Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Late-stage peptide labeling with near-infrared fluorogenic nitrobenzodiazoles by manganese-catalyzed C–H activation†
Tsuyoshi
Oyama‡
a,
Lorena
Mendive-Tapia‡
 b,
Verity
Cowell
b,
Adelina
Kopp
a,
Marc
Vendrell
b,
Verity
Cowell
b,
Adelina
Kopp
a,
Marc
Vendrell
 *b and
Lutz
Ackermann
*b and
Lutz
Ackermann
 *ac
*ac
aInstitut für Organische und Biomolekulare Chemie, Georg-August-Universität Göttingen, Tammanstraße 2, 37077, Göttingen, Germany. E-mail: lutz.ackermann@chemie.uni-goettingen.de
bCentre for Inflammation Research, The University of Edinburgh, EH16 4TJ Edinburgh, UK. E-mail: marc.vendrell@ed.ac.uk
cGerman Center for Cardiovascular Research (DZHK), Potsdamer Straße 58, 10785, Berlin, Germany
First published on 4th May 2023
Abstract
Late-stage diversification of structurally complex amino acids and peptides provides tremendous potential for drug discovery and molecular imaging. Specifically, labeling peptides with fluorescent tags is one of the most important methods for visualizing their mode of operation. Despite major recent advances in the field, direct molecular peptide labeling by C–H activation is largely limited to dyes with relatively short emission wavelengths, leading to high background signals and poor signal-to-noise ratios. In sharp contrast, here we report on the fluorescent labeling of peptides catalyzed by non-toxic manganese(I) via C(sp2)–H alkenylation in chemo- and site-selective manners, providing modular access to novel near-infrared (NIR) nitrobenzodiazole-based peptide fluorogenic probes.
Introduction
Among the vast array of pharmaceutical compounds, peptides have emerged as powerful candidates for drug development.1 Since the first medical use of insulin in 1921,1 the field of peptide-based drugs has progressively grown, currently accounting for 5% of the global pharmaceutical market and with approvals steadily increasing. Many therapeutic peptides operate by binding to cell membrane components with high affinity, triggering specific intracellular effects.2 In particular, antimicrobial peptides are emerging as a promising alternative to combat antimicrobial drug-resistance.3 In this regard, fluorogenic labeling of peptides which enables real-time imaging studies is of key importance to analyze their functional mechanism and to develop new therapeutics.4 While a variety of chemical motifs have been employed in the development of fluorescent probes, those emitting in the near-infrared (NIR) window (650–900 nm) are attracting particular attention, since they have the advantages of low autofluorescence of biomolecules as well as low toxicities due to light irradiation at relatively long excitation wavelengths.5 Nitrobenzodiazole (NBD) fluorescent dyes have been identified as powerful labeling tags because of their small size, neutral character, cell permeability and large Stokes shifts.6 In addition, Vendrell and Lee recently reported NBD derivatives with NIR fluorescent properties.7 Examples of NBD labeling peptide and protein were reported, including the modification of nucleophilic cysteine or 3-amino-alanine residues (Scheme 1a and b).8 Despite remarkable progress, the introduction of NBD is hence largely limited to the nucleophilic region, such as N-terminal groups, thiols or phenols using classical condensations or SNAr type reactions.8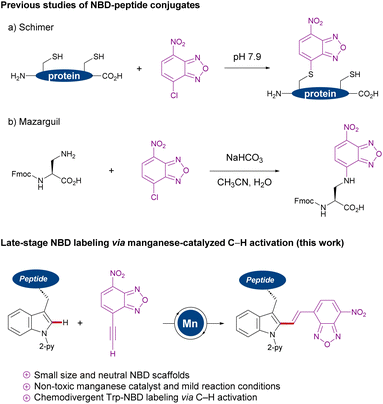 | ||
| Scheme 1 Nucleophilic NBD labeling (a and b) versus manganese(I)-catalyzed NBD labeling of tryptophans. | ||
During the past decade, transition-metal-catalyzed C–H activation9 has surfaced as a transformative platform for the direct modification of peptides,10 with major contributions from Lavilla and Albericio,11 Chen,12 Wang,13 Shi,14 Yu,15 and Ackermann,16 among others.17 While significant progress has thereby been realized, molecular peptide labeling with NIR dyes has thus proven elusive. In sharp contrast, we now have identified a strategy to stitch tryptophan with NBD fluorophores via earth-abundant and non-toxic manganese(I)-catalyzed C–H activation,18 and applied this approach for real-time imaging by using a model peptide targeting bacterial cell membranes. This strategy has the power to facilitate structure–activity relationship studies of fluorescent probes with NBD, since there is no report of direct conjugation to tryptophan residues.
Result and discussion
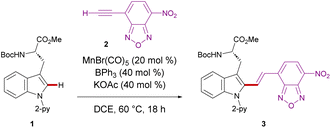
We initiated our studies by proving difficult reaction conditions with tryptophan derivative 1 and 4-ethynyl-7-nitrobenzoxadiazole (2) (Table 1). After optimization of reaction parameters (see details in the ESI Table S1†), we obtained the desired C2 functionalized product 3 in 95% yield in the presence of MnBr(CO)5, KOAc, and BPh3 (entry 1). Subsequently, we investigated the efficiency of other catalysts, such as ReBr(CO)5, which delivered product 3 only in moderate yield (entry 2). Likewise, significantly decreased catalytic activity was observed with Mn2(CO)10 (entry 3). It is noteworthy that Pd(OAc)2, [RuCl2(p-cymene)]2, [RhCp*Cl2]2, and Co(OAc)2 showed no catalytic efficiencies (entries 4–7), indicating the challenges associated with a chemo-selective NBD labeling. Interestingly, the manganese(I)-catalyzed C–H activation proceeded even at physiological temperature of 37 °C, delivering the desired product 3 in 78% yield (entry 8). Control experiments showed that manganese(I) catalyst, KOAc, and BPh3 were essential for the reaction (entries 9–11).| Entry | Deviation from the standard conditions | Yieldc (%) |
|---|---|---|
| a DCE: 1,2-dichloroethane. b Standard conditions: 1 (0.10 mmol), 2 (0.15 mmol), MnBr(CO)5 (20 mol%), BPh3 (40 mol%), KOAc (40 mol%), DCE (1.0 mL), 60 °C, 18 h. c Isolated yields. | ||
| 1 | Noneb | 95 |
| 2 | ReBr(CO)5 in place of MnBr(CO)5 | 60 |
| 3 | Mn2(CO)10 in place of MnBr(CO)5 | Trace |
| 4 | Pd(OAc)2 in place of MnBr(CO)5 | — |
| 5 | [RuCl2(p-cymene)]2 in place of MnBr(CO)5 | — |
| 6 | [RhCp*Cl2]2 in place of MnBr(CO)5 | — |
| 7 | Co(OAc)2 in place of MnBr(CO)5 | — |
| 8 | At 37 °C | 78 |
| 9 | Without MnBr(CO)5 | — |
| 10 | Without BPh3 | 4 |
| 11 | Without KOAc | Trace |
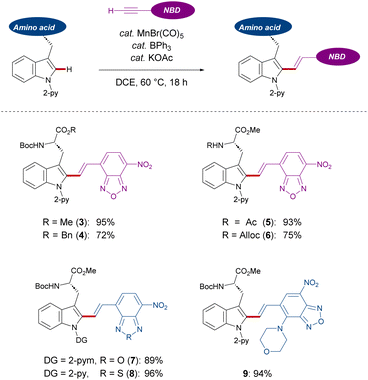
With the optimized reaction conditions for NBD-based fluorescence labeling in hand, we explored the generality of the Mn(I) catalysis (Scheme 2). The Mn(I) catalysis tolerated various tryptophans with Ac and Alloc groups at the N-terminus, generating the desired products 5 and 6 in good yields with complete diastereo-E-selectivities. It must be noted that benzyl esters at the C-terminus of tryptophan was compatible with our catalytic reaction conditions, offering the potential for further derivatization into peptides (4). In addition, NBD-alkynes bearing a different chalcogen or electron-donating group could also be converted using our strategy (8 and 9).
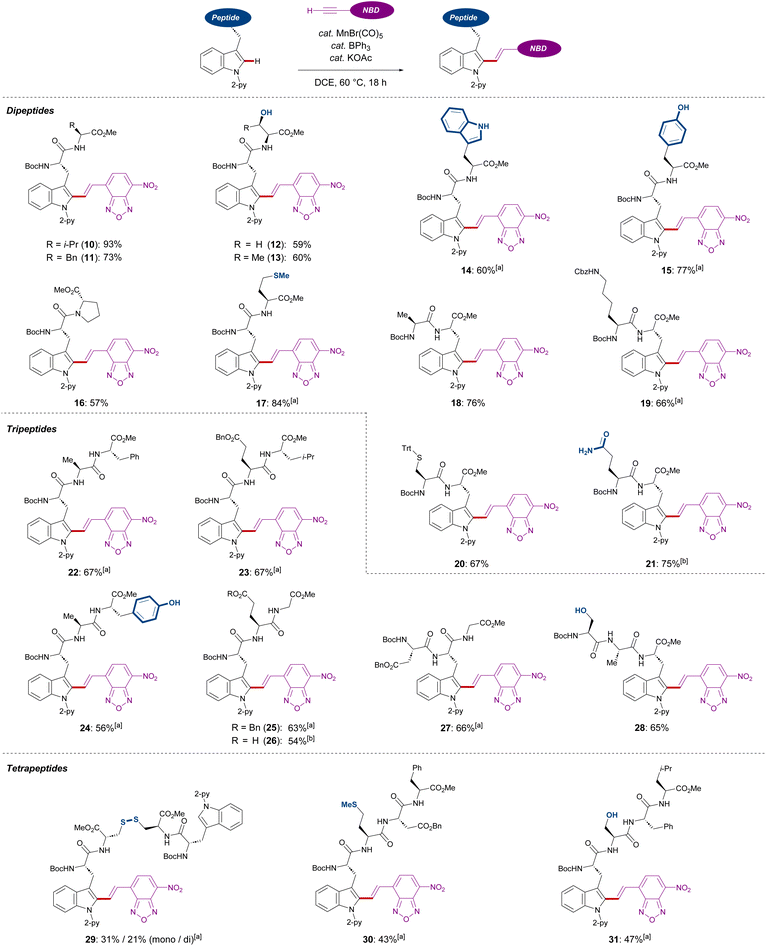
Subsequently, we investigated the late-stage diversification of dipeptides. An outstanding chemo-selectivity was demonstrated by the complete tolerance of sensitive functional groups on the amino acid side chains. For instance, successful transformation of dipeptides with alkyl side chains were observed without epimerization (10, 11, 16, and 18). Furthermore, the presence of unprotected primary and secondary alcohols or phenols did not interfere with the reaction, furnishing the desired products 12, 13, and 15 in good yields. Oxidation-sensitive groups found in methionine and protected cysteine were also tolerated (17 and 20) (Scheme 3).
Interestingly, dipeptides containing an unmodified indole moiety delivered the desired alkenylated product with excellent site selectivity (14). Lastly, good tolerance of substituted amines and unprotected amides showed the robustness of these reaction conditions (19 and 21). Triggered by the versatility of our C–H alkenylation regime, we expanded to more challenging tri- and tetra-peptides which were converted in a site-, regio-, chemo-, and diastereo-E-selective manner (22–31). The robustness of our Mn(I) catalysis was also highlighted with the successful transformation of aspartic or glutamic acid-containing substrates (23, 25–27, and 30), which are known to cyclize into 5- or 6-membered amides.19 In addition, the side formation of dehydro-alanine derivatives was not observed, which can be formed in serine-containing substrates via β-elimination (28 and 31). It must be noted that a tetrapeptide containing a disulfide bond was also amenable to our Mn(I)-catalyzed labeling process (29).
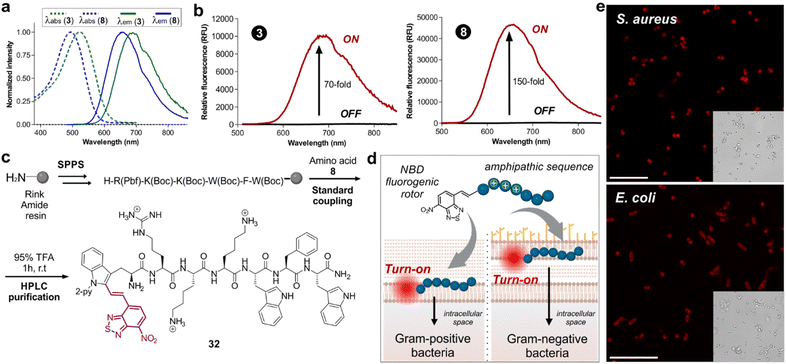
Next, we examined the fluorescence properties of the oxygen and sulfur NBD-labeled tryptophan derivatives 3 and 8, revealing absorption maxima wavelengths at 522 nm and 496 nm, and emission maxima wavelengths in the NIR-I window (688 nm and 658 nm respectively, Fig. 1a). Notably, both labeled amino acids exhibited very large Stokes shifts over 150 nm. Another important consideration is that NBD is relatively small compared to previously reported NIR-emitting scaffolds, helping to potentially retain the biological activity of the native peptides. Considering the potential molecular rotor features of our NBD-labeled tryptophan analogues, we also compared the turn-on effect of the amino acids 3 and 8 (Fig. 1b). As anticipated, both amino acids exhibited remarkable turn-on fluorescence in dioxane compared to H2O, with the S-containing compound 8 showing over 150-fold fluorescence increase in hydrophobic environments. Encouraged by the excellent fluorogenicity of the amino acid 8, we decided to use it for solid-phase peptide synthesis (SPPS) of the bacterial-targeting sequence 32 (Fig. 1c). This sequence is based on previous reports on cationic amphipathic peptides possessing both hydrophobic and hydrophilic regions, known to enable binding to the microbial cell envelope.20 Compound 8 proved compatible with standard SPPS coupling agents (e.g., Pyoxim) and cleavage conditions (e.g., trifluoroacetic acid, TFA), and we could successfully synthesize the labeled peptide 32 (Fig. 1c) in 4% yield and with high purities over 95%. Next, we exploited the turn-on features of peptide 32 for wash-free imaging of bacterial cells (Fig. 1d). First, we tested different concentrations of peptide 32 for imaging Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria under fluorescence microscopy (Fig. S1†). Importantly, the environmental sensitivity of the NBD moiety allowed direct visualization of both bacterial cells without any additional steps or washing procedures (Fig. S2†). After optimization of the labeling protocol, we employed peptide 32 in confocal fluorescence microscopy for real-time and wash-free imaging of S. aureus and E. coli bacterial cultures with high signal-to-noise ratios (Fig. 1e). Altogether, these results confirm that the NBD moiety did not alter the bacterial-targeting ability of the peptide sequence and showcase the utility of this fluorescence labeling method for NIR wash-free imaging in live cells.
Conclusions
In summary, we have developed an unprecedent late-stage labeling strategy of structurally complex peptides with NIR-emitting dyes via C(sp2)–H activation. Thereby, the assembly of fluorescent amino acids and peptides was achieved with excellent site-, chemo-, and diastereo-E-selectivity using an earth-abundant manganese(I) pre-catalyst of low toxicity. Furthermore, we identified that the NIR-labeled peptide 32 can be used for imaging of live bacteria and under wash-free conditions. Overall, this strategy features excellent properties of NBD labeled amino acids and peptides, such as large Stokes shifts, NIR emission wavelengths, and excellent fluorogenicity for real time imaging in live cells.Data availability
Data available from the authors upon reasonable request.Author contributions
T. O., and L. A. conceived the project. T. O. and A. K. performed the synthetic experiments. L. M.-T. and V. C. performed imaging experiments. T. O., L. M.-T., M. V., and L. A. wrote the manuscript with contributions from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge support from the DFG (Gottfried-Wilhelm-Leibniz award) and DZHK (PI at German Centre for Cardiovascular Research) to L. A., the European Union's Horizon 2020 research and innovation programme (Marie Skłodowska-Curie Grant Agreement No. 860762) to T. O. L. M.-T. acknowledges funding the Wellcome Trust Institutional Strategic Support Fund (ISSF) at the University of Edinburgh. M. V. acknowledges funding from an ERC Consolidator Grant (771443, DYNAFLUORS).References
- (a) L. Wang, N. Wang, W. Zhang, X. Cheng, Z. Yan, G. Shao, X. Whang and abd C. Fu, Signal Transduct Target Ther., 2022, 7, 48 CrossRef CAS PubMed; (b) M. Muttenthaler, G. F. King, D. J. Adams and P. F. Alewood, Nat. Rev. Drug Discovery, 2021, 20, 309 CrossRef CAS PubMed; (c) J. L. Lau and M. K. Dunn, Bioorg. Med. Chem., 2018, 26, 2700 CrossRef CAS PubMed; (d) W. Chiangjong, S. Chutipongtanate and S. Hongeng, Int. J. Oncol., 2020, 57, 678 CrossRef CAS PubMed.
- P. Hoppenz, S. Els-Heindl and A. G. Beck-Sickinger, Front. Chem., 2020, 8, 571 CrossRef CAS PubMed.
- (a) B. H. Gan, J. Gaynord, S. M. Rowe, T. Deingruber and D. R. Spring, Chem. Soc. Rev., 2021, 50, 7820 RSC; (b) M. Rima, M. Rima, Z. Fajloun, J.-M. Sabatier, B. Bechinger and T. Naas, Antibiotics, 2021, 10, 1095 CrossRef CAS PubMed.
- (a) Z. Cheng, E. Kuru, A. Sachdeva and M. Vendrell, Nat. Rev. Chem., 2020, 4, 275 CrossRef CAS PubMed; (b) M. Sameiro and T. Gonçalves, Chem. Rev., 2009, 109, 190 CrossRef PubMed.
- (a) Z. Guo, S. Park, J. Yoon and I. Shin, Chem. Soc. Rev., 2014, 43, 16 RSC; (b) A. L. Vahrmeijer, M. Hutteman, J. R. van der Vorst, C. J. H. van de Velde and J. V. Frangioni, Nat. Rev. Clin. Oncol., 2013, 10, 507 CrossRef CAS PubMed.
- (a) S. Benson, A. Fernandez, N. D. Barth, F. de Moliner, M. H. Horrocks, C. S. Herrington, J. L. Abad, A. Delgado, L. Kelly, Z. Chang, Y. Feng, M. Nishiura, Y. Hori, K. Kikuchi and M. Vendrell, Angew. Chem., Int. Ed., 2019, 58, 6911 CrossRef CAS PubMed; (b) D. R. Jenkinson, A. J. Cadby and S. Jones, Chem.–Eur. J., 2017, 23, 12585 CrossRef CAS PubMed; (c) Y. Shen, Y. Zhang, X. Zhang, C. Zhang, L. Zhang, J. Jin, H. Li and S. Yao, Anal. Methods, 2014, 6, 4797 RSC; (d) L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142 CrossRef CAS PubMed.
- F. de Moliner, I. Biazruchka, K. Konsewicz, S. Benson, S. Singh, J.-S. Lee and M. Vendrell, Front. Chem. Sci. Eng., 2022, 16, 128 CrossRef CAS.
- (a) S. Benson, F. de Moliner, A. Fernandez, E. Kuru, N. L. Asiimwe, J.-S. Lee, L. Hamilton, D. Sieger, I. R. Bravo, A. M. Elliot, Y. Feng and M. Vendrell, Nat. Commun., 2021, 12, 2369 CrossRef CAS PubMed; (b) V. Verdoliva, G. Digilio, M. Saviano and S. D. Luca, Catal. Sci. Technol., 2021, 11, 1067 RSC; (c) A. R. Akram, N. Avlonitis, E. Scholefield, M. Vendrell, N. McDonald, T. Aslam, T. H. Craven, C. Gray, D. S. Collie, A. J. Fisher, P. A. Corris, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Rep., 2019, 9, 8422 CrossRef PubMed; (d) I. Dufau and H. Mazarguil, Tetrahedron Lett., 2000, 41, 6063 CrossRef CAS; (e) N. C. Prices, M. Cohn and R. H. Schirmer, J. Biol. Chem., 1975, 250, 644 CrossRef; (f) P. B. Ghosh and M. W. Whitehouse, Biochem. J., 1968, 108, 155 CrossRef CAS PubMed.
- For selected reviews on C–H activation, see: (a) P. Gandeepan and L. Ackermann, Chem, 2018, 4, 199 CrossRef CAS; (b) Y. Wei, P. Hu, M. Zhang and W. Su, Chem. Rev., 2017, 117, 8864 CrossRef CAS PubMed; (c) M. M. Lorion, K. Maindan, A. R. Kapdi and L. Ackermann, Chem. Soc. Rev., 2017, 46, 7399 RSC; (d) J. A. Leitch and C. G. Frost, Chem. Soc. Rev., 2017, 46, 7145 RSC; (e) J. He, M. Wasa, K. S. L. Chan, Q. Shao and J.-Q. Yu, Chem. Rev., 2017, 117, 8754 CrossRef CAS PubMed; (f) Q.-Z. Zheng and N. Jiao, Chem. Soc. Rev., 2016, 45, 4590 RSC; (g) M. Moselage, J. Li and L. Ackermann, ACS Catal., 2016, 6, 498 CrossRef CAS; (h) B. Ye and N. Cramer, Acc. Chem. Res., 2015, 48, 1308 CrossRef CAS PubMed; (i) J. Wencel-Delord and F. Glorius, Nat. Chem., 2013, 5, 369 CrossRef CAS PubMed; (j) G. Rouquet and N. Chatani, Angew. Chem., Int. Ed., 2013, 52, 11726 CrossRef CAS PubMed; (k) S. R. Neufeldt and M. S. Sanford, Acc. Chem. Res., 2012, 45, 936 CrossRef CAS PubMed; (l) L. McMurray, F. O'Hara and M. J. Gaunt, Chem. Soc. Rev., 2011, 40, 1885 RSC; (m) L. Ackermann, R. Vicente and A. R. Kapdi, Angew. Chem., Int. Ed., 2009, 48, 9792 CrossRef CAS PubMed.
- (a) M. Jbara, S. K. Maity and A. Brik, Angew. Chem., Int. Ed., 2017, 56, 10644 CrossRef CAS PubMed; (b) A. F. M. Noisier and M. A. Brimble, Chem. Rev., 2014, 114, 8775 CrossRef CAS PubMed.
- Selected examples, see: (a) R. Subiros-Funosas, L. Mendive-Tapia, J. Sot, J. D. Pound, N. Barth, Y. Varela, F. M. Goni, M. Paterson, C. D. Gregory, F. Albericio, I. Dransfield, R. Lavilla and M. Vendrell, Chem. Commun., 2017, 53, 945 RSC; (b) S. Preciado, L. Mendive-Tapia, F. Albericio and R. Lavilla, J. Org. Chem., 2013, 78, 8129 CrossRef CAS PubMed; (c) J. Ruiz-Rodrí-guez, F. Albericio and R. Lavilla, Chem.–Eur. J., 2010, 16, 1124 CrossRef PubMed.
- Selected examples, see: (a) X. Chen, F. Ye, X. Luo, X. Liu, J. Zhao, S. Wang, Q. Zhou, G. Chen and P. Wang, J. Am. Chem. Soc., 2019, 141, 18230 CrossRef CAS PubMed; (b) X. Zhang, G. Lu, M. Sun, M. Mahankali, Y. Ma, M. Zhang, W. Hua, Y. Hu, Q. Wang, J. Chen, G. He, X. Qi, W. Shen, P. Liu and G. Chen, Nat. Chem., 2018, 10, 540 CrossRef CAS PubMed; (c) G. He, B. Wang, W. A. Nack and G. Chen, Acc. Chem. Res., 2016, 49, 635 CrossRef CAS PubMed; (d) B. Wang, W. A. Nack, G. He, S.-Y. Zhang and G. Chen, Chem. Sci., 2014, 5, 3952 RSC; (e) S.-Y. Zhang, Q. Li, G. He, W. A. Nack and G. Chen, J. Am. Chem. Soc., 2013, 135, 12135 CrossRef CAS PubMed; (f) Y. Feng and G. Chen, Angew. Chem., Int. Ed., 2010, 49, 958 CrossRef CAS PubMed.
- (a) L. Liu, X. Fan, B. Wang, H. Deng, T. Wang, J. Zheng, J. Chen, Z. Shi and H. Wang, Angew. Chem., Int. Ed., 2022, 61, e2022061 Search PubMed; (b) C. Cai, F. Wang, X. Xiao, W. Sheng, S. Liu, J. Chen, J. Zheng, R. Xie, Z. Bai and H. Wang, Chem. Commun., 2022, 58, 4861 RSC; (c) S. Liu, C. Cai, Z. Bai, W. Sheng, J. Tan and H. Wang, Org. Lett., 2021, 23, 2933 CrossRef CAS PubMed; (d) T. Long, L. Liu, Y. Tao, W. Zhang, J. Quan, J. Zheng, J. D. Hegemann, M. Uesugi, W. Yao, H. Tian and H. Wang, Angew. Chem., Int. Ed., 2021, 60, 13414 CrossRef CAS PubMed; (e) Z. Bai, Q. Chen, J. Gu, C. Cai, J. Zheng, W. Sheng, S. Yi, F. Liu and H. Wang, ACS Catal., 2021, 11, 15125 CrossRef CAS; (f) Z. Bai, C. Cai, W. Sheng, Y. Ren and H. Wang, Angew. Chem., Int. Ed., 2020, 59, 14686 CrossRef CAS PubMed.
- (a) X.-S. Yin, W.-Y. Qi and B.-F. Shi, Chem. Sci., 2021, 12, 13137 RSC; (b) L. Liu, Y.-H. Liu and B.-F. Shi, Chem. Sci., 2020, 11, 290 RSC; (c) B.-B. Zhan, J. Fan, L. Jin and B.-F. Shi, ACS Catal., 2019, 9, 3298 CrossRef CAS; (d) B. B. Zhan, Y. Li, J. W. Xu, X. L. Nie, J. Fan, L. Jin and B. F. Shi, Angew. Chem., Int. Ed., 2018, 57, 5858 CrossRef CAS PubMed; (e) G. Liao, X.-S. Yin, K. Chen, Q. Zhang, S.-Q. Zhang and B.-F. Shi, Nat. Commun., 2016, 7, 12901 CrossRef PubMed; (f) K. Chen, X. Li, S.-Q. Zhang and B.-F. Shi, Org. Chem. Front., 2016, 3, 204 RSC; (g) K. Chen, S.-Q. Zhang, J.-W. Xu, F. Hu and B.-F. Shi, Chem. Commun., 2014, 50, 13924 RSC; (h) K. Chen and B. F. Shi, Angew. Chem., Int. Ed., 2014, 53, 11950 CrossRef CAS PubMed; (i) K. Chen, F. Hu, S.-Q. Zhang and B.-F. Shi, Chem. Sci., 2013, 4, 3906 RSC.
- (a) T. Liu, J. X. Qiao, M. A. Poss and J.-Q. Yu, Angew. Chem., Int. Ed., 2017, 56, 10924 CrossRef CAS PubMed; (b) G. Chen, T. Shigenari, P. Jain, Z. Zhang, Z. Jin, J. He, S. Li, C. Mapelli, M. M. Miller, M. A. Poss, P. M. Scola, K.-S. Yeung and J.-Q. Yu, J. Am. Chem. Soc., 2015, 137, 3338 CrossRef CAS PubMed; (c) J. He, S. Li, Y. Deng, H. Fu, B. N. Laforteza, J. E. Spangler, A. Homs and J.-Q. Yu, Science, 2014, 343, 1216 CrossRef CAS PubMed; (d) W. Gong, G. Zhang, T. Liu, R. Giri and J.-Q. Yu, J. Am. Chem. Soc., 2014, 136, 16940 CrossRef CAS PubMed.
- (a) X. Hou, N. Kaplaneris, B. Yuan, J. Frey, T. Ohyama, A. M. Messinis and L. Ackermann, Chem. Sci., 2022, 13, 3461 RSC; (b) N. Kaplaneris, J. Son, L. Mendive-Tapia, A. Kopp, N. D. Barth, I. Maksso, M. Vendrell and L. Ackermann, Nat. Commun., 2021, 12, 3389 CrossRef CAS PubMed; (c) M. Stangier, A. M. Messinis, J. C. A. Oliveira, H. Yu and L. Ackermann, Nat. Commun., 2021, 12, 4736 CrossRef CAS PubMed; (d) N. Kaplaneris, F. Kaltenhӓuser, G. Sirvinskaite, S. Fan, T. D. Oliveira, L.-C. Conradi and L. Ackermann, Sci. Adv., 2021, 7, eabe6202 CrossRef CAS PubMed; (e) W. Wang, J. Wu, R. Kuniyil, A. Kopp, R. N. Lima and L. Ackermann, Chem, 2020, 6, 3428 CrossRef CAS; (f) J. Wu, N. Kaplaneris, S. Ni, F. Kaltenhäusera and L. Ackermann, Chem. Sci., 2020, 11, 6521 RSC; (g) A. Schischko, N. Kaplaneris, T. Rogge, G. Sirvinskaite, J. Son and L. Ackermann, Nat. Commun., 2019, 10, 3553 CrossRef PubMed; (h) W. Wang, P. Subramanian, O. Martinazzoli, J. Wu and L. Ackermann, Chem.–Eur. J., 2019, 25, 10585 CrossRef CAS PubMed; (i) N. Kaplaneris, T. Rogge, R. Yin, H. Wang, G. Sirvinskaite and L. Ackermann, Angew. Chem., Int. Ed., 2019, 58, 3476 CrossRef CAS PubMed; (j) M. M. Lorion, N. Kaplaneris, J. Son, R. Kuniyil and L. Ackermann, Angew. Chem., Int. Ed., 2019, 58, 1684 CrossRef CAS PubMed; (k) W. Wang, M. M. Lorion, O. Martinazzoli and L. Ackermann, Angew. Chem., Int. Ed., 2018, 57, 10554 CrossRef CAS PubMed; (l) M. Bauer, W. Wang, M. M. Lorion, C. Dong and L. Ackermann, Angew. Chem., Int. Ed., 2018, 57, 203 CrossRef CAS PubMed; (m) H. Wang, M. M. Lorion and L. Ackermann, Angew. Chem., Int. Ed., 2017, 56, 6339 CrossRef CAS PubMed; (n) A. Schischko, H. Ren, N. Kaplaneris and L. Ackermann, Angew. Chem., Int. Ed., 2017, 56, 1576 CrossRef CAS PubMed; (o) Z. Ruan, N. Sauermann, E. Manoni and L. Ackermann, Angew. Chem., Int. Ed., 2017, 56, 3172 CrossRef CAS PubMed; (p) Y. Zhu, M. Bauer and L. Ackermann, Chem.–Eur. J., 2015, 21, 9980 CrossRef CAS PubMed; (q) Y. Zhu, M. Bauer, J. Ploog and L. Ackermann, Chem.–Eur. J., 2014, 20, 13099 CrossRef CAS PubMed.
- (a) X. Wang, S. Niu, L. Xu, C. Zhang, L. Meng, X. Zhang and D. Ma, Org. Lett., 2017, 19, 246 CrossRef CAS PubMed; (b) M. San Segundo, I. Guerrero and A. Correa, Org. Lett., 2017, 19, 5288 CrossRef CAS PubMed; (c) A. J. Reay, L. A. Hammarback, J. T. W. Bray, T. Sheridan, D. Turnbull, A. C. Whitwood and I. J. S. Fairlamb, ACS Catal., 2017, 7, 5174 CrossRef CAS PubMed; (d) G. L. Tolnai, J. P. Brand and J. Waser, Beilstein J. Org. Chem., 2016, 12, 745 CrossRef CAS PubMed; (e) T. J. Osberger, D. C. Rogness, J. T. Kohrt, A. F. Stepan and M. C. White, Nature, 2016, 537, 214 CrossRef CAS PubMed; (f) B. Mondal, B. Roy and U. Kazmaier, J. Org. Chem., 2016, 81, 11646 CrossRef CAS PubMed; (g) E. Hernando, J. Villalva, Á. M. Martínez, I. Alonso, N. Rodríguez, R. Gómez Arrayás and J. C. Carretero, ACS Catal., 2016, 6, 6868 CrossRef CAS; (h) X. Wu, D. Zhang, S. Zhou, F. Gao and H. Liu, Chem. Commun., 2015, 51, 12571 RSC; (i) O. Daugulis, J. Roane and L. D. Tran, Acc. Chem. Res., 2015, 48, 1053 CrossRef CAS PubMed; (j) L. D. Tran and O. Daugulis, Angew. Chem., Int. Ed., 2012, 51, 5188 CrossRef CAS PubMed; (k) H. Dong, C. Limberakis, S. Liras, D. Price and K. James, Chem. Commun., 2012, 48, 11644 RSC.
- (a) Y. Hu and C. Wang, ChemCatChem, 2019, 11, 1167 CrossRef CAS; (b) W. Liu and L. Ackermann, ACS Catal., 2016, 6, 3743 CrossRef CAS; (c) C. Wang, Synlett, 2013, 24, 1606 CrossRef CAS.
- (a) S. A. Bernhard, A. Berger, J. H. Carter, E. Katchalski, M. Sela and Y. Shalitin, J. Am. Chem. Soc., 1962, 84, 2421 CrossRef CAS; (b) D. W. Clayton, G. W. Kenner and R. C. Sheppard, J. Chem. Soc., 1956, 371 RSC.
- (a) L. Mendive-Tapia, J. Wang and M. Vendrell, Pept. Sci., 2021, 113, e24181 CAS; (b) C. Zhao, L. Mendive-Tapia and M. Vendrell, Arch. Biochem. Biophys., 2019, 661, 187 CrossRef CAS PubMed; (c) C. Zhao, A. Fernandez, N. Avlonitis, G. V. Velde, M. Bradley, N. D. Read and M. Vendrell, ACS Comb. Sci., 2016, 18, 689 CrossRef CAS PubMed; (d) L. Mendive-Tapia, C. Zhao, A. R. Akram, S. Preciado, F. Albericio, M. Lee, A. Serrels, N. Kielland, N. D. Read, R. Lavilla and M. Vendrell, Nat. Commun., 2016, 7, 10940 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01868g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |