Open Access Article
Open Access Article3D printing for flow biocatalysis
Elena
Gkantzou
a,
Marie
Weinhart
bc and
Selin
Kara
 *ad
*ad
aInstitute of Technical Chemistry, Leibniz University Hannover, 30167 Hannover, Germany. E-mail: selin.kara@iftc.uni-hannover.de
bInstitute of Physical Chemistry and Electrochemistry, Leibniz Universität Hannover, 30167 Hannover, Germany
cInsitute of Chemistry and Biochemistry, Freie Universität Berlin, 14195 Berlin, Germany
dBiocatalysis and Bioprocessing Group, Department of Biological and Chemical Engineering, Aarhus University, 8000 Aarhus, Denmark. E-mail: selin.kara@bce.au.dk
First published on 8th August 2023
Abstract
Additive manufacturing has evolved at such a level nowadays that it follows the sustainability pathways, from applied materials to processing costs. This is a fundamental reason that more and more scientific effort is devoted to incorporating this technology in different research fields. Implementation of 3D printing technology in flow biocatalysis can be addressed at every process design level, (i) either the reactor itself, (ii) the support material for biocatalyst confinement, or (iii) the peripheral accessories that can establish a highly controlled process. 3D printing is an attractive option for enabling the development of more efficient processes, along with facile performance optimization. Moreover, the 3D printing of a biocatalyst entrapped in a protecting scaffold offers an alternative immobilization approach with promising results for a cost-effective and green process design.
Sustainability spotlightFlow systems have become part of numerous research laboratories in academia and industry, while biocatalytic systems have long been described as the sustainable alternative to classical chemical catalysis. Thus, there are increasing requirements for systems and devices that sustainably support biocatalytic flow systems. Additive manufacturing can be employed in different stages of a flow bioprocess, offering the scientists the benefit of customizing the equipment and even enhancing the system's productivity with tailor-made reactors and biocatalyst supports. The goals for sustainable consumption and production (UN-SDG12) can be addressed in this work, with literature examples of laboratory practices that promote functionality and longevity of industrially-relevant bioprocesses. An overview of recycling trends and time-to-process management is also described. |
1. Introduction
Additive manufacturing (AM), or, as it is commonly known, three-dimensional (3D) printing, connects the virtual world with the objects we get in our hands in everyday life. The AM ecosystem consists of hardware, software/modeling, and materials; in its ideal form, it is a closed-loop system. This means that nowadays, 3D printing is focusing on sustainable sources of printing materials (inks, resins, and filaments) and follows pathways for recycling and chemical circularity.1–3 The goal is to make AM a production technology whose products undergo the maximum possible reusing. At the end of their lifetime, every product either biodegrades or is recycled back into material feedstock.4 If we add the low – or extremely low in some cases – processing costs of this technology, we can talk about a sustainability diamond, ready to be employed in a vast range of distinct technological sectors, from health and chemistry to mechanics, aerospace, and environmental applications.5Additive manufacturing processes have also been implemented in the field of flow biocatalysis. This emerging application of biocatalysis is defined as the processes that take place under continuous flow conditions in specially designed reactor systems, elaborating biocatalysts either isolated, i.e., enzymes, or in their whole-cell form.6 The answer to why perform biocatalysis in flow reactors has been comprehensively answered by Tamborini et al. (2018)7 and De Santis & Meyer et al. (2020)8 in their reviews for biocatalytic process intensification. Prof. Polona Žnidaršič-Plazl has also reported on the promises and challenges of biotransformations in microflow.9 In these and several other works,10–12 it has been highlighted the substantial improvement in space-time yields (STYs) – a fundamental parameter used to describe the productivity in biocatalytic processes – when shifting from batch to continuous flow systems.13–15 So, it can be argued that flow reactors are used when continuous productivity is of utmost importance, and there is no specific need for products at large scales. However, this is not entirely true. Scale-up of flow processes can be realized utilizing parallelization and numbering-up approaches. By doing so, production on a bigger scale can be more economically feasible if we consider the significantly smaller equipment, the decrease in reaction time from hours to minutes, and the highly controlled systems that can be realized when we have to do with small dimensions.7 So, more efficient reactions and minimized waste production? industry's blessings.
Nevertheless, how 3D printing technology is incorporated into flow systems design? For flow chemistry, we need reactors and/or peripherals like pumps, sensors, reservoirs, etc. For biocatalysis, a highly controlled microenvironment in a confined space is required, that can be translated to a chemically modified scaffold to host the biocatalyst, a defined reaction chamber with sufficient nutrients or substrates supply, or a specially designed reactor that could serve special needs like light penetration in photobiocatalysis.16,17 3D printing can do all of these. 3D-printed parts have been elaborated in all stages of a continuous flow biocatalytic process: (i) the reactor itself, (ii) the support material for biocatalyst confinement, or (iii) the peripheral accessories that can establish a highly controlled process.18 Going with the 3D printing concept, rather than using already patented reactors, scaffolds, and accessories, offers the critical advantage of customizable products, fitted to the particular needs of each process. It can also be a relatively economical alternative since materials applied for 3D printers, or the 3D printers themselves, are becoming cheaper and more user-friendly. Complex geometries with high porosity can be realized, and a ‘trial-and-error’ method can be adopted by the user. Another critical aspect is that the software for 3D design is readily available to everyone, with multiple open-source programs, and a vast online community supporting the users in their endeavors.
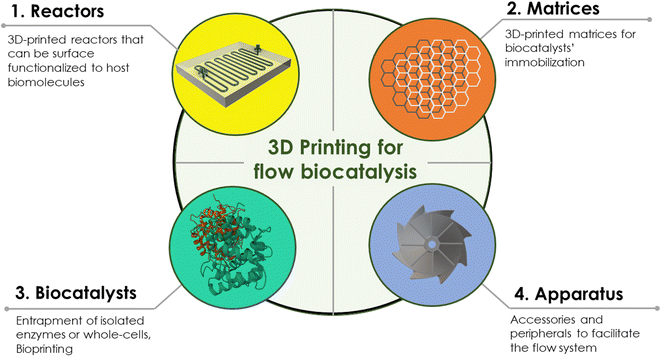
In their review, Zentel et al. (2020) explored the potential of 3D printing for chemical reaction engineering.19 They highlighted the impact of tailor-made designs and fabrication techniques as a game-changer in constructing reactors and downstreaming units in flow chemistry. Within this review article, we aim to bring together the reported knowledge in elaborating 3D printing for flow biocatalysis. We will delve into the scientific community's different approaches to utilize this flourishing technology to upgrade the continuous flow biocatalytic systems. Our approach is to divide the literature into 3D printing of reactors and reactor matrices (Section 2), 3D printing of biocatalysts (Section 3), and 3D printing of reactor apparatus (Section 4) necessary for the flow system performance (Fig. 1). This review aims to contribute to the existing literature for applying AM techniques in different fields of study. To the best of our knowledge, it is the first review with a special focus on flow biocatalysis.
2. 3D printing of reactors and reactor matrices for flow biocatalysis
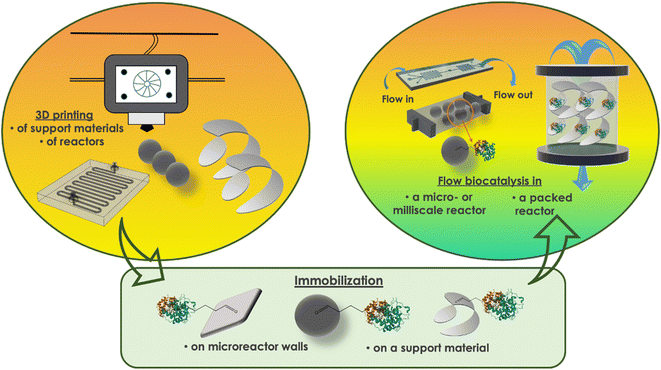
In flow biocatalysis, biotransformations typically occur within confined spaces in reactors or reactor matrices, which both can be 3D-printed. At first, it was just an appropriate scaffold, chemically inert and mechanically stable. But nowadays, AM has evolved so that chemical functionalities can easily be added to initially inert printing materials. Matrix materials, blended materials, or reactive moieties have been explored in this context, offering superior performance to the 3D-printed scaffolds.18,19 Recently, it was also reported a novel hydrogel as a reactor packing material produced with a mild 3D printing process.20 One step further, the functionalities on the 3D-printed reactor surface can be used for targeted material–biosystem interactions, with enzyme immobilization being the most common approach.21 Studies have shown that when the biocatalyst is fixed in the reactor surface, significant advantages in terms of stability and reusability arise.22 So, the process follows the route: choose the appropriate material, 3D-print the scaffold, integrate the biocatalyst, and finally connect to the overall fluidic system.Previous reports have reviewed the 3D printing techniques used for bio-related applications, including descriptions of the diverse materials that have been investigated.18,19,21,23 In the following paragraphs, we will examine some relevant examples from the literature, where 3D printing was elaborated for scaffold design and development in flow biocatalysis (Fig. 2). Table 1 summarizes the referenced literature.
| Description | Scale | Technique | Material | Enzymes | Comments | Ref. |
|---|---|---|---|---|---|---|
| Chemically surface-modified 3D-printed reactor | Milli-scale | Selective laser sintering (SLS) | Nylon 6 (fossil based) | ω-Transaminase ATA117 | • Well-plate printed of the same material as the reactor for optimization of immobilization strategy | 24 |
| • Remarkable activity and stability in flow, a total of 105 cycles of use | ||||||
| Chemically surface-modified 3D-printed reactor with integrated sphere-shaped scaffolds | Milli-scale | Fused deposition modeling (FDM) | Carbon-reinforced polylactic acid (C-PLA) (biobased) | Penicillin G acylase from Providencia rettgeri | • Different enzyme classes immobilized with the same protocol | 25 |
| Protease WQ9-2 | • Good operability proved with lactosucrose and amoxicillin synthesis under continuous flow | |||||
| Glycosidase NJEM01 | ||||||
| Lipase YCJ01 | ||||||
| Polydopamine-coated 3D-printed batch and microfluidic reactors | Milli-scale | Fused deposition modeling (FDM) | Polypropylene (fossil based) | Acetophenone reductase from Geotrichum candidum | • Novel surface modification protocol for 3D-printed materials | 26 |
| • Immobilized acetophenone reductase in microfluidic reactors produced higher (S)-1-phenylethanol than previously reported yield with free enzyme | ||||||
| Chitosan-coated 3D-printed microreactor | Micro-scale | Fused deposition modeling (FDM) | Polylactic acid (biobased) | Laccase from Trametes versicolor | • Spectroscopic characterization revealed the degree of surface enrichment | 27 |
• Total turnover number of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 386 386![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for hydroxytyrosol conversion under continuous flow 000 for hydroxytyrosol conversion under continuous flow |
||||||
| Chitosan-coated 3D-printed microreactor | Micro-scale | Fused deposition modeling (FDM) | Polylactic acid (biobased) | Lipase from Candida antarctica | • 3D-printed material compatible with deep eutectic solvents (DES) for biocatalysis in non-conventional media | 28 |
| • Exceptional stability under different conditions and incubation in DES | ||||||
| ZIF-8 modified 3D-printed micromixer | Micro-scale | Stereolithography (SLA) | Urethane methacrylate-based resin (clear resin V4) (fossil based) | Carbonic anhydrase from bovine erythrocytes | • Co-immobilization and domino immobilization approaches investigated | 29 |
| Formate dehydrogenase from Candida boidinii | • Almost 10 times increased yield for formic acid compared to conventional bubble column | |||||
| 3D-printed packing material for reactors | Milli-scale | Selective laser sintering (SLS) | Polyamide 12 (PA12) (Fossil based) | Phenolic acid decarboxylase from Mycobacterium colombiense | • Ferulic acid (FA) decarboxylation to 2-methoxy-4-vinylphenol (MVP) with in situ extraction of MVP with an organic solvent | 30 |
| • 85% conversion in the first hour of reaction, 83% of the product extracted in the first 15 minutes of operation | ||||||
| 3D-printed supports for enzyme immobilization as reactor inserts | Micro-scale | Stereolithography (SLA) | Ceramics | Phenolic acid decarboxylase from Bacillus subtilis | • Lattice structure with high surface area, reaching 58% immobilization yield and 6.02 U g−1 enzymatic activity | 31 |
| • The decarboxylation of coumaric acid to vinylphenol reached a STY of 39.21 g (L h)−1, outperforming previous reports by 8-fold | ||||||
| Adapted reactor design with incorporated textile-immobilized biocatalyst | Milli-scale | Fused deposition modeling (FDM) | Transparent polyester copolymer (fossil based) | Peroxidase MaxiBright® | • Compartmentalized membrane-type reactor with low and continuously increasing concentrations of the inhibiting substrate | 32 |
| • Yields under continuous flow improved by a factor of 30 compared to reactions in flasks | ||||||
| 3D-printed interfacial device | Milli-scale | Fused deposition modeling (FDM) | Acrylonitrile-butadiene-styrene (ABS) (Fossil based) | Carbonic anhydrase from bovine erythrocytes | • The flotation level of the floating units was easily controlled by changing the void volume of the 3D models | 33 |
| • The highest interfacial CO2 conversion was obtained when the CA fibers were located right at the air–liquid interface |
2.1 3D printing the reactors
Another example of a novel modification protocol on 3D-printed surfaces is that of Sriwong and Matsuda (2022), who applied polydopamine (PDA)-coated 3D-printed materials for continuous flow biocatalysis.26 Dopamine is an important compound in chemical biology and materials science because it shows intrinsic adhesiveness towards many different material surfaces, on which it attaches as a chemically reactive PDA priming layer. Its intrinsic reactivity towards, e.g., thiols and amines, acts as a platform for secondary reactions for specific surface functionalization. The researchers here used thermoplastic polypropylene as an inexpensive material that is chemically stable towards a range of organic solvents. The enzyme of choice was an acetophenone reductase from Geotrichum candidum (GcAPRD), and the system was applied for asymmetric ketone reduction in a continuous flow mode. Both batch and microfluidic reactors were designed, going from 1.5 cm to 1.5 mm internal diameter. The batch reactors were used to optimize the immobilization protocol, and the best-performing protocol was elaborated for microfluidics fabrication. A comparison between the batch and microreactor systems revealed that higher immobilized protein per area could be achieved with the microfluidic system providing with an efficient use of materials surface for biomolecular interactions. The versatility of the 3D printing technology offered the opportunity to improve the system performance by designing and testing different reactors, and thus investigate the optimum enzyme immobilization yield, bioreactor area per volume, and continuous flow operation. As a result, 12.5 Units of immobilized enzyme on 3D-printed microfluidic bioreactors produced 43.2 mg of (S)-1-phenylethanol with >99% ee, while the previously reported milliscale reaction gave 98 mg of product, catalyzed by 50–250 Units of free enzyme, with up to >99% ee.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 386
386![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 was achieved, indicating the viability of the designed system for repeated use of the immobilized biocatalyst. The versatility of the proposed reactor was demonstrated with the continuous flow biotransformation of several phenolic compounds from edible sources, reaching high conversion efficiencies in only one hour of residence time with mild reaction conditions.
000 was achieved, indicating the viability of the designed system for repeated use of the immobilized biocatalyst. The versatility of the proposed reactor was demonstrated with the continuous flow biotransformation of several phenolic compounds from edible sources, reaching high conversion efficiencies in only one hour of residence time with mild reaction conditions.
The same research group, in a next attempt, used this chitosan-modified 3D-printed microreactor to perform biocatalysis in deep eutectic solvents (DESs)-based media,28 that have been previously described as efficient solvents for biocatalytic processes.34 This time using lipase from Candida antarctica, the continuous flow concept was used to investigate biocatalytic characteristics of the immobilized lipase like enzyme concentration, continuous flow kinetics in the presence or absence of DESs, operational stability, and storage stability in 100 vol% DES. Notably, the modified reactors maintain their rigidity after exposure to 100 vol% DES, demonstrating the potential of biobased 3D-printed materials for use with alternative solvents. Further findings were the solvent stabilizing effect on enzymatic activity, even at 100 vol% DES concentration, the 90% residual activity of the immobilized enzyme at 4 °C after two months of storage, and that the presence of the DES betaine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol in the reaction system improved the apparent kinetic constant (KM,app) of the enzyme greatly. The enzyme-microreactor system retained 53% of its activity after 30 days of storage in 100 vol% DES at 60 °C, while it got completely deactivated when stored in buffer solution. The study also exhibits a continuous flow transesterification reaction to produce glyceryl ferulate, a hard-to-obtain natural antioxidant, showing higher productivity than the batch reaction with a commercially available immobilized enzyme preparation.
glycerol in the reaction system improved the apparent kinetic constant (KM,app) of the enzyme greatly. The enzyme-microreactor system retained 53% of its activity after 30 days of storage in 100 vol% DES at 60 °C, while it got completely deactivated when stored in buffer solution. The study also exhibits a continuous flow transesterification reaction to produce glyceryl ferulate, a hard-to-obtain natural antioxidant, showing higher productivity than the batch reaction with a commercially available immobilized enzyme preparation.
2.2 3D printing the reactor matrices
An example of 3D-printed packing material for reactors was developed in the work of Büscher et al. (2020).30 This study describes a tailor-made structure, acting as an enzyme immobilization matrix and an extraction-phase distributor at the same time. The motive of the study was the precise control of geometry that can directly affect the flow behavior of an organic extraction phase and can be readily achieved with additive manufacturing. Indeed, 3D printing allowed the researchers to investigate different reactor internal structures, and hence improve their influence on extraction relevant parameters, like the surface-to-volume ratio, the droplet size, and the residence time. Selective laser sintering (SLS) was chosen as a 3D printing approach that offers high surface roughness which can be beneficial for enzyme immobilization. Phenolic acid decarboxylase (PAD) from Mycobacterium colombiense was immobilized on a matrix material made of 3D-printed polyamide 12 located in a glass column reactor system. The aim was to perform ferulic acid decarboxylation to 2-methoxy-4-vinylphenol (MVP), implementing in situ extraction of MVP with an organic solvent. PAD has been shown to undergo product inhibition under the conditions (Ki,MVP/KM,FA = 0.06),38 so the importance of in situ product extraction is underlined by the authors. This type of reactor had been previously used for ferulic acid bioconversion. Still, it was the first time that a study focused on improving the mass transfer of the reaction product to the organic extraction phase. By doing so, 85% conversion was observed in the first hour of reaction, while 83% of the product was extracted in the first 15 minutes of operation. The newly developed matrix material also contributed to an enhanced surface area, compared to previous studies, for an increased enzyme loading that is substantial for conversion efficiency. Overall, this study is a good example of bioprocess intensification by combining biocatalysis and in situ extraction within a countercurrently operated reactor utilizing 3D-printed packing material.Also, the work by Valotta et al. (2021)31 locates in the field of 3D printing reactor inserts for continuous flow biocatalysis. PAD from Bacillus subtilis was the enzyme of choice, this time for the decarboxylation of coumaric acid to vinylphenol. The enzyme was covalently immobilized on the 3D-printed support structures after proper support functionalization to introduce reactive groups on its surface. Different geometries of the inserted structures were assessed in terms of flow behavior with a residence time distribution (RTD) approach. Ceramics were used as the 3D-printed material in a standard vat photopolymerization (VPP or commonly SLA) printing technique. The choice for ceramics, instead of standard 3D-printable polymer resins, resulted in great enzyme-carrier compatibility, demonstrated by long-term activity and stability. The ‘freedom of design’ offered by 3D printing resulted in an optimum lattice structure with a high surface area, reaching 58% immobilization yield and 6.02 U g−1 enzymatic activity. The continuous flow bioconversion was performed in an automated screening set-up for the optimization of the reaction. 3D printing was employed also for the fabrication of static mixers to provide the system with a feed stream of uniform concentration before it reaches the reactor inlet. Applying a design of experiments (DoE) approach with this setup, a space-time yield (STY) of 39.21 g (L h)−1 was reached, outperforming the previously reported by eight-fold. The authors claimed that the possibility to easily design and manufacture interchangeable inserts via 3D printing allows for rapid adaptation to the reaction needs, thus achieving optimal process conditions. This work was also a good example of realizing functional low-cost alternatives to analytical equipment that increased the level of process understanding.
3. 3D printing the biocatalysts
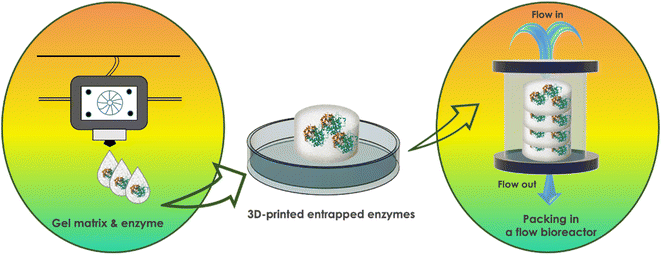
3D printing of the biocatalyst requires formulating it with support material, typically hydrogel-forming polymeric pregels, and using this mixture as a 3D-printable ink or resin (Fig. 3). Bioinks and bioresins are obtained when living cells are used as the biocatalyst and mixed in the polymeric pregel. The resulting 3D printing process is called ‘bioprinting’.39 It involves assembling cellular bio-entities via extrusion-based printing of one or multiple bioinks or, less commonly, by crosslinking the bioresin layer-by-layer via SLA- or digital light processing (DLP)-based printing to create bio-engineered living materials. In contrast, formulating active enzymes into a polymeric pregel without cells results in a bioactive and/or biocatalytic ink or resin to 3D-print bioactive scaffolds or matrixes for biomedical applications or biotechnological processes. The striking argument for 3D printing of the biocatalyst compared to a simple casting is the potential to easily control the 3D shape of the resulting biocatalytic matrix as well as the spatial distribution and/or local concentration of the biocatalyst in a modular fashion, including multiple-enzyme or cellular coculture systems for cascade biotransformations.3.1 3D printing of cells
For whole-cell biocatalysis, only a few examples have been described so far involving yeast cells to develop living materials for biocatalytic process intensification or bacteria-algae co-cultures for bioremediation processes.40–42 However, with the continuous bioprinting advancements in the field of tissue engineering and regenerative medicine, naturally focusing on mammalian cell printing, the dawning of bioprinted whole-cell catalysts can be foreseen with a shift towards prokaryotic cell printing.43 A pioneering example in that direction was reported in 2021 by Duraj-Thatte et al.,44 who established a ‘microbial ink’ and printed functional living materials from the shear-thinning hydrogel based on fibrin-inspired nanofibers produced by genetically engineered E. coli. The inks contained different genetically engineered E. coli to demonstrate their functionality and application diversity with, e.g., chemically-induced, on-demand production and extracellular secretion of azurin as a potential anti-cancer drug. Similarly, Cui et al. established the extrusion-based 3D printing of hyaluronic acid-producing Streptococcus zooepidemicus in a conventional photocrosslinkable gelatin-based matrix often used for mammalian cell printing.45 They demonstrated the benefits of 3D printing for the yield and production efficiency of the bacterial metabolite hyaluronic acid due to improved mass transfer within bacteria-containing hydrogel grid structures with optimized geometry and high surface-to-volume ratios compared to bulk gels. Because extrusion-based bioprinting has virtually the same ink requirements for eukaryotic and prokaryotic cells in terms of overall biocompatibility, mechanical integrity and stability of printed structures, and reduction of shear stress relative to the cell membrane, many of the inks previously developed for mammalian cells can also be used for prokaryotes. Besides extrusion- also SLA-based bioprinting has been demonstrated to be applicable to print bacterial cells.46In general, whole-cell biocatalysis is especially advantageous for cofactor-dependent reactions since the inherent presence of cofactors that are generated by the host cell and the ease of their recycling greatly improve the economics of the process.47 The potential to convert cheap and abundant raw materials in multistep reactions into valuable chemicals without time- and material-intensive enzyme isolation and purification, thus often simplifying both up- and downstream processing, contributes to their economic attractivity. Adverse substrate transfer limitations across the cell membrane can be overcome by printing surface-engineered cells, which display heterologous enzymes extracellularly.48 Hence, with the many powerful tools available nowadays to engineered both cells and inks, rapid advances in the field of bioprinting sustainable living materials for whole-cell biotransformations in continuous flow systems are to be expected.
3.2 3D printing of enzymes
When it comes to bioactive cell components, i.e., enzymes, we can find more literature reports, some of which also dealing with flow biocatalysis. A list of the 3D-printed biocatalysts so far and their applications is presented in Table 2. Previous review papers have also described the advances in biocatalysts incorporation in 3D-printable matrices. Focusing on the 3D-printed enzymes, the idea is based on the search for new immobilization carriers since conventional carriers, in the form of beads and granules for packed-bed reactors, do not allow for complex 3D structures with the desired shapes, sizes, and physical properties needed to produce customized reactor components for biocatalytic applications.49 Simple one-step fabrication methods incorporate enzymes in the printing material (entrapment), resulting in high enzyme activity retention due to the mild non-covalent immobilization conditions. However, careful consideration of enzyme compatibility with the raw printing materials is needed since printability and enzyme activity are directly influenced. The advantages, challenges, and future perspectives of 3D printing of enzymes entrapped in hydrogels are presented in recent reviews.49,50 An extrusion-based 3D printing technique is commonly employed for polymer-based enzyme formulations, which physically gel upon, e.g., temperature change or covalently crosslink upon irradiation of the printed structures in the presence of a suitable photoinitiator. Physical thermogelation is generally advantageous over chemical photocrosslinking in biocatalyst printing, as it does not produce radicals potentially affecting the encapsulated enzyme's structure and activity or require post-print processing steps. Furthermore, compared to living tissue printing, the requirements for printing resolution are generally lower in biocatalyst printing as it hardly affects the performance of the biocatalytic process. Thus, the often-encountered limited printing resolution of extrusion-based printers – particularly with thermogelling materials – compared to resin-based SLA printers can be neglected for most matrix-embedded biocatalyst prints. However, temperatures up to 60 °C may be needed in extrusion-based printing, again particularly with thermogelling inks, to ensure the optimal ink viscosity for an efficient printing process. Thus, for such applications, a high thermostability of the used enzymes is a prerequisite.51 In the following paragraphs, we will examine some examples from the literature that have employed this immobilization method to develop continuous flow biocatalytic reactors.| Biocatalysts | Technique | Material | Comments | Ref. |
|---|---|---|---|---|
| Alcohol dehydrogenase from Lactobacillus brevis, benzoyl formate decarboxylase from Pseudomonas putida, β-galactosidase from Aspergillus oryzae | Pneumatic extrusion-based 3D printing | Poly(ethylene glycol) diacrylate (PEG-DA) | Advantages and limitations of 3D-printed reactors with entrapped enzymes for biocatalysis | 53 |
| β-galactosidase from Aspergillus oryzae | Pneumatic extrusion-based 3D printing | Poly(ethylene glycol) diacrylate (PEG-DA) with viscosity-enhancing additives Laponite RD, Deuteron XG, Bentone MA | Development of a 3D-printable hydrogel material for long-term biocatalytic applications | 54 |
| Laccase from Trametes versicolor | Extrusion-based bioprinting | Sodium alginate-acrylamide hydrogel with hydroxyapatite additive | Biodegradation of the phenolic compound p-chlorophenol | 55 |
| β-Galactosidase | 3D jet writing | Poly(ethylene glycol) (PEG) and poly(acrylic acid) (PAA)-based hydrogels mixed with poly(ethylene glycol)diacrylate (PEGDA) | Evaluation of enzyme activity, reactants transport capacity, and long-term stability after immobilization in a hierarchically defined scaffold | 56 |
| Glucose oxidase, catalase | Extrusion-based bioprinting | Sodium alginate/polyacrylamide/hydroxyapatite (SA/PAM/HA) hybrid interpenetrating polymer network (HIPN) hydrogel | One-pot multi-enzyme immobilization for the synthesis of gluconic acid | 57 |
| Tyrosinase | Extrusion-based bioprinting | Gelatin-methacryloyl (GelMA)-collagen hydrogels | Exploration of the dual role of tyrosinase: hydrogel cross-linking and skin regeneration process | 58 |
| Cellulase | Pneumatic extrusion-based 3D printing | Cellulose nanofiber (CNF) reinforced chitosan (CHI) hydrogels | Controlled enzyme release for the inherent biodegradation of implantable biomaterials for tissue engineering | 59 |
| Yeast cells, and Bacillus subtilis co-entrapped with Chlorella vulgaris | Extrusion-based 3D printing | cucurbit[8]uril (CB[8])-mediated host–guest complex and methacrylate-based photochemically double-crosslinked dynamic bioink based on hyaluronic acid | Yeast-laden materials for the fermentation of glucose to ethanol and, bacteria-microalgae cocultured living materials system for bioremediation | 42 |
| Baker's yeast (Saccharomyces cerevisiae) | Pneumatic extrusion-based 3D printing | Polyethylene glycol dimethacrylate (PEGDA) with nanocellulose filler | Glucose fermentation | 40 |
| Yeast cells (Saccharomyces cerevisiae) | Direct-write 3D printing | F127-dimethacrylate (F127-DMA)-based hydrogel | Bioethanol production | 41 |
| Amine dehydrogenase variant (AmDH-v1), formate dehydrogenase (Cb-FDH) | Stereolithography | Agarose gel | Reductive amination of benzaldehyde in continuous flow | 60 |
| Ketoisovalerate decarboxylase (KIVD) mutant from Lactococcus lactis | Pneumatic extrusion-based 3D printing | Agarose hydrogel | Two-step biotransformation of ketoisovalerate (KIVD-catalyzed decarboxylation) into isobutanol | 52 |
| Alcohol dehydrogenase (ADH) from Alicyclobacillus acidocaldarius | ||||
| Phenacrylate decarboxylase from Enterobacter sp. | Pneumatic extrusion-based 3D printing | Agarose hydrogel | Continuous flow production of 4-hydroxystilbene from p-coumaric acid | 61 |
Maier et al. (2018) demonstrated the development of biocatalytic flow reactor cartridges composed of only the enzyme and an inexpensive agarose matrix that can be produced on-site, on-demand, and are biodegradable after use.52 Esterase (EstII) and alcohol dehydrogenase (ADH) from thermophilic organisms and an engineered decarboxylase from a mesophilic source were evaluated as the biocatalytically active ink's components. It is worth here to mention that the authors used the term ‘bioink’ for the described application of isolated enzymes, which we herein prefer to call ‘biocatalytically active inks'. After the preparation of the enzyme-containing inks by mixing the buffered agarose polymer solution with the respective purified enzyme at elevated temperature, their activity in fresh and temperature-treated gelled samples was analyzed. Therefore, the samples were incubated at 60 °C, simulating the temperature of the printer shell. Their residual EstII-catalyzed de-esterification activity towards a fluorescein derivative and their ADH-catalyzed reduction to yield isobutanol were analyzed in comparison with untreated controls. It was found that the enzymes could be incubated at 60 °C without significant activity loss – around 80% residual activity was detected after 240 min. Thus, after the 15 min printing process, the enzymatic activity of 85 ± 9% for ADH and 96 ± 12% for EstII was maintained.
The proof-of-concept under flow conditions was demonstrated with a mesophilic enzyme designed with protein engineering to broaden the applicability of the method to non-naturally thermostable variants. The enzyme of the first step of the cascade was the engineered ketoisovalerate decarboxylase (KIVD_mutant). A recovered activity of 36 ± 7% was detected after the printing process for the decarboxylation reaction of ketoisovalerate at 60 °C. Under flow conditions in the first 40 min of operation, the 3D-printed engineered enzyme reached 40% product yield in transforming ketoisovalerate to isobutyraldehyde within the agarose matrix. A flow reactor system was later used for the two-step biotransformation of ketoisovalerate into isobutanol catalyzed by KIVD_mutant and ADH 3D-printed enzyme-agarose disks in a cascade fashion. Isobutanol yield was found to be ADH dependent, as adding more ADH disks to the system increased the final product concentration. The authors argued that the 3D-printed hydrogel structures enabled the direct flow of the substrate solution inside the reactor and estimated the diffusion into the gel to reach a rate of 100 μm s−1. Data were also provided for the enhanced tolerance of the agarose-entrapped enzymes towards organic solvents, proving the agarose matrix's significant protection of the enzymes. Molecular weight-dependent enzyme leaching from the agarose gel matrix was observed and quantified with western blot analysis of the outflow. This revealed a maximum enzyme elution of 0.9% for ADH, and 0.7% for KIVD per mL flow-through. The proposed setting could withstand operational volumes of up to 70 mL of product. The study showed that this 3D printing approach allowed for the rapid prototyping of the desired structures, enabling systematic screening of construction parameters and optimization of reactor performance by modulating the shape and surface/volume aspects, utilizing inexpensive materials and simple immobilization methods.
In the subsequent research from the same group, Peng et al. (2019) demonstrated the 3D printing of biocatalytic gel modules to be integrated into a chemoenzymatic reaction cascade that would produce 4-hydroxystilbene, a pharmacologically relevant compound.61 Different phenacrylate decarboxylases were screened for their thermostability since 60 °C was needed for the extrusion-based 3D printing process (initial heating step at 60 °C for 15 min). The enzyme variants were mixed with 2-hydroxyethyl agarose solutions to formulate the biocatalytically active inks and print the gel-based reactor modules. After integration in a flow reactor system, outflow analysis revealed one enzyme variant with higher thermostability and product yield than the others. It is important to notice, though, that a time-dependent decrease in the conversion rate was observed and was related to the leaching of the encapsulated enzyme, as previous studies had similarly shown. After an optimization process, an efficiency of 98% over 40 hours of use was obtained. A scale increase by numbering-up was also demonstrated, leading to the conversion of 35 mg p-coumaric acid in a total volume of 211 mL with an average total turnover number (TTN) of 590 (4-vinyl phenol molecules per enzyme). The outflows were collected, and an isolated yield of 54% was obtained. 4-Hydroxystilbene was produced in a yield of 15% after a chemocatalytic step. The authors conclude that the proposed method can be elaborated to easily screen new enzyme classes compatible with 3D bioprinting. Also, the manufacturing process facilitated the modularity and scalability of the reactor system, introducing new enzyme classes into flow reaction systems.
Croci et al. (2022) approached the matter from a different perspective. They compared the hydrogel-based 3D-printed enzymes with conventional immobilization techniques – affinity or covalent – on commercial bead carriers in batch and continuous flow systems.60 The application focused on producing α-chiral amines, found in 40% of the commercialized optically active pharmaceutical intermediates. The reductive amination of benzaldehyde to benzylamine was performed by co-immobilizing a novel engineered amine dehydrogenase variant (AmDH) and formate dehydrogenase (FDH) as an NADH-recycling enzyme. For comparison with the commercial carriers, cation-affinity binding with EziG (preloaded with Fe3+) and Purolite (pre-loaded with Co2+) beads, or Sepabeads (EC-EP/s) and Relizyme (113/s and 403/s) for epoxide-amine covalent attachment were used for AmDH and FDH co-immobilization. Several issues were encountered with the commercial beads, from low product yields in some cases to substrate adsorption and enzyme leaching resulting in the incompatibility of the tested methods for establishing a reductive amination process in flow. Therefore, the researchers developed an ‘agarose gel reactor’ of pre-defined geometry and proposed a way to enable the enzyme entrapment at a lower temperature (40 °C) to be generally applicable also with mesophilic enzymes. For this reason, 3D-printed enzyme-agarose hydrogels were cast at 40 °C in a 3D-printed mold made of methacrylate-based resin. Different reactors and reactor channels were tested without re-printing the enzyme-containing module. Sufficient stability and reusability of the enzyme-loaded gel matrix were observed. An optimized process was performed over the course of 120 hours with 30 mM substrate concentration, and the system afforded 47% analytic product yield, of which 34% was isolated by extraction. Finally, the authors demonstrated an improved efficiency of the amination reaction in terms of product formed per amount of biocatalysts by implementing co-entrapped AmDH/FDH enzymes in a continuous flow cascade reaction. Detailed tables for a direct comparison of the amination reaction in the proposed system with previously established batch systems were also provided, highlighting a TTN of 3239 in flow versus the previously reported value of 943.
4. 3D printing of reaction apparatus
Apart from directly applied biocatalytic modules, 3D printing has also shown great potential in designing the desirable apparatus to facilitate the biocatalytic process. Researchers generally prefer to flexibly design and implement their accessories and devices to their overall system needs by modifying and adjusting in a ‘trial-and-error’ mode. In contrast, working with fixed commercial products and trying to fit them into the respective system is often process-limiting and laborious. The preferable 3D printing technique here is fused deposition modeling (FDM) because of the quick design and modular parts' mechanical robustness. Due to the vast choice of thermoplastic materials, the mechanical and chemical stability of the final additively manufactured product can easily be adjusted to and matched with the requirements of the desired application. In literature, we can find numerous examples with 3D-printed apparatus for fluidic applications:62 molds and scaffolds as an indirect approach to develop fluidic networks, complex structures and devices like microvalves, pumps, and multiplexers, cell sorters for cell biology applications, microfluidic tools for immunodetection, or even point-of-care (POC) devices manually operated. An exciting approach by Hou et al. (2021) proposed the automatic generation of 3D-printed reactionware for chemical synthesis through a computational platform that predicts and generates the specific reactor (tailored for the desired geometry, flow profile, dimensions, or also considering the material choice) fitted for the target chemical reaction.63In the field of flow biocatalysis, Wunschik et al. (2020) adopted the 3D printing technology to develop a reactor module for the enzymatic epoxidation of cyclohexene by a textile-immobilized peroxidase.32 This module would host the biocatalytic textile to realize a continuous flow process. The shift from a batch to a continuous process in this work was performed as a tool to tackle enzyme inhibition by the substrate hydrogen peroxide. At the same time, it also allowed for a modular design to fit into the specific process needs. Moreover, continuous product extraction could be performed while maintaining maximum biocatalyst activity.
For this reason, the FDM approach was adopted with a transparent polyester copolymer filament based on terephthalic acid to fabricate a two-compartment reactor. The upper chamber was filled with cyclohexene as a substrate, and the bottom part with acetonitrile as an extraction solvent. Peroxidase enzyme was immobilized on the textile and placed at the phase boundary. Cyclohexene oxidation occurred at the textile upon influx of hydrogen peroxide from the cyclohexene overlaying aqueous phase. The formed epoxide was quickly removed by extraction to the organic phase on the other compartment side of the reactor. Hence, a membrane-type reactor was developed with a low and continuously increasing concentration of the inhibiting substrate (hydrogen peroxide). Notably, the yields obtained in the developed reactor were enhanced by a factor of 30 compared to the reactions performed in flasks. The authors conclude that only a conjunction of immobilization technique and reactor design can lead to a new economic chemo-enzymatic reaction route.
In a different approach, Kim and colleagues (2020) demonstrated the usefulness of AM-enabled design freedom for an interfacial device that could facilitate the biocatalytic CO2 conversion at a gas–liquid interface.33 The enzyme of interest here was carbonic anhydrase (CA), immobilized onto electrospun fibers as a promising carrier for CA that was previously proved to enhance enzyme stability significantly.64 Although immobilized CA has already been utilized in previous studies, the advancement of this work was the interfacial positioning of the biocatalyst, resembling the natural CO2 sequestration. Such precise positioning could be effectively realized using 3D printing technology that allowed for the design of a density-adjustable platform consisting of biocatalytic and floating units. FDM printing technique with a widely used acrylonitrile-butadiene-styrene (ABS) copolymer filament was employed. The flotation level of the floating units was easily controlled by changing the void volume of the 3D models. For the bioreactor part, different modules for each step of the reaction were assembled, and the position of the immobilized CA fibers was investigated. The highest interfacial CO2 conversion was obtained when the CA fibers were located right at the air–liquid interface (flotation level 0 cm). A gradual performance decrease was observed at higher flotation levels, reaching the range of non-enzymatic processes at levels −2 and −2.5 cm. This way, it was proved that enzymatic CO2 conversion primarily occurs at the air–water interface, suggesting the importance of adequately positioning immobilized CA or other enzymes for successful interfacial biocatalysis. Adopting the proposed 3D-printed interfacial device could facilitate the control of the spatial positioning at the biphasic interface simply by adjusting the density of the floating enzyme carrier.
5. Sustainability assessment
It is worth to examining how we can address the sustainability and efficiency by the design of 3D printing for flow biocatalysis covering (i) materials and (ii) time-to-process. The advantages that flow biocatalysis can bring alone are not covered since this has been extensively reviewed before, as highlighted in the Introduction.5.1 Materials
To start with, a life cycle assessment of the materials used in the most common 3D printing technologies has been described in several review articles, while ‘case studies’ are currently being conducted to assess the recycling and reusing potential of these materials. There are some analytic reports3,4 on sustainability aspects of additive manufacturing that the reader can be redirected to. More specifically, there have been detailed references for (i) recyclable and biobased photoresins,2 (ii) recycled and biodegradable filaments,65 and (iii) sustainable hydrogels that have been very recently described in a dedicated book.66 For example, PLA (bio)degradation and recycling has been under intensive study67–70 since it is the most commonly used material in the 3D printing world. Thus, chemical,71 mechanical,72 and biological methods73,74 have already been described, while there are also companies specializing in PLA and other 3D-printed filaments recycling. Shredding and re-extruding are also widely applied by individuals and companies (Filabot, Dyze Design, etc). Table 3 reports recent examples of existing flow technologies and their materials for biocatalytic applications. These data suggest plenty of room for 3D-printed materials to invade and change the sustainability aspects of currently applied flow technologies. Commonly used polymer materials like PMMA and PTFE have already entered the market in 3D-printable forms. Regarding hydrogels for enzyme- or cell entrapment, we already highlighted in the previous sections the contribution of 3D printing that several laboratories increasingly elaborate on. When it comes to the most common material in terms of all kinds of reactors, glass is already on the market for 3D printing, and maybe such materials could already start to replace some traditionally manufactured glass microstructures.75–77| Material | Flow technology | Productivity | Ref. |
|---|---|---|---|
| Cellulose microcrystalline Avicel® PH-101 | Immobilized protein preparations packed into an Omnifit column | 0.37 gproduct L−1 h−1 | 78 |
| Agarose epoxy beads, polytetrafluoroethylene (PTFE) | Immobilized enzymes preparations packed into a custom-made PTFE reactor | 19.31 gproduct d−1 L−1 | 79 |
| Poly(methyl methacrylate) (PMMA) | Crude-enzyme hydrogel monolithic microreactor | 46.3 gproduct L−1 h−1 | 80 |
| Hydrogel comprising dimethacrylate-functionalized Pluronic F127 (F127-DMA) and sodium alginate (Alg) | Packed column | 74.6% dye degradation | 81 |
| Purolite resin beads | Immobilized enzyme beads packed into an Omnifit column | 10.6 gproduct genzyme−1 | 82 |
| Magnetic nanoparticles, PTFE | Magnetic nanoparticles loaded tube microreactor | 11.6 mmol L−1 h−1 | 83 |
| Commercially available streptavidin (STV)-coated magnetic particles (MB), PMMA | Straight channel microreactor made of PMMA packed with magnetic particles with immobilized enzyme | 28.9 mol L−1 h−1 | 83 |
| Epoxide microparticles, PTFE | Reactor chamber made of PTFE packed with microparticles with immobilized cells (all-enzyme hydrogel particles) | 18.8 gproduct L−1 d−1 | 84 |
| Perfluoroelastomer (PFE) | Segmented-flow PFE coil microreactor with whole-cells | 12.5 gproduct L−1 h−1 | 85 |
| Carbon particles, glass beads | Omnifit glass column packed with carbon and glass beads with immobilized enzymes | 11.2 gproduct L−1 h−1 | 86 |
| Alginate hydrogel, glass, silicon, Teflon | Glass-silicon-glass microreactor in a Teflon housing loaded with alginate hydrogel and enzyme mixture | 52.94% and 33.61% conversion yields for two different products | 87 |
| Perfluoroalkoxy (PFA) | Flow reactor and Y-mixer made of PFA loaded with commercially available immobilized enzyme preparations | 85.2% main product yield | 88 |
| Multi-wall carbon nanotubes, quartz | Multi-wall carbon nanotube lined quartz tubes with surface immobilized enzyme | 38.7 gProduct L−1 h−1 | 89 |
| Borosilicate glass, zinc oxide (ZnO) nanowires | Glass capillaries with in situ grown ZnO nanowires and surface immobilization of enzyme | 0.29 mgproduct min−1 mgenzyme−1 | 90 |
5.2 Time-to-process
Flow biocatalysis meets essential requirements of sustainable processes, like high reusability of the systems, time and cost-efficiency with low reaction times and small-scale equipment, and minimal waste production.7 Incorporating additive manufacturing in the establishment of biocatalytic flow systems has a strong potential to enhance these aspects further. In chemical engineering, including the field of biocatalysis in flow systems, highly sophisticated systems are typically required to establish precise process control and reassure the biocatalyst's longevity. We saw in this review examples of substantial productivity enhancements when shifting to a continuous flow regime with the help of 3D printing. It can be predicted that the easy-to-adjust trait of this technology will be increasingly elaborated in future biocatalytic applications. Fluidic systems possess modularity, so there is a great need for accordingly modular accessories and reactionware that would fit a specific process. Moreover, the opportunity to 3D-print the enzyme, protected inside its scaffold, offers an alternative immobilization approach with promising results for a cost-effective and green process design.6. Conclusion and future perspectives
In conclusion, it is foreseen that the principles of ‘quality of design’ and ‘quality by control’ could be applied since the combination of flow biocatalysis and additive manufacturing leads to automated and/or highly controlled processes. From design to implementation, the time and costs are constantly being reduced, framing not only innovative but also sustainable processes. The joined forces of materials science and bioprocess engineering will profit extensively regarding the tailor-made material design that provides the desired characteristics, including biodegradation and functionality. The targeted material–biomolecular interactions will pave the way for highly efficient continuous biocatalytic production. Additive manufacturing has undoubtedly changed scientists' way of thinking due to technology-enabled freedom of experimental design. Developing custom devices has now become an everyday practice for numerous laboratories, as the costs of this technology are getting consistently lower. It is important to note, though, that an important barrier to the industrial adoption of such processes is print-to-print inconsistency.91 Reasons for this can be found in the differences between batches of materials and the inability of the 3D printing systems to couple the deposition parameters to the dynamic material properties. Scientists are already on their way to overcoming issues like this by adopting artificial intelligence (AI) to design closed-loop control systems and achieve optimum printing consistency.92 This way, we can expect a broader implementation of highly intricate systems in chemical engineering sciences that would unlock tremendous potential for the future of biocatalytically produced compounds.Author contributions
E. Gkantzou and S. Kara conceived the conception and design of this work. S. Kara was responsible for funding acquisition and project administration. All authors contributed to the writing of the original draft, reviewed, and edited the final draft, and agreed with the final version of the submitted manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. K. thanks the Independent Research Fund Denmark, PHOTOX-f project, grant no. 9063 00031B, for the grant funding. S. K. thanks the Ministry for Science and Culture for Lower Saxony for the Holen & Halten starting grant (grant no. 12.5-76251-17-9/ 20).References
- M. Touri, F. Kabirian, M. Saadati, S. Ramakrishna and M. Mozafari, Adv. Eng. Mater., 2019, 21 Search PubMed.
- K. P. Cortés-Guzmán, A. R. Parikh, M. L. Sparacin, A. K. Remy, L. Adegoke, C. Chitrakar, M. Ecker, W. E. Voit and R. A. Smaldone, ACS Sustainable Chem. Eng., 2022, 10, 13091–13099 CrossRef.
- E. M. Maines, M. K. Porwal, C. J. Ellison and T. M. Reineke, Green Chem., 2021, 23, 6863–6897 RSC.
- E. Sanchez-Rexach, T. G. Johnston, C. Jehanno, H. Sardon and A. Nelson, Chem. Mater., 2020, 32, 7105–7119 CrossRef CAS.
- A. J. Capel, R. P. Rimington, M. P. Lewis and S. D. R. Christie, Nat. Rev. Chem., 2018, 2, 422–436 CrossRef.
- J. Britton, S. Majumdar and G. A. Weiss, Chem. Soc. Rev., 2018, 47, 5891–5918 RSC.
- L. Tamborini, P. Fernandes, F. Paradisi and F. Molinari, Trends Biotechnol., 2018, 36, 73–88 CrossRef CAS.
- P. De Santis, L.-E. Meyer and S. Kara, React. Chem. Eng., 2020, 5, 2155–2184 RSC.
- P. Žnidaršič-Plazl, Biotechnol. J., 2019, 14, 1800580 CrossRef PubMed.
- L.-E. Meyer, M. Hobisch and S. Kara, Curr. Opin. Biotechnol., 2022, 78, 102835 CrossRef CAS PubMed.
- G. Vernet, M. Hobisch and S. Kara, Curr. Opin. Green Sustainable Chem., 2022, 38, 100692 CrossRef CAS.
- S. C. Cosgrove and A. P. Mattey, Chem.–Eur. J., 2022, 28(13), e202103607 CAS.
- A. J. Tušek, A. Šalić and B. Zelić, Appl. Biochem. Biotechnol., 2017, 182, 1575–1590 CrossRef PubMed.
- A. Valotta, L. Malihan-Yap, K. Hinteregger, R. Kourist and H. Gruber-Woelfler, ChemSusChem, 2022, 15, e202201468 CAS.
- M. P. Thompson, I. Peñafiel, S. C. Cosgrove and N. J. Turner, Org. Process Res. Dev., 2018, 23, 9–18 CrossRef.
- S. N. Chanquia, A. Valotta, H. Gruber-Woelfler and S. Kara, Front. catal., 2022, 1 Search PubMed.
- V. Alphand, W. J. H. van Berkel, V. Jurkaš, S. Kara, R. Kourist, W. Kroutil, F. Mascia, M. M. Nowaczyk, C. E. Paul, S. Schmidt, J. Spasic, P. Tamagnini and C. K. Winkler, ChemPhotoChem, 2023, 7, e202200325 CrossRef CAS.
- V. Sans, Curr. Opin. Green Sustainable Chem., 2020, 25, 100367 CrossRef.
- K. M. Zentel, M. Fassbender, W. Pauer and G. A. Luinstra, Adv. Chem. Eng., 2020, 97–137, DOI:10.1016/bs.ache.2020.08.002.
- L.-E. Meyer, D. Horváth, S. Vaupel, J. Meyer, M. Alcalde and S. Kara, React. Chem. Eng., 2023, 8, 984–988 RSC.
- T. Pose-Boirazian, J. Martinez-Costas and G. Eibes, Macromol. Biosci., 2022, 22, e2200110 CrossRef PubMed.
- M. Romero-Fernandez and F. Paradisi, Curr. Opin. Chem. Biol., 2020, 55, 1–8 CrossRef CAS PubMed.
- G. Palmara, F. Frascella, I. Roppolo, A. Chiappone and A. Chiado, Biosens. Bioelectron., 2021, 175, 112849 CrossRef CAS.
- E. Peris, O. Okafor, E. Kulcinskaja, R. Goodridge, S. V. Luis, E. Garcia-Verdugo, E. O'Reilly and V. Sans, Green Chem., 2017, 19, 5345–5349 RSC.
- J. Ye, T. Chu, J. Chu, B. Gao and B. He, ACS Sustainable Chem. Eng., 2019, 7, 18048–18054 CrossRef CAS.
- K. T. Sriwong and T. Matsuda, React. Chem. Eng., 2022, 7, 1053–1060 RSC.
- E. Gkantzou, A. Skonta, A. Tsakni, A. Polydera, D. Moschovas, K. Spyrou, A. Avgeropoulos, D. Gournis, D. Houhoula and H. Stamatis, J. Biotechnol., 2022, 350, 75–85 CrossRef CAS PubMed.
- M. G. Bellou, E. Gkantzou, A. Skonta, D. Moschovas, K. Spyrou, A. Avgeropoulos, D. Gournis and H. Stamatis, Micromachines, 2022, 13(11), 1954 CrossRef PubMed.
- M. Chai, S. Razavi Bazaz, R. Daiyan, A. Razmjou, M. Ebrahimi Warkiani, R. Amal and V. Chen, Chem. Eng. J., 2021, 426 Search PubMed.
- N. Büscher, C. Spille, J. K. Kracht, G. V. Sayoga, A. W. H. Dawood, M. I. Maiwald, D. Herzog, M. Schlüter and A. Liese, Org. Process Res. Dev., 2020, 24, 1621–1628 CrossRef.
- A. Valotta, M. C. Maier, S. Soritz, M. Pauritsch, M. Koenig, D. Brouczek, M. Schwentenwein and H. Gruber-Woelfler, J. Flow Chem., 2021, 11, 675–689 CrossRef CAS PubMed.
- D. S. Wunschik, K. N. Ingenbosch, P. Suss, U. Liebelt, S. Quint, M. Dyllick-Brenzinger, R. Zuhse, U. Menyes, K. Hoffmann-Jacobsen, K. Opwis and J. S. Gutmann, Enzyme Microb. Technol., 2020, 136, 109512 CrossRef CAS PubMed.
- H. S. Kim, S.-G. Hong, J. Yang, Y. Ju, J. Ok, S.-J. Kwon, K.-M. Yeon, J. S. Dordick and J. Kim, J. CO2 Util., 2020, 38, 291–298 CrossRef CAS.
- M. Pätzold, S. Siebenhaller, S. Kara, A. Liese, C. Syldatk and D. Holtmann, Trends Biotechnol., 2019, 37, 943–959 CrossRef PubMed.
- S. R. Bazaz, A. A. Mehrizi, S. Ghorbani, S. Vasilescu, M. Asadnia and M. E. Warkiani, RSC Adv., 2018, 8, 33103–33120 RSC.
- J. Hou, G. Qian and X. Zhou, Chem. Eng. J., 2011, 167, 475–482 CrossRef CAS.
- J. Yue, G. Chen, Q. Yuan, L. Luo and Y. Gonthier, Chem. Eng. Sci., 2007, 62, 2096–2108 CrossRef CAS.
- L. Pesci, M. Baydar, S. Glueck, K. Faber, A. Liese and S. Kara, Org. Process Res. Dev., 2017, 21, 85–93 CrossRef CAS.
- A. C. Daly, M. E. Prendergast, A. J. Hughes and J. A. Burdick, Cell, 2021, 184, 18–32 CrossRef CAS PubMed.
- F. Qian, C. Zhu, J. M. Knipe, S. Ruelas, J. K. Stolaroff, J. R. DeOtte, E. B. Duoss, C. M. Spadaccini, C. A. Henard, M. T. Guarnieri and S. E. Baker, Nano Lett., 2019, 19, 5829–5835 CrossRef CAS PubMed.
- A. Saha, T. G. Johnston, R. T. Shafranek, C. J. Goodman, J. G. Zalatan, D. W. Storti, M. A. Ganter and A. Nelson, ACS Appl. Mater. Interfaces, 2018, 10, 13373–13380 CrossRef CAS PubMed.
- F. He, Y. Ou, J. Liu, Q. Huang, B. Tang, F. Xin, J. Zhang, M. Jiang, S. Chen and Z. Yu, Small, 2022, 18, 2104820 CrossRef CAS PubMed.
- Y. Liu, X. Xia, Z. Liu and M. Dong, Small, 2023, 19, 2205949 CrossRef CAS PubMed.
- A. M. Duraj-Thatte, A. Manjula-Basavanna, J. Rutledge, J. Xia, S. Hassan, A. Sourlis, A. G. Rubio, A. Lesha, M. Zenkl, A. Kan, D. A. Weitz, Y. S. Zhang and N. S. Joshi, Nat. Commun., 2021, 12 Search PubMed.
- Z. Cui, Y. Feng, F. Liu, L. Jiang and J. Yue, ACS Macro Lett., 2022, 11, 452–459 CrossRef CAS PubMed.
- K. Dubbin, Z. Dong, D. M. Park, J. Alvarado, J. Su, E. Wasson, C. Robertson, J. Jackson, A. Bose, M. L. Moya, Y. Jiao and W. F. Hynes, Nano Lett., 2021, 21, 1352–1359 CrossRef CAS PubMed.
- B. Lin and Y. Tao, Microb. Cell Fact., 2017, 16, 106 CrossRef PubMed.
- M. R. Smith, E. Khera and F. Wen, Ind. Eng. Chem. Res., 2015, 54, 4021–4032 CrossRef CAS PubMed.
- J. Shen, S. Zhang, X. Fang and S. Salmon, Gels, 2022, 8(8), 460 CrossRef CAS PubMed.
- J. Meyer, L. E. Meyer and S. Kara, Eng. Life Sci., 2022, 22, 165–177 CrossRef CAS.
- Y. Shao, Z. Liao, B. Gao and B. He, ACS Omega, 2022, 7, 11530–11543 CrossRef CAS PubMed.
- M. Maier, C. P. Radtke, J. Hubbuch, C. M. Niemeyer and K. S. Rabe, Angew. Chem., Int. Ed. Engl., 2018, 57, 5539–5543 CrossRef CAS PubMed.
- B. Schmieg, J. Dobber, F. Kirschhofer, M. Pohl and M. Franzreb, Front. Bioeng. Biotechnol., 2018, 6, 211 CrossRef PubMed.
- B. Schmieg, A. Schimek and M. Franzreb, Eng. Life Sci., 2018, 18, 659–667 CrossRef CAS PubMed.
- J. Liu, X. Shen, Z. Zheng, M. Li, X. Zhu, H. Cao and C. Cui, Int. J. Biol. Macromol., 2020, 164, 518–525 CrossRef CAS PubMed.
- A. Steier, B. Schmieg, Y. Irtel Von Brenndorff, M. Meier, H. Nirschl, M. Franzreb and J. Lahann, Macromol. Biosci., 2020, 20, 2000154 CrossRef CAS PubMed.
- X. Shen, M. Yang, C. Cui and H. Cao, Colloids Surf., A, 2019, 568, 411–418 CrossRef CAS.
- Y. Shi, T. L. Xing, H. B. Zhang, R. X. Yin, S. M. Yang, J. Wei and W. J. Zhang, Biomed. Mater., 2018, 13, 035008 CrossRef CAS.
- A. K. Tamo, T. A. Tran, I. Doench, S. Jahangir, A. Lall, L. David, C. Peniche-Covas, A. Walther and A. Osorio-Madrazo, Materials, 2022, 15, 6039 CrossRef CAS PubMed.
- F. Croci, J. Vilim, T. Adamopoulou, V. Tseliou, P. J. Schoenmakers, T. Knaus and F. G. Mutti, Chembiochem, 2022, 23, e202200549 CrossRef CAS PubMed.
- M. Peng, E. Mittmann, L. Wenger, J. Hubbuch, M. K. M. Engqvist, C. M. Niemeyer and K. S. Rabe, Chemistry, 2019, 25, 15998–16001 CrossRef CAS PubMed.
- C. K. Dixit, K. Kadimisetty and J. Rusling, Trends Anal. Chem., 2018, 106, 37–52 CrossRef CAS PubMed.
- W. Hou, A. Bubliauskas, P. J. Kitson, J.-P. Francoia, H. Powell-Davies, J. M. P. Gutierrez, P. Frei, J. S. Manzano and L. Cronin, ACS Cent. Sci., 2021, 7, 212–218 CrossRef CAS PubMed.
- S.-H. Jun, J. Yang, H. Jeon, H. S. Kim, S. P. Pack, E. Jin and J. Kim, Environ. Sci. Technol., 2020, 54, 1223–1231 CrossRef CAS PubMed.
- J. Pakkanen, D. Manfredi, P. Minetola and L. Iuliano, in Sustainable Design and Manufacturing, 2017, ch. 73, pp. 776–785 Search PubMed.
- M. B. Salehi and A. M. Moghadam, in Sustainable Hydrogels, ed. S. Thomas, B. Sharma, P. Jain and S. Shekhar, Elsevier, 2023, pp. 23–46 Search PubMed.
- P. Zhao, C. Rao, F. Gu, N. Sharmin and J. Fu, J. Cleaner Prod., 2018, 197, 1046–1055 CrossRef CAS.
- M. Dieterle and J. Ginter, Procedia CIRP, 2022, 105, 13–18 CrossRef.
- M. F. Cosate De Andrade, P. M. S. Souza, O. Cavalett and A. R. Morales, J. Polym. Environ., 2016, 24, 372–384 CrossRef CAS.
- B. Xu, Y. Chen, J. He, S. Cao, J. Liu, R. Xue, F. Xin, X. Qian, J. Zhou, W. Dong and M. Jiang, Environ. Rev., 2021, 30, 30–38 CrossRef.
- R. Yang, G. Xu, B. Dong, H. Hou and Q. Wang, Macromolecules, 2022, 55, 1726–1735 CrossRef CAS.
- J. Finnerty, S. Rowe, T. Howard, S. Connolly, C. Doran, D. M. Devine, N. M. Gately, V. Chyzna, A. Portela, G. S. N. Bezerra, P. McDonald and D. M. Colbert, J. Compos. Sci., 2023, 7, 141 CrossRef CAS.
- A. N. Mistry, B. Kachenchart, A. Wongthanaroj, A. Somwangthanaroj and E. Luepromchai, Polym. Degrad. Stab., 2022, 202, 110051 CrossRef CAS.
- J. Yu, P. D. Kim, Y. Jang, S.-K. Kim, J. Han and J. Min, Biodegradation, 2022, 33, 477–487 CrossRef CAS PubMed.
- V. Hotař, M. Stará, V. Máková and B. N. Holubová, Pure Appl. Chem., 2022, 94, 169–179 CrossRef.
- C. Liu, T. Oriekhov and M. Fokine, Front. Mater., 2022, 9, 978861 CrossRef.
- E. Gal-Or, Y. Gershoni, G. Scotti, S. M. E. Nilsson, J. Saarinen, V. Jokinen, C. J. Strachan, G. Boije af Gennäs, J. Yli-Kauhaluoma and T. Kotiaho, Anal. Methods, 2019, 11, 1802–1810 RSC.
- R. A. Rocha, L. Esquirol, V. Rolland, P. Hands, R. E. Speight and C. Scott, Enzyme Microb. Technol., 2023, 169, 110268 CrossRef CAS PubMed.
- S. Gallus, E. Mittmann, A. J. Weber, M. Peng, C. M. Niemeyer and K. S. Rabe, ChemCatChem, 2023, 15 Search PubMed.
- Q. Chen, Y. Wang and G. Luo, ChemSusChem, 2023, 16, e202201654 CAS.
- Y. Pinyakit, P. Romphophak, P. Painmanakul and V. P. Hoven, Biomacromolecules, 2023, 24, 3138–3148 CrossRef CAS PubMed.
- S. C. Cosgrove, G. J. Miller, A. Bornadel and B. Dominguez, ACS Sustainable Chem. Eng., 2023, 11, 8556–8561 CrossRef CAS.
- T. Burgahn, P. Pietrek, R. Dittmeyer, K. S. Rabe and C. M. Niemeyer, ChemCatChem, 2020, 12, 2452–2460 CrossRef CAS.
- P. Bitterwolf, A. E. Zoheir, J. Hertel, S. Kröll, K. S. Rabe and C. M. Niemeyer, Chem.–Eur. J., 2022, 28, e202202157 CrossRef CAS PubMed.
- N. Adebar, A. Nastke, J. Löwe and H. Gröger, Angew. Chem., Int. Ed., 2021, 60, 15863–15869 CrossRef CAS PubMed.
- B. Poznansky, L. A. Thompson, S. A. Warren, H. A. Reeve and K. A. Vincent, Org. Process Res. Dev., 2020, 24, 2281–2287 CrossRef CAS PubMed.
- A. Kazan, X. Hu, A. Stahl, H. Frerichs, I. Smirnova and O. Yesil-Celiktas, J. Chem. Technol. Biotechnol., 2021, 96, 3349–3357 CrossRef CAS.
- L.-H. Du, M. Xue, M.-J. Yang, Y. Pan, L.-Y. Zheng, Z.-M. Ou and X.-P. Luo, Catalysts, 2020, 10, 1419 CrossRef CAS.
- L. A. Thompson, J. S. Rowbotham, J. H. Nicholson, M. A. Ramirez, C. Zor, H. A. Reeve, N. Grobert and K. A. Vincent, ChemCatChem, 2020, 12, 3913–3918 CrossRef CAS.
- E. Gkantzou, K. Govatsi, A. V. Chatzikonstantinou, S. N. Yannopoulos and H. Stamatis, ACS Sustainable Chem. Eng., 2021, 9, 7658–7667 CrossRef CAS.
- C. Wang, X. P. Tan, S. B. Tor and C. S. Lim, Addit. Manuf., 2020, 36, 101538 Search PubMed.
- M. Piovarči, M. Foshey, J. Xu, T. Erps, V. Babaei, P. Didyk, S. Rusinkiewicz, W. Matusik and B. Bickel, ACM Trans. Graph., 2022, 41, 1–10 CrossRef.
| This journal is © The Royal Society of Chemistry 2023 |