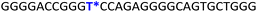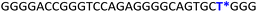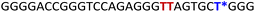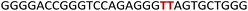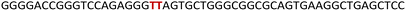Open Access Article
Open Access ArticleProbing juxtaposed G-quadruplex and hairpin motifs using a responsive nucleoside probe: a unique scaffold for chemotherapy†
Saddam Y.
Khatik
 a,
Sruthi
Sudhakar
a,
Sruthi
Sudhakar
 b,
Satyajit
Mishra
b,
Satyajit
Mishra
 c,
Jeet
Kalia
c,
Jeet
Kalia
 cd,
P. I.
Pradeepkumar
cd,
P. I.
Pradeepkumar
 b and
Seergazhi G.
Srivatsan
b and
Seergazhi G.
Srivatsan
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research (IISER), Pune, Dr Homi Bhabha Road, Pune 411008, India. E-mail: srivatsan@iiserpune.ac.in
bDepartment of Chemistry, Indian Institute of Technology Bombay, Mumbai 400076, India
cDepartment of Biological Sciences, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal Bypass Road, Bhauri, Bhopal 462066, India
dDepartment of Chemistry, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal Bypass Road, Bhauri, Bhopal 462066, India
First published on 2nd May 2023
Abstract
Paucity of efficient probes and small molecule ligands that can distinguish different G-quadruplex (GQ) topologies poses challenges not only in understanding their basic structure but also in targeting an individual GQ form from others. Alternatively, G-rich sequences that harbour unique chimeric structural motifs (e.g., GQ-duplex or GQ-hairpin junctions) are perceived as new therapeutic hotspots. In this context, the epidermal growth factor receptor (EGFR) gene, implicated in many cancers, contains a 30 nucleotide G-rich segment in the promoter region, which adopts in vitro two unique architectures each composed of a GQ topology (parallel and hybrid-type) juxtaposed with a hairpin domain. Here, we report the use of a novel dual-app probe, C5-trifluoromethyl benzofuran-modified 2′-deoxyuridine (TFBF-dU), in the systematic analysis of EGFR GQs and their interaction with small molecules by fluorescence and 19F NMR techniques. Notably, distinct fluorescence and 19F NMR signals exhibited by the probe enabled the quantification of the relative population of random, parallel and hybrid-type GQ structures under different conditions, which could not be obtained by conventional CD and 1H NMR techniques. Using the fluorescence component, we quantified ligand binding properties of GQs, whereas the 19F label enabled the assessment of ligand-induced changes in GQ dynamics. Studies also revealed that mutations in the hairpin domain affected GQ formation and stability, which was further functionally verified in polymerase stop assay. We anticipate that these findings and useful properties of the nucleoside probe could be utilized in designing and evaluating binders that jointly target both GQ and hairpin domains for enhanced selectivity and druggability.
Introduction
The epidermal growth factor receptor (EGFR) is a transmembrane protein belonging to the family of protein kinase receptors. The EGFR gene codes for a tyrosine kinase receptor, which is activated by physiological extracellular ligands initiating an important signal transduction pathway that is required for normal cell growth, differentiation and proliferation in mammalian cells.1,2 Overexpression or mutations that elevate the activity of the EGFR signalling pathway is directly linked to the progression of several cancers including that of lung, breast and glioblastoma.3,4 Presently available therapeutic strategies to counter the effects of upregulation of EGFR activity employ tyrosine kinase inhibitors or monoclonal antibodies that interfere with the binding of ligands to the extracellular receptor domain.5–7 However, their efficiency is limited by intrinsic or acquired resistance.7,8 Alternatively, we envision that targeting non-canonical nucleic acid structural elements that act as natural regulators could be a viable strategy to control the EGFR expression at replication and transcription levels.9–11 One such class of structures is a G-quadruplex (GQ), which is formed by guanine rich sequences.12,13 Four guanine bases in a sequence via Hoogsteen hydrogen bonding form a tetrad (G-tetrad) and two or more G-tetrads, stabilized by monovalent cations (K+ or Na+), stack one above the other to form GQ structures.14 Compelling experimental data indicate that GQs present in DNA and RNA serve as gene regulatory elements.15,16 Dysfunction of these elements in several genes is linked to tumour progression,17,18 and hence, small molecule ligands that stabilize these structures and down regulate gene expression are perceived as alternative therapeutic tools to mitigate cancer.19–25 Though many small molecule ligands developed so far show good selectivity between GQ and duplex structures, they seldom distinguish between different topologies of GQs as they have a similar tetrad skeleton.26 Recently, a few ligands have been developed that bind to a specific GQ topology.27–32 Nevertheless, targeting a specific GQ motif amongst others in the genome remains a major challenge. Alternatively, G-rich segments that harbour GQ-duplex or GQ-hairpin junctions are considered as druggable targets,33,34 wherein ligand scaffolds capable of simultaneously binding to GQ and duplex regions increase the specific targeting of such motifs.35–37 In the context of the EGFR gene, a 30 nucleotide G-rich sequence upstream of the transcription start site (−272 position) forms two unique GQ topologies (parallel and hybrid-type), which are stabilized by a short hairpin located at the terminal loop.38 This domain represents a new point of intervention to potentially attenuate the disease-causing activity of the gene. Therefore, it is important to first understand (i) the structural polymorphism of EGFR GQs, (ii) how the GQs interact with small molecule ligands and (iii) the functional role of GQs in a cellular process (e.g., DNA replication).GQ exhibits a high degree of structural polymorphism, which apart from environmental conditions (e.g., ionic conditions, molecular crowding and confinement) depends on the sequence composition, namely, the number of G-tracts and loop nucleotides that connect them.39,40 G-tracts composed of four or more guanine bases can form multiple dynamically interchangeable GQ conformations.41–43 The EGFR promoter region contains four G-tracts (4-3-4-3) that can support multiple GQ forms as guanine bases which can pair differently (Fig. 1A). As deduced from 1H NMR and CD analysis, this sequence adopts two main intramolecular folded structures, namely hybrid-type and parallel GQs.38 The third loop forms a hairpin junction in both the forms as a result of base-pairing between G20-C26 and C21-G25 (Fig. 1A). Notably, the hairpin structure positively contributes to GQ stability. Therefore, molecular scaffolds that can target the GQ and proximal hairpin simultaneously are envisioned to improve the druggability score of this new target.
 | ||
| Fig. 1 (A) EGFR-272 G-rich sequence folds into a mixture of hybrid and parallel GQ topologies with a hairpin structure in their third loop.38 (B) Mutations (G20 and C21 to T20 and T21) in the EGFR G-rich ON abolishes the formation of the hairpin structure in both the GQ topologies. (C) Chemical structure of the modified nucleoside TFBF-dU (1) and its corresponding phosphoramidite (2) used in the synthesis of labeled ONs. | ||
Formation of GQ structures, and their stability and interaction with small molecule ligands are commonly studied in vitro by CD, UV-thermal melting, fluorescence, NMR and X-ray techniques using unlabelled oligonucleotides (ONs) or probe-labeled ONs.44–47 Alternatively, fluorescent ligands, which show changes in spectral properties upon binding to GQ structures serve as useful sensors.26,48,49 Notably, GQ-specific antibodies and fluorescent ligands have also been developed to visualize GQs in a cellular environment.50–53 However, when multiple GQ species are present, these methods and probes rarely provide useful information on the individual topologies and their structural equilibrium as they fail to distinguishing individual topologies. In this context, single-molecule approaches using force-based (magnetic tweezers and optical tweezers) and fluorescence-based (smFRET) techniques have been somewhat successful,54–57 but these experimentations require a sophisticated instrument setup. Importantly, these methods cannot be easily used to detect lowly populated GQs and extended to cell-based analysis.
Recently, we developed a highly environment-sensitive nucleoside analog by conjugating a fluorobenzofuran moiety at the C5 position of 2′-deoxyuridine (FBF-dU).58 The heterocycle modification imparts fluorescence and endows an 19F NMR label, thereby allowing a two-channel spectroscopic analysis of the GQ structures formed by the human telomeric overhang. While this analog is highly useful, assay conditions require very high concentrations of the labeled oligonucleotides (ONs, ∼200 μM) and is not suitable for detecting lowly populated species coexisting in a dynamic equilibrium. In order to amplify the signal without compromising the sensitivity of the original probe system, we designed a second generation probe by conjugating trifluoromethyl benzofuran at the C5 position of 2′-deoxyuridine (TFBF-dU 1). This probe containing three equivalent 19F atoms significantly increased the detection limit and was used in detecting i-motif (iM) structures formed by model C-rich ONs.59 Given the ability of an EGFR promoter to adopt unique GQ architectures, whose structural polymorphism could be perturbed by environmental conditions (e.g., ionic conditions) and ligand binding, we decided to harness the true potential of TFBF-dU in studying this biologically important system in detail to validate its therapeutic potential.
Here, we report the use of TFBF-dU in probing GQ structures adopted by the wild-type and mutated EGFR promoter region, their population equilibrium and the influence of a hairpin junction in driving GQ formation. TFBF-dU incorporated into EGFR G-rich sequences provides distinct spectral readouts for the individual GQ structures, thereby enabling the quantification of their relative population under different conditions. The probe helps in identifying the preferred GQ topology in the absence and presence of small molecule ligands under intracellular ionic conditions. The utility of the probe in determining the GQ structure adopted by the ON in a cell-like environment is also demonstrated by performing 19F NMR in frog egg lysate and extract. Furthermore, using polymerase stop assay we ascertained the implication of GQs and GQs bound to ligands in the DNA replication process.
Results and discussion
Synthesis of TFBF-dU-labeled ONs to detect GQs
To gain insights into the GQ folding dynamics and interaction of GQ structures with small molecule ligands, we decided to use TFBF-dU 1 for the following reasons. This fluorescent probe containing a 19F label, when incorporated into ON sequences, is minimally perturbing and highly responsive to changes in the microenvironment as minute changes in the conformation and its interaction with neighbouring bases are manifested in the form of changes in fluorescence and 19F chemical shifts.59 100% natural abundance, high sensitivity and absence of fluorine in biological systems make 19F a good label for in-cell NMR analysis.60,61 A notable number of fluorine-labeled probes have been developed to investigate different nucleic acid structures and their interactions with ligands or proteins.62–68 The next important consideration we took into account is the placement of TFBF-dU in GQ-forming sequences. In comparison to G-tetrads, the loop residues that connect G-tetrads exhibit differences in conformation as well as interactions with adjacent bases in different GQ topologies (Fig. 1A). Hence, replacing a loop dT residue with TFBF-dU would have a very minimum impact on the GQ formation and would also provide distinct spectral signatures for different topologies by virtue of its ability to sense microenvironment changes (Fig. 1A). Before incorporation into the EGRF sequence, which forms multiple GQs, we first studied the use of the nucleoside analog by employing a hybrid-type GQ formed by a human telomeric DNA repeat ON sequence (Telo1, Table 1).69,70 For this purpose, the modified ON Telo2 in which one of the loop dTs is replaced with the nucleoside analog was synthesized by the solid-phase ON synthesis method using phosphoramidite 2 (Fig. 1C and Table 1). Similarly, in EGFR ONs 3 and 4, the nucleoside analog 1 was placed in the second loop (T11) and third loop (T27), respectively (Fig. 1A and Table 1). In addition, a mutated ON 5 (G20 and C21 to T) with a modification at T27 was synthesized to probe the role of a hairpin in the formation of GQs (Fig. 1B). All the ONs were purified by PAGE under denaturing conditions, and their purity and identity were confirmed by RP-HPLC and mass analyses, respectively (Fig. S1–S3 and Table S1†).Impact of TFBF-dU labeling on the formation of GQs
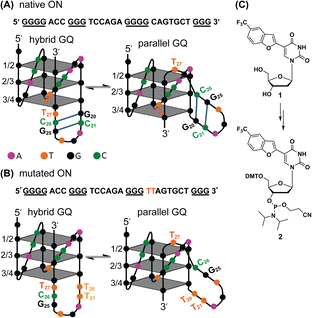
Native Telo1 and modified Telo2 ONs were annealed in 10 mM Tris·HCl buffer (pH 7.4) containing 100 mM KCl and the formation of the hybrid-type GQ structure and its stability were studied by CD and UV-thermal melting analysis. CD profiles of both control and modified ONs gave a positive band at ∼285 nm, a prominent shoulder at ∼269 nm and a negative band at ∼240 nm characteristic of a hybrid-type GQ (Fig. S4A†).71 The Tm values for the ONs were also found to be very similar (Fig. S4B and Table S2†). Furthermore, a thermal difference spectrum (TDS) obtained by subtracting the UV absorption spectrum of the folded state from that of the unfolded state gave a profile corresponding to a GQ structure (Fig. S5†).72,73 Together, these results suggest that the modification has negligible impact on the structure and stability of a Telo GQ. Similarly, EGFR ONs 3–5 and control unmodified ONs 6 and 7 were annealed in 10 mM Tris·HCl buffer (pH 7.4), and the formation of GQ structures with an increase in K+ ion concentration was monitored by CD spectroscopy. In the absence of KCl, EGFR ONs did not show a CD profile corresponding to a GQ structure (Fig. 2). In the presence of K+ ions (100 mM), control ON 6 exhibited a positive peak at 265 nm and a negative peak at 240 nm indicating the formation of a parallel GQ (Fig. 2A and B, dashed lines). In addition, a prominent shoulder at 285 nm representing a hybrid-type GQ form was observed irrespective of the K+ ion concentration (Fig. 2A, B and S6†).38,74 Modified ON 3 gave significantly less intense peaks at +265 nm and −240 nm as compared to the native ON, suggesting that the modification at position T11 hampered GQ formation (Fig. 2A, red solid line). On the other hand, ON 4 displayed a CD profile (peak at +265 nm, −240 nm, and a shoulder at 285 nm) similar to that of control ON 6 (Fig. 2B, red lines). This confirms that nucleoside 1 placed at T27 in ON 4 has negligible impact on the GQ structure formation. Mutations in the stem of the hairpin affected GQ formation of ONs 5 and 7 with a more pronounced effect on the parallel topology (Fig. 2C). Furthermore, we observed that labeled ON 4 and control ON 6 exhibited a similar profile in all K+ ion concentrations tested (50 mM to 200 mM, Fig. S6B†). Although the parallel GQ structure seemed to be higher in amounts as compared to the hybrid form, CD profiles suggested that these structures coexist with no apparent change in the structural equilibrium as a function of KCl concentration. UV-thermal melting analysis indicates that a TFBF-dU modification at T11 and T27 positions only marginally affects the stability of GQs as compared to that of unmodified control ONs (Fig. S7 and Table S2†). However, CD data indicate that a modification at T11 (ON 3) affects the GQ equilibrium, and hence for further analysis native ON 4 and mutated ON 5 containing the modification at T27 were used.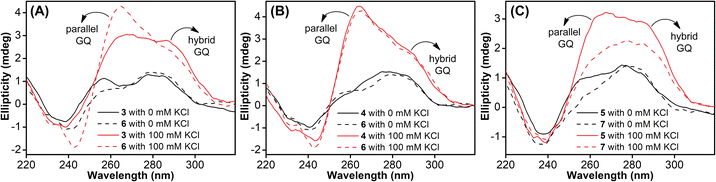 | ||
| Fig. 2 (A, B and C) CD spectra (5 μM) of modified EGFR ONs 3–5 (solid lines) and their control ONs 6 and 7 (dashed lines) under different ionic conditions. See Fig. S6† for CD profiles at different KCl concentrations (50 mM to 200 mM). | ||
Probing the GQ structure of the human telomeric repeat by fluorescence and 19F NMR
TFBF-dU modified Telo2 ON predominantly forms a mixed parallel–antiparallel stranded hybrid-type 2 GQ structure like that of control Telo1 ON (Fig. S8†).69,70 Upon excitation at 330 nm, the GQ form of Telo2 exhibited a fluorescence band with an emission maximum at ∼423 nm (Fig. 3A). When Telo2 was hybridized to its complementary C-rich strand (TeloC), it formed a duplex structure (Fig. S8†), which exhibited a very weak fluorescence band. The observed fluorescence outcome in these structures is due to the difference in the microenvironment around the probe. It is to be noted that several probes display significant quenching in fluorescence due to stacking interaction with neighbouring bases and when located near a guanine base.75,76 TFBF-dU placed at the T12 position upon duplex formation would base pair and partially stack with adjacent bases namely, G11 and T13. As a result of stacking interaction and closeness to the guanine base, the probe shows a quenched emission band. Based on the NMR structure of the hybrid 2 GQ form (PDB: 2JPZ),70 the TFBF modification at the C5-position would be projected into the groove away from the G-tetrad formed by the G11 residue (Fig. S9†). Possibly due to reduced stacking interaction the GQ form shows higher fluorescence intensity compared to the duplex structure.76 Much like the fluorescent component, the fluorine label was also found to be conformation sensitive. TFBF-dU gave a distinct 19F NMR signal for GQ and duplex structures (Fig. 3B). The formation of these structures was also confirmed by an imino proton signal appearing in GQ and duplex regions (Fig. S10†). Collectively, these results indicate that our probe is structurally non-invasive and highly responsive to nucleic acid conformations, and hence, we embarked on using TFBF-dU in the analysis of EGFR GQs. | ||
| Fig. 3 (A) Fluorescence and (B) 19F NMR spectra of telomeric GQ and duplex structures. See the ESI† for experimental conditions. | ||
Probing EGFR GQs by fluorescence
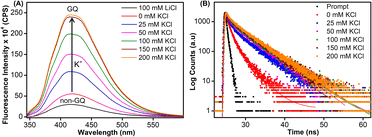
Samples of EGFR ON 4 annealed in Tris·HCl buffer (pH 7.4) containing 100 mM LiCl or varying concentrations of K+ ions were excited at 330 nm, and changes in steady-state emission were recorded. The unstructured ON in the presence of LiCl or in the absence of KCl (see CD profiles, Fig. 2B and S6B†) exhibited a very weak fluorescence band centered at around 419 nm (Fig. 4A). As the concentration of K+ ions was increased (25–200 mM), a progressive increase in the fluorescence intensity with a minor shift in emission maximum was observed as a result of formation of GQs. The fluorescence intensity of ON 4 saturated at 150 mM of KCl (∼8-fold increase as compared to the non-GQ form). In contrast, the CD profile was very similar at all K+ ion concentrations tested suggesting that there is no apparent change in the structural equilibrium (Fig. S6B†). To further probe the structures formed by EGFR ON 4, we performed time-resolved fluorescence analysis (Fig. 4B). In the absence of KCl, three decay components corresponding to lifetimes τ1 = 1.07 ns, τ2 = 2.99 ns, and τ3 = 0.11 ns were observed (Table 2). This is consistent with the literature report suggesting the possibility of three hairpin structures co-existing at zero concentration of KCl.38 At 25 mM KCl, we observed three different components with lifetimes τ1 = 1.44 ns, τ2 = 4.70 ns, and τ3 = 0.14 ns. The individual lifetime components could correspond to random coil, parallel GQ and hybrid-type GQ structures in no particular order. The longest lifetime component (τ2 = 4.70 ns) was found to be significantly more populated as compared to the other two fast decay components (τ1 and τ3). Interestingly, as we increased the concentration of KCl (25 mM to 200 mM), the population of the species corresponding to τ2 increased from 63% to 80% with a concomitant decrease in the population of τ1 and τ3 (Table 2). Lifetime analysis also indicated a saturation point at 150 mM KCl similar to steady-state analysis. It is to be noted that the fluorescence of the free nucleoside analog was not significantly affected by changes in the K+ ion concentration, indicating that the observed fluorescence is due to differences in the microenvironment of the nucleoside analog in different conformations adopted by the ON (Fig. S11A†).| KCl (mM) | τ 1 (ns) | a 1 | τ 2 (ns) | a 2 | τ 3 (ns) | a 3 |
|---|---|---|---|---|---|---|
| a Relative amplitude (a1, a2, and a3) corresponding to lifetime components (τ1, τ2, and τ3). | ||||||
| 0 | 1.07 ± 0.03 | 37% | 2.99 ± 0.05 | 21% | 0.11 ± 0.003 | 42% |
| 25 | 1.44 ± 0.14 | 15% | 4.70 ± 0.10 | 63% | 0.14 ± 0.02 | 22% |
| 50 | 1.29 ± 0.20 | 11% | 4.60 ± 0.12 | 72% | 0.11 ± 0.01 | 17% |
| 100 | 1.35 ± 0.07 | 8% | 4.59 ± 0.10 | 77% | 0.13 ± 0.02 | 14% |
| 150 | 1.27 ± 0.22 | 9% | 4.51 ± 0.02 | 79% | 0.12 ± 0.01 | 13% |
| 200 | 1.38 ± 0.10 | 8% | 4.56 ± 0.001 | 80% | 0.13 ± 0.01 | 13% |
Based on the fluorescence and CD data, and by comparing the structural models predicted for EGFR GQs in an earlier report,38 we infer the following. The CD profile of ON 4 in the presence of KCl (200 mM) shows signatures for parallel and hybrid-type GQs with a ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30.38 This trend was consistent with the fluorescence data, and hence, we assigned the longest lifetime component (τ2 = ∼4.70 ns) to the parallel GQ topology and τ1 and τ3 to either a random coil or hybrid-type GQ structure. The loop residues in a juxtaposed quadruplex and hairpin motif can orient coaxially or orthogonally depending on the GQ topology adopted by the sequence. While a coaxial orientation between the hybrid-type GQ and stem-loop results in continuous base stacking across the two structures, orthogonal interaction between the parallel GQ and hairpin does not involve base stacking between the two components.77,78 To gain further insights, models of hybrid and parallel GQ forms were built by computational analysis. The hairpin structure was found to be coaxially oriented with the hybrid-type GQ core (Fig. S12A†). In this topology, T27 was sandwiched between the hairpin domain and a G-tetrad (Fig. 5A and S12A†). Hence, TFBF-dU placed at this position may experience a similar environment, wherein it would be stacked between C26 of the hairpin and G4 of the bottom tetrad. As a result of stacking interaction, the hybrid GQ structure of ON 4 exhibits a lower fluorescence intensity and a shorter lifetime. In the case of parallel topology, the hairpin structure was found to be orthogonal to the GQ core (Fig. S12B†). In this orientation, T27 was found to partially stack with G17 that formed the top G-tetrad (Fig. 5B and S12B†). TFBF-dU placed at T27 should experience reduced stacking interaction with neighbouring bases, and hence, the parallel form of ON 4 displays higher fluorescence and a longer lifetime. For these reasons, the fluorescence intensity and contribution of τ2 increases as a function of K+ ion concentration due to a progressive shift in the equilibrium towards the formation of the more emissive parallel GQ form.
30.38 This trend was consistent with the fluorescence data, and hence, we assigned the longest lifetime component (τ2 = ∼4.70 ns) to the parallel GQ topology and τ1 and τ3 to either a random coil or hybrid-type GQ structure. The loop residues in a juxtaposed quadruplex and hairpin motif can orient coaxially or orthogonally depending on the GQ topology adopted by the sequence. While a coaxial orientation between the hybrid-type GQ and stem-loop results in continuous base stacking across the two structures, orthogonal interaction between the parallel GQ and hairpin does not involve base stacking between the two components.77,78 To gain further insights, models of hybrid and parallel GQ forms were built by computational analysis. The hairpin structure was found to be coaxially oriented with the hybrid-type GQ core (Fig. S12A†). In this topology, T27 was sandwiched between the hairpin domain and a G-tetrad (Fig. 5A and S12A†). Hence, TFBF-dU placed at this position may experience a similar environment, wherein it would be stacked between C26 of the hairpin and G4 of the bottom tetrad. As a result of stacking interaction, the hybrid GQ structure of ON 4 exhibits a lower fluorescence intensity and a shorter lifetime. In the case of parallel topology, the hairpin structure was found to be orthogonal to the GQ core (Fig. S12B†). In this orientation, T27 was found to partially stack with G17 that formed the top G-tetrad (Fig. 5B and S12B†). TFBF-dU placed at T27 should experience reduced stacking interaction with neighbouring bases, and hence, the parallel form of ON 4 displays higher fluorescence and a longer lifetime. For these reasons, the fluorescence intensity and contribution of τ2 increases as a function of K+ ion concentration due to a progressive shift in the equilibrium towards the formation of the more emissive parallel GQ form.
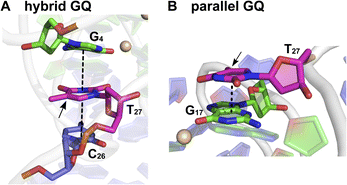 | ||
| Fig. 5 Representative structure of the major cluster of (A) hybrid and (B) parallel GQ topologies. A zoomed-in image representing the stacking interaction of T27 (in magenta) with adjacent bases is shown here. Dashed lines represent the stacking interaction. The C5 position of T27 is shown by an arrow where TFBF heterocycle modification is placed in ON 4. Carbon atoms of G-tetrads are represented in green and all other nucleotides are represented in purple. Nitrogen atoms are represented in blue, oxygen in red, and phosphate in orange. Hydrogen atoms are omitted for clarity. See the ESI† for details. | ||
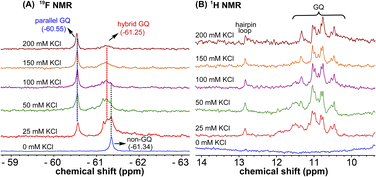
Probing EGFR GQ structural equilibrium by 19F NMR
1H NMR spectra of modified ON 4 and control ON 6 revealed broad peaks for imino protons in the GQ region (10 to 12 ppm) suggesting the presence of multiple GQ topologies in equilibrium. In addition, a peak at 12.8 ppm indicated the presence of a Watson–Crick paired hairpin structure (Fig. 6B and S13†). However, due to only minor changes in the signals, the effect of ionic conditions on the conformational equilibrium could not be deduced from 1H NMR analysis. Rewardingly, the incorporated nucleoside analog 1 gave distinct and resolved 19F signals for different structures. A systematic analysis using 19F signatures obtained under different ionic conditions allowed us to gain a deeper understanding of the GQ structural equilibrium. Notably, upon increasing the concentration of KCl, the peak at −60.55 ppm increased progressively at the expense of other peaks (Fig. 6A). At 200 mM KCl, this peak was found to be predominant. CD and fluorescence data obtained at high concentrations of K+ ions indicate the formation of a parallel GQ as the major form and a hybrid-type GQ as the minor form. So, the peak at −60.55 ppm was judiciously assigned to a parallel GQ topology and the broad peak (minor, −61.25 ppm) to hybrid GQ topology (Fig. 6A). Furthermore, integrating the 19F NMR peaks at different KCl concentrations provided valuable information on the population dynamics of parallel and hybrid GQ topologies. At 25 mM of KCl, the relative population of the parallel GQ was found to be 23%, and the hybrid topology and non-GQ structures accounted for the remaining population. Population of the parallel GQ significantly increased from 23% to 66% with increase in KCl concentration, while the population of hybrid GQ and non-GQ structures decreased to 34%, a result similar to the fluorescence data. A control experiment with nucleoside probe 1 in the presence of different concentrations of KCl did not affect the 19F NMR chemical shift similar to fluorescence (Fig. S11B†). Collectively, these results confirm that 19F signatures exhibited by modified ON 4 is due to differences in the microenvironment of the probe in different structures.Juxtaposed hairpin structure influences the GQ structural equilibrium
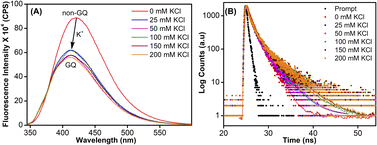
To study the role of the hairpin junction, G20 and C21 nucleotides, involved in base paring, were mutated with Ts (ON 5, Table 1). Mutated ON 5 exhibited contrasting fluorescence compared to native ON 4. The random coil form of ON 5 in the absence of KCl displayed a weak emission band centered at 419 nm (Fig. 7A). Addition of KCl (25 mM) resulted in quenching in fluorescence with a slight blue shift in the emission maximum as a result of formation of GQ structures (CD profiles also suggest the formation of GQs, Fig. 2C). Furthermore, addition of KCl did not affect the fluorescence profile of ON 5. The ON exhibited three different decay components with lifetimes τ1 = 0.95 ns, τ2 = 0.11 ns and τ3 = 2.67 ns in the presence of K+ ions (Fig. 7B and Table 3). Much like steady-state fluorescence, the population of these three components was more or less even and did not change with an increase in the KCl concentration (Table 3). The highest lifetime component (τ3 = 2.67 ns) was assigned to the parallel GQ topology as before, whereas shorter lifetimes could correspond to hybrid-type GQ and random coil structures (Table 3). Collectively, fluorescence at different K+ ion concentrations indicates that mutations in the hairpin domain significantly reduced the population of the more emissive parallel GQ topology and increased the amount of less emissive hybrid GQ topology.| KCl (mM) | τ 1 (ns) | a 1 | τ 2 (ns) | a 2 | τ 3 (ns) | a 3 |
|---|---|---|---|---|---|---|
| a Relative amplitude (a1, a2, and a3) corresponding to lifetime components (τ1, τ2, and τ3). | ||||||
| 0 | 1.69 ± 0.02 | 58% | 0.30 ± 0.01 | 42% | — | — |
| 25 | 0.95 ± 0.02 | 37% | 0.11 ± 0.01 | 34% | 2.67 ± 0.05 | 29% |
| 50 | 1.00 ± 0.09 | 37% | 0.12 ± 0.01 | 34% | 2.93 ± 0.17 | 29% |
| 100 | 1.14 ± 0.001 | 37% | 0.13 ± 0.001 | 36% | 3.39 ± 0.11 | 27% |
| 150 | 1.18 ± 0.03 | 38% | 0.13 ± 0.001 | 35% | 3.74 ± 0.003 | 27% |
| 200 | 1.04 ± 0.04 | 35% | 0.11 ± 0.002 | 34% | 3.61 ± 0.11 | 31% |
1H NMR spectra of mutated ON 5 and its corresponding unmodified ON 7 in the presence of K+ ions revealed broad imino proton peaks between 10 and 12 ppm indicating the formation of more than one GQ structure (Fig. 8B and S14†). Furthermore, the absence of a peak at 12.8 ppm confirmed that the 3rd loop did not adopt a hairpin structure. 19F NMR of 5 in the absence of KCl showed a single peak at −61.44 ppm corresponding to a non-GQ structure (Fig. 8A). In the presence of KCl, ON 5 folded into a mixture of parallel (−60.83 ppm) and hybrid GQ (−61.25 ppm) topologies. Even at high concentrations of K+ ions, the ON formed a random coil structure. The CD profile revealed the presence of a mixture of GQs along with a random coil form (Fig. 2C). As compared to native ON 4, which majorly formed a parallel GQ conformation, mutated ON 5 formed different structures that were more or less evenly populated (e.g., see peaks corresponding to parallel and hybrid-type at 100 mM KCl, Fig. S15†). These observations are also consistent with fluorescence intensity and lifetime data, alluding that both the labels behave similarly in reporting different conformations adopted by the ONs. Furthermore, the existence of a random coil structure along with GQs clearly indicates that mutations in the hairpin domain reduced the overall stability of GQ structures. A ΔTm of 4 °C between ON 4 and ON 5 also supports the above results obtained from fluorescence and NMR studies (Table S2†). Collectively, these results highlight the usefulness of our nucleoside probe in elucidating the role of a juxtaposed hairpin motif on the formation and stability of different GQs of the EGRF promoter region.
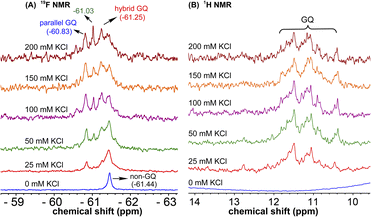 | ||
| Fig. 8 (A) 19F NMR and (B) 1H NMR spectra of mutated modified ON 5 at different KCl concentrations. Peak at −61.03 could not be assigned. | ||
Probing ligand binding by fluorescence and 19F NMR
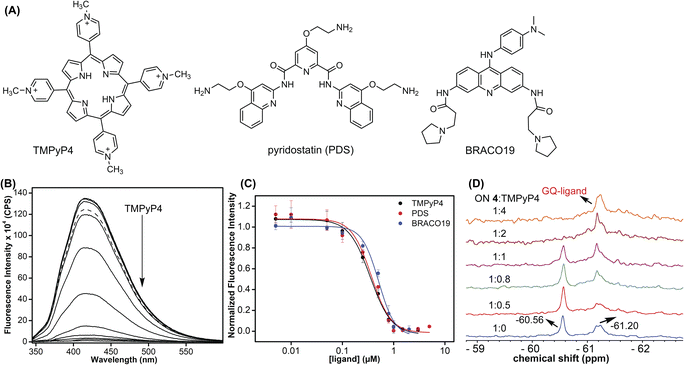
Using the dual-labeled analog, we devised assays to estimate the ligand binding to GQ structures formed by the EGFR G-rich segment. While the fluorescent label enabled the quantification of ligand binding to an ensemble of GQs, the 19F label provided information on the efficiency of ligand interaction with different co-existing GQ structures. Titration of modified ON 4 with increasing concentrations of commercially available GQ binders (TMPyP4, PDS, and BRACO-19, Fig. 9A) resulted in a dose-dependent quenching in the fluorescence intensity with no apparent change in the emission maximum (Fig. 9B and S16†). At low concentrations of ligands, we observed a slight increase in fluorescence intensity. However, the quenching effect was significant as the ligand concertation was increased further, which gave reliable Kd values. The observed fluorescence quenching is likely due to the proximity of the probe to the bound polyaromatic ligands favouring a nonradiative decay pathway.79 TMPyP4 and PDS displayed a similar affinity for an EGFR GQ with Kd values of 0.35 ± 0.02 μM and 0.37 ± 0.04 μM, respectively (Fig. 9C). However, BRACO-19 displayed a slightly lower affinity with a Kd value of 0.51 ± 0.05 μM. Furthermore, the tested ligands showed a cooperative binding with a hill coefficient of nearly 2. Since TMPyP4 binding to GQ structures exhibit a red-shifted absorption band,80 the binding of this ligand to EGFR ON 4 was also evaluated by UV-vis absorption spectroscopy. Notably, UV titration experiments gave a comparable apparent Kd value (0.14 ± 0.01 μM) for TMPyP4 binding to GQs of ON 4 (Fig. S17†).It is not straight forward to determine topology-specific binding of ligands when a sequence forms multiple structures. Ligands could bind either to a specific topology or topologies and stabilize the structures or could transform the structure, thereby redistributing the population.81 The ability of our probe to display distinct 19F signals for different GQs was put to use in detecting topology-specific binding of ligands. Modified ON 4, which folds into a mixture of parallel (−60.56 ppm) and hybrid GQ (−61.20 ppm) topologies at 100 mM KCl was titrated with TMPyP4, PDS, and BRACO-19 (Fig. 9D and S18†). Upon increasing the TMPyP4 concentration, peak intensity corresponding to the parallel GQ (−60.56 ppm) decreased, and furthermore, it disappeared at a higher concentration of TMPyP4 (ON![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand = 1
ligand = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Fig. 9D). A broad peak at around −61.20 ppm increased with increasing concentrations of the ligand. A CD spectrum revealed the presence of multiple conformations (mainly parallel and hybrid forms, Fig. S19A†). Hence, it is speculated that the signal at −61.20 ppm could be associated with different GQ–ligand complexes. In contrast, PDS and BRACO-19 showed affinity for both parallel and hybrid GQ topologies. We observed that with an increase in PDS and BRACO-19 concentration, the peaks corresponding to parallel (−60.55 ppm) and hybrid (−61.21 ppm) GQs reduced, and new peaks corresponding to different GQ–ligand complexes appeared (Fig. S18A and B†), which was confirmed by the presence of CD signatures for both parallel and hybrid GQs (Fig. S19B and C†). Furthermore, ligands are known to interact with GQs via different binding modes (external stacking, intercalation, and groove binding).79 We speculate that multiple peaks observed upon PDS and BRACO-19 binding might be due to different modes of interaction of these ligands with GQs.82,83 From our studies, we find that although ligands (TMPyP4, PDS, and BRACO-19) interact with EGFR GQs with similar Kd values (∼0.35–0.5 μM), their specificity, mode of interaction and influence on structural equilibrium are different.
2, Fig. 9D). A broad peak at around −61.20 ppm increased with increasing concentrations of the ligand. A CD spectrum revealed the presence of multiple conformations (mainly parallel and hybrid forms, Fig. S19A†). Hence, it is speculated that the signal at −61.20 ppm could be associated with different GQ–ligand complexes. In contrast, PDS and BRACO-19 showed affinity for both parallel and hybrid GQ topologies. We observed that with an increase in PDS and BRACO-19 concentration, the peaks corresponding to parallel (−60.55 ppm) and hybrid (−61.21 ppm) GQs reduced, and new peaks corresponding to different GQ–ligand complexes appeared (Fig. S18A and B†), which was confirmed by the presence of CD signatures for both parallel and hybrid GQs (Fig. S19B and C†). Furthermore, ligands are known to interact with GQs via different binding modes (external stacking, intercalation, and groove binding).79 We speculate that multiple peaks observed upon PDS and BRACO-19 binding might be due to different modes of interaction of these ligands with GQs.82,83 From our studies, we find that although ligands (TMPyP4, PDS, and BRACO-19) interact with EGFR GQs with similar Kd values (∼0.35–0.5 μM), their specificity, mode of interaction and influence on structural equilibrium are different.
Probing GQ structures in a model cellular environment by 19F NMR
Next, we studied the utility of the 19F label in detecting the formation of GQ structures in a model mimicking a cellular environment. Lysate and egg extract from frog oocytes serve as good ex vivo models to conduct NMR experiments to study the structure of biopolymers.84,85 To gain a progressive understanding of GQ structures formed by EGFR ON 4 in cell-free and cellular environments, 19F NMR experiments were performed under intraoocyte ionic conditions, oocyte clear lysate and egg extract. The clear lysate was prepared by mechanically crushing the oocytes and the suspension thus obtained was incubated at 95 °C for 10 min to denature the proteins. The lysate was obtained by centrifugation. To obtain an egg extract, the oocytes were crushed and without any further manipulations the mixture was centrifuged. The inter-phase egg extract is considered to provide a molecular crowded environment and also known to maintain most of the biological activity of a cell.8419F NMR of ON 4 recorded in a buffer mimicking intraoocyte ionic conditions (25 mM HEPES pH = 7.5, 110 mM KCl, 10.5 mM NaCl, 130 nM CaCl2, 1 mM MgCl2, 0.1 mM EDTA) gave a spectral profile reminiscent of GQ structures formed in Tris buffer containing 100 mM KCl (Fig. S20†). However, under intraoocyte ionic conditions the population of a hybrid GQ (−61.23 ppm) was found to be discernibly higher compared to that of the parallel form (−60.55 ppm), which was the major conformer in Tris buffer containing 100 mM KCl (compare with Fig. 6A). Interestingly, in both lysate and egg extract the signal corresponding to the hybrid GQ population dominated as the signal for a parallel GQ structure almost disappeared. Based on these data it appears that a hybrid topology of the EGFR ON is supported in a cellular environment. The observed signal broadening in the egg extract is not surprising as inhomogeneity and crowding can reduce the rate of tumbling and augment the relaxation process.84–86 To ascertain if the 19F signals obtained in lysate are associated with the GQ structures formed by the intact ON, the lysate sample was subjected to HPLC and mass analyses. A comparison of HPLC chromatograms of lysate, lysate plus ON 4 (after recording NMR), ON 4 and modified nucleoside analog clearly indicates that ON 4 is stable in the lysate and did not degrade during the NMR acquisition process (Fig. S21†). Furthermore, mass analysis of ON 4 isolated from the lysate sample confirmed the identity of the ON (Fig. S22†). It is important to note that the 1H NMR spectrum of ON 4 gives basic information on GQ formation till the lysate level, but it fails to provide information on the egg extract due to severe line broadening. This is a commonly observed effect and a limitation of 1H NMR use in the cell analysis.GQ formation stalls the processivity of DNA polymerase
Noncanonical DNA structures can cause genomic instability by stalling the DNA replication process.87 To study the effect of GQs, we designed three templates (T1–T3) composed of a wild-type EGFR G-rich sequence, a mutated EGFR sequence or a random non-GQ forming sequence downstream of the polymerization start site (Table 4). A 5′-FAM-labeled primer P1 was annealed with templates T1–T3 in Tris·HCl buffer containing 100 mM KCl, and the replication reaction was performed using Taq DNA polymerase at 37 °C. At different time intervals, individual reaction samples were quenched by adding denaturing gel loading buffer and flashed cooled on a dry ice bath. Reaction products were resolved by PAGE under denaturing conditions and imaged using a fluorescence scanner. Reactions with T3 majorly produced the full-length product in 1 min as a result of the absence of a GQ-forming motif in the template (Fig. 10A, blue bars and Fig. S23C,† gel image). On the other hand, T1 containing the wild-type GQ-forming sequence stalled the polymerase activity and produced largely truncated primer-extended products just above the primer band along with minor amounts of the full-length product (Fig. 10A black bars and Fig. S23A,† gel image). At a reaction time of 5 min, it produced nearly 40% of the full-length product as compared to 60% product in the presence of a control template T3. A mutated EGFR template T2 also yielded stalled products similar to template T1 (Fig. 10A, red bars and Fig. S23B,† gel image). While these results indicate that GQ formation hampers the processivity of Taq polymerase, mutations in the loop hairpin of the GQ has only a minor impact on the polymerization process. This is because mutations in the hairpin per se only destabilized the GQs by ∼4 °C (Table S2†) and do not fully hamper the formation of respective GQs of the EGFR as proved by our CD, fluorescence and 19F NMR studies (vide supra).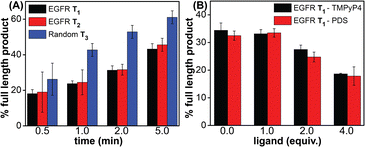 | ||
| Fig. 10 Percentage of the full-length product obtained from Taq polymerase reactions. (A) Reactions performed using templates T1–T3 at different time points. (B) Reactions performed using T1 with increasing concentrations of ligands (TMPyP4 and PDS) at 2 min. For gel images, see Fig. S23B and S24.† | ||
Next, to evaluate the effect of GQ–ligand interaction on the polymerase activity, stop assay was performed in the presence of ligands TMPyP4 and PDS. Since the primer extension with a wild-type EGFR template produced reasonable amounts of the full-length product in 2 min (∼35%), reactions with increasing concentration of ligands were performed for 2 min at 37 °C (Fig. S24†). Upon increasing the concentration of both the ligands, the intensity of the full-length product reduced significantly with a concomitant increase in the stalled products. At 4 equivalents of the ligand, a nearly two-fold decrease in the efficiency of formation of the full-length product was observed (Fig. 10B and S24†). The observed extent of inhibition of the polymerase activity as a result of ligand binding to the GQs is reasonable in the case of the EGFR because regulated EGRF expression is very important for normal cell growth, differentiation and proliferation. Hence, instead of totally abrogating the expression of the EGRF gene, it is more useful if the overexpression of this gene is suppressed so that its normal cellular functions are not affected. In addition, EGRF expression can be more specifically controlled by designing ligands that target hybrid GQ-hairpin conformations adopted by the promoter region.
Conclusions
Given the high number of putative GQ forming sequences in the human genome, non-canonical GQ-hairpin forms exhibited by the EGFR G-rich promoter DNA sequence are perceived as unique hotspots for achieving ligand-binding selectivity. To validate this notion, we utilized a new dual-purpose nucleoside analog probe (TFBF-dU) that enabled a systematic analysis of different GQ topologies formed by native and mutated EGFR promoter ONs by fluorescence and 19F NMR techniques. Distinct spectral signatures exhibited by the labels allowed the (i) detection of different GQ topologies, (ii) quantification of the GQ population equilibrium, and (iii) estimation of ligand binding to different GQs. Notably, the 19F label provided insights into the ligand-induced conformational changes in GQs, thereby suggesting that our probe could be utilized to detect topology-specific binders. Furthermore, a comparative analysis of the readouts from the probe within native and mutated ONs clearly revealed the influence of the hairpin domain in not only affecting the stability of GQs but also redistributing the population of hybrid-type and parallel GQ topologies. Notably, experiments in frog egg lysate and extract suggest that TFBF-dU could serve as a useful probe in detecting nucleic acid structures in a cellular environment. Finally, progressive stalling of the DNA polymerase activity in the absence and then in the presence of ligands, suggests that stabilization of EGFR GQs could be used as a viable approach to repress the oncogenic activity of the EGFR. Taken together, the utility of TFBF-dU as a GQ probe, which could be potentially used in the cellular milieu, and deeper insights gained on the structural and functional aspects of EGFR GQs should facilitate the development of binders that simultaneously target both GQ and hairpin domains for enhanced selectivity and druggability.Data availability
The datasets supporting this article are available in the ESI.†Author contributions
S. G. S. designed and supervised this project. S. Y. K. designed and performed the experiments. S. S. and P. I. P. built computational models of GQ structures. S. M. and J. K. isolated the frog eggs and helped in performing NMR experiments in egg lysate and extract. S. Y. K. and S. G. S. analysed the results and wrote the manuscript in consultation with all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. Y. K. acknowledges IISER Pune and India Alliance for a graduate research fellowship. S. S. acknowledges the Prime Minister's Research Fellows for the PhD fellowship. We thank SpaceTime HPC, IIT Bombay for computing facilities. This work was supported by India Alliance (IA/S/16/1/502360) and SERB (CRG/2022/000284) grants to S. G. S.References
- A. Ullrich, L. Coussens, J. S. Hayflick, T. J. Dull, A. Gray, A. W. Tam, J. Lee, Y. Yarden, T. A. Libermann, J. Schlessinger, J. Downward, E. L. V. Mayes, N. Whittle, M. D. Waterfield and P. H. Seeburg, Nature, 1984, 309, 418–425 CrossRef CAS PubMed.
- P. Blume-Jensen and T. Hunter, Nature, 2001, 411, 355–365 CrossRef CAS PubMed.
- S. Sigismund, D. Avanzato and L. Lanzetti, Mol. Oncol., 2018, 12, 3–20 CrossRef PubMed.
- N. Normanno, A. D. Luca, C. Bianco, L. Strizzi, M. Mancino, M. R. Maiello, A. Carotenuto, G. D. Feo, F. Caponigro and D. S. Salomon, Gene, 2006, 366, 2–16 CrossRef CAS PubMed.
- J.-Y. Han, K. Park, S.-W. Kim, D. H. Lee, H. Y. Kim, H. T. Kim, M. J. Ahn, T. Yun, J. S. Ahn, C. Suh, J.-S. Lee, S. J. Yoon, J. H. Han, J. W. Lee, S. J. Jo and J. S. Lee, J. Clin. Oncol., 2012, 30, 1122–1128 CrossRef CAS PubMed.
- J. R. Goffin and K. Zbuk, Clin. Ther., 2013, 35, 1282–1303 CrossRef CAS PubMed.
- A. J. Cooper, L. V. Sequist and J. J. Lin, Nat. Rev. Clin. Oncol., 2022, 19, 499–514 CrossRef CAS PubMed.
- A. N. Hata, M. J. Niederst, H. L. Archibald, M. Gomez-Caraballo, F. M. Siddiqui, H. E. Mulvey, Y. E. Maruvka, F. Ji, H. E. C. Bhang, V. K. Radhakrishna, G. Siravegna, H. Hu, S. Raoof, E. Lockerman, A. Kalsy, D. Lee, C. L. Keating, D. A. Ruddy, L. J. Damon, A. S. Crystal, C. Costa, Z. Piotrowska, A. Bardelli, A. J. Iafrate, R. I. Sadreyev, F. Stegmeier, G. Getz, L. V. Sequist, A. C. Faber and J. A. Engelman, Nat. Med., 2016, 22, 262–269 CrossRef CAS PubMed.
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC.
- A. Jain, G. Wang and K. M. Vasquez, Biochimie, 2008, 90, 1117–1130 CrossRef CAS PubMed.
- H. A. Assi, M. Garavís, C. González and M. J. Damha, Nucleic Acids Res., 2018, 46, 8038–8056 CrossRef CAS PubMed.
- G. W. Collie and G. N. Parkinson, Chem. Soc. Rev., 2011, 40, 5867–5892 RSC.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- S. Neidle and G. N. Parkinson, Curr. Opin. Struct. Biol., 2003, 13, 275–283 CrossRef CAS PubMed.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- H. Tateishi-Karimata and N. Sugimoto, Nucleic Acids Res., 2021, 49, 7839–7855 CrossRef CAS PubMed.
- C. L. Grand, T. J. Powell, R. B. Nagle, D. J. Bearss, D. Tye, M. Gleason-Guzman and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 6140–6145 CrossRef CAS PubMed.
- A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A, 2002, 99, 11593–11598 CrossRef CAS PubMed.
- J.-H. Tan, T.-M. Ou, J.-Q. Hou, Y.-J. Lu, S.-L. Huang, H.-B. Luo, J.-Y. Wu, Z.-S. Huang, K.-Y. Wong and L.-Q. Gu, J. Med. Chem., 2009, 52, 2825–2835 CrossRef CAS PubMed.
- K. I. E. McLuckie, Z. A. E. Waller, D. A. Sanders, D. Alves, R. Rodriguez, J. Dash, G. J. McKenzie, A. R. Venkitaraman and S. Balasubramanian, J. Am. Chem. Soc., 2011, 133, 2658–2663 CrossRef CAS PubMed.
- M. Micco, G. W. Collie, A. G. Dale, S. A. Ohnmacht, I. Pazitna, M. Gunaratnam, A. P. Reszka and S. Neidle, J. Med. Chem., 2013, 56, 2959–2974 CrossRef CAS PubMed.
- M. Debnath, S. Ghosh, D. Panda, I. Bessi, H. Schwalbe, K. Bhattacharyya and J. Dash, Chem. Sci., 2016, 7, 3279–3285 RSC.
- V. Dhamodharan and P. I. Pradeepkumar, ACS Chem. Biol., 2019, 14, 2102–2114 CAS.
- S. Asamitsu, S. Obata, Z. Yu, T. Bando and H. Sugiyama, Molecules, 2019, 24, 429–457 CrossRef PubMed.
- B. R. Vummidi, J. Alzeer and N. W. Luedtke, ChemBioChem, 2013, 14, 540–558 CrossRef CAS PubMed.
- S. Wu, L. Wang, N. Zhang, Y. Liu, W. Zheng, A. Chang, F. Wang, S. Li and D. Shangguan, Chem.–Eur. J., 2016, 22, 6037–6047 CrossRef CAS PubMed.
- V. Grande, C.-A. Shen, M. Deiana, M. Dudek, J. Olesiak-Banska, K. Matczyszyn and F. Würthner, Chem. Sci., 2018, 9, 8375–8382 RSC.
- Y. Ma, K. Iida, S. Sasaki, T. Hirokawa, B. Heddi, A. T. Phan and K. Nagasawa, Molecules, 2019, 24, 263–277 CrossRef PubMed.
- J.-H. Yuan, W. Shao, S.-B. Chen, Z.-S. Huang and J.-H. Tan, Biochem. Biophys. Res. Commun., 2020, 531, 18–24 CrossRef CAS PubMed.
- T. Vo, S. Oxenford, R. Angell, C. Marchetti, S. A. Ohnmacht, W. D. Wilson and S. Neidle, ACS Med. Chem. Lett., 2020, 11, 991–999 CrossRef CAS PubMed.
- S. Kumar, S. P. P. Pany, S. Sudhakar, S. B. Singh, C. S. Todankar and P. I. Pradeepkumar, Biochemistry, 2022, 61, 2546–2559 CrossRef CAS PubMed.
- K. W. Lim, P. Jenjaroenpun, Z. J. Low, Z. J. Khong, Y. S. Ng, V. A. Kuznetsov and A. T. Phan, Nucleic Acids Res., 2015, 43, 5630–5646 CrossRef CAS PubMed.
- Y. M. Vianney and K. Weisz, Nucleic Acids Res., 2022, 50, 11948–11964 CrossRef PubMed.
- T. Q. N. Nguyen, K. W. Lim and A. T. Phan, Sci. Rep., 2017, 7, 11969–11975 CrossRef PubMed.
- S. Asamitsu, S. Obata, A. T. Phan, K. Hashiya, T. Bando and H. Sugiyama, Chem.–Eur. J., 2018, 24, 4428–4435 CrossRef CAS PubMed.
- M. Yang, S. Carter, S. Parmar, D. D. Bume, D. R. Calabrese, X. Liang, K. Yazdani, M. Xu, Z. Liu, C. J. Thiele and J. S. Schneekloth Jr, Nucleic Acids Res., 2021, 49, 7856–7869 CrossRef CAS PubMed.
- M. L. Greco, A. Kotar, R. Rigo, C. Cristofari, J. Plavec and C. Sissi, Nucleic Acids Res., 2017, 45, 10132–10142 CrossRef CAS PubMed.
- J. T. Grün and H. Schwalbe, Biopolymers, 2022, 113, e23477–e23491 Search PubMed.
- P. A. Rachwal, T. Brown and K. R. Fox, Biochemistry, 2007, 46, 3036–3044 CrossRef CAS PubMed.
- H. You, J. Wu, F. Shao and J. Yan, J. Am. Chem. Soc., 2015, 137, 2424–2427 CrossRef CAS PubMed.
- J. Marquevielle, C. Robert, O. Lagrabette, M. Wahid, A. Bourdoncle, L. E. Xodo, J.-L. Mergny and G. F. Salgado, Nucleic Acids Res., 2020, 48, 9336–9345 CrossRef CAS PubMed.
- R. C. Monsen, L. DeLeeuw, W. L. Dean, R. D. Gray, T. M. Sabo, S. Chakravarthy, J. B. Chaires and J. O. Trent, Nucleic Acids Res., 2020, 48, 5720–5734 CrossRef CAS PubMed.
- M. Cheng, Y. Cheng, J. Hao, G. Jia, J. Zhou, J.-L. Mergny and C. Li, Nucleic Acids Res., 2018, 46, 9264–9275 CrossRef CAS PubMed.
- L. Lacroix, A. Séosse and J.-L. Mergny, Nucleic Acids Res., 2011, 39, e21–e31 CrossRef PubMed.
- K. N. Luu, A. T. Phan, V. Kuryavyi, L. Lacroix and D. J. Patel, J. Am. Chem. Soc., 2006, 128, 9963–9970 CrossRef CAS PubMed.
- G. N. Parkinson, M. P. H. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS PubMed.
- F. Doria, M. Nadai, M. Zuffo, R. Perrone, M. Freccero and S. N. Richter, Chem. Commun., 2017, 53, 2268–2271 RSC.
- Y. V. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- A. Henderson, Y. Wu, Y. C. Huang, E. A. Chavez, J. Platt, F. B. Johnson, R. M. Brosh Jr, D. Sen and P. M. Lansdrop, Nucleic Acids Res., 2014, 42, 860–869 CrossRef CAS PubMed.
- A. Laguerre, K. Hukezalie, P. Winckler, F. Katranji, G. Chanteloup, M. Pirrotta, J.-M. Perrier-Cornet, J. M. Y. Wong and D. Monchaud, J. Am. Chem. Soc., 2015, 137, 8521–8525 CrossRef CAS PubMed.
- A. Shivalingam, M. A. Izquierdo, A. L. Marois, A. Vyšniauskas, K. Suhling, M. K. Kuimova and R. Vilar, Nat. Commun., 2015, 6, 8178–8187 CrossRef CAS PubMed.
- Z. Yu, J. D. Schonhoft, S. Dhakal, R. Bajracharya, R. Hegde, S. Basu and H. Mao, J. Am. Chem. Soc., 2009, 131, 1876–1882 CrossRef CAS PubMed.
- H. You, X. Zeng, Y. Xu, C. J. Lim, A. K. Efremov, A. T. Phan and J. Yan, Nucleic Acids Res., 2014, 42, 8789–8795 CrossRef CAS PubMed.
- L. Ying, J. J. Green, H. Li, D. Klenerman and S. Balasubramanian, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14629–14634 CrossRef CAS PubMed.
- S. L. Noer, S. Preus, D. Gudnason, M. Aznauryan, J.-L. Mergny and V. Birkedal, Nucleic Acids Res., 2016, 44, 464–471 CrossRef CAS PubMed.
- S. Manna, D. Sarkar and S. G. Srivatsan, J. Am. Chem. Soc., 2018, 140, 12622–12633 CrossRef CAS PubMed.
- S. Y. Khatik and S. G. Srivatsan, Bioconjugate Chem., 2022, 33, 1515–1526 CrossRef CAS PubMed.
- H. Chen, S. Viel, F. Ziarelli and L. Peng, Chem. Soc. Rev., 2013, 42, 7971–7982 RSC.
- L. B. T. Pham, A. Costantino, L. Barbieri, V. Calderone, E. Luchinat and L. Banci, J. Am. Chem. Soc., 2023, 145, 1389–1399 CrossRef CAS PubMed.
- M. Himmelstoß, K. Erharter, E. Renard, E. Ennifar, C. Kreutz and R. Micura, Chem. Sci., 2020, 11, 11322–11330 RSC.
- Q. Li, J. Chen, M. Trajkovski, Y. Zhou, C. Fan, K. Lu, P. Tang, X. Su, J. Plavec, Z. Xi and C. Zhou, J. Am. Chem. Soc., 2020, 142, 4739–4748 CrossRef CAS PubMed.
- H.-L. Bao, H.-S. Liu and Y. Xu, Nucleic Acids Res., 2019, 47, 4940–4947 CrossRef CAS PubMed.
- M. R. Baranowski, M. Warminski, J. Jemielity and J. Kowalska, Nucleic Acids Res., 2020, 48, 8209–8224 CrossRef CAS PubMed.
- J. Riedl, R. Pohl, L. Rulíšek and M. Hocek, J. Org. Chem., 2012, 77, 1026–1044 CrossRef CAS PubMed.
- T. Sakamoto, D. Hasegawa and K. Fujimoto, Chem. Commun., 2015, 51, 8749–8752 RSC.
- N. B. Barhate, R. N. Barhate, P. Cekan, G. Drobny and S. Th. Sigurdsson, Org. Lett., 2008, 10, 2745–2747 CrossRef CAS PubMed.
- A. T. Phan, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2006, 34, 5715–5719 CrossRef CAS PubMed.
- J. Dai, M. Carver, C. Punchihewa, R. A. Jones and D. Yang, Nucleic Acids Res., 2007, 35, 4927–4940 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS PubMed.
- J.-L. Mergny, J. Li, L. Lacroix, S. Amrane and J. B. Chaires, Nucleic Acids Res., 2005, 33, e138–e144 CrossRef PubMed.
- L. Evans, A. Kotar, M. Valentini, A. Filloux, S. Jamshidi, J. Plavec, K. M. Rahman and R. Vilar, RSC Chem. Biol., 2023, 4, 94–100 RSC.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- S. O. Kelley and J. K. Barton, Science, 1999, 283, 375–381 CrossRef CAS PubMed.
- S. Doose, H. Neuweiler and M. Sauer, ChemPhysChem, 2009, 10, 1389–1398 CrossRef CAS PubMed.
- K. W. Lim and A. T. Phan, Angew. Chem., Int. Ed., 2013, 52, 8566–8569 CrossRef CAS PubMed.
- K. W. Lim, Z. J. Khong and A. T. Phan, Biochemistry, 2014, 53, 247–257 CrossRef CAS PubMed.
- D. D. Le, M. D. Antonio, L. K. M. Chan and S. Balasubramanian, Chem. Commun., 2015, 51, 8048–8050 RSC.
- C. Wei, G. Jia, J. Yuan, Z. Feng and C. Li, Biochemistry, 2006, 45, 6681–6691 CrossRef CAS PubMed.
- Y. Ma, K. Iida and K. Nagasawa, Biochem. Biophys. Res. Commun., 2020, 531, 3–17 CrossRef CAS PubMed.
- B. Machireddy, H.-J. Sullivan and C. Wu, Molecules, 2019, 24, 1010–1035 CrossRef CAS PubMed.
- L.-Y. Liu, T.-Z. Ma, Y.-L. Zeng, W. Liu and Z.-W. Mao, J. Am. Chem. Soc., 2022, 144, 11878–11887 CrossRef CAS PubMed.
- R. Hänsel, F. Löhr, S. Foldynová-Trantírková, E. Bamberg, L. Trantírek and V. Dötsch, Nucleic Acids Res., 2011, 39, 5768–5775 CrossRef PubMed.
- H.-L. Bao, T. Ishizuka, T. Sakamoto, K. Fujimoto, T. Uechi, N. Kenmochi and Y. Xu, Nucleic Acids Res., 2017, 45, 5501–5511 CrossRef CAS PubMed.
- Y. Ye, X. Liu, Z. Zhang, Q. Wu, B. Jiang, L. Jiang, X. Zhang, M. Liu, G. J. Pielak and C. Li, Chem.–Eur. J., 2013, 19, 12705–12710 CrossRef CAS PubMed.
- L. K. Lerner and J. E. Sale, Genes, 2019, 10, 95–119 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, CD, fluorescence, mass and NMR spectra and gel images. See DOI: https://doi.org/10.1039/d3sc00519d |
| This journal is © The Royal Society of Chemistry 2023 |