Open Access Article
Open Access ArticleDevelopment of liquid-crystalline smectic nanoporous membranes for the removal of SARS-CoV-2 and waterborne viruses†
Takeshi
Sakamoto
 *a,
Kazuhiro
Asakura
a,
Naru
Kang
a,
Riki
Kato
*a,
Kazuhiro
Asakura
a,
Naru
Kang
a,
Riki
Kato
 a,
Miaomiao
Liu
b,
Tsuyoshi
Hayashi
c,
Hiroyuki
Katayama
a,
Miaomiao
Liu
b,
Tsuyoshi
Hayashi
c,
Hiroyuki
Katayama
 *bd and
Takashi
Kato
*bd and
Takashi
Kato
 *ade
*ade
aDepartment of Chemistry and Biotechnology, School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: t_saka@chembio.t.u-tokyo.ac.jp; kato@chiral.t.u-tokyo.ac.jp
bDepartment of Urban Engineering, School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: katayama@env.t.u-tokyo.ac.jp
cDepartment of Virology II, National Institute of Infectious Diseases, Musashi-Murayama, Tokyo 208-0011, Japan
dResearch Center for Water Environment Technology, School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo 113-8656, Japan
eResearch Initiative for Supra-Materials, Shinshu University, Wakasato, Nagano 380-8553, Japan
First published on 24th August 2023
Abstract
Obtaining safe and affordable water free from bio-contaminants is a critical global issue. Filtration using polymer membranes with nanopores is a significant method for water purification. Here, we demonstrate the fabrication of water-treatment membranes with ordered nanochannels, exhibiting significant virus removal properties, by fixing the assembled structures of ionic liquid-crystalline molecules via photopolymerization. Nanostructured water-permeable membranes are prepared from ionic liquid-crystalline smectic compounds composed of a rod-shaped rigid core to form two-dimensional nanochannels. The removal of viruses, including inactivated SARS-CoV-2, in a virus cocktail solution is investigated. Tuning of the smectic assembled structures is discussed from the perspective of self-assembled molecular structures. In addition, the effects of the ionic channel morphology on water permeability are examined.
1 Introduction
Ensuring safe and affordable water supply is currently an important global issue.1–6 Water-permeable subnano-, nano- and microporous membranes are essential components of filtration systems tuned to remove hazardous targets.2–14 Various polymers, such as aromatic polyamides,15–17 cellulose esters and polysulfones, have been employed to fabricate water-treatment membranes.2–5 In addition, various materials with nano-ordered channels, such as block copolymers,18–20 carbon materials21–23 and liquid crystals,9–12 have attracted attention as new separation membrane materials.7–12Among the water-permeable membranes, reverse osmosis (RO) and nanofiltration (NF) membranes have been employed to remove small molecules and ions from water.2–7 The effective pore sizes of RO membranes are on the sub-nanometre scale, whereas those of NF membranes range from sub-nanometres to several nanometres.3,7 Currently, commonly employed RO and NF membranes are composed of crosslinked aromatic polyamides15–17 with nanopores formed during polymerization. However, obtaining well-ordered pores and pinhole-free surfaces by conventional processing of membranes is still challenging.13,24 Although current RO systems achieve virus rejection higher than 4 and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10, some degree of virus advection can occur.25,26 Novel membranes have been intensively studied for higher virus removal efficiencies.19,23,27–31
log10, some degree of virus advection can occur.25,26 Novel membranes have been intensively studied for higher virus removal efficiencies.19,23,27–31
Our intension was to develop highly efficient water-permeable membranes based on liquid-crystalline (LC) molecular order. Liquid crystals are soft materials capable of self-assembly into ordered nanometre-scale structures.10–12,28–50 The self-organization of molecules with block structures results in the formation of one-, two- and three-dimensional (1D, 2D and 3D) nanochannels depending on the molecular structures and intermolecular interactions (Fig. 1a). The nanochannels of aligned functional groups exhibit specific properties due to ionic interactions.10,11,51–54
Fixing the ordered structures of low-molecular-weight LC compounds via in situ photopolymerization is a reliable method for preparing polymer films with nano-ordered structures.10–12,28–31,39–50 Because small molecules are more fluid than polymers, pinhole-free surfaces and high ordering are possible. For membranes with a distribution of pore sizes, the efficiency of virus rejection is low owning to the existence of large pores (Fig. 1b(i)). Nano-ordered membranes arising from nanophase segregation of LC molecules exhibit excellent virus filtration properties owing to their regular nanopore sizes and low defect structures (Fig. 1b(ii)).19,28–31
The 2D nanochannels29,30,44,45 of smectic (Sm) LC phases provide a larger fraction of water-permeable paths than the 1D channels of columnar (Col) LC phases11,31,46 and the 3D channels10,11,28,31,47–49 of bicontinuous cubic (Cubbi) LC phases (Fig. 1a).32 This larger fraction of channels in Sm membranes is expected to enhance virus rejection and water permeability (Fig. 1b(iii)).29,30 For example, the ionic Sm LC polymer membranes with a thickness only 100 nm reject Aichi virus (AiV) with >7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 rejection (>99.99999%), which is considered high-performance virus removal.29 The diameter of AiV (approximately 25 nm) is smaller than that of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (approximately 120 nm). Furthermore, high-performance virus removal of Sm membranes may be useful if smaller and more harmful viruses emerge.
log10 rejection (>99.99999%), which is considered high-performance virus removal.29 The diameter of AiV (approximately 25 nm) is smaller than that of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (approximately 120 nm). Furthermore, high-performance virus removal of Sm membranes may be useful if smaller and more harmful viruses emerge.
Herein, we report the preparation of nanoporous membranes from LC molecules composed of rod-shaped rigid cores, which form 2D channels. The membranes were applied to the removal of viruses from a cocktail solution containing ultraviolet (UV)-inactivated SARS-CoV-2 and bacteriophages Qβ (Qβ). The effects of the morphologies of the 2D structures of the membranes on virus removal and water permeation are studied.
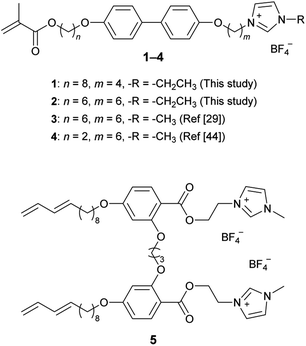
Fig. 2 shows the molecular structures of the polymerizable liquid crystals. Compounds 1 and 2 were designed in the present study. We expected compounds 1 and 2 to exhibit smectic A (SmA) phase. Compounds 3–5 were previously investigated.29,30,44 We reported the preparation and functions of nanostructured water-permeable membranes with 2D nanochannels prepared from compounds 3 (ref. 29) and 5.30 The membrane prepared from 3 contains both semi-bilayer and monolayer nanochannels.29 In contrast, compound 5 in the LC phase forms only stable semi-bilayer channels, and the membranes prepared from compound 5 exhibits improved water permeability.30
In the thermotropic LC SmA phase of molecules containing ionic hydrophilic and hydrophobic moieties, 2D nanopores form via nanophase segregation.29,30,33,44,55,56 Molecules in the SmA phases are arranged in layers such that their long axes are, on average, perpendicular to the layer plane (Fig. 3). The SmA phase can be classified into three categories according to the relationship between the molecular and periodic lengths of the layered structures: bilayer (Type 1), semi-bilayer (Type 2) and monolayer (Type 3) structures (Fig. 3). In the case of bilayers, one layer consists of two molecules, and the structure is termed SmA2 (Type 1).38,57 In a monolayer system, the layer spacing is equivalent to the molecular length, and the system is known as SmA1 (Type 3). Semi-bilayers are characterized by interdigitation, and the periodic length can be up to approximately 1.4 times the molecular length (Type 2). Semi-bilayer SmA is termed SmAd.
 | ||
| Fig. 3 Layered assemblies of molecules in the SmA phases. Type 1: bilayer structure (SmA2); Type 2: semi-bilayer structure (SmAd); Type 3: monolayer structure (SmA1). | ||
The bilayer and semi-bilayer systems yield nanoporous structures, in which only hydrophilic moieties are assembled. In the monolayer system, the molecules exhibit random head-to-tail orientations (Fig. 3). The 2D channels of the bilayer and semi-bilayer structures may be more beneficial for water-permeation owning to their hydrophilicity. Compound 5 has advantageous features for the formation of water permeable pores. In contrast, the compounds with rigid rod cores are easier to synthesize than the H-shaped molecules. In this study, we aimed to obtain molecules composed of rigid rod cores to promote the formation of bilayer or semi-bilayer Sm phase.
2 Experimental section
2.1 General
The LC phase transition behaviours were examined using a BX-53 polarizing optical microscope (POM) (Evident, Tokyo, Japan) equipped with a 10084L hot stage (Linkam, Redhill, UK). Differential scanning calorimetry (DSC) measurements were performed using a DSC 3500 Sirius system (NETZSCH, Selb, Germany) at a cooling rate of 10 °C min−1. Nuclear magnetic resonance (NMR) spectra of samples in CDCl3 were recorded using a JNM-ECX400 spectrometer (JEOL, Tokyo, Japan) operating at 400 MHz for 1H NMR and 100 MHz for 13C NMR. The chemical shifts of the 1H and 13C NMR signals were referenced to the internal standards, Me4Si (δ = 0.00) and CDCl3 (δ = 77.00), respectively. NMR results were expressed in terms of chemical shifts in ppm (δ), multiplicity, coupling constant (Hz), and relative intensity. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was performed on an Autoflex™ speed spectrometer (Bruker, Billerica, MA, USA) using dithranol as the matrix. Elemental analyses were performed using a CE-440 Elemental Analyzer (Exeter Analytical, North Chelmsford, MA, USA). X-ray diffraction (XRD) measurements were performed using a RINT-2500 system (Rigaku, Tokyo, Japan) and a SmartLab system (Rigaku). UV irradiation was performed using a UV-light-emitting diode (UV-LED) HLDL100U system (CCS, Kyoto, Japan). Thermogravimetric (TG) measurements were conducted on a TG-8122 (Rigaku) under N2 (100 cm3 min−1) in the temperature range of 25 up to 1000 °C at a heating rate of 10 °C min−1. Attenuated total reflection (ATR) Fourier transformed infrared (FTIR) spectra were obtained with a FT/IR-6100 spectrometer (JASCO, Tokyo, Japan) equipped a diamond prism system PKS-D1 (JASCO). Scanning electron microscopy (SEM) images were obtained with a JSM-7800FPRIME (JEOL) operated at 3 kV. SEM samples were coated with osmium in advance. Details of the syntheses are provided in the ESI (Schemes S1 and S2).†2.2 Materials
All reagents were purchased from Sigma-Aldrich Japan (Tokyo, Japan), Kanto Chemical (Tokyo, Japan), TCI (Tokyo, Japan) and FUJIFILM Wako Pure Chemical (Osaka, Japan) and were used as received. Unless otherwise noted, all reactions were performed under an argon atmosphere in a dry solvent purchased from Kanto Chemical. Poly(ethylene terephthalate) (PET) films coated with a poly(vinyl alcohol) (PVA) layer (SO sheet) were provided by AICELLO (Toyohashi, Japan). Flat-sheet polysulfone ultrafiltration membrane CF-30S was obtained from Nitto/Hydranautics (Osaka, Japan).2.3 Membrane fabrication
Membranes for virus filtration (P1–3 membranes) were fabricated via photopolymerization. The monomer solution was prepared by mixing the monomer (2 wt%) with photoinitiator 2,2-dimethoxy-2-phenylacetophenone (0.02 wt%) in dichloromethane.29 Transcription method using a sacrificial layer of PVA was performed to prepare the membranes.29 The monomer solution was spin-coated onto a PVA-coated PET film, which was transcribed onto a polysulfone ultrafiltration membrane by manual pressing. The membrane was heated to 100 °C and then maintained or cooled to the desired temperature (100 °C for compound 1; 60 °C for compounds 2 and 3). The membrane was irradiated with UV light (365 nm, 30 mW cm−2) using a UV-LED system for 10 min. The PVA layer was removed by immersing the sample in water after the polymerization.Polymerized films of the compounds (polymer P1–3) were used for the POM, XRD, TG, DSC and FTIR analyses of the LC polymers. The films of monomer and the photoinitiator 2,2-dimethoxy-2-phenylacetophenone (1 wt%) were heated and irradiated with UV light (365 nm, 30 mW cm−2) using a UV-LED system for 20 min in their LC state (100 °C for compound 1; 60 °C for compounds 2 and 3) on glass substrates. Details are shown in the ESI.†
2.4 Filtration experiment
Rejection of the prepared membranes was studied using 25 mm membrane filtration units equipped with an agitator (UHP-25K, Advantec, Tokyo, Japan) and connected to a reservoir (RP-2, Advantec) containing a feed solution of Milli-Q water. The membranes were submerged in Milli-Q water for 15 min before viral filtration. Pure air containing less than 0.1 ppm CO2 was used to provide a pressure of 0.3 MPa for the feed solution entering the stirred cell. Filtrate samples were collected over periods of 0–0.5, 0.5–1, 1–2 and 2–3 h. This filtration period is shorter than those of the previous studies28,29 owing to the high flux and limitation of the feed solution amount. The log reduction value (LRV) was calculated using the following equation:where Cfeed and Ceffluent denote the virus concentrations in the feed and effluent solutions, respectively.
2.5 Preparation of the feed solution containing viruses
Qβ was propagated in the laboratory using Escherichia coli (E. coli) K12 F+ (A/λ) as the host bacteria, whereas mouse hepatitis virus (MHV) and AiV were propagated using mouse brain tumor cells and Vero cells, respectively. SARS-CoV-2 [2019-nCoV/Japan/TY/WK-521/2020 strain] was propagated in VeroE6 cells overexpressing transmembrane protease serine 2 (TMPRSS2)58 in a biosafety level 3 facility at the National Institute of Infectious Diseases. Propagated SARS-CoV-2 was UV-inactivated using a TFS-20V UV transilluminator (Analytik Jena, Jena, Germany) (254 nm, 100 V, 25 W) for 15 min. The samples were then inoculated into VeroE6/TMPRSS2 cells to confirm virus inactivation.The virus suspensions were then centrifuged at 5000 × g for 15 min and filtered through cellulose acetate filters (0.2 μm, DISMIC-25CS, Advantec) to remove host bacteria/cell debris. Bacterial debris and medium components were also removed from the virus solutions using Illustra MicroSpin column S-300 HR (Merck Japan, Tokyo, Japan) to minimize their influence on the removal efficiencies of the tested membranes. For the membrane tests using a virus cocktail, Qβ, MHV, AiV and SARS-CoV-2 were mixed and spiked into the feed solution, equivalent to a dilution of 1000 times. For the membrane tests using a single virus Qβ, the purified stock was also diluted by 1000 times into the feed solution to obtain a concentration of approximately 6 × 107 copies per mL.
2.6 Quantification of viruses by reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Viral ribonucleic acid (RNA) was extracted from 140 μL of water samples (feed and effluent at each time point) using the QIAamp Mini Viral RNA Kit (QIAGEN, Venlo, Netherlands). Deoxyribonucleic acid (DNA) was obtained by reverse transcription reactions using a high-capacity cDNA reverse transcription kit with an RNase inhibitor (Applied Biosystems, Waltham, MA, USA).TaqMan-based qPCR assays for Qβ, MHV, AiV and SAR-CoV-2 were performed using a StepOnePlus™ real-time PCR system (Applied Biosystems), with the primers and probes listed in the ESI (Table S1).† The qPCR reaction mixture (25 μL) consisted of 12.5 μL of TaqMan™ gene expression master mix (Applied Biosystems), 1 μL of forward and reverse primers (10 μM), 0.5 μL of TaqMan probes (5 μM) and 5 μL of cDNA, to which UltraPure™ DNase/RNase-free distilled water (Invitrogen) was added. qPCR was performed on an ABI StepOnePlus thermocycler (Thermo Fisher, Waltham, MA, USA) using the thermal conditions described in previous reports.59–62 Standard curves were generated using 10-fold serial dilutions of predetermined plasmid DNA containing the target gene sequence ranging from 105 to 100 copies per reaction. The amplification efficiencies were >90%, and the correlation factor (R2) was >0.99.
3 Results and discussion
3.1 Molecular design and synthesis
Compounds 1–4 comprise a biphenyl core, a polymerizable methacrylate group and an imidazolium moiety, which are connected by alkylene spacers (Fig. 2). Compound 3 has alkylene spacers of the same length on both sides of the biphenyl moiety (n, m = 6) and forms the enantiotropic monolayer SmA (SmA1) phase.29 In contrast, compound 4 has different spacer lengths (n = 2; m = 6) and forms the semi-bilayer SmA (SmAd) phase, although the LC state of compound 4 is monotropic.44 In addition, the temperature range in which compound 4 exhibits the SmA phase is narrower than that of compound 3. The length of the alkylene spacers affects the assembled structures of these molecules and the thermal stabilities of the Sm phases.In the present study, compound 1 was designed as an ionic LC smectic compound composed of a rod-shaped rigid core to form the stable SmA2 or SmAd phase, thus improving water permeation. The lengths of the alkylene spacers were increased and modified (n = 8; m = 4) to achieve the assembled structures. We expected that alkylene spacers with the same spacer length on both sides of the biphenyl core would result in the formation of the SmA1 phase and that variations in the lengths of the spacers would induce the SmA2 or SmAd phase. In addition, the ethylimidazolium moiety was employed instead of the methylimidazolium moiety to reduce the temperature of transition from the less viscous isotropic liquid to the LC states, allowing efficient and stable processing during the preparation of membranes. Compound 2, with alkylene spacers of the same length (n, m = 6), was also synthesized to clarify the effects of the molecular structures on the self-assembled structures and water permeation. Compounds 1 and 2 were synthesized from 4,4′-biphenol using Mitsunobu reactions, which were similar to the processing in our previous study.29 Details are shown in the ESI.†
3.2 Assembled structures of liquid-crystalline smectic monomers and in situ polymerization
On heating, compound 1 melted at 85 °C and formed an SmA phase up to 109 °C, whereas, on cooling, it formed the SmA phase from 106 to 54 °C (Fig. 4a and S1a†). XRD patterns revealed that the layer spacing of the SmA phase was approximately 5.3 nm (Fig. 4a right). The molecular length of compound 1 was estimated to be approximately 3.9 nm, suggesting that compound 1 formed an interdigitated bilayer SmAd structure (Fig. S2a†). The hydrophobic alkyl chains with the polymerizable methacrylate moiety were likely interdigitated. Compound 2 showed an SmA phase from 80 to 25 °C on cooling (Fig. S1b†). In contrast to compound 1, the layer spacing of compound 2 was approximately 3.8 nm (Fig. S3a right†), suggesting that the assembled molecular structure of compound 2 was a monolayer (Fig. S4a†).POM observations revealed the molecular orientations of LC compounds 1 and 2 introduced between the PVA-coated glasses. Typical fan-shaped focal conic textures (Fig. 4a left and Fig. S3a left†) were observed, suggesting the formation of Sm phases in which parallel molecular arrangements were induced. The alignment of LC molecules on PVA substrates is important because our water-permeable nanostructured membranes are prepared using LC monomer films on PVA substrates.10,11,29 In contrast, between the glass substrates, molecules composed of rigid cores shown in Fig. 2, were homeotropically aligned (perpendicular to the substrate) in the SmA phase owing to the interaction between the molecules and hydrophilic surface.44 The decrease in the hydrophilicity of the substrate surface led to a change in the molecular orientation.
The transition temperatures on cooling and self-assembled structures of the Sm phases of compounds 1–4 are summarized in Table 1. Compounds 1 and 4 with asymmetric chain lengths on both sides of the rigid core, exhibited the SmAd phases. In contrast, compounds 2 and 3 with symmetric chain lengths formed the SmA1 structures. We postulated that the same spacer length on both sides of the biphenyl core site would result in the monolayer structure and that different lengths of the alkylene spacer would lead to the bilayer or semi-bilayer structures. The assembled molecular structures were consistent with our postulate.
Instead of the methylimidazolium moiety employed in previous studies,29,44 the ethylimidazolium moiety (Fig. 2) was employed in this study as the ionic moiety of the block structure to lower the isotropization temperature. The transition temperature of compound 2 was >15 °C lower than that of compound 3. The isotropization temperature of compound 1 was higher than that of compound 3 and it can be because of differences in the assembled structures and interactions between the hydrophobic groups. Further discussion about assembled molecular structures and thermal behavior is shown in the ESI (Fig. S2b and S4b).†
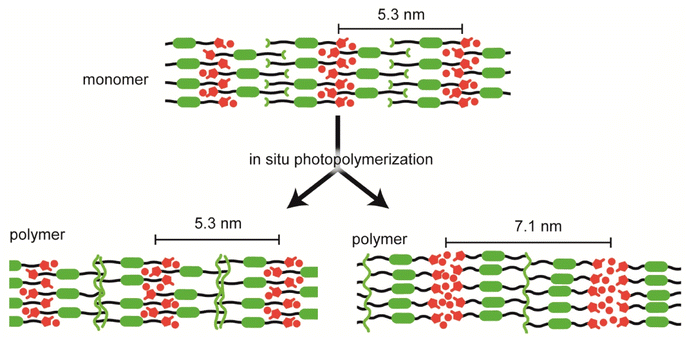
UV irradiation of mixtures containing the LC compounds and photoinitiator led to the formation of polymer films. FTIR spectra of 1 and P1 revealed that the methacrylate groups polymerized during UV irradiation (Fig. S5†). The XRD peaks of P1 were more complex than those of monomer 1 (Fig. 4a right and Fig. 4b right). This observation suggested that several ordered structures, in addition to the SmAd structures were formed during the polymerization of compound 1. Among the periodic structures observed in P1, the longest had a length of approximately 7.1 nm, which was approximately twice the molecular length of 1 (Fig. 5 and S2c†). In addition to fixation of the SmAd structure, aggregation of the polymerizable moiety likely induced structural change, from the interdigitated semi-bilayer to the bilayer structures. However, the POM images of compound 1 before and after photopolymerization were similar (Fig. 4a left and Fig. 4b left), suggesting that the molecular alignment of compound 1 was maintained during polymerization. On heating, P1 did not undergo phase transitions and decomposed at around 170 °C (Fig. S6 and S7†).
The XRD patterns of P2 suggested that during the in situ polymerization of compound 2, a periodic structure (d = 5.7 nm) was formed, in addition to the periodic structure found in the monomer (Fig. S3b right†). This is similar to the polymerization of compound 3.29 Aggregation of the methacrylate moiety likely induced the formation of the interdigitated bilayer structures (Fig. S4c†).
3.3 Virus filtration by membranes with 2D channels
Polymer membranes based on compounds 1, 2 and 3 (P1, P2 and P3 membranes, respectively) were prepared for filtration of viruses according to an established method using a PVA film as the sacrificial layer.29 We expected that the molecules in the SmA phases were aligned parallel to the PVA substrate, and the 2D channels of the LC structures were perpendicular to the substrate. The LC polymer layers were supported by polysulfone and nonwoven polyester (Fig. S8a†). These supporting layers impart mechanical toughness to the membranes.10,29 SEM images of the cross-section of P1 membrane revealed that thickness of the P1 layer on the substrate was 100–150 nm (Fig. S8b†).In the present study, a virus cocktail solution containing enveloped and non-enveloped viruses was used to compare the virus removal efficiencies. A single Qβ solution was also used to examine the filtration properties of the Sm membranes. Because Qβ is similar to typical picornavirus in size and composition, Qβ was employed as an indicator.63 AiV is representatively used to evaluate virus removal at full-scale wastewater treatment facilities.64 UV-inactivated SARS-CoV-2 and MHV were used as surrogates for SARS-CoV-2 with intact virions because both viruses belong to Betacoronavirus.
Cell culturing is the standard method used to measure the concentrations of viruses and bacteria in water, which in turn are used to determine the infection risk of pathogens in water. However, it cannot be applied to certain types of viruses because of the lack of appropriate cells. PCR is a more robust and sensitive method to enumerate viruses and measure the physical removal ratio via membrane filtration. A shortcoming of the PCR method is that it detects all viral genomes including those from inactivated viruses. However, this is not an issue in evaluating the ability of the membrane to remove viruses. Because the removal efficiency is calculated using the difference between the feed and end concentrations, although leakage of RNA/DNA may occur, it will not overestimate the removal efficiency. Therefore, PCR has been employed as a tool to investigate virus removal during various treatment steps,65,66 including membrane filtration.26 The present study also employed PCR to determine the LRVs of Qβ, demonstrating stable and sufficient removal by the polymer membranes.
The filtration properties of P1, P2 and P3 membranes were investigated using a high concentration of Qβ solution (>1.0 × 107 copies per mL). Fig. 6 shows the performance of P1, P2 and P3 membranes in filtering water containing Qβ. P1 and P2 membranes exhibited high reduction values for Qβ as well as P3 membrane. The LRVs of P2 membrane were higher than 7. For some of the filtered samples, the concentrations of the viruses were below the limit for quantitative evaluation.
The water flux through P1 membrane was higher than those through P2 and P3 membranes (Fig. 6b). In addition, P2 membrane exhibited a water flux similar to that of P3 membrane. These results indicated that the enhanced water permeability of P1 membrane did not originate in the difference between the ethylimidazolium moiety (P1 and P2) and methylimidazolium moiety (P3) but in the difference between the assembled structures of the Sm phases. As discussed earlier, the hydrophilic ionic nanochannels of the bilayer and semi-bilayer structures allow a higher water flux than the nanochannels of the monolayer structure.
The LRVs for viruses (Qβ, AiV, MHV and SARS-CoV-2) in cocktail solutions with P1 and P3 membranes are presented in Fig. 7. All viruses were simultaneously rejected with high efficiency. For all samples, the concentrations of MHV and SARS-CoV-2 after filtration were below the detection limit for quantitative evaluation. For some samples, the concentrations of Qβ and AiV after filtration were also below this limit. The different LRVs in Fig. 7 originated in the different concentrations of the feed solutions. In our previous study,31 Col and Cubbi membranes prepared from wedge-shaped molecules exhibit high rejections of viruses in cocktail solutions. The 2D channels used in this study are larger and more flexible than the Col and Cubbi channels, and thus the ion removal rates of the 2D channels are lower than those of the Col and Cubbi channels.29,30 The results shown in Fig. 7 indicated that the 2D channels of the Sm phases were effective in stopping various viruses, exhibiting a performance comparable to that of the channels of the Col and Cubbi structures.
 | ||
| Fig. 7 LRVs for viruses in cocktail solutions filtered through P1 and P3 membranes for 1 and 2 h. Error bars denote the standard deviation. Arrows show right-censoring LRVs. | ||
Studies on the fate of viruses during water treatment have mainly targeted non-enveloped viruses, such as norovirus or poliovirus, which cause waterborne diseases. The fate of SARS-CoV-2 during water treatment has attracted attention owing to its presence in wastewater since the first outbreak in December 2019.67 SARS-CoV-2 is an enveloped virus; therefore, it is challenging to determine its fate during water treatment owing to the lack of an optimized and standardized protocols.68 Wang et al. reported that membrane bioreactors show stable SARS-CoV-2 removal.69 However, studies on SARS-CoV-2 removal from water are limited. The removal of target viruses, especially SARS-CoV-2, to undetectable level by the LC nanoporous membranes indicates that these membranes are promising for the prevention of SARS-CoV-2 pollution.
4 Conclusions
We demonstrated that the 2D ionic channels of nanostructured polymer membranes prepared via the in situ polymerization of Sm monomers effectively rejected viruses of various sizes, including SARS-CoV-2, in cocktail solutions. In addition, the membrane prepared from the semi-bilayer structures of compound 1 exhibited improved water permeability compared with the membrane prepared from the monolayer structures of compound 2 and 3, although the virus removal efficiencies of the three membranes were similar. These results may promote a new strategy for the future development of water-permeable membranes to achieve clean and safe water supply.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was partially supported by JST-CREST, JPMJCR20H3, and JSPS KAKENHI (grant number: JP19H05714 and JP19H05715) (grant-in-aid for scientific research on innovative areas: aquatic functional materials, no. 6104). We thank Nitto/Hydranautics (Osaka, Japan) for providing flat-sheet polysulfone ultrafiltration membrane samples. We thank Dr Junya Uchida for helpful discussions. A part of this work was supported by “Advanced Research Infrastructure for Materials and Nanotechnology in Japan (ARIM)” of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (grant number: JPMXP1223UT-0198). We thank proof-reading service, Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.References
- J. Eliasson, Nature, 2015, 517, 6 CrossRef CAS.
- V. G. Gude, Water Res., 2016, 89, 87 CrossRef CAS PubMed.
- M. A. Shannon, P. W. Bohn, M. Elimelech, J. G. Georgiadis, B. J. Mariñas and A. M. Mayes, Nature, 2008, 452, 301 CrossRef CAS PubMed.
- M. S. Mauter, I. Zucker, F. Perreault, J. R. Werber, J. H. Kim and M. Elimelech, Nat. Sustain., 2018, 1, 166 CrossRef.
- A. Lee, J. W. Elam and S. B. Darling, Environ. Sci.: Water Res. Technol., 2016, 2, 17 RSC.
- Y. Zhao, T. Tong, X. Wang, S. Lin, E. M. Reid and Y. Chen, Environ. Sci. Technol., 2021, 55, 1359 CrossRef CAS PubMed.
- J. R. Werber, C. O. Osuji and M. Elimelech, Nat. Rev. Mater., 2016, 1, 16018 CrossRef CAS.
- I. Sadeghi, P. Kaner and A. Asatekin, Chem. Mater., 2018, 30, 7328 CrossRef CAS.
- D. L. Gin and R. D. Noble, Science, 2011, 332, 674 CrossRef CAS PubMed.
- M. Henmi, K. Nakatsuji, T. Ichikawa, H. Tomioka, T. Sakamoto, M. Yoshio and T. Kato, Adv. Mater., 2012, 24, 2238 CrossRef CAS PubMed.
- T. Sakamoto, T. Ogawa, H. Nada, K. Nakatsuji, M. Mitani, B. Soberats, K. Kawata, M. Yoshio, H. Tomioka, T. Sasaki, M. Kimura, M. Henmi and T. Kato, Adv. Sci., 2018, 5, 1700405 CrossRef.
- J. Kloos, N. Joosten, A. Schenning and K. Nijmeijer, J. Membr. Sci., 2021, 620, 118849 CrossRef CAS.
- C. Chen, L. Guo, Y. Yang, K. Oguma and L. Hou, Sci. Total Environ., 2021, 801, 149678 CrossRef CAS PubMed.
- B. N. Bhadra, L. K. Shrestha and K. Ariga, CrystEngComm, 2022, 24, 6804 RSC.
- Z. Yang, H. Guo and C. Y. Tang, J. Membr. Sci., 2019, 590, 117297 CrossRef.
- T. Kawakami, M. Nakada, H. Shimura, K. Okada and M. Kimura, Polym. J., 2018, 50, 327 CrossRef CAS.
- H. Shimura, Polym. J., 2022, 54, 767 CrossRef CAS.
- C. Lang, M. Kumar and R. J. Hickey, Soft Matter, 2021, 17, 10405 RSC.
- S. Y. Yang, I. Ryu, H. Y. Kim, J. K. Kim, S. K. Jang and T. P. Russell, Adv. Mater., 2006, 18, 709 CrossRef CAS.
- N. Hampu, J. R. Werber, W. Y. Chan, E. C. Feinberg and M. A. Hillmyer, ACS Nano, 2020, 14, 16446 CrossRef CAS PubMed.
- R. L. McGinnis, K. Reimund, J. Ren, L. Xia, M. R. Chowdhury, X. Sun, M. Abril, J. D. Moon, M. M. Merrick, J. Park, K. A. Stevens, J. R. McCutcheon and B. D. Freeman, Sci. Adv., 2018, 4, e1700938 CrossRef.
- J. Lyu, X. Wen, U. Kumar, Y. You, V. Chen and R. K. Joshi, RSC Adv., 2018, 8, 23130 RSC.
- D. Ma, H. Li, Z. Meng, C. Zhang, J. Zhou, J. Xia and Y. Wang, Environ. Sci. Technol., 2021, 55, 15206 CrossRef CAS PubMed.
- T. Urase, K. Yamamoto and S. Ohgaki, J. Membr. Sci., 1996, 115, 21 CrossRef CAS.
- M. Yasui, H. Iso, S. Torii, Y. Matsui and H. Katayama, Water Res., 2021, 206, 117735 CrossRef CAS PubMed.
- R. R. Trussell, J. D. Kenny, R. A. Tackaert, A. N. Pisarenko and R. S. Trussell, AWWA Water Sci., 2020, 2, e1206 CAS.
- O. Gustafsson, L. Manukyan and A. Mihranyan, Global chall., 2018, 2, 1800031 CrossRef PubMed.
- N. Marets, D. Kuo, J. R. Torrey, T. Sakamoto, M. Henmi, H. Katayama and T. Kato, Adv. Healthcare Mater., 2017, 6, 1700252 CrossRef PubMed.
- D. Kuo, M. Liu, K. R. S. Kumar, K. Hamaguchi, K. P. Gan, T. Sakamoto, T. Ogawa, R. Kato, N. Miyamoto, H. Nada, M. Kimura, M. Henmi, H. Katayama and T. Kato, Small, 2020, 16, 202001721 Search PubMed.
- K. Hamaguchi, R. Ichikawa, S. Kajiyama, S. Torii, Y. Hayashi, J. Kumaki, H. Katayama and T. Kato, ACS Appl. Mater. Interfaces, 2021, 13, 20598 CrossRef CAS PubMed.
- D. Kuo, T. Sakamoto, S. Torii, M. Liu, H. Katayama and T. Kato, Polym. J., 2022, 54, 821 CrossRef CAS PubMed.
- Handbook of Liquid Crystals, ed. J. W. Goodby, P. J. Collings, H. Gleeson, P. Raynes, T. Kato and C. Tschierske, Wiley-VCH, Weinheim, 2nd edn, 2014 Search PubMed.
- S. Ujiie and T. Kato, in Handbook of Liquid Crystals, ed. J. W. Goodby, P. J. Collings, H. Gleeson, P. Raynes, T. Kato and C. Tschierske, Wiley-VCH, Weinheim, 2nd edn, 2014, ch. 11, vol. 7, pp. 381–414 Search PubMed.
- M. Ishizu, K. Hisano, M. Aizawa, C. J. Barrett and A. Shishido, ACS Appl. Mater. Interfaces, 2022, 14, 48143 CrossRef CAS.
- T. Kato, M. Yoshio, T. Ichikawa, B. Soberats, H. Ohno and M. Funahashi, Nat. Rev. Mater., 2017, 2, 17001 CrossRef.
- H. K. Bisoyi and Q. Li, Chem. Rev., 2022, 122, 4887 CrossRef CAS PubMed.
- Y. Inoue, K. Takada, A. Kawamura and T. Miyata, ACS Appl. Mater. Interfaces, 2022, 14, 31513 CrossRef CAS PubMed.
- N. Kapernaum, A. Lange, M. Ebert, M. A. Grunwald, C. Haege, S. Marino, A. Zens, A. Taubert, F. Giesselmann and S. Laschat, ChemPlusChem, 2022, 87, e202100397 CrossRef CAS PubMed.
- T. Kato, J. Uchida, T. Ichikawa and B. Soberats, Polym. J., 2018, 50, 149 CrossRef CAS.
- J. Uchida, B. Soberats, M. Gupta and T. Kato, Adv. Mater., 2022, 34, 2109063 CrossRef CAS PubMed.
- T. Kato, J. Uchida, T. Ichikawa and T. Sakamoto, Angew. Chem., Int. Ed., 2018, 57, 4355 CrossRef CAS PubMed.
- D. J. Broer, C. M. W. Bastiaansen, M. G. Debije and A. P. H. J. Schenning, Angew. Chem., Int. Ed., 2012, 51, 7102 CrossRef CAS PubMed.
- D. L. Gin, X. Lu, P. R. Nemade, C. S. Pecinovsky, Y. Xu and M. Zhou, Adv. Funct. Mater., 2006, 16, 865 CrossRef CAS.
- K. Hoshino, M. Yoshio, T. Mukai, K. Kishimoto, H. Ohno and T. Kato, J. Polym. Sci., Part A: Polym. Chem., 2003, 41, 3486 CrossRef CAS.
- H. P. C. van Kuringen, G. M. Eikelboom, I. K. Shishmanova, D. J. Broer and A. P. H. J. Schenning, Adv. Funct. Mater., 2014, 24, 5045 CrossRef CAS.
- P. M. S. Roman, K. Nijmeijer and R. P. Sijbesma, J. Membr. Sci., 2022, 644, 120097 CrossRef.
- M. J. McGrath, S. H. Hardy, A. J. Basalla, G. E. Dwulet, B. C. Manubay, J. J. Malecha, Z. Shi, H. H. Funke, D. L. Gin and R. D. Noble, ACS Mater. Lett., 2019, 1, 452 CrossRef CAS.
- S. M. Dischinger, J. Rosenblum, R. D. Noble and D. L. Gin, J. Membr. Sci., 2019, 592, 117313 CrossRef.
- O. Q. Imran, P. Li, N. K. Kim, D. L. Gin and C. O. Osuji, Chem. Commun., 2021, 57, 10931 RSC.
- X. Feng, Q. Imran, Y. Zhang, L. Sixdenier, X. Lu, G. Kaufman, U. Gabinet, K. Kawabata, M. Elimelech and C. O. Osuji, Sci. Adv., 2019, 5, eaav9308 CrossRef CAS PubMed.
- Y. Ishii, N. Matubayasi, G. Watanabe, T. Kato and H. Washizu, Sci. Adv., 2021, 7, eabf0669 CrossRef CAS PubMed.
- R. Watanabe, T. Sakamoto, K. Yamazoe, J. Miyawaki, T. Kato and Y. Harada, Angew. Chem., Int. Ed., 2020, 59, 23461 CrossRef CAS PubMed.
- G. Graziano, Nat. Rev. Chem, 2020, 4, 636 CrossRef CAS PubMed.
- Y. Chen, H. Y. Chang, M. T. Lee, Z. R. Yang, C. H. Wang, K. Y. Wu, W. T. Chuang and C. L. Wang, J. Am. Chem. Soc., 2022, 144, 7768 CrossRef CAS PubMed.
- D. Högberg, B. Soberats, R. Yatagai, S. Uchida, M. Yoshio, L. Kloo, H. Segawa and T. Kato, Chem. Mater., 2016, 28, 6493 CrossRef.
- K. Hamaguchi, H. Lu, S. Okamura, S. Kajiyama, J. Uchida, S. Sato, G. Watanabe, Y. Ishii, H. Washizu, G. Ungar and T. Kato, ChemPhysChem, 2023, 24, e202200927 CrossRef CAS PubMed.
- J. W. Goodby, in Handbook of Liquid Crystals, ed. J. W. Goodby, P. J. Collings, H. Gleeson, P. Raynes, T. Kato and C. Tschierske, Wiley-VCH, Weinheim, 2nd edn, 2014, ch. 2, vol. 4, pp. 43–51 Search PubMed.
- S. Matsuyama, N. Nao, K. Shirato, M. Kawase, S. Saito, I. Takayama, N. Nagata, T. Sekizuka, H. Katoh, F. Kato, M. Sakata, M. Tahara, S. Kutsuna, N. Ohmagari, M. Kuroda, T. Suzuki, T. Kageyama and M. Takeda, Proc. Natl. Acad. Sci. U.S.A., 2020, 117, 7001 CrossRef CAS PubMed.
- S. Wolf, J. Hewitt, M. Rivera-Aban and G. E. Greening, J. Virol. Methods, 2008, 149, 123 CrossRef CAS PubMed.
- D. G. Besselsen, A. M. Wagner and J. K. Loganbill, Comp. Med., 2002, 52, 111 CAS.
- M. Kitajima, A. Hata, T. Yamashita, E. Haramoto, H. Minagawa and H. Katayama, Appl. Environ. Microbiol., 2013, 79, 3952 CrossRef CAS PubMed.
- Centers for Disease Control and Prevention, 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes, 2019, https://www.fda.gov/media/134922/download, accessed February 2023 Search PubMed.
- IAWPRC, Water Res., 1991, 25, 529 CrossRef.
- V. D. Canh, I. Kasuga, H. Furumai and H. Katayama, Food Environ. Virol., 2019, 11, 40 CrossRef CAS PubMed.
- R. Kato, T. Asami, E. Utagawa, H. Furumaim and H. Katayama, Water Res., 2018, 132, 61 CrossRef CAS PubMed.
- H. Katayama, E. Haramoto, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima and S. Ohgaki, Water Res., 2008, 42, 1441 CrossRef CAS PubMed.
- G. La Rosa, M. Iaconelli, P. Mancini, G. B. Ferraro, C. Veneri, L. Bonadonna, L. Lucentini and E. Suffredini, Sci. Total Environ., 2020, 736, 139652 CrossRef CAS PubMed.
- M. Kitajima, W. Ahmed, K. Bibby, A. Carducci, C. P. Gerba, K. A. Hamilton, E. Haramoto and J. B. Rose, Sci. Total Environ., 2020, 739, 139076 CrossRef CAS PubMed.
- R. Wang, M. Alamin, S. Tsuji, H. Hara-Yamamura, A. Hata, B. Zhao, M. Ihara and R. Honda, Sci. Total Environ., 2022, 851, 158310 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ta02705h |
| This journal is © The Royal Society of Chemistry 2023 |