 Open Access Article
Open Access ArticleDevelopment of fluorogenic substrates for colorectal tumor-related neuropeptidases for activity-based diagnosis†
Norimichi
Nagano‡
 a,
Yuki
Ichihashi‡
a,
Toru
Komatsu
*a,
Hiroyuki
Matsuzaki
b,
Keisuke
Hata
b,
Toshiaki
Watanabe
b,
Yoshihiro
Misawa
a,
Misa
Suzuki§
a,
Shingo
Sakamoto§
a,
Yu
Kagami§
a,
Ayumi
Kashiro
c,
Keiko
Takeuchi
c,
Yukihide
Kanemitsu
d,
Hiroki
Ochiai
d,
Rikiya
Watanabe
a,
Yuki
Ichihashi‡
a,
Toru
Komatsu
*a,
Hiroyuki
Matsuzaki
b,
Keisuke
Hata
b,
Toshiaki
Watanabe
b,
Yoshihiro
Misawa
a,
Misa
Suzuki§
a,
Shingo
Sakamoto§
a,
Yu
Kagami§
a,
Ayumi
Kashiro
c,
Keiko
Takeuchi
c,
Yukihide
Kanemitsu
d,
Hiroki
Ochiai
d,
Rikiya
Watanabe
 e,
Kazufumi
Honda
cf and
Yasuteru
Urano
*ag
e,
Kazufumi
Honda
cf and
Yasuteru
Urano
*ag
aGraduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. E-mail: komatsu@g.ecc.u-tokyo.ac.jp; uranokun@m.u-tokyo.ac.jp
bDepartment of Surgical Oncology, Graduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
cInstitute for Advanced Medical Sciences, Nippon Medical School, 1-1-5 Sendagi, Bunkyo-ku, Tokyo 113-0033, Japan
dNational Cancer Center Hospital, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan
eCluster for Pioneering Research, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan
fGraduate School of Medicine, Nippon Medical School, 1-1-5 Sendagi, Bunkyo-ku, Tokyo 113-8602, Japan
gGraduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
First published on 11th April 2023
Abstract
The M3 metalloproteases, neurolysin and THOP1, are neuropeptidases that are expressed in various tissues and metabolize neuropeptides, such as neurotensin. The biological roles of these enzymes are not well characterized, partially because the chemical tools to analyse their activities are not well developed. Here, we developed a fluorogenic substrate probe for neurolysin and thimet oligopeptidase 1 (THOP1), which enabled the analysis of enzymatic activity changes in tissue and plasma samples. In particular, the probe was useful for studying enzyme activities in a single-molecule enzyme assay platform, which can detect enzyme activity with high sensitivity. We detected the activity of neurolysin in plasma samples and revealed higher enzyme activity in the blood samples of patients with colorectal tumor. The result indicated that single-molecule neurolysin activity is a promising candidate for a blood biomarker for colorectal cancer diagnosis.
Introduction
In living systems, there are more than 5000 enzymatic reactions and alterations in enzymatic activity are often tightly connected to pathogenesis. Analysing these disease-related alterations is a powerful source for the detection, understanding, and treatment of diseases.1,2 Fluorogenic substrate probes are molecules that act as substrates for targeted enzymes and report their activities through fluorescence signal changes.3 They are useful tools for studying enzyme activities in physiology and pathology. Especially when they are used in single-molecule enzyme activity analysis, they offer a highly sensitive enzyme analysis methodology to study disease biomarker enzyme activities for diagnosis.4Currently, many enzymes lack selective fluorogenic substrate probes to aid in reporting their activities in complex biological systems. The M3 metalloproteases, neurolysin (EC 3.4.24.16) and thimet oligopeptidase 1 (THOP1, EC 3.4.24.15), are two of those enzymes that do not have specific fluorogenic substrates.5 We have recently identified their altered activity levels in colorectal tumor tissues; therefore, detecting their activity in tissue and blood samples will offer a useful source for activity-based disease diagnosis.6,7 Neurolysin is expressed in various tissues and is responsible for the metabolism of neurotensin, bradykinin, and substance P.8 The biological role of THOP1 is not well characterized; however, it has more than 50% sequence homology with neurolysin.9 Both peptidases are thought to act as neuropeptidases that channel neuropeptide signalling by metabolizing them.
Endopeptidase probes are designed by the fluorescence resonance energy transfer (FRET) mechanism, in which the fluorophore and quencher are attached to both sides of the substrate peptide; thus, cleavage of the linker peptides leads to fluorescence activation. Currently, fluorogenic substrates available for the M3 metalloproteases are limited to those based on the 2-aminobenzoic acid (Abz) fluorophore (λex. = 320 nm and λem. = 420 nm), such as Abz–GFSPFR–Dnp, which is derived from bradykinin;5 however, its short excitation/emission wavelength is not suitable for detecting activity in complex biological systems. The challenge in designing substrate probes for the M3 metalloproteases lies in the unique substrate specificity of the enzyme; substrate recognition is not based on specific amino acid side chains (such as the P1 position) as in most endopeptidases, but on the overall molecular structure. This is due to the presence of a long and deep substrate binding cleft (Fig. 1A).9–12 It is known that unstructured short peptides less than 20 amino acids long are preferable,5 but this knowledge was not sufficient to predict or design good substrates.
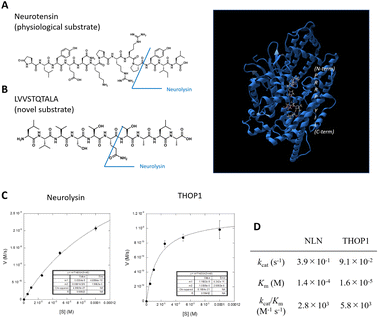 | ||
| Fig. 1 (A) Structure of neurotensin (left), a physiological substrate for the M3 metalloproteases, and structure of neurolysin (right) binding of neurotensin fragments to the active site (data are derived from PDB ID: 5LUZ). The protein backbone is shown as a ribbon diagram (blue), and the substrate is shown as a ball-and-stick model. (B) Structure of novel neurolysin substrate LVVSTQTALA peptide. (C and D) Michaelis–Menten analysis of LVVSTQTALA metabolism by recombinant neurolysin or THOP1. | ||
Results and discussion
We previously identified the altered activity of neurolysin and THOP1 in colorectal tumor tissues via the elevated metabolism of the LVVSTQTALA peptide6,13 (Fig. 1B), and this was because the peptide serves as a good substrate for these neuropeptidases (Fig. 1C, D and S1†).6 We considered its potential as an ideal source for designing novel fluorogenic substrates. We studied the structure–activity relationship of this peptide to attach fluorophore and quencher moieties to both sides of the peptides. We prepared various peptide derivatives and compared their reactivities against neurolysin by monitoring the formation of a common peptide fragment (Fig. S2 and S3†).First, we focused on N-terminal structures (Fig. 2A and S2†), where N-terminal Leu was changed to other amino acids. Leu was first changed to Tyr or acetyl-Tyr, which were both accepted. Changing Leu–Val to Gly–Gly–Gly was also accepted, so the N-terminus was relatively open to modification. However, the modification of hydroxycoumarin, a structurally small fluorophore, at the N-terminus reduced the activity, and change of the fluorophore to fluorescein, a visible light activatable fluorophore, showed an almost complete loss in the activity. We then added a fluorescence quencher Dabcyl (in the form of a linker to be incorporated into the Fmoc solid phase peptide synthesis, named aminopropyl Dabcyl (Apd)) to the N-terminus. This also reduced the reactivity. The N-terminal pocket of neurolysin has a limited length,14 so we removed several N-terminal amino acids to shorten the structure, and the shorter peptide, Apd–STQTALA, was selected as a substrate with acceptable reactivity (approximately 20% reactivity of the original substrate LVVSTQTALA).
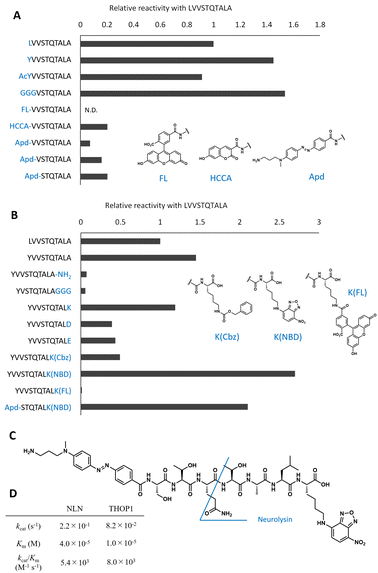 | ||
| Fig. 2 Optimization of peptide sequences to design the neurolysin probe from the original peptide. (A) Optimization of the N-terminal sequences. The relative reaction rate for recombinant neurolysin was calculated from Fig. S2.† (B) Optimization of C-terminal sequences. The relative reaction rate for recombinant neurolysin was calculated from Fig. S3.† (C) Structure of Apd–STQTALK(NBD) and expected cleavage site. (D) Michaelis–Menten analysis of Apd–STQTALK(NBD) metabolism by recombinant neurolysin (NLN) or THOP1. | ||
To optimize the C-terminal structures (Fig. 2B and S3†), we used the YVVSTQTALA peptide as the initial peptide for structural modification. Amidation of C-terminal Ala or the extension of glycine linkers after it significantly reduced the activity; therefore, it seemed inappropriate to modify the main chain of peptides at the C-terminal; hence, we tried to modify the side chain of the C-terminal amino acid. Changing the C-terminal Ala to other amino acids, such as Lys, Asp, Glu, and Lys(Cbz), was accepted, and modifying the C-terminal amino acid to Lys(NBD) enhanced its activity. In the same position, modification of fluorescein was not accepted; therefore, the use of an appropriate fluorophore was indispensable for the design. NBD is a fluorophore that exhibits visible-light absorbance and fluorescence. It was combined with the N-terminal Apd modification, and the overall peptide, NH2–Apd–STQTAL–Lys(NBD)–OH, was established as the best substrate, with the expectation that visible light fluorescence activation occurs upon cleavage of the peptide (Fig. 2C). The probe was sufficiently soluble in aqueous media to up to 50 μM (Fig. S4†). In this concentration range, Michaelis–Menten analysis was performed, and we have determined the kinetic values by non-linear regression; kcat/Km was calculated to be 5.4 × 103 (M−1 s−1) for neurolysin and 8.0 × 103 (M−1 s−1) for THOP1 (Fig. 2D and S5†). The peptide probe reacted with recombinant neurolysin to exhibit a fluorescence increase of more than 15 times (Fig. 3A), and we confirmed that the cleavage site was as designed, between Gln and Thr, for both neurolysin and THOP1 (Fig. 3B).
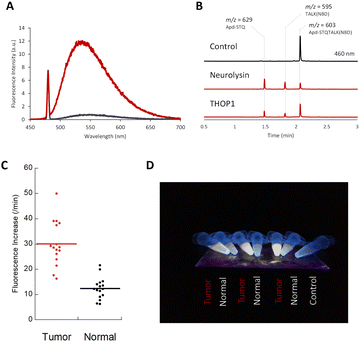 | ||
| Fig. 3 (A) Fluorescence spectra of Apd–STQTALK(NBD) (10 μM) before (black line) and after (red line) incubation with recombinant neurolysin (0.36 μg mL−1), for 4 h in DPBS (pH 7.4) containing CHAPS (0.1%). λex. = 475 nm. (B) LC-MS chromatogram (absorbance = 460 nm) of Apd–STQTALK(NBD) (50 μM) before and after incubation with recombinant neurolysin (0.1 μg mL−1) for 1 h. The representative m/z values observed for each peak are shown. (C) Fluorescence increases of Apd–STQTALK(NBD) (10 μM) after mixing with tumor and non-tumor tissue lysates (0.02 mg mL−1) from a colorectal tumor surgical specimen for 30 min. The detailed data are shown in Fig. S6.† (D) Picture of tissue lysates (tumor and non-tumor tissues), incubated with Apd–STQTALK(NBD) as shown in (C). The fluorescence was observed under irradiation, using a handy UV lamp (365 nm). | ||
We utilized a probe to detect neuropeptidase activity in colorectal tumor tissues. As reported previously, the activities of neurolysin and THOP1 were highly elevated in colorectal tumor tissues compared with that in non-tumor tissues,6,15 and we were able to detect changes in the tissue lysates from surgical specimens using a fluorometric multiwell plate reader (Fig. 3C and S6†) or by the naked eye (Fig. 3D) by fluorescence activation.
As we have successfully detected the activity of neuropeptidases in tumor tissues, we next tried to detect their activities in blood samples. It is often more challenging to detect activity of enzymes in blood samples than in tissues, because of the lower number of molecules present,16 and the activity can be modified by endogenous enzyme inhibitors in circulation.17 We have recently reported that single-molecule enzyme assays are useful for analysing enzyme activities in blood samples because of their high sensitivity and the possibility to discriminately detect the varied activities of enzyme molecules originating from enzyme subtypes and proteoforms.4,15 We applied our probe for single-molecule analysis of neuropeptidase activities in blood samples. The single-molecule enzyme activity analysis is performed by preparing a microdevice that has 105 to 106 reaction chambers with volumes of 10–100 fL and loading the diluted enzyme solution such that theoretically 0 or 1 target enzyme molecule is included in each chamber (Fig. 4A). All chambers are loaded with the fluorogenic probes and are sealed with oil. If the target single-molecule enzyme is included in the chamber, the turnover of the enzyme generates the fluorescent product, which quickly accumulates to reach the detectable concentration in the chamber. Currently, only limited classes of enzymes, phosphatases4,18,19 and glycosidases,20 have been analysed in this platform. We considered that our probe, with visible light excitation/emission and good reactivity with neuropeptidases, was suitable for the assay platform.
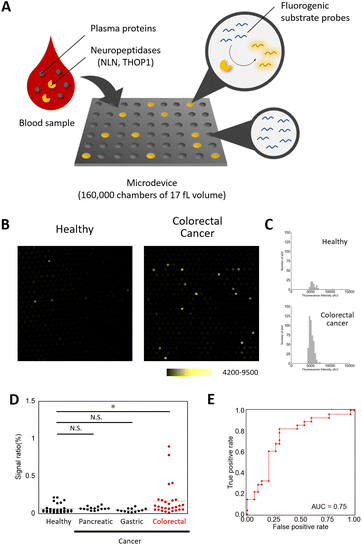 | ||
| Fig. 4 Detection of single-molecule neuropeptidase activity as a candidate for a blood-based diagnostic biomarker for colorectal cancer. (A) Schematic view of the single-molecule enzyme activity assay.4 (B) Fluorescence images of the microdevices for plasma samples of healthy subjects and colorectal tumor subjects (1/500 dilution) with Apd–STQTALK(NBD) (50 μM), after incubation at 37 °C for 4 h. (C) The histograms represent the distribution of the fluorescence increase for each sample. (D) The dot-plot diagram of the signal ratio (microchamber with the fluorescence signal/total microchamber analysed) value in each plasma sample (*P < 0.01, N. S. = not significant, Steel test). (E) Receiver operating characteristic (ROC) curve and AUC value for diagnosis of patients with colorectal cancer. The ROC curve was calculated from the number of detected points for patients with colorectal cancer versus healthy subjects. | ||
The assay was performed by loading the probe mixed with the diluted enzyme in the microdevice, and after sealing with oil and incubation, the fluorescence images were taken to detect and count the wells with the signals.21,22 We first optimized the assay conditions using the probe and the microdevice and determined that the addition of sufficient concentrations of detergents (i.e. >3 mM Triton X-100) was required for the reliable assay (Fig. S7†). Using the optimized assay conditions, the calculated detection limit of the active enzyme was 1.49 × 10−2 fM (Fig. S8†). We then tested if neuropeptidase activities were detectable in the blood samples. In the conventional fluorometric assay using a microplate reader, the activity was barely detectable (Fig. S9 and S10†), presumably because of the low concentration of active enzymes in blood. However, in single-molecule analysis, the activity spots were observed, with the fluorescence intensities comparable to that of recombinant neurolysin (Fig. S11†). Based on the knowledge that, at the tissue level, neuropeptidase activity was elevated in patients of colorectal cancer, we expected that enzyme activity in blood might also show the elevation and could be useful as a biomarker for the detection of colorectal tumors. We analysed the single-molecule activities of neuropeptidases in plasma samples from 30 healthy subjects and 28 patients with colorectal cancer (stages I–IV, Table S1†).18,19 The study was approved by the ethical committee of Nippon Medical School. As a result, we discovered that some patients showed elevated counts of the enzymes in blood samples (Fig. 4B–D and S12–S14, Table S3†), and this change was not observed in patients with other cancers (pancreatic and gastric cancer, Steel test P < 0.01). An area under the curve (AUC) of 0.75 (sensitivity = 0.82, specificity = 0.70, Table S5†) was obtained in discriminating patients with colorectal cancer from healthy subjects (Fig. 4E). The result was validated by preparing a different set of samples from 30 healthy subjects and 27 colorectal tumor patients who are treated by the National Cancer Center (Tokyo, Japan) and repeating the experiments under double-blind conditions (P < 0.05, Mann–Whitney U test, AUC = 0.65, Fig. S15 and Tables S2, S4 and S6†).
While neurolysin is known to function in potentiating immune responses, the contribution of neurolysin to tumor pathogenesis remains unknown.23,24 These results provide useful leads for further research on their involvement in colorectal tumors, such as immunologic activities around tumor areas and in circulation.25–27
Currently, the methods for early detection of colorectal tumors have not been sufficiently developed. The screening test based on a faecal occult has low specificity, and the blood biomarker test using the representative tumor markers CA19-9 and CEA is reported to have limited AUC (AUC = 0.62 for CA19-9 and 0.669 for CEA).28 Endoscopic analysis is more reliable, but it is more invasive and costly and is not easily performed. In some cases, colorectal tumors are detected at an advanced stage, which cannot be completely cured. Here, the development of novel biomarkers for colorectal tumors is highly coveted. Therefore, the blood neurolysin activity discovered in the study has promising potential for diagnosis, especially since neuropeptidase activity can be detected in plasma from a small amount of blood sample (sample requirement was less than 1 μL).
Conclusions
In summary, our study outlines the development of a fluorogenic probe for the M3 metalloproteases, neurolysin and THOP1, which enabled the detection of neuropeptidase activity in tissue lysates and blood samples. In particular, the study indicated that the activity of neurolysin was elevated in the plasma samples of patients with colorectal tumors, thus highlighting the potential of single-molecule neurolysin activity analysis as a promising candidate for a colorectal tumor biomarker.Author contributions
T. K. and Y. U. conceived the project and designed the experiments. N. N., Y. I., Y. M. and T. K. performed the experiments and data analyses. H. M., K. H., T. W., Y. K. and H. O. collected human samples. Y. I. and Y. M. synthesized the compounds. R. W. prepared microdevices. M. S., S. S., Y. K., A. K., K. T., and R. W. directed and supported the experiments. N. N. and T. K. wrote the manuscript.Conflicts of interest
S. S. and Y. K. are employees and shareholders of Cosomil, Inc. M. S. is an employee of Cosomil, Inc. K. H., R. W. and T. Komatsu are advisors and shareholders of Cosomil, Inc.Acknowledgements
We thank Prof. Ueno and Dr Nakamori for collecting the plasma samples. We thank Ms Mayano Minoda for technical support of the experiments. This work was financially supported by MEXT (15H05371, 18H04538, 19H02846, 20H04694, 21A303, and 22H02217 to T. K. and 20H05931 to R. W.), JST (PRESTO (13414915), PRESTO Network (17949814), CREST (19204926) to T. Komatsu and R. W., and START (20353017) to T. K., K. H., and R. W.), and AMED (FORCE (22581634) to T. K., K. H., and R. W., PRIME to R. W., P-CREATE (22ama221213h0001 to T. K., 22ama221401h0001 to T. K. and K. H., and 18cm0106403h0003 to K. H.), and CREST to Y. U.). The blood samples were collected with support of Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor and Welfare, and Health and Labor Sciences Research Grants from the Ministry of Health. T. K. received support from the Naito Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Chugai Foundation for Innovative Drug Discovery Science, and the MSD Life Science Foundation.Notes and references
- T. Komatsu and Y. Urano, J. Biochem., 2020, 167, 139–149 CAS.
- A. Saghatelian and B. F. Cravatt, Nat. Chem. Biol., 2005, 1(3), 130–142 CrossRef CAS PubMed.
- T. Komatsu and Y. Urano, Anal. Sci., 2015, 31, 257–265 CrossRef CAS PubMed.
- S. Sakamoto, T. Komatsu, R. Watanabe, Y. Zhang, T. Inoue, M. Kawaguchi, H. Nakagawa, T. Ueno, T. Okusaka, K. Honda, H. Noji and Y. Urano, Sci. Adv., 2020, 6, eaay0888 CrossRef CAS PubMed.
- V. Oliveira, M. Campos, R. L. Melo, E. S. Ferro, A. C. M. Camargo, M. A. Juliano and L. Juliano, Biochemistry, 2001, 40, 4417–4425 CrossRef CAS PubMed.
- J. Onagi, T. Komatsu, Y. Ichihashi, Y. Kuriki, M. Kamiya, T. Terai, T. Ueno, K. Hanaoka, H. Matsuzaki, K. Hata, T. Watanabe, T. Nagano and Y. Urano, J. Am. Chem. Soc., 2017, 139, 3465–3472 CrossRef CAS PubMed.
- Y. Ichihashi, T. Komatsu, E. Kyo, H. Matsuzaki, K. Hata, T. Watanabe, T. Ueno, K. Hanaoka and Y. Urano, Anal. Chem., 2019, 91, 11497–11501 CrossRef CAS PubMed.
- A. J. Al-Ahmad, I. Pervaiz and V. T. Karamyan, J. Neuroendocrinol., 2021, 33, e12931 CrossRef CAS PubMed.
- K. Ray, C. S. Hines and D. W. Rodgers, Protein Sci., 2002, 11, 2237 CrossRef CAS PubMed.
- P. F. Teixeira, G. Masuyer, C. M. Pinho, R. M. M. Branca, B. Kmiec, C. Wallin, S. K. T. S. Wärmländer, R. P. A. Berntsson, M. Ankarcrona, A. Gräslund, J. Lehtiö, P. Stenmark and E. Glaser, J. Mol. Biol., 2018, 430, 348–362 CrossRef CAS PubMed.
- E. J. Lim, S. Sampath, J. Coll-Rodriguez, J. Schmidt, K. Ray and D. W. Rodgers, Journal of, Biol. Chem., 2007, 282, 9722–9732 CrossRef CAS PubMed.
- C. K. Brown, K. Madauss, W. Lian, M. R. Beck, W. D. Tolbert and D. W. Rodgers, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 3127 CrossRef CAS PubMed.
- M. Polanski and N. L. Anderson, Biomarker Insights, 2006, 1, 1 CrossRef.
- C. K. Brown, K. Madauss, W. Lian, M. R. Beck, W. D. Tolbert and D. W. Rodgers, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 3127 CrossRef CAS PubMed.
- P. Mogalisetti and D. R. Walt, Methods Enzymol., 2016, 581, 541–560 CAS.
- M. Polanski and N. L. Anderson, Biomarker Insights, 2006, 1, 1 CrossRef.
- P. G. W. Gettins, Chem. Rev., 2002, 102, 4751–4804 CrossRef CAS PubMed.
- K. Honda, T. Okusaka, K. Felix, S. Nakamori, N. Sata, H. Nagai, T. Ioka, A. Tsuchida, T. Shimahara, M. Shimahara, Y. Yasunami, H. Kuwabara, T. Sakuma, Y. Otsuka, N. Ota, M. Shitashige, T. Kosuge, M. W. Büchler and T. Yamada, PLoS One, 2012, 7, e46908 CrossRef CAS PubMed.
- K. Honda, Y. Hayashida, T. Umaki, T. Okusaka, T. Kosuge, S. Kikuchi, M. Endo, A. Tsuchida, T. Aoki, T. Itoi, F. Moriyasu, S. Hirohashi and T. Yamada, Cancer Res., 2005, 65, 10613–10622 CrossRef CAS PubMed.
- Y. Rondelez, G. Tresset, K. v. Tabata, H. Arata, H. Fujita, S. Takeuchi and H. Noji, Nat. Biotechnol., 2005, 23, 361–365 CrossRef CAS PubMed.
- Y. Rondelez, G. Tresset, K. v. Tabata, H. Arata, H. Fujita, S. Takeuchi and H. Noji, Nat. Biotechnol., 2005, 23(3), 361–365 CrossRef CAS PubMed.
- R. Watanabe, N. Soga, D. Fujita, K. v. Tabata, L. Yamauchi, S. H. Kim, D. Asanuma, M. Kamiya, Y. Urano, H. Suga and H. Noji, Nat. Commun., 2014, 5(1), 1–8 Search PubMed.
- R. Goldman, Z. Bar-Shavit and D. Romeo, FEBS Lett., 1983, 159, 63–67 CrossRef CAS PubMed.
- K. Kalafatakis and K. Triantafyllou, Regul. Pept., 2011, 170, 7–17 CrossRef CAS PubMed.
- B. Magalhães, F. Trindade, A. S. Barros, J. Klein, F. Amado, R. Ferreira and R. Vitorino, Proteomics, 2018, 18, e1800187 CrossRef PubMed.
- D. M. Cavalcanti, L. M. Castro, J. C. Rosa Neto, M. Seelaender, R. X. Neves, V. Oliveira, F. L. Forti, L. K. Iwai, F. C. Gozzo, M. Todiras, I. Schadock, C. C. Barros, M. Bader and E. S. Ferro, J. Biol. Chem., 2014, 289, 15426–15440 CrossRef CAS PubMed.
- L. M. Castro, D. M. Cavalcanti, C. B. Araujo, V. Rioli, M. Y. Icimoto, F. C. Gozzo, M. Juliano, L. Juliano, V. Oliveira and E. S. Ferro, J. Proteomics, 2014, 111, 238–248 CrossRef CAS PubMed.
- Y. Yoshioka, N. Kosaka, Y. Konishi, H. Ohta, H. Okamoto, H. Sonoda, R. Nonaka, H. Yamamoto, H. Ishii, M. Mori, K. Furuta, T. Nakajima, H. Hayashi, H. Sugisaki, H. Higashimoto, T. Kato, F. Takeshita and T. Ochiya, Nat. Commun., 2014, 5(1), 1–8 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc07029d |
| ‡ These authors contributed equally. |
| § Present address: Cosomil, Inc., Entrepreneur Lab, South Clinical Research Bldg., 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8485, Japan. |
| This journal is © The Royal Society of Chemistry 2023 |
