Open Access Article
Open Access ArticleNonribosomal peptides protect Pseudomonas nunensis 4A2e from amoebal and nematodal predation†
Sebastian
Pflanze
 a,
Ruchira
Mukherji
a,
Anan
Ibrahim
a,
Ruchira
Mukherji
a,
Anan
Ibrahim
 a,
Markus
Günther
a,
Sebastian
Götze
a,
Markus
Günther
a,
Sebastian
Götze
 a,
Somak
Chowdhury
a,
Lisa
Reimer
a,
Lars
Regestein
b and
Pierre
Stallforth
a,
Somak
Chowdhury
a,
Lisa
Reimer
a,
Lars
Regestein
b and
Pierre
Stallforth
 *ac
*ac
aDepartment of Paleobiotechnology, Leibniz Institute for Natural Product Research and Infection Biology – Leibniz-HKI, Beutenbergstrasse 11a, 07745 Jena, Germany. E-mail: pierre.stallforth@leibniz-hki.de
bBio Pilot Plant, Leibniz Institute for Natural Product Research and Infection Biology – Leibniz-HKI, Beutenbergstrasse 11a, 07745 Jena, Germany
cFaculty of Chemistry and Earth Sciences, Friedrich Schiller University (FSU), Jena, Germany
First published on 2nd October 2023
Abstract
The rhizosphere is a highly competitive environment forcing bacteria to evolve strategies to oppose their enemies. The production of toxic secondary metabolites allows bacteria to counteract predators. In this study, we describe the anti-predator armamentarium of the soil-derived bacterium Pseudomonas nunensis 4A2e. Based on a genome mining approach, we identified several biosynthetic gene clusters coding for nonribosomal peptide synthetases. Generation of gene deletion mutants of the respective clusters shows a loss of defense capabilities. We isolated the novel lipopeptides keanumycin D and nunapeptins B and C, and fully elucidated their structures by a combination of in-depth mass spectrometry experiments, stable isotope labelling, and chemical synthesis. Additionally, investigation of the quorum sensing-dependent biosynthesis allowed us to elucidate parts of the underlying regulation of the biosynthetic machinery. Ecology-inspired bioassays highlight the role of these peptides as a defence strategy against protozoans and led us to find a previously unknown function against the bacterivorous nematode Oscheius myriophilus.
Introduction
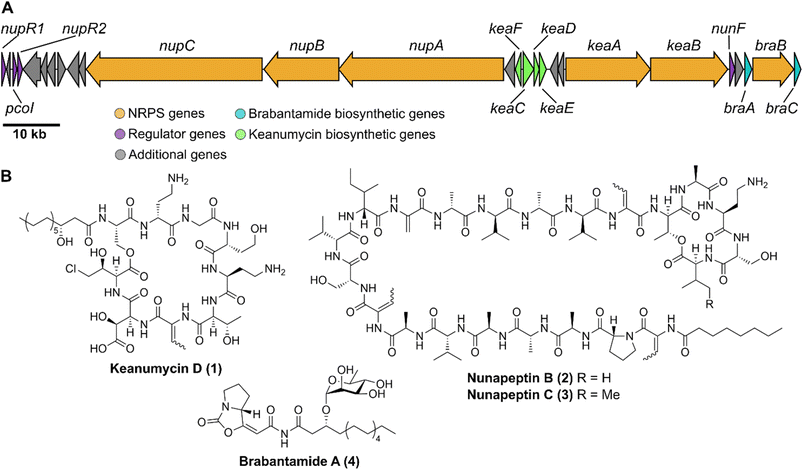
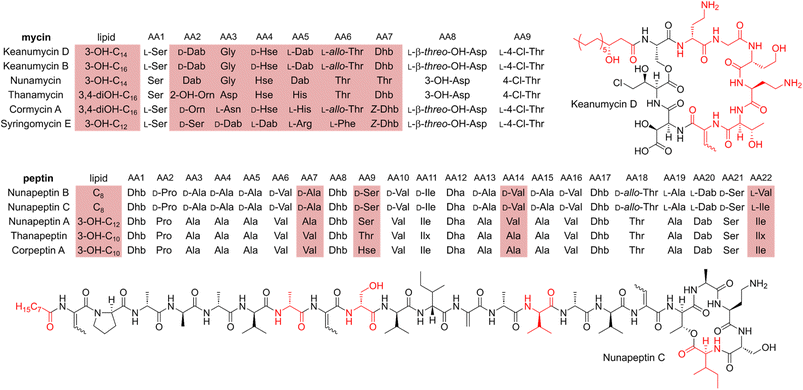
Microbial predator–prey interactions play an important role in shaping the microbial communities of virtually all ecosystems.1–3 Predation puts bacterial and fungal communities under a tremendous selection pressure, driving the evolution of traits to resist and overcome grazing. These defence strategies may rely on morphological changes including biofilm formation or increased motility.2,3 The secretion of toxic metabolites is, however, particularly effective in this regard.2,3 Especially the production of secondary metabolites including nonribosomal peptides, has been shown to be an effective way to fight various voracious predators such as unicellular eukaryotes and nematodes.2,4–6Pseudomonas spp., for instance, have been reported to produce the cyclic lipopeptides (CLPs) massetolide and viscosin to resist grazing from the soil-dwelling amoeba Naegleria americana;7 whilst the endosymbiont Candidatus Mycoavidus necroximicus protects its fungal host from the fungivorous nematode Aphelenchus avenae by the biosynthesis of anthelmintic benzolactones and a synergistically acting lipodepsipeptide.8,9 Besides grazing evasion, lipopeptides also play an important role in rhizosphere ecology, as many peptides have been found in either phytopathogenic or plant beneficial pseudomonads.10,11 One of the best studied CLPs is syringomycin, a chlorinated lipopeptide isolated from the phytopathogen Pseudomonas syringae pv. syringae.12 Other members of the syringomycin-like class of CLPs, sometimes referred to as “mycins”,10 include cormycin A from the phytopathogenic P. corrugata,13 but also thanamycin, nunamycin, and brasmycin from disease-suppressive pseudomonads.14–18 These compounds belong to the family of nonribosomal peptides (NRPs), which are generated by dedicated nonribosomal peptide synthetases (NRPSs). The incorporation of amino acids is not dependent on the ribosome, however, typically multimodular NRPSs allow for the generation of peptides of high structural diversity. Each module contains domains that select amino acids (the adenylation or A domain) and domains that catalyse the amide bond formation (the condensation or C domains). Powerful bioinformatic tools have been developed to both identify the presence of NRPS biosynthetic gene clusters (BGCs) in genomes and the nature of the incorporated amino acids. Details of NRP biosynthesis have been extensively reviewed elsewhere.19,20Interestingly, the biosynthetic gene clusters for mycin biosyntheses are usually tightly co-clustered with BGCs of another family of NRPs, the “peptins”.10 Members of the peptin family include CLPs such as syringopeptins, suppressing the phytopathogenic fungus Botrytis cinerea,21 as well as thanapeptins and nunapeptin, which have been reported to act on oomycete pathogens.15,17 Recently, we demonstrated how Pseudomonas sp. QS1027 uses a combination of structurally related CLPs, keanumycins and jessenipeptin, to overcome grazing by the model social amoeba Dictyostelium discoideum.22 Interestingly, it has been hypothesised that predation resistance is linked to phytopathogen-suppressive traits, opening potential application in agricultural systems.23,24 A good example is the work on CLPs of the amphisin family.11 While anikasin was shown to protect Pseudomonas fluorescens HKI0770 from its predator, the social amoeba Polysphondylium violaceum, a tensin-like congener supressed Streptomyces scabies, the causal organism of potato scab, significantly in planta.25,26 Therefore, to deepen our understanding of the molecular ecology of the anti-predator repertoire, we set out to investigate the interaction of pseudomonads and their predators. Here, we describe the isolation, structure elucidation, and functional characterisation of keanumycin D (1), nunapeptin B (2) and nunapeptin C (3), from the amoebicidal Pseudomonas nunensis 4A2e, which was co-isolated with the social amoeba Polysphondylium pallidum RM1 from forest soil. Bioassays with 1–3 and the co-produced lipopeptide brabantamide A (4) against a range of bacterivorous amoebae and an environmental nematode emphasise the importance of those lipopeptides against a broad spectrum of predators; whilst characterisation of gene deletion mutants of co-clustering LuxR-type proteins and the identification of a quorum sensing mediator shed light on the underlying regulation of NRP biosynthesis. We believe, that our results support our understanding of molecular microbial ecology and may help to engineer suppressive soils.27
Results and discussion
Isolation and in silico genome analysis of Pseudomonas nunensis 4A2e
In order to identify bacterial natural products with anti-predator activity, we co-isolated bacteria (prey) and social amoebae (predators) from forest soil near Jena (50.962 N 11.592 E). Co-cultivation experiments between predators and prey in a 24-well plate format allowed us to subdivide the bacterial isolates in those, which are edible or inedible for these amoebae. The latter were tested for the production of amoebicidal natural products.28–30 Further investigation of methanolic culture extracts of inedible strains led to the identification of Pseudomonas nunensis 4A2e as a producer of anti-amoebal metabolites. Genome sequencing and consecutive genome mining using antiSMASH31 revealed a region coding for three NRPS BGCs (Fig. 1A and S1†), which is highly similar to a phytopathogen-suppressive genomic island found in the biocontrol strain Pseudomonas nunensis In5 (Fig. S2 and S10†).16,32 Since recent studies have demonstrated a link between predation resistance traits and phytopathogen inhibition, we further examined these NRPS genes.22,23 The largest BGC codes for three NRPSs (NupA, NupB, NupC) and shares a protein identity of >97% with the respective NRPS genes of the nunapeptin BGC in P. nunensis In5. A smaller BGC found in the vicinity of the nunapeptin BGC displays the unique genomic architecture of the syringomycin family BGCs, coding for two NRPSs (KeaA and KeaB) and four additional proteins (KeaC–KeaF), with the latter probably being involved in the tailoring of the characteristic 4-chloro-L-threonine and L-β-threo-OH-aspartic acid motif of this natural product class.33–35 The third NRPS (BraB) contains two modules with specificity for serine and proline, and is flanked by an upstream glycosyltransferase (BraA) and a downstream FAD-dependent monooxygenase (BraC) (Fig. S6 and Tables S3 and S4†) similar to Pseudomonas sp. SH-C52, which is also the producer of thanapeptin and thanamycin.15,36Nonribosomal lipopeptides protect against amoebal predators
We proceeded by generating 4A2e mutants with inactivated biosynthetic genes using a markerless allelic replacement strategy.37 Deletion of the region coding for the adenylation (A) domain and the peptidyl carrier protein (PCP) domain in the first module of each NRPS BGC resulted in three strains: Δbra, Δnup, and Δkea. The resulting gene deletion mutants were co-cultivated with the bacterivorous amoeba Polysphondylium pallidum RM1, which was co-isolated with 4A2e. Whilst the Δbra mutant was still inedible, the Δnup and Δkea mutants lost their amoebicidal phenotype and allowed P. pallidum RM1 to form characteristic fruiting bodies (Fig. 2A and S15†). This indicated that the metabolites produced by both NRPSs are involved in the anti-predator resistance of the pseudomonad. Similar results were observed when we co-cultured the bacterial mutants with the laboratory strain Polysphondylium pallidum PN500.38 When tested against other social amoebae such as Dictyostelium discoideum28 and Dictyostelium purpureum,39 only the Δkea mutant proved to be edible. For Dictyostelium caveatum,40 however, Δnup was edible, whilst Δkea still showed no sign of fruiting body formation (Fig. 2B, S16 and S17†). These findings indicate that the presence of both NRPs is necessary to counteract a broader spectrum of protozoal predators and further drew our interest to the structural elucidation of these nonribosomal peptides.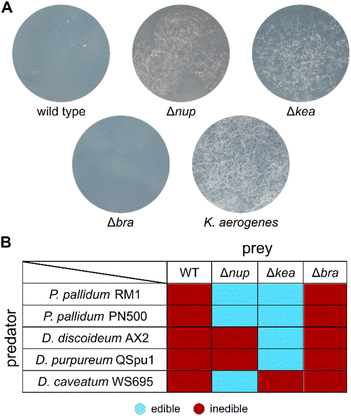 | ||
| Fig. 2 Edibility assays with selected social amoebae. (A) Co-cultivation of wild type 4A2e and mutant strains with P. pallidum RM1. Klebsiella aerogenes served as positive control. (B) Edibility of inactivation mutants with a set of social amoebae (Fig. S15–S17†). | ||
Although the kea and nup BGCs in 4A2e show a high degree of synteny with those of P. nunensis In5 (Fig. S9 and S10†), LCMS analysis of methanolic extracts of wild type 4A2e showed no match with the reported data for nunamycin or nunapeptin, however putative mycin and peptin congeners were detected and identified by comparison of wild type and mutant extracts (Fig. S11†). Due to low production titres of these candidate molecules in the initial amoeba–bacteria co-cultivation medium (SM/5), we conducted an extensive screening to optimise the culture medium for further fermentation experiments, leading to defined cultivation medium (modified Davis medium). Extraction of the supernatant with adsorbent resin and subsequent extensive chromatographic separation yielded four compounds (1–4). It is noteworthy, that careful adjustment of the phosphate concentration substantially influenced the production of 1.41,42
Structure elucidation of keanumycin D
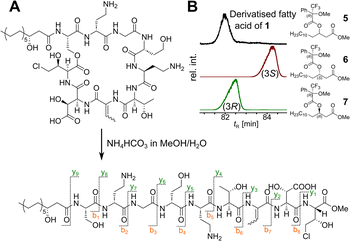
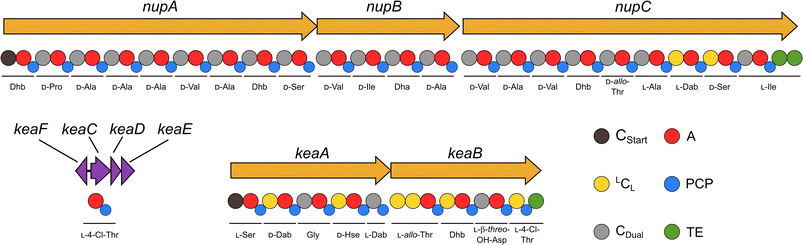
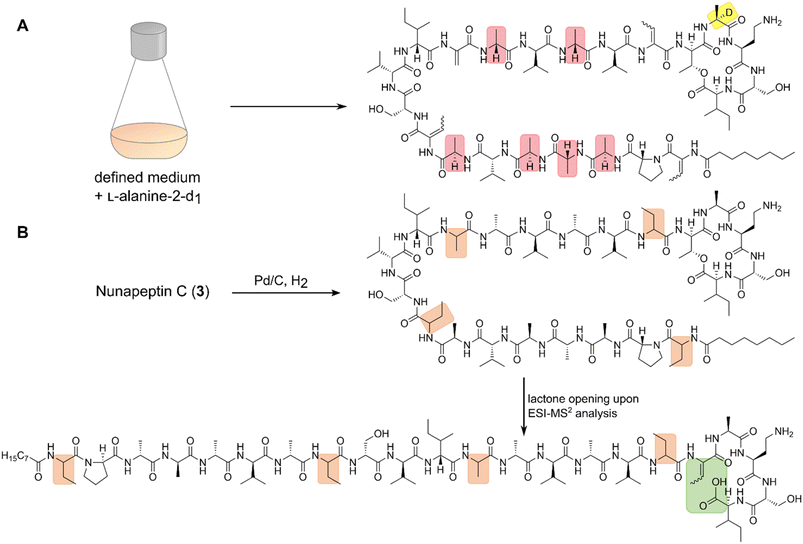
Our recent identification of keanumycins A–C proved to be helpful for structure elucidation of 1 (m/z 1122.5433 [M+H]+).22,43 Except for keanumycin C, all reported mycins are cyclic lipopeptides. Mild saponification13 of 1 in MeOH led to the formation of compound with a mass increase of 32 Da (Fig. 3A). This corresponds to the formal addition of methanol, which is in agreement with the hydrolysis of the lactone and the formation of a C-terminal methyl ester. This assumption was supported by subsequent high-resolution tandem mass spectrometry (HRMS2) analysis of the now linearised peptide and allowed us to deduce most of the amino acid sequence. Uncertainty remained about the identity of amino acids AA4 and AA6, which could not be assigned unambiguously by MS2. Both positions showed a fragment loss of 101 Da, which indicates either a homoserine (Hse) or threonine (Thr) fragment. Bioinformatic prediction of the adenylation domain specificity suggested the incorporation of threonine for module 6 (Fig. S4†), whilst homoserine was the most likely substrate of module 4 (Fig. S5†). Subsequent Marfey's analysis (Fig. S32 and S33†) further supported this assumption, as it revealed the presence of 1x L-Ser, 1x D-Dab, 1x Gly, 1x D-Hse, 1x L-Dab, 1x L-allo-Thr, 1x L-β-threo-OH-Asp, and 1x 4-Cl-L-Thr,44 which allowed the unambiguous determination of the configuration of most amino acids, except for the position of L- and D-Dab. Here, closer examination of the genomic architecture of the kea BGC suggested that the Dab of position two is D-configured due to a downstream Cdual domain, which not only condenses the following Gly to the growing peptide chain, but also epimerises the upstream Dab in the process (Fig. 4).45 The fifth module, however, is adjacent to two downstream LCL domains, that condense an L-configured amino acid to another L-configured amino acid. Therefore, an L-configuration was assigned to Dab at position five. Additionally, we found that module five is lacking an A domain, which is a characteristic of mycin BGCs of the P. mandelii group.10 It was hypothesised for the nunamycin BGC of In5, that the absent A domain in module five is compensated by the Dab-specific A domain of module two.16 After deducing the structure of the peptide backbone, we focused on the analysis of the lipid motif of the CLP. Based on our HRMS2 data, we expected the presence of an N-terminal, hydroxylated myristoyl moiety. Acidic hydrolysis, followed by esterification with trimethylsilyldiazomethane and subsequent derivatisation with (S)-Mosher's acid chloride yielded ester 5, which was then subjected to GCMS analysis. Comparison with synthetic 3-hydroxymyristic acid standards 6 and 7 allowed us to establish the identity of the lipid as (R)-3-hydroxymyristic acid (Fig. 3B and S38†). Thus, 1 was identified as a congener of the recently discovered keanumycins, differing from keanumycin B only by a slightly shorter lipid motif (Fig. 5). Interestingly, although whole-genome phylogenetic analysis shows that 4A2e and In5 share an average nucleotide identity of >96% across their entire genomes (Fig. S8†) and the mycin BGC NRPS core genes share an amino acid sequence identity of >98% (Fig. S10†), they result in slightly different peptide backbones. In contrast, even though 4A2E and QS1027 share low protein identities between KeaA and KeaB (63% and 75%, respectively), they produce CLPs with an identical amino acid sequence and configuration (Fig. 5). More detailed analyses of nonribosomal lipopeptide families have recently been published elsewhere.10,11,46Structure elucidation of nunapeptin B and C
A comparable approach was applied for structure elucidation of peptides 2 and 3. Extensive MS2 analysis allowed to determine most of the amino acid sequence of both compounds (Fig. S25 and S26†), which is highly similar to nunapeptin isolated from In5 (which we now refer to as nunapeptin A).16 As a consequence, we named our novel lipopeptides nunapeptins. However, our MS2 data indicated that 2 and 3 do not have a 3-hydroxyl fatty acid motif, which is usually observed in Pseudomonas-derived nonribosomal lipopeptides, but instead bear an N-terminal octanoic acid, which was also detected upon basic hydrolysis of 3 (Fig. S39†). Furthermore, derivatisation of the hydrolysed amino acids with Marfey's reagent yielded the following amino acid composition for 3: 1x D-Pro, 6x D-Ala, 1x L-Ala, 4x D-Val, 2x D-Ser, 1x D-Ile, 1x D-allo-Thr, 1x L-Dab, and 1x L-Ile (Fig. S29 and S31†). When we performed the same analysis with 2, we observed 1x L-Val, but no more L-Ile, which is consistent with the mass difference of 14 Da between 2 and 3. The position of the L-configured amino acids was deduced from the bioinformatic prediction of LCL condensation domains, which were predicted downstream of the Ala- and Dab-specific A domains in module 19 and 20 (Fig. 4). Therefore, we concluded that these positions must be L-configured. For L-Ala19, this assumption was later confirmed by stable isotope labelling. In NRPS biosynthesis, D-amino acids usually originate from L-amino acids, which are epimerised by specialised epimerisation (E) domains or Cdual domains with epimerisation activity. Epimerisation of an α-deuterium-labelled amino acid and exchange with a proton from the culture medium results in the loss of the label, whilst L-configured positions would retain the deuterium label, localisable in the final natural product. Hence, we cultivated 4A2e in a defined medium, supplemented with L-alanine-2-d1, and analysed peptin 3 (Fig. 6A). Comparison of the monoisotopic ion intensity and the intensity of the labelled isotopic ion for peptin 3 showed an increased relative intensity of the isotopologue's ion for the C-terminal part of the peptin sequence (Tables S10 and S11†). Thus, we deduced that an isotope enrichment must have occurred for this part of the molecule. Hence, by combining the bioinformatic prediction and isotope labelling data, we concluded that Ala19 must be L-configured. Having determined the sequence and configuration of the amino acids, we were left with the determination of the ring size of the cyclic part of the lipopeptide. Upon MS2 analysis, we observed the linearisation of the peptide resulting in the opening of the lactone to give the C-terminal carboxylic acid and a 2,3-unsaturated amino acid.26 The presence of several unsaturated amino acids in the MS2 dataset left us uncertain at which position the transformation occurred. Therefore, we masked the unsaturated amino acids in the native peptide by Pd-catalysed hydrogenation, followed by MS2 analysis of the product (Fig. 6B and S37†). Four positions (AAs 1, 8, 12, and 17) showed an increase of 2 Da as a result of olefin reduction, whilst a single position (AA18) was still identified as Dhb. We concluded that the lactone must be formed between this position (D-allo-Thr18) and the C-terminal carboxylic acid, resulting in a 16-membered ring motif for the nunapeptins (Fig. 6).Regulation of secondary metabolite biosynthesis
After elucidating the structures of the lipopeptides 1–3, our attention was drawn to putative regulators of the NRPS biosynthesis. Protein BLAST47 and InterProScan48 analysis of nearby open reading frames (ORFs) (Tables S3 and S4†) revealed the presence of three genes coding for LuxR-type regulatory proteins (NupR1, NupR2, NunF) and a gene coding for an autoinducer synthase (PcoI). For Pseudomonas sp. QS1027 and Pseudomonas corrugata CFBP 5454, proteins similar to NupR1, NupR2, and PcoI were described to be involved in the regulation of the NRPS BGCs, whilst in P. nunesis In5, NunFIn5 was reported as a regulator of nunamycin and nunapeptin biosynthesis.22,49–51 Analysis of gene deletion mutants of 4A2e showed a drastic reduction of the production of 1–3 and also brabantamide A (4), indicating that the respective BGCs are under the influence of the same regulatory network (Fig. S12†). Closer examination of the LuxR-type regulators and a search for conserved domains within the encoded protein sequences predicted an autoinducer-binding domain (Pfam PF03472) and a DNA-binding helix-turn-helix (HTH) domain (Pfam PF00196) in NupR1, whereas only one HTH domain was predicted for NupR2 and NunF (Fig. S7†). Since PcoI and NupR1 resemble a LuxI/LuxR-type quorum sensing system, we became interested in the putative signalling molecule produced by PcoI. Analysis of wild type supernatants revealed the presence of N-hexanoyl homoserine lactone, that was absent in the ΔpcoI mutant (Fig. S13†). Therefore, we sought out to re-initiate lipopeptide biosynthesis in the mutant by supplementation with synthetic N-hexanoyl-L-homoserine lactone, which was able to restore the production of 1–4 completely (Fig. S14†). Interestingly, N-hexanoyl-L-homoserine lactone has also been reported as regulator of peptin and mycin biosyntheses in CFBP 5454 and QS1027, and was also detected in extracts of In5.22,49,52,53 From an evolutionary point of view it is fascinating how the mycin and peptin NRPS genes in these pseudomonads gradually diversified, whilst the underlying regulatory mechanisms seemingly remained conserved.In contrast to NupR1, the LuxR-type proteins NupR2 and NunF lack designated autoinducer-binding domains, which makes it likely that their putative ligands are signals other than homoserine lactones, or they act in a ligand independent manner.54,55 Whilst the nature of potential ligands remains unknown, it has been demonstrated for NunFIn5 from the biocontrol strain P. nunensis In5, that NunF is strongly upregulated in response to glycerol and trehalose, which are fungal- and oomycete-associated molecules but also found in a variety of amoebae.49,50,56,57 This further strengthens the link between biocontrol traits and resistance to microbial predation,23,58,59 and has the potential to develop biocontrol formulations of plant beneficial pseudomonads for agricultural application.60,61
Bioactivity of 4A2e-derived lipopeptides
The anti-amoebal activity of purified 1–4 was determined against the model social amoeba Dictyostelium discoideum, and the human pathogens Acanthamoeba castellanii and Acanthamoeba comandoni (Table 1 and S6†).28,62 Interestingly, all peptides showed at least low micromolar activity (IC50) against the amoebae, indicating the importance of the whole genomic region to overcome protozoal grazing.| Organism | Keanumycin D (1) | Nunapeptin B (2) | Nunapeptin C (3) | Brabantamide A (4) |
|---|---|---|---|---|
| D. discoideum | 74 nM | 5.7 μM | 3.1 μM | 4.7 μM |
| A. castellanii | 22.8 μM | 3.5 μM | 2.6 μM | 14.6 μM |
| A. comandoni | 20.6 μM | 4.0 μM | 3.3 μM | 8.9 μM |
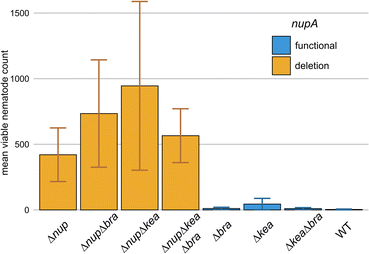
Nunapeptins suppress bacterivorous nematodes
The activity of these natural products against a variety of protozoans led us to the question, if other microfaunal predators are also affected by either one of the lipopeptides. For example, bacterivorous nematodes have been extensively studied as microbial predators,2,63–65 and the production of anthelmintic natural products was described as an effective mean to overcome their predation pressure.59,66–71 Therefore, we tested 4A2e against the bacterivorous nematode Oscheius myriophilus SP1 that was isolated from the same sampling site as 4A2e and P. pallidum RM1. Initial tests showed, that nematode cultures co-cultivated with the ΔnupΔkeaΔbra triple mutant would proliferate, whilst living worms could be hardly recovered when co-cultivated with wild type 4A2e. Encouraged by these results, we set out to determine which specific lipopeptide caused this effect by conducting co-cultivation experiments with a set of 4A2e mutants having several gene deletion combinations. Nematodes and bacteria were co-cultivated for 10 days, which would allow O. myriophilus to undergo at least two life cycles and produce offspring. At the end of the experiment, the worms were gently detached from the plates and counted (Fig. 7). Interestingly, the Δnup mutants allowed nematode cultures to thrive, whilst the number of viable worms was drastically reduced in all mutants with an intact nup BGC (ESI Table S4;† see github). This indicated, that the products of the nup BGC, the nunapeptins, interfere with the viability and propagation of O. myriophilus. Although naturally occurring nunapeptin concentrations are most likely lower, we still expect these lipopeptides to contribute to nematode repulsion, especially in combination with other secondary metabolites, such as HCN, which is also produced to some extend by 4A2e under co-cultivation conditions and in several other media (Fig. S40†).59Conclusions
In this study, we applied an ecologically motivated approach to isolate three novel nonribosomal lipopeptides from microbial predator–prey interactions. Bioinformatic as well as extensive MS2 analyses, combined with synthetic chemistry and stable isotope labelling facilitated the structure elucidation of these secondary metabolites. A selection of bioassays shed light on the broad-spectrum activity of these metabolites against several microfaunal predators. Eventually, we were able to gain an insight in the intraspecific regulation of these metabolites by a LuxR/LuxI-type quorum sensing system. The structural similarity of these peptides with secondary metabolites that have been described in studies of phytopathogen suppression as well as the high degree of synteny of the respective BGCs raise the question regarding the role such soil-dwelling pseudomonads may play in environmental communities, how they shape the corresponding microbiomes, and how we can use them to engineer suppressive soils.Data availability
Whole genome sequence data of Pseudomonas nunensis 4A2e (bioproject accession no. PRJNA956857), the 16S rDNA sequence of P. nunensis 4A2e (OQ830499), and the 18S rDNA sequence data of P. pallidum RM1 (OQ842737) and O. myriophilus SP1 (OQ832783) have been deposited in Genbank. The codes used to generate the genomic analysis and their corresponding ESI tables can be found in github (https://github.com/Darcy220606/NRPS-Pnunensis4A2e).Author contributions
Sebastian Pflanze: investigation, methodology, conceptualisation, formal analysis, visualisation, writing – original draft. Ruchira Mukherji: investigation, formal analysis, writing – review & editing. Anan Ibrahim: formal analysis, data curation, writing – review & editing. Markus Günther: formal analysis, visualisation, writing – review & editing. Sebastian Götze: investigation, writing – review & editing. Somak Chowdhury: formal analysis, data curation, writing – review & editing. Lisa Reimer: investigation, validation. Lars Regestein: resources. Pierre Stallforth: conceptualisation, funding acquisition, project administration, supervision, writing – original draft and review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Andrea Perner (HRMS2), Heike Heinecke (NMR), Patrick Berthel, Michael Cyrulies, Jan Schönemann, Matthias Steinacker, Karsten Willing (fermentation), and Mandy Mlotek (GCMS) for their excellent technical support. We would like to thank Ron Hermenau for his generous gift of racemic β-threo-OH-Asp. This research was supported by the Werner Siemens Foundation, the Leibniz Association, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy EXC 2051 (Project-ID 390713860, “Balance of the Microverse”), and Dr Illing Stiftung.References
- R. R. Nair, M. Vasse, S. Wielgoss, L. Sun, Y.-T. N. Yu and G. J. Velicer, Nat. Commun., 2019, 10, 4301 CrossRef PubMed.
- A. Jousset, Environ. Microbiol., 2012, 14, 1830–1843 CrossRef PubMed.
- C. Matz and S. Kjelleberg, Trends Microbiol., 2005, 13, 302–307 CrossRef CAS PubMed.
- C. Matz, J. S. Webb, P. J. Schupp, S. Y. Phang, A. Penesyan, S. Egan, P. Steinberg and S. Kjelleberg, PLoS One, 2008, 3, e2744 CrossRef PubMed.
- A. Jousset, E. Lara, L. G. Wall and C. Valverde, Appl. Environ. Microbiol., 2006, 72, 7083–7090 CrossRef CAS PubMed.
- M. Künzler, PLoS Pathog., 2018, 14, e1007184 CrossRef PubMed.
- M. Mazzola, I. de Bruijn, M. F. Cohen and J. M. Raaijmakers, Appl. Environ. Microbiol., 2009, 75, 6804–6811 CrossRef CAS PubMed.
- H. Büttner, S. P. Niehs, K. Vandelannoote, Z. Cseresnyés, B. Dose, I. Richter, R. Gerst, M. T. Figge, T. P. Stinear, S. J. Pidot and C. Hertweck, Proc. Natl. Acad. Sci. U.S.A., 2021, 118, e2110669118 CrossRef PubMed.
- H. Büttner, S. J. Pidot, K. Scherlach and C. Hertweck, Chem. Sci., 2023, 14, 103–112 RSC.
- L. Girard, M. Höfte and R. D. Mot, Crit. Rev. Microbiol., 2020, 46, 397–419 CrossRef PubMed.
- C. Cesa-Luna, N. Geudens, L. Girard, V. De Roo, H. R. Maklad, J. C. Martins, M. Höfte and R. De Mot, mSystems, 2023, 8, e00988 CrossRef PubMed.
- N. Fukuchi, A. Isogai, J. Nakayama, S. Takayama, S. Yamashita, K. Suyama, J. Y. Takemoto and A. Suzuki, J. Chem. Soc., Perkin Trans., 1992, 1, 1149–1157 RSC.
- A. Scaloni, M. Dalla Serra, P. Amodeo, L. Mannina, R. M. Vitale, A. L. Segre, O. Cruciani, F. Lodovichetti, M. L. Greco, A. Fiore, M. Gallo, C. D'Ambrosio, M. Coraiola, G. Menestrina, A. Graniti and V. Fogliano, Biochem. J., 2004, 384, 25–36 CrossRef CAS PubMed.
- R. Mendes, M. Kruijt, I. de Bruijn, E. Dekkers, M. van der Voort, J. H. M. Schneider, Y. M. Piceno, T. Z. DeSantis, G. L. Andersen, P. A. H. M. Bakker and J. M. Raaijmakers, Science, 2011, 332, 1097–1100 CrossRef CAS PubMed.
- M. Van Der Voort, H. J. G. Meijer, Y. Schmidt, J. Watrous, E. Dekkers, R. Mendes, P. C. Dorrestein, H. Gross and J. M. Raaijmakers, Front. Microbiol., 2015, 6, 693 Search PubMed.
- C. F. Michelsen, J. Watrous, M. A. Glaring, R. Kersten, N. Koyama, P. C. Dorrestein and P. Stougaard, mBio, 2015, 6, e0079 CrossRef PubMed.
- C. F. Michelsen, H. Jensen, V. J. Venditto, R. C. Hennessy and P. Stougaard, PeerJ, 2015, 3, e1476 CrossRef PubMed.
- H. Zhao, Y.-P. Liu and L.-Q. Zhang, Front. Microbiol., 2019, 10, 544 CrossRef PubMed.
- R. D. Süssmuth and A. Mainz, Angew. Chem., Int. Ed., 2017, 56, 3770–3821 CrossRef PubMed.
- S. L. Wenski, S. Thiengmag and E. J. N. Helfrich, Synth. Syst. Biotechnol., 2022, 7, 631–647 CrossRef CAS PubMed.
- P. Lavermicocca, N. Sante Iacobellis, M. Simmaco and A. Graniti, Physiol. Mol. Plant Pathol., 1997, 50, 129–140 CrossRef CAS.
- S. Götze, R. Vij, K. Burow, N. Thome, L. Urbat, N. Schlosser, S. Pflanze, R. Müller, V. G. Hänsch, K. Schlabach, L. Fazlikhani, G. Walther, H.-M. Dahse, L. Regestein, S. Brunke, B. Hube, C. Hertweck, P. Franken and P. Stallforth, J. Am. Chem. Soc., 2023, 145, 2342–2353 CrossRef PubMed.
- N. Amacker, Z. Gao, B. C. Agaras, E. Latz, G. A. Kowalchuk, C. F. Valverde, A. Jousset and S. Weidner, Front. Microbiol., 2020, 11, 614194 CrossRef PubMed.
- S. Weidner, E. Latz, B. Agaras, C. Valverde and A. Jousset, Plant Soil, 2017, 410, 509–515 CrossRef CAS.
- S. Götze, R. Herbst-Irmer, M. Klapper, H. Görls, K. R. A. Schneider, R. Barnett, T. Burks, U. Neu and P. Stallforth, ACS Chem. Biol., 2017, 12, 2498–2502 CrossRef PubMed.
- A. Pacheco-Moreno, F. L. Stefanato, J. J. Ford, C. Trippel, S. Uszkoreit, L. Ferrafiat, L. Grenga, R. Dickens, N. Kelly, A. D. Kingdon, L. Ambrosetti, S. A. Nepogodiev, K. C. Findlay, J. Cheema, M. Trick, G. Chandra, G. Tomalin, J. G. Malone and A. W. Truman, eLife, 2021, 10, e71900 CrossRef CAS PubMed.
- R. Gómez Expósito, I. de Bruijn, J. Postma and J. M. Raaijmakers, Front. Microbiol., 2017, 8, 2529 CrossRef PubMed.
- R. Froquet, E. Lelong, A. Marchetti and P. Cosson, Nat. Protoc., 2009, 4, 25–30 CrossRef CAS PubMed.
- M. Klapper, S. Götze, R. Barnett, K. Willing and P. Stallforth, Angew. Chem., Int. Ed., 2016, 55, 8944–8947 CrossRef CAS PubMed.
- J. Arp, S. Götze, R. Mukherji, D. J. Mattern, M. García-Altares, M. Klapper, D. A. Brock, A. A. Brakhage, J. E. Strassmann, D. C. Queller, B. Bardl, K. Willing, G. Peschel and P. Stallforth, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 3758–3763 CrossRef CAS PubMed.
- K. Blin, S. Shaw, A. M. Kloosterman, Z. Charlop-Powers, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, W29–W35 CrossRef CAS PubMed.
- F. Ntana, R. C. Hennessy, A. Zervas and P. Stougaard, Int. J. Syst. Evol. Microbiol., 2023, 73, 005700 CAS.
- F. H. Vaillancourt, J. Yin and C. T. Walsh, Proc. Natl. Acad. Sci. U.S.A., 2005, 102, 10111–10116 CrossRef CAS PubMed.
- G. M. Singh, F. H. Vaillancourt, J. Yin and C. T. Walsh, Chem. Biol., 2007, 14, 31–40 CrossRef CAS PubMed.
- G. M. Singh, P. D. Fortin, A. Koglin and C. T. Walsh, Biochemistry, 2008, 47, 11310–11320 CrossRef CAS PubMed.
- Y. Schmidt, M. van der Voort, M. Crüsemann, J. Piel, M. Josten, H.-G. Sahl, H. Miess, J. M. Raaijmakers and H. Gross, ChemBioChem, 2014, 15, 259–266 CrossRef CAS PubMed.
- L. R. Hmelo, B. R. Borlee, H. Almblad, M. E. Love, T. E. Randall, B. S. Tseng, C. Lin, Y. Irie, K. M. Storek, J. J. Yang, R. J. Siehnel, P. L. Howell, P. K. Singh, T. Tolker-Nielsen, M. R. Parsek, H. P. Schweizer and J. J. Harrison, Nat. Protoc., 2015, 10, 1820–1841 CrossRef CAS PubMed.
- A. J. Heidel, H. M. Lawal, M. Felder, C. Schilde, N. R. Helps, B. Tunggal, F. Rivero, U. John, M. Schleicher, L. Eichinger, M. Platzer, A. A. Noegel, P. Schaap and G. Glöckner, Genome Res., 2011, 21, 1882–1891 CrossRef CAS PubMed.
- S. Basu, P. Fey, Y. Pandit, R. Dodson, W. A. Kibbe and R. L. Chisholm, Nucleic Acids Res., 2012, 41, D676–D683 CrossRef PubMed.
- C. Schilde, H. M. Lawal, K. Kin, I. Shibano-Hayakawa, K. Inouye and P. Schaap, Mol. Phylogenet. Evol., 2019, 134, 66–73 CrossRef PubMed.
- D. C. Gross, J. Appl. Bacteriol., 1985, 58, 167–174 CrossRef CAS PubMed.
- J. H. Zhang, N. B. Quigley and D. C. Gross, Appl. Environ. Microbiol., 1997, 63, 2771–2778 CrossRef CAS PubMed.
- S. Götze and P. Stallforth, Org. Biomol. Chem., 2020, 18, 1710–1727 RSC.
- H. K. Webb and R. G. Matthews, J. Biol. Chem., 1995, 270, 17204–17209 CrossRef CAS PubMed.
- S. Dekimpe and J. Masschelein, Nat. Prod. Rep., 2021, 38, 1910–1937 RSC.
- S. Götze and P. Stallforth, Nat. Prod. Rep., 2020, 37, 29–54 RSC.
- C. Camacho, G. Coulouris, V. Avagyan, N. Ma, J. Papadopoulos, K. Bealer and T. L. Madden, BMC Bioinf., 2009, 10, 421 CrossRef PubMed.
- T. Paysan-Lafosse, M. Blum, S. Chuguransky, T. Grego, B. L. Pinto, G. A. Salazar, M. L. Bileschi, P. Bork, A. Bridge, L. Colwell, J. Gough, D. H. Haft, I. Letunić, A. Marchler-Bauer, H. Mi, D. A. Natale, C. A. Orengo, A. P. Pandurangan, C. Rivoire, C. J. A. Sigrist, I. Sillitoe, N. Thanki, P. D. Thomas, S. C. E. Tosatto, C. H. Wu and A. Bateman, Nucleic Acids Res., 2023, 51, D418–D427 CrossRef CAS PubMed.
- R. C. Hennessy, C. B. W. Phippen, K. F. Nielsen, S. Olsson and P. Stougaard, MicrobiologyOpen, 2017, 6, e00516 CrossRef PubMed.
- L. Christiansen, K. S. Alanin, C. B. W. Phippen, S. Olsson, P. Stougaard and R. C. Hennessy, Appl. Environ. Microbiol., 2020, 86, e01284 CrossRef CAS PubMed.
- G. Licciardello, A. Caruso, P. Bella, R. Gheleri, C. P. Strano, A. Anzalone, E. A. Trantas, P. F. Sarris, N. F. Almeida and V. Catara, Front. Microbiol., 2018, 9, 521 CrossRef PubMed.
- G. Licciardello, I. Bertani, L. Steindler, P. Bella, V. Venturi and V. Catara, FEMS Microbiol. Rev., 2007, 61, 222–234 CAS.
- G. Licciardello, C. P. Strano, I. Bertani, P. Bella, A. Fiore, V. Fogliano, V. Venturi and V. Catara, J. Biotechnol., 2012, 159, 274–282 CrossRef CAS PubMed.
- C. Bez, S. Covaceuszach, I. Bertani, K. S. Choudhary and V. Venturi, mSphere, 2021, 6, e01322 CAS.
- C. Bez, A. M. Geller, A. Levy and V. Venturi, mSystems, 2023, e01039 Search PubMed.
- L. A. Temesvari, G. Klein and D. A. Cotter, Mycologia, 1996, 88, 819–830 CrossRef.
- R. Deslauriers, H. C. Jarrell, R. A. Byrd and I. C. P. Smith, FEBS Lett., 1980, 118, 185–190 CrossRef CAS PubMed.
- M. S. Müller, S. Scheu and A. Jousset, PLoS One, 2013, 8, e66200 CrossRef PubMed.
- N. Neidig, R. J. Paul, S. Scheu and A. Jousset, Microb. Ecol., 2011, 61, 853–859 CrossRef CAS PubMed.
- S. Tienda, C. Vida, E. Lagendijk, S. de Weert, I. Linares, J. González-Fernández, E. Guirado, A. de Vicente and F. M. Cazorla, Front. Microbiol., 2020, 11, 1874 CrossRef PubMed.
- A. Bejarano and G. Puopolo, in How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases, ed. A. De Cal, P. Melgarejo and N. Magan, Springer International Publishing, Cham, 2020, vol. 21, pp. 275–293 Search PubMed.
- M. R. Mungroo, R. Siddiqui and N. A. Khan, Folia Microbiol., 2021, 66, 689–699 CrossRef CAS PubMed.
- N. Neidig, A. Jousset, F. Nunes, M. Bonkowski, R. J. Paul and S. Scheu, Funct. Ecol., 2010, 24, 1133–1138 CrossRef.
- J. Zheng, F. Dini-Andreote, L. Luan, S. Geisen, J. Xue, H. Li, B. Sun and Y. Jiang, mBio, 2022, 13, e03293 Search PubMed.
- L. K. Carta, J. Nematol., 2000, 32, 362–369 CAS.
- M. Nandi, C. Selin, A. K. C. Brassinga, M. F. Belmonte, W. G. D. Fernando, P. C. Loewen and T. R. de Kievit, PLoS One, 2015, 10, e0123184 CrossRef PubMed.
- P. Burlinson, D. Studholme, J. Cambray-Young, D. Heavens, J. Rathjen, J. Hodgkin and G. M. Preston, ISME J., 2013, 7, 1126–1138 CrossRef CAS PubMed.
- W. Zhou, J. Vergis and T. Mahmud, J. Nat. Prod., 2022, 85, 590–598 CrossRef CAS PubMed.
- E. Pradel, Y. Zhang, N. Pujol, T. Matsuyama, C. I. Bargmann and J. J. Ewbank, Proc. Natl. Acad. Sci. U.S.A., 2007, 104, 2295–2300 CrossRef CAS PubMed.
- F. Ballestriero, J. Nappi, G. Zampi, P. Bazzicalupo, E. Di Schiavi and S. Egan, Sci. Rep., 2016, 6, 29284 CrossRef PubMed.
- J. E. E. U. Hellberg, M. A. Matilla and G. P. C. Salmond, Front. Microbiol., 2015, 6, 137 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03335j |
| This journal is © The Royal Society of Chemistry 2023 |