Open Access Article
Open Access ArticleEngineered vitamin E-tethered non-immunogenic facial lipopeptide for developing improved siRNA based combination therapy against metastatic breast cancer†
Argha Mario
Mallick
 a,
Abhijit
Biswas‡
b,
Sukumar
Mishra‡
a,
Sonali
Jadhav
c,
Kasturee
Chakraborty
a,
Abhijit
Biswas‡
b,
Sukumar
Mishra‡
a,
Sonali
Jadhav
c,
Kasturee
Chakraborty
 a,
Archana
Tripathi
a,
Arnab
Mukherjee
a,
Archana
Tripathi
a,
Arnab
Mukherjee
 *c and
Rituparna Sinha
Roy
*c and
Rituparna Sinha
Roy
 *ade
*ade
aDepartment of Biological Sciences, Indian Institute of Science Education and Research Kolkata, Mohanpur, 741246, India. E-mail: rituparna@iiserkol.ac.in
bDepartment of Chemical Sciences, Indian Institute of Science Education and Research Kolkata, Mohanpur, 741246, India
cDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Pune, 411008, India. E-mail: arnab.mukherjee@iiserpune.ac.in
dCentre for Advanced Functional Materials, Indian Institute of Science Education and Research Kolkata, Mohanpur, 741246, India
eCentre for Climate and Environmental Studies, Indian Institute of Science Education and Research Kolkata, Mohanpur, 741246, India
First published on 3rd July 2023
Abstract
RNA interference based therapeutic gene silencing is an emerging platform for managing highly metastatic breast cancer. Cytosolic delivery of functional siRNA remains the key obstacle for efficient RNAi therapy. To overcome the challenges of siRNA delivery, we have engineered a vitamin E-tethered, short, optimum protease stabilized facial lipopeptide based non-immunogenic, biocompatible siRNA transporter to facilitate the clinical translation in future. Our designed lipopeptide has an Arginine-Sarcosine-Arginine segment for providing optimum protease-stability, minimizing adjacent arginine–arginine repulsion and reducing intermolecular aggregation and α-tocopherol as the lipidic moiety for facilitating cellular permeabilization. Interestingly, our designed non-immunogenic siRNA transporter has exhibited significantly better long term transfection efficiency than HiPerFect and can transfect hard to transfect primary cell line, HUVEC. Our engineered siRNA therapeutics demonstrated high efficacy in managing metastasis against triple negative breast cancer by disrupting the crosstalk of endothelial cells and MDA-MB-231 and reduced stemness and metastatic markers, as evidenced by downregulating critical oncogenic pathways. Our study aimed at silencing Notch1 signalling to achieve “multi-targeted” therapy with a single putative molecular medicine. We have further developed mechanistically rational combination therapy combining Notch1 silencing with a repurposed drug m-TOR inhibitor, metformin, which demonstrated synergistic interaction and enhanced antitumor efficacy against cancer metastasis.
1. Introduction
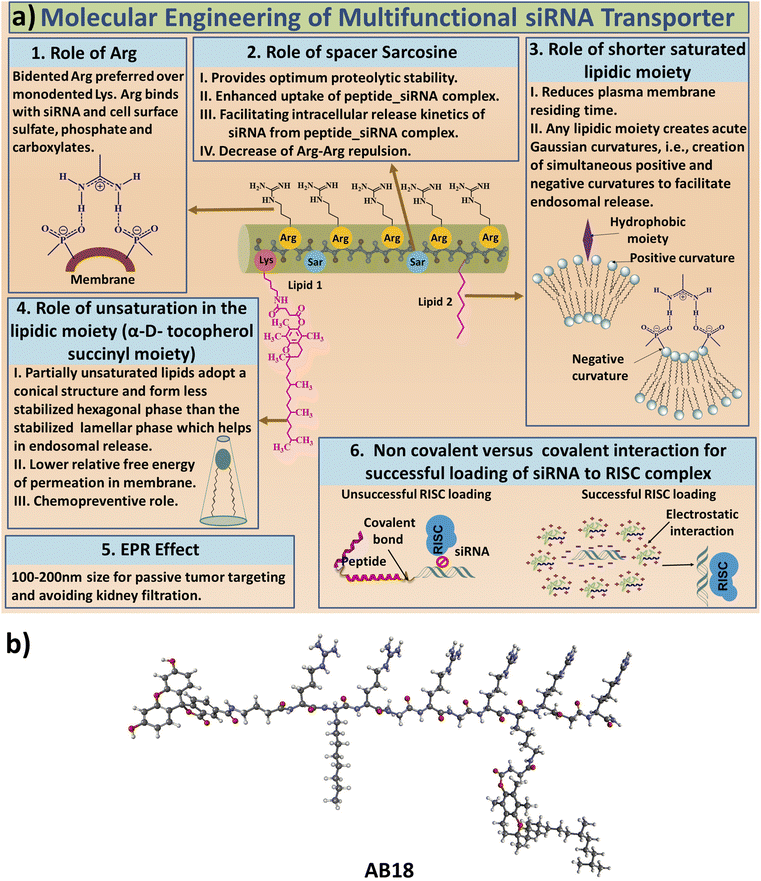
RNA interference (RNAi) has emerged as a potential therapy for treating many incurable diseases having undruggable targets. FDA has approved the first RNAi therapy called ONPATTRO™ (Patisiran) in August 2018 to treat transthyretin-mediated amyloidosis.1 Only about twenty siRNA based therapies are under clinical trial.2 siRNA is a double-stranded non-coding RNA with around 21–23 base pairs having two nucleotide overhangs on both 3′ sides.3,4 RNAi machinery faces the formidable challenge of efficient and safe intracellular delivery of functional siRNA due to its large molecular weight (≈14.5 kDa) and high anionic charge of the siRNA.5 To enable intracellular entry and cytosolic delivery, siRNA needs to (i) cross the endothelial membrane and avoid kidney filtration, (ii) penetrate the extracellular matrix, (iii) enter the targeted cell membrane, (iv) overcome the endocytic barrier inside the cell and (v) remain protected from different intracellular RNases while retaining its functional state to access the RNAi machinery.4,6 siRNA delivery vehicles include synthetic polymers, lipids, cationic peptides and viral delivery strategies.4,7 The delivery vehicle has to be biocompatible with optimum size and charge to ensure cellular penetration avoiding rapid kidney filtration. Another key hurdle for developing effective siRNA therapy is providing long term gene silencing. Towards this goal, cationic and amphipathic peptides have emerged as potent siRNA transporters.8 Bidentate cationic amino acid residue, arginine, is advantageous over monodentate lysine residue since arginine residue can interact more efficiently with cell surface carboxylate, sulfate and phosphate, thus facilitating increased cellular uptake.4,9–11 Wender et al. have demonstrated that a polyarginine peptide with 9 Arg residues shows optimum cellular internalization.12 Rothbard et al. claimed that the usage of an Arg-Xxx-Arg moiety in Arg enriched peptide sequences is preferred and cell permeability increases for linear spacer residue between two adjacent arginine residues from (Xxx = Gly; CH2 = 1) to spacer residue (Xxx = 6-amino caproic acid; CH2 = 5).13 Our group has reported the use of N-methyl glycine and D-histidine as spacer residues in linear facial siRNA transporters having an Arg-Xxx-Arg moiety.14,15 We have also demonstrated that the usage of an unsaturated lipid is preferred over the saturated one in facial lipopeptide based siRNA transporters.4,15 Parang and his group designed homochiral, cyclic arginine and tryptophan-enriched amphipathic peptide-based nuclear transporters.16 Cyclization enables the static presentation of guanidinium groups between Arg residues and enhances intracellular permeability17 and constraining the conformation of a peptide backbone also increases its bioactivity, bioavailability and cellular uptake.19,20 Dowdy and co-workers designed an ∼65 residue peptide-based siRNA transporter, which has exhibited high transfection efficiency against three different types of hard to transfect primary cell lines.21Cancer progression and acquiring resistance are greatly enhanced by the elevated levels of reactive oxygen species (ROS).22 Interestingly, naturally occurring vitamin E can scavenge ROS from cells.23 Peptides having larger hydrophobic moieties have higher binding affinity to the cell membranes and also aid in endocytosis by enabling budding formation in the cell membrane.24 Unsaturated lipid moieties may have favourable free energies of insertion at the bilayer interfaces.25 Interestingly, constrained cell penetrating peptides (CPPs) bind to the endosomal membrane with greater affinity and also exit the endosome more easily than linear CPPs due to their entropic advantage.26 Introducing backbone or side chain constrains in a peptide enables rapid endosomal escape by vesicle budding and collapse and also probably by membrane disruption.26,27 Vesicle budding requires the formation of acute Gaussian curvatures, which simultaneously have both positive and negative curvatures in orthogonal directions at the budding neck.28,29 Introducing a constraint into CPPs leads to the insertion of hydrophobic groups in between phospholipid molecules generating a positive curvature.28,29 The guanidinium groups of arginine residues in the CPPs form a bidentate hydrogen bond with the phosphate group of lipids inducing a simultaneous negative curvature, facilitating budding formation.28,29
Activated Notch signalling is a hallmark feature of triple negative breast cancer (TNBC).30 Notch1 oncogene is responsible for cancer initiation, progression, metastasis, maintaining stemness and chemoresistance leading to very poor overall survival in TNBC patients.30 Targeting the Notch signalling pathway is considered as an approach to achieve “multi-targeted” therapy with a single putative molecular medicine. Notch upregulation also helps in immune-suppression and chemoresistance of TNBC through the m-TOR pathway.31,32 Combination therapy has become the corner stone in cancer care.33 Inhibiting the m-TOR signalling pathway gives rise to a Notch dependent drug resistant cancer stem cell (CSC) population in TNBC.34 However, combination strategies targeting both Notch and m-TOR signalling can target CSCs in TNBC.35 Moreover, targeting different molecular pathways by multiple drugs leads to delayed oncogenic mutations and adaption of cancer cells.33 The anti-diabetic drug metformin can improve cancer prognosis by inhibiting the m-TOR signalling pathway.36
In this study, we have engineered clinically safe, short, peptide based siRNA transporter to provide long term gene silencing and tackling cancer metastasis by mechanistically inspired combination therapy. The interesting feature of our engineered siRNA transporter is its ability to transfect hard-to-transfect primary cells and successfully preventing metastasis as monotherapy and combination therapy in highly metastatic breast cancer cells. Additionally, we have determined the therapeutic outcome of combination drugs involving long term Notch1 silencing and m-TOR inhibitor metformin.
2. Results and discussion
2.1 Design of lipopeptides having constrained lipidic moieties as siRNA transporters
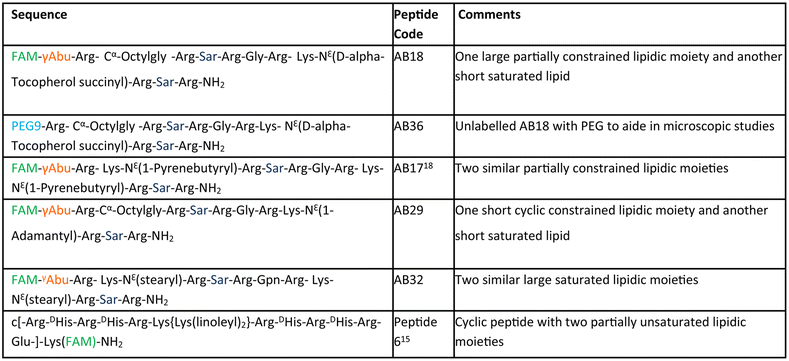
The aim of our study is to engineer short, efficient, optimum protease stabilized peptide based siRNA transporter to enable intracellular delivery of siRNA in a biologically active form. Towards this goal, we have engineered a library of short facial lipopeptides having an Arg-Xxx-Arg backbone and constrained and unconstrained lipidic moieties (Tables 1, S1,†Fig. 1a, b, S1 and S2†) by employing the Fmoc solid phase peptide synthesis strategy (Fig. S3†). Fig. 1a describes our detailed molecular engineering strategy of the siRNA transporter, peptide AB18. Such lipopeptides having an Arg-Sarcosine-Arg moiety provides optimum protease stability, reduced intermolecular aggregation and minimized adjacent arginine–arginine repulsion. Lipidic residues were incorporated to enhance hydrophobicity and promote vesicle budding through the formation of acute Gaussian curvatures.29 Octyl glycine, stearyl, 1-pyrenebutyryl, alpha-D-tocopherol succinyl and 1-adamantyl were used as lipidic moieties to examine the effect of constrained and unconstrained lipids on siRNA transporter designing. Alpha-D-tocopherol can be easily internalized in cells by uptake facilitated by lipase, lipid transfer proteins and also by receptor-mediated lipoprotein endocytosis, which further helps in its cellular internalization.37 FDA approved alpha-D-tocopherol succinate was selected as the lipidic moiety to make a clinically safe siRNA transporter. Alpha-D-tocopherol (constituent of vitamin E) has shown an antitumor effect on breast cancer by demonstrating cell cycle arrest.38,39 Higher levels of reactive oxygen species (ROS) have a significant role in breast cancer progression, metastasis and acquiring resistance in cancer.40 Alpha-D-tocopherol has the potential to effectively scavenge intracellular reactive oxygen.23 Vitamin E-tethered siRNA transporter lipopeptides, AB18 and AB36 may have additional chemopreventive potential activity against breast cancer. Unsaturated lipidic moieties are used in lipopeptide designing, as unsaturation facilitates fusogenicity and endosomal escape.7,41,42 Gamma-aminobutyric acid (γAbu) was used as a spacer between the fluorophore (FAM) and peptide backbone. Peptides were conjugated with FAM dye to aid in studying cellular internalization by flow cytometry and visualization under a microscope. Unlabelled lipopeptide AB36 was used to evaluate protein expression monitoring the TRITC signal by immunofluorescence studies. Peptide 6 (Table 1) was used as a positive control and for comparison.15 The mass of the lipopeptides was examined by MALDI-TOF mass spectra (Fig. S4–S8†).| a Please note that lipopeptides AB18 and AB36 have similar sequences. Lipopeptide AB18 is attached to fluorophore FAM, whereas lipopeptide AB36 is not attached to any fluorophore for the convenience in microscopic studies. Peptide 6 is reported in ref. 15 and is used as a positive control for comparison. FAM is 5(6)-carboxyfluorescein dye. The residue Gabapentin is abbreviated as ‘Gpn’. N-Methyl glycine or Sarcosine residue is abbreviated as ‘Sar’. PEG residue used in this study is designated as PEG9. 8-Amino-3,6-dioxaoctanoyl is abbreviated as PEG9. The structures of PEG9 and Gpn are illustrated in Table S1. |
|---|
|
 | ||
| Fig. 1 (a) Engineering strategy to design lipopeptide AB18. (b) Schematic diagram of lipopeptide AB18. | ||
2.2 Physicochemical characterization of lipopeptides and lipopeptide_siRNA nanocomplexes
For each lipopeptide a negative CD band was observed at around 200 nm, indicating that the synthesized lipopeptides remain unstructured in aqueous solution (Fig. S9†). The hydrodynamic radius of lipopeptides AB17, AB18, AB29 and AB32 was found to be 236 ± 11 nm, 111 ± 10 nm, 65 ± 0.8 nm and 92 ± 13 nm respectively, in cell culture media (DMEM supplemented with 10% FBS) from DLS studies (Fig. S10†). Cell culture media with FBS were used to mimic physiological conditions and the hydrodynamic radius was measured considering the protein corona effect. The size of the lipopeptide_siRNA complex was also determined by FE-SEM and cryo-TEM studies. The average diameter of the self-assembled complexes obtained from FE-SEM of lipopeptides AB17_siRNA, AB18_siRNA, AB29_siRNA and AB32_siRNA complexes was 133 ± 22 nm, 182 ± 20 nm, 144 ± 23 nm, and 100 ± 14 nm, respectively (Fig. S11†). The average diameter of the self-assembled lipopeptide_siRNA complexes AB17_siRNA, AB18_siRNA, AB29_siRNA and AB32_siRNA complexes obtained from cryo-TEM data was 132 ± 24 nm, 168 ± 28 nm, 99 ± 20 nm and 59 ± 7.5 nm, respectively (Fig. 2a and S12†). All the nanoparticles have a spherical morphology and their sizes are ideal for the lipopeptide siRNA complex to reach and to be retained at the tumor site by enhanced retention and permeability (EPR) effect and avoid kidney filtration. The hydrodynamic radius of lipopeptide-fluorophore unlabelled siRNA (SignalSilence control siRNA) complexes in cell culture media at different molar ratios (MRs) ranging from 5 to 100 is given in Table S2† and Fig. 2b. The zeta potential of AB17_siRNA and AB18_siRNA complexes from MR 5–100 is plotted in Fig. 2c and their values along with that of the AB36_siRNA complex are given in Table S3.† The zeta potential of the synthesized lipopeptide_siRNA complexes at MR 5, 10, 20, 50 and 100 showed net positive charge at MR50 and above. For cellular internalization, it is necessary that the negative charge of the siRNA needs to be completely masked and the lipopeptide_siRNA complexes need to have a net positive charge. Upon increasing the molar ratio of lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA, the MR turns positive but too high positive charge of the nanocomplex is also not desirable as it may impair the release of siRNA from the lipopeptide_siRNA complexes. Moreover, a gel retardation study confirms that the siRNA is completely masked by the lipopeptide at higher than MR 35 (Fig. S13†). So, the optimum MR 50 is chosen for all biological studies.
siRNA, the MR turns positive but too high positive charge of the nanocomplex is also not desirable as it may impair the release of siRNA from the lipopeptide_siRNA complexes. Moreover, a gel retardation study confirms that the siRNA is completely masked by the lipopeptide at higher than MR 35 (Fig. S13†). So, the optimum MR 50 is chosen for all biological studies.
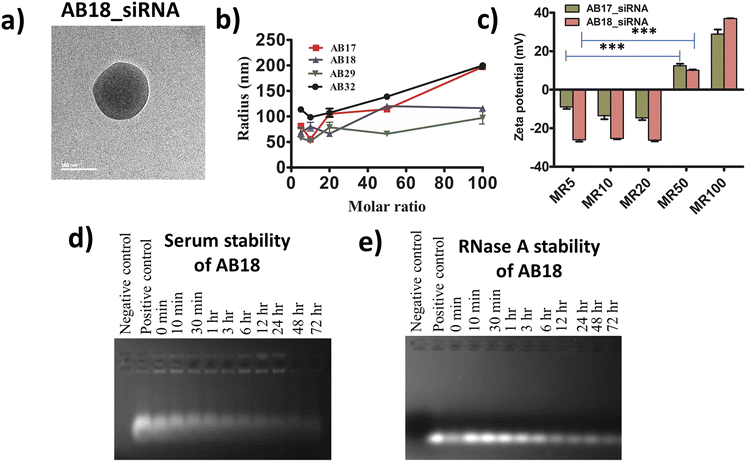 | ||
| Fig. 2 (a) Cryo-TEM images of representative lipopeptide AB18_siRNA complexes showing the spherical nanostructure of self-assembled nanocomplexes at MR 50. (b) Hydrodynamic radius of lipopeptide_fluorophore unlabelled siRNA complexes in culture media at different MRs ranging from 5 to 100, and their values are mentioned in ESI Table S2.† Cell culture media with 10% FBS were used to mimic physiological conditions and the hydrodynamic radius was measured considering the protein corona effect. (c) Zeta potential of the lipopeptide AB17_siRNA and lipopeptide AB18_siRNA complexes at different molar ratios. Their values are given in ESI Table S3.† Error bars indicate SEM from three separate replicates. (d and e) Stability of lipopeptide AB18_siRNA complexes in the presence of (d) serum and (e) RNase A up to a time point of 72 h. | ||
2.3 Stability of siRNA in lipopeptide_siRNA nanocomplexes against serum and ribonuclease A
Free siRNA gets degraded upon the addition of serum and RNase A. Protease stabilized lipopeptide AB18 could protect unmodified siRNA from degradation in the presence of both serum (Fig. 2d) and RNase A (Fig. 2e) till 72 h. This ensures that the siRNA remains protected under in vivo conditions by the synthesized lipopeptide AB18.2.4 Protease stability of lipopeptides
The optimum protease stability of siRNA transporters ensures higher internalization of lipopeptides, lipopeptide_siRNA complexes and disassembly of internalized lipopeptide_siRNA complexes. siRNA transporters need to be cleared off from the in vivo system failing which will lead to bio-accumulation and toxicity. The Arg-Sarcosine-Arg template is preferred for providing optimum protease stability, since Arg-DHis-Arg is resistant to proteolytic cleavage.15 AB36 in the presence of 0.1 μg trypsin starts to degrade from 1–3 h and is completely degraded at 6 h ensuring that the lipopeptide acts as an efficient siRNA transporter without any bio-accumulation (Fig. S14†).2.5 Cell viability in the presence of lipopeptides and lipopeptide_siRNA nanocomplexes
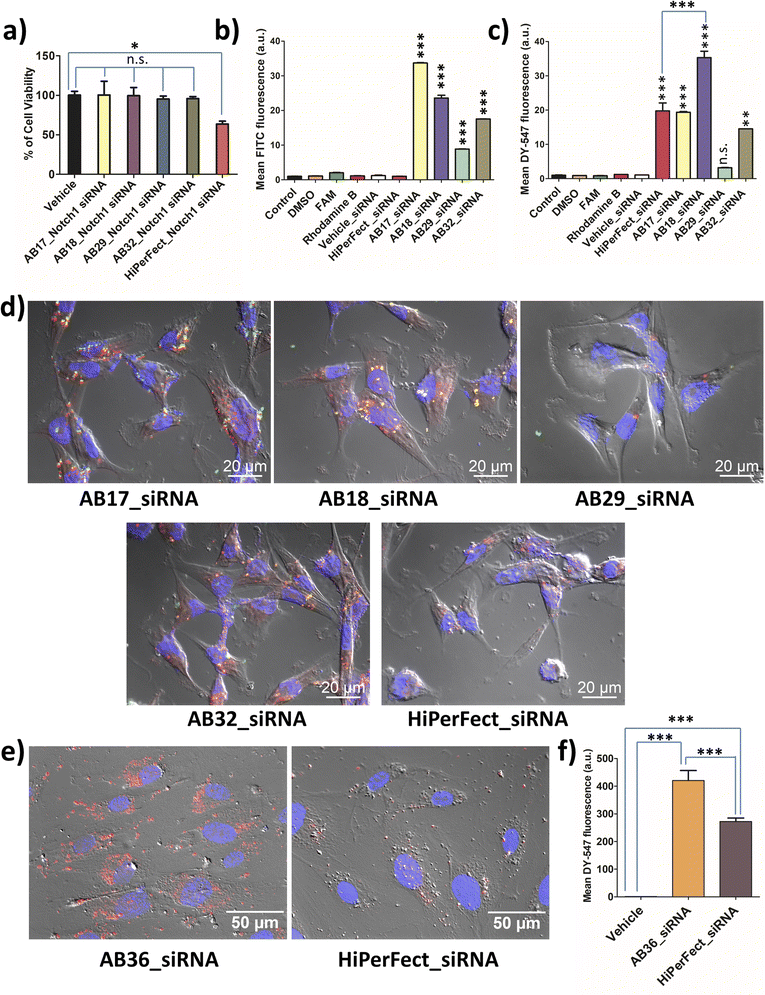
MTT assay was carried out for all the synthesized lipopeptides at 24 h, 48 h and 72 h time points for TNBC cell lines MDA-MB-231 (human) and 4T1 (murine) and also for non-cancer cell lines, human fibroblast cell line and human embryonic kidney cell line, HEK-293. All the synthesized lipopeptides were non-toxic till the 72 h time point at up to 2.5 μM concentration in all the cell lines (Fig. S15–S18†). All biological studies were done at 1.25 μM concentration of the lipopeptides, which is half the dose of 2.5 μM used in MTT assay. MTT assay of the lipopeptide_Notch1 silencing siRNA complex was compared to that of the HiPerFect_siRNA complex at 48 h and 72 h time points. It is observed that the synthesized lipopeptide_Notch1 silencing siRNA complex was not toxic, whereas the HiPerFect_siRNA complex showed a significant amount of toxicity at the 48 h time point (Fig. S19†) and 72 h time point (Fig. 3a). The optimum protease stability of the synthesized N-methylated lipopeptides prevents prolonged bioaccumulation of cationic lipopeptides, ensuring that the lipopeptides are non-toxic to the cells.2.6 Cellular uptake studies of lipopeptide_siRNA nanocomplexes
Lipopeptides were FAM labelled and siRNA was DY-547 labelled, which can be detected using FITC (green) and TRITC (red) channels, respectively. The lipopeptide is internalized most in the case of the AB17_siRNA complex at the 1 h time point (Fig. 3b). Flow cytometry data (1 h incubation) demonstrate that the highest siRNA internalization was achieved by lipopeptide AB18 and was 1.79 times higher than that by HiPerFect_siRNA in the TNBC cell line MDA-MB-231 as quantified by flow cytometry analysis (Fig. 3c). Lipopeptide AB29 internalized siRNA with much lower efficiency compared to lipopeptide AB18 (Fig. 3c).At the 6 h time point, the co-localization of lipopeptide_siRNA complexes was observed in MDA-MB-231 cells from microscopic studies (Fig. 3d). Apotome based microscopic data reveal that lipopeptide AB18 showed higher levels of cytosolic distribution of siRNA and a healthier cell morphology than the commercially used transfecting agent HiPerFect (Fig. 3d). To understand the difference in internalization inside the cellular membrane, we performed computational studies with lipidic portions of lipopeptide AB18 and lipopeptide AB29 interacting with the cancer cell mimetic model membrane. Cancer metastasis is facilitated in the presence of HUVEC cells. Primary cell lines like HUVEC are “notoriously difficult to transfect” as they are highly sensitive to transfection reagents which are toxic and degrade exogenous nucleic acids readily.43 Lipopeptide AB36 (the unlabelled version of lipopeptide AB18 with PEG) exhibited significantly higher (1.53 times more) cellular internalization and cytoplasmic distribution of siRNA compared to HiPerFect in HUVEC cells (Fig. 3e and f).
The presence of a large partially constrained lipidic moiety and another short saturated lipidic moiety in lipopeptide AB36 results in its improved cellular internalization with less residing time in the plasma membrane thus ensuring high cytoplasmic distribution of the lipopeptide_siRNA nanocomplexes.4,41 Moreover, the optimum protease stability of the synthesized lipopeptide allows maximum internalization of the nanocomplex and also de-complexation of lipopeptide_siRNA nanocomplexes and subsequently the release of siRNA in the cytoplasm.
2.7 Computer simulation studies
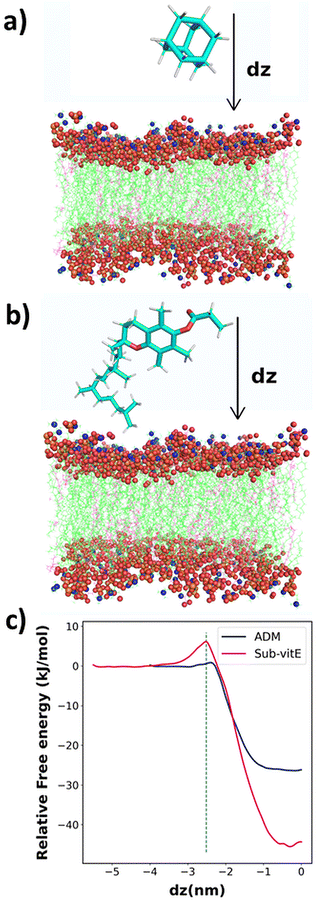
From cellular uptake studies of lipopeptide_siRNA nanocomplexes we observed that the highest level of siRNA is internalized by lipopeptide AB18 and the lowest amount of siRNA by lipopeptide AB29, among the designed lipopeptides (Fig. 3c). Interestingly, lipopeptides AB18 and AB29 differ only by a single lipidic moiety attached to the same peptide backbone. We used computational modelling to understand the reason of the difference in the permeability of these lipopeptides through the membrane. However, these lipopeptides are too large, and therefore computationally extremely expensive for large-scale atomistic modelling and simulations. Since these lipopeptides differ only in the substituent adamantane (ADM) and substituted vitamin-E (Sub-vitE) in the side chain of a particular residue, we used only these substituents to study the permeability. Fig. S20a and S20b† show the chemical structures of the lipopeptides and the substituents of interest with which computational studies are performed inside a brown box.The free energy profiles for ADM and Sub-vitE permeation were constructed from umbrella sampling simulations. The free energy profiles show that the ADM has to cross a small barrier of 0.92 kJ mol−1 to enter into the membrane. After crossing the barrier, the free energy decreases and reaches the minimum in the region between dz = −0.30 and dz = −0.09. When ADM goes further along the coordinate, passing the centre into the bilayer (i.e., dz = 0), the free energy slightly increases, as expected. Overall ADM is stable by 26.0 kJ mol−1 (6.21 kcal mol−1) at the centre of the membrane (dz = 0) compared to that in water. The free energy barrier to get into the membrane for Sub-vitE is larger (6 kJ mol−1). However, it reaches the minimum at dz = −0.30, with a free energy stability of −44.1 kJ mol−1 (10.6 kcal mol−1) compared to that in water. The atomistic model of the membrane and the permeants along with the reaction coordinate dz is shown in Fig. 4a and b. The comparison of the free energy profiles of permeation of both ADM and Sub-vitE indicates that Sub-vitE will be more stable in the bilayer and therefore will have a greater chance of permeation. The stability of ADM is poor in the membrane and therefore its chance of permeation will also be less (Fig. 4c).
2.8 Cellular trafficking pathways of lipopeptide_siRNA complexes
To investigate the cellular trafficking pathways of lipopeptide_siRNA complexes, we have used different endocytosis inhibitors and agents which cause energy depletion. The inhibitors of endocytosis and their dose were chosen from well-documented literature.14,16,44–47 The internalization pathways of cargo depend on the size of the cargo, physiochemical properties of peptides used as transporters, dose of cargo used, and type of cell lines.46 Cells were treated with different endocytic inhibitors such as dynasore – a dynamin – dependent pathway inhibitor, chlorpromazine (abbreviated as CPZ) – a clathrin inhibitor, methyl-β-cyclodextrin (abbreviated as MβCD) – a caveolae and lipid raft inhibitor, 5-(N-ethyl-N-isopropyl)amiloride (abbreviated as EIPA) – a macropinocytosis inhibitor and cytochalasin D (abbreviated as Cyto D) – a actin polymerization inhibitor. It was observed that the siRNA internalization shows a marked decrease in the presence of 80 μM dynasore and 3.5 mM methyl-β-cyclodextrin (Fig. S21†), suggesting that the lipopeptide_siRNA complexes are most likely internalized by both clathrin and caveolae dependent endocytosis.2.9 Endosome mimetic GUV poration assay
Crossing the endosomal membrane is one of the important physiological barriers a good siRNA transporter needs to ensure for cytosolic delivery of the siRNA. Here, we have synthesized giant unilamellar vesicles or GUVs by the gel swelling technique and have engineered the composition of GUVs such that they mimic the membrane composition of the early endosome (phosphatidylcholine (PC)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphatidylethanolamine (PE) = 65
phosphatidylethanolamine (PE) = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) and late endosome (bis(monoacylglycro)phosphate (BMP)
15) and late endosome (bis(monoacylglycro)phosphate (BMP)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PC
PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE = 77
PE = 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
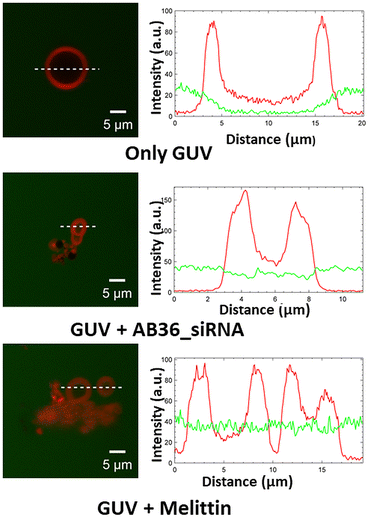
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4).48,49 We have performed a leakage study to investigate the efficiency of our synthesized lipopeptide_siRNA complex to pass through the endosomal membrane and qualify for endosomal escape for efficient cytosolic distribution of siRNA to be loaded to the RISC complex.50 The synthesized GUVs were CM-Dil labelled (red colour) and were kept in a buffer containing 5(6)-carboxyfluorescein (abbreviated as FAM, green colour). Optical sectioning of confocal microscopy was done such that it allowed visualization of the dark background within the GUV, even in the presence of soluble dye outside. The addition of the lipopeptide AB36_Notch1 siRNA complex to GUVs for 1 h resulted in the influx of the FAM containing green coloured buffer inside both the GUVs mimicking early endosome and late endosome (Fig. 5 and S22†). This was possible due to the fact that the lipopeptide AB36_Notch1 siRNA complex could form pores in both early endosome and late endosome mimicking GUVs. Melittin (5 μM), a well characterized, pore forming membrane active peptide derived from bee venom, was taken as a positive control and it also induced poration of both types of GUVs.51 Lipopeptide AB36 having the vitamin E lipidic moiety promotes fusion of the lipid bilayer. Our synthesized lipopeptide has an advantage over the cell penetrating peptide (CPP) TAT, as TAT can perform endosomal escape only in the late endosome in the presence of BMP49 but our synthesized lipopeptide AB36_siRNA nanocomplex can escape from both early and late endosomes (Fig. 5 and S22†). Escape from the early endosome is an important criterion as in the late endosome the pH turns more acidic which can be detrimental to maintaining the functionality of the siRNA. Lipopeptide AB36 was chosen as the siRNA transporter in this study as it is the unlabelled version of lipopeptide AB18 (without fluorophore FAM), which exhibited the maximum siRNA internalization from flow cytometry studies (Fig. 3c). Both lipopeptide AB18 and buffer contain FAM (green colour), so to avoid any false positive result, we have used unlabelled lipopeptide AB36. The presence of a large unsaturated hydrophobic group in lipopeptide AB36 possibly helps in adapting a conical-like structure in the membrane.4,7 The insertion of such lipidic moieties having unsaturation in a biological membrane favours a less stable hexagonal phase compared to a more stable lamellar phase.4,7 This phenomenon possibly also results in the disruption of the endosomal membrane by the lipopeptide AB36 encapsulated siRNA complex.
4).48,49 We have performed a leakage study to investigate the efficiency of our synthesized lipopeptide_siRNA complex to pass through the endosomal membrane and qualify for endosomal escape for efficient cytosolic distribution of siRNA to be loaded to the RISC complex.50 The synthesized GUVs were CM-Dil labelled (red colour) and were kept in a buffer containing 5(6)-carboxyfluorescein (abbreviated as FAM, green colour). Optical sectioning of confocal microscopy was done such that it allowed visualization of the dark background within the GUV, even in the presence of soluble dye outside. The addition of the lipopeptide AB36_Notch1 siRNA complex to GUVs for 1 h resulted in the influx of the FAM containing green coloured buffer inside both the GUVs mimicking early endosome and late endosome (Fig. 5 and S22†). This was possible due to the fact that the lipopeptide AB36_Notch1 siRNA complex could form pores in both early endosome and late endosome mimicking GUVs. Melittin (5 μM), a well characterized, pore forming membrane active peptide derived from bee venom, was taken as a positive control and it also induced poration of both types of GUVs.51 Lipopeptide AB36 having the vitamin E lipidic moiety promotes fusion of the lipid bilayer. Our synthesized lipopeptide has an advantage over the cell penetrating peptide (CPP) TAT, as TAT can perform endosomal escape only in the late endosome in the presence of BMP49 but our synthesized lipopeptide AB36_siRNA nanocomplex can escape from both early and late endosomes (Fig. 5 and S22†). Escape from the early endosome is an important criterion as in the late endosome the pH turns more acidic which can be detrimental to maintaining the functionality of the siRNA. Lipopeptide AB36 was chosen as the siRNA transporter in this study as it is the unlabelled version of lipopeptide AB18 (without fluorophore FAM), which exhibited the maximum siRNA internalization from flow cytometry studies (Fig. 3c). Both lipopeptide AB18 and buffer contain FAM (green colour), so to avoid any false positive result, we have used unlabelled lipopeptide AB36. The presence of a large unsaturated hydrophobic group in lipopeptide AB36 possibly helps in adapting a conical-like structure in the membrane.4,7 The insertion of such lipidic moieties having unsaturation in a biological membrane favours a less stable hexagonal phase compared to a more stable lamellar phase.4,7 This phenomenon possibly also results in the disruption of the endosomal membrane by the lipopeptide AB36 encapsulated siRNA complex.
2.10 Knockdown of Notch1, Erk1 and Erk2 genes by lipopeptide_siRNA nanocomplexes
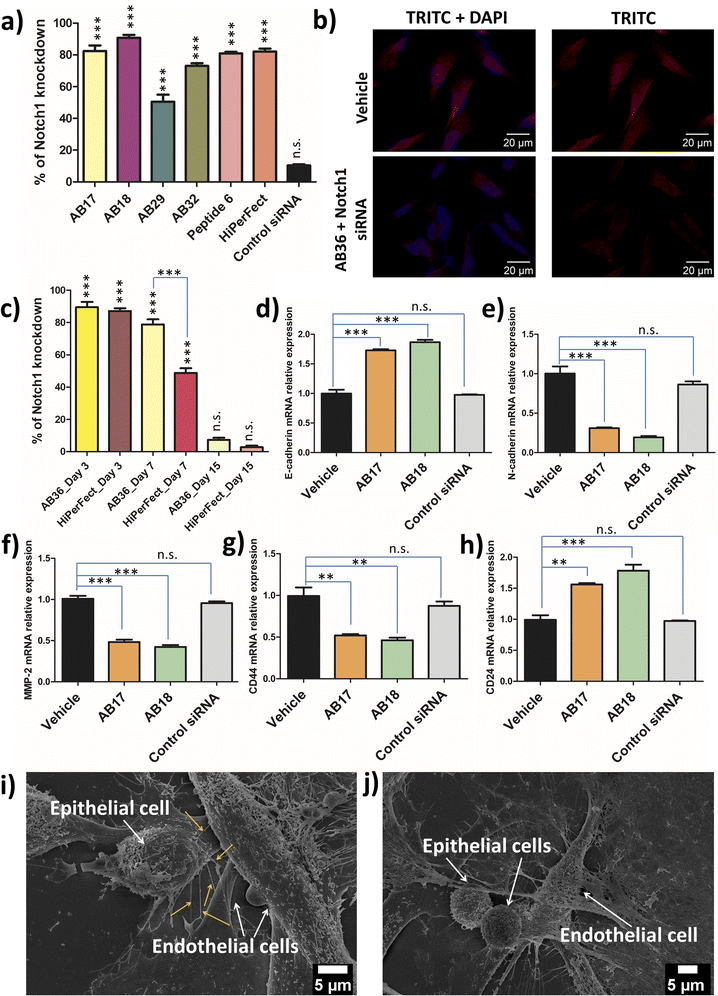
Notch1 and Erk1/2 genes are abruptly overexpressed in the TNBC cell line MDA-MB-231.52,53 TNBC has a very high mortality rate and patients with TNBC are hard to treat as TNBC is highly metastatic. Notch pathways act as a key player in tumour malignancy by employing epithelial mesenchymal transition.54 Erk1/2 genes play a major role in cell proliferation, angiogenesis, cellular differentiation and cell survivability and the Notch1 gene is mostly responsible for promoting metastasis, stemness and chemoresistance in TNBC.52 The synthesized lipopeptide_siRNA complexes were able to silence Notch1, Erk1 and Erk2 genes effectively and thus can have high therapeutic potential.Real time PCR was done to study the silencing of the Erk1 gene at 48 h and 72 h time points by the synthesized lipopeptide_siRNA complexes at MR50 and compared to the HiPerFect_siRNA complex. At 48 h the synthesized lipopeptide_siRNA complex showed lower transfection efficiency as compared to HiPerFect (Fig. S23†) but at the 72 h time point the transfection efficiency of the synthesized lipopeptide AB17_siRNA and lipopeptide AB18_siRNA complexes was comparable to the silencing done by the HiPerFect_siRNA complex for all the investigated oncogenes, i.e., Erk1, Erk2 and Notch1 genes (Fig. 6a, S24 and S25†). The knockdown percentage of different breast cancer oncogenes, namely the Erk1 gene at 48 h (Fig. S23†) and 72 h (Fig. S24†), the Erk2 gene at 72 h (Fig. S25†) and the Notch1 gene at 72 h incubation (Fig. 6a), is mentioned in Table 2.
| Oncogene | Incubation time | siRNA transporter | Knockdown percentage |
|---|---|---|---|
| Erk1 | 48 h | AB17 | 67.9 ± 7.6% |
| AB18 | 62.6 ± 12.5% | ||
| AB29 | 52.6 ± 10.2% | ||
| AB32 | 51.6 ± 7.4% | ||
| Peptide6 | 31.2 ± 2.2% | ||
| HiPerFect | 88.7 ± 1.3% | ||
| 72 h | AB17 | 85.9 ± 8.8% | |
| AB18 | 89.1 ± 6.3% | ||
| AB29 | 58.8 ± 10.9% | ||
| AB32 | 70.6 ± 2.3% | ||
| Peptide6 | 78.1 ± 2.0% | ||
| HiPerFect | 86.5 ± 9.6% | ||
| Erk2 | 72 h | AB17 | 75.8 ± 5.1% |
| AB18 | 85.9 ± 16.7% | ||
| AB29 | 59.8 ± 7.2% | ||
| AB32 | 60.3 ± 3.2% | ||
| Peptide6 | 71.6 ± 9.4% | ||
| HiPerFect | 83.6 ± 2.9% | ||
| Notch1 | 72 h | AB17 | 82.5 ± 5.6% |
| AB18 | 90.8 ± 3.06% | ||
| AB29 | 50.5 ± 7.7% | ||
| AB32 | 73.1 ± 2.8% | ||
| Peptide6 | 80.9 ± 1.6% | ||
| HiPerFect | 82.2 ± 3.0% |
All the knockdown studies were done at MR50 and SignalSilence control siRNA was used as a negative control. The lipopeptide AB17_siRNA complex and lipopeptide AB18_siRNA complex showed a high level of knockdown for all the oncogenes examined with comparable knockdown efficiency to that of the HiPerFect_siRNA complex. Real time PCR analysis reveals lipopeptide mediated cytosolic delivery of functional siRNA, which can access the RNAi machinery.
The immunostaining of Notch1 and Erk1/2 proteins by the lipopeptide AB36_ERK1/2 silencing siRNA complex at MR50 for 72 h showed a significant level of protein downregulation compared to HiPerFect_ERK1/2 silencing siRNA (Fig. 6b, S26 and S27†). Notch1 and Erk1/2 proteins were secondarily labelled by the AlexaFour 568 secondary antibody, which is visible in the TRITC channel. In the case of both Notch1 and Erk1/2 knockdown, the TRITC signal is significantly lowered compared to the untreated vehicle, inferring that both Notch1 (Fig. 6b) and Erk1/2 (Fig. S26†) proteins are significantly lowered at the protein level at 72 h incubation. Our real time-PCR and ApoTome data infer that lipopeptide AB18 and its variant AB36 having one large partially constrained lipidic moiety (alpha-D-tocopherol succinyl) and another short saturated lipid (octyl chain) attached to the peptide backbone have demonstrated the most efficient oncogene silencing at both gene and protein levels.
Among the components of siRNA transporters, our experimental data demonstrate that the introduction of a constrained or unsaturated lipidic moiety into siRNA transporters possibly facilitates the internalization of the lipopeptide encapsulated siRNA by reducing the residing time of the nanocomplex in the plasma membrane and also possibly facilitates endosomal escape. Additionally, the optimum proteolytic stability of the peptide backbone (Arg-Sar-Arg template) enables enhanced cellular uptake and possibly successful decomplexation of lipopeptide_siRNA nanocomplexes. These characteristics facilitate increased gene knockdown efficacy of an efficient siRNA transporter.
2.11 Long term gene silencing efficacy of the synthesized lipopeptide_siRNA nanocomplex
As RNA interference is an invasive procedure, it is desirable that the treatment regime exhibits long term oncogene silencing for effective cancer treatment. To examine the long term knockdown efficacy, MDA-MB-231 cells were treated with AB36_Notch1 siRNA and HiPerFect_Notch1 siRNA and their knockdown efficiency was checked on day 3, day 7 and day 15 post transfection (Fig. 6c). On day 3, siRNA transporter lipopeptide AB36 and HiPerfect showed similar gene knockdown efficiency. Interestingly on day 7, siRNA transporter lipopeptide AB36 exhibited 1.6 times more gene knockdown efficiency than HiPerFect. The Notch1 gene knockdown efficiency by AB36_Notch1 siRNA and HiPerFect_Notch1 siRNA on day 7 is 78.7 ± 5.6% and 48.7 ± 5%, respectively. Superior long term gene silencing efficacy by siRNA transporter AB36 may support its great therapeutic importance.2.12 Relative expression of genes related to metastasis and stemness in Notch1 silenced MDA-MB-231
After successful Notch1 down regulation we wanted to investigate the therapeutic outcomes of Notch1 silenced TNBC cell line MDA-MB-231 cells by the lipopeptide AB18_Notch1 siRNA complex. The Notch1 signalling pathway drive cells towards EMT.55 “Cadherin switching” involves the changes of isoforms of cadherins at adherens junctions of cells.55 This occurs in the normal developmental process and allows cell types to segregate from one another. During tumor metastasis, cadherin switching is recapitulated and this allows the tumor tissue to leave its primary site and metastasize in a distant site. During EMT, cells shed their epithelial markers like E-cadherin and acquire features of mesenchymal cells, and mesenchymal markers like N-cadherin and vimentin.56 This leads the tumour cells to lose their strong affinity with epithelial cells and instead associate weakly with mesenchymal cells, facilitating cell migration and invasion, leading to metastasis.56 Shao et al. have exhibited that the silencing of Notch1 in the TNBC cell line MDA-MB-231 resulted in decreased expression of mesenchymal markers and subsequently increased the expression of epithelial markers.57 The lipopeptide AB18_Notch1 siRNA complex upregulated E-cadherin 1.87 times and down regulated N-cadherin 5.18 times with respect to the vehicle (Fig. 6d and e) indicating inhibition of EMT and metastasis.The high rate of mortality occurring in the aggressive TNBC cell line is mainly due to the TNBC cells metastasizing to lungs and the pleural effusions.58 As a matter of fact, MDA-MB-231 is derived from pleural effusions.59 Nearly 40% of TNBC patients have lung metastasis and these patients have a median survival of only around 22.5 months with systemic chemotherapy.60,61 One of the crucial genes associated with aggressive forms of breast cancer metastasizing in lung and pleural effusions is the MMP-2 gene (gelatinase A).59 Notch1 upregulation leads to the upregulation of both NF-κB and Snail proteins, both of which upregulates MMP-2 protein.56,62,63 Rise in MMP-2 leads to the degradation of extracellular matrix proteins, which facilitates increased invasion of cancer cells. We examined the MMP-2 gene level in Notch1 silenced MDA-MB-231 cells by the lipopeptide AB18_Notch1 siRNA complex by real time PCR analysis (Fig. 6f). The lipopeptide AB18_Notch1 siRNA complex lowered the MMP-2 gene level 2.38 times with respect to untreated MDA-MB-231 cells marked as a vehicle (Fig. 6f), inferring decreased metastasis in Notch1 silenced cells.
TNBC tumors have a high population of cancer stem cells (CSCs).64 CD44+/CD24− is considered an important biomarker for CSCs.65 Fillmore and Kuperwasser claim that more than 90% of TNBC cell line MDA-MB-231 has this CSC phenotype.66 CSCs are capable of self-renewal, has tumor heterogeneity and are held responsible for tumorigenesis, the prolific rate of metastasis, tumor recurrence and drug resistance.67 The presence of such a high population of CSCs may account for a high rate of metastasis, resistance to chemotherapy and eventually high mortality rate of TNBC. Elevated Notch1 signalling leads to increased CD44 and decreased CD24 gene expression in breast cancer.68 Chang et al. have demonstrated that down regulating the CD44 gene led to the inhibition of lung metastasis of MDA-MB-231 cells and also reduced breast tumor outgrowth in a mice model.69
In this study, we were interested to investigate the stem cell phenotypes in Notch1 silenced MDA-MB-231 cells by real time PCR analysis (Fig. 6g and h). CD44 expression was decreased 2.15 fold and CD24 expression was increased 1.8 fold by the lipopeptide AB18_Notch1 siRNA complex (Fig. 6g and h), indicating reduced CSC properties of MDA-MB-231 cells and making TNBC cells less invasive and more responsive to chemotherapy.
2.13 Disruption of nanoscale conduit-mediated communication between tumor cells and endothelial cells
Angiogenesis and metastasis are two hallmark features of cancer progression. Both these phenomena involve communication between the epithelial tumor cell and endothelial cells of blood vessels. Communications are mainly mediated by paracrine signalling by growth factors and by the exosome packed with tumor secreted micro-RNAs (miRNAs).70,71 Recently Sengupta and co-workers have shown that metastatic breast cancer cells develop physical nanoscale conduits or nanobridges which transfer miRNAs from tumour cells to endothelial cells.72 Generally miRNA-132 is transferred from the tumor cells to the endothelial cells and makes the blood vessel pathological by creating a pre-metastatic niche in the blood vessel, thus aiding in tumor metastasis.72 The same group has shown that cancer cells deploy physical nanotubes which connect with immune cells and “hijack” the mitochondria from immune cells.73 This event metabolically empowers the cancer cell and depletes the immune cells thus providing a strategy for immune escape of the tumour cells. In our present study we have examined the presence of nanobridge formation in the 3D co-culture of epithelial (MDA-MB-231 cells) and primary endothelial cells (HUVEC) in matrigel![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS (1
PBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Our studies confirm the presence of nanobridges arising from the epithelial cells and connecting the endothelial cells (Fig. 6i). These nanobridges selectively arises from the side of the MDA-MB-231 cells facing the HUVEC and only from MDA-MB-231 cells which are in close vicinity to HUVEC cells (Fig. 6i). The nanobridges are of 185 ± 40 nm in the short axis and 9.86 ± 1.4 μm in the long axis are very different from other cellular protrusions like lamellipodia.72 Interestingly, Notch1 silenced MDA-MB-231 in close proximity to HUVEC did not exhibit any nanobridge formation, indicating the prevention of metastasis and combating the immunosuppression of TNBC (Fig. 6j).
1). Our studies confirm the presence of nanobridges arising from the epithelial cells and connecting the endothelial cells (Fig. 6i). These nanobridges selectively arises from the side of the MDA-MB-231 cells facing the HUVEC and only from MDA-MB-231 cells which are in close vicinity to HUVEC cells (Fig. 6i). The nanobridges are of 185 ± 40 nm in the short axis and 9.86 ± 1.4 μm in the long axis are very different from other cellular protrusions like lamellipodia.72 Interestingly, Notch1 silenced MDA-MB-231 in close proximity to HUVEC did not exhibit any nanobridge formation, indicating the prevention of metastasis and combating the immunosuppression of TNBC (Fig. 6j).
2.14 Designing combination therapy and evaluation of its effect on the metastasis and stemness of TNBC
Combination therapy has become a corner stone for cancer therapy since it involves combining more than one therapeutic agent such that it results in enhanced therapeutic potential, reduced toxicity and prevention of drug resistance.33 Towards this goal we have selected the “miracle drug” metformin in combination with Notch1 silencing by the lipopeptide AB36_Notch1 siRNA complex against the TNBC cell line MDA-MB-231. Metformin has been used widely as an anti-diabetic and anti-ageing agent.74 We have repurposed this FDA approved drug for combination mediated cancer therapy. Metformin acts a mTOR inhibitor by inhibiting mTORC1 (a catalytic subunit of mTOR) via IGF1 and the insulin signalling pathway.36 High Notch1 signalling in TNBC leads to pro-survival signalling by mTOR-dependent PI3K/Akt inhibition of tumor suppressor protein p53.32 MDA-MB-231 has a high level of mutant p53,75 so restoring the native p53 level is critically important for therapy. Activated Akt signalling in cooperation with Notch signalling leads to self-renewal of cancer stem cells.76 High Akt signalling leads to chemoresistance in breast cancer and inhibition of m-TOR sensitizes cells to cytotoxic agents.77 Notch also upregulates PD-L1 in MDA-MB-231 cells through the m-TOR pathway thus helping in immunosuppression of cancer cells.31 Metformin induces autophagy, causes cell cycle arrest and is also associated with the metabolo-epigenetic link in cancer.78,79 So for all these reasons, we have chosen a m-TOR blocker metformin in combination with siRNA mediated Notch1 silencing.2.15 Evaluation of the drug combination effect of metformin and Notch1 silencing by the lipopeptide AB36_Notch1 siRNA complex and calculation of the combination index (CI)
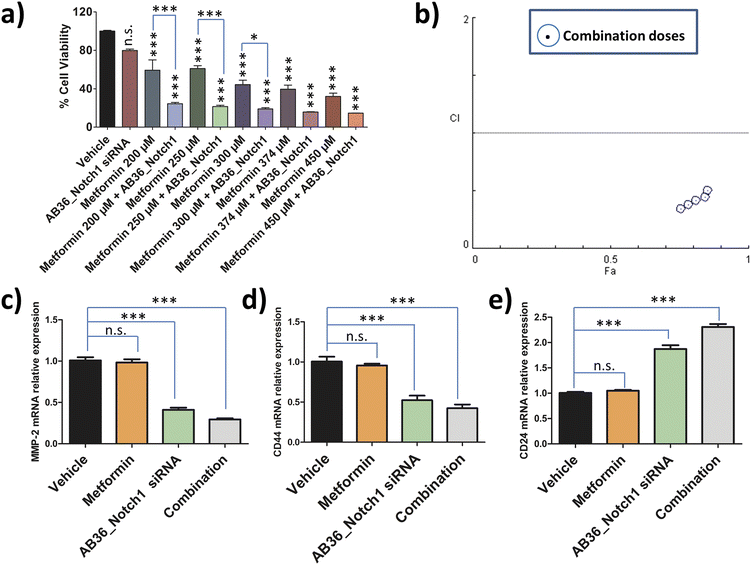
To assess the combination effect of metformin and Notch1 silencing by the lipopeptide AB36_Notch1 siRNA complex in MDA-MB-231 cells, we first determined the IC50 value of metformin against MDA-MB-231 cells at the 72 h time point. MTT assay was performed with a range of doses and the IC50 value was determined by using CompuSyn software (data not shown). The IC50 value of metformin against MDA-MB-231 at 72 h was determined to be 374 μM. For combination studies, we have used a non-fixed ratio.80 Non-fixed ratios are employed when one drug is much more potent than another drug.80 Here, metformin is much more cytotoxic than the lipopeptide AB36_Notch1 siRNA complex which shows negligible cytotoxicity at 72 h. MTT assay was performed for the drug combinations at 72 h keeping the dose of lipopeptide AB36_Notch1 siRNA fixed and varying the metformin dose. The metformin doses were selected near the IC50 value of metformin (200 μM, 250 μM, 300 μM, 374 μM and 450 μM) as it is recommended in combinations with a non-fixed ratio (Fig. 7a).80 The CI of the combination was determined by the Chou–Talalay method using CompuSyn software.81Fig. 7b depicts the CI vs. effect (effect is abbreviated as “Fa”) of all doses used in combination to determine drug interactions which is acquired from CompuSyn software. In general, a CI value > 1 refers to an antagonistic effect, CI = 1 refers to an additive effect and CI < 1 refers to a synergistic effect.81 All the doses in combination yielded a synergistic value with CI < 0.7. The strongest synergistic value was obtained with the dose of 200 μM metformin (CI = 0.34) and thus we have chosen this dose of metformin along with Notch1 silencing by the lipopeptide AB36_Notch1 siRNA complex while applying combination therapy to all other in vitro and in vivo experiments in this study. Please note that both lipopeptide AB18 and lipopeptide AB36 have similar sequences. To avoid the interference from fluorophore FAM, instead of lipopeptide AB18, lipopeptide AB36 was used. These in vitro data of combination therapy were used for designing in vivo experiments in zebrafish, where avoiding the interference of the FAM fluorophore is critically needed.2.16 Relative expression of genes related to metastasis and stemness after applying combination therapy
The relative expression of genes like MMP-2, CD24 and CD44 related to metastasis and stemness were checked by RT-PCR at 72 h after applying combination therapy and compared to that of only Notch1 silenced MDA-MB-231 cells. Notch1 silencing was done by the lipopeptide AB36_Notch1 siRNA complex at 72 h and combination therapy was done by co-administering the lipopeptide AB36_Notch1 siRNA silencing complex and metformin (200 μM) on MDA-MB-231 cells for 72 h. For all the genes investigated combination yielded better therapeutic results compared to only Notch1 silencing of MDA-MB-231 cells.The relative expression of the MMP-2 gene in combination drug treated MDA-MB-231 cells was 0.29 compared to 0.41 in only Notch1 silenced MDA-MB-231 (Fig. 7c). The relative expression of the CD44 gene in combination drug treated MDA-MB-231 cells was 0.42 compared to 0.52 in only Notch1 silenced MDA-MB-231 cells (Fig. 7d) and the relative expression of the CD24 gene in combination drug treated MDA-MB-231 cells was 2.31 compared to 1.87 in only Notch1 silenced MDA-MB-231 cells (Fig. 7e). In our present study, we can infer that the combination of Notch1 silencing along with metformin administration could further decrease metastasis and stemness compared to only Notch1 silenced in MDA-MB-231 cells as seen from the relative gene expression levels of MMP-2, CD24 and CD44 genes. The treatment of metformin (200 μM) alone did not show any changes in the relative expression of any genes. So, we can infer that metformin (200 μM) did not show any changes in gene expression in MMP-2, CD24 and CD44 genes but when used along with Notch1 silencing siRNA, it can further reduce metastasis and stemness in the TNBC cell line MDA-MB-231.
2.17 Role of Notch1 silencing and combination therapy in preventing metastasis and cell proliferation in an in vivo zebrafish model
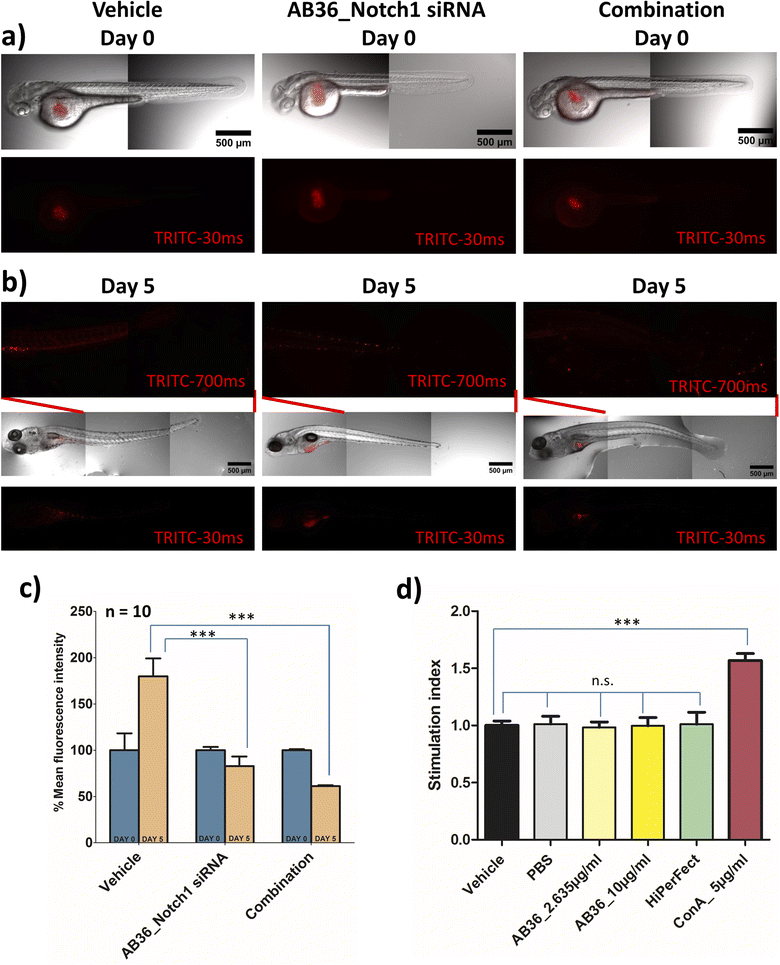
The in vivo experimental procedures used in this study were approved by the Institutional Animal Ethics Committee (IAEC, Animal Use Protocol No.: IISERK/IAEC/AP/2022/83). After achieving successful therapeutic effects under in vitro conditions, we further wanted to check the efficacy of the siRNA transporter and designed combination therapy in an in vivo zebrafish model. In vivo models provide us insights into the molecular mechanisms and signalling pathways involved in the progression of cancer and play a great role in developing different therapeutic agents. Murine xenograft models are considered benchmark in an in vivo breast cancer model but this model suffers from drawbacks like a high cost of intense care for immunocompromised mice, longer tumor generation time and requirement of a large number of cells to be injected for successful tumor inoculation in mice.82 To avoid these complications we have used a zebrafish (Danio rerio) xenograft in vivo model for our study. The tiny zebrafish is making big waves in the field of cancer research, being used in developing xenograft models of various human cancers and for testing of various therapeutic drugs.83,84 Zebrafish shares genes of 71% of all proteins and 82% of disease-causing protein with that of humans.85 Zebrafish models have benefits like (i) transparency of embryos can be obtained by 1-phenyl 2-thiourea (PTU) treatment leading to easy visualization of tumor development and angiogenesis, (ii) cost effectiveness in rearing zebrafish and a large number of embryos can be obtained in a single day, (iii) the absence of a fully developed immune system in zebrafish larvae till 11 days post fertilization allows successful xenograft of different human cancer cells and (iv) zebrafish embryos and larvae can be cultured in small places like in 96 well format and requires a very small amount of drug for testing its pharmacological efficiency.18,82,86,87 All these reasons make zebrafish a great platform for studying cancer development and for testing the efficacy of anticancer drugs. As we synthesize the lipopeptides in our lab and due to the huge cost of anti-Notch1 specific siRNA we have selected the zebrafish model which will require a less quantity of drug over the mice model.In this study, we have developed a zebrafish xenograft model of human breast cancer. Towards the goal of examining the efficiency of Notch1 silencing and combination therapy, we first silenced the Notch1 gene in in vitro MDA-MB-231 cells with the lipopeptide AB36_Notch1 silencing complex for 72 h and co-administered metformin with the lipopeptide AB36_Notch1 silencing complex for 72 h in in vitro MDA-MB-231 cells for combination. These cells were labelled with CM-Dil and injected into perivitelline space of 48 h post fertilization (hpf) zebrafish larvae. Untreated labelled MDA-MB-231 cells were also injected and considered as a vehicle. It is reported that the injection of MDA-MB-231 cells into zebrafish larvae results in cellular proliferation and micro-metastasis to a secondary site generally towards the tail-fin region.88,89 We studied the proliferation and micro-metastasis of MDA-MB-231 cells to the tail region at 0 day post injection (dpi) (Fig. 8a) and 5 days post injection (dpi) (Fig. 8b) by capturing images by epifluorescence microscopy. Untreated MDA-MB-231 cells exhibited active cell proliferation and cell migration towards the tail region at 5 dpi in the case of larvae injected with untreated MDA-MB-231 cells. Larvae injected with Notch1 silenced MDA-MB-231 cells by lipopeptide AB36_Notch1 showed significantly less cellular proliferation and micro-metastasis of MDA-MB-231 cells to the tail region compared to larvae injected with untreated MDA-MB-231 cells at 5 dpi (Fig. 8b). Further less cellular proliferation and negligible micro-metastasis were observed in larvae injected with MDA-MB-231 cells treated with combination of metformin (200 μM) and the lipopeptide AB36_Notch1 silencing complex for 72 h (Fig. 8b). Cellular proliferation was quantified by measuring the fluorescence of CM-Dil labelled MDA-MB-231 cells (Fig. 8c). Untreated MDA-MB-231 cells showed nearly 1.8 times more cell population at 5 dpi compared to 0 dpi. Interestingly, Notch1 silenced MDA-MB-231 cells and MDA-MB-231 cells treated with combination drugs showed 1.2 times and 1.63 times less cell population at 5 dpi compared to 0 dpi, respectively (Fig. 8c). To demonstrate micro-metastasis, the region from the mid-section to the tail section in zebrafish larvae was imaged by epifluorescence microscopy with higher exposure (700 ms) (Fig. 8b). Negligible metastasis was observed in the case of both Notch1 silenced MDA-MB-231 cells and MDA-MB-231 cells treated with combination drugs. The comparatively big red dots in the insets of middle and extreme right sections in Fig. 8b are some residual pigments in zebrafish larvae and artefacts during imaging which are not from MDA-MB-231 cells.
Our study infers that Notch1 silenced MDA-MB-231 cells by the lipopeptide AB36_Notch1 silencing complex does not proliferate and shows negligible micro-metastasis in the in vivo zebrafish model. MDA-MB-231 cells treated with combination of metformin and Notch1 silencing siRNA enhances the therapeutic outcome in the in vivo zebrafish model by mitigating cell proliferation and further reducing micro-metastasis in zebrafish larvae.
2.18 Immunogenicity of synthesized lipopeptides
One of the key criteria for a clinically safe peptide based drug and other health care materials is that it must be non-immunogenic so that it does not elicit any abrupt immune response. We have checked the immunogenicity of our synthesized lipopeptide AB36 and HiPerFect by PBMC proliferation assay (Fig. 8d). PBMC was isolated from goat blood. Concanavalin A (5 μg ml−1), a T cell mitogen was used as a positive control.90 Lipopeptide AB36 was used at its working concentration for all experiments (1.25 μM or 2.635 μg ml−1) and at a much higher concentration (10 μg ml−1) to check its immunogenic nature and HiPerFect was used at a dose as recommended in the user manual for 96 well plates. PBS was used as a negative control. Both lipopeptide AB36 and HiPerFect did not show any mitogenic activity on PBMC cells at the 72 h time point, which infers that the synthesized lipopeptide AB36 and HiPerFect are non-immunogenic (Fig. 8d).Cartoon representation of the key point summary of this work is illustrated in Fig. 9. Briefly the lipopeptide encapsulated Notch1 silencing siRNA complexes are internalized by clathrin and caveolae mediated endocytosis. The size of these nanocomplexes qualify for the EPR effect, thus passively targeting the tumor site. The nanocomplexes can escape from both early and late endosomes and silence the Notch1 mRNA in the cytoplasm. Notch1 is highly expressed in triple negative breast cancer and is responsible for its aggressive metastasis and chemoresistance.30 Notch1 silencing in combination with metformin (mTOR inhibitor) exhibits synergistic drug interaction against TNBC.
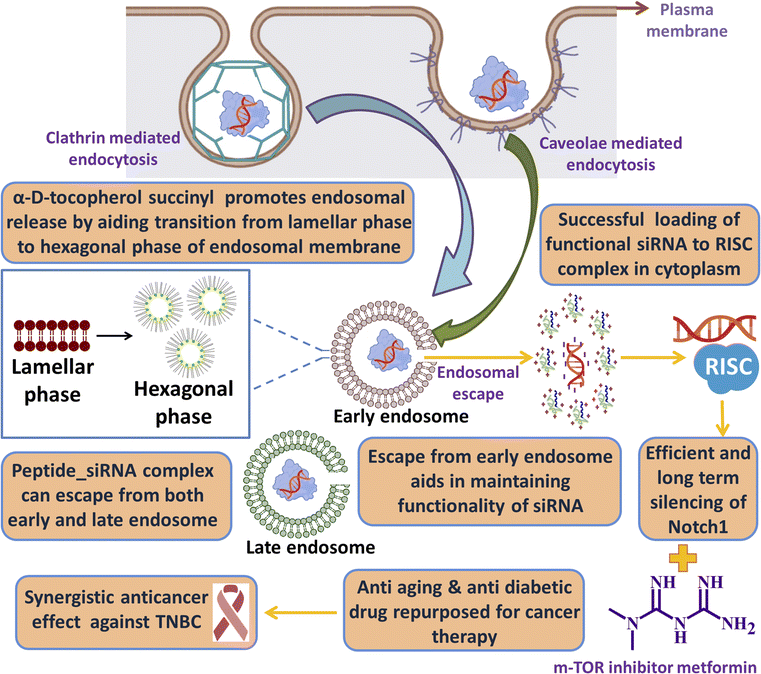 | ||
| Fig. 9 Cartoon representation of the key point summary of this work. Image was created with the help of https://www.biorender.com/. | ||
3. Experimental section
3.1 Materials
Resin and Fmoc protected amino acids were purchased from Novabiochem and GL Biochem and employed in synthesis without further purification. Coupling reagents HATU (1-[bis(dimethylamino)methylene]-H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate), HBTU (2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) and PyBOP (benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate) were obtained from Novabiochem and hydroxybenzotriazole (HOBt) was procured from Sisco Research Laboratories Pvt. Ltd (SRL). Anhydrous dimethylformamide (DMF) and dichloromethane (DCM) used in the coupling reaction were obtained from Acros Organics and N,N-diisopropylethylamine (DIPEA) from TCI Chemicals. Stearic acid, pyrene butyric acid, D-alpha-tocopherol succinate, adamantanecarboxylic acid, 2,4,6-trinitrobenzenesulfonic acid (TNBS), thioanisol, phenol, and piperidine were purchased from Sigma-Aldrich and EDT was obtained from Acros Organics. The washing solvents DMF and DCM were purchased from Merck India. 5(6)-Carboxyfluorescein (FAM) dye was procured from Invitrogen. HiPerFect was purchased from Qiagen. siGLO Red Transfection reagent (DY-547-labelled siRNA) (Cat. No. # D-001630-02-20) was obtained from Dharmacon. Fetal bovine serum (FBS), 0.25% trypsin–ethylenediaminetetraacetic acid (EDTA) (1X), Dulbecco's Modified Eagle's Medium (DMEM) and Opti-MEM were bought from Gibco, Life Technologies. RPMI-1640 and colourless RPMI-1640 were procured from HiMedia. Fluoroshield, Fluoroshield with 4′,6-diamidino-2-phenylindole (DAPI) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent and cholesterol were procured from Sigma-Aldrich. POPC (Cat. No.: 850457P), 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–18
0–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PE (Cat. No.: 850757P) and 18
1 PE (Cat. No.: 850757P) and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 BMP (S,S) (Cat. No.: 857135C) for constructing GUVs were procured from Avanti Polar Lipids. Notch1 siRNA (Cat. No.: sc-36095) was acquired from Santacruz Biotechnology. SignalSilence p44/42 MAPK (Erk1/2) siRNA (Cat. No. #6560) and SignalSilence control siRNA (unconjugated) (Cat No. #6568), and p44/42MAPK (Erk1/2) antibody (Cat. No. #9102) were purchased from Cell Signaling Technology. Goat anti-rabbit IgG (H + L) secondary antibody and Alexa Fluor 568 (Cat. No. A-11036) were procured from Thermo Fisher Scientific.
1 BMP (S,S) (Cat. No.: 857135C) for constructing GUVs were procured from Avanti Polar Lipids. Notch1 siRNA (Cat. No.: sc-36095) was acquired from Santacruz Biotechnology. SignalSilence p44/42 MAPK (Erk1/2) siRNA (Cat. No. #6560) and SignalSilence control siRNA (unconjugated) (Cat No. #6568), and p44/42MAPK (Erk1/2) antibody (Cat. No. #9102) were purchased from Cell Signaling Technology. Goat anti-rabbit IgG (H + L) secondary antibody and Alexa Fluor 568 (Cat. No. A-11036) were procured from Thermo Fisher Scientific.
3.2 Lipopeptide synthesis
The Fmoc solid phase peptide synthesis strategy was utilized to synthesize lipopeptides manually. The lipidic moieties of pyrene, tocopherol, adamantane, and stearic acid coupling was done on the Lys side chain after deprotection of the mild acid labile Mtt group (4-methyl trityl) from the Lys(Mtt) moiety. The Mtt group deprotection was done with a 1% trifluoroacetic acid (TFA) + 5% TIS + DCM mixture for 1 h after 5 minutes interval. The coupling of the lipidic moiety was done using 6 equiv PyBop + 6 equiv HOBt + DIPEA in DMF for 12 h. This coupling was done twice for complete conjugation. The fluorophore 5,6-FAM was tagged at the N terminus of the peptide and the rest of the procedures were followed as described in ref. 14. The lipopeptide purification was done by HPLC using a C18 column using ACN as solvent and confirmed by MALDI-TOF mass spectra (Fig. S4–S8†). The general synthesis scheme of FAM-labelled lipopeptides having constrained lipidic moieties is shown in Fig. S3.†3.3 Circular dichroism
Circular dichroism was carried out as described in ref. 14. Spectra were recorded in a range of 195–300 nm and the spectra were obtained by accumulating three scans with a data pitch of 0.2 nm, scan speed of 100 nm min−1, and bandwidth of 1.0 nm. After smoothing and baseline correction, final spectra were generated. All spectra were plotted using OriginPro 8 software.3.4 Dynamic light scattering
Dynamic light scattering experiments were carried out as mentioned in ref. 15. For the lipopeptide_fluorophore unlabelled siRNA complex, the hydrodynamic size in cell culture media (DMEM + 10% FBS) and the zeta potential in aqueous solution were measured at a molar ratio (MR) [lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA] ranging from 5
siRNA] ranging from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 100
1 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 respectively. To form the lipopeptide_siRNA complex, the lipopeptide and siRNA were mixed at a definite concentration and allowed to rest at room temperature for 30 min. All data were measured in triplicate. OriginPro 8 and GraphPad Prism 5 software were used to plot all data, and the values are represented as mean ± standard error of the mean (SEM) of three independent experiments.
1 respectively. To form the lipopeptide_siRNA complex, the lipopeptide and siRNA were mixed at a definite concentration and allowed to rest at room temperature for 30 min. All data were measured in triplicate. OriginPro 8 and GraphPad Prism 5 software were used to plot all data, and the values are represented as mean ± standard error of the mean (SEM) of three independent experiments.
3.5 Scanning electron microscopy
Field emission scanning electron microscopy (FE-SEM) studies for lipopeptide_siRNA complexes were performed at MR50 as described in ref. 14.3.6 Cryogenic transmission electron microscopy
Cryogenic transmission electron microscopy (Cryo-TEM) studies for lipopeptide_siRNA complexes were performed at MR50 as described in ref. 14.3.7 Gel retardation assay
To determine the formation of the lipopeptide_siRNA complex at different MRs, gel retardation assay was performed as reported in ref. 14. The range of MR taken was from MR 0–MR 100.3.8 Protease stability of lipopeptides
The protease stability of lipopeptide AB36 was examined in the presence of trypsin as reported in ref. 14.3.9 Serum and ribonuclease A protection assay
The ability of the synthesized lipopeptides to protect the siRNA against serum and RNase A was determined using the protocol as done in ref. 14. The time period for which the lipopeptide_siRNA complexes at MR 50 were exposed to serum and RNase A was 0 min, 10 min, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h, 48 h and 72 h.3.10 Computational studies
Two heterogenous POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS lipid bilayer systems were constructed using CHARMM-GUI membrane builder91,92 one for the adamantane (ADM) and other for substituted vitamin-E (Sub-vitE). The lipid bilayers in both the systems had 160 POPC and 40 POPS lipid molecules per leaflet mimicking the cell membrane of cancer cells. Based on the hydration number the lipid bilayers were solvated using 9999 TIP3P93 water molecules for the ADM system and 15
POPS lipid bilayer systems were constructed using CHARMM-GUI membrane builder91,92 one for the adamantane (ADM) and other for substituted vitamin-E (Sub-vitE). The lipid bilayers in both the systems had 160 POPC and 40 POPS lipid molecules per leaflet mimicking the cell membrane of cancer cells. Based on the hydration number the lipid bilayers were solvated using 9999 TIP3P93 water molecules for the ADM system and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 999 TIP3P water molecules for the substituted vitE system. The systems were neutralized by adding 150 mM KCl salt. The systems had dimensions of 7.74 nm × 7.74 nm × 9.00 nm and 7.80 nm × 7.80 nm × 11.76 nm for ADM and Sub-vitE systems, respectively. The periodic boundary conditions were enforced along all directions. For both systems, we used CHARMM36 (ref. 94) for the lipids and CHARMM general force field (CGenFF)95 for the permeants; the latter were randomly inserted into the boxes containing the lipid bilayer and TIP3P water molecules using the insert-molecules module implemented in GROMACS which replaces some water molecules from the boxes to accommodate the permeants.
999 TIP3P water molecules for the substituted vitE system. The systems were neutralized by adding 150 mM KCl salt. The systems had dimensions of 7.74 nm × 7.74 nm × 9.00 nm and 7.80 nm × 7.80 nm × 11.76 nm for ADM and Sub-vitE systems, respectively. The periodic boundary conditions were enforced along all directions. For both systems, we used CHARMM36 (ref. 94) for the lipids and CHARMM general force field (CGenFF)95 for the permeants; the latter were randomly inserted into the boxes containing the lipid bilayer and TIP3P water molecules using the insert-molecules module implemented in GROMACS which replaces some water molecules from the boxes to accommodate the permeants.
The systems were energy minimized using the steepest descent algorithm,96 followed by equilibration at a temperature of 310 K using a V-rescale thermostat97 for 3 ns with a coupling constant of 0.1 ps. After NVT equilibration, the systems were subjected to NPT equilibration for 7 ns at a temperature of 310 K and pressure of 1 bar using a Nosé–Hoover thermostat98,99 and Parrinello–Rahman barostat.100 The coupling constants for temperature and pressure coupling were 0.5 ps and 5 ps, respectively.
In the final production run the system was equilibrated in an NPT ensemble for 20 ns using a Nosé–Hoover thermostat and Parrinello–Rahman barostat with coupling constants 0.5 ps and 2 ps respectively. Particle-Mesh-Ewald101 was used to calculate long-range electrostatic interactions. Bonds to all atoms were constrained using a Linear Constraint Solver (LINCS)101 and the MD integration time step was 2 fs. A cut-off distance of 1.2 nm was used for the short-range neighbour list and the neighbour list was updated every 10 fs. The MD simulations were performed using GROMACS-2021.4.102
3.11 Free energy calculation
Free energy for permeation of adamantane (ADM) and substituted vit-E (Sub-vitE) was determined using the umbrella sampling method.103,104 Umbrella sampling simulations were performed using PLUMED-2.7.4 (ref. 105) patched with GROMACS-2021.4.102 The bilayer was aligned to have its surface normal along the Z direction. Therefore, the collective variable for the umbrella sampling was chosen as the z-coordinate of the distance separating the centre of mass (COM) of the all atoms of the lipid bilayer and the COM of the permeants. The atomistic model of the membrane and the permeants along with the reaction coordinate dz is shown in Fig. 4a and b.Since the bilayer is symmetric, we calculated the free energy of the permeants from the solvent up to the COM of the bilayer. For the adamantane system, the length of the box in the Z-direction was 8 nm. Therefore, the adamantane was brought from z = −4 (surface) to z = 0 (COM of the membrane), with a window spacing of 0.0625 nm, using successive umbrella sampling simulations. For the Sub-vitE system, the windows were placed from z = −5.5 nm to z = 0 nm with a window spacing of 0.0625 nm as the box size in the Z-direction was 11 nm.
After several trials, we used 300–500 kJ mol−1 nm−2 force constants for each window in our umbrella sampling simulations. Based on the convergence criteria (Fig. S28†), the length of the simulations for each window was 30 ns. The final free energy profile for ADM and Sub-vitE systems was obtained by combining 64 and 88 no. of simulations respectively using the WHAM program from Grossfield's lab.106,107
3.12 Cell culture
Triple negative human breast adenocarcinoma cells (MDA-MB-231), the human fibroblast cell line and the human embryonic kidney cell line HEK-293 were cultured in Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum, 1% penicillin–streptomycin and 0.1% amphotericin B. Murine triple negative breast cancer cell line 4T1 cells were cultured in RPMI-1640 with 10% bovine serum and 1% penicillin–streptomycin. Primary cells HUVEC were cultured using complete EGM2 media (Lonza) and were seeded in plates coated with 0.1% gelatin.3.13 Cell viability assay in the presence of cell-penetrating lipopeptides and lipopeptide_siRNA complexes
MTT assay was performed on MDA-MB-231, 4T1, the human fibroblast cell line and the HEK-293 cell line as mentioned in ref. 14. The concentrations of lipopeptides used were 500 nM, 1 μM and 2.5 μM for a time point of 24 h, 48 h and 72 h. For the cytotoxicity assay of the lipopeptide_siRNA complex MR50 was used with 1.25 μM lipopeptide and 25 nM Notch1 siRNA was used along with the HiPerFect_siRNA complex with a dose as mentioned in the HiPerFect usage manual for 25 nM siRNA at time points of 48 h and 72 h. Data were plotted using GraphPad prism software and all values are represented here as mean ± SEM of three independent experiments.3.14 Cellular uptake studies of lipopeptide_siRNA complexes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were analyzed for each sample and samples were taken in duplicate while performing the experiment. The flow cytometer instrument (BD FACSAria) was used to investigate the uptake of the FAM-labelled lipopeptide_DY-547-labelled siRNA complex employing fluorescein isothiocyanate (FITC) and polyethylene (PE) channels. The FAM-labelled lipopeptide at 1.25 μM and the unlabelled lipopeptide encased 25 nM DY-547-labelled siRNA complex were used for FITC and PE signal compensation, respectively. Data were analyzed with FACSDiva software for lipopeptide_siRNA uptake analysis and plotted using GraphPad Prism 5 software as mean ± SEM of two replicates.
000 cells were analyzed for each sample and samples were taken in duplicate while performing the experiment. The flow cytometer instrument (BD FACSAria) was used to investigate the uptake of the FAM-labelled lipopeptide_DY-547-labelled siRNA complex employing fluorescein isothiocyanate (FITC) and polyethylene (PE) channels. The FAM-labelled lipopeptide at 1.25 μM and the unlabelled lipopeptide encased 25 nM DY-547-labelled siRNA complex were used for FITC and PE signal compensation, respectively. Data were analyzed with FACSDiva software for lipopeptide_siRNA uptake analysis and plotted using GraphPad Prism 5 software as mean ± SEM of two replicates.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were grown on coverslips overnight and HUVEC cells (50
000 cells per well) were grown on coverslips overnight and HUVEC cells (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were cultured on coverslips pre coated with 0.1% gelatin overnight, followed by incubation with 1.25 μM FAM-labelled lipopeptide_25 nM DY-547-labelled siRNA complexes (MR50) at 37 °C for 6 h. The rest of the protocol was followed as discussed in ref. 15. Images were acquired at 63× magnification for MDA-MB-231 cells and 40× magnification for HUVEC cells using AXIO Observer Z1 and ApoTome systems. Image acquisition and processing were performed using Zen 2 Pro software.
000 cells per well) were cultured on coverslips pre coated with 0.1% gelatin overnight, followed by incubation with 1.25 μM FAM-labelled lipopeptide_25 nM DY-547-labelled siRNA complexes (MR50) at 37 °C for 6 h. The rest of the protocol was followed as discussed in ref. 15. Images were acquired at 63× magnification for MDA-MB-231 cells and 40× magnification for HUVEC cells using AXIO Observer Z1 and ApoTome systems. Image acquisition and processing were performed using Zen 2 Pro software.
3.15 Early and late endosome mimetic GUV preparation and nanoparticle mediated GUV membrane poration assay
GUVs were prepared by the gel swelling method as discussed in ref. 48 with minor modifications. Briefly, etched cover slips were lysine coated, UV exposed followed by coating with 5% (w/v) polyvinylalcohol (PVA) and dried overnight in an oven. To mimic the early endosome, phosphatidylcholine (PC)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphatidylethanolamine (PE) = 65
phosphatidylethanolamine (PE) = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 lipid composition was prepared.49 Likewise, to mimic the late endosome, bis(monoacylglycro)phosphate (BMP)
15 lipid composition was prepared.49 Likewise, to mimic the late endosome, bis(monoacylglycro)phosphate (BMP)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PC
PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE = 77
PE = 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 lipid composition was prepared.49 To the lipid mixture, 10 μL of 5 μM CM-Dil dye was added. The solution was vortexed to prevent incomplete mixing and then added dropwise gently on a 5% (w/v) PVA coated cover slip and the organic solvent was completely evaporated with a gentle stream of nitrogen. The cover slips were kept in a high vacuum for 1 h. Approximately, 200 μL of PIPES buffer was added to the cover slip and incubated in the dark at room temperature for 30 min for gel swelling. 100 μL of such solution was incubated for 30 min at 37 °C. To this solution, 200 μL of PIPES buffer (200 mM glucose) and 200 μL of FAM (in 6 μM PIPES buffer with 200 mM sucrose) were added. To this GUV solution, lipopeptide_siRNA nanocomplexes were incubated at a concentration of lipopeptide AB36 of 1.25 μM and 25 nM siRNA at 37 °C for 1 h. The samples were gently drop cast on a passivated glass slide (using 5% BSA solution in ddH2O) and visualized under a confocal microscope. 5 μM melittin was used as a positive control.
4 lipid composition was prepared.49 To the lipid mixture, 10 μL of 5 μM CM-Dil dye was added. The solution was vortexed to prevent incomplete mixing and then added dropwise gently on a 5% (w/v) PVA coated cover slip and the organic solvent was completely evaporated with a gentle stream of nitrogen. The cover slips were kept in a high vacuum for 1 h. Approximately, 200 μL of PIPES buffer was added to the cover slip and incubated in the dark at room temperature for 30 min for gel swelling. 100 μL of such solution was incubated for 30 min at 37 °C. To this solution, 200 μL of PIPES buffer (200 mM glucose) and 200 μL of FAM (in 6 μM PIPES buffer with 200 mM sucrose) were added. To this GUV solution, lipopeptide_siRNA nanocomplexes were incubated at a concentration of lipopeptide AB36 of 1.25 μM and 25 nM siRNA at 37 °C for 1 h. The samples were gently drop cast on a passivated glass slide (using 5% BSA solution in ddH2O) and visualized under a confocal microscope. 5 μM melittin was used as a positive control.
3.16 In vitro gene silencing studies by real-time PCR
Knockdown of Notch1, Erk1 and Erk2 at 48 h and 72 h was carried out in MDA-MB-231 cells, and the gene levels of Notch1, Erk1 and Erk2 were analyzed by real-time PCR. MDA-MB-231 cells were transfected with 1.25 μM lipopeptides_25 nM silencing siRNA of Erk1/2 or Notch1. Also Notch1 silenced MDA-MB-231 cells were tested for their relative expression of E-cadherin, N-cadherin, MMP-2, CD44 and CD24 genes. The detailed protocol followed, the primers used and the calculation of gene expression level were as in ref. 15. The primer used for Notch1 is: (F) 5′-GTCAACGCCGTAGATGACC-3′, (R) 5′-TTGTTAGCCCCGTTCTTCAG-3′, E-cadherin: (F) 5′-ATTCTGATTCTGCTGCTCTTG-3′; (R) 5′-AGTCCTGGTCCTCTTCTCC-3′; N-cadherin: (F) 5′-ATGGTGTATGCCGTGAGAAG -3′, (R) 5′-TGTGCTTACTGAATTGTCTTGG-3′; MMP-2: (F) 5′-CGCTCAGATCCGTGGTGAG-3′, (R) 5′-TTGTCACGTGGCGTCACAG-3′; CD44: (F) 5′-GATGGAGAAAGCTCTGAGCATC-3′, (R) 5′-CACCTTCTTCGACTGTTGAC-3′; CD24: (F) 5′-TTCTCCAAGCACCCAGCA-3′, (R) 5′-TGGAATAAATCTGCGTGGGTA-3′. The percentage of knockdown was calculated by using the formula knockdown (%) = [100 − (2−ΔΔCt × 100)]%. In a separate experiment to evaluate the duration of gene silencing RT-PCR was performed after day 3, day 7 and day 15. All primer synthesis was carried out by Eurofins Genomics India Pvt. Ltd. The results were plotted as the mean ± SEM of three replicates using GraphPad Prism 5 software.3.17 Immunostaining of Notch1 and ERK proteins
MDA-MB-231 cells (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well for 72 h incubation) were seeded on etched coverslips in a 24-well plate and incubated overnight. On the following day, the cells were transfected with 1.25 μM lipopeptide AB36 (unlabelled) complexed to 25 nM Erk1/2 or Notch1 silencing siRNAs in separate experiments (MR 50). Transfection with HiPerFect complexed with erk1/2 silencing siRNA was also carried out for comparative studies. The protocol followed is as given in ref. 15.
000 cells per well for 72 h incubation) were seeded on etched coverslips in a 24-well plate and incubated overnight. On the following day, the cells were transfected with 1.25 μM lipopeptide AB36 (unlabelled) complexed to 25 nM Erk1/2 or Notch1 silencing siRNAs in separate experiments (MR 50). Transfection with HiPerFect complexed with erk1/2 silencing siRNA was also carried out for comparative studies. The protocol followed is as given in ref. 15.
3.18 3D Co-culture of epithelial and endothelial cells to study the inhibition of physical nanoscale conduit formation by scanning electron microscopy
Matrigel (BD Biosciences) was diluted with PBS in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and applied on etched, lysine coated cover slips and kept undisturbed at room temperature for 10 min and at 37 °C for around 30 min. Endothelial cells (HUVEC) (30
1 ratio and applied on etched, lysine coated cover slips and kept undisturbed at room temperature for 10 min and at 37 °C for around 30 min. Endothelial cells (HUVEC) (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) were added on the cover slip with 1
000 cells) were added on the cover slip with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Matrigel
1 Matrigel![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS and incubated for 6 h in EGM2 media. Then triple negative breast cancer epithelial cells (MDA-MB-231) were seeded in the same cover slip in a 1
PBS and incubated for 6 h in EGM2 media. Then triple negative breast cancer epithelial cells (MDA-MB-231) were seeded in the same cover slip in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (HUVEC
1 ratio (HUVEC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MDA-MB-231) and incubated in 1
MDA-MB-231) and incubated in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMEM
1 DMEM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EGM2 media for 24 h and a 3D co-culture was established. Normal MDA-MB-231 and Notch1 silenced MDA-MB-231 by 1.25 μM AB36 complexed to 25 nM Notch1 silencing siRNA were added to matrigel containing HUVEC in different sets. The 3D co-culture was fixed with 2% glutaraldehyde overnight and a FE-SEM experiment was performed as described in ref. 108 to visualize nanobridges formed from epithelial cells to endothelial cells.
EGM2 media for 24 h and a 3D co-culture was established. Normal MDA-MB-231 and Notch1 silenced MDA-MB-231 by 1.25 μM AB36 complexed to 25 nM Notch1 silencing siRNA were added to matrigel containing HUVEC in different sets. The 3D co-culture was fixed with 2% glutaraldehyde overnight and a FE-SEM experiment was performed as described in ref. 108 to visualize nanobridges formed from epithelial cells to endothelial cells.
3.19 Combination studies
3.20 Zebrafish xenograft
The in vivo experimental procedures used in this study were approved by the Institutional Animal Ethics Committee (IAEC, Animal Use Protocol No.: IISERK/IAEC/AP/2022/83). Briefly untreated MDA-MB-231 cells, MDA-MB-231 cells transfected with the lipopeptide AB36_Notch1 silencing siRNA complex and MDA-MB-231 cells transfected with the lipopeptide AB36_Notch1 silencing siRNA complex in combination with metformin (200 μM) which exhibited the highest synergistic interaction were stained with 3.8 μM CM-DiI (Invitrogen) according to the product's instruction manual. These stained cells were trypsinized and were injected in 48 hpf PTU treated zebrafish larvae by using an Eppendorf FemtoJet4x nanoinjector keeping the larvae on a 1% agarose injecting chamber made with a microinjection mould. PTU treatment prevented the formation of pigments. Around 400 cells of MDA-MB-231 cells were injected into the perivitelline space of each embryo. Larvae with the correct site of injection and a similar number of cells were screened by using a fluorescence stereomicroscope (Olympus Szx16) and were considered as successfully injected. The screened larvae were imaged at 0 dpi with a fluorescence stereomicroscope and epifluorescence microscope (Olympus IX81). These larvae were incubated at 33 °C at which both the MDA-MB-231 cells and the larvae can grow. At 5 dpi these xenograft larvae were imaged by using a fluorescence stereomicroscope and epifluorescence microscope and were evaluated for the growth of tumor mass and micrometastasis of MDA-MB-231 cells to the tail portion. During microinjection and image accusation, the larvae were kept in tricaine to immobilize them. The images of both 0 dpi and 5 dpi were captured using the same microscope settings (exposure, intensity and threshold). After acquiring images, the zebrafish larvae xenografts were sacrificed by keeping the anesthetized zebrafish larvae in iced cold water. Image stitching and quantification of fluorescence intensity were carried out using ImageJ software and plotted as percentage mean fluorescence intensity taking 0 dpi readings as the baseline.3.21 Evaluation of immunogenicity of the designed lipopeptides
To evaluate if the designed lipopeptides elicit any immune response in peripheral blood mononuclear cells (PBMCs) immunogenicity assay was performed. Goat blood was collected from a local butcher shop in Alsever's solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). PBMC was isolated in Histopaque®-1077 gradient (Sigma) according to the product instruction manual. The PBMC isolated was dissolved in colourless RPMI-1640 media. 4 × 105 PMBC cells were seeded into each well of a 96 well plate with colourless RPMI-1640 media supplemented with 10% FBS, 0.1% amphotericin B and 1% penicillin–streptomycin and incubated overnight. MTT assay was performed after treatment of lipopeptide AB36 (2.635 μg ml−1 and 10 μg ml−1) and compared with HiPerFect at the 72 h time point. PBS and concanavalin A or ConA (Himedia) were taken as negative and positive controls respectively. The stimulation index (SI) was calculated using the following formula.109
5). PBMC was isolated in Histopaque®-1077 gradient (Sigma) according to the product instruction manual. The PBMC isolated was dissolved in colourless RPMI-1640 media. 4 × 105 PMBC cells were seeded into each well of a 96 well plate with colourless RPMI-1640 media supplemented with 10% FBS, 0.1% amphotericin B and 1% penicillin–streptomycin and incubated overnight. MTT assay was performed after treatment of lipopeptide AB36 (2.635 μg ml−1 and 10 μg ml−1) and compared with HiPerFect at the 72 h time point. PBS and concanavalin A or ConA (Himedia) were taken as negative and positive controls respectively. The stimulation index (SI) was calculated using the following formula.109
SI = (mean OD570 of stimulated PBMCs − mean OD570 of blank)/mean OD570 of unstimulated PBMCs.
4. Conclusions
We have engineered short, clinically safe, non-immunogenic, optimum protease resistant peptide based siRNA transporters for efficient knockdown of oncogenes upregulated in TNBC. Lipopeptide encased Notch1 silencing siRNA nanocomplexes have successfully disrupted the crosstalk of HUVEC and MDA-MB-231 cell mediated pre-metastatic niche aiding in metastasis. Mechanistically inspired combination therapy involving a m-TOR inhibitor, repurposed drug, metformin and Notch1 silencing has demonstrated a synergistic effect. Engineered siRNA therapeutics has reduced metastasis and stemness markers both in vitro and in vivo in a zebrafish model. Our siRNA-based long term gene silencing strategy and combination therapy hold promise for designing pharmaceutically feasible new clinical trials, clinical translation and personalized therapy of cancer patients. Additionally, siRNA-based gene silencing in organelles may broaden the potential for treating hereditary genetic disorder, antimicrobial resistance and development of mRNA based vaccines.Data availability
Schemetic diagrams of the lipopeptides, general synthesis scheme, MALDI-TOF mass spectra and other supportive figures and tables (circular dichroism spectra, FE-SEM and cryo-TEM data, cell viability data, flowcytometry data, RT-PCR and immunofluorescence data, etc.) are available in the ESI.†Author Contributions
A. B. and S. M. are shared second authors. R. S. R. and A. M. M. designed the experiments. A. M. M. performed all cell-based assays. S. M., K. C. and A. T. helped A. M. M. in cell biology work. A. M. M. and S. M. performed all zebrafish-related experiments. A. B. synthesized peptides. A. B. and A. M. M. performed the physicochemical characterization. Computational studies were performed by A. M. and S. J and the computational part was written by A. M. and S. J. A. M. M. and R. S. R. contributed to writing the entire manuscript except the computational part.Conflicts of interest
The authors declare the following competing financial interests: we have filed Indian patent entitled “Lipopeptides having constrained lipidic moieties for transfection and siRNA delivery to cancer cells” on peptide-based siRNA transporters (patent application no.: 201931001645). RSR, Prof. Dhananjay Bhattacharyya, AMM, AB, KC and Dr Sanchita Mukherjee are inventors in the patent application no: 201931001645.Acknowledgements
This study is funded by DBT Nanobiotechnology (BT/PR22086/NNT/28/1222/2017) and by SERB POWER (Grant No: SPF/2021/000024) (partially). The computational part of the project was partially supported by the Department of Biotechnology, India (BT/PR34215/AI/133/22/2019). AMM thanks CSIR and DBT Nanobiotechnology (BT/PR27059/NNT/28/1543/2017) for fellowship. AB is thankful to UGC for fellowship, SM thanks CSIR for fellowship, SJ thanks UGC for fellowship, KC thanks CSIR and DST-SERB (EMR/2016/003829) and AT thanks IISER Kolkata for fellowship. RSR thanks SERB for SERB POWER fellowship (Grant No: SPF/2021/000024). The authors thank Dr Kausik Chakraborty (CSIR-IGIB, India), Dr Krishanu Ray (TIFR, India) and Dr Sourav Haldar (CDRI-Lucknow, India), for their insightful discussions. The authors sincerely thank Dr Rajeeb K. Swain (ILS Bhubaneswar, India) for his help with zebrafish experimental training. The authors are grateful to Prof. Surajit Sinha and Dr Chandra Bose (IACS, Kolkata, India) for their help with the needle puller instrumentation facility. The authors acknowledge the support and the resources provided by the ‘PARAM Brahma Facility’ under the National Supercomputing Mission, Government of India at the Indian Institute of Science Education and Research, Pune. The authors thank Prof. Amitabha Bandyopadhyay (IIT Kanpur, India) and Prof. Jonaki Sen (IIT Kanpur, India) for providing nano-injection needles. The authors express their sincere gratitude to Dr Sandeep Singh (NIBMG, India) for helping us with flow cytometry experiments. The authors thank Mr Amarendra Nath Biswas, (Bose Institute, Kolkata, India,) for mass spectroscopy data, Mr Kashinath Sahu (IISER-K SEM facility) for SEM data, Mr Ritabrata Ghosh (IISER-K imaging facility) for ApoTome and confocal microscopy data, Mr Tamal Ghosh (IISER-K Flow Cytometry facility) and Mrs Bhaswati Tarafdar (NIBMG, India) for flow cytometry data, and Mr Sumanta Moi (IISER-K TEM facility) for TEM data. The authors sincerely thank Mr Somnath Jan for helping in CD experiments, Ms Asmita Mukherjee and Dr Arun Bahadur Gurung for their help during manuscript preparation.References
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope and T. D. Madden, The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs, Nat. Nanotechnol., 2019, 14, 1084–1087 CrossRef CAS.
- Y. Dong, D. J. Siegwart and D. G. Anderson, Strategies, design, and chemistry in siRNA delivery systems, Adv. Drug Delivery Rev., 2019, 144, 133–147 CrossRef CAS PubMed.
- H. Dana, G. M. Chalbatani, H. Mahmoodzadeh, R. Karimloo, O. Rezaiean, A. Moradzadeh, N. Mehmandoost, F. Moazzen, A. Mazraeh and V. Marmari, Molecular mechanisms and biological functions of siRNA, Int. J. Biomed. Sci., 2017, 13, 48 CrossRef.
- A. M. Mallick, A. Tripathi, S. Mishra, A. Mukherjee, C. Dutta, A. Chatterjee and R. Sinha Roy, Emerging Approaches for Enabling RNAi Therapeutics, Chem. – Asian J., 2022, 17, e202200451 CrossRef CAS PubMed.
- J. Wang, Z. Lu, M. G. Wientjes and J. L.-S. Au, Delivery of siRNA therapeutics: barriers and carriers, AAPS J., 2010, 12, 492–503 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Knocking down barriers: advances in siRNA delivery, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Delivery materials for siRNA therapeutics, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- W. Tai and X. Gao, Functional peptides for siRNA delivery, Adv. Drug Delivery Rev., 2017, 110, 157–168 CrossRef PubMed.
- J. B. Rothbard, T. C. Jessop and P. A. Wender, Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells, Adv. Drug Delivery Rev., 2005, 57, 495–504 CrossRef CAS.
- I. Nakase, H. Akita, K. Kogure, A. Graslund, U. Langel, H. Harashima and S. Futaki, Efficient intracellular delivery of nucleic acid pharmaceuticals using cell-penetrating peptides, Acc. Chem. Res., 2012, 45, 1132–1139 CrossRef CAS PubMed.
- M. Gooding, L. P. Browne, F. M. Quinteiro and D. L. Selwood, siRNA delivery: from lipids to cell-penetrating peptides and their mimics, Chem. Biol. Drug Des., 2012, 80, 787–809 CrossRef CAS.
- P. A. Wender, D. J. Mitchell, K. Pattabiraman, E. T. Pelkey, L. Steinman and J. B. Rothbard, The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters, Proc. Natl. Acad. Sci. U.S.A., 2000, 97, 13003–13008 CrossRef CAS PubMed.
- J. B. Rothbard, E. Kreider, C. L. VanDeusen, L. Wright, B. L. Wylie and P. A. Wender, Arginine-rich molecular transporters for drug delivery: role of backbone spacing in cellular uptake, J. Med. Chem., 2002, 45, 3612–3618 CrossRef CAS.
- C. Dutta, K. Chakraborty and R. Sinha Roy, Engineered nanostructured facial lipopeptide as highly efficient molecular transporter, ACS Appl. Mater. Interfaces, 2015, 7, 18397–18405 CrossRef CAS.
- A. Biswas, K. Chakraborty, C. Dutta, S. Mukherjee, P. Gayen, S. Jan, A. M. Mallick, D. Bhattacharyya and R. Sinha Roy, Engineered histidine-enriched facial Lipopeptides for enhanced intracellular delivery of functional siRNA to triple negative breast cancer cells, ACS Appl. Mater. Interfaces, 2019, 11, 4719–4736 CrossRef CAS PubMed.
- D. Mandal, A. Nasrolahi Shirazi and K. Parang, Cell-penetrating homochiral cyclic peptides as nuclear-targeting molecular transporters, Angew. Chem., Int. Ed., 2011, 50, 9633–9637 CrossRef CAS.
- G. Lättig-Tünnemann, M. Prinz, D. Hoffmann, J. Behlke, C. Palm-Apergi, I. Morano, H. D. Herce and M. C. Cardoso, Backbone rigidity and static presentation of guanidinium groups increases cellular uptake of arginine-rich cell-penetrating peptides, Nat. Commun., 2011, 2, 1–6 Search PubMed.
- K. Chakraborty, A. Biswas, S. Mishra, A. M. Mallick, A. Tripathi, S. Jan and R. Sinha Roy, Harnessing Peptide-Functionalized Multivalent Gold Nanorods for Promoting Enhanced Gene Silencing and Managing Breast Cancer Metastasis, ACS Appl. Bio Mater., 2023, 6, 458–472 CrossRef CAS PubMed.
- J. E. Bock, J. Gavenonis and J. A. Kritzer, Getting in shape: controlling peptide bioactivity and bioavailability using conformational constraints, ACS Chem. Biol., 2013, 8, 488–499 CrossRef CAS PubMed.
- N. S. Robertson and D. R. Spring, Using peptidomimetics and constrained peptides as valuable tools for inhibiting protein–protein interactions, Molecules, 2018, 23, 959 CrossRef PubMed.
- A. Eguchi, B. R. Meade, Y.-C. Chang, C. T. Fredrickson, K. Willert, N. Puri and S. F. Dowdy, Efficient siRNA delivery into primary cells by a peptide transduction domain–dsRNA binding domain fusion protein, Nat. Biotechnol., 2009, 27, 567–571 CrossRef CAS PubMed.
- S. Kumari, A. K. Badana and R. Malla, Reactive oxygen species: a key constituent in cancer survival, Biomarker Insights, 2018, 13, 1177271918755391 CrossRef PubMed.
- J. Tucker and D. Townsend, Alpha-tocopherol: roles in prevention and therapy of human disease, Biomed. Pharmacother., 2005, 59, 380–387 CrossRef CAS PubMed.
- Y. Tian, X. Zeng, J. Li, Y. Jiang, H. Zhao, D. Wang, X. Huang and Z. Li, Achieving enhanced cell penetration of short conformationally constrained peptides through amphiphilicity tuning, Chem. Sci., 2017, 8, 7576–7581 RSC.
- W. C. Wimley and S. H. White, Experimentally determined hydrophobicity scale for proteins at membrane interfaces, Nat. Struct. Biol., 1996, 3, 842–848 CrossRef CAS PubMed.
- Z. Qian, A. Martyna, R. L. Hard, J. Wang, G. Appiah-Kubi, C. Coss, M. A. Phelps, J. S. Rossman and D. Pei, Discovery and mechanism of highly efficient cyclic cell-penetrating peptides, Biochemistry, 2016, 55, 2601–2612 CrossRef CAS PubMed.
- D. Pei and M. Buyanova, Overcoming endosomal entrapment in drug delivery, Bioconjugate Chem., 2018, 30, 273–283 CrossRef.
- A. Sahni, Z. Qian and D. Pei, Cell-penetrating peptides escape the endosome by inducing vesicle budding and collapse, ACS Chem. Biol., 2020, 15, 2485–2492 CrossRef CAS.
- P. G. Dougherty, A. Sahni and D. Pei, Understanding cell penetration of cyclic peptides, Chem. Rev., 2019, 119, 10241–10287 CrossRef CAS.
- M. Giuli, E. Giuliani, I. Screpanti, D. Bellavia and S. Checquolo, Notch signaling activation as a hallmark for triple-negative breast cancer subtype, J. Oncol., 2019, 2019 Search PubMed.
- F. A. Mansour, A. Al-Mazrou, F. Al-Mohanna, M. Al-Alwan and H. Ghebeh, PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis, Oncoimmunology, 2020, 9, 1729299 CrossRef PubMed.
- S. K. Mungamuri, X. Yang, A. D. Thor and K. Somasundaram, Survival signaling by Notch1: mammalian target of rapamycin (mTOR)–dependent inhibition of p53, Cancer Res., 2006, 66, 4715–4724 CrossRef CAS PubMed.
- K. Chakraborty, A. Tripathi, S. Mishra, A. M. Mallick and R. S. Roy, Emerging concepts in designing next-generation multifunctional nanomedicine for cancer treatment, Biosci. Rep., 2022, 42, BSR20212051 CrossRef CAS PubMed.
- N. E. Bhola, V. M. Jansen, J. P. Koch, H. Li, L. Formisano, J. A. Williams, J. R. Grandis and C. L. Arteaga, Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell PopulationTargeting TORC1/2 and Notch1 Abrogates Tumor Initiation, Cancer Res., 2016, 76, 440–452 CrossRef CAS.
- F. Hossain, C. Sorrentino, D. A. Ucar, Y. Peng, M. Matossian, D. Wyczechowska, J. Crabtree, J. Zabaleta, S. Morello and L. Del Valle, Notch signaling regulates mitochondrial metabolism and NF-κB activity in triple-negative breast cancer cells via IKKα-dependent non-canonical pathways, Front. Oncol., 2018, 8, 575 CrossRef PubMed.
- S. Amin, A. Lux and F. O'Callaghan, The journey of metformin from glycaemic control to mTOR inhibition and the suppression of tumour growth, Br. J. Clin. Pharmacol., 2019, 85, 37–46 CrossRef CAS PubMed.
- P. Mardones and A. Rigotti, Cellular mechanisms of vitamin E uptake: relevance in α-tocopherol metabolism and potential implications for disease, J. Nutr. Biochem., 2004, 15, 252–260 CrossRef CAS PubMed.
- K.-W. Tam, C.-T. Ho, S.-H. Tu, W.-J. Lee, C.-S. Huang, C.-S. Chen, C.-H. Wu, C.-H. Lee and Y.-S. Ho, α-Tocopherol succinate enhances pterostilbene anti-tumor activity in human breast cancer cells in vivo and in vitro, Oncotarget, 2018, 9, 4593 CrossRef PubMed.
- A. K. Smolarek and N. Suh, Chemopreventive activity of vitamin E in breast cancer: a focus on γ-and δ-tocopherol, Nutrients, 2011, 3, 962–986 CrossRef CAS PubMed.
- F. Hecht, C. F. Pessoa, L. B. Gentile, D. Rosenthal, D. P. Carvalho and R. S. Fortunato, The role of oxidative stress on breast cancer development and therapy, Tumor Biol., 2016, 37, 4281–4291 CrossRef CAS.
- J. Heyes, L. Palmer, K. Bremner and I. MacLachlan, Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids, J. Controlled Release, 2005, 107, 276–287 CrossRef CAS PubMed.
- S. Leekumjorn, H. J. Cho, Y. Wu, N. T. Wright, A. K. Sum and C. Chan, The role of fatty acid unsaturation in minimizing biophysical changes on the structure and local effects of bilayer membranes, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 1508–1516 CrossRef CAS PubMed.
- M. A. Hunt, M. J. Currie, B. A. Robinson and G. U. Dachs, Optimizing transfection of primary human umbilical vein endothelial cells using commercially available chemical transfection reagents, J. Biomol. Tech., 2010, 21, 66 Search PubMed.
- J. S. Appelbaum, J. R. LaRochelle, B. A. Smith, D. M. Balkin, J. M. Holub and A. Schepartz, Arginine topology controls escape of minimally cationic proteins from early endosomes to the cytoplasm, Chem. Biol., 2012, 19, 819–830 CrossRef CAS PubMed.
- K. L. J. Leong, M. M.-L. Ng and J. J. H. Chu, The essential role of clathrin-mediated endocytosis in the infectious entry of human enterovirus 71, J. Biol. Chem., 2011, 286, 309–321 CrossRef PubMed.
- F. Duchardt, M. Fotin-Mleczek, H. Schwarz, R. Fischer and R. Brock, A comprehensive model for the cellular uptake of cationic cell-penetrating peptides, Traffic, 2007, 8, 848–866 CrossRef CAS PubMed.
- I. Nakase, M. Niwa, T. Takeuchi, K. Sonomura, N. Kawabata, Y. Koike, M. Takehashi, S. Tanaka, K. Ueda and J. C. Simpson, Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement, Mol. Ther., 2004, 10, 1011–1022 CrossRef CAS PubMed.
- A. Weinberger, F.-C. Tsai, G. H. Koenderink, T. F. Schmidt, R. Itri, W. Meier, T. Schmatko, A. Schröder and C. Marques, Gel-assisted formation of giant unilamellar vesicles, Biophys. J., 2013, 105, 154–164 CrossRef CAS.
- A. Erazo-Oliveras, K. Najjar, D. Truong, T.-Y. Wang, D. J. Brock, A. R. Prater and J.-P. Pellois, The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos, Cell Chem. Biol., 2016, 23, 598–607 CrossRef CAS PubMed.
- S. Haldar, E. Mekhedov, C. D. McCormick, P. S. Blank and J. Zimmerberg, Lipid-dependence of target membrane stability during influenza viral fusion, J. Cell Sci., 2019, 132, jcs218321 CAS.
- M.-T. Lee, T.-L. Sun, W.-C. Hung and H. W. Huang, Process of inducing pores in membranes by melittin, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, 14243–14248 CrossRef CAS.
- C. Bartholomeusz, A. M. Gonzalez-Angulo, P. Liu, N. Hayashi, A. Lluch, J. Ferrer-Lozano and G. N. Hortobágyi, High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients, Oncologist, 2012, 17, 766–774 CrossRef CAS.
- X. X. Lai, G. Li, B. Lin and H. Yang, Interference of Notch 1 inhibits the proliferation and invasion of breast cancer cells: involvement of the β-catenin signaling pathway, Mol. Med. Rep., 2018, 17, 2472–2478 CAS.
- J. J. Speiser, Ç. Erşahin and C. Osipo, The functional role of Notch signaling in triple-negative breast cancer, Vitam. Horm., 2013, 93, 277–306 CAS.
- R. Kar, N. K. Jha, S. K. Jha, A. Sharma, S. Dholpuria, N. Asthana, K. Chaurasiya, V. K. Singh, S. Burgee and P. Nand, A “NOTCH” deeper into the epithelial-to-mesenchymal transition (EMT) program in breast cancer, Genes, 2019, 10, 961 CrossRef CAS PubMed.
- S. Lamouille, J. Xu and R. Derynck, Molecular mechanisms of epithelial–mesenchymal transition, Nat. Rev. Mol. Cell Biol., 2014, 15, 178–196 CrossRef CAS PubMed.
- S. Shao, X. Zhao, X. Zhang, M. Luo, X. Zuo, S. Huang, Y. Wang, S. Gu and X. Zhao, Notch1 signaling regulates the epithelial–mesenchymal transition and invasion of breast cancer in a Slug-dependent manner, Mol. Cancer, 2015, 14, 1–17 CrossRef PubMed.
- L. Jin, B. Han, E. Siegel, Y. Cui, A. Giuliano and X. Cui, Breast cancer lung metastasis: Molecular biology and therapeutic implications, Cancer Biol. Ther., 2018, 19, 858–868 CrossRef CAS.
- A. J. Minn, G. P. Gupta, P. M. Siegel, P. D. Bos, W. Shu, D. D. Giri, A. Viale, A. B. Olshen, W. L. Gerald and J. Massagué, Genes that mediate breast cancer metastasis to lung, Nature, 2005, 436, 518–524 CrossRef CAS PubMed.
- W. D. Foulkes, I. E. Smith and J. S. Reis-Filho, Triple-negative breast cancer, N. Engl. J. Med., 2010, 363, 1938–1948 CrossRef CAS PubMed.
- H. Y. Yhim, S. W. Han, D. Y. Oh, W. Han, S. A. Im, T. Y. Kim, Y. T. Kim, D. Y. Noh, E. K. Chie and S. W. Ha, Prognostic factors for recurrent breast cancer patients with an isolated, limited number of lung metastases and implications for pulmonary metastasectomy, Cancer, 2010, 116, 2890–2901 CrossRef PubMed.
- H. Kim and G. Jung, Notch1 increases Snail expression under high reactive oxygen species conditions in hepatocellular carcinoma cells, Free Radical Res., 2014, 48, 806–813 CrossRef CAS PubMed.
- L. Li, F. Zhao, J. Lu, T. Li, H. Yang, C. Wu and Y. Liu, Notch-1 signaling promotes the malignant features of human breast cancer through NF-κB activation, PLoS One, 2014, 9, e95912 CrossRef.
- N. Fultang, M. Chakraborty and B. Peethambaran, Regulation of cancer stem cells in triple negative breast cancer, Cancer Drug Resist., 2021, 4, 321 CAS.
- M. Al-Hajj, M. S. Wicha, A. Benito-Hernandez, S. J. Morrison and M. F. Clarke, Prospective identification of tumorigenic breast cancer cells, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 3983–3988 CrossRef CAS.
- C. M. Fillmore and C. Kuperwasser, Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy, Breast Cancer Res., 2008, 10, 1–13 CrossRef PubMed.
- L. T. H. Phi, I. N. Sari, Y.-G. Yang, S.-H. Lee, N. Jun, K. S. Kim, Y. K. Lee and H. Y. Kwon, Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment, Stem Cells Int., 2018, 2018, 5416923 Search PubMed.
- X. Zhang, X. Zhao, S. Shao, X. Zuo, Q. Ning, M. Luo, S. Gu and X. Zhao, Notch1 induces epithelial-mesenchymal transition and the cancer stem cell phenotype in breast cancer cells and STAT3 plays a key role, Int. J. Oncol., 2015, 46, 1141–1148 CrossRef CAS.
- G. Chang, J. Wang, H. Zhang, Y. Zhang, C. Wang, H. Xu, Y. Lin, L. Ma, Q. Li and T. Pang, CD44 targets Na+/H+ exchanger 1 to mediate MDA-MB-231 cells' metastasis via the regulation of ERK1/2, Br. J. Cancer, 2014, 110, 916–927 CrossRef CAS.
- Y. Cao, J. Arbiser, R. J. D'Amato, P. A. D'Amore, D. E. Ingber, R. Kerbel, M. Klagsbrun, S. Lim, M. A. Moses and B. Zetter, Forty-year journey of angiogenesis translational research, Sci. Transl. Med., 2011, 3, 114rv113 Search PubMed.
- W. Zhou, M. Y. Fong, Y. Min, G. Somlo, L. Liu, M. R. Palomares, Y. Yu, A. Chow, S. T. F. O'Connor and A. R. Chin, Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis, Cancer Cell, 2014, 25, 501–515 CrossRef CAS PubMed.
- Y. Connor, S. Tekleab, S. Nandakumar, C. Walls, Y. Tekleab, A. Husain, O. Gadish, V. Sabbisetti, S. Kaushik, S. Sehrawat, A. Kulkarni, H. Dvorak, B. Zetter, E. Edelman and S. Sengupta, Physical nanoscale conduit-mediated communication between tumour cells and the endothelium modulates endothelial phenotype, Nat. Commun., 2015, 6, 1–14 Search PubMed.
- T. Saha, C. Dash, R. Jayabalan, S. Khiste, A. Kulkarni, K. Kurmi, J. Mondal, P. K. Majumder, A. Bardia and H. L. Jang, Intercellular nanotubes mediate mitochondrial trafficking between cancer and immune cells, Nat. Nanotechnol., 2022, 17, 98–106 CrossRef CAS PubMed.
- V. N. Anisimov, Metformin: do we finally have an anti-aging drug?, Cell Cycle, 2013, 12, 3483–3489 CrossRef CAS.
- M. Olivier, R. Eeles, M. Hollstein, M. A. Khan, C. C. Harris and P. Hainaut, The IARC TP53 database: new online mutation analysis and recommendations to users, Hum. Mutat., 2002, 19, 607–614 CrossRef CAS PubMed.
- K. S. Papadakos, M. Bartoschek, C. Rodriguez, C. Gialeli, S.-B. Jin, U. Lendahl, K. Pietras and A. M. Blom, Cartilage oligomeric matrix protein initiates cancer stem cells through activation of Jagged1-Notch3 signaling, Matrix Biol., 2019, 81, 107–121 CrossRef CAS PubMed.
- C. Dong, J. Wu, Y. Chen, J. Nie and C. Chen, Activation of PI3K/AKT/mTOR pathway causes drug resistance in breast cancer, Front. Pharmacol., 2021, 12, 628690 CrossRef CAS PubMed.
- Y. Wang, W. Xu, Z. Yan, W. Zhao, J. Mi, J. Li and H. Yan, Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways, J. Exp. Clin. Cancer Res., 2018, 37, 1–12 CrossRef PubMed.
- E. Cuyàs, S. Verdura, B. Martin-Castillo and J. A. Menendez, Metformin: Targeting the Metabolo-Epigenetic Link in Cancer Biology, Front. Oncol., 2021, 10, 620641 CrossRef PubMed.
- I. V. Bijnsdorp, E. Giovannetti and G. J. Peters, in Cancer cell culture, Springer, 2011, pp. 421–434 Search PubMed.
- T.-C. Chou, Drug Combination Studies and Their Synergy Quantification Using the Chou–Talalay MethodSynergy Quantification Method, Cancer Res., 2010, 70, 440–446 CrossRef CAS.
- M. Konantz, T. B. Balci, U. F. Hartwig, G. Dellaire, M. C. André, J. N. Berman and C. Lengerke, Zebrafish xenografts as a tool for in vivo studies on human cancer, Ann. N.Y. Acad. Sci., 2012, 1266, 124–137 CrossRef PubMed.
- T. Dhawan, Developing Zebrafish as an In Vivo Model to Screen Compounds for Anti-Cancer Activity in Human Breast Cancer, 2019 Search PubMed.
- S. Berghmans, C. Jette, D. Langenau, K. Hsu, R. Stewart, T. Look and J. P. Kanki, Making waves in cancer research: new models in the zebrafish, Biotechniques, 2005, 39, 227–237 CrossRef CAS PubMed.
- D. G. Howe, Y. M. Bradford, A. Eagle, D. Fashena, K. Frazer, P. Kalita, P. Mani, R. Martin, S. T. Moxon and H. Paddock, The Zebrafish Model Organism Database: new support for human disease models, mutation details, gene expression phenotypes and searching, Nucleic Acids Res., 2017, 45, D758–D768 CrossRef CAS PubMed.
- A. Eguiara, O. Holgado, I. Beloqui, L. Abalde, Y. Sanchez, C. Callol and A. G. Martin, Xenografts in zebrafish embryos as a rapid functional assay for breast cancer stem-like cell identification, Cell Cycle, 2011, 10, 3751–3757 CrossRef CAS.
- L. Truong, S. L. Harper and R. L. Tanguay, in Drug safety evaluation, Springer, 2011, pp. 271–279 Search PubMed.
- J. Ren, S. Liu, C. Cui and P. Ten Dijke, Invasive behavior of human breast cancer cells in embryonic zebrafish, J. Visualized Exp., 2017, e55459 Search PubMed.
- H. K. Brown, K. Schiavone, S. Tazzyman, D. Heymann and T. J. Chico, Zebrafish xenograft models of cancer and metastasis for drug discovery, Expert Opin. Drug Discovery, 2017, 12, 379–389 CrossRef CAS PubMed.
- Y. Ando, C. Yasuoka, T. Mishima, T. Ikematsu, T. Uede, T. Matsunaga and M. Inobe, Concanavalin A-mediated T cell proliferation is regulated by herpes virus entry mediator costimulatory molecule, In Vitro Cell. Dev. Biol.: Anim., 2014, 50, 313–320 CrossRef CAS PubMed.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan and R. M. Venable, CHARMM-GUI membrane builder toward realistic biological membrane simulations, J. Comput. Chem., 2014, 35, 1997–2004 CrossRef CAS PubMed.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes, Biophys. J., 2009, 97, 50–58 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O'Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell Jr and R. W. Pastor, Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types, J. Phys. Chem. B, 2010, 114, 7830–7843 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes and I. Vorobyov, CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comput. Chem., 2010, 31, 671–690 CAS.
- W. H. Press, S. A. Teukolsky, W. T. Vetterling and B. P. Flannery, Numerical recipes: The Art of Scientific Computing, Cambridge university press, 3rd edn, 2007 Search PubMed.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- G. J. Martyna, M. L. Klein and M. Tuckerman, Nosé–Hoover chains: the canonical ensemble via continuous dynamics, J. Chem. Phys., 1992, 97, 2635–2643 CrossRef.
- S. Nosé, A molecular dynamics method for simulations in the canonical ensemble, Mol. Phys., 1984, 52, 255–268 CrossRef.
- M. Parrinello and A. Rahman, Polymorphic transitions in single crystals: a new molecular dynamics method, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, LINCS: a linear constraint solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- H. J. Berendsen, D. van der Spoel and R. van Drunen, GROMACS: a message-passing parallel molecular dynamics implementation, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- G. M. Torrie and J. P. Valleau, Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- D. Bochicchio, E. Panizon, R. Ferrando, L. Monticelli and G. Rossi, Calculating the free energy of transfer of small solutes into a model lipid membrane: comparison between metadynamics and umbrella sampling, J. Chem. Phys., 2015, 143, 10B612_611 CrossRef.
- G. A. Tribello, M. Bonomi, D. Branduardi, C. Camilloni and G. Bussi, PLUMED 2: new feathers for an old bird, Comput. Phys. Commun., 2014, 185, 604–613 CrossRef CAS.
- S. Kumar, J. M. Rosenberg, D. Bouzida, R. H. Swendsen and P. A. Kollman, The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method, J. Comput. Chem., 1992, 13, 1011–1021 CrossRef CAS.
- A. Grossfield, WHAM: the weighted histogram analysis method, version 2.0. 9, available at membrane, 2013, https://www.google.com/search?q=urmc.+rochester.+edu%2Fcontent%2Fwham%26rlz=1C1GCEA_enIN1051IN1051%26oq=urmc.+rochester.+edu%2Fcontent%2Fwham%26aqs=chrome..69i57j33i160l3.941j0j9%26sourceid=chrome%26ie=UTF-8, Accessed November, 2013, 15 Search PubMed.
- S. Ghosh, P. Gayen, S. Jan, A. V. Kishore, V. Kumar, A. M. Mallick, A. Mukherjee, S. K. Nandi and R. Sinha Roy, Bioinspired Non-Immunogenic Multifunctional Sealant for Efficient Blood Clotting and Suture-Free Wound Closure, ACS Biomater. Sci. Eng., 2020, 6, 6378–6393 CrossRef CAS PubMed.
- N. Molaae, G. Mosayebi, A. Pishdadian, M. Ejtehadifar and A. Ganji, Evaluating the proliferation of human PeripheralBlood mononuclear cells using MTT assay, Int. J. Basic Sci. Med., 2017, 2, 25–28 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01071f |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |