 Open Access Article
Open Access ArticleSurface-enhanced Raman scattering sensing for detection and mapping of key cellular biomarkers
Yuanjiao
Yang†
,
Shan
Wu†
,
Yunlong
Chen
 * and
Huangxian
Ju
* and
Huangxian
Ju
 *
*
State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: ylc@nju.edu.cn; hxju@nju.edu.cn
First published on 25th October 2023
Abstract
Cellular biomarkers mainly contain proteins, nucleic acids, glycans and many small molecules including small biomolecule metabolites, reactive oxygen species and other cellular chemical entities. The detection and mapping of the key cellular biomarkers can effectively help us to understand important cellular mechanisms associated with physiological and pathological processes, which greatly promote the development of clinical diagnosis and disease treatment. Surface-enhanced Raman scattering (SERS) possesses high sensitivity and is free from the influence of strong self-fluorescence in living systems as well as the photobleaching of the dyes. It exhibits rich and narrow chemical fingerprint spectra for multiplexed detection, and has become a powerful tool to detect and map cellular biomarkers. In this review, we present an overview of recent advances in the detection and mapping of different classes of cellular biomarkers based on SERS sensing. These advances fully confirm that the SERS-based sensors and sensing methods have great potential for the exploration of biological mechanisms and clinical applications. Additionally, we also discuss the limitations of present research and the future developments of the SERS technology in this field.
Introduction
The identification and monitoring of biomarkers have attracted considerable attention in the early screening and diagnosis of diseases. These biomarkers including proteins,1–3 nucleic acids,4–6 glycans7,8 and many small molecules,9,10 such as small biomolecule metabolites and reactive oxygen species (ROS), and their expression, distribution, transformation and interaction in body fluids, organisms, tissues and cells are closely related to physiological and pathological processes. The abnormal excess or deficiency in expression of these biomarkers generally indicates the occurrence and deterioration of diseases. Therefore, the dynamic monitoring of the variant progress of biomarkers related to a particular disease is extremely significant and helpful for clinical diagnosis and therapy for diseases. The biggest challenge in the detection of these biomarkers is their ultralow expression levels at the early stages of the disease.11,12 Establishing highly sensitive, specific and reliable detection strategies of biomarkers is highly demanded for early disease diagnosis.A wide range of methods based on electrochemical analysis, mass spectrometry, and optical techniques have been developed for the detection of biomarkers in fluid samples.13–15 Among them, fluorescence and Raman technologies can be equipped with a microscope or confocal microscope to provide the species, morphology and distribution of the target molecules at the cellular level. Thus, they possess great potential in the detection and mapping of biomarkers at single cell and in vivo levels.16,17 Compared to fluorescence microscopy, Raman microscopy can avoid the interference of strong self-fluorescence in living systems and the photobleaching of the dyes, and is more suitable for the sensing applications in biological systems.18 Owing to the narrow fingerprint vibrational spectrum, Raman or surface-enhanced Raman scattering (SERS) microscopy has been extensively used in multiplexed detection.19
Since the SERS effect was first discovered in 1974,20 different enhancement mechanisms, such as electromagnetic enhancement (EM) and chemical enhancement (CE), have been proposed. The EM mechanism exhibits an amplification of Raman signals by 104 to 1011 times, while the CE mechanism only enhances the signals by 10–100 times. Therefore, the EM-based SERS effect greatly promotes the development of highly sensitive sensing and bioanalytical technology by integrating it with nanotechnology, particularly noble metal nanoparticles,21,22 and SERS methods have become powerful tools for the detection and mapping of key cellular biomarkers in complex living systems. Generally, the SERS sensing of living systems can be classified into direct and indirect methods. The direct methods are based on the analysis of SERS signals emitted by the target molecules themselves, thereby revealing the inherent Raman characteristics as well as important information, such as conformation and orientation during adsorption in a simple, label-free manner. However, the SERS signals of the target molecules are easily interfered with or even overwhelmed in complex living systems. To address this problem, indirect analysis based on nanoprobes functionalized with Raman reporters and targeting groups has been developed, which promises indirect methods with selectivity and practicality in multiplexed detection. Nevertheless, the nanoprobe-based SERS methods lose the rich structural and molecular information of the targets. Thus, the SERS sensing strategies for key cellular biomarkers still face some challenges in life science research and clinical diagnosis and therapy of diseases.
Recent research in this field mainly focuses on the highly sensitive visualization and real-time accurate quantification of important biomarkers in vivo. Besides, many SERS sensing methods have been widely applied in the detection and mapping of various biomarkers in cellular processes, which dig out rich and reliable biological information. Some review articles have been published to introduce the SERS-based methodology for multi-scale biomedical imaging and SERS sensing of molecular biomarkers in cellular processes.23–26 In this review, we comprehensively discuss the recent advances in SERS sensing for the detection and mapping of key biomarkers, including small biomolecules, proteins, nucleic acids, and especially glycans, with a particular focus on the latest consideration to improve the sensing performance including the efficiency, sensitivity, and selectivity and applications. We divide this topic into three categories: (1) the detection and mapping of small molecular biomarkers such as pH, ROS and small biomolecule metabolites, (2) the detection and mapping of biomacromolecules like proteins, nucleic acids and glycans, and (3) multi-modal analysis related SERS sensing (Fig. 1). The mechanisms of these sensing strategies along with the biological backgrounds, probe preparation and practical application are also presented. We hope this review will be favourable for researchers to design more SERS sensing methods for the analysis of key cellular biomarkers in single cells as well as complex living systems, which may eventually promote the exploration of vital mechanisms and the development of clinical diagnosis and treatment of diseases.
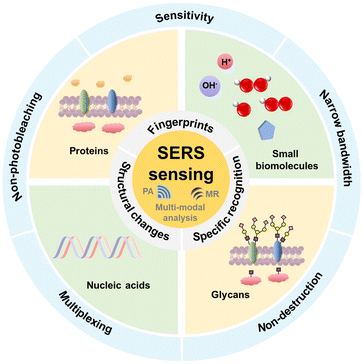 | ||
| Fig. 1 Schematic of the principles and advantages of SERS sensing strategies for detection or mapping of key cellular biomarkers including multi-modal analysis. | ||
Small biomolecules
pH
The environmental pH decides the protonation and deprotonation of some molecules. Both intracellular and extra-cellular pH values are crucial to regulate the functions and physiological activities of cells, which further influence many cell processes and cause malignant diseases.27,28 Thus, the real-time and spatial change in pH inside or outside cells is vital, and can be precisely and stably measured by SERS sensing with protonated or deprotonated Raman reporters responding to different pH environments.Raman reporters with protonation or deprotonation features are commercially available, including 4-mercaptopyridine (4-MPy),29 4-mercaptobenzoic acid (4-MBA)30 and 4-mercaptobenzonitrile (4-MBN). For example, Ren's group designed a 4-MPy modified gold nanoparticle (AuNP)-assembled SERS substrate to in situ sense the extracellular pH during cell apoptosis (Fig. 2a).31 They further used 4-MPy or 4-MBA to functionalize AuNPs with different morphologies to reveal pH-related pathological processes, including gradual alkalization, rapid acidification, and equilibrium pH recovery in the cell cycle.32
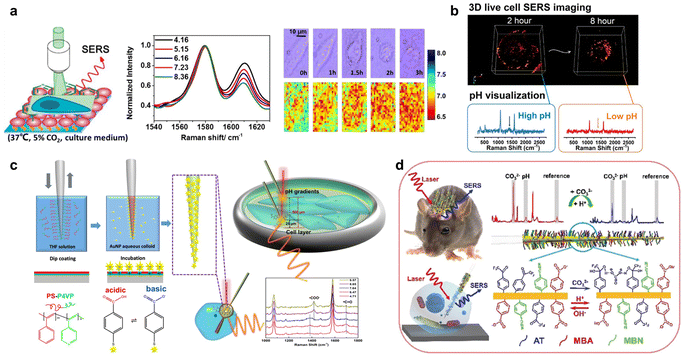 | ||
| Fig. 2 (a) In situ imaging of live-cell extracellular pH during cell apoptosis by SERS sensing. Reprinted with permission from ref. 31. Copyright 2018, American Chemical Society. (b) Live-cell three-dimensional SERS imaging of intracellular pH. Reprinted with permission from ref. 35. Copyright 2020, American Chemical Society. (c) The fabrication procedure of the SERS nanosensor and its application for measuring the pH of cells and pH gradients in cell culture environments. Reprinted with permission from ref. 36. Copyright 2020, American Chemical Society. (d) The developed SERS probe for the simultaneous biosensing of the CO32− concentration and pH value in live brains and single neurons. Reprinted with permission from ref. 40. Copyright 2019, Wiley-VCH. | ||
In acidic lysosomes, pH influences the degradation of many proteins related to lysosomal functions and diseases.33 Li et al. developed a SERS method to monitor the decrease and increase of pH in lysosomes corresponding to the autophagy and apoptosis processes with 4-MPy and BSA conjugated Au nanostars (AuNSs).34 To expand the observation of pH change from limited cell dimensions to whole-cell dimensions, Liz-Marzán's group proposed three dimensional (3D) SERS imaging of pH change during the endocytosis of AuNPs using 4-MBA and polyarginine modified AuNSs, which was promising to detect pH variations up to the single-particle level during further endocytosis of the nanoparticles (Fig. 2b).35
The stability and uneven distribution of the constructed SERS probe can influence the reproducibility of the SERS sensing of pH. Masson's group used 4-MBA functionalized AuNSs to uniformly decorate nanofibers and produce intrinsic hotspots. By inserting the SERS nanofibers into cells or placing the SERS nanofibers near (25 μm) or far (500 μm) from monolayer cancer cells, the intracellular and extracellular pH gradients of cells were measured with high sensitivity and spatial resolution (Fig. 2c).36
In addition to small molecules, some cytosine-rich ssDNA sequences can reversibly and tunably respond to pH through the formation of a DNA tetraplex, an i-motif structure. Zhang et al. designed cytosine-rich ssDNA and 4-MBN modified AuNPs as the SERS probe to dynamically trace intracellular pH through the nanostructure change of ssDNA sequences to the i-motif, then triggering the aggregation of AuNPs to produce an enhanced SERS signal of 4-MBN.37 Significantly, 4-MBN has often been used in cellular SERS detection because its C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N group can provide a Raman signal within the cellular Raman-silent region.
N group can provide a Raman signal within the cellular Raman-silent region.
From the cellular level to the in vivo level, the SERS probes for pH change are usually inserted into samples with minimal invasion to sense the pH. Dong's group designed 4-MBA attached gold nanoshells to cover the tip of a 260 μm-diameter stainless-steel acupuncture needle, which was inserted into a rat joint to sense pH and pulled out within 5 min to record the SERS spectra in vitro.38 Subsequently, to protect SERS probes from being removed during the insertion process, this group developed a SERS-active optical fibre with an inclined end and a 4-MBA attached gold nanoshell integrated hole near the inclined end, which could dynamically sense the pH changes in the muscle and brain of a living rat in situ without pulling out the fiber.39 For the SERS mapping and simultaneous quantification of pH in vivo, Tian's group assembled 4-MBA and 4-MBN on a layer of rough gold film coated quartz taper to achieve ratiometric SERS imaging and quantification of pH in a live brain through a plot of the peak intensity ratio of 4-MBN to 4-MBA versus the corresponding pH value in vitro. The SERS spectrum of 4-MBN in the cellular Raman-silent region was used as the inner reference for correcting the SERS spectrum of 4-MBA. In addition, they performed pH sensing in a single neuron through a SERS nanoprobe with a 200 nm tip to further reveal the mechanism of neuron death on the single-cell level (Fig. 2d).40
Small biomolecule metabolites
Small biomolecule metabolites are an important class of metabolites related to life processes and disease diagnosis, such as ATP, glucose and lactate. Some small biomolecule metabolites of cells and bacteria possess a strong SERS effect and can always be label-free detected according to their characteristic SERS fingerprints on SERS substrates.Lussier et al. placed a patch-clamp nanopipette decorated with AuNPs near the living cells and simultaneously monitored the content variety of secreted metabolites at the single molecule level such as pyruvate, lactate, ATP, and urea in the extracellular medium through the change in the corresponding SERS signal of small biomolecule metabolites (Fig. 3a).41 Liz-Marzán's group developed a plasmonic substrate formed with a superlattice of AuNPs on a glass cover slip to sense kynurenine, tryptophan and their ratio for assessing prognosis by SERS, and image the accumulation of purine derivatives in the cellular stress induced cancer cell death model.42 Despite cellular small biomolecule metabolites, Ju's group designed a SERS sandwich sticky note by wrapping AuNSs between hexagonal boron nitride layers, which was pasted on biofilms for real-time imaging and monitoring of pyocyanin secretion from Pseudomonas aeruginosa (Fig. 3b).43
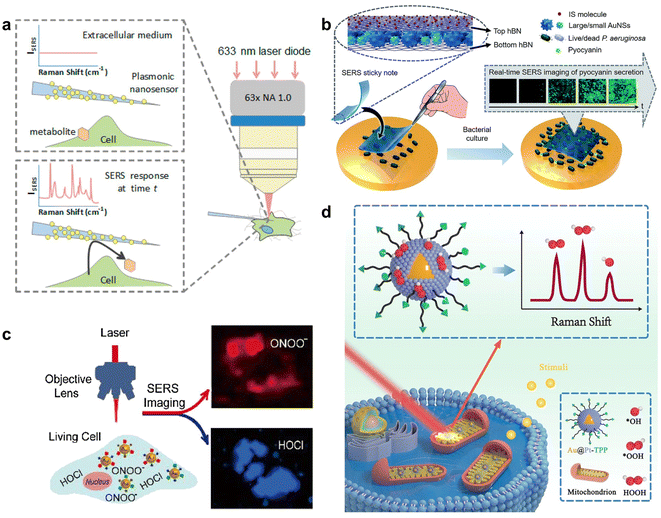 | ||
| Fig. 3 (a) SERS nanosensor to monitor cellular secretion. Reprinted with permission from ref. 41. Copyright 2018, American Chemical Society. (b) The sandwich type structure of the SERS sticky note and its attachment on live P. aeruginosa for SERS imaging of pyocyanin secretion. Reprinted with permission from ref. 43. Copyright 2020, The Royal Society of Chemistry. (c) Simultaneous SERS imaging of endogenous HOCl and ONOO− in a single living cell with AuNPs/MMP/MBAPE nanoprobes. Reprinted with permission from ref. 48. Copyright 2018, Elsevier B.V. (d) SERS borrowing strategy based on Au@Pt nanoprobes for direct spectroscopic identification of multiple mitochondrial ROS in living cells. Reprinted with permission from ref. 50. Copyright 2022, Wiley-VCH. | ||
In addition to the above label-free SERS detection of the secreted extracellular small biomolecule metabolites with AuNPs decorated substrates, Zhang et al. utilized the dimers of DTNB labelled DNA-Au Janus nanoparticles induced by ATP to generate an enhanced SERS signal of DTNB, and proposed a method for in situ sensing of intracellular ATP.44
ROS
ROS including superoxide anions (O2−), hydrogen peroxide (H2O2) and hydroxyl radicals (˙OH) are important to many diseases like cancer, diabetes and cardiovascular diseases.45 The sensing of ROS in living cells can be realized through indirect or direct measurements, which respectively utilize the ROS-induced structural transition of specific Raman reporters labelled on SERS probes or the fingerprint peaks of ROS. Kumar et al. designed a Raman probe with myoglobin modified core-satellite AuNPs to respond to the conversion of Fe(III)–OH2 of myoglobin to Fe(IV)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in the presence of ROS, which could be used to detect non-specific ROS in the concentration range from 10−2 to 10−10 M.46 Except for the non-specific and general reaction of Fe(III)–OH2 with ROS, some groups also used special reactions of sensing molecules on SERS probes to monitor a specific ROS type. Li et al. prepared 2-mercapto-4-methoxy-phenol (MMP)47 and 4-mercaptophenyl boronic acid pinacol ester (MBAPE) co-modified AuNPs, which exhibited a transition of the Raman spectrum from hydroxyl and methoxyl moieties to diquinone moieties and phenolic hydroxyl groups upon specific reaction with HOCl and ONOO−, enabling the highly selective detection and imaging of HOCl and ONOO− in living cells due to the specific probing reactions (Fig. 3c).48 Tang's group prepared seleno-phenylboronic acid pinacol ester modified AuNPs through the stable Au–Se interaction, which were used to detect H2O2 based on H2O2-specific recognition and quantify H2O2 in mitochondria via the characteristic peak ratio of B–O at 993 cm−1 to the constant internal standard (IS) peak of C–H at 1071 cm−1.49
O in the presence of ROS, which could be used to detect non-specific ROS in the concentration range from 10−2 to 10−10 M.46 Except for the non-specific and general reaction of Fe(III)–OH2 with ROS, some groups also used special reactions of sensing molecules on SERS probes to monitor a specific ROS type. Li et al. prepared 2-mercapto-4-methoxy-phenol (MMP)47 and 4-mercaptophenyl boronic acid pinacol ester (MBAPE) co-modified AuNPs, which exhibited a transition of the Raman spectrum from hydroxyl and methoxyl moieties to diquinone moieties and phenolic hydroxyl groups upon specific reaction with HOCl and ONOO−, enabling the highly selective detection and imaging of HOCl and ONOO− in living cells due to the specific probing reactions (Fig. 3c).48 Tang's group prepared seleno-phenylboronic acid pinacol ester modified AuNPs through the stable Au–Se interaction, which were used to detect H2O2 based on H2O2-specific recognition and quantify H2O2 in mitochondria via the characteristic peak ratio of B–O at 993 cm−1 to the constant internal standard (IS) peak of C–H at 1071 cm−1.49
As for direct Raman detection of multiple ROS in the mitochondria of living cells, Zhu's group developed Au@Pt core–shell NPs to capture ROS via the strong interaction between Pt and O, and simultaneously obtained the enhanced Raman fingerprint peaks of different mitochondrial multiple ROS in living cells (Fig. 3d).50
In summary, the SERS sensing of small molecular biomarkers in cellular processes can be performed with both the direct analysis of SERS signals of the target biomarker molecules and the indirect analysis of suitable SERS probes prepared by immobilizing small molecular biomarker-responding Raman reporters on SERS substrates. The pH-responding Raman reporters possess protonation or deprotonation features, and thus their SERS spectra are sensitive to the change in pH environments. The approach of small biomolecule metabolites on SERS substrates can greatly enhance their sensing signals, which leads to sensitive SERS sensing methods based on the inherent fingerprint spectra of these metabolites. Besides, metabolite-induced aggregation of Raman reporter labelled AuNPs, and ROS-responding structural transition of Raman reporters on AuNPs provide the possibility to detect multiple intracellular small molecular biomarkers with the SERS sensing platforms.
Proteins
Cellular proteins are highly crucial in regulating many processes in biological events such as intercellular communication, material transfer, cell migration, and cell apoptosis.51–53 The overexpression, inappropriate morphological changes and abnormal aggregation of cellular proteins are often closely related to the occurrence and deterioration of diseases. By detecting relevant membrane protein expression levels on cells, different cell lines can be distinguished accurately and efficiently, which is very essential to promote the diagnosis and prognosis of diseases. Owing to the inadequate information om single biomarker analysis, the development of highly sensitive simultaneous detection methods for multiple cancer biomarkers has become appealing for diagnostic and therapeutic strategies. To address this unmet need, many SERS-based strategies have been conducted in a multiplex manner using multiple Raman reporters and by combining a traditional or novel multi-channel device such as the paper-based flow assay and microfluidic platforms. Matt et al. developed a microfluidic platform combined with SERS-based spectral encoding for rapid, specific, sensitive and multiplexed detection of cancer protein biomarkers overexpressed in cancer cells (Fig. 4a).54 This platform could not only simultaneously capture target biomarkers from complex biological samples, but also enabled specific and sensitive detection including that of human epidermal growth factor receptor 2 (HER2), cell surface associated mucin 1 (MUC1), epidermal growth factor receptor (EGFR), and cell surface associated mucin 16 (MUC16) with a limit of detection (LOD) of 10 fg mL−1 in patient serum samples. Li et al. also proposed a SERS-based sandwich-type platform combined with novel Au–Ag alloy NBs utilizing the nanoyeast-scFvs as mAb alternatives for multiplex detection of soluble cancer protein biomarkers (sPD-1, sPD-L1 and sEGFR) (Fig. 4b).55 The LOD for sPD-1, sPD-L1, and sEGFR was 6.17, 0.68, and 69.86 pg mL−1, respectively. Zhang et al. constructed a lateral flow assay (LFA) based on core–shell SERS nanotags for multiplex detection of three cardiac biomarkers that play important roles in the early diagnosis of acute myocardial infarction (AMI).56 By utilizing silver–gold core–shell bimetallic nanotags, this method realized quantitative detection with ultrahigh sensitivity. The LODs for myoglobin, cardiac troponin I, and creatine kinase-MB isoenzymes were calculated to be below the clinical cut-off values, which were 3.2, 0.44, and 0.55 pg mL−1, respectively. In order to avoid cross-reactivity and false-negative results due to the hook effect of LFA and achieve more accurate and high-throughput quantitative analysis, Su et al. designed a paper-based SERS-vertical flow biosensor, named iREX (integrated Raman spectroscopic EXOsomes) biosensor, to quantitatively detect multiple exosomal proteins in clinical serum samples of patients (Fig. 4c).57 Through profiling of the discriminative expression of MUC1, HER2 and CEA in the EXOsome (EXO) samples from different breast cancer cell subtypes, the cancer molecular subtypes were identified noninvasively, which allowed accurate clinical diagnosis and efficient treatment of breast cancer. These studies indicate that SERS-based multiple detection has been gradually expanded to the analysis of multiple biomarkers for clinical diagnosis. Future progress in this field should persistently expand the capability of multiplexing, sensitivity and specificity with the help of algorithms and novel microfluidic devices.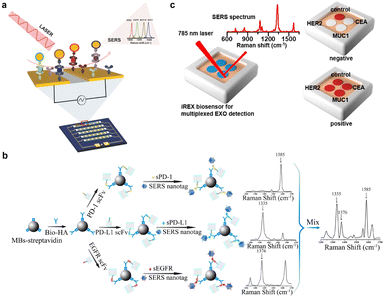 | ||
| Fig. 4 (a) Schematic illustration of multiplexed protein biomarker detection using ac-EHD-induced SERS-immunoassay. Reprinted with permission from ref. 54. Copyright 2016, Wiley-VCH. (b) Schematic illustration of the detection of sPD-1, sPD-L1, and sEGFR. Reprinted with permission from ref. 55. Copyright 2018, American Chemical Society. (c) Schematic of the iREX biosensor based multiplexed detection of exosomal proteins in the presence of no EXO and multiple EXOs. Reprinted with permission from ref. 57. Copyright 2023, American Chemical Society. | ||
Apart from in vitro direct detection of biomarkers, many SERS sensing strategies have been developed to visualize protein activities at the cellular level. This field has been expanded from the traditional methods of simple identification and detection at the cellular level to the current numerous ratiometric sensing strategies. These strategies commonly involve the utilization of two distinct types of Raman reporters modified on SERS probes. One is designed to specifically respond to the target analytes, and the other served as an IS, which largely diminishes the effects of heterogeneous distribution of nanoprobes. Zhong et al. designed a sensitive ratio-type SERS nanoprobe for imaging of the protease activity in single living cells of different cancer cells subtypes (Fig. 5a).58 They functionalized both rhodamine B (Rh B)-conjugated peptide substrates as a specific matrix metalloproteinase-2 (MMP-2) recognizer and 2-naphthalenethiol (2-NT) as an IS molecule on the plasmon-active AuNP surface, which exhibited sensitive and stable intensity. With the specific cleavage of substrate peptides by MMP-2, the SERS emission of RhB decreased or even disappeared whereas the emission of 2-NT was unaffected, which achieved the detection of MMP-2 activity in biological samples. Their results showed that the SERS nanoprobe enabled optical visualization of MMP-2 activity in normal breast epithelial cells and demonstrated the differential expression of the MMP-2 protein in different breast cancer subtypes. Li et al. also realized the quantitative ratiometric discrimination of two different types of breast cancer cell lines using a ratiometric SERS strategy (Fig. 5b).59 Two SERS nanoprobes modified with different Raman molecules were coupled to the urokinase plasminogen activated receptor (uPAR) and EGFR targeting peptides, respectively. To verify this ratiometric strategy, they examined the in vitro SERS imaging ability of these two nanoprobes in living cells, which showed that this imaging method could simultaneously quantify the expression of the uPAR and EGFR in MDA-MB-231 and MCF-7 cells. Additionally, Wang et al. developed a DNA colocalization-dependent system (Co-DNA-Locker) integrated ratiometric sensing strategy using controlled self-assembly of gold nanorod (AuNR) arrays for ultrasensitive detection of exosomes specifically secreted by SK-Br-3 cells.60 The AuNRs uniformly arranged in a hexagonal shape provided dense hot spots and highly improved the sensitivity of detection. Combined with the ratiometric strategy, the three-signal input switch pf DNA colocalization enabled the detection of exosomes specifically and accurately. This sensor realized a wide detection range (1.0 × 104–5.0 × 106 particles mL−1) and a low LOD (5.3 × 103 particles mL−1), which illustrated promising applications in clinical disease diagnosis and therapy. These ratiometric SERS strategies not only exclude the background interference such as influencing factors in a complex biological environment, but also significantly improve the signal-to-noise ratio of targeted cells in SERS imaging. At the same time, by combining them with the merits of high sensitivity and powerful multiplexing capability of SERS, as well as the accuracy of ratio identification without an external reference, accurate diagnosis and screening of breast cancer cells can be realized. In the future, the stability and sensitivity of SERS probes in vivo need to be further improved for their clinical applications.
 | ||
| Fig. 5 (a) Schematic illustration of the visualization of intracellular MMP-2 activity with the ratiometric SERS nanoprobe. Reprinted with permission from ref. 58. Copyright 2022, Elsevier B.V. (b) Schematic illustration of simultaneous SERS imaging of uPAR and EGFR biomarkers expressed on cells using uPAR- and EGFR-SERS nanoprobes encoded with distinct Raman signatures. Reprinted with permission from ref. 59. Copyright 2019, The Royal Society of Chemistry. | ||
Besides in vitro detection and in situ visualization of proteins associated with the recognition and enzymatic functions, some research studies to sensitively visualize the proteins that play important roles in the regulation of cellular plasma membrane repairing processes have also been reported. The SERS probes are usually constructed with Raman reporters and targeting groups for specific recognition. Ju’s group designed a pore-forming protein-induced SERS strategy for visualizing the membrane repairing process and further revealing its mechanism (Fig. 6a).61 Through conjugating the pore-forming protein streptolysin O (SLO) with AuNSs modified with Raman reporters, the SERS signal could be induced and obviously decreased with the initiation of membrane repair owing to the departure of the conjugated SLO. Thus, the results presented the visualization information of the complete cell membrane repair and illustrated the exact mechanism of plasma membrane repair, that is, the SLO endocytosis- and exocytosis-mediated repairing effect. This strategy holds great potential for screening the timely repair-blocking process to provide new insights into the design of the precision therapeutic schedule.
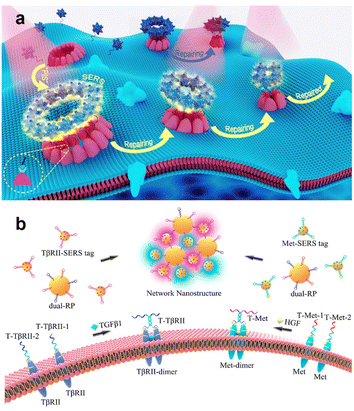 | ||
| Fig. 6 (a) Schematic illustration of the pore-forming protein-induced SERS strategy for dynamic monitoring of cell membrane repair. Reprinted with permission from ref. 61. Copyright 2017, Elsevier B.V. (b) Schematic illustration of simultaneous SERS imaging of the dimers of Met and TβRII proteins on a live cell membrane. Reprinted with permission from ref. 64. Copyright 2022, American Chemical Society. | ||
Transmembrane proteins can experience extracellular ligand binding processes that enable conformational changes to be induced so as to promote signal transduction and stimulate signalling pathways in important intracellular processes.62,63 However, it remains a major challenge to detect the protein dimerization state and in situ visualize the occurrence of dimerization as well as the subsequent activation of the cellular pathway. SERS sensing has the potential to provide very reliable solutions to address these limitations. Dong et al. established a highly sensitive and specific SERS imaging strategy for simultaneous imaging of the dual homodimerizations of membrane proteins based on the networking of AuNP-based dual-recognition probes and SERS tags with a proximity ligation-assisted catalytic hairpin assembly (CHA) (Fig. 6b).64 They synthesized dual-RPs by co-functionalizing hairpin-structured ssDNAs H1-Met (mesenchymal epithelial transition factor) and H1-TβRII (transforming growth factor-β type II receptor) on one 50 nm AuNP and hairpin-structured single-stranded DNAs H2-Met or H2-TβRII on 15 nm AuNPs, respectively, which were labelled with two sensitive Raman molecules on membrane proteins Met and TβRII. They confirmed that this SERS imaging strategy could simultaneously image dual dimerizations (i.e., Met-dimerizations and TβRII dimerizations) without any mutual interference by controlling the aggregation and isolation of Met and TβRII proteins. Subsequently, they further visualized the signal transduction between cancer cells and stromal cells or stem cells by imaging homodimerization of the dual membrane proteins.
The studies described above demonstrate that effective SERS imaging methods can serve as effective methods for identifying single or multiple signalling pathways accurately through the detection of membrane protein dimerization, and have the potential to monitor multiple intercellular signal transductions in both natural and complex cellular environments simultaneously. The future advancement of in vivo protein detection with SERS sensing platforms requires the sensitivity and specificity of SERS sensing strategies to be further enhanced for facilitating clinical applications.
Nucleic acids
Nucleic acids, mainly including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), play central roles in various biological events such as gene storage, gene encoding or decoding, gene transmission and gene expression.65,66 It is worth developing highly sensitive methods to detect the sequences and concentrations of DNA and RNA that can serve as biomarkers for clinical diagnosis and research on biological events. Among nucleic acids, microRNAs (miRNAs) are a group of small non-coding RNA molecules (19–25 nucleotides), and play significant roles in many important cellular processes such as cell proliferation and apoptosis.67–69 The vital regulatory role of miRNAs in the pathogenesis of different diseases like cancers suggests the potential application of the highly sensitive detection of miRNAs as biomarkers in cancer diagnosis and prevention.70–72 At present, abundant SERS sensing strategies have been well-established for miRNA detection due to the excellent characteristics of SERS, some of which also allow satisfactory quantitative determination. To overcome the limitation of traditional SERS substrates made of plasmonic metals with simple structures having low sensitivity and poor binding affinity with the targets for gaining more accurate detection, many research teams have begun to focus on designing a new SERS substrate by adjusting the structure of Au-based nanomaterials for essentially enhancing the detection efficiency. Wang's group designed an AuNP-bridge array to improve the DNA hybridization efficiency at the solid–liquid interface and realized the highly sensitive detection of miRNA-21 (Fig. 7a).73 They prepared this AuNP-bridge array by self-assembly of AuNPs in the gap of a SiO2 opal array, which promoted the formation of vertical nanochannels and microcavities. By inducing permeability and the micro-vortex effect, they improved the mass transfer efficiency of the target analyte and the probability of collision between the target analyte and acceptor at the solid–liquid interface consequently. They found that the DNA hybridization efficiency of the Au NP-bridge array was 15-fold higher than that of the planar array, and the constructed sensor for miRNA-21 showed faster DNA hybridization kinetics compared with previous sensing matrices, demonstrating potential in the implementation of various biosensor chips for genes, drugs, and disease markers.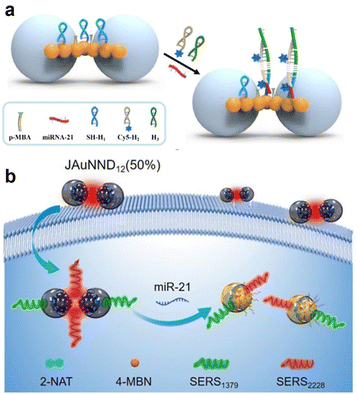 | ||
| Fig. 7 (a) Schematic illustration of the principle of microRNA-21 detection on an AuNP-bridge array. Reprinted with permission from ref. 73. Copyright 2022, American Chemical Society. (b) Schematic illustration of SERS-based ratiometric detection of miR-21 using JAuNND12 (50%). Reprinted with permission from ref. 74. Copyright 2021, Wiley-VCH. | ||
Meanwhile, the visualization of miRNA in living cells and in vivo has also been reported. Yang's group designed asymmetric core–shell AuNPs for SERS ratiometric detection of miRNA (Fig. 7b).74 They synthesized a series of Janus nanogap AuNPs (JAuNNPs) with varying proportions of Au shell coverage of ca. 100/75/50/25% and self-assembled the shell-to-shell and core-to-core JAuNNDs (50%) from amphiphilic JAuNNPs (50%) by modifying the hydrophilic and hydrophobic polymer properties on the Au core or shell to construct the JAuNND12 (50%) nanoprobe for miRNA detection and visualization by functionalizing specific DNA sequences with different Raman signal molecules. The sensitive JAuNND12 (50%) nanoprobe enabled specific detection and visualization of miRNA-21 both in living cells and in vivo without suffering from the physiological background and external interference. These studies respectively designed new types of SERS sensing substrates to improve the efficiency of DNA hybridization, which essentially cemented the interaction between the targets and the adjusted structure of the SERS substrate to realize more sensitive and accurate analysis of nucleic acids.
In addition, the identification and characterization of genetic DNA modifications and structural changes are vital for diagnosis, personalized therapy, and prognosis assessment of diseases such as cancer.75 As SERS sensing can provide intrinsic fingerprint information of analytes, a lot of direct and label-free detection strategies for unmodified DNAs have been developed, though some methods cannot provide reliable detection results due to the presence of impurity signals caused by modified layers of nanoparticles and the weak interaction between DNA and the metal surface. Recently, some nanoparticles with positive charge and aggregating agents or interfacial agents have been used to effectively improve the SERS sensing performance. Guerrini et al. reported an ultrasensitive label-free SERS method to detect unmodified dsDNA using positively charged Ag colloids with a spermine coating (AgNP@Sp) (Fig. 8a).76 Owing to the electrostatic interaction, the negatively charged DNA could adhere to AgNP@Sp, resulting in the stable clusters formed by nanoparticles aggregation, which generated reproducible and responsible SERS spectra. This simple strategy could not only quantify the hybridization events but also monitor a slight change in the single base in duplexes. Ren's group developed a reliable method for highly sensitive detection of single- and double-stand DNA based on iodide-modified Ag nanoparticles using a phosphate backbone signal as an IS, which obtains the absolute signal of each base.77 Guo's group also reported a universal SERS-based strategy for quantification of A/G bases in single stranded DNAs based on a dual-ion (I− and Al3+) modified silver nanoparticle with a novel interfacial agent, dichloromethane. These methods achieved the capability of detecting DNAs with longer chains and more complex structures.78 Liu et al. reported a desirable method by combining SERS and the polymerase chain reaction with multivariate statistical analysis to detect and classify BRAF wild type and V600E mutant genes. They synthesized plasmonic nanostructures with the control of the positive charge on their surface to facilitate high-affinity binding between target DNAs and nanostructures.79 Qi et al. designed a novel and uniform plasmonic sunflower-like gold nanostructure with high-density gap-plasmon “hot spots” and achieved reliable SERS detection in real-time monitoring of structural changes in cellular DNAs at the base level.80 These studies showed that the interaction between the detection object and SERS substrate can be greatly improved through charge modification of the SERS substrate, which further enhances the stability, sensitivity and repeatability of the label-free SERS sensing strategies.
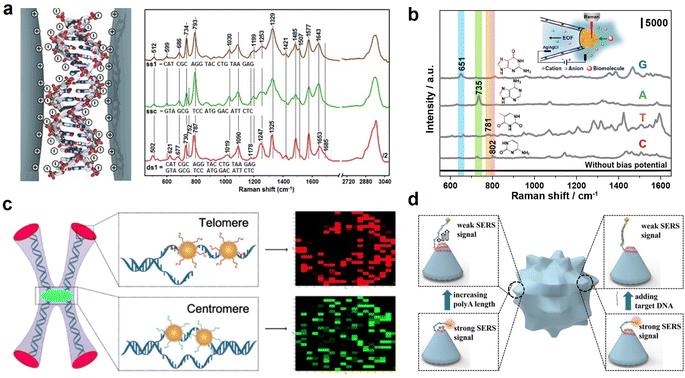 | ||
| Fig. 8 (a) Schematic illustration of spermine-modified Ag NPs for DNA detection. Reprinted with permission from ref. 76. Copyright 2015, Wiley-VCH. (b) Schematic illustration of detection of the four nucleobases by using gold plasmonic nanopores. Reprinted with permission from ref. 81. Copyright 2019, American Chemical Society. (c) Schematic illustration of the hybridization between the SERS nanoprobes and genome DNA. Reprinted with permission from ref. 82. Copyright 2016, American Chemical Society. (d) Schematic illustration of DNA conformation modulation with the polyA length on AuNSs and in situ SERS monitoring of conformation transformation. Reprinted with permission from ref. 83. Copyright 2017, Wiley-VCH. | ||
Some researchers integrated SERS sensing with the nanopore technique to perform the comprehensive analysis of the DNA structure. For example, Wang's group prepared gold plasmonic nanoprobes (GPNs) by in situ reduction of gold at the tip of a glass nanopipette (Fig. 8b).81 Under a biased electric field, DNA and amino acids could be driven through the nanopore, which provided a lot of information on the biomolecular structures by SERS. They found that GPNs could detect translocation of 10−9 M adenine after applying a bias potential and distinguished the difference in oligonucleotides at the single base level. In addition to exploring the structure of DNA, it was also of great significance for the regulation and characterization of its conformational changes.
Besides, both the DNA sequence and conformation-related information can be analysed in a more specialized manner. Zong et al. presented a SERS-based in situ hybridization method to measure the telomere length, which utilized two kinds of SERS nanoprobes, a telomere specific telo-probe and centromere specific centro probe, to hybridize with genome DNAs in situ (Fig. 8c).82 The SERS signal intensity of the telo-probe gradually increased with increasing telomere length, while that of the centro probe remained unchanged, which was used as the inner control signal. Therefore, the telomere length could be evaluated by virtue of the redefined T/C ratio. Ju's group reported a facile method for the modulation of DNA conformation from the “lie-down” to the “stand-up” conformation on the surface of multibranched AuNPs, which was regulated by the length of polyadenine connected with the DNA sequence and the hybridization of the target DNA sequence (Fig. 8d).83 This strategy could specifically recognize the DNA targets, and had been exploited to detect DNA sequences related to severe acute respiratory syndrome (SARS), demonstrating its great significance for the investigation of DNA-related cell physiological processes. To sum up, current nucleic acid sensing based on SERS not only enables ultrasensitive, selective detection and in situ imaging of nucleic acids in living cells, but also facilitates the exploration of nucleic acid sequences and variation of conformation, which provides more accurate results for the exploration of the vital mechanisms at the genetic level.
Glycans
Glycans are covalent assemblies of oligosaccharides and polysaccharides that can exist in the free form or as covalent glycoconjugates with proteins or lipids,84 and play key roles in regulating fundamental molecular and cell biology processes, tumor proliferation, invasion, as well as cell communication and interactions.85 They are distributed both on the cell surface and within the cells. The Raman signal of natural glycans is mainly composed of C–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational stretches, which is easily overwhelmed by the strong Raman signals of other biomolecules in cells. Therefore, the Raman sensing of glycans usually uses bioorthogonal Raman reporters or glycan-specific recognised groups like the boronic group to label glycans for SERS detection.
O vibrational stretches, which is easily overwhelmed by the strong Raman signals of other biomolecules in cells. Therefore, the Raman sensing of glycans usually uses bioorthogonal Raman reporters or glycan-specific recognised groups like the boronic group to label glycans for SERS detection.
Chen's group metabolically incorporated different bioorthogonal Raman reporters, including azide, alkyne, and nitrile groups, into glycans on cell surfaces, which produced the Raman signals in the cellular Raman-silent region for the direct detection or imaging of glycans by SERS (Fig. 9a).86 Then, they developed arrays of AuNPs or AgNPs on silicon wafers or glass slides as versatile SERS imaging platforms for visualizing various biomolecules like proteins, glycans, and lipids metabolically incorporated by biorthogonal Raman reporters on living cell surfaces.87
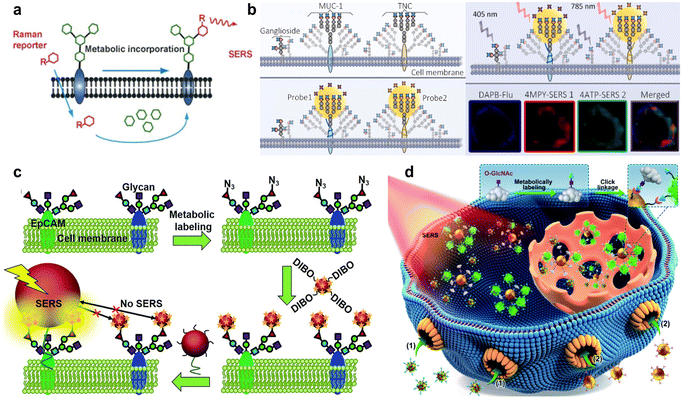 | ||
| Fig. 9 (a) Bioorthogonal Raman reporter strategy for SERS detection of glycans on live cells. Reprinted with permission from ref. 86. Copyright 2013, Wiley-VCH. (b) Classification process for the profiling of cell-surface SA subgroups. Reprinted with permission from ref. 94. Copyright 2021, Elsevier B.V. (c) Zone-controllable SERS effect for imaging of protein-specific glycans on the cell surface. Reprinted with permission from ref. 95. Copyright 2016, The Royal Society of Chemistry. (d) Quantitative SERS imaging strategy for O-GlcNAcylation mapping of single living cells. Reprinted with permission from ref. 97. Copyright 2022, The Royal Society of Chemistry. | ||
The termini of glycans are generally occupied by a negatively charged nine-carbon monosaccharide, sialic acid (SA), which is a critical biomarker84 related to malignant and metastatic phenotypes of cancer.88 SA can be specifically recognized and bound by phenylboronic acid (PBA) via strong esterification and the production of stable cyclic structures at physiological pH 7.4.89 Therefore, various SERS strategies have used PBA as a SA-recognized moiety as well as a Raman reporter to sense SA on the cell surface. Tabatabaei et al. imaged distinct SA expression of different position controlled cells on 4-mercaptophenylboronic acid (4-MPBA) functionalized plasmonic platforms by SERS.90 Ju group's prepared 3-MPBA and Raman reporter DTNB co-functionalized gold nanoflowers and poly(N-acetylneuraminic acid) modified AuNPs to form a single-core-multi-satellite nanostructure on cells, which could produce a strong SERS signal to image SA on living cells.91 Subsequently, they developed a similar PBA-SA system to form nano-conjugates for the detection of SA in the tumor region of the xenografted mice by SERS.92 In addition to the ordinary Raman reporter functionalized AuNPs, Liu's group proposed alkyne-bridged dimer AuNPs for precise imaging of SA expression of cancer cells.93
Except for the imaging of the glycan on the whole cells, glycan on specific proteins can provide more information related to protein glycosylation. Lv et al. used dansylamino phenylboronic acid (DAPB), a fluorescent boronic acid derivative that can recognize SA, and an MUC-1-recognizing aptamer or tenascin C (TNC)-recognizing aptamer to modify AuNPs pre-labelled with Raman reporters 4MPY and 4ATP. Combined with the fluorescence and SERS images, the merging of the fluorescence and SERS signals could indicate the SA distribution in the distinct space domains of MUC-1 or TNC on living cells (Fig. 9b).94 Ju's group also designed a zone-controllable SERS strategy to image the protein-specific glycosylation on the cell surface. Instead of labelling SA with PBA, they used dibenzocyclooctyne (DIBO) to label the azide-tagged SA through the bioorthogonal reaction on DTNB modified 10 nm AuNPs, and meanwhile utilized 30 nm or 40 nm AuNPs modified with the EpCAM aptamer to construct different SERS effect zones on the target protein. Only the SA on the target protein could bring DTNB on the 10 nm AuNP probe closer to the efficient SERS zone of the 30 nm or 40 nm AuNP probe, which exhibited a SERS signal for the imaging of EpCAM-specific SA on the living cell surface (Fig. 9c).95
In addition to glycans on the cell surface, the in situ detection of glycans inside cells remains a great challenge. O-GlcNAcylation connects single N-acetylglucosamine (GlcNAc) residues to O-linkages (serine or threonine) on proteins, which is abundant in cells and regulates many cancer processes.96 Ju's group proposed an in situ quantitative SERS imaging strategy for quantifying O-GlcNAcylation in living cells, which used azide labelled O-GlcNAcylated compounds (OGCs) and azide and DTNB co-labelled 15 nm-AuNPs to competitively bind with dibenzocyclooctyne labelled 40 nm-AuNPs to produce the OGC-negatively correlated SERS signal. Combined with an in vitro calibration curve, they successfully achieved the quantitative SERS imaging of OGCs inside cells (Fig. 9d).97
In summary, the SERS sensing strategies for glycans are usually performed through glycans labelled with bioorthogonal Raman reporters to produce Raman signals in the cellular Raman-silent region or Raman reporters modified SERS probes to produce enhanced Raman signals. These SERS sensing strategies successfully realize the sensing of extracellular and intracellular glycans as well as the glycans on specific proteins, and thus provide valuable information for the exploration of glycan-related life mechanism.
Multi-modal analysis
When the SERS sensing for the detection and imaging of multiple cell biomarkers has been achieved in vitro and in vivo, these developed methods have some problems, including abnormal aggregation, uneven distribution, and unnecessary accumulation of SERS tags in tissues and in vivo, which lead to unreliable signals.98,99 Multi-modal analysis is a practical solution for these problems, and has been developed by combining SERS sensing with other sensing technologies such as photoacoustic (PA) imaging, preoperative magnetic resonance imaging (MRI), and near infrared II (NIR II) imaging.100,101 In fact, multi-modal nanoprobes have been exploited for in vivo imaging of tumours to provide extremely rich, sensitive and real-time visual information at a greater tissue depth than previous research.SERS sensing can sensitively track the fingerprint signals of Raman reporters in tumors, and thus, can be used to distinguish tumors from the surrounding normal tissue.102 To avoid the interference of a complex tumor microenvironment, the exploration of a highly efficient enhancing substrate is of great significance in improving the reproducibility and stability of the SERS signal. Thus, the synthesis of novel nanostructures and assembled substrates of plasmonic mental nanomaterials with other materials has attracted increasing interest in recent years, and enables better biocompatibility and physiological stability for the SERS sensing of tumors. Wang's group established a highly effective theranostic strategy by preparing gold nanoframeworks (AuNFs) with large mesopores, which enabled the combination of Raman-photoacoustic multi-modal imaging and photothermal chemotherapy of tumours in the NIR-II biowindow.103 The AuNFs exhibited great absorbance in the NIR-II region, showing excellent ability of PA imaging and photothermal therapy (PTT) for deeper tumours. In order to target CD44-overexpressed tumor cells (MDA-MB-231), the AuNFs were modified with hyaluronic acid (HA), which was further embedded with gatekeeping doxorubicin (DOX) to evaluate the drug-loading efficiency and functionalized with a Raman reporter to realize SERS imaging. The synthesized conjugates had powerful efficacy for Raman-PA dual-modal imaging and NIR-II photo-chemotherapy to completely eradicate the tumour. Sun group's designed a pH-responsive SERS-based nanosystem that enabled the timely tracking of the drug release process and monitoring of the chemo-phototherapeutic effect (Fig. 10a).104 They deposited Fe3O4@Au@Ag on graphene oxide (GO) nanocomposites and further attached the Raman reporter 4MPBA and DOX to form the SERS probe, which could track the DOX dynamic release according to the real-time change in the 4-MPBA SERS intensity. Moreover, the tumour-targeting delivery of these nanocomposites could be realized through the specific binding of PBA moieties to SA overexpressed on the surface of tumour cells. Their results demonstrated that these SERS probes exhibited effective suppression of tumour xenografts by means of the release of DOX to induce the GO-mediated PTT killing effect. Song's group reported a dual-ratiometric core–satellite gold nanostructure combined with SERS and PA imaging for real-time and quantitative detection of the H2O2 produced in inflammation and tumours in vivo (Fig. 10b).105 For SERS and PA imaging, they constructed an amide bond between the mesoporous silica-coated nanogapped gold nanorods, which were equipped with ammonia and AuNPs co-functionalized with 4-mercaptobenzoboric acid and D-(+)-galactose. The combination of PA and SERS imaging enabled the accurate quantification of H2O2 produced in inflammation and tumours, as well as the monitoring of the anti-inflammation treatment progress in real-time.
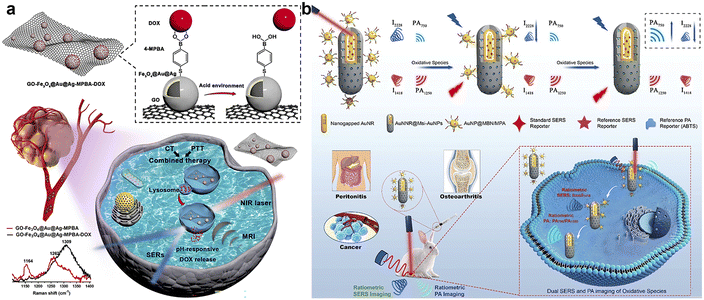 | ||
| Fig. 10 (a) Schematic illustration of real-time monitoring of pH-responsive DOX release for SERS/MR imaging-guided chemo-phototherapy. Reprinted with permission from ref. 104. Copyright 2023, Elsevier B.V. (b) Schematic illustration of the core–satellite nanostructure with dual ratiometric SERS and PA imaging signals for the detection of oxidative species (e.g. H2O2) produced in inflammation, tumors and osteoarthritis in rabbits with high precision in real-time. Reprinted with permission from ref. 105. Copyright 2020, Wiley-VCH. | ||
The combination of SERS sensing with other technologies is an attractive theranostic platform to acquire multiplex information in a biological environment, including the tracking of nanomedicine-based drug delivery/release and the monitoring of the therapeutic process, which provides new insights into the design of multi-modal strategies for responsible disease diagnosis. Moreover, nanoprobes equipped with NIR II imaging modes also exhibit the same advantages and can further exert the therapeutic effect. Thus, developing more appealing multi-modal strategies is of great importance for expanding the application of SERS sensing in disease diagnosis and therapy.
Summary and outlook
The detection and mapping of key cellular biomarkers are a long-term issue, and have been developed by numerous research groups for many years. This review provides an overview of recent advances of various SERS-based sensors and strategies for the sensitive detection and imaging of small molecules, proteins, nucleic acids, and glycans at cellular and in vivo levels. We specifically review several representative strategies for different classes of cellular biomarkers.Most SERS sensing strategies are designed with multifunctional SERS substrates, which are cleverly integrated with multiple functional groups to improve the recognition ability of targeted objects and achieve highly specific detection. Combining the multiplexing ability of SERS sensing with specific recognition in microfluid systems can realize the simultaneous analysis and screening of multiple biomarkers with high sensitivity and specificity. The sensitivity and accuracy of SERS sensing can be further improved by using the asymmetric or special array structure as the SERS substrate and introducing the ratiometric strategy to reduce the interference of the background signal. By optimizing the interaction between the targeted group and SERS substrate, label-free SERS sensing methods can offer the intrinsic fingerprint information of biomarkers in a more sensitive and reproducible manner. In addition, SERS sensing has been combined with other technologies to conduct multi-modal analysis, and has achieved excellent imaging and therapy monitoring of tumors. These multi-modal strategies exhibit the possibility of clinical application and increasing alternative to the previous single-modal techniques.
Despite the great improvement of SERS sensing performance, more comprehensive, universal and practical strategies for the detection and mapping of biomarkers to reveal significant biological mechanisms are still an urgent demand. The SERS sensing of biomarkers first need to overcome the interference of other species in complex biological samples. The search for novel SERS substrates that can provide more hot spots and improve the aggregation of detection targets in a maximally enhanced region is also urgent. The stability and biocompatibility of SERS probes should meet the requirements of signal reliability, reproducibility and detection accuracy. Moreover, the temporal resolution for monitoring important disease-related cellular physiological processes triggered by target biomarkers needs to be further improved for obtaining more information.
We predict that SERS sensing strategies can be designed to develop the practical applications from the following aspects: (1) using novel recognition mechanisms and microfluid sensors to improve the capability of extracting, identifying and screening of the target biomarkers; (2) improving SERS substrate fabrication following the guidance of physical enhancement mechanisms or by combining it with other tools such as DNA origami and nanopores; (3) attempting to assemble nanoparticles with novel materials for optimizing the stability and biocompatibility of SERS probes; (4) expanding the multiplexing capability with the assistance of algorithms to achieve high diagnostic accuracy, including machine learning and artificial intelligence; (5) upgrading confocal Raman microscopy and integrating it with other technologies such as super-resolution imaging and photoacoustic imaging to improve the spatial resolution and tissue depth of in vivo SERS sensing from the perspective of multi-modal analysis. We believe that SERS sensing strategies will achieve more sensitive and effective detection and mapping of more cellular biomarkers to offer more biological information and play essential roles in clinical diagnosis and therapy of diseases.
Author contributions
All authors contributed to the writing of the manuscript and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21974063, 21827812 and 21890741) and the program B for Outstanding PhD candidate of Nanjing University.References
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. LaBean, Science, 2003, 301, 1884–1886 CrossRef PubMed.
- C. L. Sun, H. W. Wang, M. Hayashi, L. C. Chen and K. H. Chen, J. Am. Chem. Soc., 2006, 128, 8378–8379 CrossRef.
- K. Morimoto, M. Toya, J. Fukuda and H. Suzuki, Anal. Chem., 2008, 80, 5616–5621 CrossRef PubMed.
- R. Elghanian, J. J. Storhoff, R. C. Mucic, R. L. Letsinger and C. A. Mirkin, Science, 1997, 277, 1078–1081 CrossRef CAS PubMed.
- T. A. Taton, C. A. Mirkin and R. L. Letsinger, Science, 2000, 289, 1757–1760 CrossRef CAS PubMed.
- K. Warncke and J. M. Canfield, J. Am. Chem. Soc., 2004, 126, 5932–5933 CrossRef PubMed.
- M. N. Christiansen, Proteomics, 2014, 14, 525–546 CrossRef CAS PubMed.
- K. Ueda, Proteom.–Clin. Appl., 2013, 7, 607–617 CrossRef CAS.
- B. A. Webb, M. Chimenti, M. P. Jacobson and D. L. Barber, Nat. Rev. Cancer, 2011, 11, 671–677 CrossRef CAS PubMed.
- S. S. Sabharwal and P. T. Schumacker, Nat. Rev. Cancer, 2014, 14, 709–721 CrossRef CAS PubMed.
- A. M. Lutz, J. K. Willmann, F. V. Cochran, P. Ray and S. S. Gambhir, PLoS Med., 2008, 5, e170 CrossRef PubMed.
- V. S. P. K. S. A. Jayanthi, A. B. Das and U. Saxena, Biosens. Bioelectron., 2017, 91, 15–23 CrossRef CAS PubMed.
- A. P. Black, H. Liang, C. A. West, M. Wang, H. P. Herrera, B. B. Haab, P. M. Angel, R. R. Drake and A. S. Mehta, Anal. Chem., 2019, 91, 8429–8435 CrossRef CAS PubMed.
- L. Ning, X. Li, D. Yang, P. Miao, Z. Ye and G. Li, Anal. Chem., 2014, 86, 8042–8047 CrossRef CAS PubMed.
- R. Yang, F. Li, W. Zhang, W. Shen, D. Yang, Z. Bian and H. Cui, Anal. Chem., 2019, 91, 13006–13013 CrossRef CAS.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS PubMed.
- X. Qian, X. H. Peng, D. Ansari, Q. Yin-Goen, G. Z. Chen, D. M. Shin, A. N. Young, M. D. Wang and S. Nie, Nat. Biotechnol., 2008, 26, 83–90 CrossRef CAS PubMed.
- Q. Xiong, C. Y. Lin, J. Ren, J. Zhou, K. Pu, M. B. Chan-Park, H. Mao, Y. C. Lam and H. Duan, Nat. Commun., 2018, 9, 1743 CrossRef PubMed.
- Y. Qiu, Y. Zhang, M. Li, G. Chen, C. Fan, K. Cui, J. B. Wan, A. Han, J. Ye and Z. Xiao, ACS Nano, 2018, 12, 7974–7985 CrossRef CAS PubMed.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- Y. Wang, B. Yan and L. Chen, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- J. Langer, J. D. Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, K. Faulds, F. J. G. Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, R. Itoh, M. Kall, J. Kneipp, N. A. Kotov, H. Kuang, E. C. L. Ru, H. K. Lee, J. F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J. Nam, S. M. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L. Tay, K. G. Thomas, Z. Tian, R. P. V. Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzan, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- J. S. Lin, X. D. Tian, G. Li, F. L. Zhang, Y. Wang and J. F. Li, Chem. Soc. Rev., 2022, 51, 9445–9468 RSC.
- D. Wu, Y. Chen, S. Hou, W. Fang and H. Duan, ChemBioChem, 2019, 20, 2432–2441 CrossRef CAS PubMed.
- H. Chen, Z. Cheng, X. Zhou, R. Wang and F. Yao, Anal. Chem., 2022, 94, 143–164 CrossRef CAS PubMed.
- Y. Hang, J. Boryczka and N. Wu, Chem. Soc. Rev., 2022, 51, 329–375 RSC.
- B. A. Webb, M. Chimenti, M. P. Jacobson and D. L. Barber, Nat. Rev. Cancer, 2011, 11, 671–677 CrossRef CAS PubMed.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- X.-S. Zheng, P. Hu, Y. Cui, C. Zong, J.-M. Feng, X. Wang and B. Ren, Anal. Chem., 2014, 86, 12250–12257 CrossRef CAS PubMed.
- J. E. Park, N. Yonet-Tanyeri, E. V. Ende, A.-I. Henry, B. E. P. White, M. Mrksich and R. P. V. Duyne, Nano Lett., 2019, 19, 6862–6868 CrossRef CAS PubMed.
- M. X. Xu, X. Ma, T. Wei, Z.-X. Lu and B. Ren, Anal. Chem., 2018, 90, 13922–13928 CrossRef CAS PubMed.
- X.-S. Zheng, C. Zong, X. Wang and B. Ren, Anal. Chem., 2019, 91, 8383–8389 CrossRef CAS PubMed.
- F. R. Maxfield, J. M. Willard and S. Lu, Lysosomes: Biology, Diseases, and Therapeutics, John Wiley & Sons, New York, 2016 Search PubMed.
- S.-S. Li, M. Zhang, J.-H. Wang, F. Yang, B. Kang, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2019, 91, 8398–8405 CrossRef CAS PubMed.
- Y. Z. Zhang, D. J. de Aberasturi, M. Henriksen-Lacey, J. Langer and L. M. Liz-Marzań, ACS Sens., 2020, 5, 3194–3206 CrossRef CAS PubMed.
- X. J. Zhao, S. Campbell, G. Q. Wallace, A. Claing, C. G. Bazuin and J.-F. Masson, ACS Sens., 2020, 5, 2155–2167 CrossRef CAS PubMed.
- Y. Zhang, Q. Liu, M. T. Liu, X. N. Zhang, X. Y. Li, L. H. Dai, M. Meng, D. B. Liu, Y. M. Yin and R. M. Xi, Sens. Actuators, B, 2022, 353, 131162 CrossRef CAS.
- C. Y. Pan, X. C. Li, J. Sun, Z. Li, L. Zhang, W. P. Qian, P. M. Wang and J. Dong, ACS Appl. Bio Mater., 2019, 2, 2102–2108 CrossRef CAS PubMed.
- C. Y. Pan, S. Y. Zhang, X. L. Xiong, Z. Li, B. W. Ai, W. P. Qian and J. Dong, Adv. Mater. Interfaces, 2022, 9, 2200328 CrossRef CAS.
- W. K. Wang, F. Zhao, M. Z. Li, C. P. Zhang, Y. H. Shao and Y. Tian, Angew. Chem., Int. Ed., 2019, 58, 5256–5260 CrossRef CAS PubMed.
- F. Lussier, T. Brulé, M. Vishwakarma, T. Das, J. P. Spatz and J.-F. Masson, Nano Lett., 2016, 16, 3866–3871 CrossRef CAS PubMed.
- J. Plou, I. García, M. Charconnet, I. Astobiza, C. García-Astrain, C. Matricardi, A. Mihi, A. Carracedo and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 1910335 CrossRef CAS.
- J. X. Guo, Y. Liu, Y. L. Chen, J. Q. Li and H. X. Ju, Chem. Sci., 2018, 9, 5906–5911 RSC.
- X. R. Zhang, Y. H. Ge, M. H. Liu, Y. J. Pei, P. He, W. L. Song and S. S. Zhang, Anal. Chem., 2022, 94, 7823–7832 CrossRef CAS PubMed.
- C. C. Winterbourn, Nat. Chem. Biol., 2008, 4, 278–286 CrossRef CAS PubMed.
- S. Kumar, A. Kumar, G.-H. Kim, W.-K. Rhim, K. L. Hartman and J.-M. Nam, Small, 2017, 13, 1701584 CrossRef PubMed.
- D.-W. Li, J.-J. Sun, Z.-F. Gan, H.-Y. Chen and D. Guo, Anal. Chim. Acta, 2018, 1018, 104–110 CrossRef CAS PubMed.
- D.-W. Lia, H.-Y. Chena, Z.-F. Gana, J.-J. Suna, D. Guoa and L.-L. Qu, Sens. Actuators, B, 2018, 227, 8–13 CrossRef.
- X. X. Li, X. Y. Duan, P. Yang, L. Li and B. Tang, Anal. Chem., 2021, 93, 4059–4065 CrossRef CAS PubMed.
- S. C. Lin, H. J. Ze, X.-G. Zhang, Y.-J. Zhang, J. Song, H. M. Zhang, H.-L. Zhong, Z.-L. Yang, C. Y. Yang, J.-F. Li and Z. Zhu, Angew. Chem., Int. Ed., 2022, 61, e202203511 CrossRef CAS PubMed.
- A. K. Dunker, C. J. Brown, J. D. Lawson, L. M. Iakoucheva and Z. Obradović, Biochemistry, 2002, 41, 6573–6582 CrossRef CAS PubMed.
- P. E. Wright and H. J. Dyson, J. Mol. Biol., 1999, 293, 321–331 CrossRef CAS PubMed.
- I. Rousalova and E. Krepela, Int. J. Oncol., 2010, 37, 1361–1378 CAS.
- K. K. Reza, J. Wang, R. Vaidyanathan, S. Dey, Y. Wang and M. Trau, Small, 2017, 13, 1602902 CrossRef PubMed.
- J. Li, J. Wang, Y. S. Grewal, C. B. Howard, L. J. Raftery, S. Mahler, Y. Wang and M. Trau, Anal. Chem., 2018, 90, 10377–10384 CrossRef CAS PubMed.
- D. Zhang, L. Huang, B. Liu, H. Ni, L. Sun, E. Su, H. Chen, Z. Gu and X. Zhao, Biosens. Bioelectron., 2018, 106, 204–211 CrossRef CAS PubMed.
- X. Su, X. Liu, Y. Xie, M. Chen, C. Zheng, H. Zhong and M. Li, ACS Nano, 2023, 17, 4077–4088 CrossRef CAS.
- Q. Zhong, K. Zhang, X. Huang, Y. Lu, J. Zhao, Y. He and B. Liu, Biosens. Bioelectron., 2022, 207, 114194 CrossRef CAS PubMed.
- L. Li, M. Liao, Y. Chen, B. Shan and M. Li, J. Mater. Chem. B, 2019, 7, 815–822 RSC.
- J. Wang, H. Xie and C. Ding, ACS Appl. Mater. Interfaces, 2021, 13, 32837–32844 CrossRef CAS PubMed.
- Y. J. Yang, Y. L. Chen, J. X. Guo, H. P. Liu and H. X. Ju, iScience, 2021, 24, 102980 CrossRef CAS PubMed.
- P. J. Chitwood, S. Juszkiewicz, A. Guna, S. Shao and R. S. Hegde, Cell, 2018, 175, 1507–1519 CrossRef CAS.
- P. Leippe, J. Broichhagen, K. Cailliau, A. Mougel, M. Morel, C. Dissous, D. Trauner and J. Vicogne, Angew. Chem., Int. Ed., 2020, 59, 6720–6723 CrossRef CAS PubMed.
- C. Dong, X. Fang, J. Xiong, J. Zhang, H. Gan, C. Song and L. Wang, ACS Nano, 2022, 16, 14055–14065 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- S. Minchin and J. Lodge, Essays Biochem., 2019, 63, 433–456 CrossRef CAS PubMed.
- G. Bertoli, C. Cava and I. Castiglioni, Theranostics, 2015, 5, 1122–1143 CrossRef CAS PubMed.
- T. W. Nilsen, Trends Genet., 2007, 23, 243–249 CrossRef CAS.
- G. Stefani and F. J. Slack, Nature, 2008, 9, 219–230 CAS.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS.
- E. K. Lim, T. Kim, S. Paik, S. Haam, Y. M. Huh and K. Lee, Chem. Rev., 2015, 115, 327–394 CrossRef CAS PubMed.
- A. Esquela-Kerscher and F. J. Slack, Nat. Rev. Cancer, 2006, 6, 259–269 CrossRef CAS.
- X. Huang, W. Zhao, X. Chen, J. Li, H. Ye, C. Li, X. Yin, X. Zhao, X. Qiao, Z. Xue and T. Wang, J. Am. Chem. Soc., 2022, 144, 17533–17539 CrossRef CAS.
- R. Zhu, H. Feng, Q. Li, L. Su, Q. Fu, J. Li, J. Song and H. Yang, Angew. Chem., Int. Ed., 2021, 133, 12668–12676 CrossRef.
- E. Garcia-Rico, R. A. Alvarez-Puebla and L. Guerrini, Chem. Soc. Rev., 2018, 47, 4909–4923 RSC.
- L. Guerrini, Z. Krpetic, D. Lierop, R. A. Alvarez-Puebla and D. Graham, Angew. Chem., Int. Ed., 2015, 54, 1144–1148 CrossRef CAS PubMed.
- L. J. Xu, Z. C. Lei, J. Li, C. Zong, C. J. Yang and B. Ren, J. Am. Chem. Soc., 2015, 137, 5149–5154 CrossRef CAS.
- Y. Li, T. Gao, G. Xu, X. Xiang, B. Zhao, X. X. Han and X. Guo, Anal. Chem., 2019, 91, 7980–7984 CrossRef CAS.
- Y. Liu, N. Lyu, V. K. Rajendran, J. Piper, A. Rodger and Y. Wang, Anal. Chem., 2020, 92, 5708–5716 CrossRef CAS PubMed.
- G. Qi, D. Wang, C. Li, K. Ma, Y. Zhang and Y. Jin, Anal. Chem., 2020, 92, 11755–11762 CrossRef CAS PubMed.
- J. M. Yang, L. Jin, Z. Q. Pan, Y. Zhou, H. L. Liu, L. N. Ji, X. H. Xia and K. Wang, Anal. Chem., 2019, 91, 6275–6280 CrossRef CAS.
- S. Zong, C. Chen, Z. Wang, Y. Zhang and Y. Cui, ACS Nano, 2016, 10, 2950–2959 CrossRef CAS PubMed.
- J. X. Guo, Y. L. Chen, Y. J. Jiang and H. X. Ju, Chem. Eur. J., 2017, 23, 9332–9337 CrossRef CAS PubMed.
- M. M. Fuster and J. D. Esko, Nat. Rev. Cancer, 2005, 5, 526–542 CrossRef CAS PubMed.
- S. S. Pinho and C. A. Reis, Nat. Rev. Cancer, 2015, 15, 540–555 CrossRef CAS PubMed.
- L. Lin, X. D. Tian, S. L. Hong, P. Dai, Q. C. You, R. Y. Wang, L. S. Feng, C. Xie, Z.-Q. Tian and X. Chen, Angew. Chem., Int. Ed., 2013, 52, 7266–7271 CrossRef CAS PubMed.
- M. Xiao, L. Lin, Z. F. Li, J. Liu, S. L. Hong, Y. Y. Li, M. L. Zheng, X. M. Duan and X. Chen, Chem.–Asian J., 2014, 9, 2040–2044 CrossRef CAS PubMed.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS PubMed.
- S. Deshayes, H. Cabral, T. Ishii, Y. Miura, S. Kobayashi, T. Yamashita, A. Matsumoto, Y. Miyahara, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2013, 135, 15501–15507 CrossRef CAS PubMed.
- M. Tabatabaei, G. Q. Wallace, F. A. Caetano, E. R. Gillies, S. S. G. Fergusonb and F. Lagugńe-Labarthet, Chem. Sci., 2016, 7, 575–582 RSC.
- W. Y. Song, L. Ding, Y. L. Chen and H. X. Ju, Chem. Commun., 2016, 52, 10640–10643 RSC.
- Y. R. Wang, Y. J. Yang, J. X. Guo, H. X. Ju and Y. L. Chen, Chem. Sci., 2023, 14, 923–927 RSC.
- H. X. Di, H. Q. Liu, M. M. Li, J. Li and D. B. Liu, Anal. Chem., 2017, 89, 5874–5881 CrossRef CAS PubMed.
- J. Lv, S. Chang, X. Y. Wang, Z. R. Zhou, B. B. Chen, R. C. Qian and D. W. Li, Sens. Actuators, B, 2022, 351, 130877 CrossRef CAS.
- Y. L. Chen, L. Ding, W. Y. Song, M. Yang and H. X. Ju, Chem. Sci., 2016, 7, 569–574 RSC.
- G. W. Hart, C. Slawson, G. Ramirez-Correa and O. Lagerlof, Annu. Rev. Biochem., 2011, 80, 825–858 CrossRef CAS PubMed.
- Y. J. Yang, Y. L. Chen, S. Y. Zhao, H. P. Liu, J. X. Guo and H. X. Ju, Chem. Sci., 2022, 13, 9701–9705 RSC.
- W. K. Wang, F. Zhao, M. Z. Li, C. P. Zhang, Y. H. Shao and Y. Tian, Angew. Chem., Int. Ed., 2019, 58, 5256–5260 CrossRef CAS PubMed.
- W. Wang, F. Zhao, M. Li, C. Zhang, Y. Shao and Y. Tian, Angew. Chem., Int. Ed., 2019, 131, 5310–5314 CrossRef.
- M. V. Yigit, L. Y. Zhu, M. A. Ifediba, Y. Zhang, K. Carr, A. Moore and Z. Medarova, ACS Nano, 2011, 5, 1056–1066 CrossRef CAS PubMed.
- J. He, S. Y. Hua, D. X. Zhang, K. Wang, X. Y. Chen and M. Zhou, Adv. Funct. Mater., 2022, 32, 2208028 CrossRef CAS.
- U. S. Dinish, Z. Song, C. J. H. Ho, G. Balasundaram, A. B. E. Attia, X. Lu, B. Z. Tang, B. Liu and M. Olivo, Adv. Funct. Mater., 2015, 25, 2316–2325 CrossRef CAS.
- J. P. Wang, J. Y. Sun, Y. H. Wang, T. Chou, Q. Zhang, B. L. Zhang, L. Ren and H. J. Wang, Adv. Funct. Mater., 2020, 30, 1908825 CrossRef CAS.
- X. Q. Huang, B. B. Sheng, H. M. Tian, Q. X. Chen, Y. Q. Yang, B. Bui, J. Pi, H. H. Cai, S. Chen, J. L. Zhang, W. Chen, H. Zhou and P. H. Sun, Acta Pharm. Sin. B, 2023, 13, 1303–1317 CrossRef CAS.
- Q. Q. Li, X. G. Ge, J. M. Ye, Z. Li, L. C. Su, Y. Wu, H. H. Yang and J. B. Song, Angew. Chem., Int. Ed., 2021, 60, 7323–7332 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |
