Open Access Article
Open Access ArticleEngineering fluorescent protein chromophores with an internal reference for high-fidelity ratiometric G4 imaging in living cells†
Jiao-Na
Han
,
Caijun
Zhong
,
Mingmin
Ge
,
Shi
Kuang
 * and
Zhou
Nie
* and
Zhou
Nie
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology, Hunan University, Changsha, 410082, P. R. China. E-mail: niezhou.hnu@gmail.com; kuangshi@hnu.edu.cn
First published on 29th March 2023
Abstract
G-quadruplexes (G4s) are significant nucleic acid secondary structures formed by guanine-rich sequences. Many single-emission G4 fluorescent probes that are lit up by inhibiting intramolecular rotation have been reported. However, they are non-fluorescent unless structurally rigidified, making them sensitive to other intracellular crowding and confinement environments in the cell, like viscosity. Ratiometric measurements provide built-in self-calibration for signal correction, enabling more sensitive and reliable detection. Herein, we structurally modulate green fluorescent protein (GFP)-like chromophores by integrating the imidazolidinone scaffold of the GFP chromophore and coumarin 6H, obtaining a G4 responsive dual-emission chromophore, called NHCouI. The red emission signal of NHCouI can specifically respond to parallel G4s, while its green emission signal is inert and acts as an internal reference signal. NHCouI–G4 complexes feature high fluorescence quantum yield and excellent anti-photobleaching properties. NHCouI can self-calibrate the signal and avoid viscosity disturbances within the range of major subcellular organelles during G4 imaging in living cells. It is also applied to reflect the difference between apoptosis and ferroptosis via tracking G4s. To the best of our knowledge, NHCouI is the first small molecule G4 probe enabled by internal reference correction capability, opening up new avenues for dual-emission chromophore development and high-fidelity and reliable analysis in G4 imaging research.
Introduction
G-quadruplexes (G4s) are noncanonical nucleic acid secondary structures formed by guanine-rich sequences and contain two or more stacked G-quartets, which are stabilized by Hoogsteen hydrogen bonding.1–5 G4s play significant roles in multiple biological processes, such as DNA replication regulation,6,7 gene expression and stability,8–10 and telomere biology.11,12 Thus, developing a high-fidelity and reliable G4 visualization approach is highly significant.Recently, many single-emission fluorescent probes for G4s have been reported that are lit up by inhibiting intramolecular rotation.13–19 Typically, these probes are non-fluorescent. After binding with G4s, the molecular structure of G4 probes is rigidified, which reduces the nonradiative decay caused by twisted intramolecular charge transfer (TICT) and activates the fluorescence emission.20,21 However, other intracellular crowding and confinement effects like viscosity can also cause molecular rotation inhibition, which results in nonspecific signals in living cell imaging.
Compared with single-emission chromophores, dual-emission chromophores are less affected by target concentration-independent experimental or physiological factors and allow for absolute intensity-independent signal readout via self-calibration using the ratio between two emission wavelengths.22,23 Notably, ratiometric probes with one internal reference signal, in which one signal can specifically respond to the target of interest, and the other one is target-insensitive and acts as an internal reference, offer a fidelity and reliable approach for ratiometric sensing and imaging.26 They allow for precise monitoring of both the target of interest and the distribution of probes in live cells.24–27
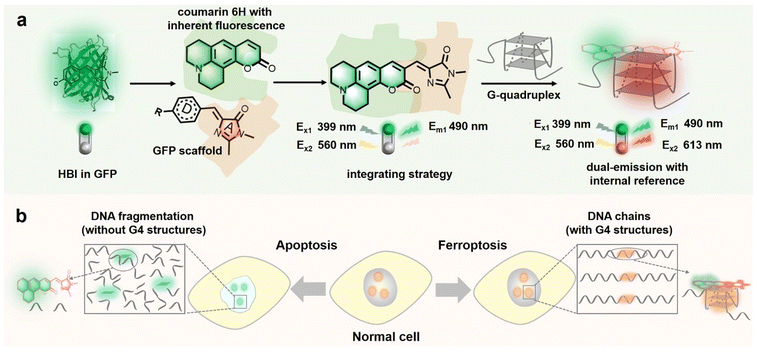
Herein, based on the characteristics of fluorescent protein (FP) chromophores, including (1) a highly modular molecular structure and being easy to integrate,28–31 (2) a rich spectral palette32,33 and (3) responsiveness to G4s,13,14 we rationally design and integrate coumarin 6H having bright fluorescence34,35 with the imidazolinone scaffold of the FP chromophore (Scheme 1) to produce a novel ratiometric G4 probe with an internal reference, named NHCouI, which can precisely probe the G4s and track the distribution of probes in live cells. The intrinsic emission of NHCouI at 490 nm originates from coumarin 6H, acting as an internal reference, while the fluorogenic signal at 613 nm is significantly enhanced upon interaction with G-quarts of G4s. Particularly, benefiting from the internal reference at 490 nm, the ratio (F613/F490) of NHCouI is free from background interference from the local concentration of the chromophore. Also, it can correct the viscosity interference within the viscosity range of major subcellular organelles (50–130 cP).36 Besides, NHCouI exhibits robust topological selectivity towards parallel G4s rather than other topological structures. Furthermore, we apply this integration strategy to other molecular frameworks, demonstrating its universality. Following the unique qualities of NHCouI, it enables self-calibrated imaging of G4s in living cells, remarkably improving imaging fidelity and tracking the distribution of the probe in live cells. Finally, NHCouI is applicable for tracking G4 changes with self-calibration in apoptosis and ferroptosis cell death pathways. Overall, this study reports the first small molecule G4 probe with internal reference correction capability and that can be applied for high-fidelity and reliable analysis in G4 imaging research.
Results and discussion
Constructing the ratiometric probe from the GFP-Like chromophore via step-by-step structural modification
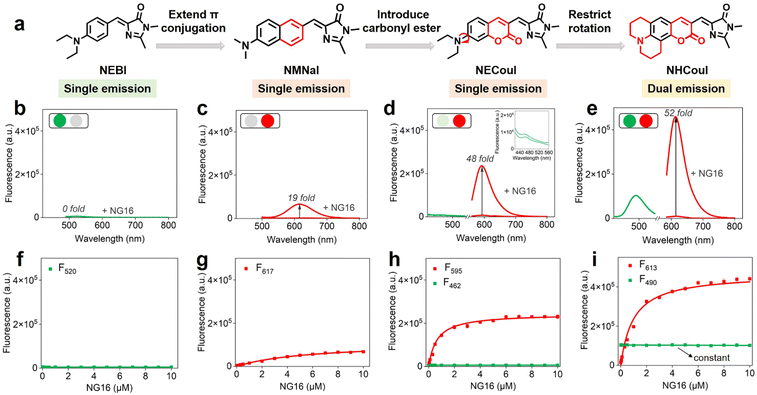
Recently, due to the modularity and tunability of GFP chromophores,37–39 a series of fluorescent protein chromophore derivatives were constructed to light up the G4s by forming the G4 mimics of FPs and dramatically enriching the FPs' spectral palette.15,16 The GFP chromophore-HBI (4-hydroxy benzyl imidazolinone) featured a phenolic moiety, making HBI susceptible to protonation. Thus, we first substituted the phenolic moiety with the diethylamino group, obtaining NEBI (N,N-(diethylamino)benzylidene-imidazolinone) and investigated its response to the G4 structure. Here, we chose NG16 as a template since it can form a typical parallel G4 topology (Fig. S19†). As the NG16 concentration increased (0–10 μM), both absorption and emission exhibited a negligible change at 456 nm (Fig. S20†) and 522 nm (Fig. 1a and b), respectively. These results indicated no significant interaction between NEBI and G4 (Fig. 1f). They might result from the relatively small aromatic ring planar structure of NEBI, which prevented its π–π stacking efficiency with the G4 structure. Subsequently, to investigate the feasibility of extending the π-conjugate system for increasing the π–π stacking efficiency and G4 binding potency, we replaced N,N-diethylaniline with N,N-dimethyl-naphthalene-2-amine. And a small and compact conjugate structure with a donor–acceptor (D–A) molecular configuration, NMNaI, was constructed. As expected, a 19-fold fluorescence enhancement can be observed at 617 nm with the addition of NG16 (Fig. 1c and g). Besides, the NMNaI/NG16 complex displayed a 95 nm red-shifted compared with the NEBI/NG16 complex, owing to the robust electron-donating D group. These results verified our notion that expanding the conjugate plane of the benzene group was beneficial to the π–π stacking of the chromophore with G4 structures and wavelength redshift of the chromophore.To further optimize the photophysical properties, we integrated a coumarin analog, which is a widely used fluorophore known for its bright fluorescence and ease of modification and regulation.40–42 As shown in Fig. 1d and h, N,N-(diethyl) coumarin-imidazolidinone (NECouI) exhibited a significant fluorescence enhancement (48-fold) at 595 nm upon mixing with NG16. Meanwhile, the Kd value for NECouI/NG16 is 1.37 ± 0.18 μM, which was twice as low as NMNaI/NG16 (2.79 ± 0.12 μM), corresponding to a higher binding affinity (Table S1 and S2†). Interestingly, an extremely weak emission at 462 nm, using 399 nm excitation, can be observed, which interested us in constructing the ratiometric probe. For this one emitter with two emitting states system, a possible method to increase the radiative efficacy is to decrease the nonradiative decay by inhibiting the rotator.43 Here, coumarin 6H with a more rigid structure was integrated into the FP chromophore backbone to design NHCouI. As shown in Fig. 1e, i, and S21,† the finally obtained NHCouI displayed a remarkable fluorescence enhancement (52-fold) at 613 nm upon mixing with NG16 (with a Kd value 1.48 ± 0.15 μM). As shown in Fig. S20,† the chromophore-NG16 complexes exhibited excellent absorption bathochromic shifts (25 nm, 15 nm, and 14 nm for NMNaI, NECouI, and NHCouI, respectively), revealing that the spectral properties of the chromophores can change with strong interactions with G4s. Moreover, NHCouI was sensitive to the polarity of the environment, exhibiting positive solvatochromism in organic solvents (Fig. S22†). Surprisingly, NHCouI had a moderate-intensity fluorescence at 490 nm, which kept constant and independent of the NG16 concentration (Fig. 1i). This unique feature inspired us to construct an HBI-like ratiometric G4 probe with internal reference correction capability. Next, we evaluated the ratiometric response properties of NHCouI by dividing the fluorescence intensity at 613 nm by 490 nm. As shown in Fig. S23,† the concentration-dependent titration experiment demonstrated that NHCouI had an excellent linear response in a range of NG16 concentrations (0.03–0.5 μM) with the lowest detection value of 0.0168 μM. Taken together, NHCouI had a constant emission at 490 nm and an NG16 concentration-dependent fluorescence enhancement at 613 nm, which allowed NHCouI to ratiometrically detect G4 structures with superior G4 binding ability and internal reference correction capability.
Evaluating the photophysical properties of NHCouI to bind with G4
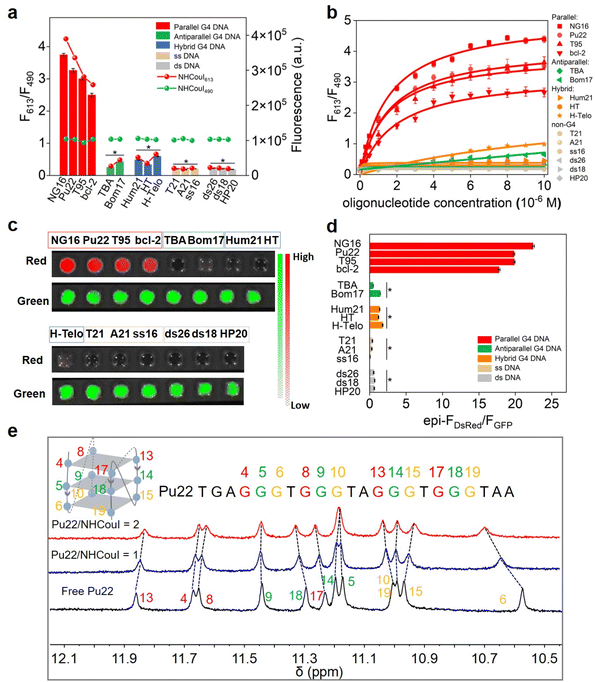
Because the topology of G4s is polymorphic, we initially investigated the fluorescence properties of NHCouI with different G4s and non-G4 oligonucleotides by fluorescence spectroscopy. The sequences of the oligonucleotides used are seen in Table S3,† and the CD spectra of G4 oligonucleotides are characterized in Fig. S24† to verify the topology structures. As shown in Fig. 2a, the addition of parallel G4s (NG16, Pu22, T95, and bcl-2) significantly enhanced the red emission signal (35–52 fold), while the green fluorescence intensity remained unchanged. In contrast, when tested with other G4 topology structures, including anti-parallel G4s (TBA and Bom17), hybrid G4s (Hum21, HT and H-Telo), single-stranded DNAs (T21, A21 and ss16), and double-stranded DNAs (ds26, ds18, and HP20), the green fluorescence remained with the same intensity towards parallel G4s, while red fluorescence signals didn't show enhancement. Besides, the cross-sectional study results between NEBI, NMNaI, NECouI, and NHCouI (Fig. S25†) further highlighted NHCouI's outstanding response performance towards parallel G4s. Next, the titration experiment of parallel G4s with NHCouI gave a Kd value ranging from 1.47 ± 0.15 μM to 2.59 ± 0.19 μM (Fig. 2b and S26 and Table S4†), similar to NMNaI and NECouI (Fig. S27†). Beyond that the NHCouI-NG16 complex had a relatively higher fluorescence quantum yield (Q.Y. = 0.62) than most conventional fluorescent proteins (i.e., TagRFP-1, mRuby and mCherry), which are 0.22–0.41.44–46 It is reported that the Pb2+ can drive G4 from a parallel to antiparallel conformational transition,47 making it an ideal model to investigate the topology specificity of NHCouI. As we expected, the F613/F490 ratio of NHCouI showed a significant decrease with the addition of Pb2+, corresponding to the Pb2+-induced antiparallel G4 conformational transition. Next, the introduction of EDTA, a Pb2+ chelator, which can trap the Pb2+ from the Pb2+–G4 complex and result in the reformulation of parallel G4s by rebinding to K+, rapidly recovered the F613/F490 ratio of NHCouI (Fig. S28†). These results indicated that using the red emission, NHCouI could dynamically recognize parallel G4s with high fluorescence quantum yield (Table S5†), while the green emission is inert to different nucleic acid secondary structures.Next, the influence of NHCouI on the formation of the G4 structures was investigated using circular dichroism (CD) with NG16 as an example. In the presence of KCl, the addition of NHCouI had little effect on the formation of the parallel topology of NG16. The subsequent increase in the amount of NHCouI did not affect its topology. In the absence of KCl, it was difficult for NG16 to form a parallel G4 topology, and NHCouI slightly facilitated the folding of NG16 (Fig. S19†). Besides, the CD melting experiments showed that the melting temperature (Tm) of the NHCouI–NG16 complex was 4.7 °C higher than that of NG16 alone in the presence of KCl, while the Tm value was 2.6 °C higher than in the absence of KCl (Fig. S29†). Compared with a significant increase in ΔTm (26.4 °C) induced by K+, these slight effects (2–4 °C) from NHCouI suggested that the probe hardly interfered with the stability and folding kinetics of the G4 structure, possessing the potential to detect natural G4s in cells. Subsequently, we explored the possibility of NHCouI to achieve direct dual-color visualization of G4 oligonucleotides by using the IVIS Lumina XR small animal optical in vivo imaging system with the GFP and DsRed channel. In this experiment, the green emission at 490 nm was received by the GFP channel, and the red emission at 613 nm was received by the DsRed channel. As shown in Fig. 2c and S30,† the radiance of the GFP channel kept within a range of 1.10 ± 0.15 × 107 p sec−1 cm−2 sr−1 and independent of the concentration gradients of NG16. In contrast, the signal from the DsRed channel increased considerably with the NG16 concentration. These results demonstrated that NHCouI had a selective and rapid visualization effect on parallel G4s. Besides, the visualization ratio results (Fig. 2d) were consistent with those of the fluorescence ratio (F613/F490) of NHCouI responsive oligonucleotides in Fig. 2a, demonstrating the potential of NHCouI as a direct visualization tool for parallel G4s.
Binding mechanism of the NHCouI–G4 complexes and theoretical calculation
First, the binding mode of NHCouI with NG16 in solution was demonstrated to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 through Job's plot analysis (Fig. S31†). To further understand the interaction mechanism between NHCouI and the G4 structure, molecular docking (MD) simulations were performed by using AutoDock, including parallel G4 DNA NG16 (PDB ID: 2LXV), anti-parallel G4 DNA TBA (PDB ID: 5MJX), and hybrid parallel G4 DNA HT (PDB ID: 2GKU) (Fig. S32†). The docking experiments results proved that NHCouI had a coplanar conformation with NG16, but a non-planar conformation with TBA and HT, respectively. These non-planar conformations suggested that NHCouI stacked on the G-quartets of both HT and TBA insufficiently, which can't inhibit the TICT effect and was not favorable for fluorescence generation. More directly, NHCouI bound more strongly to NG16 (−8.01 kcal mol−1) than to TBA (−5.27 kcal mol−1) and HT (−5.78 kcal mol−1), which was attributed to the synergy of these two factors, demonstrating why NHCouI had a higher affinity towards parallel G4s than other topology structures.
1 through Job's plot analysis (Fig. S31†). To further understand the interaction mechanism between NHCouI and the G4 structure, molecular docking (MD) simulations were performed by using AutoDock, including parallel G4 DNA NG16 (PDB ID: 2LXV), anti-parallel G4 DNA TBA (PDB ID: 5MJX), and hybrid parallel G4 DNA HT (PDB ID: 2GKU) (Fig. S32†). The docking experiments results proved that NHCouI had a coplanar conformation with NG16, but a non-planar conformation with TBA and HT, respectively. These non-planar conformations suggested that NHCouI stacked on the G-quartets of both HT and TBA insufficiently, which can't inhibit the TICT effect and was not favorable for fluorescence generation. More directly, NHCouI bound more strongly to NG16 (−8.01 kcal mol−1) than to TBA (−5.27 kcal mol−1) and HT (−5.78 kcal mol−1), which was attributed to the synergy of these two factors, demonstrating why NHCouI had a higher affinity towards parallel G4s than other topology structures.
To obtain more intuitively structure information and understand the dynamic binding mode of the interaction between NHCouI and DNA G4 in solution, the 1H NMR spectrum of the NHCouI–G4 complex was obtained. Herein, a well-characterized Pu22 DNA from the oncogene Myc promoter was utilized (Fig. 2e).48 Pu22-G4-forming guanine imine protons exhibited characteristic chemical shifts in the 10–12 ppm region.49 With increasing concentration of NHCouI, the imine proton peaks of G6, G10, G13, G15, G17 and G19 received significant interference. These guanines come from the end of the Pu22 G-quartet, suggesting that NHCouI may stack onto the terminal side of the G-quartet. The terminal stacking mode of NHCouI was further supported by the competitive inhibition experiment of 5,10,15,20-tetra (N-methyl-4-pyridyl) porphyrin (TMPyP4), a known G4 terminal stacking ligand with an ultra-high binding affinity (Kd = 0.011–0.31 μM for oncogene Myc promoter G4s) (Fig. S33†).50 The experimental results showed that the F613/F490 ratio of the NHCouI–NG16 complex decreased greatly with the increase of TMPyP4 concentration, indicating that TMPyP4 was able to replace the NHCouI molecule in the NHCouI–NG16 complex. Taken together, the results of the molecular docking and NMR analysis supported our solution experiment findings that NHCouI selectively bound with the parallel G4s and further demonstrated that it tended to bind with the terminal G-quartet of parallel G4s in a stacking pattern.
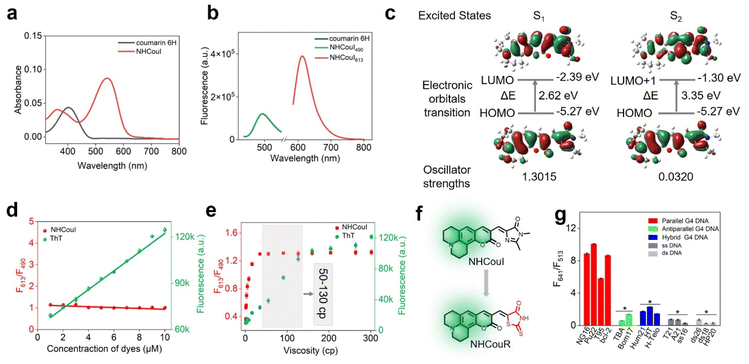
Additionally, theoretical calculations were performed to investigate the mechanism for ratiometric imaging of NHCouI. The molecule geometry of NHCouI was optimized by the density functional theory (DFT) method at the B3LYP/6–31 + g(d) level, and the results showed that the NHCouI molecule existed in a chair conformation in the ground state (Fig. S34 and Table S6†). Two absorption peaks were found (λab = 363 nm, 542 nm), representing two independent excitation pathways for NHCouI. Among them, the absorption at 363 nm was more consistent with the absorption of coumarin 6H (401 nm) (Fig. 3a). The emission of NHCouI at 490 nm was in perfect agreement with the emission of coumarin 6H from its locally excited states, supporting the notion that the internal reference signal at 490 nm was derived from the coumarin 6H moiety (Fig. 3b). Based on the optimized structure, the electronic transitions were further calculated by the time-dependent density functional theory (TDDFT) calculations at the B3LYP/6–31 + g(d) level. As shown in Fig. 3c, S0 → S1 had a higher oscillator strength (f = 1.3015) and lower energy gap (2.62 eV), corresponding to the red emission at 613 nm. And S0 → S2 had a lower oscillator strength (f = 0.0320) and a higher energy gap (3.56 eV), corresponding to the green emission at 490 nm, which was similar to the energy gap of coumarin 6H (Fig. S35†). Additionally, we visualized the frontier orbitals of NHCouI (Fig. 3c). It was found that the HUMO orbitals were mainly concentrated on the coumarin 6H moiety at S0. In contrast, under irradiation, the LUMO orbitals were primarily focused on the imidazolinone ring. It was therefore inferred that NHCouI had a robust intramolecular charge transfer process in the excited state, involving the rationale of the observed inhibiting TICT-induced red emission.
Lighting up G4s using ratiometric probe NHCouI with internal reference correction capability
The fluorescence signal from single-emission probes is not only determined by the concentration of the target of interest but also by the local concentration of the probe and the power of the excitation light source, which can affect the imaging accuracy and the sample contrast. Additionally, while imaging living cells using the probes that are lit by the restriction of intramolecular rotation, the influence of viscosity fluctuations should be considered. Notably, the ratiometric probe with an internal reference can effectively avoid these disadvantages. Thioflavin T (ThT) is a commercially available single-emission probe widely used for G4 imaging.51,52 In this section, we chose ThT as the control group. As shown in Fig. 3d, in the presence of 0.5 μM NG16, the signal from ThT remarkably increased (6.8-folds) with a ThT concentration rise. However, the F613/F490 ratio of NHCouI showed a probe concentration (1–10 μM) independent response, which ranges in value from 1.05 to 1.17. Additionally, in the absence of NG16, the fluorescence of ThT increased with its concentration, while the fluorescence ratio of NHCouI remained almost unchanged (Fig. S36†). It was proved that the NHCouI could detect G4s by the ratio signal with the assistance of the calibration of the internal reference. This property protected NHCouI from the effect of its probe local concentration and made NHCouI linearly respond to the concentration of G4. Next, the excitation laser intensity-dependent experiment was performed using confocal laser scanning microscopy (CLSM) to evaluate the self-calibration capability of NHCouI under different excitation light source intensities. The results showed that while the laser intensity increased 10-fold, the Fred/Fgreen ratio of NHCouI was almost invariable (11.25 ± 1.06), while the fluorescence intensity of ThT increased 7.26 times (Fig. S37†).Next, to evaluate whether the probe will be affected by viscosity, we explored the effect of viscosity on the fluorescence intensity of NHCouI. Here, we utilized the viscosity sensitivity (χ) to describe the relationship between the fluorescence intensity of the probe and viscosity. We measured the fluorescence intensities of these probes in binary mixtures of water and glycerol of known viscosity. We also calculated χ from the plot of the logarithm of the viscosity and the logarithm of the fluorescence intensity based on the Föster–Hoffmann equation (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Φ = χ
Φ = χ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) η + C).53 The viscosity sensitivities of NHCouI at red emission (λem = 613 nm) and ThT (λem = 484 nm) in the system of water and glycerol were 0.46 and 0.71, respectively. Significantly, the viscosity sensitivity of NHCouI at green emission (λem = 490 nm) in the system of water and glycerol was 0.15. This low χ value might explain why the green emission can act as the inter reference. Additionally, the χ value of NHCouI at green emission (λem = 490 nm) was consistent with the viscosity sensitivity of coumarin 6H (χ = 0.13), a more rigid coumarin (Fig. S38†). It was reported that the viscosity of the major organelles within the cell was between 50 and 130 cP.36 Encouragingly, within this range, the F613/F490 ratio of NHCouI kept at 1.29–1.31, which changed less than 2% and was much lower than the response to parallel G4s (F613/F490 = 2.67–4.39). In contrast, the fluorescence intensity of ThT showed a 2.2-fold change in the same viscosity range (Fig. 3e). These data suggested that the NHCouI with internal reference correction capability could detect G4 with a high degree of fidelity and reliability, independent of probe local concentration and viscosity.
η + C).53 The viscosity sensitivities of NHCouI at red emission (λem = 613 nm) and ThT (λem = 484 nm) in the system of water and glycerol were 0.46 and 0.71, respectively. Significantly, the viscosity sensitivity of NHCouI at green emission (λem = 490 nm) in the system of water and glycerol was 0.15. This low χ value might explain why the green emission can act as the inter reference. Additionally, the χ value of NHCouI at green emission (λem = 490 nm) was consistent with the viscosity sensitivity of coumarin 6H (χ = 0.13), a more rigid coumarin (Fig. S38†). It was reported that the viscosity of the major organelles within the cell was between 50 and 130 cP.36 Encouragingly, within this range, the F613/F490 ratio of NHCouI kept at 1.29–1.31, which changed less than 2% and was much lower than the response to parallel G4s (F613/F490 = 2.67–4.39). In contrast, the fluorescence intensity of ThT showed a 2.2-fold change in the same viscosity range (Fig. 3e). These data suggested that the NHCouI with internal reference correction capability could detect G4 with a high degree of fidelity and reliability, independent of probe local concentration and viscosity.
The general applicability of the integration strategy for developing ratiometric G4 probes
Encouraged by these lines of evidence, we assume that the coumarin 6H is crucial to achieving GFP chromophore-like ratiometric G4 probes with internal reference correction capability. Recently, rhodamine derivatives have been developed as protein chromophores, which can selectively bind the target protein and produce intense fluorescence, acting as an activating protein tag.54–56 We thus explored the feasibility of our integration strategy on the rhodamine skeleton, which was named NHCouR. As shown in Fig. 3f, upon the addition of NG16, the fluorescence of NHCouR was strongly enhanced at 641 nm while remaining unchanged at 513 nm, demonstrating the versatility of this “integrating strategy” for constructing ratiometric G4 probes (Fig. S39†). The theoretical calculation of NHCouR further revealed its dual-emission mechanism consistent with NHCouI (Fig. S40 and Table S7†). Besides, the spectrophotometrically studied results showed that NHCouR was still specific toward parallel G4s (Fig. 3g), as well as the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric end-stacking binding mode (Fig. S41†). NHCouR expressed a significant absorption bathochromic shift of 14 nm when adding NG16, suggesting that the spectral properties of NHCouR can change with strong interactions with G4s (Fig. S42†). Subsequently, the effect of viscosity on the fluorescence intensity of NHCouR was also explored. The viscosity sensitivity of NHCouR was 0.05 at 513 nm and 0.68 at 641 nm, respectively, similar to those of NHCouI (Fig. S43†). Due to the difference in the octanol−water partition coefficients of NHCouI (log
1 stoichiometric end-stacking binding mode (Fig. S41†). NHCouR expressed a significant absorption bathochromic shift of 14 nm when adding NG16, suggesting that the spectral properties of NHCouR can change with strong interactions with G4s (Fig. S42†). Subsequently, the effect of viscosity on the fluorescence intensity of NHCouR was also explored. The viscosity sensitivity of NHCouR was 0.05 at 513 nm and 0.68 at 641 nm, respectively, similar to those of NHCouI (Fig. S43†). Due to the difference in the octanol−water partition coefficients of NHCouI (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.26) and NHCouR (log
P = 1.26) and NHCouR (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.14), we hypothesized that it was more challenging for NHCouR with fewer alkyl substituents and nitrogen atoms to penetrate biological membranes than NHCouI. The research on NHCouR hinted at the versatility of the integrating strategy for designing ratiometric G4 probes. Due to the superior properties (i.e., high fluorescence quantum yield and excellent affinity for G4s), the subsequent research efforts have been focused on NHCouI.
P = 1.14), we hypothesized that it was more challenging for NHCouR with fewer alkyl substituents and nitrogen atoms to penetrate biological membranes than NHCouI. The research on NHCouR hinted at the versatility of the integrating strategy for designing ratiometric G4 probes. Due to the superior properties (i.e., high fluorescence quantum yield and excellent affinity for G4s), the subsequent research efforts have been focused on NHCouI.
Tracking G4s in cells viaNHCouI with the self-calibration capability
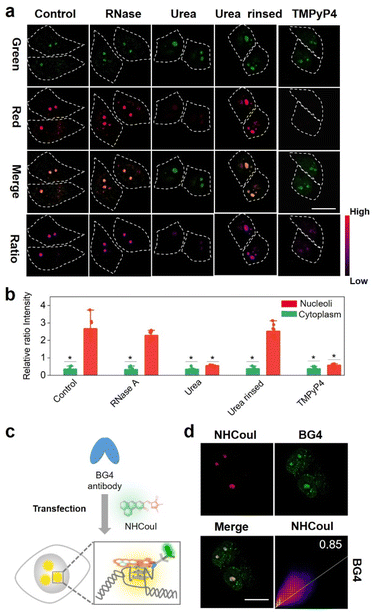
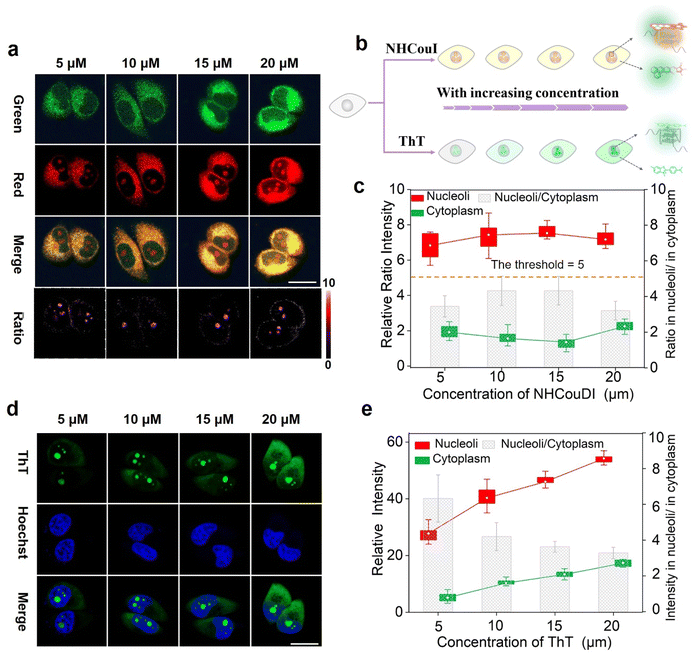
After collecting these promising results from the solution experiments described above, we subsequently determined the specificity of NHCouI for G4 in cells. The initial studies were conducted using fixed HeLa cells and recorded with a confocal laser scanning microscope (CLSM) to determine the feasibility. As shown in Fig. 4a, treatment with NHCouI (5 μM) for 120 min resulted in the appearance of bright green and red fluorescent spots in the nucleoli of fixed HeLa cells, with weak fluorescence in the cytoplasm. The signal intensity and the Fred/Fgreen ratio in the nucleoli had no significant difference before and after RNase treatment, which was 2.38 and 2.29, respectively, approximately 6.8 and 6.5 times higher than those of the cytoplasm (Fig. 4b). Next, fixed HeLa cells were incubated with urea to denature the secondary structure of the nucleic acids.57 In the presence of urea, the red channel fluorescence foci disappeared in the nucleoli, but they recovered after simple washing, while the green channel fluorescence remained unchanged, which supported the selective binding of NHCouI with the secondary structure of nucleic acids. In ligand competition experiments using TMPyP4,50 the red fluorescence foci in the nucleoli were significantly reduced with the addition of TMPyP4, and the green fluorescence remained largely unchanged, demonstrating the staining of G4s by NHCouI. Additionally, we performed immunofluorescence experiments, using the highly specific antibody against G4 structures (BG4), to compare its colocalization with the red fluorescence of NHCouI (Fig. 4c). As shown in Fig. 4d, the red foci of NHCouI colocalized well with the staining of BG4 in the nucleoli of HeLa cells (Pearson's correlation coefficient = 0.85). These results suggested that the red fluorescence of NHCouI can be lit-up by G4-specificity. In contrast, the green fluorescence of NHCouI remained constant after different treatments. Our results significantly demonstrated the potential of NHCouI for ratiometric G4 imaging with internal reference correction capability in cells.The ratiometric fluorescence of NHCouI was not affected by its concentration, avoiding some false positive signals caused by the increase of NHCouI local concentration and enabling high-fidelity imaging of G4s. The photobleaching resistance and biocompatibility were investigated to evaluate its G4 detection ability, which helped track G4 complexes in living cells (Fig. S44 and S45†). Then, we explored whether dual-emission NHCouI could track G4s with self-calibration capability in living cells and trace the probe distribution. The HeLa cells were incubated with 5 μM, 10 μM, 15 μM and 20 μM NHCouI for 2 hours (Fig. 5a and S46†). The fluorescence signal from the red and green channels increased with the NHCouI concentration. Notably, from the inert green channel, we can trace the NHCouI subcellular distribution easily. It was found that NHCouI is evenly distributed in the nucleus or cytoplasm, but there is much less NHCouI in the nucleus than in the cytoplasm. However, the Fred/Fgreen ratio in the nucleoli was 6.68 to 7.44, which was 3.86-fold higher than the ratio in the cytoplasm (ranging from 1.52 to 2.26). Notably, the data allowed us to set the Fred/Fgreen ratio at five as a threshold to eliminate background interference, making it possible to achieve high-precision imaging G4s in living cells (Fig. 5c). In contrast, with the identical ThT concentration incubation for 2 hours, the signals in both the nucleoli and cytoplasm were enhanced by the increase of its concentration, indicating that the signal bound to G4s was affected by its concentration, causing some signal distortion (Fig. 5d and e). These results fully demonstrated the concentration self-calibration properties of NHCouI, which can present the G4 distribution more objectively. The subcellular distribution of NHCouI was performed by comparison with commercially available subcellular organelle localization dyes. Confocal fluorescence images showed that the green fluorescence signals of NHCouI did not colocalize with those from Lyso-Tracker red and Mito-Tracker red, and the red fluorescence signals of NHCouI did not colocalize with those from Lyso-Tracker green and Mito-Tracker green (Fig. S47 and S48†). These results implied that NHCouI was distributed in the cytoplasm without subcellular organelle targeting ability and the green signal is sufficient to act as the internal reference. Besides, the colocalization analysis of coumarin 6H with Lyso-Tracker red and Mito-Tracker red shows its random cytoplasm distribution (Fig. S49†). In general, the findings of these experiments demonstrated that NHCouI could distinguish the targeting G4 signals from the signal of the distribution of the probe in live cells, providing a possible approach for high-fidelity imaging of G4s in cells.
Tracking G4 formation during different cell death pathways
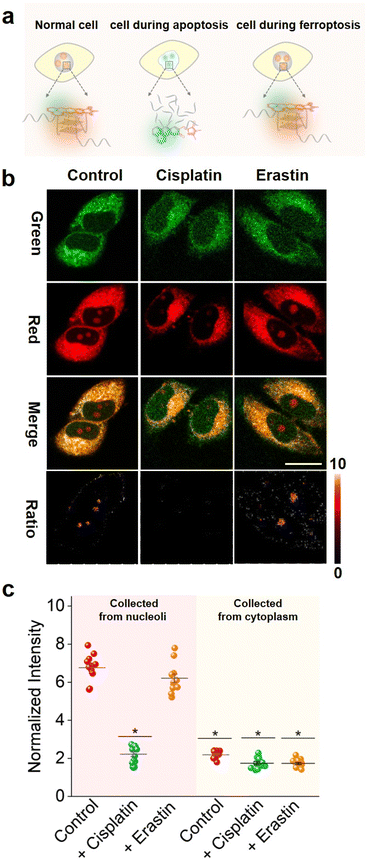
Recently, extensive pathways to regulate cell death have been discovered. Among them apoptosis58,59 and ferroptosis60,61 are two commonly utilized mechanisms. Apoptosis commonly induces the activation of caspases, an aspartic cysteine-specific protease, resulting in the cleavage of caspase-activated DNase inhibitors and subsequent release of DNase, which catalyzes DNA degradation.62 Ferroptosis induces cell death by inhibiting the cystine-glutamate antiporter or directly inactivating glutathione peroxidase 4 (GPX4) activity, resulting in the loss of cellular redox homeostasis.60 Interestingly, apoptosis involves nuclear changes (such as chromatin condensation, nuclear shrinkage, DNA fragmentation and apoptotic body formation), whereas ferroptosis does not. Significantly, DNA fragmentation induced by apoptosis causes endonucleolytic DNA cleavage at multiple internucleosomal sites, which overlaps with the G4-riched sequence.63,64Hence, we hypothesized that it is possible to test the DNA fragmentation by tracing G4s and then distinguish the apoptosis and ferroptosis cell death pathways.65 To this end, we first explored whether NHCouI could track the differences in DNA G4s in the nucleoli during apoptosis and ferroptosis(Fig. 6a). HeLa cells were incubated with cisplatin for 12 hours to induce apoptosis59,66 and then treated with NHCouI (5 μM). As shown in Fig. 6b, after treatment with cisplatin, the Fred/Fgreen ratio signal in the nucleoli was significantly decreased (Fig. 6c and S50†), which was reduced by 67% compared with the control group. Besides, for the cells incubated with ThT, the fluorescence in the nucleoli was also significantly reduced after cisplatin treatment, consistent with the imaging results of NHCouI (Fig. S51†). Next, the G4 formation during ferroptosis was investigated in the HeLa cells that were treated with erastin, a widely used ferroptosis inducer.65 As shown in Fig. 6b, the erastin treatment would not change the Fred/Fgreen ratio signal, consistent with the imaging results of ThT. Additionally, the TUNEL assay is able to identify the DNA breaks formed when DNA fragmentation occurs in the last phase of apoptosis. It relies on the enzyme terminal deoxynucleotide transferase (TdT), which attaches deoxynucleotides to the 3′-hydroxyl terminus of DNA breaks.67 Thus, we performed the TUNEL assay to evaluate the DNA fragmentation in HeLa cells after the aforementioned cell death pathway inducing pretreatments. The results showed that there was no distinct fluorescence change in the negative control group and erastin pretreatment group, while the fluorescence was significantly enhanced in the cisplatin pretreatment group, which was consistent with the results of the positive control group after DNase pretreatment, implying significant DNA fragmentation in HeLa cells after cisplatin pretreatment (Fig. S52†). Compared with the TUNEL assays, NHCouI allowed direct and quick evaluation of DNA fragmentation in living cell nucleoli. Besides, the flow cytometry, utilizing Propidium Iodide (PI) to stain DNA, proved that the DNA damage was more intense during apoptosis and might reduce G4 formation relative to control and ferroptosis-treated cells (Fig. S53†). Overall, this study demonstrated the potential of NHCouI to visualize the difference between apoptosis and ferroptosis via tracking G4s, which could provide a valuable tool for studying the mechanisms underlying different types of cell death.
Conclusions
In summary, we developed the first ratiometric G4s probe with internal reference correction capability, named NHCouI, by integrating the GFP chromophore building block with coumarin 6H. NHCouI had the absolute intensity-independent, viscosity-independent, local probe concentration-independent, and excitation laser power-independent signal readout. It overcame the interference from the viscosity disturbances in major subcellular organelles (50–130 cP) and the probe's uneven subcellular distribution. We have also demonstrated the generalizability of the integration strategy with other molecular backbones. NHCouI exhibits high specificity for parallel G4s than other topological structures. Additionally, NHCouI was applied to track the difference between apoptosis and ferroptosis by imaging G4s for the first time and offered a new perspective for studying nuclear changes during cell death. Given the high modularity of this strategy, it is expected to be used for precise quantification of G4s in cells via further tuning the photophysical properties of this probe. Overall, we envision that the ratiometric probes with internal reference correction capability would afford a promising toolkit for precision imaging of G4s and tracing the probe distribution. This work opens up new avenues for high-fidelity and reliable analysis in G4 imaging research.Data availability
Experimental procedures, compound characterization, NMR spectra, and additional data are located within the ESI.†Author contributions
J.-N. Han, S. Kuang, and N. Zhou proposed the idea, designed the experiments, and revised the manuscript. J.-N. Han carried out experimental work and wrote the manuscript. C. Zhong and M. Ge assisted in the synthesis and characterization of chromophores. All authors contributed to the manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFA0910100), the National Natural Science Foundation of China (22034002, 21725503, and 22204047), and the Fundamental Research Funds for the Central Universities.Notes and references
- M. Gellert, M. N. Lipsett and D. R. Davies, Proc. Natl. Acad. Sci. U. S. A., 1962, 48, 2013–2018 CrossRef CAS PubMed
.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed
.
- Y. Xu, Chem. Soc. Rev., 2011, 40, 2719–2740 RSC
.
- J. Spiegel, S. Adhikari and S. Balasubramanian, Trends Chem., 2020, 2, 123–136 CrossRef CAS PubMed
.
- J. Chen, M. Cheng, P. Stadlbauer, J. Šponer, J.-L. Mergny, H. Ju and J. Zhou, CCS Chem., 2022, 4, 3036–3050 CrossRef CAS
.
- P. Sarkies, C. Reams, L. J. Simpson and J. E. Sale, Mol. Cell, 2010, 40, 703–713 CrossRef CAS PubMed
.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed
.
- R. Hänsel-Hertsch, M. Di Antonio and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2017, 18, 279–284 CrossRef PubMed
.
- X. Liu, S. Cui, Q. Qi, H. Lei, Y. Zhang, W. Shen, F. Fu, T. Tian and X. Zhou, Nucleic Acids Res., 2022, 50, 11387–11400 CrossRef PubMed
.
- T. Chang, W. Li, Z. Ding, S. Cheng, K. Liang, X. Liu, T. Bing and D. Shangguan, Anal. Chem., 2017, 89, 8162–8169 CrossRef CAS PubMed
.
- Z. Tan, J. Tang, Z.-Y. Kan and Y.-H. Hao, Curr. Top. Med. Chem., 2015, 15, 1940–1946 CrossRef CAS PubMed
.
- Y. Sun, C. Zhao, T. Cui, H. Qin, J. Niu, J. Ren and X. Qu, Nucleic Acids Res., 2020, 48, 9986–9994 CrossRef CAS PubMed
.
- G. Feng, C. Luo, H. Yi, L. Yuan, B. Lin, X. Luo, X. Hu, H. Wang, C. Lei and Z. Nie, Nucleic Acids Res., 2017, 45, 10380–10392 CrossRef CAS PubMed
.
- J. Zhang, H. Li, B. Lin, X. Luo, P. Yin, T. Yi, B. Xue, X.-L. Zhang, H. Zhu and Z. Nie, J. Am. Chem. Soc., 2021, 143, 19317–19329 CrossRef CAS PubMed
.
- M. Di Antonio, A. Ponjavic, A. Radzevičius, R. T. Ranasinghe, M. Catalano, X. Zhang, J. Shen, L.-M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed
.
- X.-C. Chen, G.-X. Tang, W.-H. Luo, W. Shao, J. Dai, S.-T. Zeng, Z.-S. Huang, S.-B. Chen and J.-H. Tan, J. Am. Chem. Soc., 2021, 143, 20779–20791 CrossRef CAS PubMed
.
- X.-C. Chen, S.-B. Chen, J. Dai, J.-H. Yuan, T.-M. Ou, Z.-S. Huang and J.-H. Tan, Angew. Chem., Int. Ed., 2018, 57, 4702–4706 CrossRef CAS PubMed
.
- R. Kumar, K. Chand, S. Bhowmik, R. N. Das, S. Bhattacharjee, M. Hedenström and E. Chorell, Nucleic Acids Res., 2020, 48, 1108–1119 CrossRef CAS PubMed
.
- B. Prasad, M. Doimo, M. Andréasson, V. L'Hôte, E. Chorell and S. Wanrooij, Chem. Sci., 2022, 13, 2347–2354 RSC
.
- A. Baldridge, S. R. Samanta, N. Jayaraj, V. Ramamurthy and L. M. Tolbert, J. Am. Chem. Soc., 2011, 133, 712–715 CrossRef CAS PubMed
.
- M. S. Baranov, K. A. Lukyanov, A. O. Borissova, J. Shamir, D. Kosenkov, L. V. Slipchenko, L. M. Tolbert, I. V. Yampolsky and K. M. Solntsev, J. Am. Chem. Soc., 2012, 134, 6025–6032 CrossRef CAS PubMed
.
- T. Peng, G. M. Bonamy, E. Glory-Afshar, D. R. Rines, S. K. Chanda and R. F. Murphy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2944–2949 CrossRef CAS PubMed
.
- Q. Chen, H. Fang, X. Shao, Z. Tian, S. Geng, Y. Zhang, H. Fan, P. Xiang, J. Zhang and X. Tian, Nat. Commun., 2020, 11, 1–10 CrossRef PubMed
.
- J. Li, X. Yin, B. Li, X. Li, Y. Pan, J. Li and Y. Guo, Anal. Chem., 2019, 91, 5354–5361 CrossRef CAS PubMed
.
- X. Mu, F. Wu, R. Wang, Z. Huang, T. Lv, Y. Lu, B. Liu and X. Zhou, Sens. Actuators B: Chem., 2021, 338, 129842 CrossRef CAS
.
- L. Shi, C. Yan, Z. Guo, W. Chi, J. Wei, W. Liu, X. Liu, H. Tian and W.-H. Zhu, Nat. Commun., 2020, 11, 793 CrossRef CAS PubMed
.
- Y. Yao, Y. Zhang, C. Yan, W.-H. Zhu and Z. Guo, Chem. Sci., 2021, 12, 9885–9894 RSC
.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS PubMed
.
- X. Li, J. Wu and S. R. Jaffrey, Angew. Chem., Int. Ed., 2021, 60, 24153–24161 CrossRef CAS PubMed
.
- Y. Liu, C. H. Wolstenholme, G. C. Carter, H. Liu, H. Hu, L. S. Grainger, K. Miao, M. Fares, C. A. Hoelzel, H. P. Yennawar, G. Ning, M. Du, L. Bai, X. Li and X. Zhang, J. Am. Chem. Soc., 2018, 140, 7381–7384 CrossRef CAS PubMed
.
- W. Wan, Y. Huang, Q. Xia, Y. Bai, Y. Chen, W. Jin, M. Wang, D. Shen, H. Lyu, Y. Tang, X. Dong, Z. Gao, Q. Zhao, L. Zhang and Y. Liu, Angew. Chem., Int. Ed., 2021, 60, 11335–11343 CrossRef CAS PubMed
.
- S. Ye, H. Zhang, J. Fei, C. H. Wolstenholme and X. Zhang, Angew. Chem., Int. Ed., 2021, 60, 1339–1346 CrossRef CAS PubMed
.
- C. H. Wolstenholme, H. Hu, S. Ye, B. E. Funk, D. Jain, C.-H. Hsiung, G. Ning, Y. Liu, X. Li and X. Zhang, J. Am. Chem. Soc., 2020, 142, 17515–17523 CrossRef CAS PubMed
.
- M. Tasior, I. Deperasińska, K. Morawska, M. Banasiewicz, O. Vakuliuk, B. Kozankiewicz and D. T. Gryko, Phys. Chem. Chem. Phys., 2014, 16, 18268–18275 RSC
.
- J. L. Vázquez, I. Velazco-Cabral, E. Alvarado-Méndez, M. Trejo-Durán, M. Flores-Alamo, E. Peña-Cabrera, M. A. García-Revilla and M. A. Vázquez, Phys. Chem. Chem. Phys., 2021, 23, 22466–22475 RSC
.
- D. Su, C. L. Teoh, L. Wang, X. Liu and Y.-T. Chang, Chem. Soc. Rev., 2017, 46, 4833–4844 RSC
.
- C. L. Walker, K. A. Lukyanov, I. V. Yampolsky, A. S. Mishin, A. S. Bommarius, A. M. Duraj-Thatte, B. Azizi, L. M. Tolbert and K. M. Solntsev, Curr. Opin. Chem. Biol., 2015, 27, 64–74 CrossRef CAS PubMed
.
- E. A. Rodriguez, R. E. Campbell, J. Y. Lin, M. Z. Lin, A. Miyawaki, A. E. Palmer, X. Shu, J. Zhang and R. Y. Tsien, Trends Biochem. Sci., 2017, 42, 111–129 CrossRef CAS PubMed
.
- W. B. Frommer, M. W. Davidson and R. E. Campbell, Chem. Soc. Rev., 2009, 38, 2833–2841 RSC
.
- D. Cao, Z. Liu, P. Verwilst, S. Koo, P. Jangjili, J. S. Kim and W. Lin, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS PubMed
.
- N. Jiang, J. Fan, F. Xu, X. Peng, H. Mu, J. Wang and X. Xiong, Angew. Chem., Int. Ed., 2015, 54, 2510–2514 CrossRef CAS PubMed
.
- S. S. Matikonda, J. Ivanic, M. Gomez, G. Hammersley and M. J. Schnermann, Chem. Sci., 2020, 11, 7302–7307 RSC
.
- S. K. Behera, S. Y. Park and J. Gierschner, Angew. Chem., Int. Ed., 2021, 60, 22624–22638 CrossRef CAS PubMed
.
- N. C. Shaner, M. Z. Lin, M. R. McKeown, P. A. Steinbach, K. L. Hazelwood, M. W. Davidson and R. Y. Tsien, Nat. Methods, 2008, 5, 545–551 CrossRef CAS PubMed
.
- N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567–1572 CrossRef CAS PubMed
.
- S. Kredel, F. Oswald, K. Nienhaus, K. Deuschle, C. Röcker, M. Wolff, R. Heilker, G. U. Nienhaus and J. Wiedenmann, PLoS One, 2009, 4, e4391 CrossRef PubMed
.
- T. Li, E. Wang and S. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 CrossRef CAS PubMed
.
- J. Dickerhoff, J. Dai and D. Yang, Nucleic Acids Res., 2021, 49, 5905–5915 CrossRef CAS PubMed
.
- J. Dai, M. Carver, L. H. Hurley and D. Yang, J. Am. Chem. Soc., 2011, 133, 17673–17680 CrossRef CAS PubMed
.
- D. D. Le, M. Di Antonio, L. K. M. Chan and S. Balasubramanian, Chem. Commun., 2015, 51, 8048–8050 RSC
.
- S. Zhang, H. Sun, H. Chen, Q. Li, A. Guan, L. Wang, Y. Shi, S. Xu, M. Liu and Y. Tang, Biochim. Biophys. Acta - Gen. Subj., 2018, 1862, 1101–1106 CrossRef CAS PubMed
.
- X. Luo, B. Xue, G. Feng, J. Zhang, B. Lin, P. Zeng, H. Li, H. Yi, X.-L. Zhang, H. Zhu and Z. Nie, J. Am. Chem. Soc., 2019, 141, 5182–5191 CrossRef CAS PubMed
.
- T. Förster and G. Hoffmann, Z. Phys. Chem., 1971, 75, 63–76 CrossRef
.
- M.-A. Plamont, E. Billon-Denis, S. Maurin, C. Gauron, F. M. Pimenta, C. G. Specht, J. Shi, J. Quérard, B. Pan, J. Rossignol, K. Moncoq, N. Morellet, M. Volovitch, E. Lescop, Y. Chen, A. Triller, S. Vriz, T. L. Saux, L. Jullien and A. Gautier, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 497–502 CrossRef CAS PubMed
.
- P. Chen, R. Wang, K. Wang, J.-N. Han, S. Kuang, Z. Nie and Y. Huang, Chem. Sci., 2022, 13, 12187–12197 RSC
.
- K. S. Mineev, S. A. Goncharuk, M. V. Goncharuk, N. V. Povarova, A. I. Sokolov, N. S. Baleeva, A. Y. Smirnov, I. N. Myasnyanko, D. A. Ruchkin, S. Bukhdruker, A. Remeeva, A. Mishin, V. Borshchevskiy, V. Gordeliy, A. S. Arseniev, D. A. Gorbachev, A. S. Gavrikov, A. S. Mishin and M. S. Baranov, Chem. Sci., 2021, 12, 6719–6725 RSC
.
- X. Lu, X. Wu, S. Kuang, C. Lei and Z. Nie, Anal. Chem., 2022, 94, 10283–10290 CrossRef CAS PubMed
.
- E. A. Prokhorova, A. V. Zamaraev, G. S. Kopeina, B. Zhivotovsky and I. N. Lavrik, Cell. Mol. Life Sci., 2015, 72, 4593–4612 CrossRef CAS PubMed
.
- J. Gong, A. Costanzo, H.-Q. Yang, G. Melino, W. G. Kaelin, M. Levrero and J. Y. J. Wang, Nature, 1999, 399, 806–809 CrossRef CAS PubMed
.
- B. R. Stockwell, J. P. F. Angeli, H. Bayir, A. I. Bush, M. Conrad, S. J. Dixon, S. Fulda, S. Gascón, S. K. Hatzios, V. E. Kagan, K. Noel, X. Jiang, A. Linkermann, M. E. Murphy, M. Overholtzer, A. Oyagi, G. C. Pagnussat, J. Park, Q. Ran, C. S. Rosenfeld, K. Salnikow, D. Tang, F. M. Torti, S. V. Torti, S. Toyokuni, K. A. Woerpel and D. D. Zhang, Cell, 2017, 171, 273–285 CrossRef CAS PubMed
.
- J. Yi, A. M. Minikes and X. Jiang, Cell Chem. Biol., 2019, 26, 621–622 CrossRef CAS PubMed
.
- K. Vermeulen, D. R. Van Bockstaele and Z. N. Berneman, Cell Prolif., 2003, 36, 131–149 CrossRef CAS PubMed
.
- C. Liang, X. Zhang, M. Yang and X. Dong, Adv. Mater., 2019, 31, 1904197 CrossRef CAS PubMed
.
- S. Nagata, H. Nagase, K. Kawane, N. Mukae and H. Fukuyama, Cell Death Differ., 2003, 10, 108–116 CrossRef CAS PubMed
.
- K. N. Wang, L. Y. Liu, D. Mao, S. Xu, C. P. Tan, Q. Cao, Z. W. Mao and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 15095–15100 CrossRef CAS PubMed
.
- A. H. Lau, Kidney Int., 1999, 56, 1295–1298 CrossRef CAS PubMed
.
- Y. Gavrieli, Y. Sherman and S. A. Ben-Sasson, J. Cell Biol., 1992, 119, 493–501 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc00022b |
| This journal is © The Royal Society of Chemistry 2023 |