Open Access Article
Open Access ArticleNative mass spectrometry of proteoliposomes containing integral and peripheral membrane proteins†
Yun
Zhu‡
,
Sangho D.
Yun‡
 ,
Tianqi
Zhang
,
Jing-Yuan
Chang
,
Lauren
Stover
and
Arthur
Laganowsky
,
Tianqi
Zhang
,
Jing-Yuan
Chang
,
Lauren
Stover
and
Arthur
Laganowsky
 *
*
Department of Chemistry, Texas A&M University, College Station, TX 77843, USA. E-mail: ALaganowsky@chem.tamu.edu
First published on 21st November 2023
Abstract
Cellular membranes are critical to the function of membrane proteins, whether they are associated (peripheral) or embedded (integral) within the bilayer. While detergents have contributed to our understanding of membrane protein structure and function, there remains challenges in characterizing protein–lipid interactions within the context of an intact membrane. Here, we developed a method to prepare proteoliposomes for native mass spectrometry (MS) studies. We first use native MS to detect the encapsulation of soluble proteins within liposomes. We then find the peripheral Gβ1γ2 complex associated with the membrane can be ejected and analyzed using native MS. Four different integral membrane proteins (AmtB, AqpZ, TRAAK, and TREK2), all of which have previously been characterized in detergent, eject from the proteoliposomes as intact complexes bound to lipids that have been shown to tightly associate in detergent, drawing a correlation between the two approaches. We also show the utility of more complex lipid environments, such as a brain polar lipid extract, and show TRAAK ejects from liposomes of this extract bound to lipids. These findings underscore the capability to eject protein complexes from membranes bound to both lipids and metal ions, and this approach will be instrumental in the identification of key protein–lipid interactions.
Introduction
Cell membranes are made up of an intricate and heterogeneous mixture of membrane-associated proteins (MAPs) and lipids that collectively carry out various biological tasks. Constituting about a third of human proteome, MAPs can be categorized into two types based on their association with the biological membrane: integral membrane proteins and peripheral membrane proteins.1,2 These two types are distinct in their function, structural properties, and interactions with cellular membrane. Integral membrane proteins contain one or more transmembrane helices, and part or whole is embedded in the bilayer. They also carry out a number of essential biological processes, such as transport of ions and molecules across the bilayer, and they represent one of the largest targets for drug discovery.3 Peripheral membrane proteins associate with the surface of the bilayer through electrostatic interactions, e.g. polybasic domains, and from post-translational modifications, such as prenylation.4,5 More generally, the lipid environment is known to modulate the structure and function of MAPs. For example, the activities of integral membrane proteins, such as the bacterial ammonia channel (AmtB) and the two-pore domain potassium channel (TRAAK), have been shown to be regulated by specific lipids.6–9 For peripheral membrane proteins, post-translational modification with membrane anchors are known to play a critical role in various biological processes.10 A well-known example is prenylation of the C-terminus of K-Ras, which has been shown to be important for nanoclustering on the membrane surface.11,12Native mass spectrometry (MS) commonly uses detergents to solubilize membrane protein complexes for analysis. There are some advantages of studying membrane protein complexes in detergent environments. One is the ability to preserve non-covalent interactions.13 Another is the capability to distinguish and analyze the binding of individual ligands to complexes.14 This approach is especially effective for understanding membrane protein–lipid interactions, leading to the discovery of lipids pivotal for membrane protein function. More specifically, over the past decade, native MS has shown that certain protein–lipid interactions can have an allosteric effect on other protein interactions, as well as those with lipids, and small molecule drugs, and identify lipids important for function, like PI(4,5)P2's role in G-protein-coupled receptor initiation and G-protein specificity.13–22
Beyond detergent, other approaches have been employed to introduce membrane proteins from more native-like environments into the mass spectrometer. One strategy involves the incorporation of membrane proteins into nanodiscs or bicelles, both of which employ small, discoidal arrangements of phospholipid bilayers.23 These strategies have been shown to better preserve the oligomeric state of membrane protein complexes.24,25 MS analysis often involves ejection of membrane proteins from nanodiscs.24,26 Notably, membrane proteins eject from nanodisc as intact complexes bound to lipids. This has been useful for ejecting membrane proteins complexes from heterogenous lipid environments, providing insight into lipid binding to AmtB and the bacterial water channel (AqpZ).27 Recent work has utilized super charging molecules, which dramatically increases the charge states of protein complexes ionized using electrospray ionization.28 The addition of supercharging molecules enables measurements of the intact mass of membrane proteins embedded in nanodiscs, circumventing the need to activate and eject the embedded membrane protein.29 One particular drawback of using bicelles and nanodisc is the inability to generate an electrochemical gradient, which may be required to measure protein activity, such as for a potassium flux assay.
Another strategy involves the use of membrane vesicles or proteoliposomes. Robinson and co-workers developed a method that makes use of sonication to disrupt and destabilize membrane vesicles prior to introduction30 into the mass spectrometer.31,32 Like nanodiscs, MS analysis requires the use of collisional activation to eject the associated membrane protein complexes from the destabilized vesicles. However, the inherent heterogeneous nature of cell membranes hinders the ability to study targeted, specific interactions. Russell and co-workers developed a method that involves the freeze drying of vesicles and dissolution in isobutanol prior to MS analysis.33,34 This approach has been used to capture the lipid-dependence of Gramicidin A conformers. More recently, Schmidt and co-workers developed an approach that makes use of incorporating proteins into defined lipid environments,35 and Gupta and co-workers have refined the approach to use a mixture of synthetic lipids to mimic cellular membranes.30 Similar to nanodiscs, the addition of super charging molecules aids the ejection of membrane protein complexes from these liposomes and the ejected complexes retain bound lipids.29,30 This approach has been particularly useful to study how the lipid environment influences the monomer–dimer equilibrium of semisweet.30
The integration of integral and peripheral membrane proteins with liposomes of defined composition is an attractive route for functional assays,36,37 cryoEM studies,38–40 and MS analysis of MAPs. Here, we report a simplified method for the preparation of proteoliposomes for native MS analysis. Using this method, we demonstrate the encapsulation of soluble proteins within liposomes to mimic a confined microenvironment, and incorporation of peripheral and integral membrane proteins into proteoliposomes. MS analysis shows the soluble and MAPs can readily be ejected and detected. The integral membrane proteins eject from proteoliposomes bound to lipids, and the bound lipids align with the results obtained from studies of the detergent-solubilized complexes.
Results
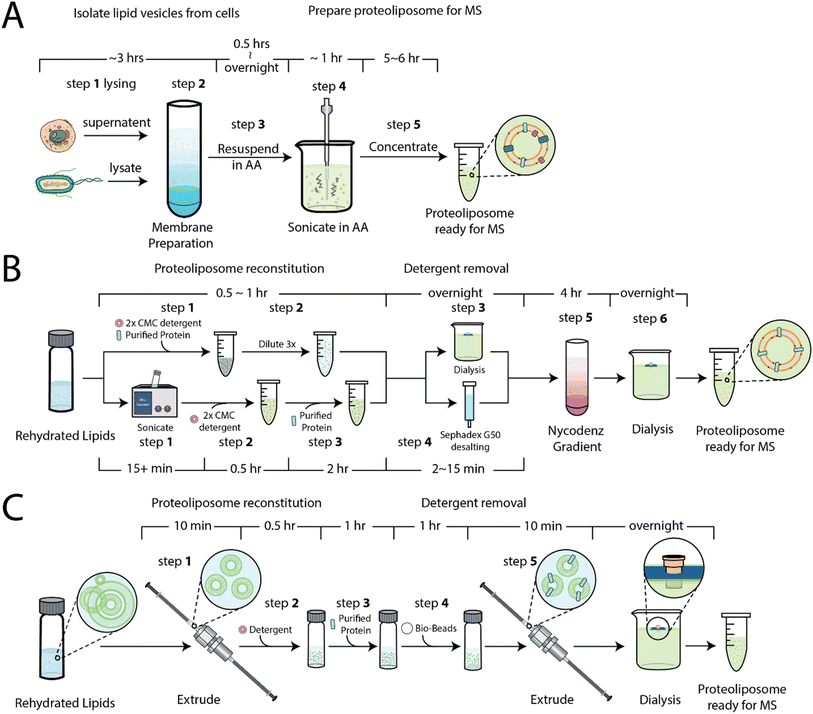
A simplified method for proteoliposome preparation for native MS
Our first objective was to develop simplified method for the preparation of proteoliposomes for native MS. As noted above, one approach31 involves the purification of membranes, in some cases containing overexpressed membrane proteins, followed by resuspension and subjected to a series of sonication pulses (Fig. 1A), which can inadvertently lead to sample heating. This approach produces non-uniform vesicles and has recently been used to capture insight into rhodopsin signaling in native membranes.41 A more recent approach30,35 involves the preparation of the proteoliposomes using a combination of size exclusion chromatography or dialysis to remove detergents38 and a floatation step that involves a nycodenz (or sucrose) gradient, which requires ultracentrifugation over several hours (Fig. 1B). To simplify the procedure, we sought to develop a method (Fig. 1C and S1†) that starts with small unilamellar liposomes (SULs) of a defined lipid composition prepared by the extrusion method. SULs (20–100 nm) and large unilamellar liposomes (100–1000 nm) are commonly used in membrane protein functional assays.36,42 Purified soluble proteins or detergent-solubilized membrane proteins are incubated with SULs in the presence of detergent. The excess detergent is removed through absorption to beads (see methods) to facilitate incorporation and generation of proteoliposomes. The proteoliposomes are then extruded and, like other methods, dialyzed against ammonium acetate, a volatile buffer that facilitates the MS analysis of ejected proteins by reducing adducts, such as those from sodium or potassium chloride salts.Encapsulation soluble proteins within liposomes
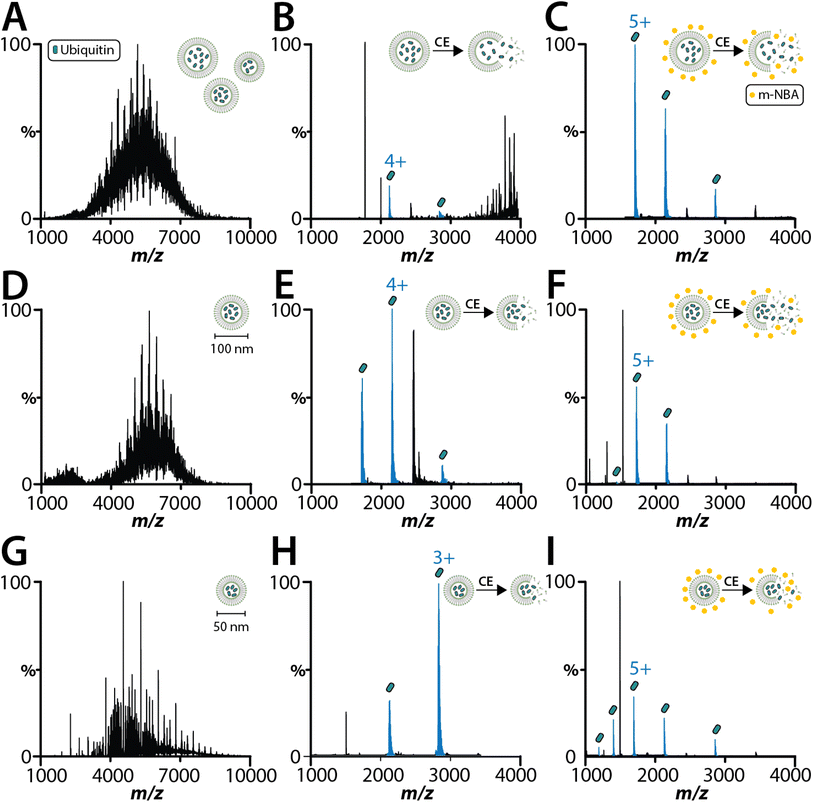
As interest increases in encasing biological samples in lipid-based solutions like lipid nanoparticles and liposomes for drug delivery,43 native MS analysis of proteoliposomes containing soluble biologics might offer insights into the final integrity of the encapsulated cargo. As proof of concept, we encapsulated two soluble proteins (lysozyme and ubiquitin) within 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) SULs. As the size of the proteoliposome has been reported to be important for native MS analysis and cryoEM studies,30,38 we analyzed ubiquitin encapsulated in SULs before and after extrusion using filter membranes with pore sizes of 50 and 100 nm (Fig. 2). With minimal activation energy, the encapsulated ubiquitin is not detected (Fig. 2A, D and G), confirming that the protein is fully encapsulated within the liposome. Applying collisional energy to disrupt the non-extruded liposomes yields signal for ubiquitin (Fig. 2B). Extruded SULs displayed significantly improved spectra for encapsulated ubiquitin and lysozyme after disrupting the SULs by collisional activation (Fig. 2B, E, H and S3A†). Notably, these samples did not require the addition of a super-charging molecule to detect the encapsulated cargo. The most abundant charge states for ubiquitin and lysozyme encapsulated in SULs was 3+ and 5+, respectively. These charge states were lower (by two charges) compared to samples analyzed in solution (Fig. S2 and S3B†). The encapsulation of cargo shields it from picking up excess charge and the detection of cargo requires collisional activation of liposomes, which occurs in the mass spectrometer. Interestingly, ubiquitin ejected out of the SULs in the presence of m-nitrobenzyl alcohol (m-NBA), a super-charging molecule,28 displayed charge states like that observed for the unencapsulated protein (Fig. 2C, F and I). Taken together, native MS can be used to analyze the encapsulated cargo of liposomes.Proteoliposomes containing peripheral membrane proteins
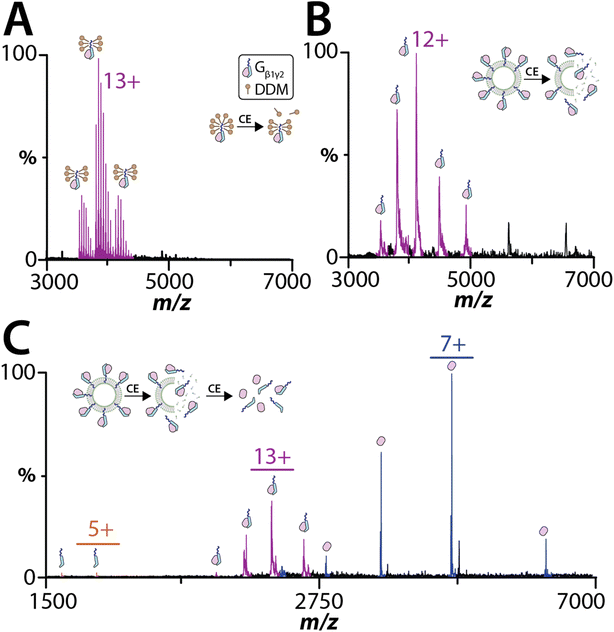
Peripheral membrane proteins play a key role in the activation of many signaling pathways, such as the heterotrimeric G protein complex that consists of α, β, and γ subunits.44,45 Here, we investigated the heterodimeric Gβ1γ2 complex, which the C-terminus of the Gγ2 subunit is post-translationally modified by geranylgeranylation. Gβ1γ2 complex purified from membranes and solubilized in n-dodecyl-beta-maltoside (DDM) was subjected to native MS analysis (Fig. 3A). Under the mild instrument conditions (CE of 20 eV and CID of 10 eV), the dimeric complex remains intact and bound to several DDM molecules. The mass of complex is consistent with geranylgeranylation and processing of the C-terminal CaaX motif (Tables S5 and S6†).46 Increasing the collision activation energy to CE of 80 eV and CID of 60 eV resulted in dissociation of the DDM adducts and Gβ1γ2 complex, confirming the stoichiometry of the heterodimeric complex (Fig. S4†). To eject and detect the Gβ1γ2 complex from proteoliposomes required the addition of m-NBA and required less energy (CE of 30 eV and CID of 30 eV) than that needed to dissociate the Gβ1γ2 complex in DDM (Fig. 3B). The charge state distribution on the ejected Gβ1γ2 complex was comparable to the complex solubilized in DDM. Unlike the sample analyzed in DDM, Gβ1γ2 complex ejected from proteoliposome did not display any adducts. Additionally, an increase in activation energy to CE of 30 eV and CID of 80 eV also resulted in dissociation of the ejected Gβ1γ2 complex into individual subunits (Fig. 3C). These results indicate peripheral membrane proteins associated with liposomes can be analyzed by native MS.Model bacterial integral membrane proteins embedded in SULs
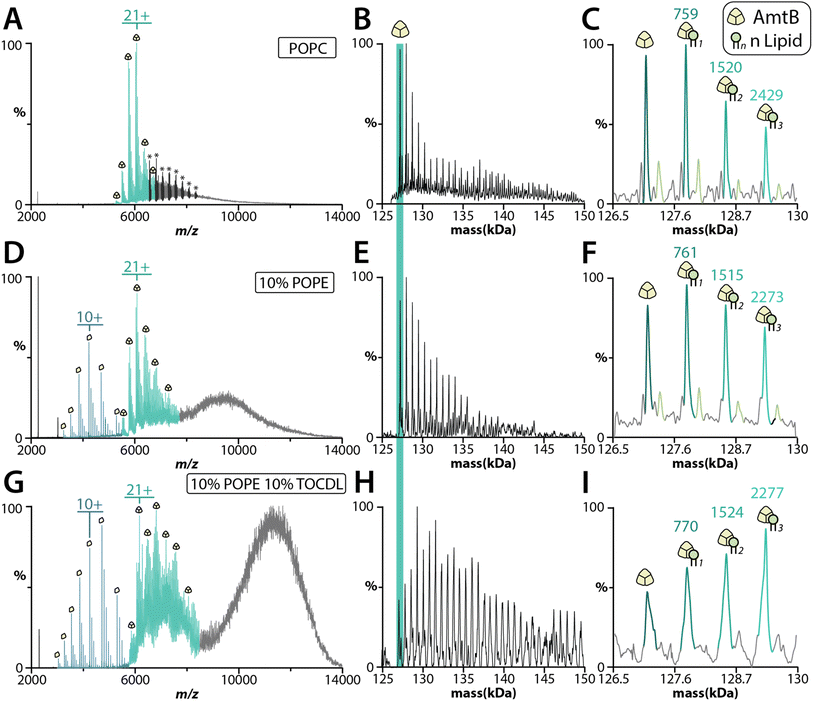
The bacterial trimeric AmtB and tetrameric AqpZ channels solubilized in detergent have been extensively studied using native MS,13,47–55 and represent model membrane proteins to benchmark the proteoliposome method. We first reconstituted AmtB into POPC SULs. MS analysis revealed that extrusion and the addition of m-NBA improved the quality of the mass spectra for the ejected AmtB complex (Fig. S5†). AmtB was then reconstituted into SULs containing different compositions: POPC; 90% POPC and 10% POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine); and 80% POPC, 10% POPE and 10% 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (TOCDL). Proteoliposomes containing POPE and TOCDL were selected because previous work has shown PE enhances AmtB–CDL interactions.53,56 AmtB could readily be ejected from the proteoliposomes containing different lipids in the presence of m-NBA (Fig. 4). Interestingly, AmtB reconstituted in POPC protoeliposomes required less energy to eject the complex compared to the other proteoliposome formulations (See Fig. 4 legend). The proteoliposomes containing either POPE or POPE and TOCDL showed ejection of the trimeric AmtB complex along with signal for the dissociated subunit (Fig. 4D–G). Significantly more lipids were bound the ejected AmtB complex for the proteoliposomes containing POPE and TOCDL (Fig. 4G–I). This observation is consistent with previous work showing that PE enhances AmtB–CDL interactions.53,56 Moreover, the intensities of AmtB-lipid bound species is complex and the irregularity indicates these may likely represent mixed lipid bound states. TOCDL is about twice the mass of POPE, and the broadness of the adduct peaks makes it difficult to assign with confidence. It is also worth noting proteoliposomes containing POPE also showed more lipids bound to AmtB compared to proteoliposomes containing only POPC. The dissociated monomers retained adducts with a mass in the ballpark of either POPE or POPC. We also incorporated AqpZ with C-terminal fusion to the green fluorescent protein (GFP) into POPC SULs. The tetrameric complex could also be ejected from proteoliposomes in the presence of m-MBA (Fig. S6A†). The ejected complex also retained several bound POPC molecules (Fig. S6B†).Human two-pore domain potassium channels reconstituted in different lipid environments
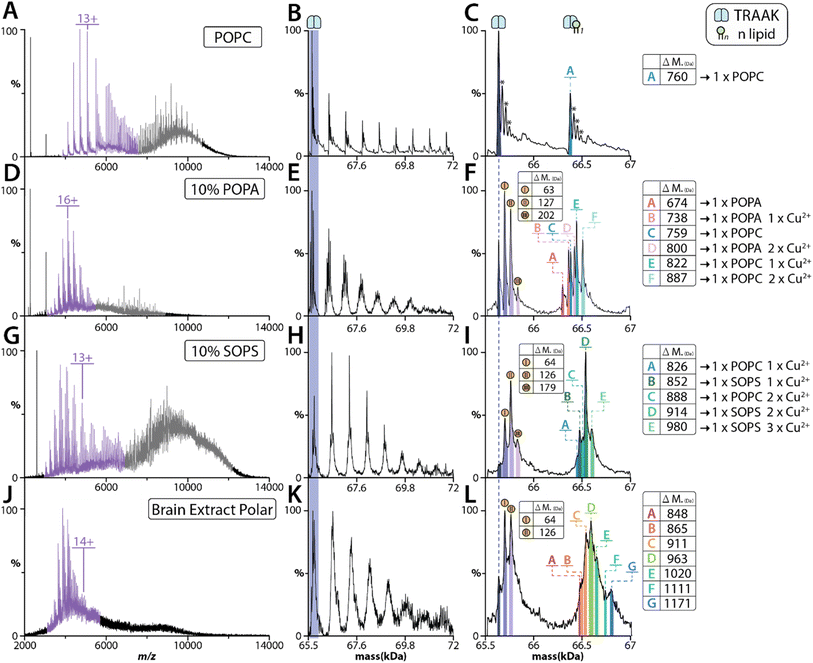
Given the results for AmtB and AqpZ, we next focused our investigation on the human two-pore domain potassium channels, TRAAK and TREK2. Both potassium channels carry out essential physiological and pathological processes, such as regulating membranes resting potential, pain-perception, and neuroprotection.57–59 We first performed MS analysis of TRAAK reconstituted in POPC proteoliposomes, which showed the dimeric complex could be ejected (Fig. 5A–C). However, a significant decrease in the number of POPC bound to the ejected complex was observed with increasing charge state i.e., compare charge states centered around 6000 versus 5000 m/z (Fig. 5A). This pattern is consistent with the higher charge state ions experiencing higher collision energies, leading to dissociation of the bound POPC lipids. As previous work from our group has revealed that TRAAK selectively binds POPA,60 we reconstituted the channel into proteoliposomes containing 10% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA). TRAAK ejected from these proteoliposomes displayed a markedly different retention of lipid species compared to POPC alone (Fig. 5D–F). More specifically, signals for both POPC and POPA bound to TRAAK were observed in addition to other species containing ∼63 Da adducts (Fig. 5F). In our recent work, we have discovered that copper(II) binds TRAAK,8 and the adducts observed here are likely copper(II) binding to the channel.We previously have shown that copper(II) specifically modulates TRAAK-phosphatidylserine interactions. Therefore, we reconstituted TRAAK and TREK2 into proteoliposomes containing POPC and 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (SOPS). SOPS (788 Da) was selected over POPS (761 Da) to enhance the mass difference with POPC (760 Da). MS analysis of proteoliposomes containing SOPS revealed TRAAK ejected predominantly with SOPS and copper(II) bound species (Fig. 5G–I). As a comparison, we performed similar experiments for TREK2 reconstituted into proteoliposomes of the same lipid composition (Fig. S7†). Although TREK2 shares ∼45% sequence identity with TRAAK,59,61 copper(II) does not modulate TREK2–lipid interactions.62 The ejected TREK2 complexes were bound to a near equal abundance of POPC and SOPS (Fig. S7†). Unlike TRAAK, there was no signal corresponding to copper(II) binding (Fig. S7D†). In short, these findings highlight the utility of retaining key membrane protein–lipid interactions, including those modulated by metal ion binding.
We have recently developed an approach that combines native MS and lipidomic approaches to identify specific protein–lipids interactions from natural sources, such as a brain polar lipid extract56 (Fig. S9†). This inspired us to investigate whether the method described herein could be extended to more complex lipid mixtures. TRAAK was reconstituted into a brain polar lipid extract showed a subset of species ejecting with the dimeric complex (Fig. 3G). The heterogenous nature of the natural lipid extract as well as the presence of potential Cu2+ adducts makes it difficult to assign the different adducts with high confidence. The two highest abundant adducts (783 Da and 835 Da, assuming these are bound to two Cu2+ ions) ejected with TRAAK are consistent with those identified using a protocol56 to enrich lipids from brain polar extract in detergent (Fig. S9†). These lipids likely correspond to phosphatidylserine. Taken together, these results demonstrate that complex lipid extracts can be used and TRAAK ejects with specific lipids.
Applicability to functional assays
We next examined if the proteoliposomes prepared for native MS studies are suitable for functional assays. After several modifications (see methods) to an established cell-free potassium flux assay,36 the same proteoliposomes used for native MS studies could be used to measure potassium flux (Fig. S8†). Other membrane mimetics, such as bicelles and nanodiscs, do not permit the generation of an electrochemical gradient that is often necessary for functional assays. While we focus on potassium transport, we envisage that other functional assays can be implemented to test activity of the reconstituted membrane protein.Discussion
In this work, we developed an improved and streamlined procedure for the preparation of unilamellar proteoliposomes for native MS studies. We find SULs used for encapsulation of cargo or reconstitution of MAPs prepared by extrusion produce samples that are most amenable for native MS analyses. We also observed ejection of protein complexes for larger size vesicles, such as 100 nm. However, the smaller liposomes (50 nm) resulted in improved mass spectra of ejected protein complexes, especially for membrane proteins bound to lipids. Detection of encapsulated cargo was less dependent on liposome size and m-NBA is not required but facilitates activation of liposomes. Presumably the smaller liposome size enables more efficient collisional activation, which is necessary to eject membrane protein complexes. Here, we demonstrate broad utility of the method, from detection of encapsulated cargo to peripheral and membrane proteins embedded in proteoliposomes. The simplified procedure requires less preparation time and fewer steps compared to previously established protocols.30,31 Proteoliposomes were prepared at a protein–lipid ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. Any empty liposomes will not interfere with the analysis of ejected proteins from proteoliposomes as these signals are unique. We also show that proteoliposomes prepared for native MS studies can also be used for functional assays.
1000. Any empty liposomes will not interfere with the analysis of ejected proteins from proteoliposomes as these signals are unique. We also show that proteoliposomes prepared for native MS studies can also be used for functional assays.
A better understanding of MAP–lipid interactions will undoubtably require studies that include intact lipid bilayers. Here, we explored the utility of intact bilayers using Gβ1γ2, a peripheral membrane protein complex involved in cell signaling, and four integral membrane proteins complexes, two of which are human two-pore domain potassium channels. The Gβ1γ2 complex ejects as an intact complex from proteoliposomes. Increasing the collisional energy dissociates the complex into subunits, further validating the assignment of the heterodimeric complex. Unlike Gβ1γ2 and encapsulated soluble proteins, integral membrane proteins required the addition of m-NBA, a result consistent with previous reports.29,30 Other supercharging agents, such as sulfolane,63 had a similar effect.
We have previously determined the lipid binding affinity of AmtB and TRAAK in detergent, providing the unique opportunity to directly compare the lipids that remain bound to the proteins ejected from proteolioposomes. In general, proteoliposomes containing only POPC were easier to eject membrane protein complexes compared to those containing mixed lipids. In all cases, the addition of a lipid known to display high affinity for the membrane protein complex in detergent ejected with the protein. This is of particular importance as the composition of mixed lipids proteoliposomes used here contain 80–90% POPC. More specifically, AmtB ejected from proteoliposomes containing POPE and TOCDL showed an overall increase in lipid bound species. This result is consistent with previous findings that PE enhances AmtB–CDL interactions.48,56 TRAAK reconstituted in 10% POPA ejected bound to POPA and POPC, and in combination with copper(II). The observation of POPA bound, despite being only 10% in composition, is consistent with results obtained in detergent, where POPA binds TRAAK with about four-fold higher affinity over POPC. Moreover, TRAAK ejected from proteoliposomes containing 10% SOPS revealed an abundance of SOPS and copper(II) bound to TRAAK. TREK2 ejected from the same lipid composition showed near equal binding of POPC and SOPS, and no copper(II) binding. These results further corroborate our observations for TRAAK and TREK2 solubilized in detergent.62 More complex lipid environments can be explored, such as TRAAK ejected from proteoliposomes formed from a brain polar lipid extract. However, future directions will be the development of methods to further dissect adduct heterogeneity. These results demonstrate that membrane protein complexes can not only eject with lipids but also metal ions, which maybe important for regulating specific interactions and function.
In summary, we have developed a simplified method for preparing proteoliposomes for native MS studies. We show the encapsulation of cargo in SULs can be analyzed by native MS, presenting opportunities to better understand how confined environments, or molecular crowding, influence protein stability, aggregation, and molecular interactions. The capacity to employ native MS to release and study both peripheral and integral membrane complexes from specific lipid environments offers promising avenues to delve deeper into how lipids regulate protein structure and function. Moreover, the use of natural lipid extracts can potentially lead to the identification of pivotal protein–lipid interactions. In short, the ability to interrogate the retention of membrane-protein lipids post ejection from the membrane and in conjunction with functional assay opens new and exciting opportunities.
Material and methods
Protein expression and purification
Guanine nucleotide-binding protein subunits beta-1 (Gβ1, UniProt P62873, residues 2–340) and gamma-2 (Gγ2, UniProt P59768, residues 1–71) were subcloned into a pACEBac1 (Geneva Biotech) plasmid modified to co-express Gβ1 with a N-terminal tobacco etch virus (TEV) protease cleavable His6 tag and Gγ2 with a N-terminal TEV protease cleavable Strep-tag II. The plasmid was transformed into DH10EMBacY cells (Geneva Biotech) following the manufactures protocol, and the recombinant bacmid was purified and used to transfect Spodoptera frugiperda (Sf9) cells using PEI Max (Polysciences) transfection reagent to generate P1 virus.64 Gβ1 and Gγ2 were expressed in Trichoplusia ni (Tni) cell with the addition of baculovirus for 72 h at 27 °C while shaking. Cells were harvested by centrifugation at 4000 × g for 10 min and stored at −80 °C. Cell pellets were resuspended in KCl lysis buffer (200 mM KCl, 50 mM Tris, pH 7.4 at room temperature) and lysed using a Microfluidics M-110P microfluidizer operating at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi. The cell debris was removed by centrifugation at 20
000 psi. The cell debris was removed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 25 min at 4 °C. Membranes were pelleted through ultracentrifugation at 100
000 × g for 25 min at 4 °C. Membranes were pelleted through ultracentrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 hours at 4 °C. The pelleted membranes were resuspended in KCl-resuspension buffer (150 mM KCl, 50 mM Tris, pH 7.4 at room temperature) and 1% (w/v) n-dodecyl-beta-maltoside (DDM) was added for 2 hours to extract the Gβ1γ2 complex. The extracted proteins were centrifugated at 20
000 × g for 2 hours at 4 °C. The pelleted membranes were resuspended in KCl-resuspension buffer (150 mM KCl, 50 mM Tris, pH 7.4 at room temperature) and 1% (w/v) n-dodecyl-beta-maltoside (DDM) was added for 2 hours to extract the Gβ1γ2 complex. The extracted proteins were centrifugated at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 10 min and supernatant was filtered with a 0.45-micron syringe filter (Pall Corp.). The cleared extract was loaded onto a 2 mL bed volume of Ni-NTA Superflow resin (Qiagen) gravity-flow column pre-equilibrated in KRA-DDM (150 mM KCl, 50 mM Tris, 20 mM imidazole, 0.025% DDM, pH 7.4 at room temperature), washed with KRA-DDM, and the Gβγ complex was then eluted with KRB-DDM (KRA-DDM with 500 mM imidazole). Eluted complexes were diluted 20x volume with KRA-DDM and were further purified with a gravity-flow column packed with 1 mL bed volume of Strep-tactin Sepharose resin (IBA-Lifesciences) in KRA-DDM. After several column washes using KRA-DDM, the purified complex was eluted with KRA-DDM supplemented with 2.5 mM desthiobiotin. The eluent was concentrated using a 30 kDa MWCO concentrator (MilliporeSigma), aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C for future use.
000 × g at 4 °C for 10 min and supernatant was filtered with a 0.45-micron syringe filter (Pall Corp.). The cleared extract was loaded onto a 2 mL bed volume of Ni-NTA Superflow resin (Qiagen) gravity-flow column pre-equilibrated in KRA-DDM (150 mM KCl, 50 mM Tris, 20 mM imidazole, 0.025% DDM, pH 7.4 at room temperature), washed with KRA-DDM, and the Gβγ complex was then eluted with KRB-DDM (KRA-DDM with 500 mM imidazole). Eluted complexes were diluted 20x volume with KRA-DDM and were further purified with a gravity-flow column packed with 1 mL bed volume of Strep-tactin Sepharose resin (IBA-Lifesciences) in KRA-DDM. After several column washes using KRA-DDM, the purified complex was eluted with KRA-DDM supplemented with 2.5 mM desthiobiotin. The eluent was concentrated using a 30 kDa MWCO concentrator (MilliporeSigma), aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C for future use.
AmtB from Escherichia coli (UniProt P69681) were expressed and purified as previously descried.65 In brief, E. coli AmtB was expressed in E. coli C43 (DE3) (Lucigen) as a N-terminal HRV3C protease cleavable 10x His-tag and maltose binding protein (MBP) in TB (IBI Scientific). The cells were induced with 0.5 mM IPTG at an OD600 reached between 0.6–0.8 and expressed overnight at 20 °C while shaking. Cells were harvested by centrifugation (4000 × g for 10 min, 4 °C), resuspended in TBS buffer (150 mM NaCl, 50 mM Tris, pH 7.4 at room temperature), and were lysed by passing through a Microfluidics M-110P microfluidizer operating at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi. Insoluble material was pelleted by centrifugation (20
000 psi. Insoluble material was pelleted by centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 minutes at 4 °C), and the supernatant was subjected to ultracentrifugation (100
000 × g for 20 minutes at 4 °C), and the supernatant was subjected to ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 h, 4 °C) to pellet membrane. The purified membranes were resuspended in resuspension buffer (100 mM NaCl, 20 mM Tris, 20% glycerol, 5 mM βME (2-mercaptoethanol), pH 7.4 at room temperature) and was extracted overnight with 5% octyl glucoside (OG) at 4 °C. Extracted proteins were centrifuged to remove insoluble debris (40
000 × g for 2 h, 4 °C) to pellet membrane. The purified membranes were resuspended in resuspension buffer (100 mM NaCl, 20 mM Tris, 20% glycerol, 5 mM βME (2-mercaptoethanol), pH 7.4 at room temperature) and was extracted overnight with 5% octyl glucoside (OG) at 4 °C. Extracted proteins were centrifuged to remove insoluble debris (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min at 4 °C) and the supernatant was filtered (0.45 μm, Cytiva Whatman) prior to loading onto a HisTrap HP column (Cytiva) pre-equilibrated in NHA-DDM buffer (200 mM NaCl, 20 mM Tris, 20 mM imidazole, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature). The immobilized protein was eluted with NHB-DDM buffer (100 mM NaCl, 20 mM Tris, 500 mM imidazole, 10% glycerol, 0.025% DDM; pH 7.4 at room temperature). The eluent was then loaded onto a 5 mL MBPTrap HP column (Cytiva) pre-equilibrated in the MBP-loading buffer (100 mM NaCl, 20 mM Tris, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature) and eluted with MBP-elution buffer (MBP-loading buffer supplement with 10 mM maltose). Eluted protein was treated with HRV3C protease at a 100
000 × g for 20 min at 4 °C) and the supernatant was filtered (0.45 μm, Cytiva Whatman) prior to loading onto a HisTrap HP column (Cytiva) pre-equilibrated in NHA-DDM buffer (200 mM NaCl, 20 mM Tris, 20 mM imidazole, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature). The immobilized protein was eluted with NHB-DDM buffer (100 mM NaCl, 20 mM Tris, 500 mM imidazole, 10% glycerol, 0.025% DDM; pH 7.4 at room temperature). The eluent was then loaded onto a 5 mL MBPTrap HP column (Cytiva) pre-equilibrated in the MBP-loading buffer (100 mM NaCl, 20 mM Tris, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature) and eluted with MBP-elution buffer (MBP-loading buffer supplement with 10 mM maltose). Eluted protein was treated with HRV3C protease at a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 protein-to-protease ratio overnight at 4 °C. The sample was then passed over a HisTrap HP and flow through containing the tag-less protein was collected, concentrated, and purified by Superdex 200 Increase 10/300 GL column (Cytiva) with DDM-GF buffer (100 mM NaCl, 20 mM Tris, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature). Fractions containing AmtB were pooled, aliquoted, flash frozen and stored at −80 °C for future use.
1 protein-to-protease ratio overnight at 4 °C. The sample was then passed over a HisTrap HP and flow through containing the tag-less protein was collected, concentrated, and purified by Superdex 200 Increase 10/300 GL column (Cytiva) with DDM-GF buffer (100 mM NaCl, 20 mM Tris, 10% glycerol, 0.025% DDM, pH 7.4 at room temperature). Fractions containing AmtB were pooled, aliquoted, flash frozen and stored at −80 °C for future use.
AqpZ from Escherichia coli (UniProt P60844) was expressed and purified as a C-terminal TEV protease cleavable fusion to super folder green fluorescent protein66 and His6 tag as previously described.13 In brief, the AqpZ-GFP plasmid was transformed into BL21-A1 (Invitrogen). Expression of AqpZ-GFP was induced with the addition of 0.2% arabinose when the OD600 reached 0.6–0.8 and incubated overnight at 20 °C while shaking. The purification of AqpZ-GFP was identical to that described for AmtB with the exception that TEV protease was used.
TRAAK (K2p4.1b, Addgene #158744, residues 1–290 with N104Q and N108Q mutations to remove predicted N-linked glycosylation sites, Uniprot Q9NYG8-2) was expressed and purified as previously described.6 In brief, the TRAAK plasmid was introduced into the Glycoswitch Komagataella phaffii SuperMan5 (och1-Δ1, GAP-mannosidaseHDEL, pep4-Δ1, prb1-Δ1) strain (BioGrammatics Inc.) by electroporation (ECM 630, BTX) following the manufacturer's protocol. High-expression colonies were selected using a 96-well high throughput screen67 and were scaled up for protein expression. Selected colony in YPD from 96-well screen was used to inoculate a 50 mL YPD (1% yeast extract, 2% tryptone, 2% dextrose) and was allowed to grow overnight at 30 °C while shaking. The overnight cultured was used to inoculate 600 mL BMGY in baffled flasks and was grown for 24 h at 30 °C while shaking. The cells were spun down (2000 × g for 5 min) and were media exchanged into BMMY (0.5% MeOH) for expression at 30 °C. Anti-foaming agent was added to a final concentration of 0.01% polypropylene glycol 2000. TRAAK was expressed for 48 h before being harvested by centrifugation (2000 × g for 5 min). Cells were resuspended in KCl-lysis buffer and were lysed with a microfluidizer at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi. Cell lysate was clarified by centrifugation (20
000 psi. Cell lysate was clarified by centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min at 4 °C) and the supernatant was subjected to ultracentrifugation (100
000 × g for 20 min at 4 °C) and the supernatant was subjected to ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 h at 4 °C) to pellet the membrane. Membrane was resuspended in KCl-lysis buffer and was extracted with 1% DDM overnight at 4 °C. Insoluble debris were removed by centrifugation (20
000 × g for 2 h at 4 °C) to pellet the membrane. Membrane was resuspended in KCl-lysis buffer and was extracted with 1% DDM overnight at 4 °C. Insoluble debris were removed by centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min) followed by syringe filtration. Clarified supernatant was loaded onto a HisTrap HP column pre-equilibrated in KHA-DM buffer (200 mM KCl, 50 mM Tris, 10% glycerol, 0.2% DM(N-decyl-β-D-maltopyranoside), pH = 7.8 at room temperature) supplemented with 20 mM imidazole and the protein was eluted through gradient elution with KHB-DM (KHA-DM with 500 mM imidazole). Eluted protein was loaded onto a StrepTag-affinity column (4 mL bed volume, prepared in-house with Strep-Tactin Sepharose resin, iba) equilibrated in KHA-DM buffer and was eluted with KHA-DM supplemented with 3 mM desthiobiotin. Desalting of the eluates was performed on a HiPrep 26/10 Desalting column (Cytiva) with KHA-DM. TEV protease was added at a 1
000 × g for 10 min) followed by syringe filtration. Clarified supernatant was loaded onto a HisTrap HP column pre-equilibrated in KHA-DM buffer (200 mM KCl, 50 mM Tris, 10% glycerol, 0.2% DM(N-decyl-β-D-maltopyranoside), pH = 7.8 at room temperature) supplemented with 20 mM imidazole and the protein was eluted through gradient elution with KHB-DM (KHA-DM with 500 mM imidazole). Eluted protein was loaded onto a StrepTag-affinity column (4 mL bed volume, prepared in-house with Strep-Tactin Sepharose resin, iba) equilibrated in KHA-DM buffer and was eluted with KHA-DM supplemented with 3 mM desthiobiotin. Desalting of the eluates was performed on a HiPrep 26/10 Desalting column (Cytiva) with KHA-DM. TEV protease was added at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 protease to TRAAK molar ratio to cleave the tag overnight at room temperature. Cut-tags were removed by reverse IMAC and the flow through was further purified on a Superdex 200 Increase 10/300 GL column in KHA-DM. Fractions containing TRAAK were pooled and stored in a similar manner as AmtB.
10 protease to TRAAK molar ratio to cleave the tag overnight at room temperature. Cut-tags were removed by reverse IMAC and the flow through was further purified on a Superdex 200 Increase 10/300 GL column in KHA-DM. Fractions containing TRAAK were pooled and stored in a similar manner as AmtB.
TREK2 (Uniprot P57789-1, residues 55–335 with T58A mutation) was expressed and purified as previously described.8 Generation of recombinant baculovirus and expression of TREK2 was identical to that described for Gβ1γ2 with the exception that 5 μM kifunensine was added to the culture during expression.
Purified membranes containing overexpressed TREK2 were resuspended in KCl-lysis buffer and proteins were extracted by the addition of 1.2% OGNG (octyl glucose neopentyl glycol) for 2 h at 4 °C. The extracted protein was clarified by centrifugation (21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min) followed by syringe filtration. The supernatant was loaded onto a gravity-flow StrepTag-affinity column equilibrated in KHA-OGNG (KHA with 0.12% OGNG). Immobilized protein was washed with KHA-OGNG followed by KHA-C10E5 (KHA with 0.062% C10E5 (pentaethylene glycol monodecyl ether)) and was eluted with KHA-C10E5-DTB (KHA-C10E5 with 3 mM desthiobiotin). Eluted protein was desalted using HiPrep 26/10 Desalting column in KHA-C10E5 and was treated with EndoH to reduce the glycan heterogeneity. Strep-affinity column was used to further purify the treated proteins. TREK2 were desalted and were stored similarly as AmtB.
000 × g for 5 min) followed by syringe filtration. The supernatant was loaded onto a gravity-flow StrepTag-affinity column equilibrated in KHA-OGNG (KHA with 0.12% OGNG). Immobilized protein was washed with KHA-OGNG followed by KHA-C10E5 (KHA with 0.062% C10E5 (pentaethylene glycol monodecyl ether)) and was eluted with KHA-C10E5-DTB (KHA-C10E5 with 3 mM desthiobiotin). Eluted protein was desalted using HiPrep 26/10 Desalting column in KHA-C10E5 and was treated with EndoH to reduce the glycan heterogeneity. Strep-affinity column was used to further purify the treated proteins. TREK2 were desalted and were stored similarly as AmtB.
Preparation of proteoliposomes for native mass spectrometry analysis
A detailed flowchart of the method adapted from 6,30 with modification is provided in Fig. S1.† Lipid stocks (Avanti Polar Lipids) in chloroform were dried under nitrogen stream, washed with pentane, and dried overnight in a vacuum chamber overnight to form lipid films. Lipid films were rehydrated in water to a final concentration of 20 mM. For single-lipid-component liposomes, lipid stocks were diluted to 10 mM lipid concentration with appropriate buffer (final buffer concentration of 20 mM Tris, 150 mM NaCl or KCl at pH 7.4). For liposomes containing mixed lipids, lipid stocks were mixed to achieve the desired molar ratios. For example, 10% POPA in POPC would have 2 mM POPA and 18 mM POPC before being diluted to 10 mM lipid concentration. Lipids in buffer of choice from above were extruded using 50 nm membrane filter (Cytiva) with multiple passages, or until the solution turned translucent, to form large unilamellar vesicles (LUVs). To solubilize the LUVs, 2-20x critical micelle concentration (CMC) of detergent was added and the lipid-detergent mixture was equilibrated at 4 °C while rotating for 1 h or until the mixture became transparent/translucent. Peripheral and integral membrane protein samples were added to the solubilized lipids at a protein to lipid molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. The liposome and protein mixture was diluted with buffer to reach a final lipid concentration of 5 mM. The protein–lipid mixture was equilibrated for 1–2 h at 4 °C while rotating. Detergents were removed by the addition of bio-beads (Bio-Rad) prepared by washing with MeOH, five times with water, and three times with buffer. Bio-beads were added to fill roughly half of the solution volume and were incubated at 4 °C while rotating for 1–2 h or until mixture turned opaque. Proteoliposome mixture was remove by pipetting with attention to avoid bio-beads and extruded using 50 nm pore size membrane (Cytiva) to generate unilamellar proteoliposomes (PUVs). For native MS studies, proteoliposomes were dialyzed using a 10 kDa MWCO dialysis membrane overnight against 1 L of 200 mM ammonium acetate pH 7 at 4 °C.
1000. The liposome and protein mixture was diluted with buffer to reach a final lipid concentration of 5 mM. The protein–lipid mixture was equilibrated for 1–2 h at 4 °C while rotating. Detergents were removed by the addition of bio-beads (Bio-Rad) prepared by washing with MeOH, five times with water, and three times with buffer. Bio-beads were added to fill roughly half of the solution volume and were incubated at 4 °C while rotating for 1–2 h or until mixture turned opaque. Proteoliposome mixture was remove by pipetting with attention to avoid bio-beads and extruded using 50 nm pore size membrane (Cytiva) to generate unilamellar proteoliposomes (PUVs). For native MS studies, proteoliposomes were dialyzed using a 10 kDa MWCO dialysis membrane overnight against 1 L of 200 mM ammonium acetate pH 7 at 4 °C.
Ubiquitin and lysozyme egg white were dissolved in MiliQ water to a final concentration of 20 μM. The protein samples were filtered with a 0.45-micron syringe filter (Pall Corp.). Proteins were flash-frozen in liquid nitrogen and were stored at −80 °C for future use. For encapsulation of soluble proteins, the protocol described above was used with the exception that proteins were incorporated into the liposomes at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 protein to lipid molar ratio. Upon extrusion, samples were dialyzed against 1 L 200 mM ammonium acetate (pH = 7) at 4 °C overnight. 20 kDa MWCO dialysis membranes were specifically used to dialyze out any soluble proteins that are not incorporated into the liposome.
500 protein to lipid molar ratio. Upon extrusion, samples were dialyzed against 1 L 200 mM ammonium acetate (pH = 7) at 4 °C overnight. 20 kDa MWCO dialysis membranes were specifically used to dialyze out any soluble proteins that are not incorporated into the liposome.
Native mass spectrometry
Proteins for native MS study were prepared as previously reported.50 In brief, proteins were buffer exchanged into 200 mM ammonium acetate (2xCMC detergent was added for membrane proteins) with a centrifugal desalting column (Micro Bio-Spin 6, Bio-Rad). For Gβ1γ2 complex, 2xCMC DDM was used. For AmtB and AqpZ-GFP, 2xCMC C8E4 (tetraethylene glycol monooctyl ether) was used. For TRAAK and TREK2, 2x CMC C10E5 (pentaethylene glycol monodecyl ether) was used. Native MS data were collected on a Thermo Exactive Plus with an Extended Mass Range Orbitrap mass spectrometer (Thermo Scientific). The tuning parameters for each protein are listed in Tables S1–4.† Proteoliposome samples dialyzed against 200 mM ammonium acetate were collected on the same mass spectrometer with the tuning parameters listed in Tables S1–4.† For proteoliposomes containing peripheral and integral membrane proteins, 1–5% m-NBA (3-Nitrobenzyl alcohol, CAS no.:619-25-0, Sigma-Aldrich), a supercharging agent, was mixed with proteoliposome prior to introduction into the mass spectrometer. Native MS data were deconvoluted and analyzed using UniDec.68Fluorescence flux assay for TRAAK and TREK2
The liposome flux assay was performed as previously described60 with modification. The fluorescence was measured on a CLARIOstar (BMG LabTech) microplate reader with the excitation wavelength (410 nm with 10 nm window) and record the emission at 490 nm with a 10 nm. In brief, 10 μL of proteoliposome in 150 mM potassium acetate (pH = 7.4) was mixed with 190 μL of 150 mM sodium acetate (pH = 7.4) followed by 5 μL of ACMA solution (9-amino-6-chloro-2-methoxyacridine, 65 μM in sodium acetate). The baselined was measure for 1 min for every 5 s. CCCP (10 μL, 1.6 μM CCCP in sodium acetate) was added to initiate fluorescence decay and was recorded for 7 min every 5 s. To establish the endline, 1 μL of valinomycin (9 μM in DMSO) was added and the fluorescence was read for 5 min every 5 s.Data availability
Data associated with this manuscript is available upon reasonable request.Author contributions
Y. Z., S. Y., and A. L. designed the research. Y. Z., S. Y., T. Z., J. C., and L. S. expressed and purified proteins, and performed experiments. Y. Z., S. Y., and A. L. drafted the manuscript with input from the other authors.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by Welch Foundation (A-2106-20220331) and NIH (R01GM138863, R01GM139876 and RM1GM1454316) awarded to A.L., and instrumentation support from NIH (RM1GM149374).References
- K. R. Vinothkumar and R. Henderson, Structures of membrane proteins, Q. Rev. Biophys., 2010, 43, 65–158 CrossRef CAS PubMed.
- J. Martin and A. Sawyer, Elucidating the structure of membrane proteins, Biotechniques, 2019, 66, 167–170 CrossRef CAS PubMed.
- J. P. Overington, B. Al-Lazikani and A. L. Hopkins, How many drug targets are there?, Nat. Rev. Drug Discovery, 2006, 5, 993 CrossRef CAS PubMed.
- Y. Atsmon-Raz and D. P. Tieleman, Parameterization of Palmitoylated Cysteine, Farnesylated Cysteine, Geranylgeranylated Cysteine, and Myristoylated Glycine for the Martini Force Field, J. Phys. Chem. B, 2017, 121, 11132–11143 CrossRef CAS PubMed.
- E. Ozkirimli and C. B. Post, Src kinase activation: A switched electrostatic network, Protein Sci., 2006, 15, 1051–1062 CrossRef CAS PubMed.
- S. Schrecke, Y. Zhu, J. W. McCabe, M. Bartz, C. Packianathan, M. Zhao, M. Zhou, D. Russell and A. Laganowsky, Selective regulation of human TRAAK channels by biologically active phospholipids, Nat. Chem. Biol., 2021, 17(1), 89–95 CrossRef CAS PubMed.
- X. Cong, Y. Liu, W. Liu, X. Liang, D. H. Russell and A. Laganowsky, Determining Membrane Protein-Lipid Binding Thermodynamics Using Native Mass Spectrometry, J. Am. Chem. Soc., 2016, 138, 4346–4349 CrossRef CAS PubMed.
- Y. Zhu, S. Schrecke, S. Tang, M. T. Odenkirk, T. Walker, L. Stover, J. Lyu, T. Zhang, D. Russell, E. S. Baker, X. Yan and A. Laganowsky, Cupric Ions Selectively Modulate TRAAK–Phosphatidylserine Interactions, J. Am. Chem. Soc., 2022, 144, 7048–7053 CrossRef CAS PubMed.
- X. Cong, Y. Liu, W. Liu, X. Liang and A. Laganowsky, Allosteric modulation of protein-protein interactions by individual lipid binding events, Nat. Commun., 2017, 8, 2203 CrossRef PubMed.
- D. R. Lowy and B. M. Willumsen, Function and regulation of ras, Annu. Rev. Biochem., 1993, 62, 851–891 CrossRef CAS PubMed.
- Y. Zhou and J. F. Hancock, Lipid Profiles of RAS Nanoclusters Regulate RAS Function, Biomolecules, 2021, 11(10), 1439 CrossRef CAS PubMed.
- Y. Zhou, P. S. Prakash, H. Liang, A. A. Gorfe and J. F. Hancock, The KRAS and other prenylated polybasic domain membrane anchors recognize phosphatidylserine acyl chain structure, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(6), e2014605118 CrossRef CAS PubMed.
- A. Laganowsky, E. Reading, T. M. Allison, M. B. Ulmschneider, M. T. Degiacomi, A. J. Baldwin and C. V. Robinson, Membrane proteins bind lipids selectively to modulate their structure and function, Nature, 2014, 510, 172–175 CrossRef CAS PubMed.
- J. Gault, J. A. Donlan, I. Liko, J. T. Hopper, K. Gupta, N. G. Housden, W. B. Struwe, M. T. Marty, T. Mize, C. Bechara, Y. Zhu, B. Wu, C. Kleanthous, M. Belov, E. Damoc, A. Makarov and C. V. Robinson, High-resolution mass spectrometry of small molecules bound to membrane proteins, Nat. Methods, 2016, 13, 333–336 CrossRef PubMed.
- T. M. Allison, E. Reading, I. Liko, A. J. Baldwin, A. Laganowsky and C. V. Robinson, Quantifying the stabilizing effects of protein-ligand interactions in the gas phase, Nat. Commun., 2015, 6, 8551 CrossRef CAS PubMed.
- S. M. Fantin, K. F. Parson, S. Niu, J. Liu, D. A. Polasky, S. M. Dixit, S. M. Ferguson-Miller and B. T. Ruotolo, Collision Induced Unfolding Classifies Ligands Bound to the Integral Membrane Translocator Protein, Anal. Chem., 2019, 91, 15469–15476 CrossRef CAS PubMed.
- N. P. Barrera, N. Di Bartolo, P. J. Booth and C. V. Robinson, Micelles protect membrane complexes from solution to vacuum, Science, 2008, 321, 243–246 CrossRef CAS PubMed.
- J. Marcoux, S. C. Wang, A. Politis, E. Reading, J. Ma, P. C. Biggin, M. Zhou, H. Tao, Q. Zhang, G. Chang, N. Morgner and C. V. Robinson, Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9704–9709 CrossRef CAS PubMed.
- H. Y. Yen, J. T. S. Hopper, I. Liko, T. M. Allison, Y. Zhu, D. Wang, M. Stegmann, S. Mohammed, B. Wu and C. V. Robinson, Ligand binding to a G protein-coupled receptor captured in a mass spectrometer, Sci. Adv., 2017, 3, e1701016 CrossRef PubMed.
- J. R. Bolla, J. B. Sauer, D. Wu, S. Mehmood, T. M. Allison and C. V. Robinson, Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ, Nat. Chem., 2018, 10, 363–371 CrossRef CAS PubMed.
- J. Marcoux and C. V. Robinson, Twenty years of gas phase structural biology, Structure, 2013, 21, 1541–1550 CrossRef CAS PubMed.
- H. Y. Yen, K. K. Hoi, I. Liko, G. Hedger, M. R. Horrell, W. Song, D. Wu, P. Heine, T. Warne, Y. Lee, B. Carpenter, A. Pluckthun, C. G. Tate, M. S. P. Sansom and C. V. Robinson, PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling, Nature, 2018, 559, 423–427 CrossRef CAS PubMed.
- J. T. S. Hopper, Y. T.-C. Yu, D. Li, A. Raymond, M. Bostock, I. Liko, V. Mikhailov, A. Laganowsky, J. L. P. Benesch, M. Caffrey, D. Nietlispach and C. V. Robinson, Detergent-free mass spectrometry of membrane protein complexes, Nat. Methods, 2013, 10, 1206–1208 CrossRef CAS PubMed.
- J. E. Keener, G. Zhang and M. T. Marty, Native Mass Spectrometry of Membrane Proteins, Anal. Chem., 2021, 93, 583–597 CrossRef CAS PubMed.
- S. M. Fantin, K. F. Parson, P. Yadav, B. Juliano, G. C. Li, C. R. Sanders, M. D. Ohi and B. T. Ruotolo, Ion mobility-mass spectrometry reveals the role of peripheral myelin protein dimers in peripheral neuropathy, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(17), e2015331118 CrossRef CAS PubMed.
- M. T. Marty, K. K. Hoi, J. Gault and C. V. Robinson, Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs, Angew Chem. Int. Ed. Engl., 2016, 55, 550–554 CrossRef CAS PubMed.
- J. E. Keener, H. S. Jayasekera and M. T. Marty, Investigating the Lipid Selectivity of Membrane Proteins in Heterogeneous Nanodiscs, Anal. Chem., 2022, 94, 8497–8505 CrossRef CAS PubMed.
- A. T. Iavarone, J. C. Jurchen and E. R. Williams, Supercharged protein and peptide ions formed by electrospray ionization, Anal. Chem., 2001, 73, 1455–1460 CrossRef CAS PubMed.
- J. E. Keener, D. E. Zambrano, G. Zhang, C. K. Zak, D. J. Reid, B. S. Deodhar, J. E. Pemberton, J. S. Prell and M. T. Marty, Chemical Additives Enable Native Mass Spectrometry Measurement of Membrane Protein Oligomeric State within Intact Nanodiscs, J. Am. Chem. Soc., 2019, 141, 1054–1061 CrossRef CAS PubMed.
- A. Panda, F. Giska, A. L. Duncan, A. J. Welch, C. Brown, R. McAllister, P. Hariharan, J. N. D. Goder, J. Coleman, S. Ramakrishnan, F. Pincet, L. Guan, S. Krishnakumar, J. E. Rothman and K. Gupta, Direct determination of oligomeric organization of integral membrane proteins and lipids from intact customizable bilayer, Nat. Methods, 2023, 20, 891–897 CrossRef CAS PubMed.
- D. S. Chorev, H. Tang, S. L. Rouse, J. R. Bolla, A. von Kügelgen, L. A. Baker, D. Wu, J. Gault, K. Grünewald, T. A. M. Bharat, S. J. Matthews and C. V. Robinson, The use of sonicated lipid vesicles for mass spectrometry of membrane protein complexes, Nat. Protoc., 2020, 15, 1690–1706 CrossRef CAS PubMed.
- D. S. Chorev, L. A. Baker, D. Wu, V. Beilsten-Edmands, S. L. Rouse, T. Zeev-Ben-Mordehai, C. Jiko, F. Samsudin, C. Gerle, S. Khalid, A. G. Stewart, S. J. Matthews, K. Grunewald and C. V. Robinson, Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry, Science, 2018, 362, 829–834 CrossRef CAS PubMed.
- J. W. Patrick, R. C. Gamez and D. H. Russell, The Influence of Lipid Bilayer Physicochemical Properties on Gramicidin A Conformer Preferences, Biophys. J., 2016, 110, 1826–1835 CrossRef CAS PubMed.
- J. W. Patrick, R. C. Gamez and D. H. Russell, Elucidation of conformer preferences for a hydrophobic antimicrobial peptide by vesicle capture-freeze-drying: a preparatory method coupled to ion mobility-mass spectrometry, Anal. Chem., 2015, 87, 578–583 CrossRef CAS PubMed.
- M. Frick, C. Schwieger and C. Schmidt, Liposomes as Carriers of Membrane-Associated Proteins and Peptides for Mass Spectrometric Analysis, Angew. Chem., Int. Ed., 2021, 60, 11523–11530 CrossRef CAS PubMed.
- Z. Su, E. C. Brown, W. Wang and R. MacKinnon, Novel cell-free high-throughput screening method for pharmacological tools targeting K+ channels, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5748–5753 CrossRef CAS PubMed.
- A. M. Amati, S. Graf, S. Deutschmann, N. Dolder and C. von Ballmoos, Current problems and future avenues in proteoliposome research, Biochem. Soc. Trans., 2020, 48, 1473–1492 CrossRef CAS PubMed.
- X. Yao, X. Fan and N. Yan, Cryo-EM analysis of a membrane protein embedded in the liposome, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18497–18503 CrossRef CAS PubMed.
- V. S. Mandala and R. MacKinnon, Voltage-sensor movements in the Eag Kv channel under an applied electric field, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2214151119 CrossRef PubMed.
- V. S. Mandala and R. MacKinnon, The membrane electric field regulates the PIP(2)-binding site to gate the KCNQ1 channel, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2301985120 CrossRef CAS PubMed.
- S. Chen, T. Getter, D. Salom, D. Wu, D. Quetschlich, D. S. Chorev, K. Palczewski and C. V. Robinson, Capturing a rhodopsin receptor signalling cascade across a native membrane, Nature, 2022, 604, 384–390 CrossRef CAS PubMed.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Liposomes and polymersomes: a comparative review towards cell mimicking, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, Lipid Nanoparticles horizontal line From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- W. M. Oldham and H. E. Hamm, Heterotrimeric G protein activation by G-protein-coupled receptors, Nat. Rev. Mol. Cell Biol., 2008, 9, 60–71 CrossRef CAS PubMed.
- D. Wootten, A. Christopoulos, M. Marti-Solano, M. M. Babu and P. M. Sexton, Mechanisms of signalling and biased agonism in G protein-coupled receptors, Nat. Rev. Mol. Cell Biol., 2018, 19, 638–653 CrossRef CAS PubMed.
- M. A. Lindorfer, N. E. Sherman, K. A. Woodfork, J. E. Fletcher, D. F. Hunt and J. C. Garrison, G protein gamma subunits with altered prenylation sequences are properly modified when expressed in Sf9 cells, J. Biol. Chem., 1996, 271, 18582–18587 CrossRef CAS PubMed.
- T. M. Allison, E. Reading, I. Liko, A. J. Baldwin, A. Laganowsky and C. V. Robinson, Quantifying the stabilizing effects of protein–ligand interactions in the gas phase, Nat. Commun., 2015, 6, 8551 CrossRef CAS PubMed.
- X. Cong, Y. Liu, W. Liu, X. Liang and A. Laganowsky, Allosteric modulation of protein-protein interactions by individual lipid binding events, Nat. Commun., 2017, 8, 2203 CrossRef PubMed.
- X. Cong, Y. Liu, W. Liu, X. Liang, D. H. Russell and A. Laganowsky, Determining Membrane Protein–Lipid Binding Thermodynamics Using Native Mass Spectrometry, J. Am. Chem. Soc., 2016, 138, 4346–4349 CrossRef CAS PubMed.
- A. Laganowsky, E. Reading, J. T. Hopper and C. V. Robinson, Mass spectrometry of intact membrane protein complexes, Nat. Protoc., 2013, 8, 639–651 CrossRef CAS PubMed.
- Y. Liu, X. Cong, W. Liu and A. Laganowsky, Characterization of Membrane Protein–Lipid Interactions by Mass Spectrometry Ion Mobility Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2016, 28, 579–586 CrossRef PubMed.
- J. Lyu, Y. Liu, J. W. McCabe, S. Schrecke, L. Fang, D. H. Russell and A. Laganowsky, Discovery of Potent Charge-Reducing Molecules for Native Ion Mobility Mass Spectrometry Studies, Anal. Chem., 2020, 92, 11242–11249 CrossRef CAS PubMed.
- J. W. Patrick, C. D. Boone, W. Liu, G. M. Conover, Y. Liu, X. Cong and A. Laganowsky, Allostery revealed within lipid binding events to membrane proteins, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 2976–2981 CrossRef CAS PubMed.
- J. W. Patrick and A. Laganowsky, Generation of Charge-Reduced Ions of Membrane Protein Complexes for Native Ion Mobility Mass Spectrometry Studies, J. Am. Soc. Mass Spectrom., 2019, 30(5), 886–892 CrossRef CAS PubMed.
- S. N. Sipe, J. W. Patrick, A. Laganowsky and J. S. Brodbelt, Enhanced Characterization of Membrane Protein Complexes by Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem., 2020, 92, 899–907 CrossRef CAS PubMed.
- Y. Zhu, M. T. Odenkirk, P. Qiao, T. Zhang, S. Schrecke, M. Zhou, M. T. Marty, E. S. Baker and A. Laganowsky, Combining native mass spectrometry and lipidomics to uncover specific membrane protein–lipid interactions from natural lipid sources, Chem. Sci., 2023, 14, 8570–8582 RSC.
- J. Noel, G. Sandoz and F. Lesage, Molecular regulations governing TREK and TRAAK channel functions, Channels, 2011, 5, 402–409 CrossRef CAS PubMed.
- S. G. Brohawn, W. Wang, A. Handler, E. B. Campbell, J. R. Schwarz and R. MacKinnon, The mechanosensitive ion channel TRAAK is localized to the mammalian node of Ranvier, eLife, 2019, 8, e50403 CrossRef PubMed.
- M. Fink, F. Lesage, F. Duprat, C. Heurteaux, R. Reyes, M. Fosset and M. Lazdunski, A neuronal two P domain K+ channel stimulated by arachidonic acid and polyunsaturated fatty acids, EMBO J., 1998, 17, 3297–3308 CrossRef CAS PubMed.
- S. Schrecke, Y. Zhu, J. W. McCabe, M. Bartz, C. Packianathan, M. Zhao, M. Zhou, D. Russell and A. Laganowsky, Selective regulation of human TRAAK channels by biologically active phospholipids, Nat. Chem. Biol., 2021, 17, 89–95 CrossRef CAS PubMed.
- H. Bang, Y. Kim and D. Kim, TREK-2, a new member of the mechanosensitive tandem-pore K+ channel family, J. Biol. Chem., 2000, 275, 17412–17419 CrossRef CAS PubMed.
- Y. Zhu, S. Schrecke, S. Tang, M. T. Odenkirk, T. Walker, L. Stover, J. Lyu, T. Zhang, D. Russell, E. S. Baker, X. Yan and A. Laganowsky, Cupric Ions Selectively Modulate TRAAK-Phosphatidylserine Interactions, J. Am. Chem. Soc., 2022, 144, 7048–7053 CrossRef CAS PubMed.
- D. A. Abaye, I. A. Agbo and B. V. Nielsen, Current perspectives on supercharging reagents in electrospray ionization mass spectrometry, RSC Adv., 2021, 11, 20355–20369 RSC.
- J. Scholz and S. Suppmann, A new single-step protocol for rapid baculovirus-driven protein production in insect cells, BMC Biotechnol., 2017, 17, 83 CrossRef CAS PubMed.
- S. Kumar, Y. Zhu, L. Stover and A. Laganowsky, Step toward Probing the Nonannular Belt of Membrane Proteins, Anal. Chem., 2022, 94, 13906–13912 CrossRef CAS PubMed.
- J. D. Pedelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Engineering and characterization of a superfolder green fluorescent protein, Nat. Biotechnol., 2006, 24, 79–88 CrossRef CAS PubMed.
- R. Weis, High-Throughput Screening and Selection of Pichia pastoris Strains, Methods Mol. Biol., 2019, 1923, 169–185 CrossRef CAS PubMed.
- M. T. Marty, A. J. Baldwin, E. G. Marklund, G. K. A. Hochberg, J. L. P. Benesch and C. V. Robinson, Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles, Anal. Chem., 2015, 87, 4370–4376 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed flowchart of the proteoliposome preparation steps. Native mass spectra of ubiquitin, lysozyme and Gβ1γ2 complex in solution. Native mass spectra of AmtB, AqpZ, and lysozyme ejected from POPC liposome. TRAAK and TREK2 ejected from 10% SOPS, 90% POPC liposome. Functional assay with TRAAK incorporated in liposomes. Table values of mass spectrometer parameters used for analysis. Theoretical and measured masses of Gβ1γ2 complex obtained via native MS. See DOI: https://doi.org/10.1039/d3sc04938h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |