fig. 4B). BMSCs in each group formed orange-red calcium nodules on the 21st and 28th day. Calcium nodules in the rGO/GelMA group produced more calcium nodules compared to the control and pure GelMA groups (Fig. 4C). Chlorohexadecylpyridine was used to dissolve calcium nodules for further quantitative analysis (Fig. 4D). To further explore the osteogenic differentiation ability, we detected the relative mRNA expression levels of RUNX2, ALP, Col-1 and OCN by qRT-PCR at the gene level. The expression of osteogenesis-related genes was significantly enhanced in the rGO/GelMA group after 14 days of osteogenic induction (Fig. 4E). As far as we know, graphene derivatives have adsorption effects on dexamethasone and β-glycerophosphate.54 As a member of glucocorticoid, Dex can participate in the expression of some signal molecules in the process of osteogenesis.55 Osteoblasts cultured on graphene exhibited higher levels of mineralization and significantly upregulated osteogenic genes and proteins, including Runx2, Col-1 and OCN.56Myelination of SC is a prerequisite during nerve regeneration.57 Among them, neuronal cell adhesion molecule (NCAM) is an indicator of SCs myelination. It is only expressed in immature SCs and is reduced in myelination of SCs. In contrast, the expression levels of NGF, PMP22, and Krox20 were increased in myelinated SCs.58 As shown in Fig. 4F, NCAM gene expression was significantly decreased on rGO/GelMA scaffolds compared to GelMA scaffolds, while NGF, PMP22 and Krox20 gene expression levels were significantly increased (p < 0.05). The results indicated that the presence of rGO promoted SCs myelination.
3.4. In vivo experiments – subcutaneous heterotopic transplantation in rats
At present, ideal bone inducing materials loaded with stem cells should have three key characteristics, including successful transplantation; promote differentiation and heterotopic osteogenesis.59 To evaluate the viability and function of transplanted cells and the biological characteristics of biomaterials, scaffolds loaded with two types of cells (BMSCs and SCs) were transplanted ectopically subcutaneously (Fig. S1, ESI†). According to previous research reports, BMSCs co-cultured with SCs showed higher proliferation ability and osteogenic differentiation potential. Its related mechanism of action is mainly realized through the interaction between NGF, BDNF, TrkA, S100 and other cell signaling molecules.1 As shown, the in vivo experiments were divided into 5 groups (Fig. 5A). In group I, the groups in which cells were directly injected into the subcutaneous tissue (BMSCs and SCs groups) did not show new bone and fewer blood vessel formation due to low cell retention. For the material-only groups (GelMA and rGO/GelMA), the scaffolds resulted in the formation of an ordered arrangement of tissues without obvious inflammatory response, confirming the excellent biocompatibility of GelMA and rGO (Fig. S2, ESI†). Compared with the scaffold group not loaded with cells, the scaffolds loaded with BMSCs and SCs were surrounded by a large number of collagen fibers, which were stained blue under Masson staining indicating the excellent osteogenic ability of the scaffolds (Fig. 5B). Collagen fibers (97%) are the most abundant protein in the bone extracellular matrix. In the process of osteogenesis, hydroxyapatite crystals are mainly arranged in a periodic and staggered manner along collagen fibers, thereby realizing bone repair and regeneration.60 Then we further used immunofluorescence(IF) staining to observe the trend or situation of ectopic bone formation in vivo. The key to determining the biomaterial for stem cell transplantation is the survival rate and osteogenic differentiation of the transplanted cells. When the biomaterial is transplanted into the body, the stem cells can successfully complete the proliferation and differentiation into osteoblasts. During this period, osteoinductive materials serve space for the growth of BMSCs, absorb endogenous growth factors, and promote the proliferation and osteogenic differentiation of MSCs.61 Here, immunofluorescence staining showed that 8 weeks after implantation in the Cell-rGO/GelMA group, the transplanted stem cells clearly expressed NGF, Runx2 and Col-1 (markers of osteoblasts/Schwann cells) in the implanted area (Fig. 6). At the same time, the vascularized CD31 molecule also showed high expression(Fig. S3, ESI†). The lower concentration of rGO can enhance the migration of endothelial cells and show the characteristics of promoting angiogenesis.62 New blood vessels form into regenerated peripheral nerves to provide nutritional support. Meanwhile, graphene oxide derivatives have also been shown to accelerate osteogenic differentiation by enhancing the expression of nestin, β-tubulin III and MAP2 to promote neuronal differentiation, adhesion, and proliferation. Activation of the mechanosensitive integrin-FAK axis also plays an important role in osteogenic induction.63 Immunohistochemical staining was performed to further assess neuralized bone formation (Fig. 5B). The expression of S100β and OCN was weaker in the cell-only group and the material-only group. In contrast, strong expression of OCN was found in the BMSCs and SCs-loaded scaffold groups, which was attributed to the transplanted BMSCs. At the same time, s100β was also highly expressed, which was attributed to the transplanted SCs. The synergy of BMSCs and SCs is thought to make a significant contribution to regenerating neuralized osteoid tissue. Overall, the rGO-added hydrogel scaffolds showed good cell adhesion compared to the pure GelMA scaffolds. In terms of in vivo biosafety, we extracted the heart, liver, spleen, lung and kidney organs of experimental animals for tissue sections, and found no obvious abnormalities or residues of graphene derivatives (Fig. S4, ESI†). Taken together, compared with GelMA, rGO/GelMA has a stronger ability to promote the osteogenic differentiation of BMSCs, which is consistent with our previous in vitro results.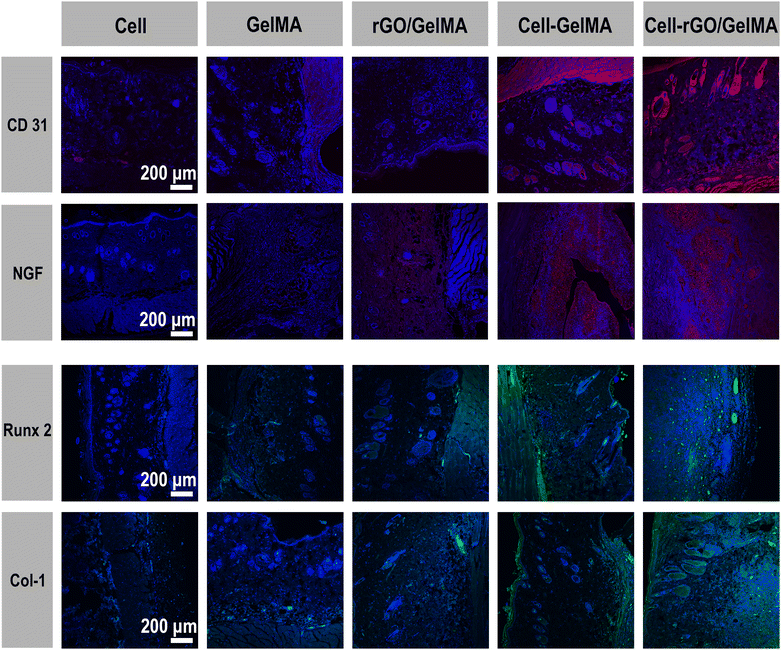 | ||
| Fig. 6 Immunofluorescence staining of tissue sections (CD31, NGF, Runx2 and Col-1) after 8 weeks of in vivo experiments. | ||
4. Conclusion
In conclusion, we fabricated rGO/GelMA scaffolds by applying 3D printing technology. The scaffolds with suitable pore sizes showed excellent biocompatibility and proliferation ability. The 0.05% rGO/GelMA scaffold has an excellent pore size of 26.35 ± 0.5 μm and good mechanical properties, which can promote the adhesion of BMSCs and SCs, and maintain high dryness and continuous proliferation within 7 days. In vitro experiments, alizarin red and alkaline phosphatase staining demonstrated its excellent osteogenic induction ability. In rats, 0.05% rGO/GelMA scaffolds loaded with both BMSCs and SCs achieved osteogenesis and neurogenesis 2 months after transplantation. This is attributed to the high adhesion capacity of the scaffold material and the potential for osteogenic/neural differentiation. Due to their flexibility as bioinks in 3D printing, GelMA hydrogels mixed with rGO are expected to have broad applications in multicellular delivery and functional tissue regeneration.Author contributions
Xinwei Zhang: writing – original draft, conceptualization, methodology, data curation. Hao Zhang: visualization, editing. Yi Zhang: data curation, investigation. Huimin Huangfu: data curation, investigation. Yixin Yang: software, validation. Qiuyue Qin: software. Yidi Zhang: conceptualization, methodology, formal analysis, project administration. Yanmin Zhou: conceptualization, project administration, supervision, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 82071152), the Development and Reform Commission of Jilin Province (Grant No. 2021C042-1), and the Jilin Provincial Science and Technology Department (Grant No. 20200201302JC).References
- R. L. Meng Zhang, X. Wang, J. Xue, C. Deng, C. Feng, H. Zhuang, J. Ma, C. Qin, L. Wan, J. Chang and C. Wu, Sci. Adv., 2020, 6, eaaz6725 CrossRef PubMed.
- M. Qu, C. Wang, X. Zhou, A. Libanori, X. Jiang, W. Xu, S. Zhu, Q. Chen, W. Sun and A. Khademhosseini, Adv. Healthcare Mater., 2021, 10, e2001986 CrossRef PubMed.
- T. Y. L. Alessandra Marrella and D. H. Lee, Mater. Today, 2017, 10, 1016 Search PubMed.
- F. X. Li, F. Xu, X. Lin, F. Wu, J. Y. Zhong, Y. Wang, B. Guo, M. H. Zheng, S. K. Shan and L. Q. Yuan, Front. Endocrinol., 2020, 11, 77 CrossRef PubMed.
- S. Grässel, Grässel Arthritis Res. Ther., 2014, 6, 485 CrossRef PubMed.
- M. A. Fernandez-Yague, S. A. Abbah, L. McNamara, D. I. Zeugolis, A. Pandit and M. J. Biggs, Adv. Drug Delivery Rev., 2015, 84, 1–29 CrossRef CAS PubMed.
- J. Cao, S. Zhang, A. Gupta, Z. Du, D. Lei, L. Wang and X. Wang, Int. J. Med. Sci., 2019, 16, 831–837 CrossRef CAS PubMed.
- R. E. Tomlinson, B. A. Christiansen, A. A. Giannone and D. C. Genetos, Front. Endocrinol., 2020, 11, 646 CrossRef PubMed.
- C. Y. Yu and P. V. Abbott, Aust. Dent. J., 2016, 61, 39–58 CrossRef PubMed.
- M. Zhang, C. Qin, Y. Wang, X. Hu, J. Ma, H. Zhuang, J. Xue, L. Wan, J. Chang, W. Zou and C. Wu, Addit. Manuf., 2022, 54, 102721 CAS.
- X. Xue, Y. Hu, Y. Deng and J. Su, Adv. Funct. Mater., 2021, 31, 2009432 CrossRef CAS.
- C. C.-S. Claudio Bordignon and M. P. Colombo, Haematologica, 1999, 84, 1110–1149 Search PubMed.
- L. S. Neves, M. T. Rodrigues, R. L. Reis and M. E. Gomes, Expert Rev. Precis. Med. Drug Dev., 2016, 1, 93–108 CrossRef.
- H. Liu, Q. Zhang, S. Wang, W. Weng, Y. Jing and J. Su, Bioact. Mater., 2022, 14, 169–181 CrossRef CAS PubMed.
- Y. Han, X. Li, Y. Zhang, Y. Han, F. Chang and J. Ding, Cells, 2019, 8, 886 CrossRef CAS PubMed.
- I. R. Avanzi, J. R. Parisi, A. Souza, M. A. Cruz, C. C. S. Martignago, D. A. Ribeiro, A. R. C. Braga and A. C. Renno, J. Biomed. Mater. Res., Part B, 2022, 1–17 Search PubMed.
- K. M. Wong, E. Babetto and B. Beirowski, Neural Regener. Res., 2017, 12, 518–524 CrossRef PubMed.
- R. Li, D. Li, C. Wu, L. Ye, Y. Wu, Y. Yuan, S. Yang, L. Xie, Y. Mao, T. Jiang, Y. Li, J. Wang, H. Zhang, X. Li and J. Xiao, Theranostics, 2020, 10, 1649–1677 CrossRef CAS PubMed.
- S. Shimizu, M. Kitada, H. Ishikawa, Y. Itokazu, S. Wakao and M. Dezawa, Biochem. Biophys. Res. Commun., 2007, 359, 915–920 CrossRef CAS PubMed.
- J. Wang, W. Zheng, L. Chen, T. Zhu, W. Shen, C. Fan, H. Wang and X. Mo, ACS Biomater. Sci. Eng., 2019, 5, 2444–2456 CrossRef CAS PubMed.
- K. Zhihui and D. Min, ACS Biomater. Sci. Eng., 2022, 8, 2849–2857 CrossRef PubMed.
- C. Liao, Y. Li and S. C. Tjong, Int. J. Mol. Sci., 2018, 19, 3564 CrossRef PubMed.
- X. P. Li, K. Y. Qu, B. Zhou, F. Zhang, Y. Y. Wang, O. D. Abodunrin, Z. Zhu and N. P. Huang, Colloids Surf., B, 2021, 205, 111844 CrossRef CAS PubMed.
- K. Zhou, P. Yu, X. Shi, T. Ling, W. Zeng, A. Chen, W. Yang and Z. Zhou, ACS Nano, 2019, 13, 9595–9606 CrossRef CAS PubMed.
- Z. Huang, M. Sun, Y. Li, Z. Guo and H. Li, J. Mater. Chem. B, 2021, 9, 2656–2665 RSC.
- S. R. Shin, C. Zihlmann, M. Akbari, P. Assawes, L. Cheung, K. Zhang, V. Manoharan, Y. S. Zhang, M. Yuksekkaya, K. T. Wan, M. Nikkhah, M. R. Dokmeci, X. S. Tang and A. Khademhosseini, Small, 2016, 12, 3677–3689 CrossRef CAS PubMed.
- H. Li, C. Tan and L. Li, Mater. Des., 2018, 159, 20–38 CrossRef CAS.
- A. Schwab, R. Levato, M. D'Este, S. Piluso, D. Eglin and J. Malda, Chem. Rev., 2020, 120, 11028–11055 CrossRef CAS PubMed.
- Z. Chen, D. Zhao, B. Liu, G. Nian, X. Li, J. Yin, S. Qu and W. Yang, Adv. Funct. Mater., 2019, 29, 1900971 CrossRef.
- N. Paxton, W. Smolan, T. Bock, F. Melchels, J. Groll and T. Jungst, Biofabrication, 2017, 9, 044107 CrossRef PubMed.
- X. Cui, J. Li, Y. Hartanto, M. Durham, J. Tang, H. Zhang, G. Hooper, K. Lim and T. Woodfield, Adv. Healthcare Mater., 2020, 9, e1901648 CrossRef PubMed.
- C. K. Kuo and P. X. Ma, J. Biomed. Mater. Res., Part A, 2008, 84, 899–907 CrossRef PubMed.
- L. Dong, Z. Bu, Y. Xiong, H. Zhang, J. Fang, H. Hu, Z. Liu and X. Li, Int. J. Biol. Macromol., 2021, 188, 72–81 CrossRef CAS PubMed.
- X. Xue, Y. Hu, S. Wang, X. Chen, Y. Jiang and J. Su, Bioact. Mater., 2022, 12, 327–339 CrossRef CAS PubMed.
- X. Bai, M. Gao, S. Syed, J. Zhuang, X. Xu and X. Q. Zhang, Bioact. Mater., 2018, 3, 401–417 CrossRef PubMed.
- A. Gutierrez-Fernandez, M. Inada, M. Balbin, A. Fueyo, A. S. Pitiot, A. Astudillo, K. Hirose, M. Hirata, S. D. Shapiro, A. Noel, Z. Werb, S. M. Krane, C. Lopez-Otin and X. S. Puente, FASEB J., 2007, 21, 2580–2591 CrossRef CAS PubMed.
- J. Liu, M. Qu, C. Wang, Y. Xue, H. Huang, Q. Chen, W. Sun, X. Zhou, G. Xu and X. Jiang, Small, 2022, 18, e2106172 CrossRef PubMed.
- M. D. Ranieri Cancedda, G. Bianchi, A. Derubeis and R. Quarto, Stem Cells, 2003, 21, 610–619 CrossRef PubMed.
- B. N. Kharbikar, P. Mohindra and T. A. Desai, Cell Stem Cell, 2022, 29, 692–721 CrossRef CAS PubMed.
- J. Y. Ng, S. Obuobi, M. L. Chua, C. Zhang, S. Hong, Y. Kumar, R. Gokhale and P. L. R. Ee, Carbohydr. Polym., 2020, 241, 116345 CrossRef CAS PubMed.
- Y. Wang, Y. Xiao, G. Gao, J. Chen, R. Hou, Q. Wang, L. Liu and J. Fu, J. Mater. Chem. B, 2017, 5, 511–516 RSC.
- W. Guo, S. Wang, X. Yu, J. Qiu, J. Li, W. Tang, Z. Li, X. Mou, H. Liu and Z. Wang, Nanoscale, 2016, 8, 1897–1904 RSC.
- S. E. D’Souza, Elsevier Science Publisher, 1991, 91, pp. 0376–5067.
- Y. Dong, P. Li, C.-B. Chen, Z.-H. Wang, P. Ma and G.-Q. Chen, Biomaterials, 2010, 31, 8921–8930 CrossRef CAS PubMed.
- J. Yan, R. Wu, S. Liao, M. Jiang and Y. Qian, Front. Bioeng. Biotechnol., 2020, 8, 590998 CrossRef PubMed.
- L. Zhang, L. Xu, G. Li and Y. Yang, Colloids Surf., B, 2019, 173, 689–697 CrossRef CAS PubMed.
- Y. Qian, X. Zhao, Q. Han, W. Chen, H. Li and W. Yuan, Nat. Commun., 2018, 9, 323 CrossRef PubMed.
- S. Hosseinpour, L. J. Walsh and C. Xu, J. Mater. Chem. B, 2020, 8, 9863–9876 RSC.
- X. Ye, Z. He, Y. Liu, X. Liu, R. He, G. Deng, Z. Peng, J. Liu, Z. Luo, X. He, X. Wang, J. Wu, X. Huang, J. Zhang and C. Wang, Int. J. Mol. Sci., 2022, 23, 9722 CrossRef CAS PubMed.
- Y. Zhang, T. Lin, H. Meng, X. Wang, H. Peng, G. Liu, S. Wei, Q. Lu, Y. Wang, A. Wang, W. Xu, H. Shao and J. Peng, J. Orthop. Transl., 2022, 33, 13–23 Search PubMed.
- D. Gan, Y. Jiang, Y. Hu, X. Wang, Q. Wang, K. Wang, C. Xie, L. Han and X. Lu, J. Orthop. Transl., 2022, 33, 120–131 Search PubMed.
- A. Raslan, L. Saenz Del Burgo, J. Ciriza and J. L. Pedraz, Int. J. Pharm., 2020, 580, 119226 CrossRef CAS PubMed.
- J. An, H. Yang, Q. Zhang, C. Liu, J. Zhao, L. Zhang and B. Chen, Life Sci., 2016, 147, 46–58 CrossRef CAS PubMed.
- W. C. Lee, C. H. Y. X. Lim, H. Shi, L. A. L. Tang, Y. Wang, C. T. Lim and K. P. Loh, ACS Nano, 2022, 5, 7334–7341 CrossRef PubMed.
- P. B. A. Scutt and M. Brautigam, Calcif. Tissue Int., 1996, 59, 154–162 CrossRef PubMed.
- D. Mohammadrezaei, H. Golzar, M. Rezai Rad, M. Omidi, H. Rashedi, F. Yazdian, A. Khojasteh and L. Tayebi, J. Biomed. Mater. Res., Part A, 2018, 106, 2284–2343 CrossRef CAS PubMed.
- J. A. Pereira, F. Lebrun-Julien and U. Suter, Trends Neurosci., 2012, 35, 123–134 CrossRef CAS PubMed.
- G. Nocera and C. Jacob, Cell. Mol. Life Sci., 2020, 77, 3977–3989 CrossRef CAS PubMed.
- Q. Zhao, M. Shi, C. Yin, Z. Zhao, J. Zhang, J. Wang, K. Shen, L. Zhang, H. Tang, Y. Xiao and Y. Zhang, Nano-Micro Lett., 2020, 13, 28 CrossRef PubMed.
- Y. Liu, D. Luo and T. Wang, Small, 2016, 12, 4611–4632 CrossRef CAS PubMed.
- R. J. Miron and Y. F. Zhang, J. Dent. Res., 2012, 736–744 CrossRef CAS PubMed.
- S. Mukherjee, P. Sriram, A. K. Barui, S. K. Nethi, V. Veeriah, S. Chatterjee, K. I. Suresh and C. R. Patra, Adv. Healthcare Mater., 2015, 1722–1732 CrossRef CAS PubMed.
- X. Yao, Z. Yan, X. Wang, H. Jiang, Y. Qian and C. Fan, Regener. Biomater., 2021, 1–9 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb01979e |
| This journal is © The Royal Society of Chemistry 2023 |