Open Access Article
Open Access ArticleMetal-coding assisted serological multi-omics profiling deciphers the role of selenium in COVID-19 immunity†
Ying
Zhou‡
 a,
Shuofeng
Yuan‡
bcd,
Fan
Xiao‡
e,
Hongyan
Li
a,
Ziwei
Ye
f,
Tianfan
Cheng
g,
Cuiting
Luo
b,
Kaiming
Tang
b,
Jianpiao
Cai
b,
Jianwen
Situ
b,
Siddharth
Sridhar
bcdh,
Wing-Ming
Chu
i,
Anthony Raymond
Tam
i,
Hin
Chu
b,
Chi-Ming
Che
a,
Shuofeng
Yuan‡
bcd,
Fan
Xiao‡
e,
Hongyan
Li
a,
Ziwei
Ye
f,
Tianfan
Cheng
g,
Cuiting
Luo
b,
Kaiming
Tang
b,
Jianpiao
Cai
b,
Jianwen
Situ
b,
Siddharth
Sridhar
bcdh,
Wing-Ming
Chu
i,
Anthony Raymond
Tam
i,
Hin
Chu
b,
Chi-Ming
Che
 a,
Lijian
Jin
a,
Lijian
Jin
 g,
Ivan Fan-Ngai
Hung
ci,
Liwei
Lu
e,
Jasper Fuk-Woo
Chan
g,
Ivan Fan-Ngai
Hung
ci,
Liwei
Lu
e,
Jasper Fuk-Woo
Chan
 *bcdhjk and
Hongzhe
Sun
*bcdhjk and
Hongzhe
Sun
 *a
*a
aDepartment of Chemistry, State Key Laboratory of Synthetic Chemistry, CAS-HKU Joint Laboratory of Metallomics on Health and Environment, The University of Hong Kong, Pokfulam, Hong Kong SAR, China. E-mail: hsun@hku.hk
bState Key Laboratory of Emerging Infectious Diseases, Carol Yu Centre for Infection, Department of Microbiology, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong Kong SAR, China. E-mail: jfwchan@hku.hk
cDepartment of Infectious Diseases and Microbiology, The University of Hong Kong-Shenzhen Hospital, Shenzhen, Guangdong, China
dCentre for Virology, Vaccinology and Therapeutics, Hong Kong Science and Technology Park, Hong Kong SAR, China
eDepartment of Pathology, Shenzhen Institute of Research and Innovation, The University of Hong Kong, Hong Kong SAR, China
fSchool of Biomedical Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
gFaculty of Dentistry, The University of Hong Kong, Pokfulam, Hong Kong SAR, Hong Kong, China
hDepartment of Microbiology, Queen Mary Hospital, Pokfulam, Hong Kong SAR, China
iDivision of Infectious Diseases, Department of Medicine, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
jAcademician Workstation of Hainan Province, Hainan Medical University-The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
kGuangzhou Laboratory, Guangdong Province, China
First published on 18th September 2023
Abstract
Uncovering how host metal(loid)s mediate the immune response against invading pathogens is critical for better understanding the pathogenesis mechanism of infectious disease. Clinical data show that imbalance of host metal(loid)s is closely associated with the severity and mortality of COVID-19. However, it remains elusive how metal(loid)s, which are essential elements for all forms of life and closely associated with multiple diseases if dysregulated, are involved in COVID-19 pathophysiology and immunopathology. Herein, we built up a metal-coding assisted multiplexed serological metallome and immunoproteome profiling system to characterize the links of metallome with COVID-19 pathogenesis and immunity. We found distinct metallome features in COVID-19 patients compared with non-infected control subjects, which may serve as a biomarker for disease diagnosis. Moreover, we generated the first correlation network between the host metallome and immunity mediators, and unbiasedly uncovered a strong association of selenium with interleukin-10 (IL-10). Supplementation of selenium to immune cells resulted in enhanced IL-10 expression in B cells and reduced induction of proinflammatory cytokines in B and CD4+ T cells. The selenium-enhanced IL-10 production in B cells was confirmed to be attributable to the activation of ERK and Akt pathways. We further validated our cellular data in SARS-CoV-2-infected K18-hACE2 mice, and found that selenium supplementation alleviated SARS-CoV-2-induced lung damage characterized by decreased alveolar inflammatory infiltrates through restoration of virus-repressed selenoproteins to alleviate oxidative stress. Our approach can be readily extended to other diseases to understand how the host defends against invading pathogens through regulation of metallome.
Introduction
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a huge threat to public health worldwide.1,2 Severe COVID-19 disease is characterized by immunopathology caused by cytokine dysregulation in addition to direct virus-induced damage.3 Recent studies have revealed a number of factors that drive COVID-19 severity including the host genetic code,4 and impaired metabolic pathways.5 By using different omics techniques, we and others have also identified other host factors such as lipids, proteins, and metabolites that play biologically important roles in the pathogenesis of COVID-19.6–10 However, the pathogenesis and potential host targets that can serve as intervention strategies remain largely unclear.Metal(loid)s are essential components of all forms of life, and serve as catalytic cofactors in almost half of all known enzymes.11,12 They are also closely associated with multiple diseases if dysregulated.13–15 Increasing evidence shows that metals and metalloids, e.g. Zn, Mn, K, Fe, Ca, and Se, are also involved in both the innate and adaptive immunity of the host including inflammatory response and antiviral immunity.16–24 On the other hand, metal ions are also critical factors for the survival and replication of viruses. For example, Zn2+, Fe2+/3+ and Mn2+ are required for proper functions of several non-structural proteins in SARS-CoV-2.25–28 Certain nutritional metal ions (e.g. Zn2+) were also reported to exert direct anti-viral activity against COVID-19 through inhibition of its replication.26
Moreover, it is increasingly recognized that serum metal(loid)s levels such as Zn, Se and Ca are closely related to the severity and even mortality of COVID-19 patients, and may serve as a risk factor of COVID-19 severity.3,29–31 However, the underlying mechanisms are largely unknown. A comprehensive understanding on how SARS-CoV-2 infection impairs the host metallome and how metal(loid)s are involved in COVID-19 pathophysiology and immunopathology may facilitate therapeutic intervention for COVID-19. Herein, we perform comprehensive serological profiling of metallome and immunoproteome using our recently established metal-coding assisted multiplex proteome assay platform32 (Scheme 1) to uncover the roles that metal(loid)s play in COVID-19 immunity, the disease prognosis, intervention and mechanistic insights.
 | ||
| Scheme 1 Schematic chart for metal-coding assisted serological multi-omics profiling for deciphering the metallomic and immunoproteomic features of COVID-19. | ||
Results
Metal fingerprint-based serological metallomic and proteomic profiling deciphers the metallome and immune response to SARS-CoV-2 infections
To investigate how the host metallome and immune system respond to SARS-CoV-2 infection, the serum levels of different metal(loid)s (i.e., elements) and immunoproteome were quantified using inductively coupled plasma-mass spectrometry (ICP-MS) by counting metal readouts as we described previously.32To define the metallome response to SARS-CoV-2, we compared the contents of different metal(loid)s, including selenium (Se), zinc (Zn), calcium (Ca), magnesium (Mg), potassium (K), chromium (Cr), iron (Fe), copper (Cu), molybdenum (Mo), and iodine (I), in the sera of non-infected controls and COVID-19 patients with different levels of disease severity. We observed extensive variations in the levels of multiple elements (Se, Zn, Ca, K, Cu, Cr, and I) after SARS-CoV-2 infection and with different levels of disease severities (Fig. 1A and S1†). Notably, the serum levels of Se, Zn and Cr were significantly lower in COVID-19 patients than those in non-infected controls, particularly in moderate to severe COVID-19 patients.
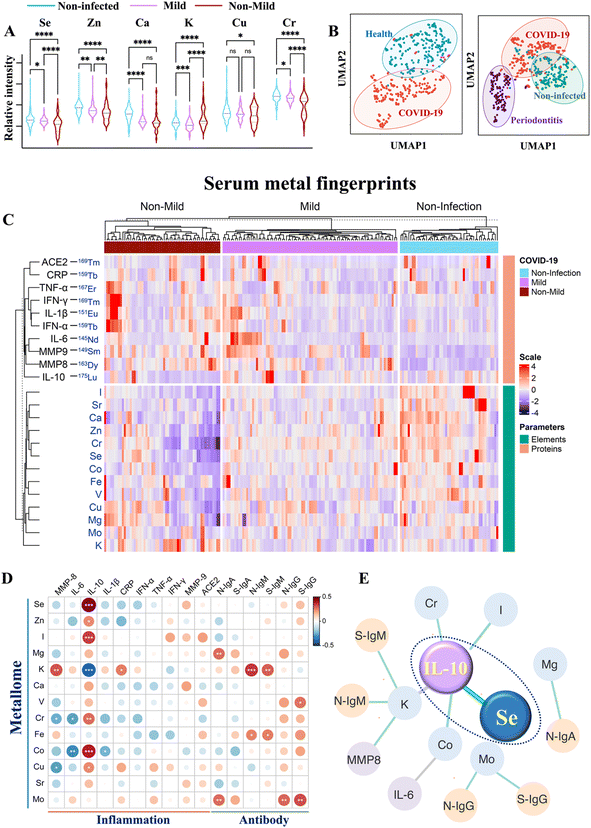 | ||
| Fig. 1 Multiplexed serological profiling of metallome and immunoproteome using ICP-MS. (A) Comparison of different metal(loid)s among the sera from non-infected individuals and COVID-19 patients with different levels of severities. The dashed line denotes the median value of each parameter across all samples used in the plot. (B) UMAP map for classification of COVID-19 patients, periodontitis patients and non-infected controls. The red, purple, and green circles represent the gates for the discrimination of COVID-19 patients, periodontitis patients and non-infected controls, respectively. (C) Metal fingerprints showing the metallome and immunoproteome pattern in the serum of each individual. Heatmap (D) and networks (E) of Spearman rho coefficients for the serum metallome with different types of antibodies and inflammation mediators. Statistical significance levels are presented with asterisks. Only significant correlations (P < 0.01) are included for the construction of correlation networks. The correlation coefficient and directionality of the correlation are pictured through line thickness and color, respectively. The green and gray lines represent the positive and negative correlation, respectively. The blue, yellow and purple of the nodes indicate metallome, antibody response, and inflammation mediators, respectively. The antibody information used partial data from our earlier study.32P values were calculated by the ANOVA test to evaluate the statistical significance of variations among groups. 0.01 < (*)P < 0.05, 0.001 < (**)P < 0.01, 0.0001 < (***)P < 0.001, (****)P < 0.0001. | ||
We next used uniform manifold approximation and projection (UMAP), an efficient dimensionality reduction approach that has been widely applied in single-cell studies,33 to evaluate whether the metallome modulation could facilitate the discrimination of COVID-19 patients from non-infected controls. All the metal(loid)s variables included in this study were used to construct a UMAP map. As shown in Fig. 1B (left), most of the COVID-19 patients were distinctly distributed (red dots) away from the non-infected controls (blue dots), suggesting that the distinct metallome pattern efficiently differentiated the two groups. Moreover, to evaluate whether the host metallome pattern could be incorporated to acute and chronic infections diagnosis, we compared the metallome-generated UMAP of non-infected controls, COVID-19 patients, and another 47 patients with generalized severe periodontitis, a severe gum infection. The results indicated that each group exhibited unique serological metallome signatures, facilitating their differentiation (Fig. 1B, right). The results indicate that the unique metallome pattern of different kinds of disease could be possibly incorporated into clinics to facilitate the diagnosis of specific disease.
It is well recognized that the homeostatic imbalance of different metal(loid)s tends to disrupt various biological pathways.15 To investigate whether SARS-CoV-2 infection affects metal(loid) homeostasis and vice versa, we examined the intrinsic correlations among each metal(loid) in non-infected controls and COVID-19 patients by Spearman's rank correlation test.34 The value of Spearman's rank correlation coefficient (ρ) was applied to assess the level of association between pairwise parameters. We observed evident alterations of the interplay of intrinsic metal(loid)s–metal(loid)s after SARS-CoV-2 infection and in COVID-19 patients with different levels of disease severity (Fig. S2A and B†). The most pronounced variations were found in the correlations of K with Mg [ρcontrol = 0.41, P < 0.0001; ρcovid-19 (ns)], Ca [ρcontrol = 0.54, P < 0.0001; ρcovid-19 (ns)], Zn [ρcontrol = 0.37, P < 0.0001; ρcovid-19 (ns)], and Cr [ρcontrol = 0.48, P < 0.0001; ρcovid-19 (ns)], and the correlations of Ca with Mg [ρcontrol = 0.60, P < 0.0001; ρcovid-19 (ns)] and Fe [ρcontrol = 0.41, P < 0.0001; ρcovid-19 (ns)]. Despite multiple metal(loid)–metal(loid) correlations being disrupted, we observed an evident increase in certain metal(loid)–metal(loid) correlations, e.g. Se and Zn (ρnon-mild = 0.77, P < 0.0001; ρcontrol = 0.56, P < 0.0001), in COVID-19 patients with more severe disease (Fig. S2†), indicative of collaborative participation of these metal(loid)s in COVID-19 disease progression. As both Se and Zn are major antioxidant essential elements, the evident increase in Se–Zn interplay in more severe COVID-19 patients may be attributable to their roles in counterbalancing of the negative effect of free radicals induced by virus infection.35
To define the serum immunological features of each COVID-19 patient, the immune-related proteins, mainly inflammation mediators [interleukin 6 (IL-6), interleukin 10 (IL-10), interferon α (IFN-α), interferon γ (IFN-γ), tumor necrosis factor-alpha (TNF-α), interleukin 1β (IL-1β), C-reactive protein (CRP), matrix metalloproteinase-8 (MMP8), and matrix metalloproteinase-9 (MMP9)], were quantified by introducing metal-reporters. Generally, the serum proteins were enriched and immunoprecipitated with their corresponding magnetic bead-conjugated capture antibodies. Subsequently, the captured proteins were magnetically separated and interacted with lanthanide-conjugated detection antibodies (metal-coding reporter antibodies). The metal-labeled detector antibodies were then eluted to the supernatant under acidic conditions, and quantified by ICP-MS. Based on this method, a holistic view of the serum metal fingerprint pattern conveying the metallome and immunoproteome information of each individual was achieved (Fig. 1C). By comparing the metal intensity corresponding to serum levels of different elements and immune-related parameters among non-infected controls and COVID-19 patients with different levels of severities (Table S1†), the host metallome and immune response to SARS-CoV-2 infections and disease progression could be characterized. The detailed information of antibodies used in the serological proteomic profiling and the ICP-MS setting parameters for metallome and proteome quantification is summarized in Tables S2–S5.†
We observed significantly elevated IL-6 (P < 0.01), IL-1β (P < 0.05), IFN-α (P < 0.0001), TNF-α (P < 0.01), CRP (P < 0.01), and MMP8 (P < 0.01) in the sera of moderate to severe than mild COVID-19 patients (Fig. S3†). Notably, IL-10, an essential anti-inflammatory cytokine, was downregulated in moderate to severe COVID-19 patients (P < 0.0001). Importantly, the IL-6/IL-10 ratio elevated more profoundly (IL-6/IL-10non-mild/mild = 2.31) than either IL-6 (IL-6non-mild/mild = 1.27) or IL-10 (IL-10mild/non-mild = 1.73) alone in moderate to severe than mild COVID-19 patients, suggesting that the IL-6/IL-10 ratio may serve as a more sensitive marker than IL-6 or IL-10 alone for disease severity stratification.
Integration of serological metallome and immunoproteome uncovers the links of metal(loid)s with COVID-19 immunity
To explore the role of metal(loid)s in COVID-19 immunity, the correlation of the host metallome with the inflammatory and antibody responses was further evaluated by integrating the metallome and immunoproteome information. The correlation of metal(loid)s with different immunity-associated parameters was generated by Spearman's rank correlation test. The intuitive association of each metal(loid) with different types of antibodies (IgA, IgM, and IgG antibodies specific to nucleocapsid (N) and spike proteins (S) of SARS-CoV-2) and inflammation mediators (IL-6, IL-10, IL-6/IL-10, IL-1β, TNF-α, MMP8, MMP9, IFN-α, IFN-γ, and CRP) was shown as a heatmap (Fig. 1D) and correlation network (Fig. 1E) based on the value of Spearman rho coefficients. In terms of antibody response, significant positive correlations between Mg and Mo with anti-SARS-CoV-2-S and -N IgA antibodies were found. Moreover, an association of K with anti-SARS-CoV-2-S and -N IgM antibodies was also observed (Fig. 1D and S4†). Apart from the correlation with IgA, Mo was also positively correlated with anti-SARS-CoV-2-S and -N IgG antibodies. These results confirm the potential involvement of metal(loid)s in the adaptive immunity of COVID-19. Notably, IL-10, a crucial anti-inflammatory cytokine, was significantly correlated with multiple metal(loid)s, including Se, Zn, I, Cr, Cu, Co, and K, among which the positive correlation of IL-10 with Se was the most pronounced (Fig. 1E and S4†). Collectively, the metal(loid)-immunity network established from the integrated multi-omics data provided direct evidence of the involvement of metal(loid)s in the inflammatory and antibody responses in COVID-19.Selenium elevates IL-10 expression in B cells and inhibits proinflammatory cytokine induction in both B and CD4+ T cells
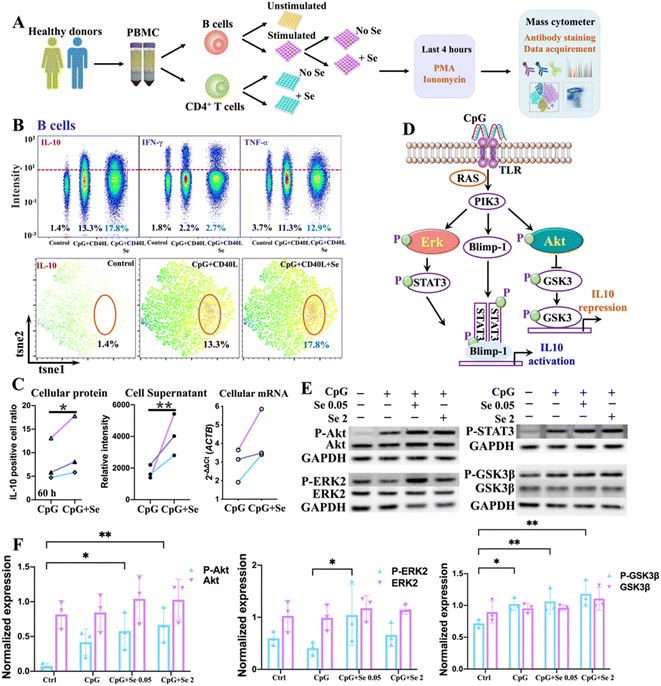
We next sought to validate the role of metal(loid)s in COVID-19 immunity. Given the strong association of Se with IL-10, we selected Se as an illustrative example and investigated its immune regulatory role in immune cells. To select the appropriate immune cell subtypes that have distinct features between COVID-19 patients and non-infected controls, we first characterized the immune cell phenotypes in peripheral blood mononuclear cells (PBMCs) of COVID-19 patients and non-infected controls by mass cytometry combined with t-distributed stochastic neighbor embedding (tSNE) to compare cell population densities according to the infection status. A panel of antibodies including seven cell surface markers, i.e., CD45, CD20, CD3, CD4, CD8, CD14, and PD1, were used (Table S6†), allowing the identification of 7 main subtypes of immune cells in both COVID-19 patients and non-infected controls, i.e., B cells, T cells, CD4+ T cells, CD8+ T cells, CD4+CD8+ T cells, CD4−CD8− T cells, and monocytes, as represented in the t-SNE plots (Fig. S5 and S6A†). The tSNE representation and differentiated cell counts showed a notable decrease in the density of B cells, and an evident increase in CD4+CD8+ T cell ratios in COVID-19 patients in comparison to non-infected controls (Fig. S6B†). Furthermore, the percentages of monocytes and CD4/CD8 showed a moderately decreasing trend in COVID-19 patients (Fig. S6B†). Notably, there was a significant increase in the expression level of programmed cell death protein 1 (PD1), an inhibitory receptor that is expressed by all activated T cells and during T cell exhaustion, in CD4+ T cells, but not in CD8+ T cells (Fig. S6C and D†), indicating that SARS-CoV-2 potentially drives T cell exhaustion in COVID-19 patients. We further assessed the expression of selenoprotein P (SEPP1), one of the most important selenoproteins responsible for Se transportation, storage and functions, in all the immune cells of COVID-19 patients and non-infected controls. Interestingly, we noted a significantly enhanced expression of selenoprotein P (SEPP1) in all types of immune cells of COVID-19 patients compared to those of the controls (Fig. S7A–D†), further confirming the involvement of Se in the cellular immune response of COVID-19.As a significant reduction of B cell ratios and the overexpressed exhaustion marker in the CD4+ T cells of PBMC from COVID-19 patients were noted, we next examined whether and how exogenous Se affects the functions of these two types of immune cells. We performed ex vivo stimulation of B cells, using CpG and CD40L in combination with supplementation of different concentrations of Se, followed by intracellular staining to monitor the cytokine response in the activated B cells by mass cytometry (Fig. 2A). We observed significantly enhanced production of IL-10 in B cells treated with a relatively lower concentration of Se (0.05 μM, 60 h) (Fig. 2B, C and S8A†). Such effects may partly account for the positive association between serum Se and IL-10 in COVID-19 patients observed in our study. Alongside the increase of IL-10 expression, we also observed enhanced expressions of IL-6 and TNF-α, both at the mRNA and protein levels, after Se treatment (Fig. S8B and C†), which is consistent with a previous report that IL-10+ B cells often co-express the pro-inflammatory cytokines (IL-6 and TNF-α).36 Additionally, the increased mRNA expression of some key selenoproteins, including SELK, GPX4, SEPP1, TXNRD1, and GPX1, was shown in the Se-treated group (0.05 μM, 60 h) (Fig. S8D†), which may participate in the Se-modulated B cell functions.
As several signaling pathways, including MAPK, PI-3K, and STAT3 pathways, are responsible for IL-10 production of B cells,37 we therefore investigated the effects of Se on these signaling pathways in B cells. We first examined the mRNA levels of key signaling molecules (MAPK1, STAT3, GSK3B, AKT1, TLR7, TLR8, TLR9, CD40, RAF1, PIK3CA, PRDM1) involved in the MAPK, PI-3K, and STAT3 pathways,37 and found significant elevation of MAPK1, STAT3, GSK3B, AKT1, RAF1, PIK3CA, and PRDM1 in CpG stimulated B cells. However, the addition of Se did not cause significant alternation of these proteins at mRNA levels (Fig. S9†). We further examined whether Se might influence the activation of signaling molecules involved in these pathways with a focus on extracellular signal-regulated kinase (ERK), STAT3, protein kinase B (Akt), and Gsk3β, given their critical roles in IL-10 production and repression of B cells (Fig. 2D).38 After pre-treatment of B cells with or without Se, the cells were stimulated with CpG for 3 hours, and the total and phosphorylated ERK, Akt, Gsk3β, and STAT3 were compared (Fig. S10†). As shown in Fig. 2E and F, the total proteins for ERK, Akt, Gsk3β, and STAT3 remained almost unchanged in the B cells stimulated with CpG and treated with Se. However, the phosphorylation levels of Akt and Gsk3β were enhanced in CpG-stimulated B cells, and further significantly increased after Se treatment. The significant enhancement of phosphorylated ERK by a low level of Se was also observed in the CpG-activated B cells. Given the positive regulatory role of phosphorylated Akt and ERK in the IL-10 expression of B cells, the ability of Se in activation of Akt and ERK may account for the Se-enhanced IL-10 expression of B cells.
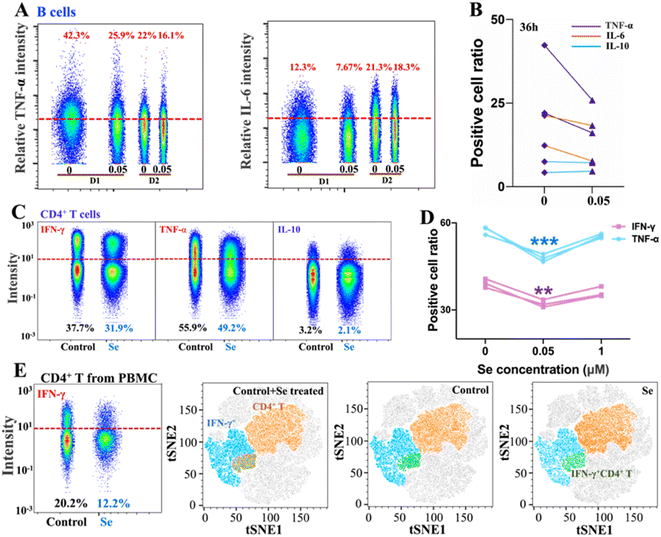
Intriguingly, alongside the enhancement of IL-10 production of B cells induced by Se, we also observed the inhibition of proinflammatory cytokines (IL-6 and TNF-α) in B cells upon a shorter period of treatment with Se (0.05 μM, 36 h) (Fig. 3A and B). Moreover, we further assessed the effects of exogenous Se on CD4+ T cells (Fig. 2A). In line with the inhibition of proinflammation cytokines of B cells, we observed markedly reduced expression of TNF-α and IFN-γ in CD4+ T cells treated with a relatively low concentration of Se (0.05 μM, 60 h) (Fig. 3C and D). Such inhibition of TNF-α and IFN-γ expression in CD4+ T cells by Se could also be observed from the mixed PBMC cells (Fig. 3E). Taken together, we demonstrate that a low level of Se (0.05 μM) inhibits the production of proinflammatory cytokines (TNF-α, IL-6, and IFN-γ) by B cell and CD4+ T cells, and stimulates anti-inflammatory cytokine (IL-10) production of B cells through activation of Akt and ERK pathways, suggesting a potential anti-inflammatory role of Se.
Selenium alleviates SARS-CoV-2-induced lung damage in K18-hACE2 mice through enhancing the expression of protective selenoproteins
Se has been regarded as an essential micronutrient and its central role in modulating the outcome of different infectious diseases has been highlighted in recent studies.39,40 Here we evaluated the in vivo immune-modulated role of Se in the K18-human angiotensin-converting enzyme II-transgenic (K18-hACE2) mouse model of COVID-19 by feeding the mice with Se (sodium selenite, 0.2 mg kg−1) for 10 days before SARS-CoV-2 infection (Fig. 4A). At 4 days post-infection, we examined the lung damage and found that Se treatment significantly decreased alveolar inflammatory infiltrates as observed in the vehicle group (Fig. 4B). We further compared the cytokine (IL-6, TNF-α, IFN-γ, and IL-10) levels in the virus-infected mouse serum with or without Se pretreatment (Fig. 4C and S11A†). The Se pretreatment could significantly suppress the enhanced pro-inflammatory cytokines, i.e., IL-6 and IFN-γ, induced by virus infection (Fig. 4C). Additionally, we compared the mRNA levels of cytokines (IL-6, TNF-α and IL-10) and chemokines (CCL2, CCL3, CXCL10, and CCL22) in the lung tissues of SARS-CoV-2-infected mice with or without Se supplement. Consistently, the mRNA levels of Ccl2 (P < 0.01), Ccl3 (P < 0.05), Il-6 (P < 0.05), and Tnf (P < 0.05) were remarkably downregulated in the mice pretreated with Se, which further corroborated an anti-inflammatory role of Se in COVID-19 (Fig. S11B†).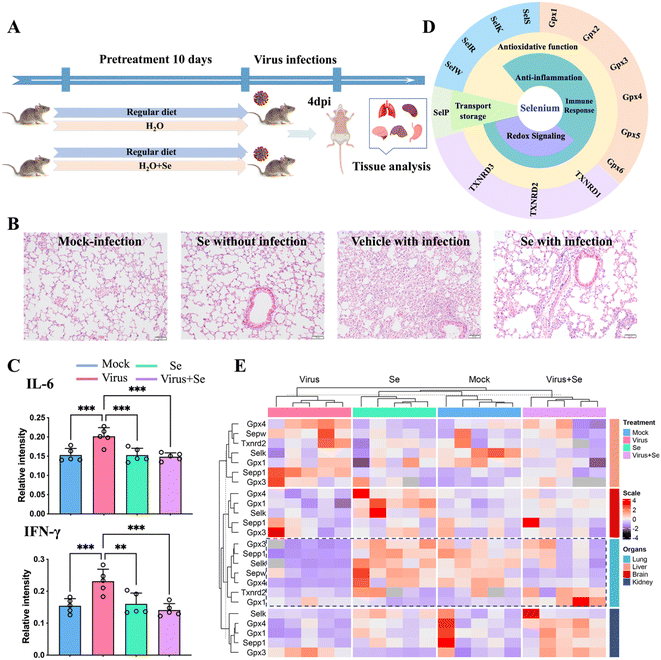 | ||
| Fig. 4 Se supplement improves SARS-CoV-2 infection outcomes of K18-hACE2 mice. (A) Experimental diagram of studies in mice. (B) Representative haematoxylin and eosin-stained sections of the collected mouse lungs at 4 days post-infection. (C) Comparison of the cytokine (IL-6 and IFN-γ) level in the mouse serum under different treatment conditions. (D) Classification of selenoproteins associated with infectious disease and their putative functions. (E) Heatmap of the expression level of Se-related genes in four organs of mice (lung, brain, kidney, and liver) under different treatment conditions [mock: non-infected control; virus: virus infection without Se treatment; Se: treatment with 0.2 mg kg−1 selenium without virus infection and virus + Se: virus infection with pre-supplement of 0.2 mg kg−1 selenium]. The mRNA levels were analyzed by the 2−ΔΔCt method. All the primer information of this study is shown in Table S7.†P values were calculated by the ANOVA test to evaluate the statistical significance of variations among groups. (*) 0.01 < P < 0.05, (**) 0.001 < P < 0.01, (***) 0.0001 < P < 0.001, (****) P < 0.0001, (ns) not significant. | ||
The biological functions of Se are mainly achieved through incorporating Se into multiple selenoproteins, which are critical to maintaining redox homeostasis and modulating the immune response against pathogens.40,41 All of these proteins are important determinants of the outcome of infections. The putative selenoproteins involved in infections include thioredoxin reductases (TXNRDs: TXNRD1, TXNRD2, and TXNRD3), glutathione peroxidases (GPXs: GPX1, GPX2, GPX3, GPX4, and GPX5) and selenoproteins (S, K, R, W, P, etc.)40,41 (Fig. 4D). To address whether SARS-CoV-2 infection also modulates the expression of selenoproteins, we further examined the mRNA level of several selenoproteins in the lungs and other organs of K18-hACE2 mice with or without SARS-CoV-2 infection, and with or without Se pretreatment. We found that SARS-CoV-2 infection induced an organ-specific reduction of selenoprotein genes, i.e., Gpx4 (P < 0.001), Gpx3 (P < 0.05), Sepp1 (P < 0.001), Sepw (P < 0.05) and Selk (P < 0.001), in the lung (Fig. 4E and S12†). Given that GPX4, GPX3, SEPP1, SEPW and SELK play central roles in antioxidant stress, the anti-inflammatory process, and activation of the immune response, such an evident reduction of these selenoproteins may account for the widely reported virus-induced oxidative stress, which may be associated with more severe disease.40,41 Importantly, pretreatment of the mice with Se could partially restore some of the repressed selenoprotein genes, i.e., Sepp1, Gpx3, Gpx4, and Selk (Fig. 4E and S12†). Additionally, Se also enhanced the expression of Gpx1 and Txnrd2 in SARS-CoV-2-infected mice. Our data indicated that the beneficial effect of Se on SARS-CoV-2-infected mice is likely attributable to the Se-enhanced expression of protective selenoproteins. Collectively, we demonstrate an in vivo anti-inflammatory role of Se in COVID-19 through rescuing virus-repressed selenoproteins to compensate for the increased oxidative stress and to alleviate the overwhelming cytokine storm.
Discussion
In the past decades, metal(loid)s have been increasingly recognized as critical modulators of the immune system.15–17 Dysregulation of metal(loid)s ions may result in an immunocompromised state, which increases the severity of infection diseases.19–24 Though there were reports and clinical data highlighting the role of metal(loid)s in COVID-19 disease,42–45 the immunomodulation of metal(loid)s in COVID-19 disease remains elusive. In this study, we systemically explore the host serological metallome and immunoproteome features in COVID-19 patients and uncover their relevance by using our developed metal-aided multiplexed proteome profiling method,32 providing a holistic view of the serological landscape and identifying a direct link between the host serum metallome and COVID-19 immunity.After being infected with SARS-CoV-2, we observed an evident decrease of multiple metal(loid)s (Se, Zn, Ca, Mg, K, I, Cr and Mo) in COVID-19 patients, among which some metal(loid)s e.g., Se, Zn, even show significant variations with disease severity. It is reported that multiple metal ions, e.g., Zn2+,27,28 are critical cofactors of many viral enzymes, proteases and polymerases that aid in viral replication and are required for proper functions of several non-structural proteins in SARS-CoV-2. The disturbance of the metabolism of these metals may result from the competition of essential elements between virus and hosts. Moreover, the impaired homeostasis of host serological metal(loid)s by SARS-CoV-2 infection may also be associated with redox disorders, electrolytes imbalance, etc.15,46–48
By integrating the metallome and immunoproteome features of COVID-19 patients with different levels of severity, we unbiasedly identified the strong correlation between Se and IL-10. IL-10 is an anti-inflammatory cytokine that plays a central role in maintaining the balance of immune response towards infections, alleviating tissue damage caused by exaggerated inflammatory response.49 The tight positive association of Se with IL-10 may contribute to the anti-inflammatory role of Se in COVID-19 disease.
Our cellular data confirm that Se could regulate immune cell functions by enhancing IL-10 expression in B cells and repressing proinflammatory cytokines in B and CD4+ T cells. The Se enhanced IL-10 production in B cells was partly attributable to the Se-mediated activation of ERK and Akt, which are crucial signaling molecules involved in IL-10 production of B cells (Fig. 5).38 It was reported previously that Se also inhibits the transcription factor (NF-kB)50 and modulates other immune cells (e.g. NK cells, macrophages, T cells), which may also account for its anti-inflammatory role in SARS-CoV-2 infection.40,51 Such an anti-inflammatory role of Se was further validated in vivo. In line with our observation, selenium deficiency was frequently observed in death cases of COVID-19 patients.30 Significant clinical benefits of selenium supplementation have also been demonstrated in other viral infections via potential immunomodulatory effects as well as its capability of affecting viral mutation and evolution.50,52 However, particular caution should be taken for Se supplementation since it may only benefit individuals with low Se status.15 Thus prior to Se treatment of COVID-19 patients, Se status should be evaluated.
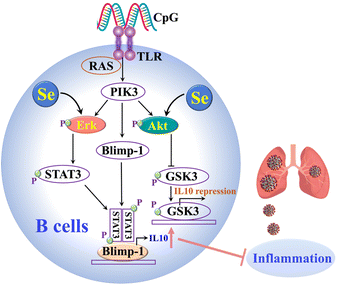 | ||
| Fig. 5 The proposed mechanisms of the anti-inflammatory role of Se in SARS-CoV-2 infections through enhancing the IL-10 expression of B cells. | ||
We further demonstrate that SARS-CoV-2 infection significantly represses several critical selenoproteins, which are involved in multiple biological pathways including oxidative stress response, redox homeostasis, endoplasmic reticulum (ER) stress, systemic inflammation and immune responses.40,41 We observed the repression of gene expression of multiple selenoproteins in lungs of virus-infected mice, including GPX4, GPX3, SELENOK and SELENOP, which may account for the widely reported viral-induced oxidative stress, resulting in a higher risk of viral genome mutation.35,50,52 The suppression of gene expression of GPX4, SELENOF, SELENOM, SELENOK and SELENOS, TXNRD3 has also been found in the virus infected cell model.53 Exogenous Se supplement restores virus-repressed selenoproteins, thus partially compensates the virus-induced oxidative stress, and alleviates the overwhelming cytokine storm and tissue damage. Collectively, the observed beneficial effect of Se on COVID-19 disease outcomes is attributable to its anti-oxidative functions, anti-inflammatory capability, and its immune modulation roles through mediating immune cell functions.
Conclusions
In conclusion, by integrating the metallome and immunoproteome, we unveiled the critical role of Se in the immune response of COVID-19 disease. We further validated that the Se-mediated enhancement of IL-10 production in B cells is achieved by stimulation of ERK and Akt activation, as well as suppression of proinflammatory cytokines in both B and CD4+ T cells. The beneficial effect of Se on improving the outcome of SARS-CoV-2-infected mice provides a fundamental basis for using Se as an auxiliary treatment of COVID-19 disease. The integrative multi-omics approach can be readily extended to long COVID, and other diseases to understand how metallomes are involved in both innate and adaptive immunity.Data availability
All detailed procedures and experimental data are available in the ESI.† Data not included there (e.g. mass cytometry data) can be provided on request.Author contributions
Y. Z., S. Y., F. X., J. F.-W. C., and H. S. conceived and designed the study. Y. Z., S. Y., F. X., H. L., Z. W. Y., T. F. C., C. T. L., K. M. T., J. P. C., J. W. S., S. S., W.-M. C., A. R. T., H. C., C.-M. C., L. J., I. F.-N. H., L. W. L., J. F.-W. C. and H. S. designed and/or performed experiments. J. F.-W. C. and H. S. acquired funding and supervised the study. Y. Z. and T. F. C. analyzed data. Y. Z., H. L. and H. S. wrote the manuscript, and all authors reviewed and edited the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was partly supported by funding from the Research Grants Council (C7060-21G, C7034-20EF, T11-709/21-N, 17318322, 2122-7S04, 17306323); the Health and Medical Research Fund (20190572, COVID1903010 – Project 7, and 07210107), the Food and Health Bureau; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government; the National Natural Science Foundation of China General Program (82272337); the National Program on Key Research Project of China (2020YFA0707500 and 2020YFA0707504); the Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; Emergency Collaborative Project (EKPG22-01) of Guangzhou Laboratory; the Emergency COVID-19 Project (2021YFC0866100), Major Projects on Public Security, National Key Research and Development Program; the Major Science and Technology Program of Hainan Province (ZDKJ202003); the Research Project of Hainan Academician Innovation Platform (YSPTZX202004); the Hainan Talent Development Project (SRC200003); and donations from the Shaw Foundation Hong Kong, Richard Yu and Carol Yu, Michael Seak-Kan Tong, May Tam Mak Mei Yin, Lee Wan Keung Charity Foundation Limited, Hong Kong Sanatorium & Hospital, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, Lo Ying Shek Chi Wai Foundation, and The University of Hong Kong (URC and Norman & Cecilia Yip Foundation). The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report. We thank the staff at the Centre for Comparative Medicine Research of the University of Hong Kong for their facilitation of the study, and Prof. Jiazuan Ni (Shenzhen University) for stimulating discussion.References
- J. F. W. Chan, et al. , Lancet, 2020, 395, 514–523 CrossRef CAS PubMed.
- K. K. W. To, et al. , Emerging Microbes Infect., 2021, 10, 507–535 CrossRef CAS PubMed.
- M. Merad, et al. , Science, 2022, 375, 1122–1127 CrossRef CAS PubMed.
- H. Namkoong, et al. , Nature, 2022, 609, 754–760 CrossRef CAS PubMed.
- F. Karagiannis, et al. , Nature, 2022, 609, 801–807 CrossRef CAS PubMed.
- S. Yuan, et al. , Cell Discovery, 2021, 7, 1–13 Search PubMed.
- A. Stukalov, et al. , Nature, 2021, 594, 246–252 CrossRef CAS PubMed.
- D. Bojkova, et al. , Nature, 2020, 583, 469–472 CrossRef CAS PubMed.
- Y. Zhou, et al. , Nat. Biotechnol., 2022, 374, 1–12 Search PubMed.
- T. T. T. Yuen, et al. , Int. J. Biol. Sci., 2022, 18, 4714–4730 CrossRef CAS PubMed.
- C. C. Murdoch and E. P. Skaar, Nat. Rev. Microbiol., 2022, 20, 657–670 CrossRef CAS PubMed.
- Y. Zhou, et al. , Annu. Rev. Biochem., 2022, 91, 449–473 CrossRef PubMed.
- D. Budinger, et al. , Lancet Neurol., 2021, 20, 956–968 CrossRef CAS PubMed.
- E. J. Ge, et al. , Nat. Rev. Cancer, 2022, 22, 102–113 CrossRef CAS PubMed.
- M. P. Rayman, Lancet, 2012, 379, 1256–1268 CrossRef CAS PubMed.
- C. Yang, et al. , J. Med. Virol., 2021, 93, 1639–1651 CrossRef CAS PubMed.
- R. Eil, et al. , Nature, 2016, 537, 539–543 CrossRef CAS PubMed.
- G. S. Lee, et al. , Nature, 2012, 492, 123–127 CrossRef CAS PubMed.
- B. Kim, et al. , Sci. Signaling, 2022, 15, eabi7400 CrossRef CAS PubMed.
- C. Wang, et al. , Adv. Immunol., 2020, 145, 187–241 CrossRef CAS PubMed.
- A. S. McKee and A. P. Fontenot, Curr. Opin. Immunol., 2016, 42, 25–30 CrossRef CAS PubMed.
- F. Baixauli, M. Villa and E. L. Pearce, Science, 2019, 363, 1395–1396 CrossRef CAS PubMed.
- T. Fukada, et al. , Nat. Immunol., 2019, 20, 248–250 CrossRef CAS PubMed.
- M. Vig and J. P. Kinet, Nat. Immunol., 2009, 10, 21–27 CrossRef CAS PubMed.
- H. Li, et al. , Chem. Commun., 2022, 58, 7466–7482 RSC.
- N. Maio, et al. , Science, 2021, 373, 236–241 CrossRef CAS PubMed.
- C. Andreini, F. Arnesano and A. Rosato, Metallomics, 2022, 14, mfac047 CrossRef PubMed.
- P. Krafcikova, et al. , Nat. Commun., 2020, 11, 3717 CrossRef CAS PubMed.
- A. V. Skalny, et al. , Metabolites, 2021, 11, 244 CrossRef CAS PubMed.
- A. Moghaddam, et al. , Nutrients, 2020, 12, 2098 CrossRef CAS PubMed.
- J. L. Domingo and M. Marquès, Food Chem. Toxicol., 2021, 152, 112161 CrossRef CAS PubMed.
- Y. Zhou, et al. , Chem. Sci., 2022, 13, 3216–3226 RSC.
- E. Becht, et al. , Nat. Biotechnol., 2019, 37, 38–44 CrossRef CAS PubMed.
- P. Sedgwick, Br. Med. J., 2014, 349, g7327 CrossRef PubMed.
- M. Laforge, et al. , Nat. Rev. Immunol., 2020, 20, 515–516 CrossRef CAS PubMed.
- M. C. Glass, et al. , Cell Rep., 2022, 39, 110728 CrossRef CAS PubMed.
- M. Michee-Cospolite, et al. , Front. Immunol., 2022, 13, 818814 CrossRef CAS PubMed.
- J. A. Bibby, et al. , Nat. Commun., 2020, 11, 3412 CrossRef CAS PubMed.
- Y. Yao, et al. , Nat. Immunol., 2021, 22, 1127–1139 CrossRef CAS PubMed.
- J. Zhang, et al. , Redox Biol., 2020, 37, 101715 CrossRef CAS PubMed.
- L. Schomburg, Nat. Rev. Endocrinol., 2012, 8, 160–171 CrossRef CAS PubMed.
- D. Jesus, et al. , Metallomics, 2020, 12, 1912–1930 CrossRef PubMed.
- I. Al-Saleh, et al. , BioMetals, 2022, 35, 125–145 CrossRef CAS PubMed.
- K. Sargsyan, et al. , Chem. Sci., 2020, 11, 9904–9909 RSC.
- J. Zhang, et al. , Am. J. Clin. Nutr., 2020, 111, 1297–1299 CrossRef PubMed.
- K. Amporndanai, et al. , Nat. Commun., 2021, 12, 3061 CrossRef CAS PubMed.
- A. Gupta, et al. , Nat. Med., 2020, 26, 1017–1032 CrossRef CAS PubMed.
- M. Noori, et al. , Rev. Med. Virol., 2022, 32, e2262 CrossRef CAS PubMed.
- S. S. Iyer and G. Cheng, Crit. Rev. Immunol., 2012, 32, 23–63 CrossRef CAS PubMed.
- M. P. Rayman, E. W. Taylor and J. Zhang, Proc. Nutr. Soc., 2023, 82, 1–12 CrossRef CAS PubMed.
- L. Huang, et al. , Immunity, 2021, 54, 1728–1744 CrossRef CAS PubMed.
- L. Hiffler and B. Rakotoambinina, Front. Nutr., 2020, 7, 164 CrossRef PubMed.
- Y. Wang, et al. , Food Chem. Toxicol., 2021, 153, 112286 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03345g |
| ‡ These authors contributed equally as co-first authors. |
| This journal is © The Royal Society of Chemistry 2023 |