 Open Access Article
Open Access ArticleHighly sensitive solid-state nanopore aptasensor based on target-induced strand displacement for okadaic acid detection from shellfish samples†
Mohamed Amin
Elaguech
 abc,
Yajie
Yin
ab,
Yunjiao
Wang
abc,
Yajie
Yin
ab,
Yunjiao
Wang
 ab,
Bing
Shao
ab,
Bing
Shao
 *d,
Chaker
Tlili
*d,
Chaker
Tlili
 *ab and
Deqiang
Wang
*ab and
Deqiang
Wang
 *abc
*abc
aChongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing, 400714, PR China. E-mail: chakertlili@cigit.ac.cn; dqwang@cigit.ac.cn
bChongqing School, University of Chinese Academy of Sciences (UCAS), Chongqing, 400714, PR China
cUniversity of Chinese Academy of Sciences (UCAS), Beijing, 100049 PR China
dDepartment of Veterinary Pharmacology and Toxicology, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China. E-mail: shaobingch@sina.com
First published on 13th October 2023
Abstract
Okadaic acid (OA) is a marine toxin that is frequently found in a wide variety of shellfish and can cause major health complications. A sensitive and selective nanopore-based aptasensor for detecting OA in shellfish is presented. The nanosensor, which is based on target-induced strand displacement reactions, is primarily composed of an OA-specific biotinylated truncated aptamer immobilized onto streptavidin-coated magnetic beads and its predesigned complementary signaling probe (cDNA) to form a dsDNA duplex. The aptamer tends to form a more stable OA–aptamer complex in the presence of OA, resulting in the displacement of the signaling probe (cDNA) from the dsDNA duplex. Using the optimal conditions, the nanosensor displayed a broad detection range of 1.0 pg mL−1 to 1.0 × 102 ng mL−1 with a limit of detection of 3.0 × 10−2 pg mL−1, showing that this method has higher sensitivity than most reported methods in the literature for OA detection and exhibited good selectivity against interfering small molecules. More significantly, the proposed nanosensor showed tremendous potential for OA detection from real samples; it was effectively used on shellfish samples with recoveries ranging from 95.33% to 110.03%. Overall, our findings suggest that the proposed nanosensor can pave the way for the development of high-performance sensing methodologies for marine biotoxins.
1. Introduction
Marine algae are among the most vital organisms supporting thriving marine ecosystems. Okadaic acid (OA) is a common by-product of harmful algal blooms (HABs) in the marine environment. OA is typically produced by algal genera such as Prorocentrum, Phalacroma, and Dinophysis.1 Numerous shellfish, including mussels, oysters, and clams, have been found to be contaminated with OA, most likely from consuming toxin-producing algae.2 Human consumption of these contaminated shellfish can result in severe health problems. High intake of OA can cause gastro-abdominal disturbances, such as diarrhea, vomiting, and nausea.3,4 Furthermore, OA may affect gene expression and normal cell activity, and even lead to tumor formation.5 Therefore, to minimize the risks associated with these toxins, several international food committees have established a maximum allowed intake of 1.6 × 102 μg kg−1.6As a consequence of strict regulations, various quantification approaches for OA determination have been developed to fulfill international requirements. Until 2011, the mouse bioassay (MBA) was considered as the gold-standard detection method for the detection of OA. However, this technique has a number of drawbacks, such as ethical concerns, lack of specificity to marine toxins, and low accuracy.7 Since then, conventional liquid chromatography coupled with tandem mass spectroscopy (LC-MS/MS) has been adopted as a viable alternative to MBA for routine OA quantification.8 More recently, with the development of biosensing techniques, various other detection strategies have been proposed. Biosensors can be an excellent alternative to the currently used techniques for OA detection due to their low cost, small size, sharp response time, and affordability. For example, Campbell and co-workers demonstrated the detection of OA on carbon black screen-printed electrodes decorated with an OA-specific protein-conjugate.9 Their result shows that the biosensor exhibited a low limit of detection (LOD) of 1.8 × 102 ng mL−1. Similarly, colorimetric immunoassay based on a double catalysis enhancement strategy has proven to be an effective detection method for OA, achieving results that are comparable to those of LC-MS.10 Additionally, it has been reported that a quantum dot nanobead-based fluorescence sensor has been successfully utilized for the detection of OA molecules.11 Based on their findings, this method offers a low LOD of 2.0 × 101 ng mL−1 and broad linear range.
Although conventional biosensors based on antibodies as a recognition element have shown outstanding performance, their high cost and complexity remain a bottleneck.12 To overcome these drawbacks, aptamers with high binding affinity toward OA have been identified. Aptamers are short synthetic oligonucleotide sequences with valuable properties,6 such as simplicity of synthesis, high binding affinity, thermal stability,13 and conformational changes upon binding with the target molecule, which allows for the development of direct sensing strategies.14 Recent studies have synthesized OA-specific aptamers with dissociation constants ranging from 2.77 nM to 3.8 × 102 nM and have integrated them into various sensing systems, including microfluidic aptasensors,12,15 microcantilever-array biosensors,14 fluorescence sensors,16 piezoelectric aptasensors17 and electrochemical sensors.18 For example, S. Eissa et al. synthesized a set of OA-specific aptamers with dissociation constant Kd values ranging from 7.7 × 101 nM to 3.8 × 102 nM.13 H. Gu et al. used the SELEX technique to synthesize different aptamers sequences for OA detection with Kd values ranging from 4.2 × 101 nM to 8.8 × 101 nM.2 More recently, Chinnappan et al. presented a truncated OA-specific aptamer with a low Kd value of 2.77 nM.19 Overall, aptamer-based sensors have shown satisfactory results in terms of sensitivity, selectivity, and limit of detection and have achieved international requirements.
During recent years, solid-state nanopore sensors have emerged as a promising alternative to existing biosensors. Nanopore sensors are label-free single molecule detection techniques that have gained tremendous attention due to their high sensitivity, cost-effectiveness, and stability in harsh chemical environments.20 Typically, a nanopore sensor consists of a nanometric orifice drilled in a free-standing membrane that allows for the translocation of target molecules through the pore under a transmembrane voltage, thereby providing valuable information about the molecules' sizes, charge polarity, and interaction with their chemical environment.21 Lately, nanopore-based sensors have demonstrated exceptional sensitivity down to the fM level,22 and have been successfully used for small molecule sensing through direct and indirect detection strategies.23,24 However, although nanopore aptasensors have been extensively applied for small molecule sensing, they have not yet been extensively applied for marine toxin detection, despite the importance of marine biotoxins.
In this context, a target-induced strand displacement assay combined with solid-state nanopores for screening OA in shellfish is described. First, a highly OA-sensitive truncated aptamer was immobilized on magnetic beads (MBs), and then a complementary signaling DNA (cDNA) was hybridized with the aptamer to form a dsDNA duplex. The addition of OA molecules will result in the melting of this dsDNA duplex, which will then allow the cDNA to be released into the solution. Subsequently, the cDNA-containing solution was translocated through a nanopore sensor. On the other hand, the cDNA sequences were tailed with poly(A), and the length of both cDNA and the poly(A) tail were tuned to boost the nanopore signal count. By employing this displacement-based signaling mechanism, the aptasensor's performance was significantly enhanced. Overall, the proposed aptasensor demonstrated outstanding performance without the need of any sensor functionalization, sample pre-treatments, or enzyme amplification.
2. Experimental section
2.1. Reagents
Nanopore chips (NBPT001YZ-HR) were purchased from Norcada. The chips used in this work are composed of a 20 ± 3 nm thick SiNx membrane suspended over a 200 μm Si substrate. Okadaic acid (OA) and microcystins MC-LR, MC-RR and MC-YR (purity >97%) were ordered from Taiwan Algal Science Inc. Dinophysistoxin-1 (DTX-1) (purity >97%) was purchased from the National Research Council of Canada. Streptavidin-decorated MBs (4 mg mL−1) were ordered from New England Biolabs Beijing, China. KCl, NaCl, NaOH, MgCl2, methanol, ethylenediaminetetraacetic acid (EDTA) and tris(hydroxymethyl)aminomethane (Tris) were all purchased from Sigma Aldrich, Shanghai. Tris–HCl was ordered from Biosharp China. Tween 20 was ordered from Sigma Life Science. Polyethylene glycol 4000 (PEG-4000) was purchased from Adamas Reagent Co., Ltd. The cDNA and aptamer sequences used in this work presented in Table S1† were synthesized by Sangon, Shanghai. The solutions used in this work were prepared using HPLC-grade purified water obtained from a Molecular 1850D purification system and filtered through 0.22 μm filters.2.2. Nanopore fabrication using dielectric breakdown technique
Prior to initiating the dielectric breakdown procedure, the nanopore chips underwent several cleaning steps to ensure their cleanliness and remove any organic contamination, following a previously reported protocol.25 Briefly, the process involved immersing the nanopore chip in ethanol at room temperature (25 °C) for 20 minutes, then transferring it to an 80 °C piranha solution (H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2 = 3
H2O2 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 60 minutes, followed by rinsing in three separate hot water baths for 10 minutes each. The cleaned chip was loaded into a custom-made PMMA flow cell with two chambers (cis and trans) filled with 200 mL of a translocation buffer (TB) consisting of 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8. Before initiating the controlled breakdown (CBD) process, the flow cell was positioned in a Faraday cage and connected to a Keithley 2450 source meter via two Ag/AgCl electrodes. During this process, the membrane's resistance, leakage current, and trans-membrane voltage of the SiNx membrane were continuously monitored while an initial current of 80 nA was applied, with subsequent increases of 5 nA every 5 seconds. To prevent multiple nanopores from forming accidentally, the transmembrane voltage was limited to a maximum of 0.8 V nm−1.26 The nanopore formation was detected through an abrupt increase in leakage current and decrease in membrane resistance when the applied current reached a critical value, typically around 270 pA. Finally, the nanopore size was estimated theoretically using an equation proposed by M. Wanunu27 and later adopted by W. Kowalczyk.28
1) for 60 minutes, followed by rinsing in three separate hot water baths for 10 minutes each. The cleaned chip was loaded into a custom-made PMMA flow cell with two chambers (cis and trans) filled with 200 mL of a translocation buffer (TB) consisting of 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8. Before initiating the controlled breakdown (CBD) process, the flow cell was positioned in a Faraday cage and connected to a Keithley 2450 source meter via two Ag/AgCl electrodes. During this process, the membrane's resistance, leakage current, and trans-membrane voltage of the SiNx membrane were continuously monitored while an initial current of 80 nA was applied, with subsequent increases of 5 nA every 5 seconds. To prevent multiple nanopores from forming accidentally, the transmembrane voltage was limited to a maximum of 0.8 V nm−1.26 The nanopore formation was detected through an abrupt increase in leakage current and decrease in membrane resistance when the applied current reached a critical value, typically around 270 pA. Finally, the nanopore size was estimated theoretically using an equation proposed by M. Wanunu27 and later adopted by W. Kowalczyk.28| G = σ[(4L/πd2) + (1/d)]−1 |
2.3. Preparation of the OA-induced displacement assay
First, streptavidin-decorated magnetic beads (MBs) were washed three times with magnetic bead buffer (1× MB buffer) consisting of 20.0 mM Tris–HCl (pH 8.0), 0.5 M NaCl, and 1.0 mM EDTA to remove any impurities. Prior to conjugation with the MBs, the aptamers were pre-heated at 95 °C for 5 minutes to remove any potential cross-linking. The aptamers were then incubated with the MBs in MB buffer for 20 minutes at 37 °C to permit their immobilization via the streptavidin–biotin affinity. The final ratio of MBs and aptamer concentration was adjusted to 20.0 μg per 1.0 μM. After immobilization, the MB–aptamer conjugate was washed three times for 3 minutes in MB buffer, deionized water, and Tris-Tween (TT) buffer (250.0 mM Tris–HCl pH 8.0, 0.1% Tween 20). To remove any physio-adsorbed molecules and weak streptavidin–biotin linkages, the solution was then washed with 150.0 mM NaOH for 10 minutes at room temperature, followed by an additional wash for 15 minutes at 70 °C with TT buffer. The cDNA was then diluted in a binding buffer containing 50.0 mM Tris–HCl (pH 7.5), 5.0 mM MgCl2, and 150.0 mM NaCl and heated to 95 °C for 5 min. Finally, the MB–aptamer conjugate and cDNA were incubated in binding buffer for 2 hours at 37 °C to form the aptamer–cDNA duplex.29,30After the aptamer–cDNA duplex formation on the magnetic beads (MBs), the binding buffer was removed using a permanent magnet and replaced with a translocation buffer (TB) containing 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8. The resulting MB–dsDNA solution, consisting of 20.0 μg MBs and 1.0 μM aptamer, was incubated with OA at concentrations ranging from 1.0 pg mL−1 to 1.0 × 102 ng mL−1, for 90 minutes at 37 °C. The cDNA-containing solution was then collected using a permanent magnet and translocated through the nanopore.
2.4. Okadaic acid extraction and detection from shellfish
Choromytilus meridionalis (mussels) and Mercenaria (clams) were procured from a local market in Chongqing, China. After removing the tissues from the shells, they were thoroughly rinsed with deionized water and strained using filter paper. The sample preparation is based on a previous report with some modifications.19 Briefly, 1 g of each tissue was homogenized using a mortar and pestle and then dispersed in a centrifugation tube containing 10 mL of methanol. The mixture was vigorously shaken and then centrifuged at 8000g for 10 min. The resulting supernatant solution (5 ml) was collected and then stored at 4 °C until further use. In order to determine the sensor's recovery, different concentrations of OA (3.0, 7.0 × 102, and 5.0 × 103 pg mL−1) were spiked into the shellfish extracts. Ultimately, all extracts were subjected to incubation with MB–dsDNA conjugate and subsequently translocated through the nanopore for 10 min.2.5. Apparatuses and data processing
The nanopore fabrication was conducted using a Keithley 2450 source meter, controlled by an in-house LabView program. DNA translocation and I–V measurements were performed on a Axopatch 200B patch-clamp amplifier (Molecular Devices, Inc. Sunnyvale, CA), with a fixed sampling rate of 100 kHz and a low-pass Bessel filter of 5 kHz. All electrical measurements were recorded over a period of 10 minutes. The physical size of the nanopore was determined through transmission electron microscopy imaging (TEM, Tecnai F20, FEI). The LC–MS spectrometer utilized was an 6530 Q-TOF LC/MS from Agilent Technologies. The translocation events' characteristics, including frequency, amplitude, and dwell time, were extracted using Clampfit software, and the data were analyzed and plotted using OriginPro 9.1 software.3. Results and discussion
3.1. Principle of the nanopore aptasensors based on target-induced strand displacement for OA detection
The ability of DNA and RNA aptamers to hybridize by Watson–Crick base pairing with complementary strands and their conformational mechanism of target binding allow for sophisticated biosensing schemes with improved capabilities and analytical performance. These aptamers are primarily engineered on the basis of several strategies that have been proposed, whereby a normal aptamer can endure a large target-binding-induced flexibility or conformation change necessary for a high signal gain.31 Among them, target-induced strand-displacement assays are the most commonly reported method for quantifying the target-binding affinity and specificity of small-molecule-binding aptamers. This technique uses a short cDNA strand that is complementary to a portion of the aptamer to generate structure-switching aptamers without using sequence engineering.32 The strategy for the nanopore aptasensors based on the target-induced strand displacement assay to detect OA is illustrated in Scheme 1. Briefly, as shown in Scheme 1, the truncated OA-binding aptamer was firstly coupled to the magnetic microbeads via a streptavidin/biotin interaction. Then, a complementary target (cDNA) tailed with poly(A) sequence was hybridized with the aptamer modified-magnetic beads. In the presence of the OA molecules, the binding between OA and its aptamers shifts the equilibrium towards dehybridization, inducing the displacement of the cDNA. The released cDNAs are positively correlated with OA concentration, therefore the supernatant after magnetic separation is used as the detection solution for the nanopore devices. Finally, by recording the changes in the frequency of cDNA translocation events, OA target concentration can be readily quantified. | ||
| Scheme 1 Graphical representation of the okadaic acid detection principle using the nanopore sensor and target-induced strand displacement assay. | ||
3.2. Optimization of the reaction conditions
To enhance the nanopore's signal count, several optimization steps were performed. The medium in which the displacement assay occurs can affect the release rate of the cDNA, and/or the conformation change of the aptamer upon binding with the OA molecule. The literature shows that the OA-mediated displacement assay has been performed in different solutions. Therefore, initially the aptamer–cDNA duplex decorated MBs were incubated with a concentration of 1.0 μg mL−1 OA molecules in a solution composed of 50.0 mM Tris–HCl (pH 8), 5.0 mM MgCl2, and 150.0 mM NaCl. Then, the supernatant solution was translocated through the nanopore. Under a transmembrane voltage of 300 mV, the open pore current of a 3.5 nm nanopore was approximately 200 pA, and the event rate was negligible. With the increase of the applied voltage from 300 mV to 600 mV, the event count increased as the applied voltage was increased from 300 mV to 600 mV. However, the nanopore has enlarged quickly as a direct result of the relatively high voltage. The buffer solution was then adjusted to 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA pH 8. Under an applied voltage of 150 mV, the open pore current of the same 3.5 nm nanopore was around 600 pA when subjected to an applied voltage of 150 mV. Fig. 1C and D reveal an unexpected finding that the MgCl2-containing solution had a much lower signal count than the KCl-containing solution. This behavior was mainly attributed to one (or both) of these following reasons: (i) the release rate of the cDNA is higher in the KCl-containing solution than in MgCl2-containing solution, and/or (ii) the net charge of the released cDNA strand strongly depends on the amount of counter-ions in the surrounding solution. In the case of the MgCl2-containing solution, Mg2+ cations will be attracted to the negatively charged phosphate groups of the DNA backbones, hence reducing the net charge of the released cDNA molecules.33 This charge reduction will eventually enhance the tendency of cDNA molecules to adhere to negatively charged SiNx surfaces.34 In addition to this, it was demonstrated that at pH 8 in an MgCl2-containing solution, DNA molecules can bind together forming a new and bigger conformation.33 Whereas, in a monovalent salt solution (KCl-containing solution) the attraction between the adjacent DNA molecules is weak and negligible.33 Because of these changes caused by the Mg2+ ions, the ability to drive a DNA molecule to cross the nanopore under relatively low applied voltages becomes harder.3.3. Optimization of the cDNA sequence
In the last few years, several OA-specific aptamers have been selected. One highly specific aptamer, developed by M. Zourob's team, has been widely used by other research groups.13 In this study, an aptamer DNA probe (OA6T2) synthesized in Zourob's lab, which has a dissociation constant of 2.77 nM (ref. 19) was used. The original complementary sequence to OA6T2 was proposed by Chinnappan et al. This 12-base ssDNA was selected over other sequences due to its strong fluorescence emission in the presence of OA molecules as reported previously.19In target-induced strand displacement assays, it is crucial to approximate the exact binding site between the aptamer and its target molecule. This helps in designing an effective complementary DNA sequence (cDNA) that reduces background signals in the absence of the target molecule while increasing the release rate in the presence of the target molecules. In order to obtain optimal results in the nanopore measurements, a series of A-tailed cDNAs of varying length was designed, based on the sequence originally proposed by Chinnappan et al.19 (see Table S1†). To investigate the impact of varying the length of the cDNA, a poly(A) tail consisting of 20 bases to the cDNA sequences was appended. In this case, 1.0 μM of each of these A-tailed cDNAs was incubated with the as-prepared MB–aptamer conjugate, and the supernatant solution was translocated through the nanopore. As shown in Fig. 1B, the washing process successfully removed almost all of the excess and weakly bound aptamers and cDNA molecules. Without the addition of OA molecules, the background signals of all cDNA sequences were negligible. After adding the same concentration of OA (1.0 μg mL−1) to the solution containing the aptamer–cDNA duplexes, the supernatant was collected and translocated through the nanopore sensor for 10 minutes. The event rates have increased dramatically for all cDNA sequences, proving the feasibility of this target-induced strand displacement assay. As shown in Fig. 2A, among the five A-rich cDNA sequences, the shortest cDNA sequence (Comp5(A20)) exhibited the highest event rate, suggesting that the binding site of OA molecules with the probe DNA OA6T2 is likely from G18 to C23. Fig. S2† presents the scatter plot of the translocation of the different cDNA sequences.
Furthermore, to investigate the effect of the A-tail length, additional cDNA sequences with different tail lengths (A10, A15, A25, and A30) were designed based on the cDNA Comp 5 template (without a tail). As depicted in Fig. 2B, the A-tail has a remarkable effect on the signal frequency, enhancing the sensor's sensitivity without the need for any complicated enzymatic amplification strategies. In addition, due to the added tail, the tailed cDNAs exhibited deeper current drops (140 ± 11 pA) compared to the non-tailed sequence (83 ± 32 pA), which helps in distinguishing the translocation signals from the noise signals. After the Poly(A20) tail was added, the signal count jumped from 0.8 events per s without a tail to 1.75 events per s with an A20 tail. Comp 5(A20) and Comp 5(A25) exhibited the highest signal frequency, with a slight difference between them, whereas the signal count dropped for Comp 5(A30). Furthermore, other studies showed the benefits of tailing the cDNA sequence on the signal count but further studies are needed to more explain and understand the origins of this effect.35 To reduce the cost of the assay, Comp 5(A20) was used throughout the rest of the experiments. Fig. 2C shows the scatter plot and the amplitude and dwell time histograms of the translocation of cDNA Comp 5(A20) through the nanopore, revealing that Comp 5(A20) has a current amplitude of 140 ± 11 pA dwell time of 2.33 ± 0.82 ms. Fig. S3† shows the translocation current traces of the control and cDNA Comp 5(A20) containing the supernatant solution through the nanopore.
3.4. Quantification of okadaic acid in buffer solution
Using the aforementioned experimental conditions, the developed aptasensor was applied for the detection of OA molecules at a concentration ranging from 1.0 pg mL−1 to 1.0 × 102 ng mL−1. The OA concentration in the solution was quantified by counting the nanopore signal (f) induced by the translocation of the cDNA-containing solution. The frequency (f) can be expressed as f = kon [Comp 5(A20)], where [Comp 5(A20)] is the concentration of the OA-released cDNA Comp 5(A20), and kon is the Comp 5(A20) occurrence rate constant.36 The sensitivity and limit of detection of the sensors were determined through the frequency (f). In this nanopore-based displacement assay, the event's frequency of the OA-released cDNA showed an increase upon incubating the as-prepared aptamer–cDNA duplex-modified MBs with different concentrations of OA molecules from 1.0 pg mL−1 to 1.0 × 102 ng mL−1 (Fig. 3A). For instance, the event rate jumped from 0.59 ± 0.01 events per second at 1.0 pg mL−1 OA to 1.05 ± 0.02 events per second at 1.0 × 102 ng mL−1 OA. To assess the sensor's reproducibility, three independent measurements were performed using different nanopore chips.Further investigation of the nanopore response as a function of the added OA concentration during the displacement assay revealed that the cDNA events frequency exhibited a wide dynamic range and a correlation coefficient R2 = 0.99. Furthermore, the regression equation describing the relationship between the nanopore response (f) and the OA concentration can be written as follows: f = 0.09![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10([OA]) + 0.59 (as shown in Fig. 3B). The limit of detection (LOD) was estimated to be 3.0 × 10−2 pg mL−1 in a total detection time of 10 min, with a linear dynamic range between 1.0 pg mL−1 to 1.0 × 102 ng mL−1. The limit of detection was calculated based on the method reported in ref. 37, the blue dashed line in Fig. S4† depicts 3× the noise level from the background noise (0.33 events per second), and the LOD was derived from the signal count that is three times higher than the background signal. It is worth mentioning that the calibration curve did not exhibit a linear relationship between the nanopore response and the OA concentration when plotted in decimal form, hence, in order to demonstrate the linear response of the sensor, the OA concentration was converted to a logarithmic form (log10) (inset Fig. 3B). The novel nanopore bioassay largely satisfies the international regulation, which strictly limits the amount of OA in food products to a maximum of 199.25 nM. As indicated in Table 1, the results of this nanopore aptasensor showed a better LOD than the gold-standard HPLC (LOD = 1.15 × 10−2 μg mL−1).38 In addition to that, the performance of the prepared sensor can compete or even outperform previously published works that require multiple reagents and rigorous preparation such as fluorescence sensors,11 colorimetric assays39 and electrochemical sensors,10,18 and even better than some amplification-based sensors. As an example, Huajie Gu et al. used a rolling circle amplification strategy for the detection of OA; although this strategy showed an excellent LOD, it requires complicated steps and is a time-consuming strategy.1 In another work, gold nanoparticles were used to amplify the OA detection signals based on a piezoelectric aptasensor; this strategy yielded to a LOD of 2.5 × 10−1 ng mL−1.17 It is important to highlight that in comparison to other techniques, the superiority of the proposed sensor resides in its simplicity, ease of use, and the fact that neither labeling nor amplification are necessary. In addition to that, the sensor exhibited excellent stability over an extended period of time during the assay runs. This stability is clearly demonstrated by the consistent current traces observed in Fig. 3A, indicating the robustness of the nanopore. Nevertheless, a notable constraint of this methodology pertains to the fabrication process of the nanopores. Despite that the CBD technique is widely recognized as a reliable method for nanopore fabrication, it is important to note that the shape of the nanopore cannot be precisely controlled using this technique. This lack of control could have an effect on the translocation of cDNA molecules.
log10([OA]) + 0.59 (as shown in Fig. 3B). The limit of detection (LOD) was estimated to be 3.0 × 10−2 pg mL−1 in a total detection time of 10 min, with a linear dynamic range between 1.0 pg mL−1 to 1.0 × 102 ng mL−1. The limit of detection was calculated based on the method reported in ref. 37, the blue dashed line in Fig. S4† depicts 3× the noise level from the background noise (0.33 events per second), and the LOD was derived from the signal count that is three times higher than the background signal. It is worth mentioning that the calibration curve did not exhibit a linear relationship between the nanopore response and the OA concentration when plotted in decimal form, hence, in order to demonstrate the linear response of the sensor, the OA concentration was converted to a logarithmic form (log10) (inset Fig. 3B). The novel nanopore bioassay largely satisfies the international regulation, which strictly limits the amount of OA in food products to a maximum of 199.25 nM. As indicated in Table 1, the results of this nanopore aptasensor showed a better LOD than the gold-standard HPLC (LOD = 1.15 × 10−2 μg mL−1).38 In addition to that, the performance of the prepared sensor can compete or even outperform previously published works that require multiple reagents and rigorous preparation such as fluorescence sensors,11 colorimetric assays39 and electrochemical sensors,10,18 and even better than some amplification-based sensors. As an example, Huajie Gu et al. used a rolling circle amplification strategy for the detection of OA; although this strategy showed an excellent LOD, it requires complicated steps and is a time-consuming strategy.1 In another work, gold nanoparticles were used to amplify the OA detection signals based on a piezoelectric aptasensor; this strategy yielded to a LOD of 2.5 × 10−1 ng mL−1.17 It is important to highlight that in comparison to other techniques, the superiority of the proposed sensor resides in its simplicity, ease of use, and the fact that neither labeling nor amplification are necessary. In addition to that, the sensor exhibited excellent stability over an extended period of time during the assay runs. This stability is clearly demonstrated by the consistent current traces observed in Fig. 3A, indicating the robustness of the nanopore. Nevertheless, a notable constraint of this methodology pertains to the fabrication process of the nanopores. Despite that the CBD technique is widely recognized as a reliable method for nanopore fabrication, it is important to note that the shape of the nanopore cannot be precisely controlled using this technique. This lack of control could have an effect on the translocation of cDNA molecules.
| Detection technique | Recognition element | Linear range | LOD | Ref. |
|---|---|---|---|---|
| Electrochemical sensor | OA protein-conjugates | 0.27–3.3 ng mL−1 | 0.15 ng mL−1 | 18 |
| Immunosensor | Antibody | 0.02–33.6 ng mL−1 | 0.02 ng mL−1 | 40 |
| Colorimetric sensor | OA-specific aptamer | 8.0–966 ng mL−1 | 0.33 ng mL−1 | 39 |
| Fluorescence sensor | Monoclonal antibody | 0.62–20 ng mL−1 | 0.62 ng mL−1 | 11 |
| Love wave sensors | Anti-BSA antibody | 10–150 ng mL−1 | 5.5 ng mL−1 | 41 |
| ELISA | Monoclonal antibody | 20–750 pg mL−1 | 12 pg mL−1 | 42 |
| Microfluidic sensor | Phosphorene-gold nanocomposite | 8–201 ng mL−1 | 6.44 pg mL−1 | 6 |
| HPLC | — | 0.4–62.67 μg mL−1 | 0.015 μg mL−1 | 38 |
| LC–MS/MS | 0.56–62.07 μg mL−1 | 0.045 μg mL−1 | ||
| Nanopore sensor | OA-specific aptamer | 1.0 pg mL−1 to 1.0 × 102 ng mL−1 | 3.0 × 10−2 pg mL−1 | This work |
3.5. Assessment of the sensor's selectivity and reproducibility
Encouraged by the outstanding sensitivity and detection range of the proposed aptasensor, the selectivity was further examined by using other marine toxin molecules. To evaluate the selectivity of the novel aptasensor, the following were used: three different microcystins (MC-LR, MC-RR, and MC-YR), aflatoxin B1 (AFB1), malachite green (MG), and DTX1 which have an analogue structure to OA. For this test, a concentration of 1.0 × 102 ng mL−1 of each interferon molecule was incubated with the already prepared aptamer–cDNA duplex decorated MBs. The effect of these molecules on the cDNA release rate was evaluated by comparing the event's frequency produced by each assay to the signal generated by 1.0 × 102 ng mL−1 of the OA target, as shown in Fig. 4A. Furthermore, Fig. 4B shows that these molecules have no significant effect on the signal frequency and their signals are comparable to the blank signal (background noise): 0.14, 0.125, 0.075, 0.09 and 0.13 events per s for MC-LR, MC-RR, MC-YR, AFB, and MG, respectively. With the exception of the DTX1 signals, which exhibit a signal frequency count of 0.415 events per s, noticeably surpassing that of the other interfering molecules, this is due to the high similarity of the OA and DTX1 structures. Fig. 4C highlights the only difference between the two chemicals where OA has a hydrogen atom at the R-group while DXT-1 has a methyl group. Moreover, further statistical analysis proved that the response of the novel aptasensor to all these interfering substances is significantly different compared to the signal of OA molecules (p < 0.05, n = 3). These results demonstrated the high specificity of the aptamer probe used in this work, and further approve the superiority of the presented approach. In addition, the sensor's reproducibility was further assessed by analyzing three independently fabricated nanopore sensors toward 1.0 × 102 ng mL−1 okadaic acid (OA) and a relative standard deviation (RSD) of 4% was obtained, demonstrating a satisfactory reproducibility of the assay. These results indicate that the nanopore aptasensor platform can be used for the accurate quantification of OA.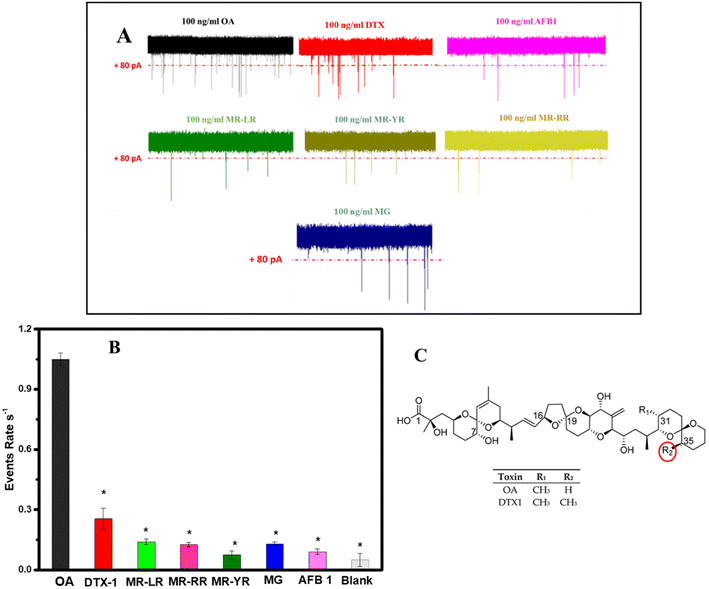 | ||
| Fig. 4 Selectivity of the proposed nanopore aptasensor has been evaluated for the detection of 1.0 × 102 ng mL−1 of OA molecules, as well as various marine toxin substances. A) Current traces induced by 1.0 × 102 ng mL−1 of each molecule. B) Effect of the interferon molecules on the event rate. All data were obtained from 3 different measurements. Level of significance: *P < 0.05. C). The chemical structure of OA and DTX-1. Graph inspired from a previous work.43 | ||
3.6. Practical application of the sensor
To further test the reproducibility and the accuracy of the proposed nanopore aptasensor, the sensor was applied to two different types of shellfish (Mytilus edilis and Mercenaria). The signal frequency of the non-spiked extract translocation was insignificant (Fig. S5†), which indicated that the shellfish has no traces of OA or that the concentration is below our sensor's detection limit. In contrast, the signal count of the spiked extracts exhibited a notable increase in comparison to the non-spiked samples. The recovery of this sensor was calculated by inserting the signal frequency of the spiked extract in the regression equation of the calibration curve and the results were summarized in Table 2. As a result, the average recovery of spiked OA in the shellfish samples was between 95.33% and 110.3%, indicating that the interferents in the shellfish sample do not significantly affect the detection of OA molecules. Moreover, the LC–MS/MS technique was used to detect OA molecules in the spiked shellfish extract. Fig. S6† shows the calibration curve of the LC–MS/MS technique; the area of signal peak exhibited a linear correlation to the OA concentration ranging from 5.0 pg mL−1 to 1.0 × 101 ng mL−1, with a regression coefficient of R2 = 0.99. The recovery levels were comparable to the recovery levels of the developed nanopore aptasensor (Table 2). As shown in Fig. S7,† the correlation analysis of the two methods' results showed a linear regression plots with a y-intercept and slope value close to the ideal values of 0 and 1 respectively, which further confirm the good agreement between the two methods, hence proving the accuracy of our nanopore aptasensor. Unlike fluorescence sensors which suffer from the auto-fluorescence noise of the biological samples,19 the proposed aptasensor does not suffer from any noise coming from the real sample extracts. Thus, this technique shows great promise as a practical application for the detection of okadaic acid directly from shellfish samples.| Shellfish species | Spiked concentration (pg mL−1) | Nanopore assay | LC–MS/MS | ||
|---|---|---|---|---|---|
| Found (pg mL−1)n | Recovery (%)n | Found (pg mL−1) | Recovery (%) | ||
| n = 3; nd: not detected or under the limit of detection. | |||||
| Mytilus edilis | 0 | nd | — | nd | — |
| 3 | 2.86 ± 0.13 | 95.33 | nd | — | |
| 7.0 × 102 | 733 ± 20 | 104.72 | 711 ± 26 | 99.11 | |
| 5.0 × 103 | 5104 ± 53 | 102.06 | 4640 ± 100 | 92.8 | |
| Mercenaria | 0 | nd | — | nd | — |
| 3 | 3.12 ± 0.06 | 104 | nd | — | |
| 7.0 × 102 | 670 ± 20 | 95.66 | 680 ± 21 | 105.7 | |
| 5.0 × 103 | 5515 ± 250 | 110.30 | 5420 ± 151 | 110.4 | |
4. Conclusion
In this work, a new amplification-free ultra-sensitive nanopore-based aptasensor was successfully developed for the detection of okadaic acid in shellfish samples. The detection strategy relied on the target-induced strand displacement on an assay between the okadaic acid molecules and the signaling probe (cDNA). The length of the poly(A) tail on the cDNA was optimized and the results were evaluated in terms of the sensor's sensitivity. Moreover, the effect of the buffer composition on the nanopore signals during the displacement assay was further optimized. Subsequently, the optimized conditions were used to detect OA molecules in both controlled solutions and in shellfish extracts, revealing the high selectivity of the aptamer used in this work (OA6T2). The proposed nanopore aptasensor outperforms existing sensors in terms of dynamic range, limit of detection, selectivity, and accuracy. However, the event count remains a bit low because of the low concentrations of OA. Additionally, compared to other methods, the proposed sensing technology is easy to use, and does not require any additional pre-treatment steps or functionalization of the nanopore sensor. Finally, to address the limitations of this method, more research on nanopore production and a more selective and high yield aptamer probe is needed.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors thank the University of Chinese Academy of Sciences (UCAS), China for supporting this work. This work was supported by the Youth Innovation Promotion Association (No. 2022388) of the Chinese Academy of Science, the Natural Science Foundation of Chongqing, China (CSTB2023NSCQ-MSX0071), and the Science Foundation of CIGIT, China (E355400201).References
- H. Gu, L. Hao, N. Duan, S. Wu, Y. Xia, X. Ma and Z. Wang, Microchim. Acta, 2017, 184, 2893–2899 CrossRef CAS.
- H. Gu, N. Duan, S. Wu, L. Hao, Y. Xia, X. Ma and Z. Wang, Sci. Rep., 2016, 6, 21665 CrossRef CAS PubMed.
- P. Zhao, H. Liu, P. Zhu, S. Ge, L. Zhang, Y. Zhang and J. Yu, Sens. Actuators, B, 2021, 344, 130174 CrossRef CAS.
- L. L. Fu, X. Y. Zhao, L. D. Ji and J. Xu, Toxicon, 2019, 160, 1–7 CrossRef CAS PubMed.
- Z. Zhang, X. Yu, J. Zhao, X. Shi, A. Sun, H. Jiao, T. Xiao, D. Li and J. Chen, Chemosphere, 2020, 246, 125622 CrossRef CAS PubMed.
- S. Ramalingam, R. Chand, C. B. Singh and A. Singh, Biosens. Bioelectron., 2019, 135, 14–21 CrossRef CAS PubMed.
- H. Suzuki and Y. Okada, J. Vet. Med. Sci., 2018, 80, 616–619 CrossRef CAS PubMed.
- A. Sassolas, A. Hayat, G. Catanante and J. L. Marty, Talanta, 2013, 105, 306–316 CrossRef CAS PubMed.
- J. L. D. Nelis, D. Migliorelli, L. Mühlebach, S. Generelli, L. Stewart, C. T. Elliott and K. Campbell, Talanta, 2021, 228, 122215 CrossRef CAS PubMed.
- Y. Tian, L. Yuan, M. Zhang, Y. He and X. Lin, Anal. Methods, 2022, 14, 1261–1267 RSC.
- H. Wang, H. Lin, R. Pan, H. Yan, S. Li, Y. Zheng, Q. Cao, Z. Xu, L. Zeng and S. Liu, Food Anal. Methods, 2022, 15, 2470–2478 CrossRef.
- S. Y. Toh, M. Citartan, S. C. B. Gopinath and T. H. Tang, Biosens. Bioelectron., 2015, 64, 392–403 CrossRef CAS PubMed.
- S. Eissa, A. Ng, M. Siaj, A. C. Tavares and M. Zourob, Anal. Chem., 2013, 85, 11794–11801 CrossRef CAS PubMed.
- Y. Wang, D. Rao, X. Wu, Q. Zhang and S. Wu, Microchem. J., 2021, 160, 105644 CrossRef CAS.
- A. Castanheira, M. B. Dos Santos, L. Rodriguez-Lorenzo, R. Queirós and B. Espiña, Analyst, 2021, 146, 2638–2645 RSC.
- W. Shan, J. Sun, R. Liu, W. Xu and B. Shao, Sens. Actuators, B, 2022, 352, 131035 CrossRef CAS.
- Y. Tian, P. Zhu, Y. Chen, X. Bai, L. Du and W. Chen, Sens. Actuators, B, 2021, 346, 130446 CrossRef CAS.
- J. L. D. Nelis, D. Migliorelli, L. Mühlebach, S. Generelli, L. Stewart, C. T. Elliott and K. Campbell, Talanta, 2021, 228, 122215 CrossRef CAS PubMed.
- R. Chinnappan, R. AlZabn, T. A. Mir, M. Bader and M. Zourob, Microchim. Acta, 2019, 186, 406 CrossRef PubMed.
- L. Xue, H. Yamazaki, R. Ren, M. Wanunu, A. P. Ivanov and J. B. Edel, Nat. Rev. Mater., 2020, 5, 931–951 CrossRef CAS.
- J. Wu, S. Wang, L. Liang, C. Zhao, Y. Yin, T. Weng, B. Yin, L. Wang and D. Wang, Sens. Actuators, B, 2021, 348, 130712 CrossRef CAS.
- K. Chuah, Y. Wu, S. R. C. Vivekchand, K. Gaus, P. J. Reece, A. P. Micolich and J. J. Gooding, Nat. Commun., 2019, 10, 1–9 CrossRef CAS PubMed.
- M. A. Elaguech, M. Bahri, K. Djebbi, D. Zhou, B. Shi, L. Liang, N. Komarova, A. Kuznetsov, C. Tlili and D. Wang, Food Chem., 2022, 389, 133051 CrossRef CAS PubMed.
- S. Rauf, L. Zhang, A. Ali, Y. Liu and J. Li, ACS Sens., 2017, 2, 227–234 CrossRef CAS PubMed.
- J. Wu, L. Liang, M. Zhang, R. Zhu, Z. Wang, Y. Yin, B. Yin, T. Weng, S. Fang, W. Xie, L. Wang and D. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 12077–12088 CrossRef CAS PubMed.
- M. Dong, Z. Tang, X. He and W. Guan, ACS Appl. Electron. Mater., 2020, 2, 2954–2960 CrossRef CAS.
- M. Wanunu, T. Dadosh, V. Ray, J. Jin, L. McReynolds and M. Drndić, Nat. Nanotechnol., 2010, 5, 807–814 CrossRef CAS PubMed.
- S. W. Kowalczyk, A. Y. Grosberg, Y. Rabin and C. Dekker, Nanotechnology, 2011, 22, 315101 CrossRef PubMed.
- K. Djebbi, M. Bahri, M. A. Elaguech, R. Tian, S. Biao, C. Tlili and D. Wang, in Smart Sensors, Measurement and Instrumentation, 2021, vol. 38, pp. 137–164 Search PubMed.
- K. Djebbi, J. Xing, T. Weng, M. Bahri, M. A. Elaguech, C. Du, B. Shi, L. Hu, S. He, P. Liao, C. Tlili and D. Wang, Anal. Chim. Acta, 2022, 1208, 339778 CrossRef CAS PubMed.
- H. Yu, O. Alkhamis, J. Canoura, Y. Liu and Y. Xiao, Angew. Chem., Int. Ed., 2021, 60, 16800–16823 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- Y. Zhang, L. Liu, J. Sha, Z. Ni, H. Yi and Y. Chen, Nanoscale Res. Lett., 2013, 8, 1–8 CrossRef PubMed.
- S. W. Kowalczyk, D. B. Wells, A. Aksimentiev and C. Dekker, Nano Lett., 2012, 12, 1038–1044 CrossRef CAS PubMed.
- T. Li, Z. Su, Y. Li, L. Xi and G. Li, Talanta, 2022, 248, 123619 CrossRef CAS PubMed.
- Y. Lin, Y. L. Ying, X. Shi, S. C. Liu and Y. T. Long, Chem. Commun., 2017, 53, 11564–11567 RSC.
- M. Bahri, S. Biao, M. A. Elaguech, K. Djebbi, D. Zhou, L. Liang, C. Tlili and D. Wang, ACS Appl. Nano Mater., 2022, 5, 5035–5044 CrossRef CAS.
- A. P. Louppis, A. V. Badeka, P. Katikou, E. K. Paleologos and M. G. Kontominas, Toxicon, 2010, 55, 724–733 CrossRef CAS PubMed.
- X. Yan, X. Qi, Y. Zhao, L. Li, R. Ma, L. Wang, S. Wang and X. Mao, J. Ocean Univ. China, 2022, 21, 400–408 CrossRef CAS.
- X. Li, Y. Cheng, R. Xu, Z. Zhang, X. Qi, L. Chen and M. Zhu, Talanta, 2022, 247, 123567 CrossRef CAS PubMed.
- L. Zou, Y. Tian, X. Zhang, J. Fang, N. Hu and P. Wang, Sens. Actuators, B, 2017, 238, 1173–1180 CrossRef CAS.
- R. Wang, L. Zeng, H. Yang, Y. Zhong, J. Wang, S. Ling, A. F. Saeed, J. Yuan and S. Wang, J. Hazard. Mater., 2017, 339, 154–160 CrossRef CAS PubMed.
- J. Kilcoyne, S. Burrell, C. Nulty, R. Salas, E. J. Wright, I. Rajotte and C. O. Miles, Mar. Drugs, 2020, 18, 1–14 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00199g |
| This journal is © The Royal Society of Chemistry 2023 |



