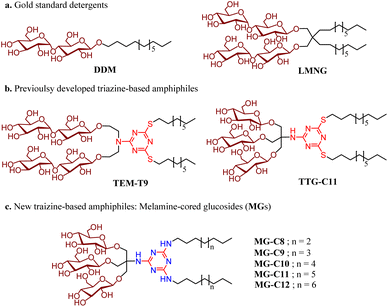
Open Access Article
Open Access ArticleMelamine-cored glucosides for membrane protein solubilization and stabilization: importance of water-mediated intermolecular hydrogen bonding in detergent performance†
Lubna
Ghani‡§
a,
Seonghoon
Kim‡
 c,
Muhammad
Ehsan‡
c,
Muhammad
Ehsan‡
 a,
Baoliang
Lan
d,
Ida H.
Poulsen
a,
Baoliang
Lan
d,
Ida H.
Poulsen
 e,
Chandra
Dev
f,
Satoshi
Katsube
e,
Chandra
Dev
f,
Satoshi
Katsube
 f,
Bernadette
Byrne
f,
Bernadette
Byrne
 g,
Lan
Guan
g,
Lan
Guan
 f,
Claus J.
Loland
f,
Claus J.
Loland
 e,
Xiangyu
Liu
*d,
Wonpil
Im
e,
Xiangyu
Liu
*d,
Wonpil
Im
 *b and
Pil Seok
Chae
*b and
Pil Seok
Chae
 *a
*a
aDepartment of Bionano Engineering, Hanyang University, Ansan 155-88, South Korea. E-mail: pchae@hanyang.ac.kr
bDepartment of Biological Sciences, Chemistry, and Bioengineering Lehigh University, Bethlehem, PA 18015, USA. E-mail: wonpil@lehigh.edu
cSchool of Computational Sciences, Korea Institute for Advanced Study, Seoul, 024-55, South Korea
dTsinghua-Peking Center for Life Sciences, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, School of Medicine, School of Pharmaceutical Sciences, Tsinghua University, Beijing 100084, China. E-mail: liu_xy@tsinghua.edu.cn
eDepartment of Neuroscience, University of Copenhagen, Copenhagen, DK-2200, Denmark
fDepartment of Cell Physiology and Molecular Biophysics, Center for Membrane Protein Research, School of Medicine, Texas Tech University Health Sciences Center, Lubbock, Texas 79430, USA
gDepartment of Life Sciences, Imperial College London, London SW7 2AZ, UK
First published on 23rd October 2023
Abstract
Membrane proteins play essential roles in a number of biological processes, and their structures are important in elucidating such processes at the molecular level and also for rational drug design and development. Membrane protein structure determination is notoriously challenging compared to that of soluble proteins, due largely to the inherent instability of their structures in non-lipid environments. Micelles formed by conventional detergents have been widely used for membrane protein manipulation, but they are suboptimal for long-term stability of membrane proteins, making downstream characterization difficult. Hence, there is an unmet need for the development of new amphipathic agents with enhanced efficacy for membrane protein stabilization. In this study, we designed and synthesized a set of glucoside amphiphiles with a melamine core, denoted melamine-cored glucosides (MGs). When evaluated with four membrane proteins (two transporters and two G protein-coupled receptors), MG-C11 conferred notably enhanced stability compared to the commonly used detergents, DDM and LMNG. These promising findings are mainly attributed to a unique feature of the MGs, i.e., the ability to form dynamic water-mediated hydrogen-bond networks between detergent molecules, as supported by molecular dynamics simulations. Thus, MG-C11 is the first example of a non-peptide amphiphile capable of forming intermolecular hydrogen bonds within a protein–detergent complex environment. Detergent micelles formed via a hydrogen-bond network could represent the next generation of highly effective membrane-mimetic systems useful for membrane protein structural studies.
Introduction
Membrane proteins are ubiquitous bio-macromolecules that reside within multifaceted cellular membranes. Approximately a quarter of the human proteome corresponds to membrane proteins.1 These bio-macromolecules play essential roles in a variety of cellular functions and are implicated in a number of diseases such as asthma, cancers, viral infections and neuro-degenerative diseases.2 Owing to their central roles in a number of physiological processes, membrane proteins constitute around 60% of approved drug targets and, therefore, their three dimensional structures are eagerly sought to assist in structure-based drug design. Despite significant and considerable recent improvements, isolation of functional membrane proteins in sufficient amounts and of the requisite quality necessary for their functional and structural studies is still challenging.3,4 Structure determination of membrane proteins has historically been and, to a certain extent, remains technically difficult, as demonstrated by the fact that membrane proteins with a known structure comprise a low fraction of all protein structures deposited in the Protein Data Bank (PDB). These challenges can arise at any point in the workflow from gene expression to structure determination, as exemplified by inadequate protein expression, poor protein extraction from native membranes, limited long-term protein stability, and recalcitrant protein crystallization. While these challenges have severely hindered membrane protein structure determination, technological advances in expression systems, solubilisation and purification techniques, and structure determination methods such as X-ray crystallography, single-particle cryogenic-electron microscopy (cryo-EM) and nuclear magnetic resonance (NMR) spectroscopy over the last decade have led to an ever-growing number of deposited membrane protein structures.5–7Detergents serve as essential tools for membrane protein solubilization, purification and structural study. These amphipathic compounds are widely used not only to extract membrane proteins from the membranes, but also to maintain protein integrity during purification and downstream characterization.8–10 OG (n-octyl-β-D-glucoside), DM (n-decyl-β-D-maltoside), and DDM (n-dodecyl-β-D-maltoside) are among the most widely used detergents for structural studies of membrane proteins.9 These conventional detergents have been successfully used for structural studies of relatively stable membrane proteins, but they tend to be inadequate to handle more challenging eukaryotic membrane proteins and multisubunit membrane protein complexes. Due to their canonical architecture of single head and tail groups, micelles formed by these detergents are markedly more dynamic than cell membranes.11 In addition, detergent micelles, particularly those formed by harsh detergents, can remove closely associated lipid molecules important for membrane protein structure and function during protein extraction. Therefore, it is necessary to develop novel amphiphiles able to both form micelles with reduced dynamics and retain lipid molecules essential for protein function.
Over the past few decades, several membrane-mimetic systems have been developed including bicelles,12 membrane scaffold protein (MSP)-based nanodiscs (NDs),13 polymeric amphiphiles [amphipols (Apols)14,15 and styrene-maleic acid copolymers (SMAs)],16,17 and peptide-based amphiphiles [lipopeptide detergents (LPDs),18 β-peptides (BPs),19 and Saposin A].20 Among these membrane-mimetic systems, nano-assemblies formed by MSP, SMA and Saposin A, namely NDs, SMA lipo-particles (SMALPs) and Salipro, respectively, are particularly interesting as they include lipids that are either added to the isolated protein or directly extracted from the native membrane along with the protein, providing a more physiological environment. While successfully used for structural studies of membrane proteins, particularly via cryo-EM, many of these systems are ineffective at membrane protein extraction from the membranes and are not suitable for membrane protein crystallization. As additional alternatives to conventional detergents, small amphiphiles have been developed, as exemplified by neopentyl glycol-based amphiphiles [e.g., maltose neopentyl glycols (MNGs), glucose neopentyl glycols (GNGs), and neopentyl glycol-derived triglucosides (NDTs)],21–23 rigid hydrophobic group-bearing amphiphiles [e.g., chobimalt and glyco-diosgenin (GDN)],24,25 carbohydrate-cored amphiphiles [e.g., mannitol-based amphiphiles (MNAs)26 and scyllo-inositol glycosides (SIGs)]27 and facial amphiphiles (FAs).28,29 Among these small detergent molecules, two amphiphiles (LMNG and GDN) are particularly notable as they have been used for structural studies of more than 450 membrane proteins during the past 10 years.30 These biochemical tools are particularly effective for structural elucidation of biologically and pharmaceutically important bio-macromolecules such as G protein-coupled receptors (GPCRs) and ATP-binding cassette (ABC) transporters, highlighting significant contributions of novel detergents to membrane protein structure determination. Very recently, we reported a couple of classes of 1,3,5-triazine-based detergents including triazine-based di-maltosides (TEMs),31 tris(hydroxymethyl)methanamine (TRIS)-bearing triazine-based glucosides (TTGs),32 and triazine-based tetra-maltosides (TZMs)33 for membrane protein stability. In this study, we report another class of 1,3,5-triazine-based amphiphiles containing a melamine unit (2,4,6-triamino-1,3,5-triazine) in the central region, denoted as melamine-cored glucosides (MGs). Due to the presence of multiple hydrogen-bonding donors and acceptors in the central melamine unit, we hypothesized that these amphiphiles form a hydrogen-bond network within the micellar structure when assembled around membrane protein surfaces and that this network increases detergent–detergent interactions which in turn enhances membrane protein stability. When tested with four model membrane proteins [two transporters (leucine transporter (LeuT) and melibiose permease (MelB)) and two G protein-coupled receptors (β2 adrenergic receptor (β2AR) and μ-opioid receptor (MOR))], MG-C11 proved highly effective at stabilizing the membrane proteins compared to two gold standard detergents (DDM and LMNG). Molecular dynamics (MD) simulations show that MG-C11 tends to form (dynamic) water-mediated hydrogen-bonding with other detergent molecules in the protein–detergent complexes (PDCs), which explains its remarkable protein stabilizing efficacy. Thus, MG-C11 not only has potential as a useful tool for membrane protein manipulation, but also provides a new strategy for the design of novel detergents optimal for membrane protein stability.
Results and discussion
Detergent design, synthesis and physical characterization studies
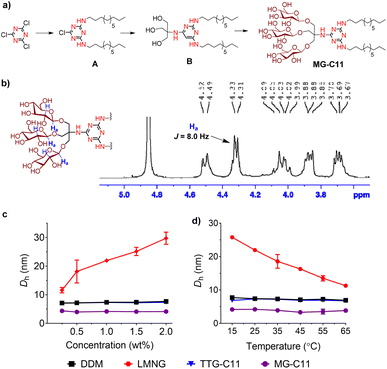
New amphiphiles containing two alkyl chains and three glucose units as the hydrophobic and hydrophilic groups were synthesised (Fig. 1). The alkyl chains were conjugated into the triazine ring via an amino linkage, while the three glucose head groups were introduced into the same ring via a tris(hydroxymethyl)aminomethane (TRIS) linker. Due to the presence of strongly nucleophilic amine functional groups, we could efficiently attach both the alkyl chain and TRIS linker into the triazine ring via nucleophilic aromatic substitution reactions. The resulting triazine-based glucoside detergents contain a core structure of 2,4,6-triamino-1,3,5-triazine (A.K.A, melamine), and are thus denoted as melamine-cored glucosides (MGs). This core unit has a unique structure distinct from that of other detergent scaffolds since it contains multiple hydrogen bond donors and acceptors. The secondary amino (NH) and tertiary amino (N) groups presented in an alternating pattern around/on the heterocyclic ring are typical hydrogen-bonding donors and acceptors, respectively (Fig. 1). This distinctive feature has facilitated the wide use of melamine-bearing compounds in supramolecular assemblies.34–37 Of note, the MGs differ from previously developed triazine-based amphiphiles (TEMs and TTGs). Both MGs and TTGs have a glucoside head group, while the TEMs have a maltoside head group. The MGs differ from the TTGs in terms of the functional group used to introduce the two alkyl chains into the triazine ring (thioether (TTGs) vs. amino (MGs)). The MGs are structurally similar to the TTGs, but greatly differ in terms of the way they interact in micelles given the presence of hydrogen bond networks formed between the amino groups in the melamine core. The alkyl chains of the MGs varied from C8 to C12 to allow identification of the optimal alkyl chain length for membrane protein study. Detergent alkyl chain length is a critical factor determining balance between detergent hydrophobicity and hydrophilicity (i.e., hydrophilic–lipophilic balance (HLB)). HLB values of the individual MGs were obtained using Griffin's method and are summarized in Table S1†.38 All MGs were found to give high HLB values (13.6–15.2) compared to DDM, LMNG and previously developed TTG-C11. For example, the HLB value of MG-C11 (13.9) is higher than those of its thioether version (TTG-C11; 12.7), DDM (13.4) and LMNG (13.6), reflecting the rather hydrophilic nature of the new detergents.The MGs were prepared in four efficient synthetic steps starting from 2,4,6-trichloro-1,3,5-triazine, comprising dialkylation, TRIS coupling, glycosylation and global deprotection (Fig. 2a). Briefly, an alkyl amine (R–NH2) and TRIS were successively reacted with 2,4,6-trichloro-1,3,5-triazine in the presence of diisopropylethylamine as a base to produce the dialkylated triazine-based triol derivative (B). The resulting compound is subjected to β-selective glycosylation to give the corresponding glycosylated product in ∼85% yield, followed by complete removal of the benzoyl protecting group using sodium methoxide (NaOMe) (see the ESI† for details). The ease of synthesis along with high synthetic yields made it possible to synthesize the designed amphiphiles in multi-gram quantities. NMR spectra corroborate the β-stereochemistry of the glycosidic bonds of the MGs. The NMR spectrum of MG-C11 is shown in Fig. 2b, as an example. The anomeric proton peak appears at 4.32 ppm with a vicinal coupling constant (3Jaa) of 8.0 Hz, which is in good agreement with the formation of the β-glycosidic bond (Fig. 2b). An anomeric proton with an α-glycosidic linkage gives higher chemical shift (5.15–5.20 ppm) and a smaller coupling constant (4.0 Hz) than the β-anomeric proton.
The individual MGs were soluble to at least 10%, with the exception of MG-A12 which had a lower water solubility of ∼5%. The solutions containing these detergents were stable at room temperature for a month. The tendency of the MGs to form self-assemblies was investigated by measuring their critical micelle concentrations (CMCs), and the size of the self-assemblies formed by these detergents was estimated via hydrodynamic radii (Rh). Detergent CMC and Rh were obtained by utilizing encapsulation of the water-insoluble dye, diphenylhexatriene (DPH), in the micelle interiors and dynamic light scattering (DLS) experiments, respectively.39 The results are summarized in Table 1. The CMCs of the MGs follow a typical trend; their CMCs decrease with increasing alkyl chain length. This trend is likely due to the fact that detergent hydrophobicity is proportional to the alkyl chain length. The MG CMCs were estimated to be in the range of 40 to 600 μM, significantly higher than those of other two alkyl chain-bearing detergents such as LMNG (∼10 μM) and TTG-C11 (∼4 μM).
| Detergent | M.W.a | CMC (mM) | CMC (wt%) | R h (nm)b | Solubility |
|---|---|---|---|---|---|
| a Molecular weight of detergents. b Hydrodynamic radius of detergents measured at 1.0 wt% by dynamic light scattering. | |||||
| MG-C8 | 941.0 | ∼0.6 | ∼0.06 | 2.1 ± 0.5 | ∼10 |
| MG-C9 | 969.1 | ∼0.5 | ∼0.05 | 2.2 ± 0.2 | ∼10 |
| MG-C10 | 997.1 | ∼0.3 | ∼0.03 | 2.3 ± 0.1 | ∼10 |
| MG-C11 | 1025.2 | ∼0.15 | ∼0.015 | 2.4 ± 0.1 | ∼10 |
| MG-C12 | 1053.3 | ∼0.04 | ∼0.004 | 2.5 ± 0.1 | ∼5 |
| TTG-C11 | 1059.3 | ∼0.004 | ∼0.0004 | 3.7 ± 0.1 | ∼10 |
| LMNG | 1005.2 | ∼0.01 | ∼0.001 | 9.8 ± 0.2 | ∼10 |
| DDM | 510.6 | 0.17 | 0.0087 | 3.4 ± 0.1 | ∼10 |
For example, the CMC of MG-C11 is roughly 40 times higher than that of TTG-C11 (150 vs. 4 μM) although these two detergents have the same head and tail groups. Such a large CMC difference likely originates from the polarity difference between the two functional groups (amino vs. thioether) used to incorporate the alkyl chain into the detergent scaffold; the amino group is highly polar, while the thioether group is slightly polar. As a result, double alkyl-chained MG-C11 gave a surprisingly high CMC comparable to single alkyl-chained DDM (150 vs. 170 μM). Notably, most novel detergents developed to date including LMNG (10 μM) and NDT-C11 (6 μM) have lower CMCs than DDM. The high CMCs suggest that the MGs have a reduced tendency to self-assemble which may facilitate detergent exchange, often necessary for membrane protein structural study.40 The MGs are also different from the TTGs and LMNG in terms of micelle sizes. The Rh of MG-C11 was estimated to be 2.4 nm, substantially smaller than those of TTG-C11 (3.7 nm) and LMNG (9.8 nm) (Table 1). The micelle sizes of the MGs are comparable to conventional detergents known to form small micelles, as exemplified by OG (2.6 nm) and LDAO (1.9 nm).30,41 It is worth mentioning that small micelle-forming detergents are beneficial for membrane protein research, as excess detergent micelles can be efficiently reduced from protein samples through molecular weight cut-off (MWCO) filters. This property is also considered beneficial for membrane protein structural study via X-ray crystallography, cryo-EM and NMR spectroscopy.42,43 Interestingly, the sizes of the micelles formed by the MGs only slightly varied with increasing detergent alkyl chain length. The Rh of detergent micelles increases by only 0.1 nm with every additional methylene unit to the alkyl chain, smaller than in other classes of amphiphiles.
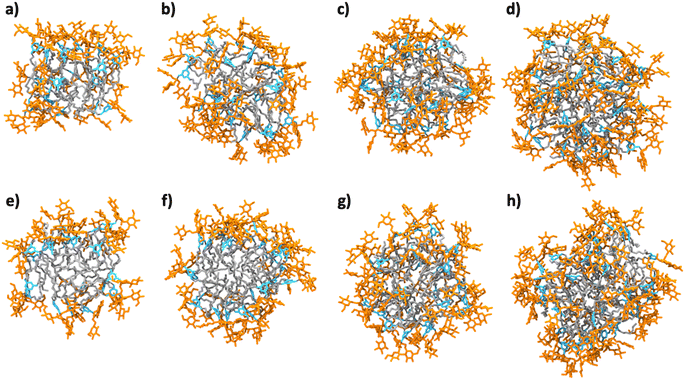
Micellar structures formed by MG-C11 and MG-C12 were calculated via MD simulations. The calculations were carried out assuming that the aggregation number (AN) of detergent micelles is either 20, 25, 30 or 40. These ANs were selected based on the experimentally determined Rh of micelles formed by MG-C11 or MG-C12. The calculation results are visualized in Fig. 3 and their micellar properties are summarized in Tables S2 and S3.† Interestingly, we observed little difference in the distances of individual detergent components (glucoside (Glu), TRIS-triazine (Tt) and alkyl tail (R)) from the micellar centre when the detergent alkyl chain length increases from C11 (MG-C11) to C12 (MG-C12) (Table S2†). In addition, the radii of gyration (Rg) of micelles increase by 0.2–0.3 Å with the same variation in the alkyl chain length of the MGs (Table S3†). The small increase in Rh of the MGs with increasing the alkyl chain length is consistent with this trend (Table 1). This result indicates that the micelle size is mainly determined by the hydrophilic group (i.e., glucoside-TRIS-triazine) rather than the alkyl chain of the MGs. Favourable interactions between the detergent hydrophilic groups may be responsible for this observation. Of note, the calculated Rg values of both MG-C11 and MG-C12 micelles were smaller than the experimentally determined Rh of these detergents (2.4 and 2.5 nm, respectively), typical for spherical micelles. With detergent AN increasing from 20 to 40, the Rg values of detergent micelles increase only 1.2 times for both MGs, indicating that new detergent molecules incorporated into MG micelles are mainly positioned to decrease the empty spaces or micelle defects in the micelle interiors rather than to increase micelle volumes (Table S3†). This indication was further supported by the substantial decreases in the solvent accessible surface areas (SASAs) of MG-C11 and MG-C12 micelles with increasing detergent AN. Interestingly, the SASA of MG-C11 micelles is substantially smaller than that of MG-C12 micelles with the same AN, indicating that MG-C11 could form more tightly packed micelles than MG-C12. Based on the distances of the glucoside head group from the micelle centre (Table S2 and Fig. S2†), the ANs of MG-C11 and MG-C12 micelles are estimated to be in the range of 25 to 30, significantly smaller than those of DDM (80–150) and LMNG (∼400).44 The dynamics of the detergents in their micelles were estimated by calculating root mean square deviation (RMSD) of detergent alkyl chains. With the detergent AN increasing from 20 to 40, the RSMDs of MG-C11 and MG-C12 were reduced from 10.3 to 7.3 Å and 10.1 to 8.0 Å, respectively (Table S4†). These RMSD values are significantly smaller (14.1 Å) than that obtained from micelles formed by 130 DDM molecules. This result indicates that MG-C11 forms micelles that are less dynamic than those formed by DDM, partly due to dynamic hydrogen-bond formation between MG-C11 molecules (vide infra).
The small micelle size of MG-C11 compared to those of DDM, LMNG, and TTG-C11 is effectively maintained with variation in detergent concentration or solution temperature (Fig. 2c and d). The small micelle size formed by MG-C11 varied little with increasing detergent concentration from 0.3 to 2.0 wt% or increasing solution temperature from 15 to 65 °C. In contrast, self-assemblies formed by LMNG increased or decreased with the same variations of detergent concentration and solution temperature. This result indicates that MG-C11 forms small micelles with a globular/elliptical shape, while micelles formed by LMNG are cylindrical.45 The MGs were further investigated in terms of their micelle size distributions. All the MGs showed unimodal size distributions of their micelles in the number- or volume-weighted DLS profiles, suggesting homogenous populations of their micelles (Fig. S3a and S3b†). Large aggregates shown in the intensity-weighted DLS profiles of the MGs are not due to size heterogeneity of detergent micelles, but result from the highly sensitive nature of light scattering depending on the aggregate size (Fig. S3c†).
Detergent evaluation with membrane proteins
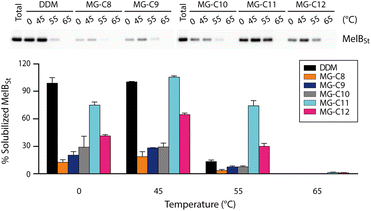
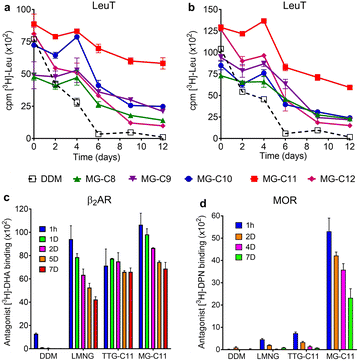
Detergent evaluation started with melibiose permease from Salmonella typhimurium (MelBSt).46a–e The permease overexpressed in E. coli membranes was extracted using 1.5 wt% of the individual MGs at 0 °C for 90 min. The extracts were further incubated at an elevated temperature (45, 55, or 65 °C) for another 90 min. The amounts of soluble MelBSt were determined using Western blot analysis following ultracentrifugation and represented as percentages (%) of the total MelBSt present in the untreated membranes. The amount of soluble MelBSt obtained from the low temperature experiment (0 °C) gives information about detergent efficiency for protein extraction, while those from the temperature variation experiments (45, 55, or 65 °C) are directly correlated with detergent efficacy for protein stabilization. Thus, this protocol provides information about both protein extraction efficiency and protein stabilization efficacy of the tested detergents. When MelBSt was extracted using the individual MGs or DDM (as a control) at 0 °C, DDM was most efficient at extracting and solubilizing MelBSt (Fig. 4). The MGs were inferior to DDM in this context, but MG-C11, the best of the five MGs, extracted MelBSt from the membranes in ∼75% yield. When the 0 °C-extracts were additionally incubated at 45 °C, the amounts of soluble MelBSt increased substantially in the case of the MG-treated samples. For instance, MG-C11 and MG-C12 gave amounts of soluble MelBSt increasing from ∼75% to 100% and from ∼40% to ∼65%, respectively. Increased detergent water-solubility and/or enhanced membrane dynamics induced by the elevated temperature are likely responsible for increased MelBSt solubilization. Detergent behaviour between the MGs and DDM was differentiated in the thermo-solubility experiment at 55 °C. At this high temperature, a small amount of the transporter (∼10%) remains in a soluble state following thermal treatment, indicating that most of the DDM-solubilized MelBSt undergoes protein denaturation and/or aggregation. In contrast, MG-C11 effectively retained protein solubility at this high temperature, resulting in ∼75% soluble MelBSt. The other MGs, except MG-C12, were ineffective at preserving MelBSt in a soluble state. The same experiment at 65 °C resulted in a complete loss in soluble MelBSt in all tested detergents. This result indicates that MG-C11 is moderately efficient at MelBSt extraction and notably effective at stabilizing the transporter.The MGs were further evaluated with another transporter, the bacterial leucine transporter (LeuT) Aquifex aeolicus.47 Protein stability was assessed by measuring the ability to bind a radioactive substrate ([3H]-leucine (Leu)) via scintillation proximity assay (SPA).48 LeuT purified in DDM was subjected to detergent exchange via sample dilution and the final concentrations of the MGs in the sample solutions were CMCs + 0.04/0.2 wt%. [3H]-Leu binding ability of the transporter in the individual MGs was monitored over a 12 day incubation period at room temperature (Fig. 5a and b). LeuT in DDM initially showed a good ability to bind Leu, but rapidly lost binding ability over the course of the incubation. After 6 days of incubation, LeuT in DDM almost completely lost its substrate binding ability. LeuT in the individual MGs, despite variation in their initial abilities to bind the substrate, was more effective at maintaining its stability than in DDM, with the best performance observed for MG-C11. The overall trend of detergent efficacy for LeuT stabilization was similar when the MGs were used at either CMCs + 0.04 wt% or 0.2 wt% (Fig. 5a and b). Along with the MelBSt results (Fig. 4), these findings indicate that MG-C11 is highly effective at stabilizing membrane transporters.
We selected MG-C11 for the next evaluation as this detergent showed the most promising results for solubilisation and/or long-term stabilization of two transporters (MelBSt and LeuT). As a model membrane protein for this evaluation, we turned to a G protein-coupled receptor (GPCR), the human β2 adrenergic receptor (β2AR).49 First, we investigated detergent efficiency for receptor solubilisation. For this purpose, we used 1.0 wt% MG-C11 to extract the receptor from the membranes at two different temperatures (4 or 25 °C) (Fig. S4†). DDM and LMNG were used as controls as these two detergents are widely used for GPCR extraction.50 When the receptor was extracted from the membranes at 4 °C, LMNG of the tested detergents was most efficient at receptor extraction, with MG-C11 yielding a similar amount of soluble β2AR to DDM. When the solubilisation experiment was carried out at 25 °C, all the tested detergents yielded comparable amounts of the soluble receptor. These results indicate that MG-C11 can be used as efficiently as DDM for β2AR extraction. Interestingly, in marked distinction from DDM and LMNG, the receptor solubilized by MG-C11 yielded two bands (Fig. S4†). These bands are likely to correspond to the monomeric and dimeric forms of the receptor with molecular weights of ∼46 and ∼92 kDa, respectively. It is known that there is a substantial amount of dimeric β2AR in physiological cell membranes that plays a critical role in receptor and cellular function.51 The marked ability of MG-C11 to stabilize membrane protein complexes may allow effective isolation of dimeric β2AR. This result is important as structural study of dimeric/oligomeric GPCRs is limited by detergent-mediated dissociation into monomers. Thus, MG-C11 may open the window to readily access dimeric GPCRs. Next, we turned to study MG-C11 efficacy for receptor stabilization. In this evaluation, we included TTG-C11 as a positive control, in addition to DDM and LMNG. LMNG is a widely used novel detergent particularly for GPCR structural study, while TTG-C11 is a previously developed triazine-based glucoside. Notably, LMNG has been used for successful determination of more than 145 GPCR structures over the past 10 years.30 β2AR purified in LMNG was diluted in a buffer solution containing LMNG, TTG-C11, or MG-C11 to reach a final detergent concentration of 0.1 wt%. The resulting protein samples were then incubated for 7 days at room temperature and receptor stability was monitored at regular intervals during the incubation (Fig. 5c). The ability of the receptor to bind a radioactive antagonist ([3H]-dihydroalprenolol (DHA)) was used to assess β2AR stability.52–54 As expected, β2AR in DDM showed low affinity for the radiolabelled ligand upon detergent exchange and lost its ligand binding ability rapidly. β2AR in LMNG showed markedly enhanced DHA binding ability upon detergent exchange, as expected from the wide use of this NG detergent for GPCR structural studies. However, this initial binding ability of LMNG gradually decreased during the 7 day incubation. TTG-C11 was overall comparable to LMNG in this regard. When MG-C11 was used to encapsulate the receptor, this detergent showed higher initial ligand binding of the receptor than LMNG and TTG-C11, and this initial capability was effectively preserved over the course of the 7 day incubation.
Based on the encouraging results of MG-C11 for β2AR stability, we further evaluated this detergent for another GPCR, the mouse μ-opioid receptor (MOR).55 This GPCR is particularly challenging to stabilize in detergent micelles. The LMNG-extracted receptor was purified in the same detergent and the resulting LMNG-purified MOR was exchanged from LMNG to the respective test detergent (DDM, LMNG, TTG-C11, or MG-C11) via sample dilution. The final detergent concentration for detergent efficacy comparison was 0.1 wt%. MOR stability was assessed via a similar method used for β2AR stability using a receptor-specific antagonist (i.e., [3H]-diprenorphine (DPN)) instead of [3H]-DHA. MOR in DDM completely lost ligand binding ability upon detergent exchange. LMNG and TTG-C11 were better than DDM, consistent with the results of β2AR with these detergents, but the receptor in these novel detergents still suffered from rapid loss of DPN binding over the 7 day incubation at 4 °C (Fig. 5d). In contrast, upon detergent exchange from LMNG to MG-C11, MOR showed markedly enhanced ability to bind DPN compared to the receptor in LMNG or TTG-C11; the DPN binding increased by 11 and 7 times, respectively, when MG-C11 was used compared to LMNG and TTG-C11. Even after the 7 day incubation at 4 °C, the DPN binding ability of the receptor in MG-C11 was five/three times higher than the initial ability of the receptor in LMNG and TTG-C11. Indeed, this detergent is the most effective of recently developed novel detergents at stabilizing MOR.56–59
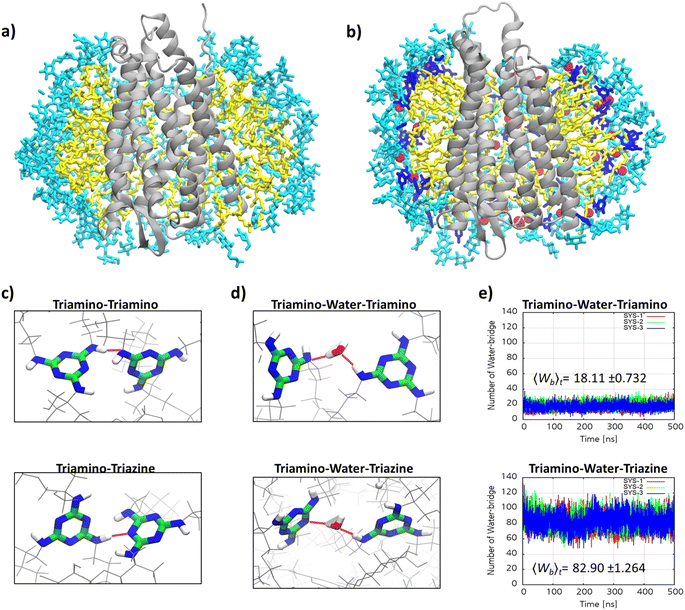
The remarkable efficacy of MG-C11 for MOR stabilization compared to LMNG prompted us to explore molecular interactions in the MOR-detergent complex via MD simulations. 500 ns-long MD simulations were performed using MOR (PDB: 6DDE) complexed with 96 molecules of MG-C11 or LMNG.60 The simulation results for complex formation of LMNG/MG-C11 with the receptor show effective encapsulation of the hydrophobic surface of the receptor by LMNG or MG-C11 molecules (Fig. S5†). When protein–detergent interactions were analysed according to the amino-acid residues of the receptor, we found that two transmembrane helices (α5 and α6) interact in a higher frequency with the alkyl chains of MG-C11 than those of LMNG (Fig. S6†). The other helices showed little noticeable difference between these two detergents. In addition, the numbers of LMNG and MG-C11 molecules in contact with the receptor surfaces were calculated from the MD simulations (Fig. S7†). When using the last 200 ns trajectory data, MG-C11 gave an average contact number of ∼79, substantially larger than that of LMNG (∼72). A similar result was obtained for comparison of the atom numbers of the detergent alkyl chains interacting with the receptor. The average number of detergent contact atoms increases from 677 (LMNG) to 723 (MG-C11). This increased frequency/contact number means a stronger interaction of MG-C11 with the receptor surface compared to that of LMNG, a feature favourably associated with enhanced receptor stability. Next, we investigated detergent–detergent interactions by calculating the numbers of intermolecular hydrogen-bonding within detergent micelles surrounding the receptor. Due to the presence of hydrogen bond donors and acceptors in the core structure, MG-C11 is likely to form hydrogen bonds with nearby molecules. Rather surprisingly, hydrogen-bonding that persists for more than 1 ns is absent in the simulated complex between MG-C11 and MOR. Thus, we next focused on dynamic hydrogen-bonding in the complex that constantly undergoes bond formation and breakage during the simulation trajectory (Fig. 6a and b). The extent of such dynamic intermolecular interactions was quantified by counting the average number of hydrogen bonds present in the complex at a given time in the last 200 nm trajectory. Direct hydrogen-bonding between detergent molecules in the protein–detergent complexes (PDCs), denoted as triamino (NH)-triamino (NH) or triamino (NH)-triazine (N) interaction, is found to be negligible. This is likely due to the fact that the presence of the bulky head group (i.e., TRIS-triglucoside) hinders these hydrogen-bonding pairs from approaching each other (Fig. 6c). When mediated by a water molecule, however, substantially larger numbers are obtained for the two types of hydrogen-bonding interactions of this MG, denoted as triamino (NH)-water-triamino (NH) and triamino (NH)-water-triazine (N) interactions, respectively (Fig. 6d). The average number of triamino (NH)-water-triamino (NH) hydrogen-bonding is ∼18 during the last 200 ns trajectory (Fig. 6e). This number dramatically increases to ∼83 for the hydrogen-bonding between the triamino and triazine units of MG-C11 (triamino (NH)-water-triazine (N)). Thus, at any given instant, the total number of water-mediated hydrogen bonds between detergent molecules in the complex reaches 101, higher than the number of detergent molecules (96) employed to build the PDCs in the simulations. This result indicates that every MG-C11 molecule, at least, is competent to form a single dynamic hydrogen bond with another detergent molecule in the PDC environment, with the assistance of water molecules that penetrate into the triazine layer of the detergent micelles. It is likely that this hydrogen-bonding network in micelles formed by MG-C11, although dynamic and water-mediated, contributes to enhanced MOR stability via increased detergent–detergent interactions. It is noteworthy that hydrogen-bonding is much stronger than van der Waals interactions and thus hydrogen-bonding between detergent molecules, even if present in a small number, could make a big difference with respect to detergent behaviour toward membrane protein stability. Almost every detergent (e.g., DDM or LMNG) can only harness relatively weak van der Waals interactions for assembly or micelle formations.
Discussion
Detergent efficacy for protein stabilization is dependent on the detergent alkyl chain length, which is closely associated with the detergent HLB and hydrophobic length. This is a reason why we typically find an optimal alkyl chain length in detergent studies for membrane protein stability. The exact alkyl chain length most effective for protein stability differs from one class of detergents to another and also varies depending on the identity of the target membrane proteins, but typically falls in in the range of C10 to C13. This narrow range of optimal alkyl chain length is mainly due to small variations in the hydrophobic width of membrane proteins (28 to 32 Å).61 The compatibility of the hydrophobic dimensions of proteins and detergents is necessary for energetically favourable protein–detergent interactions. As for the MGs, the optimal alkyl chain length was observed to be C11, the same alkyl chain length found in other detergent studies.23,32,59,62,63 MG-C11 was more effective than DDM at stabilizing all tested membrane proteins. Furthermore, this detergent conferred enhanced stability to two GPCRs (β2AR and MOR) compared to LMNG and TTG-C11. In addition, this detergent showed a reasonable extraction efficiency with both MelBSt and β2AR. Small micelle formation, with an Rh of 2.4 nm, and the rather high CMC (0.15 mM) are additional aspects of MG-C11 that are favourable for membrane protein study, distinctive from those of other novel detergents reported to date. Taken together, these results indicate that MG-C11 holds significant potential for extraction, purification and structural study of GPCRs. Notably, this detergent allowed solubilisation of the dimeric form of β2AR when the receptor was extracted from the membranes, implying a profound impact of this detergent on structural study of GPCR dimers.It is difficult to pinpoint which structural feature of MG-C11 is mainly responsible for enhanced membrane protein stability observed here since protein stability is determined by a combination of multiple factors. One important factor determining detergent efficacy for protein stabilization is the hydrophobicity of the detergent hydrophobic group.32 The presence of a small number of polar groups (e.g., amino and hydroxyl groups) in the detergent lipophilic region could be detrimental for membrane protein stability, as these weaken detergent interactions with membrane protein surfaces (i.e., protein–detergent interactions). As a result, detergents including amino functional group(s) in the detergent core or lipophilic region are rarely used for membrane protein study. The presence of relatively nonpolar groups, as exemplified by ether and thioether groups, represent a slight compromise in detergent efficacy for membrane protein stabilization. In the MGs, each alkyl chain was conjugated to the triazine core via a polar amino linkage. Thus, the lipophilic groups of the MGs are relatively polar compared to those of LMNG and TTG-C11 where the corresponding alkyl chains are introduced into the detergent cores directly (LMNG) or via a thioether linkage (TTG-C11). The high CMCs and large HLBs of the MGs relative to those of LMNG and TTG-C11 support the relatively high polarity of the MG lipophilic groups. Given the possession of high polarity, it is remarkable that MG-C11 was superior to LMNG and TTG-C11 in membrane protein stability. In order to explore the reasons for this surprisingly enhanced stability, we investigated the molecular interactions within the MOR-MG-C11 complex via MD simulations and compared them with the simulation results obtained for the MOR-LMNG complex. We found that MG-C11 interacts with the receptor more frequently and extensively than LMNG, thereby increasing protein–detergent interactions. More importantly, MG-C11 micelles around MOR form rather extensive water-mediated hydrogen-bond networks between the melamine units. Therefore, the increased detergent–detergent interaction is likely to significantly contribute to the enhanced protein stabilization efficacy of MG-C11 compared to LMNG.
Development of detergents capable of forming a hydrogen-bonding network is remarkable for a couple of reasons. There is only one class of detergents (i.e., β-peptides (BPs)) reported to form hydrogen-bonding networks in detergent micelles.19 While these peptide-based detergents showed promising results in the initial evaluation with multiple membrane proteins, they exhibit poor water-solubility and are difficult to synthesize on a large scale. In contrast, MG-C11, the first example of a non-peptide-based detergent with the ability to form hydrogen-bond networks in PDCs, is convenient for large scale synthesis (requiring only four synthetic steps) and is highly water-soluble (>10%). Detergent design capable of forming intermolecular hydrogen-bonding is challenging as hydrogen-bonding can only be attained under strict restrictions in terms of orientation and distance between the hydrogen bond donor and acceptor. This challenge is particularly difficult to achieve in micellar architecture with a large curvature. Consequently, a simple insertion of a hydrogen-bonding motif into a detergent scaffold is unlikely to induce the realization of hydrogen-bond formation between detergent molecules. For instance, there is no plausible evidence that tripod amphiphiles (TPAs) and pre-assembled detergents (PADs) containing an amide linkage and triazole unit in the detergent core, respectively, form hydrogen bonds in a micellar or PDC state.64,65 It is remarkable that MG-C11, a small micelle-forming detergent, is able to form intermolecular hydrogen bonding mediated by water molecules in micelle interiors. Due to their strong yet reversible nature, various bio-macromolecules such as ribonucleic acids, proteins and carbohydrates have evolved to widely utilize hydrogen-bonding interactions for their individual functions. Thanks to the ability to form dynamic intermolecular hydrogen-bonds, MG-C11 can form stable assemblies around membrane proteins, resulting in the markedly enhanced protein stability demonstrated by this study. The hydrogen-bond network achieved by MG-C11 may not be optimal as the bonds are both dynamic and water-mediated. Thus, the introduction of a more extensive hydrogen-bonding network into detergent micelles may further enhance protein stability. We will pursue these directions to further improve detergent efficacy for protein stabilization. It will be also interesting to investigate the utility of this detergent for in vitro reconstitution of membrane proteins into proteoliposomes as only a few detergents were shown to be effective for this application.66 Thus, new detergents capable of forming extensive and strong hydrogen-bonding could represent the next generation of novel detergents and hold significant potential for future membrane protein structural studies.
Conclusions
We have developed a set of glucoside amphiphiles with a melamine core. Of these MGs, MG-C11 showed the most effective protein stabilization and was superior to three gold standards (DDM, LMNG and TTG-C11). The MOR stability achieved by MG-C11 is particularly remarkable, indicating that this MG may be suited for structural studies of particularly unstable GPCRs. In addition, β2AR extraction by using this detergent yielded a dimeric receptor, a quaternary arrangement challenging to access using other detergents. MG-C11 possesses several favorable features for membrane protein study including synthetic accessibility, small protein–detergent complex formation, reasonable efficiency for protein extraction and marked protein stabilization efficacy. Thus, the new detergent is likely to find wide use in membrane protein structural study. As indicated by our MD simulation study, MG-C11 is capable of forming dynamic hydrogen-bonding between detergent molecules when associated with membrane proteins, distinct from other detergents. The resulting increase in detergent–detergent interactions is likely to be mainly responsible for the enhanced membrane protein stability observed in this study. Therefore, the current study not only provides a detergent tool potentially useful for membrane protein study, but also a new strategy for the design of novel detergents that facilitate membrane protein research.Data availability
The data that support the findings of this study are available in the ESI† of this article.Author contributions
P. S. C. designed the new amphiphiles. L. G. and M. E. synthesized the amphiphiles. L. G., S. K., M. E., B. L., I. H. P., C. D. and S. K. designed and performed the research and interpreted the data. P. S. C., W. I., X. L., C. J. L., L. G. and B. B. contributed to experimental design and data interpretation. P. S. C. and L. G. wrote the manuscript, with oversight from all the other authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare the following competing financial interest(s): P. S. C. and L. G. are inventors on a patent application that covers the MGs.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) (2021R1A2C2006067 and 2018R1A6A1A03024231 to P. S. C.), the National Institutes of Health (NIH) (R01GM122759 to L. G.) and the National Science Foundation (NSF) (MCB-2111728 to W. I.). This work was also supported by the Samsung Science and Technology Foundation under project number SSTF-BA2202-07.Notes and references
- L. Fagerberg, K. Jonasson, G. von Heijne, M. Uhlén and L. Berglund, Proteomics, 2010, 10, 1141–1149 CrossRef CAS PubMed.
- (a) C. R. Sanders and J. K. Myers, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 25–51 CrossRef CAS PubMed; (b) D. P. Ng, B. E. Poulsen and C. M. Deber, Biochim. Biophys. Acta, 2012, 1818, 1115–1122 CrossRef CAS PubMed.
- J. Drews, Science, 2000, 287, 1960–1964 CrossRef CAS PubMed.
- J. P. Overington, B. Al-Lazikani and A. L. Hopkins, Nat. Rev. Drug Discovery, 2006, 5, 993–996 CrossRef CAS PubMed.
- K. Shimizu, W. Cao, G. Saad, M. Shoji and T. Terada, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1077–1091 CrossRef CAS PubMed.
- J. P. Allen, F1000Research, 2019, 8 Search PubMed.
- Y. He, K. Wang and N. Yan, Protein Cell, 2014, 5, 658–672 CrossRef CAS PubMed.
- S. Newstead, S. Ferrandon and S. Iwata, Protein Sci., 2008, 17, 466–472 CrossRef CAS PubMed.
- S. Newstead, J. Hobbs, D. Jordan, E. P. Carpenter and S. Iwata, Mol. Membr. Biol., 2008, 25, 631–638 CrossRef CAS PubMed.
- R. M. Garavito and S. Ferguson-Miller, J. Biol. Chem., 2001, 276, 32403–32406 CrossRef CAS PubMed.
- M. Kieber, T. Ono, R. C. Oliver, S. B. Nyenhuis, D. P. Tieleman and L. Columbus, Biophys. J., 2019, 116, 1682–1691 CrossRef CAS PubMed.
- S. Faham and J. U. Bowie, J. Mol. Biol., 2002, 316, 1–6 CrossRef CAS PubMed.
- A. Nath, W. M. Atkins and S. G. Sligar, Biochemistry, 2007, 46, 2059–2069 CrossRef CAS PubMed.
- C. Tribet, R. Audebert and J.-L. Popot, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 15047–15050 CrossRef CAS PubMed.
- J.-L. Popot, T. Althoff, D. Bagnard, J.-L. Banères, P. Bazzacco, E. Billon-Denis, L. J. Catoire, P. Champeil, D. Charvolin and M. J. Cocco, Annu. Rev. Biophys., 2011, 40, 379–408 CrossRef CAS PubMed.
- M. Orwick-Rydmark, J. E. Lovett, A. Graziadei, L. Lindholm, M. R. Hicks and A. Watts, Nano Lett., 2012, 12, 4687–4692 CrossRef CAS PubMed.
- J. Broecker, B. T. Eger and O. P. Ernst, Structure, 2017, 25, 384–392 CrossRef CAS PubMed.
- C.-L. McGregor, L. Chen, N. C. Pomroy, P. Hwang, S. Go, A. Chakrabartty and G. G. Privé, Nat. Biotechnol., 2003, 21, 171–176 CrossRef CAS PubMed.
- H. Tao, S. C. Lee, A. Moeller, R. S. Roy, F. Y. Siu, J. Zimmermann, R. C. Stevens, C. S. Potter, B. Carragher and Q. Zhang, Nat. Methods, 2013, 10, 759–761 CrossRef CAS PubMed.
- J. Frauenfeld, R. Löving, J.-P. Armache, A. F. Sonnen, F. Guettou, P. Moberg, L. Zhu, C. Jegerschöld, A. Flayhan and J. A. Briggs, Nat. Methods, 2016, 13, 345–351 CrossRef CAS PubMed.
- P. S. Chae, S. G. Rasmussen, R. R. Rana, K. Gotfryd, R. Chandra, M. A. Goren, A. C. Kruse, S. Nurva, C. J. Loland and Y. Pierre, Nat. Methods, 2010, 7, 1003–1008 CrossRef CAS PubMed.
- P. S. Chae, R. R. Rana, K. Gotfryd, S. G. Rasmussen, A. C. Kruse, K. H. Cho, S. Capaldi, E. Carlsson, B. Kobilka and C. J. Loland, Chem. Commun., 2013, 49, 2287–2289 RSC.
- A. Sadaf, J. S. Mortensen, S. Capaldi, E. Tikhonova, P. Hariharan, O. Ribeiro, C. J. Loland, L. Guan, B. Byrne and P. S. Chae, Chem. Sci., 2016, 7, 1933–1939 RSC.
- S. C. Howell, R. Mittal, L. Huang, B. Travis, R. M. Breyer and C. R. Sanders, Biochemistry, 2010, 49, 9572–9583 CrossRef CAS PubMed.
- P. S. Chae, S. G. Rasmussen, R. R. Rana, K. Gotfryd, A. C. Kruse, A. Manglik, K. H. Cho, S. Nurva, U. Gether and L. Guan, Chem.–Eur. J., 2012, 18, 9485–9490 CrossRef CAS PubMed.
- H. Hussain, Y. Du, N. J. Scull, J. S. Mortensen, J. Tarrasch, H. E. Bae, C. J. Loland, B. Byrne, B. K. Kobilka and P. S. Chae, Chem.–Eur. J., 2016, 22, 7068–7073 CrossRef CAS PubMed.
- A. Sadaf, M. Ramos, J. S. Mortensen, Y. Du, H. E. Bae, C. F. Munk, P. Hariharan, B. Byrne, B. K. Kobilka and C. J. Loland, ACS Chem. Biol., 2019, 14, 1717–1726 CrossRef CAS PubMed.
- Q. Zhang, X. Ma, A. Ward, W. X. Hong, V. P. Jaakola, R. C. Stevens, M. e. G. Finn and G. Chang, Angew Chem. Int. Ed. Engl., 2007, 119, 7153–7155 CrossRef.
- M. Das, Y. Du, J. S. Mortensen, H. E. Bae, B. Byrne, C. J. Loland, B. K. Kobilka and P. S. Chae, Chem.–Eur. J., 2018, 24, 9860–9868 CrossRef CAS PubMed.
- H. J. Lee, H. S. Lee, T. Youn, B. Byrne and P. S. Chae, Chem, 2022, 8, 980–1013 CAS.
- L. Ghani, C. F. Munk, X. Zhang, S. Katsube, Y. Du, C. Cecchetti, W. Huang, H. E. Bae, S. Saouros and M. Ehsan, J. Am. Chem. Soc., 2019, 141, 19677–19687 CrossRef CAS PubMed.
- L. Ghani, X. Zhang, C. F. Munk, P. Hariharan, B. Lan, H. S. Yun, B. Byrne, L. Guan, C. J. Loland and X. Liu, Bioconjug. Chem., 2023, 34, 739–747 CrossRef CAS PubMed.
- L. Ghani, S. Kim, H. Wang, H. S. Lee, J. S. Mortensen, S. Katsube, Y. Du, A. Sadaf, W. Ahmed and B. Byrne, Chem.–Eur. J., 2022, 28, e202200116 CrossRef CAS PubMed.
- M. Ma and D. Bong, Langmuir, 2011, 27, 8841–8853 CrossRef CAS PubMed.
- A. Ranganathan, V. Pedireddi and C. Rao, Trends In Chemistry Of Materials: Selected Research Papers of CNR Rao, 2008, pp. 430–431 Search PubMed.
- H.-F. Ji and X. Xu, Langmuir, 2010, 26, 4620–4622 CrossRef CAS PubMed.
- P. Timmerman and L. J. Prins, Eur. J. Org Chem., 2001, 2001, 3191–3205 CrossRef.
- W. C. Griffin, J. Soc. Cosmet. Chem., 1954, 5, 249–256 Search PubMed.
- A. Chattopadhyay and E. London, Anal. Biochem., 1984, 139, 408–412 CrossRef CAS PubMed.
- L. Liu, Z. Zhu, F. Zhou, D. Xue, T. Hu, W. Luo, Y. Qiu, D. Wu, F. Zhao and Z. Le, ACS Omega, 2021, 6, 21087–21093 CrossRef CAS PubMed.
- L. Li, S. Nachtergaele, A. M. Seddon, V. Tereshko, N. Ponomarenko and R. F. Ismagilov, J. Am. Chem. Soc., 2008, 130, 14324–14328 CrossRef CAS PubMed.
- Q. Zhang, H. Tao and W.-X. Hong, Methods, 2011, 55, 318–323 CrossRef CAS PubMed.
- W. Luo, M. Yang, Y. Zhao, H. Wang, X. Yang, W. Zhang, F. Zhao, S. Zhao and H. Tao, Chem.–Eur. J., 2022, 28, e202202242 CrossRef CAS PubMed.
- A. Stetsenko and A. Guskov, Crystals, 2017, 7, 197 CrossRef.
- C. Breyton, W. Javed, A. Vermot, C.-A. Arnaud, C. Hajjar, J. Dupuy, I. Petit-Hartlein, A. Le Roy, A. Martel and M. Thépaut, Biochim. Biophys. Acta, Biomembr., 2019, 1861, 939–957 CrossRef CAS PubMed.
- (a) L. Guan, S. Nurva and S. P. Ankeshwarapu, J. Biol. Chem., 2011, 286, 6367–6374 CrossRef CAS PubMed; (b) A. S. Ethayathulla, M. S. Yousef, A. Amin, G. Leblanc, H. R. Kaback and L. Guan, Nat. Commun., 2014, 5, 3009 CrossRef PubMed; (c) A. Amin, A. S. Ethayathulla and L. Guan, J. Bacteriol., 2014, 196, 3134–3139 CrossRef PubMed; (d) A. Amin, P. Hariharan, P. S. Chae and L. Guan, Biochemistry, 2015, 54, 5849–5855 CrossRef CAS PubMed; (e) E. Cordat, I. Mus-Veteau and G. Leblanc, J. Biol. Chem., 1998, 273, 33198–33202 CrossRef CAS PubMed.
- G. Deckert, P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller and M. Aujay, Nature, 1998, 392, 353–358 CrossRef CAS PubMed.
- M. Quick and J. A. Javitch, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3603–3608 CrossRef CAS PubMed.
- D. M. Rosenbaum, V. Cherezov, M. A. Hanson, S. G. Rasmussen, F. S. Thian, T. S. Kobilka, H.-J. Choi, X.-J. Yao, W. I. Weis and R. C. Stevens, Science, 2007, 318, 1266–1273 CrossRef CAS PubMed.
- S. Lavington and A. Watts, Biophys. Rev., 2020, 12, 1287–1302 CrossRef CAS PubMed.
- (a) X. Li, M. Zhou, W. Huang and H. Yang, FEBS J., 2017, 284, 2004–2018 CrossRef CAS PubMed; (b) Y. Kwon, D. H. Kim, M. G. Jeong, M. T. Hong, S. Park, Y. Chang, K. Zhou, S. Y. Park, J. Zhang and S. H. Ryu, Cell Chem. Biol., 2022, 29, 1532–1540 CrossRef CAS PubMed.
- S. E. Mansoor, H. S. Mchaourab and D. L. Farrens, Biochemistry, 2002, 41, 2475–2484 CrossRef CAS PubMed.
- X. Yao, C. Parnot, X. Deupi, V. R. Ratnala, G. Swaminath, D. Farrens and B. Kobilka, Nat. Chem. Biol., 2006, 2, 417–422 CrossRef CAS PubMed.
- G. Swaminath, J. Steenhuis, B. Kobilka and T. W. Lee, Mol. Pharmacol., 2002, 61, 65–72 CrossRef CAS PubMed.
- A. Manglik, A. C. Kruse, T. S. Kobilka, F. S. Thian, J. M. Mathiesen, R. K. Sunahara, L. Pardo, W. I. Weis, B. K. Kobilka and S. Granier, Nature, 2012, 485, 321–326 CrossRef CAS PubMed.
- H. E. Bae, C. Cecchetti, Y. Du, S. Katsube, J. S. Mortensen, W. Huang, S. Rehan, H. J. Lee, C. J. Loland and L. Guan, Acta Biomater., 2020, 112, 250–261 CrossRef CAS PubMed.
- A. Sadaf, S. Kim, H. E. Bae, H. Wang, A. Nygaard, Y. Uegaki, Y. Du, C. F. Munk, S. Katsube and H. S. Lee, Acta Biomater., 2021, 128, 393–407 CrossRef CAS PubMed.
- M. Ehsan, H. Wang, C. Cecchetti, J. S. Mortensen, Y. Du, P. Hariharan, A. Nygaard, H. J. Lee, L. Ghani and L. Guan, ACS Chem. Biol., 2021, 16, 1779–1790 CrossRef CAS PubMed.
- H. J. Lee, M. Ehsan, X. Zhang, S. Katsube, C. F. Munk, H. Wang, W. Ahmed, A. Kumar, B. Byrne and C. J. Loland, Chem. Sci., 2022, 13, 5750–5759 RSC.
- S. Lee, S. Ghosh, S. Jana, N. Robertson, C. G. Tate and N. Vaidehi, Biochemistry, 2020, 59, 2125–2134 CrossRef CAS PubMed.
- (a) A. G. Lee, Biomembranes, 2003, 1612, 1–40 CrossRef CAS PubMed; (b) T. Nugent and D. T. Jones, BMC Bioinf., 2013, 14, 276 CrossRef PubMed.
- M. Ehsan, Y. Du, N. J. Scull, E. Tikhonova, J. Tarrasch, J. S. Mortensen, C. J. Loland, G. Skiniotis, L. Guan, B. Byrne, B. K. Kobilka and P. S. Chae, J. Am. Chem. Soc., 2016, 138, 3789–3796 CrossRef CAS PubMed.
- M. Das, Y. Du, O. Ribeiro, P. Hariharan, J. S. Mortensen, D. Patra, G. Skiniotis, C. J. Loland, L. Guan, B. K. Kobilka, B. Byrne and P. S. Chae, J. Am. Chem. Soc., 2017, 139, 3072–3081 CrossRef CAS PubMed.
- P. S. Chae, A. C. Kruse, K. Gotfryd, R. R. Rana, K. H. Cho, S. G. Rasmussen, H. E. Bae, R. Chandra, U. Gether and L. Guan, Chem.–Eur. J., 2013, 19, 15645–15651 CrossRef CAS PubMed.
- F. Zhao, Z. Zhu, L. Xie, F. Luo, H. Wang, Y. Qiu, W. Luo, F. Zhou, D. Xue and Z. Zhang, Chem.–Eur. J., 2022, 28, e202201388 CrossRef CAS PubMed.
- A. Godoy-Hernandez, A. H. Asseri, A. J. Purugganan, C. Jiko, C. de Ram, H. Lill, M. Pabst, K. Mitsuoka, C. Gerle, D. Bald and D. G. G. McMillan, ACS Cent. Sci., 2023, 9, 494–507 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03543c |
| ‡ These authors have contributed equally to this work. |
| § Department of Chemistry, Women University of Azad Jammu & Kashmir Bagh, (WUAJK), Bagh-12500, AJK, Pakistan. |
| This journal is © The Royal Society of Chemistry 2023 |