Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Diagnostics and analysis of SARS-CoV-2: current status, recent advances, challenges and perspectives
Tao
Dong
ab,
Mingyang
Wang
a,
Junchong
Liu
a,
Pengxin
Ma
a,
Shuang
Pang
a,
Wanjian
Liu
c and
Aihua
Liu
 *a
*a
aInstitute for Chemical Biology & Biosensing, College of Life Sciences, Qingdao University, 308 Ningxia Road, Qingdao 266071, China. E-mail: liuah@qdu.edu.cn
bSchool of Pharmacy, Medical College, Qingdao University, 308 Ningxia Road, Qingdao 266071, China
cQingdao Hightop Biotech Co., Ltd, 369 Hedong Road, Hi-tech Industrial Development Zone, Qingdao 266112, China
First published on 3rd May 2023
Abstract
The disastrous spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has induced severe public healthcare issues and weakened the global economy significantly. Although SARS-CoV-2 infection is not as fatal as the initial outbreak, many infected victims suffer from long COVID. Therefore, rapid and large-scale testing is critical in managing patients and alleviating its transmission. Herein, we review the recent advances in techniques to detect SARS-CoV-2. The sensing principles are detailed together with their application domains and analytical performances. In addition, the advantages and limits of each method are discussed and analyzed. Besides molecular diagnostics and antigen and antibody tests, we also review neutralizing antibodies and emerging SARS-CoV-2 variants. Further, the characteristics of the mutational locations in the different variants with epidemiological features are summarized. Finally, the challenges and possible strategies are prospected to develop new assays to meet different diagnostic needs. Thus, this comprehensive and systematic review of SARS-CoV-2 detection technologies may provide insightful guidance and direction for developing tools for the diagnosis and analysis of SARS-CoV-2 to support public healthcare and effective long-term pandemic management and control.
1. Introduction
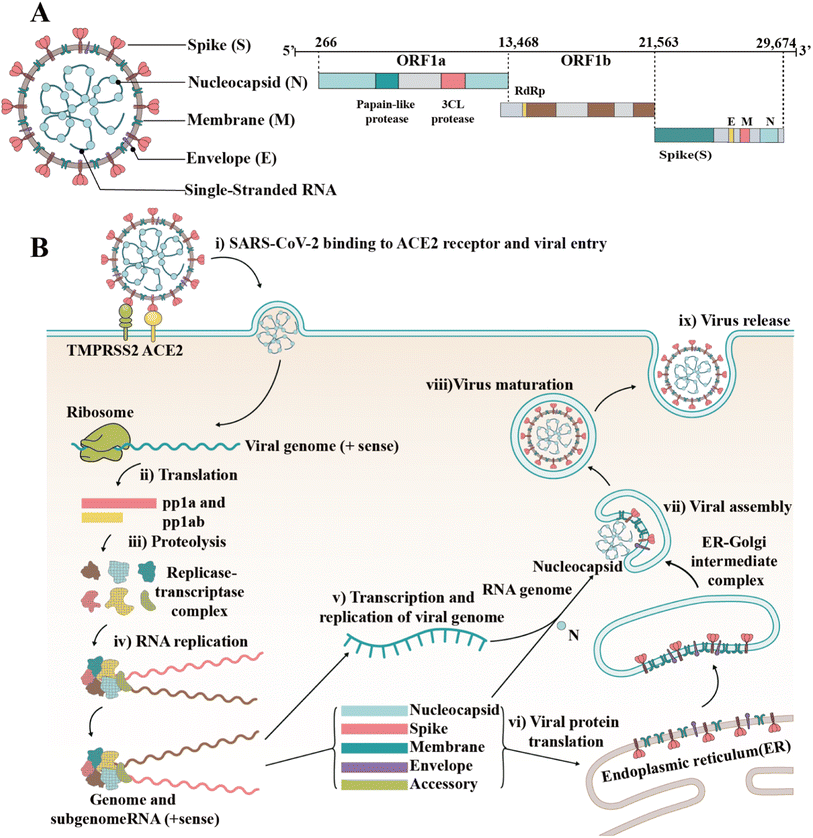
Since the outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in December 2019, this virus has spread rapidly around the world.1,2 Consequently, the rampant COVID-19 has induced severe public health problems, threatening the lives and security of people around the world and putting stress on national healthcare systems. As of March 2023, over 6.8 million people died of COVID-19 globally, according to a report by the World Health Organization (WHO). In addition, SARS-CoV-2 shows the tendency of long-term coexistence with humans and continues to mutate, generating new variants and subvariants, which induce enhanced immune escape and rapid transmission, according to the reports by the US Centers for Disease Control and Prevention (CDC) (https://www.cdc.gov/). The shutdown and delayed resumption of production caused by the epidemic have seriously weakened the global economy and led to various social “after-effects” (reduced social workforce and enlarged wealth gap).SARS-CoV-2 is an enveloped single-stranded RNA virus.3,4 Its genome length is 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 nucleotides (nt) (GenBank number MN908947). The gene fragments express the structural and nonstructural proteins, where the spike (S), envelope (E), membrane (M) and nucleocapsid (N) genes encode the structural proteins of the S protein (SP), E protein (EP), M protein (MP), and N protein (NP), respectively, while the open reading frame (ORF) region encodes nonstructural proteins, such as 3-chymotrypsin-like protease, papain-like protease and RNA-dependent RNA polymerase (RdRP) (Fig. 1A).5,6 As a type I fusion protein, SP plays a crucial role in the process of virus infection and pathogenesis.7 SP consists of subunit S1 and subunit S2, of which subunit S1 is composed of the N-terminal domain (NTD) and receptor-binding domain (RBD), while subunit S2 mediates the fusion between the virus and the cell membrane. The RBD of the S protein can bind to the cell receptor of angiotensin-converting enzyme 2 (ACE2), which is the key to the viral invasion of the body.8,9 When S proteins bind to the receptor, transmembrane serine protease 2 (TMPRSS2), located on the host cell membrane, facilitates virus entry in the cell by activating S proteins.10 Once the virus enters the cell, it releases its RNA, which begins to replicate in the host cell. Replication and transcription of the viral RNA genome occur through protein cleavage and assembly of the replicase–transcriptase complex. After replication of viral RNA, the synthesis of structural proteins, assembly and packaging in the host cell, the viral particles are released (Fig. 1B).11,12
881 nucleotides (nt) (GenBank number MN908947). The gene fragments express the structural and nonstructural proteins, where the spike (S), envelope (E), membrane (M) and nucleocapsid (N) genes encode the structural proteins of the S protein (SP), E protein (EP), M protein (MP), and N protein (NP), respectively, while the open reading frame (ORF) region encodes nonstructural proteins, such as 3-chymotrypsin-like protease, papain-like protease and RNA-dependent RNA polymerase (RdRP) (Fig. 1A).5,6 As a type I fusion protein, SP plays a crucial role in the process of virus infection and pathogenesis.7 SP consists of subunit S1 and subunit S2, of which subunit S1 is composed of the N-terminal domain (NTD) and receptor-binding domain (RBD), while subunit S2 mediates the fusion between the virus and the cell membrane. The RBD of the S protein can bind to the cell receptor of angiotensin-converting enzyme 2 (ACE2), which is the key to the viral invasion of the body.8,9 When S proteins bind to the receptor, transmembrane serine protease 2 (TMPRSS2), located on the host cell membrane, facilitates virus entry in the cell by activating S proteins.10 Once the virus enters the cell, it releases its RNA, which begins to replicate in the host cell. Replication and transcription of the viral RNA genome occur through protein cleavage and assembly of the replicase–transcriptase complex. After replication of viral RNA, the synthesis of structural proteins, assembly and packaging in the host cell, the viral particles are released (Fig. 1B).11,12
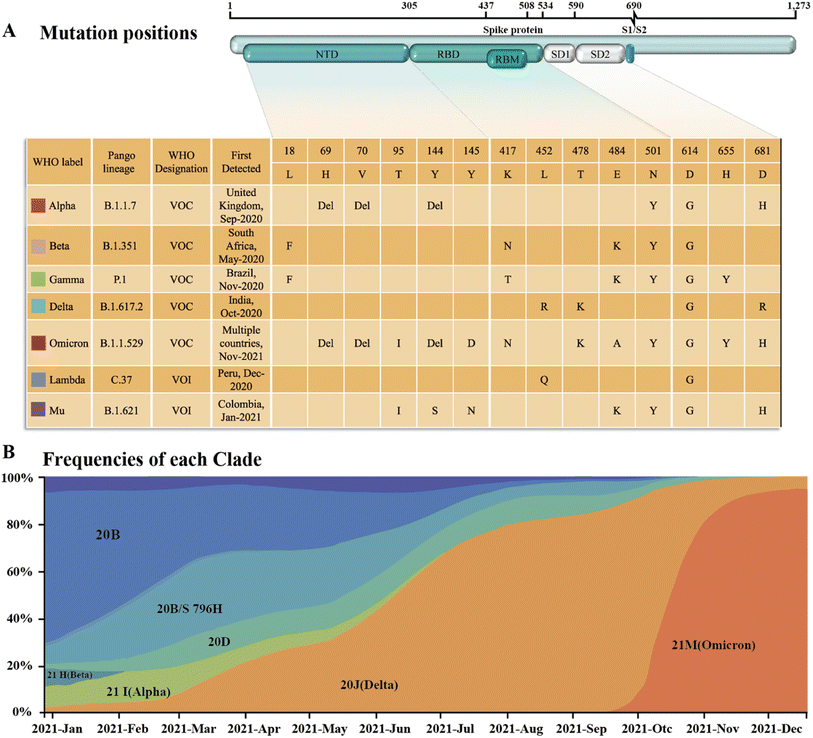
Since SARS-CoV-2 became popular at the end of 2019, many different subtypes or branches have evolved and spread globally. The mutation rate of this virus is about 2 nt per month, which is much lower than the influenza (4 nt per month) or human immunodeficiency virus (HIV) (8 nt per month).13 Accordingly, with the discovery of new variants of SARS-CoV-2, it is necessary to periodically reconfigure diagnostic tests for this virus.14 Among the SARS-CoV-2 mutant strains that have been detected and isolated, the main types of mutations were found to occur in RBD of the S protein.15,16 Mutations in this structure may increase the affinity with the receptor,17,18 weaken the effect of neutralizing antibodies,19,20 or cause the virus to escape immunity.21,22 Although mutations are also present in the N protein, compared to the S protein, it is highly conserved and often used as a rapid detection marker for SARS-CoV-2. Since December 2020, four rapidly spreading virus lineages have been identified as variants of concern (VOCs), marking the entry of the pandemic into a new phase. The four VOCs are the Alpha, Beta, Gamma and Delta variants, while many of the other variants that have subsequently emerged possess some of the mutational characteristics of these four variants. Currently, the B.1.1.529 Pango lineage (Omicron variant), including BA.1, BA.2, BA.3, BA.4, BA.5, and its descendent lineages, are prevalent. The WHO has emphasized that these descendant lineages should be monitored, and their virus characteristics should be examined.
The incubation period of COVID-19 ranges from 1 to 14 days, mostly 3 to 7 days with the main symptoms of fever, dry cough and fatigue.23–25 Some patients have diminished or lost sense of smell and taste as the first symptoms. A few patients have nasal congestion, runny nose, sore throat, conjunctivitis, myalgia and diarrhea.26–28 Severe patients usually develop respiratory distress and/or hypoxemia one week after the onset of the disease. Furthermore, they can rapidly progress to acute respiratory distress syndrome, septic shock, difficult-to-correct metabolic acidosis, coagulation dysfunction, organ failure, etc.29 Notably, many infected individuals may not have significant clinical symptoms due to the decreased virulence of this virus.30,31 Although SARS-CoV-2 infection will not be as fatal as its initial outbreak, studies have shown that many infected people suffered from severe sequelae.32–34 According to the CDC, over 80% of the nearly 24 million adults in the United States with long COVID experience difficulties in daily activities.2 Moreover, a report by the Brookings Institution also indicates that approximately 4 million people in the United States are unemployed because of long COVID symptoms.35
The spread of SARS-CoV-2 and the shocking infection data caused the world to seek effective prevention, control, and treatment methods. Thus, to help researchers in different fields have a more comprehensive understanding of the SARS-CoV-2 assays and promote the development of new assay technologies, herein, we review the articles related to SARS-CoV-2 detection technologies from 2020 to March 2023. Compared with the previous reviews, we not only elaborate on the principles of each method but also summarize its utilization, advantages and limits. In response to the simplicity and broadness of the published reviews on SARS-CoV-2 antigen detection, we overview this type of technique in terms of various antigenic subunits and analyze its applications in clinical settings. This makes our article more systematic, insightful and practically meaningful. In addition to the in-depth insight into the three main types of assays (molecular assays, serological assays, and antigen assays), we also review the corresponding countermeasure strategies for SARS-CoV-2 variants. Further, the mutational location characteristics and the epidemiological features of various variants are summarized. Meanwhile, related detection and countermeasure strategies for different mutation locations are also proposed, which are of great significance for diagnosing and preventing mutant strains. Finally, the challenges and possible strategies are prospected to explore new assays to meet different diagnostic needs.
2. Molecular assays for the detection of viral nucleic acids
SARS-CoV-2 molecular assays are the most direct and sensitive detection methods. Currently, more than 500 commercial kits are available worldwide for the molecular detection of SARS-CoV-2 RNA, and the emergence of these kits has played a critical role in the containment of the SARS-CoV-2 outbreak (Table 1).| Manufacturer | Test name | Method | Target gene | Specimen type | LODb | Sensitivity | Specificity | Statusc | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| a H: laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (chemiluminescence immunoassay, CLIA), 42 U.S.C. §263a, that meet requirements to perform high complexity tests. M: laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform moderate complexity tests. W: patient care settings operating under a CLIA Certificate of Waiver. b LOD, Limit of detection. c Status, Authorized settings include the following. | |||||||||
| LumiraDx UK Ltd | LumiraDx SARS-CoV-2 RNA STAR Complete | RT, qSTAR amplification, home collection, screening, pooling | N gene | Nasal swabs | 32 TCID50 per mL | 97% | 96% | H | 36 |
| Abbott Molecular Inc. | Abbott RealTime SARS-CoV-2 Assay | RT-PCR | N and RdRP genes | Nasopharyngeal and oropharyngeal swabs | 100 copies per mL | 95.9% | 100% | H | 37 |
| Meridian Bioscience, Inc. | Revogene SARS-CoV-2 | RT-PCR | N gene | Nasopharyngeal, oropharyngeal, anterior nasal, and mid-turbinate nasal swab specimens | 7 TCID50 per mL | 97.7% | 97.7% | H, M | 15 |
| Talis Biomedical Corporation | Talis One COVID-19 Test System | Reverse transcriptase isothermal amplification | ORF1ab gene, N gene | Nasal mid-turbinate swabs | 500 copies per mL | 100% | 100% | H, M, W | 38 |
| Cepheid | Xpert Xpress CoV-2/Flu/RSV plus | rRT-PCR | N gene, E gene, RdRP genes | Nasopharyngeal swabs, anterior nasal swabs, nasal wash/aspirate specimens | 138 copies per mL | 100% | 100% | H, M, W | 39 |
| Life Technologies Corporation (a part of Thermo Fisher Scientific Inc.) | TaqPath COVID-19 Fast PCR Combo Kit 2.0 | rRT-PCR | N gene, ORF1a, ORF1b | Saliva | 1000 GCE per mL | 97.1% | 97.6% | H | 40 |
| Detect, Inc. | Detect Covid-19 Test | RT-LAMP | ORF1ab | Anterior nasal swabs | 800 copies per mL | 90.9% | 97.5% | Home, H, M, W | 41 |
| PerkinElmer, Inc. | PKamp Respiratory SARS-CoV-2 RT-PCR Panel 1 | rRT-PCR | N gene, ORF1a gene | Nasopharyngeal swabs, anterior nasal swabs, and mid-turbinate swabs | SARS-CoV-2: 0.11 TCID50 per mL | 100% | 100% | H | 20 |
| LMSI, LLC (dba Lighthouse Lab Services) | CovidNow SARS-CoV-2 Assay | rRT-PCR | N gene | Upper respiratory specimens, self-collected anterior nasal swab specimens | 800 copies per mL | 96% | 100% | H | 21 |
| Applied DNA Sciences Inc. | Linea COVID-19 Assay Kit | rRT-PCR | S gene (S1 and S2 regions) | Nasopharyngeal swabs and oropharyngeal swabs | 1250 copies per mL | 98% | 92% | H | 22 |
| Enzo Life Sciences, Inc. | AMPIPROBE SARS-CoV-2 Test System | rRT-PCR | N gene and RdRP gene | Nasal, mid-turbinate, nasopharyngeal, and oropharyngeal swab specimens | 280 copies per mL | 96.2% | 98% | H | 42 |
| Thermo Fisher Scientific Inc. | TaqPath COVID-19 RNase P Combo Kit 2.0 | rRT-PCR | N gene, ORF1a, ORF1b | Nasopharyngeal, anterior, and mid-turbinate nasal swabs | 75 GCE per mL | 96.7% | 95% | H | 43 |
| Twist Bioscience Corporation | SARS-CoV-2 NGS Assay | NGS | Several | Nasopharyngeal swabs, oropharyngeal swabs, anterior nasal swabs | 800 copies per mL | 96.7% | 100% | H | 44 |
| BioGX, Inc. | BioGX Xfree COVID-19 Direct RT-PCR | rRT-PCR | N1 gene | Nasopharyngeal, anterior nasal, mid-turbinate | 1000–3000 copies per mL (depending on the platform used) | 95% | 100% | H | 26 |
| OSANG Healthcare | GeneFinder COVID-19 Plus RealAmp Kit | rRT-PCR | RdRP, E gene, N gene | Alveolar lavage fluid, throat swab, sputum | 10 copies per 20 μL reaction | 100% | 100% | H | 45 |
| OPTI Medical Systems | OPTI SARS-CoV-2 RT PCR Test | rRT-PCR, pooling | N gene (N1 and N2) | Anterior nasal swabs, mid-turbinate nasal swabs, nasopharyngeal swabs | 700–900 copies per mL | 100% | 100% | H | 46 |
| PathogenDx, Inc. | DetectX-Rv | rRT-PCR, DNA microarray hybridization | N gene (N1 and N2 regions) | Nasopharyngeal swabs, oropharyngeal swabs, mid-turbinate swabs, anterior nasal swabs | 1000 copies per mL | 95% | 100% | H | 47 |
| Hologic, Inc. | Aptima SARS-CoV-2 Assay | Real-time TMA, dual kinetic assay | ORF1ab (2 targets) | Nasopharyngeal, nasal, mid-turbinate, and oropharyngeal swab specimens | 212 copies per mL within a sample (83 copies per mL within the tested aliquot) | 100% | 98% | H | 48 |
| LumiraDx UK Ltd | LumiraDx SARS-CoV-2 RNA STAR Complete | RT-qSTAR amplification | ORF1a | Mid-turbinate, nasopharyngeal, and oropharyngeal swabs | 7500 copies per mL | 97.8% | 98.5% | H | 49 |
| Quidel Corporation | Solana SARS-CoV-2 Assay | Molecular isothermal Reverse Transcriptase – Helicase-Dependent Amplification (RT-HDA) | ORF1ab | Nasopharyngeal and nasal swab specimens | 1.16 ×104 copies per mL | 90.3% | 97.8% | H, M | 32 |
| Cue Health Inc. | Cue COVID-19 Test for Home and Over the Counter Use | Isothermal amplification, home collection | N gene | Anterior nares swabs | 2700 copies per mL | 96% (symptomatic), (100% asymptomatic) | 98% (symptomatic), (100% asymptomatic) | Home, H, M, W | 50 |
| Grifols Diagnostic Solutions Inc. | Procleix SARS-CoV-2 Assay | Transcription-mediated nucleic acid amplification (TMA) | Not stated | Anterior nasal and mid-turbinate nasal swabs, nasopharyngeal | 60 copies per mL | 100% | 100% | H | 51 |
| Agena Bioscience, Inc. | MassARRAY SARS-CoV-2 Panel | rRT-PCR, MALDI-TOF | N gene (N1, N2, and N3 regions), ORF1, and ORF1ab | Nasopharyngeal swabs, oropharyngeal swabs, nasal and mid-turbinate swabs | 310–2500 copies per mL | 98.9% | 100% | H | 35 |
| Cepheid | Xpert® Omni SARS-CoV-2 | rRT-PCR | E gene and N gene (N2 region) | Nasopharyngeal, oropharyngeal, nasal, or mid-turbinate swab specimens | 400 copies per mL | 100% | 100% | H, M, W | 52 |
| Lucira Health, Inc. | Lucira COVID-19 All-In-One Test Kit | RT-LAMP | N gene (two regions) | Nasal swab (self-collection), nasal swab (healthcare collection) | 2700 copies per swab (or 900 copies per mL if the sample is 100% lysed and eluted) | 94% | 98% | Home, H, M, W | 53 |
| GenMark Diagnostics, Inc. | ePlex Respiratory Pathogen Panel 2 | rRT-PCR, electrochemical detection | Not stated | Nasopharyngeal swabs | 0.01TCID50 per mL | 100% | 100% | H, M | 54 |
| Seasun Biomaterials, Inc. | AQ-TOP COVID-19 Rapid Detection Kit PLUS | RT-LAMP | ORF1ab and N genes | Oropharyngeal and nasopharyngeal swabs, anterior nasal and mid-turbinate nasal swabs | 1000 copies per mL | 100% | 100% | H | 55 |
| Clear Labs, Inc. | Clear Dx SARS-CoV-2 Test | rRT-PCR, NGS | Several | Nasopharyngeal swab, oropharyngeal swab, anterior nasal swab | 2000 copies per mL | 100% | 100% | H | 56 |
| Abbott Diagnostics Scarborough | ID NOW COVID-19 | Isothermal amplification | RdRP | Direct nasal secretions, nasopharyngeal or throat swabs | 125 copies per mL | 100% | 100% | H, M, W | 57 |
| Cue Health Inc. | Cue COVID-19 Test | Isothermal amplification | N gene | Nasal swab | 1300 copies per mL | 100% | 92% | Home, H, M, W | 58 |
2.1 PCR-based SARS-CoV-2 detection
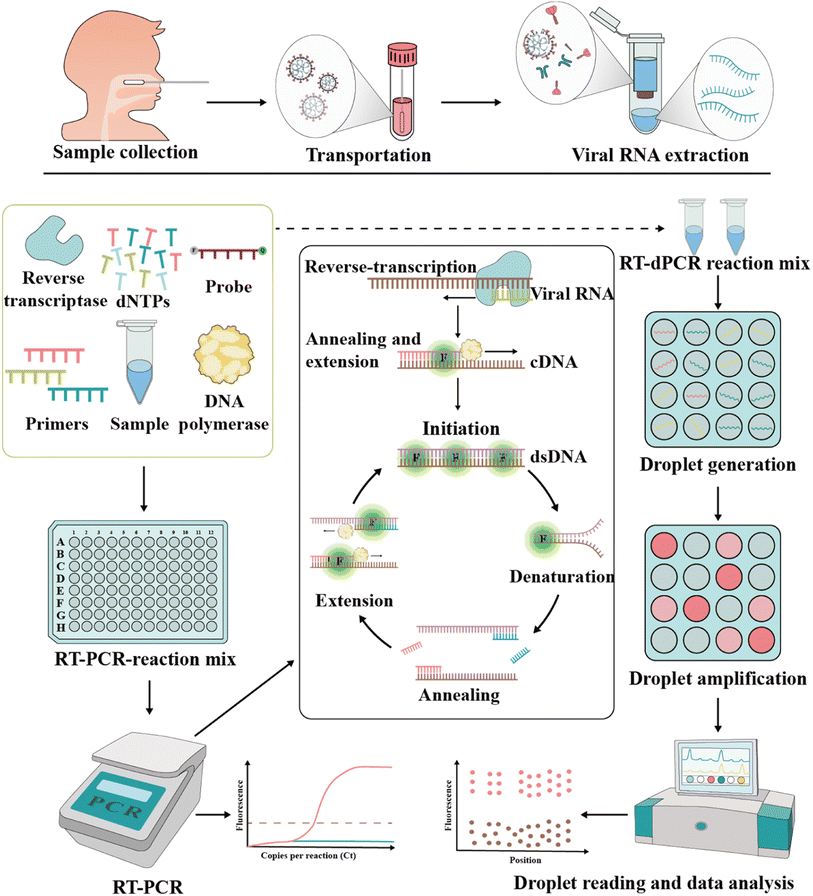
The foundation of molecular diagnosis depends on identifying and amplifying viral genetic material from suspected individual specimens. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is the gold standard for identifying SARS-CoV-2, powered by its ability to amplify minuscule amounts of viral genetic information.59,60The RT-PCR assay can target different genes72 such as RdRp gene, N gene, E gene, S gene and ORF1b or ORF8 region of the SARS-CoV-2 genome.73 Vogels et al. compared seven sets of primers provided by the WHO and found that they all had similar performances.74 Notably, Corman et al. indicated that different targets in the SARS-CoV-2 genome have different properties, affecting the sensitivity and specificity.75 Recently, many studies showed that primer probes for the detection of the N and E genes are more sensitive than that targeting the RdRp gene.74,76–78 This merit may be attributed to the mismatch of reverse primers used in detecting RdRp genes.79 To improve the diagnostic efficiency and reliability, two or more genes should be included in the RT-qPCR reaction to enhance the identification of true positives. In addition, mutations in target genes may affect the sensitivity of the assay, which may lead to false negative results.80–82 According to the sequence analysis of the SARS-CoV-2 genome by the Global Initiative of Sharing All Influenza Data (GISAID) database and China CDC, the N primer region has a higher rate of viral mutations compared to other primer sets.83 Although this does not imply that the primers cannot bind, it reveals the variability of the target region. A report showed that the deletion of S gene locations 69 and 70 in VOC B.1.1.7 failed at least one RT-PCR-based diagnostic kit for S gene detection.61 These findings highlight the importance of the independent evaluation of the primer-probe sets used in SARS-CoV-2 RT-PCR assays.
The RT-PCR assays of SARS-CoV-2 mainly use samples collected from the upper respiratory via swabs. Among the samples, sputum and nasal swabs have higher viral loads within seven days after symptom onset, whereas pharyngeal swabs are unreliable at eight days after symptom onset,84–86 given that viral loads are lower in samples after the 8th day. Generally, sputum samples have higher viral loads than pharyngeal swab samples, while urine or stool samples have lower viral RNA loads.84,86–88 After systematic evaluation of five studies, Woloshin et al. found that RT-PCR had a high rate of false negatives (2% to 29%).89 This phenomenon may be contributed from improper sampling techniques, low viral load in the sampling area, or mutations in the viral genome.90–93 Thus, to improve the sensitivity, safety and rapidity of RT-PCR assays, Erster et al. used a lysis buffer supplemented with nucleic acid stabilization and lysis buffer (NSLB) instead of the traditional viral transfer medium (VTM) for better sample preservation.94
Currently, the RT-PCR technique is the most commonly used and effective method for SARS-CoV-2 detection; however, this test often leads to population aggregation with the risk of causing virus transmission. Therefore, RT-PCR-based mixed-sample mass screening to control the COVID-19 pandemic is not optimal.
Recently, COVID-19 patients have been reported to revert to SARS-CoV-2 positivity;73,107,108 however, there are no specific clinical features to distinguish them from fully recovered patients.86,109 This has provoked public concern about the current standards for patient discharge.110 Therefore, this situation requires more sensitive detection methods compared to RT-PCR. In this case, RT-dPCR is a suitable technique with good sensitivity. Lu and team found that the LOD of RT-dPCR was 10-fold lower than that of RT-PCR.111 Alteri et al. used RT-dPCR to detect 55 suspected COVID-19 patients who tested negative by RT-PCR; however, 35% of these individuals tested positive by RT-dPCR for the N gene.112 Given that RT-dPCR is more sensitive in detecting the virus in low concentrations, it is a promising and reliable method to examine asymptomatic carriers discharged from hospitals. In addition, RT-dPCR can also be used to detect SARS-CoV-2 RNA in the air.113 It was reported that some toilets used by medical workers and patients contained a high viral load.93,114 This also indicates the importance of hygienic treatment and room ventilation to restrict COVID-19 transmission.
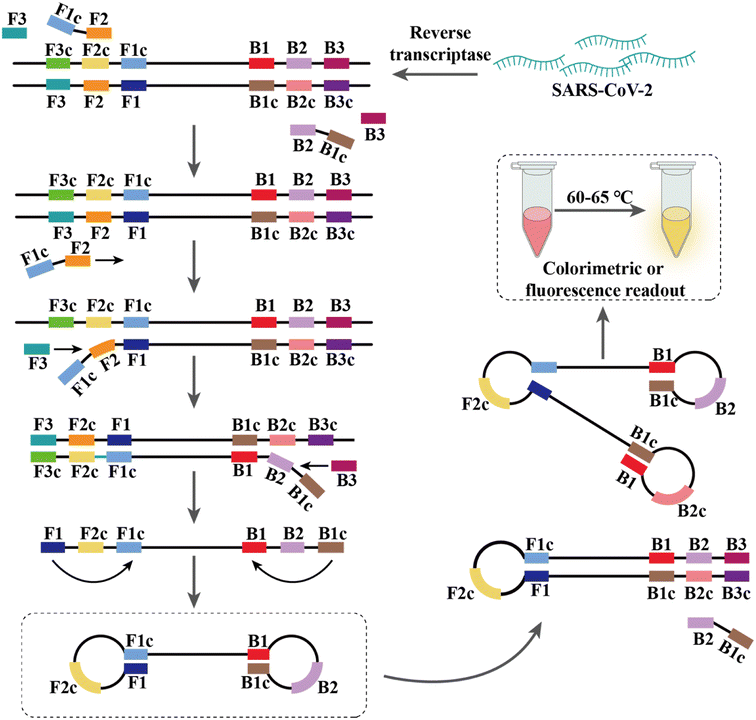
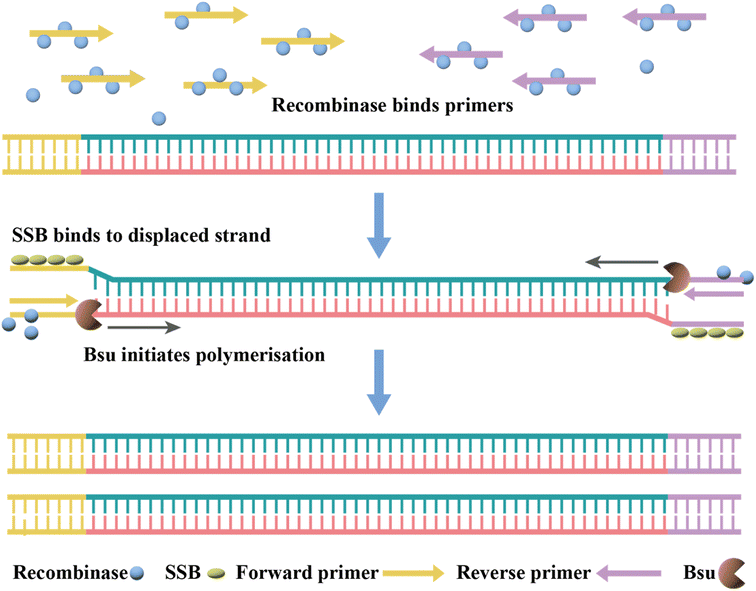
2.2 Isothermal nucleic acid amplification
PCR-based nucleic acid testing protocols are complicated, which require expensive instrumentation and professionals. In addition, these methods require multiple temperature changes for each cycle and involve complex thermo-cycling equipment.115,116 Isothermal nucleic acid amplification is another detection strategy that allows amplification without a thermal cycler at thermostatic temperature.In addition, many modified RT-LAMP procedures have been developed. For example, Zhang et al. combined the clustered regularly interspaced short palindromic repeat (CRISPR)/associated endonuclease (Cas) 12a system with RT-LAMP, and simultaneously achieved visual detection by using gold nanoparticles (AuNPs).126 Moreover, this method could be directly applied to 96-well plates for high-throughput screening.127 Thi et al. developed LAMP sequencing using barcode primers, which is scalable and can potentially analyze thousands of samples in parallel.128 Rohaim et al. introduced artificial intelligence (AI) algorithms in RT-LAMP and developed a handheld intelligent diagnostic device called AI-LAMP, which not only improved the performance of RT-LAMP assays but also reduced their operation time and subjectivity.129
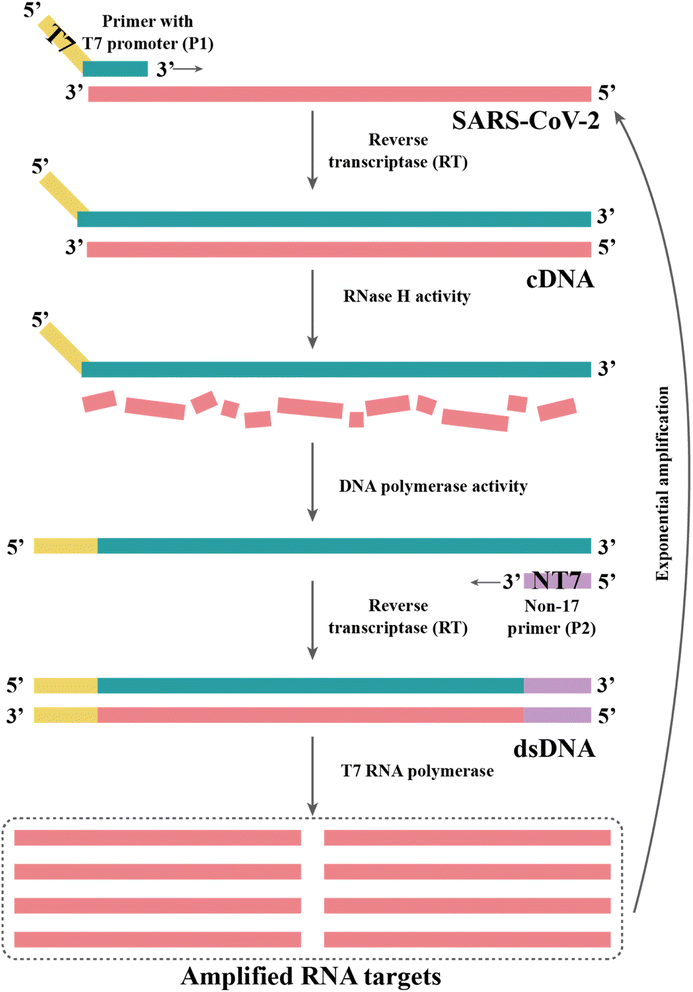
Currently, RT-RPA can be combined with other techniques such as fluorescence resonance energy transfer (FRET) or CRISPR-Cas technology.143–146 El Wahed et al. developed RT-RPA suitcase laboratory, a device that enables COVID-19 detection in resource-poor settings. The sensitivity of RdRP, E and N gene detection for SARS-CoV-2 can reach 2, 15, and 15 RNA molecules, respectively, in 15 min.147 A customized isothermal amplification integrated lateral flow strip (LFS) platform enabled rapid, simultaneous visual screening of SARS-CoV-2 and influenza viruses (influenza A and influenza B) without cross-reactivity, false positives and false negatives.148 The other applications of RT-RPA for SARS-CoV-2 detection involve CRISPR-Cas technology, which are described in the following section.
| Method | Required enzyme | Probes number | Temperature (°C) | Time (min) | Target | LOD (copies per μL) | Sensitivity | Advantagesa | Disadvantagesa | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| a Advantages and disadvantages are compared to RT-qPCR methods. | ||||||||||
| NEAR | Nicking endonuclease, strand-displacing DNA | 2 | 60–65 | 5–15 | RdRp gene | 0.125 | 48–71.7% | Fast testing, universally in various environments | Low sensitivity | 149 |
| NASBA | RNase H, reverse transcriptase, T7 DNA-dependent RNA polymerase | 2 | 41 | 90–120 | S gene N gene | 0.5 | 98.15–100% | High sensitivity | Required RNase inhibitors | 150 |
| Non-enzymatic isothermal strand displacement and amplification | Restriction endonucleases, strand-substituting DNA polymerases | 3 | 42 | <30 | RdRp gene, N gene | 10 | 96.77% | No RNA reverse transcription step, fast testing | Non-specific reactions/false positives | 151 |
| Rolling circle amplification | DNA ligase, DNA polymerase | 8 | 30–37 | <120 | N gene S gene | 1 | — | High specificity | False negatives and false positives | 5 |
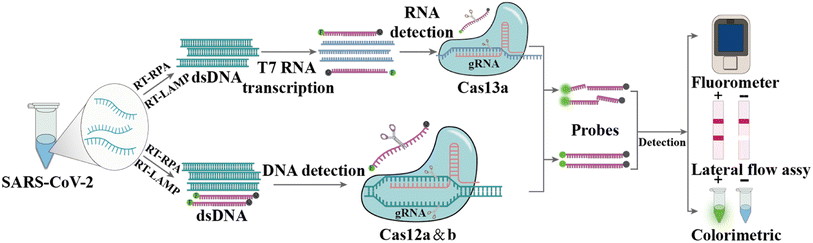
2.3 CRISPR-based diagnosis
CRISPR and CRISPR-Cas are prokaryotic defense systems that safeguard organisms against exogenous nucleic acids.152,153 After specific binding between guide RNA (gRNA) or CRISPR RNA (crRNA) and the target sequence (DNA or RNA) located next to a proto-spacer adjacent motif, the Cas enzyme is activated, which can exhibit local DNase or RNase activity, leading to local cleavage of the target DNA or RNA (cis-cleavage) as well as to collateral damage to adjacent single-stranded DNA (ssDNA) or RNA (trans cleavage) (Fig. 6), respectively.154,155 Nowadays, many different Cas enzymes have been identified, such as Cas9, Cas12, and Cas13, in which certain enzymes can be programmed for the detection of RNA viruses.156–158 The Cas12a-based system DNA endonuclease-targeted CRISPR trans reporter (DETECTR) and Cas13-based system called Specific High sensitivity Enzymatic Reporter UnLock (SHERLOCK) are two representative CRISPR technologies for infection pathogen detection.159–161 Since the COVID-19 outbreak, the SHERLOCK and DETECTR platforms have been used for SARS-CoV-2 detection and authorized for Emergency Use Authorization (EUA) by the FDA.162,163Patchsung et al. performed a two-step CRISPR-Cas13a-based SHERLOCK assay targeting the S, N and ORF1ab genes of SARS-CoV-2. The target sequences were reverse transcribed and amplified using RT-RPA, followed by transcribing into RNA using T7 RNA polymerase, and then identifying and detecting based on CRISPR-Cas13a with an LOD of 42 copies per reaction (Fig. 6).164 Broughton et al. developed and validated the DETECTR platform for detecting the N and S genes of SARS-CoV-2, which used RT-LAMP to amplify target genes, followed by Cas12a-based specific sequence cleavage. The resultant readout is also available by fluorescence or lateral flow. The estimated LOD of this platform is 10 copies per μL, with 95% and 100% agreement for positive and negative predictions, respectively.165 To fulfill COVID-19 diagnostic demands, researchers have developed many variants of CRISPR-Cas12 and CRISPR-Cas13 systems, for example, the one-pot assay iSCAN and STOPCovid.v2 (simplifying the operational steps and shortening the detection time),166–168 AIOD-CRISPR (avoiding aerosol contamination from opening the cap),169 CRISPR-FDS with the high-throughput format,170 ITP-CRISPR with point-of-care testing (POCT) potential in combination with microfluidics,171 SHERLOCK-HUDSON for direct sample detection without nucleic acid extraction172 and SHINEv.2 for the detection of multiple variants of SARS-CoV-2.173
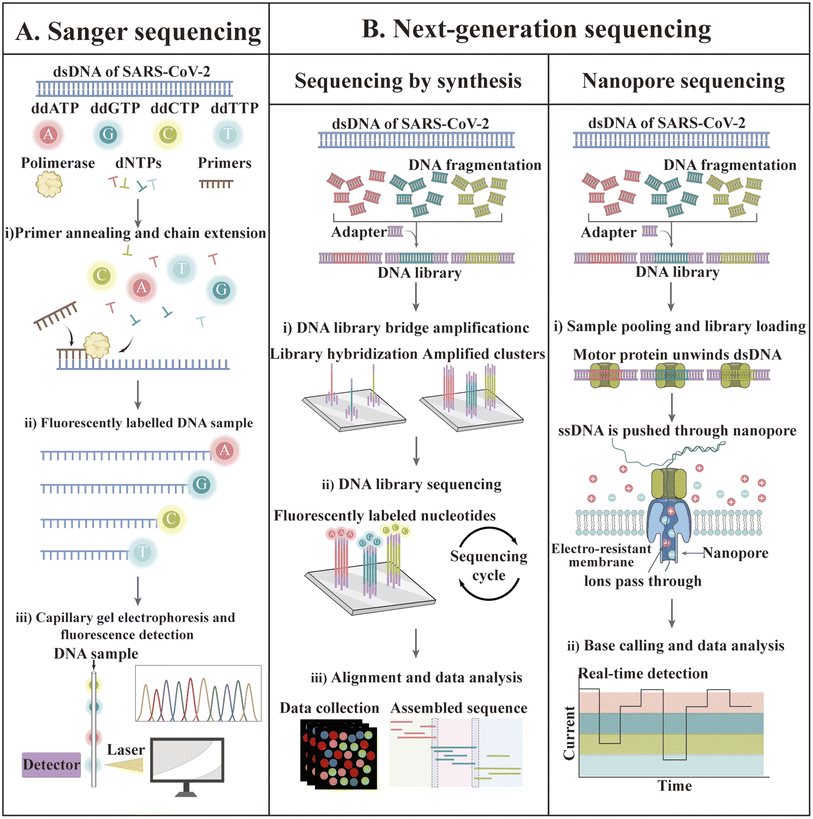
2.4 Sequencing-based technology
In recent decades, gene sequencing technology has witnessed unprecedented developments based on Sanger sequencing technology. The first SARS-CoV-2 genome was obtained by next-generation sequencing methods in less than one month after disease identification.174 Currently, GISAID shares more than 13 million SARS-CoV-2 genome sequences.175 The available sequencing technologies are Sanger sequencing as well as next-generation sequencing (NGS) technologies such as synthetic by sequencing (SBS), ion semiconductor sequencing, nanosphere sequencing, and nanopore sequencing.176–179 The principles of some of these techniques are illustrated in Fig. 7.Although most SARS-CoV-2 genome sequencing is performed by NGS, traditional Sanger sequencing can still be used for viral genome sequencing.180–183 qSanger-COVID-19, a rapid detection kit for COVID-19 developed by BillionToOne (BTO), is based on Sanger sequencing technology, which allows direct sample detection without nucleic acid extraction and offers comparable sensitivity to RT-PCR (10–20 genomic copy equivalents); nevertheless, the throughput became 30-times faster and 20-times higher.184 Despite the fact that the sample processing step is eliminated, this protocol is still highly manual given that it requires setting up multiple reactions. In addition, the final sequence comparison of sequencing results must be manually checked and identified by a trained professional, making post-sequencing analysis a challenge. Unlike Sanger sequencing, which is designed to generate consistent sequences for a single target amplicon, NGS technology allows the sequencing of millions to billions of DNA strands in parallel during a single run.185,186 EUA has licensed four kits for the targeted testing of SARS-CoV-2 RNA based on Illumina's sequencing by synthesis technology.161 The details of these kits are summarized in Table 3.
| Diagnostic | Entity | Authorized setting | Attributes | Sequencing platform | Detection target | Duration | Throughput | LOD (copies per mL) |
|---|---|---|---|---|---|---|---|---|
| a H, Laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high complexity tests. | ||||||||
| Illumina COVIDSeq Test | Illumina, Inc. | Ha | SBS | NovaSeq 6000 | 98 target genes | 12 h | 3072 | 500 |
| Helix COVID-19 NGS Test | Helix OpCo LLC (dba Helix) | Ha | SBS | NovaSeq 6000 | S and RPP30 | — | 384 | 125 |
| UCLA SwabSeq COVID-19 Diagnostic Platform | The University of California, Los Angeles (UCLA) | Ha | SBS | Illumina MiSeq | S2, synthetic internal control S2 gene and RPP30 | — | 384 | 250 |
| SARS-CoV-2 NGS Assay | Twist Bioscience Corporation | Ha | SBS | Illumina NextSeq 550 | Target the entire viral genome | — | 96 | 800 |
To date, only a few sequencing studies have been explored for SARS-CoV-2 detection compared to the frequently used assays such as RT-PCR.184,187–190 Bhoyar sequenced 752 replicate processed clinical specimens using Illumina's COVIDSeq method, which shows good sensitivity, precision and accuracy compared to RT-PCR. Moreover, the diagnostic positivity of COVIDSeq increased by 5.7% (27 positives were detected from cases diagnosed as negative and indeterminate by RT-PCR).189 Bloom et al. also compared NGS (SwabSeq) with RT-PCR and found that this method had similar or better sensitivity and specificity.191 In addition, this SwabSeq method also has potential to detect 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 samples simultaneously.191
000 samples simultaneously.191
The Clear Dx SARS-CoV-2 test (Clear Labs) is the only EUA-authorized protocol for detecting SARS-CoV-2 using Oxford Nanopore sequencing technology. Compared to the short read length of SBS technology, Oxford Nanopore sequencing yielded longer read lengths without amplification artifacts and bias at a lower cost.192,193 Wang et al. performed nanopore target sequencing of SARS-CoV-2 and other pathogens from respiratory specimens within 6–10 h. This technique displayed considerable sensitivity and specificity, with an LOD of 10 copies per mL.194
3. Serological antibody test
Diverse antibody assays, characterized by high-throughput and low workload are playing a more crucial role in supplementing the nucleic acid test. For patients who are highly suspected of COVID-19 but negative by the molecular test, serological antibody testing may be helpful for their diagnosis.195 Moreover, antibody tests are essential to diagnose whether an individual has been previously infected and may also help to confirm the presence of current infection, given that a large proportion of SARS-CoV-2 infections is asymptomatic.196 Due to the importance of serology testing, academia and industry have developed various platforms for serological diagnosis. These tests can be used in laboratories or wherever the patient is in the hospital or at home (point-of-care). Many immunoassays, referred to as total antibody assays, have been constructed to detect levels of all isotypes simultaneously.3.1 Antibody types and dynamics
Antibodies are formed by the body's immune system in response to infections, which can be detected in whole blood, plasma or serum. Three types of antibodies are created in response to infection, i.e., immunoglobulin A (IgA), IgG and IgM, which rise or fall at different times after the onset of infection.197–201 IgG is used in most antibody tests given that it persists for the longest time and may reflect longer-term immunity, while IgM typically rises quickly with infection and declines soon after infection is cleared. The early seroconversion of IgM against SARS-CoV-2 was reported at 3–5 days, and the level can last for more than 1 month, while IgG against SARS-CoV-2 may be detected after 8 days.202,203 Notably, many reports on serum antibody levels in SARS-CoV-2 patients indicate that IgM expression was observed concurrently with IgG expression.204–207 Long et al. conducted a large multicenter study and found that the median seroconversion of both isotypes was recorded on day 13.208 Udugama et al. found that antibody responses to infection took days to weeks to be reliably detectable. Then, the levels of those antibodies decreased over time.209 Although IgA has rarely been studied, as a sensitive marker of infection, IgA levels correlate with disease severity and neutralizing activity. Okba et al. used a commercial enzyme-linked immunosorbent assay (ELISA) to detect IgA and IgG and found that the specificity of the IgG and IgA assays were similar, but the IgA ELISA had a superior sensitivity over IgG ELISA. Fig. 8 shows that the RT-PCR molecular diagnostics as well as IgM and IgG antibody assays may vary over time.210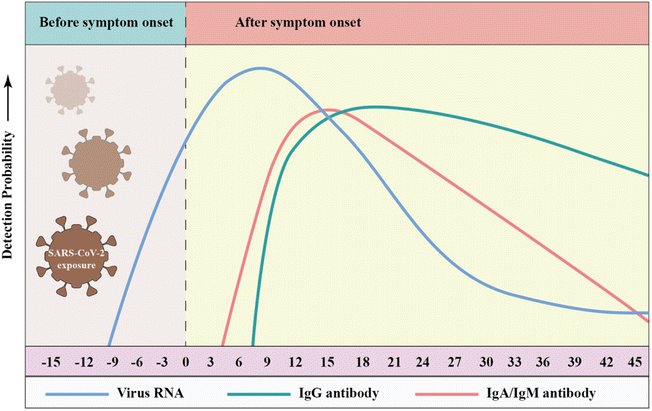 | ||
| Fig. 8 Detection probability of viral RNA or antibody (IgA, IgM, and IgG) against SARS-CoV-2 during infection (relative to symptom onset). The testing windows of nucleic acid amplification tests (RT-PCR, blue) and serology tests (antibody, green) are indicated. Reproduced with permission from Lancet Respir. Med., 2020, 8, 717–725, 10.1016/S2213-2600(20)30230-7.211 | ||
Thus far, the Foundation for Innovative Diagnostics (FIND) has listed more than 400 antibody tests, most of which are produced by commercial companies and are available on the market. Here, these assays are classified into the following categories and their properties are summarized according to the testing platforms (Table 4). It is critical to understand the strengths and limitations of these assays in various clinical and research scenarios before utilizing antibody testing for SARS-CoV-2.
| Method | Developer | Test | Sensitivity (%) | Specificity (%) | Antigen | Antibody | Time (min) | Specimen and volume | Advantages | Disadvantages | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgG | TAa | IgM | IgG | TA | IgM | |||||||||
| a TA, total antibodies. b NM, not mentioned. c ECD, extracellular domain. | ||||||||||||||
| LFIA | PRIMA Lab SA | COVID-19 IgG/IgM Rapid Test | 95.7% | 97.8% | NP | IgG, IgM | 10 | Whole blood, serum and plasma, 10 μL | Easy, fast, flexible, and accurate testing with low cost for POC testing | False positives, qualitative non-quantitative, heterogeneous presentation, the limited overall sensitivity of qualitative results only in the acute phase of the disease | ||||
| Prognosis Biotech | Rapid Test 2019-Ncov Total Immunoglobulins | 98.7% | 94.8% | NP | IgG, IgM, IgA | 10 | Serum, plasma or whole blood, 10 μL | |||||||
| Sugentech | SGTi-flex COVID-19 IgM/IgG | 90% | 94.4% | 95% | 9% | 98.3% | 100% | NP and S-RBD | IgG, IgM | 10 | Serum, plasma or whole blood, 10 μL | |||
| SG Diagnostics Pte Ltd | SG Diagnostics COVID-19 Immunity Rapid Test Kit | 96.4% | 98% | S-RBD | IgG | 10 | Serum, plasma or whole blood, 10 μL | |||||||
| Zhongshan Bio-Tech Co Ltd | SARS-CoV-2 IgM-IgG (GICA) | 96.7% | 96.7% | 50% | 85.0% | 83.8% | 97.5% | SP | IgG, IgM | 10 | Serum, plasma, or whole blood, 10 μL | |||
| Self-diagnostics | SARS CoV-2 IgM/IgG Antibody Assay Kit | 88.0% | 99.0% | Antigen | IgG, IgM | 15 | Serum, plasma, whole blood, or fingertip blood, 10 μL | |||||||
| PRIME4DIA Co., Ltd | P4DETECT COVID-19 IGM/IGG | 97.7% | 98.1% | NMb | IgG, IgM | 10 | Serum, plasma, whole blood, 10 μL | |||||||
| Nirmidas Biotech, Inc | MidaSpot™ COVID-19 Antibody Combo Detection Kit | 96.7% | 100% | 100% | 97.5% | 100% | 98.8% | NMb | IgG, IgM | 10 | Human serum, plasma, 10 μL | |||
| Beijing Wantai Biological Pharmacy Enterprise Co., Ltd | WANTAI SARS-Cov-2 Ab Rapid Test | 100% | 98.8% | S-RBD | IgG, IgM | 10 | Serum, plasma, whole blood, 10 μL | |||||||
| Biocan Diagnostics Inc | Novel Coronavirus (COVID-19) IgG/IgM Antibody Test | 92% | 100% | Antigen | IgG, IgM | 10 | Serum, plasma, and whole blood, 10 μL | |||||||
| ELSA | Plexense, Inc. | ACCEL ELISA® COVID-19 Kit | 94.4% | 100% | NP | IgG, IgM, IgA | 45 | Blood (serum), 50 μL | Less time-consuming, low sample consumption, some analyses yield quantitative results, higher overall sensitivity compared to LFIA, suitable for high throughput and automation | Not suitable for rapid testing, requires trained laboratory personnel to check in batches during the laboratory process, endogenous interference, poor repeatability | ||||
| Quansys Biosciences | Q-Plex SARS-CoV-2 Human IgG (4-Plex) | 100% | 100% | SP | IgG | 120 | Human serum, plasma, 2 μL | |||||||
| AB Diagnostic Systems GmbH | Abia SARS-CoV-2 IgG/IgM | 40.9%∼100% | 91.1% ∼100% | Antigen | IgG, IgM | 120 | Serum or plasma | |||||||
| Actim Oy | Actim ® ELISA SARS-CoV-2 IgG | 96% | 98% | S1 protein | IgG | 120 | Serum or plasma | |||||||
| AMEDA Labordiagnostik GmbH | AMP ELISA Test SARS-CoV-2 Ab | 95.9% | 100% | 100% | 98.4% | 99.5% | 99.8% | NP, S1-RBD and S2-ECDc | IgG, IgM | 70 | Serum, plasma, 100 μL | |||
| Adversis Pharma GmbH | SARS-CoV-2 IgG ELISA | 100% | 99.4% | NP | IgG | 120 | Serum, plasma, 100 μL | |||||||
| Prognosis Biotech | Bio-Shield 2019-nCoV Total Immunoglobulins | 97.3% | 98.6% | 82.6% | 97.4% | 100% | 92.3% | NMb | IgG, IgM, IgA | 120 | Serum, plasma | |||
| Diatheva | COVID-19 ELISA IgG DIATHEVA Kit | 98.8% | 98.2% | NM | IgG | 150 | Serum, plasma | |||||||
| DIA.PRO Diagnostic Bioprobes S.r.l. | COVID-19 Spike 1&2 IgG – ELISA | 98% | 98% | S1 and S2 protein | IgG | 120 | Serum, plasma | |||||||
| Icosagen AS | Serological COVID-19 ELISA Kit | 87.8% | 98.9% | SP and NP | IgG, IgM | 120 | Serum, plasma | |||||||
| CLIA | Beckman Coulter Inc. | Access SARS-CoV-2 IgG | 100% | 99.8% | S1-RBD | IgG | 60 | Serum or plasma, 20 μL | Simple operation, high sensitivity, suitable for large population detection | Expensive instruments, and well-trained professionals | ||||
| Snibe Co., Ltd | MAGLUMI 2019-nCoV IgM/IgG | 92.1% | 93.9% | 75.7% | 96% | SP and NP | IgG, IgM | 60–120 | Serum, 160 μL | |||||
| Mindray | SARS-CoV-2 IgG(CLIA) | 82.2% | 94.9% | NM | IgG | 60–120 | Serum, 10 μL | |||||||
| Inova Diagnostics, Inc. | QUANTA Flash® SARS-CoV-2 IgG | 100% (15+ days) | 99.9% | SP or NP | IgG | 60–120 | Serum or plasma, 20 μL | |||||||
| Ortho Clinical Diagnostics | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack | 100% (8+ days) | 100% | S1 protein | IgG, IgA, and IgM | ∼85 | Serum or plasma, 80 μL | |||||||
| Immunodiagnostic Systems Ltd | IDS SARS-CoV-2 IgG | 97.6% (15+ days) | 100% | NP and SP | IgG | About 60 ∼ 120 | Serum or plasma | |||||||
| Beckman Coulter | Access SARS-CoV-2 IgM | 98.3%(15+ days) | 99.9% | S-RBD | IgM | About 60 ∼ 120 | Serum or plasma, 10 μL | |||||||
| Siemens Healthcare Diagnostics Inc. | Dimension Vista SARS-CoV-2 Total Antibody Assay (COV2T) | 100% (15+ days) | 99.8% | S-RBD | IgG, IgM | About 60 ∼ 120 | Serum or plasma, 10 μL | |||||||
| Ortho-Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack | 100% (15 days+) | 100% | SP | IgG | About 85 | Serum or plasma, 20 μL | |||||||
| DiaSorin Inc. | LIAISON® SARS-CoV-2 S1/S2 IgG | 97% (15+ days) | 98% | S1 and S2 | IgG | Serum or plasma, 20 μL | ||||||||
3.2 Lateral flow immunoassay (LFIA)
LFIA is a rapid immunochromatography-based method that utilizes a cassette to inject patient samples.212–215 Usually, it requires only a few drops of whole blood from a finger prick placed on the test strip, with a band appearing as positive or negative for antibody detection. If the sample contains SARS-CoV-2-specific antibodies, they will attach to the virus antigen bound to AuNPs or another detection system, where the complex migrates along the membrane towards the test line containing a secondary antibody against the immune complex to result in a visible band.216 LFIA is a POC serology test, which can be used in emergencies with the advantages of time-saving (∼15 min), easy-operation, and easy-result-interpretation. Guedez-López et al. evaluated three commercially available antibody detection kits and found that their sensitivities were relatively low in the early stage (1–7 days from the onset of symptoms) of SARS-CoV-2 infection and gradually increased in the intermediate stage (8–14 days from the onset of symptoms) and peaked at a late stage (more than 15 days).217 After systematic analysis of 5016 references and 40 studies, Bastos et al. also concluded that LFIA had lower sensitivity in the early stage of symptom onset.218 Although the sensitivity of the assays was affected by the timing of sample acquisition, a higher overall sensitivity was consistently observed with the use of total antibody detection. Li et al. developed an LFIA for the simultaneous detection of IgG and IgM and found a significant increase in sensitivity compared to the detection of IgG or IgM, respectively (Fig. 9).219 Other studies reported the sensitivity and specificity of LFIA for SARS-CoV-2-specific total antibodies using recombinant antigens of NPs, which were 94.6% (84.9–98.9, CI 95%) and 100% (95.75–100, CI 95%) as early as 7 days post confirmation of positivity, respectively.220 In addition, an immunochromatographic fluorescence assay was developed by combining fluorescent nanotags with LFIA to ensure high sensitivity and reliability of the assay. These fluorescent nanotags such as silica-core@dual QD-shell nanocomposites (SiO2@DQD),221 fluorescent microspheres222 and selenium nanoparticles206 showed excellent accuracy when labeled with NP or SP of SARS-CoV-2. Notably, Chen et al. designed aggregation-induced emission (AIE) nanoparticles (emission wavelength, 810 nm)-labeled LFIA for the early detection of antibodies against SARS-CoV-2 and the sensitivity of the proposed test strip for detecting IgM and IgG was 78% and 95%, respectively (172 serum samples).223 Importantly, the detection of IgM or IgG in sequential clinical samples was 1–7 days after symptom onset. Recently, Liu and coworkers developed a surface-enhanced Raman scattering (SERS)-based LFIA biosensor,221 which used dual-layer Raman molecule-loaded Ag-coated SiO2 nanoparticles as advanced SERS tags. Subsequently, the SERS tag was conjugated with the S protein for the simultaneous detection of IgG/IgM antibodies with the LOD of 1 ng mL−1 S−1-protein antibody.221 Further the LOD was 1 pg mL−1 for clinical samples, which is 1000-times lower than the visualization results. Thus, the SERS-LFIA technique was proposed for rapid screening and bulk diagnosis at ultra-low detection levels when other commonly used methods are not available.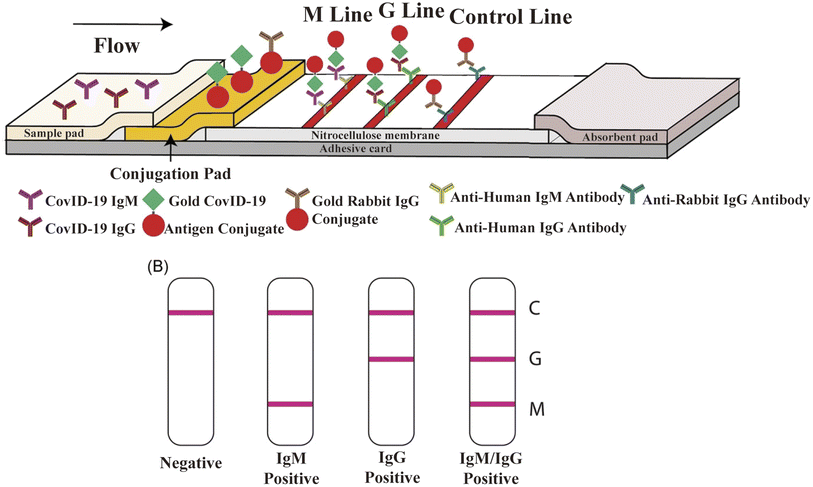 | ||
| Fig. 9 Schematic illustration of rapid SARS-CoV-2 IgM-IgG combined antibody test. (A) Schematic diagram of the detection device. (B) Illustration of different testing results, C: control line; G: IgG line; and M: IgM line. Reproduced with permission from J. Med. Virol., 2020, 92, 1518–1524, 10.1002/jmv.25727.224 | ||
3.3 Indirect enzyme-linked immunosorbent assay
Indirect ELISA is a plate-based assay, in which the microtiter wells are coated with antigens. After adding the sample, the antibodies will specifically recognize the immobilized antigens. After washing, a secondary antibody conjugate is introduced, which specifically binds to the antigen/antibody complex. Then, the color is developed, and the absorbance is related to the number of antibodies in the sample.225–227 Indirect ELISA is easily adaptable to automation for high throughput. Most indirect ELISAs utilize N or S proteins alone or in combination with antigens from the ORF1ab region. Okba et al. produced an array of different ELISA to examine antibody reactivity to SP, S1, and NP proteins, and S-RBD antigens210 and found that the sensitivity of the SP-based ELISA is higher than that of the S1-based ELISA. However, the specificity of S1-based ELISA is higher, because SP-based ELISA is cross-reactive with SARS-CoV and MERS-CoV. In a similar study, the same group found that compared to S-RBD-based ELISA, S-based ELISA is more sensitive,228 which can be expected considering that SP contains subunits S1 and S2. Lopandić et al. produced and purified recombinant protein M, which can specifically bind to antibodies from the sera of COVID-19 convalescents, and in M-based ELISA for the detection of IgG and IgM, the sensitivities were 96% and 93%, respectively.229 In general, all these antigens showed comparable sensitivities when assayed. Recently, numerous magnetic ELISA assays have been developed by combining magnetic bead systems with ELISA. Cai et al. reported that biotinylated synthetic peptides comprising different parts of SARS-CoV-2 proteins were bound to streptavidin-coated magnetic beads, providing 81.5% sensitivity for COVID-19 IgG and IgM detection in about 30 min.230 Following this, the authors used this method to detect cases of COVID-19 antibodies in RT-qPCR-negative infected asymptomatic patients, highlighting the importance of antibody detection. Subsequently, Huergo et al. immobilized recombinant His-tagged SARS-CoV-2 NP on the surface of Ni2+ magnetic beads and challenged whole blood samples obtained from controls or COVID-19 cases.231 The method only required 2 μL of whole blood, and the detection procedure could be accomplished in 12 min. Ultimately, the naked eye could evaluate the results without sophisticated instruments. Presently, indirect ELISA-based methods may offer the advantages of measuring antibody titers, high throughput and selective isotype detection, but they are labor-intensive and unsuitable for POC testing. Thus, to address these issues, microfluidic ELISA was established. Gong et al. developed a microfluidic platform that collected serum by a pulling-force spinning top and paper-based microfluidic ELISA for quantitative IgA/IgM/IgG measurements in an instrument-free manner.232 This method could isolate the serum from whole blood and provide an affordable, rapid and user-friendly way to diagnose COVID-19. Furthermore, it had higher sensitivity for detecting total antibodies compared to conventional methods (99.7% vs. 95.6%). Liu et al. solved this dilemma by presenting a reciprocating-flowing immunobinding (RF-immunobinding) strategy, which enabled the antibodies in the fluid to come into contact with the corresponding immobilized antigens on substrate repeatedly during continuous reciprocating-flowing to achieve adequate immunobinding within 60 s.233 This strategy was further developed into an immunoassay method for the serological detection of 13 suspected COVID-19 patients and obtained a 100% true negative and true positive.3.4 Chemiluminescence immunoassay (CLIA)
CLIA offers significant advantages over traditional assay detection methods, especially in the quantification of antibodies. Its principle is that the recombinant protein of SARS-CoV-2 is labeled by magnetic beads and used to bind the antibody (IgG, IgM, or IgA) in the sample, followed by the use of a secondary antibody coupled with chemiluminescent agents to recognize the antibody. Finally, a luminous signal will be generated when the substrate is added.234,235 Typically, CLIA results are obtained in 0.5–2 h. Similar to ELISA, CLIA is a high-throughput assay with high accuracy, low signal-to-noise ratio, and increased stability of reagents. Many studies utilized CLIA to address the differences in patient populations and time of onset of symptoms vs. serologic results. Grossberg et al. used a multiplex CLIA test for COVID-19 in an otherwise healthy cohort of adults and children in Colorado, and found that IgM antibodies against SARS-CoV-2 were generally detectable in the blood several days after initial infection, and the IgM levels and IgG levels were both elevated early (0–30 days following symptom onset). The IgM levels declined 30 days following symptom onset, while the IgG levels remained elevated for up to 60 days following the onset of symptoms.236 In another study, Long et al. utilized CLIA to test 285 patients with COVID-19 and found that within 19 days after the onset of symptoms, 100% of patients were tested positive for IgG. Because seroconversion for IgG and IgM occurred simultaneously or sequentially, both IgG and IgM titers plateaued within 6 days after seroconversion.237The quantitative index is significant as a biomarker, reflecting the severity of the clinical manifestations of patients. Kong et al. investigated the level of serologic IgM and IgG antibodies and compared the results of CLIA with nucleic acid test (NAT). Among 88 patients, 95.45% were confirmed as positive by the combination of NAT and CLIA, which was significantly higher than by single NAT (73.86%) or CLIA (65.91%).238 They also found that the seroconversion started on day 5 after disease onset, and the IgG level rose earlier than IgM. The comparison between patients with different disease severity suggests that early seroconversion and high antibody titer are linked with less severe clinical symptoms. These results support the combination of CLIA and NAT in routine COVID-19 diagnosis. It is worth noting that this result is opposite to a study that reported a positive correlation between clinical severity and antibody titer.238 Different quantification methods, especially the precision, range and linearity of quantification may be responsible for these differences. Besides the above-mentioned three major categories of methods, there are some other detection techniques such as fluorescent microsphere immunoassays,239 photonic ring immunoassays240 and photometric immunoassays.241
With the popularity of vaccination, it is increasingly accepted that serological antibody tests may serve to evaluate vaccine protectiveness. However, the concept of antibody-based immunization passports did not succeed. The FDA states that antibody testing cannot be used to determine immunity or protection against COVID-19, especially after individuals receive the COVID-19 vaccine.242 Although serological tests demonstrate high sensitivity and specificity, some tests detect antibodies that may be only produced after natural infection. Depending on the assay, people who were not previously infected could test negative for antibodies despite the fact that they have vaccine-induced immunity. Nowadays, antibody tests can play an important role in identifying individuals who may have been exposed to the SARS-CoV-2 virus and may have developed an adaptive immune response.
4. Neutralizing test
To prevent the spread of SARS-CoV-2, many countries are diligently promoting vaccination to achieve herd immunity as soon as possible. Vaccine protection efficacy is the most critical parameter in evaluating vaccine clinical effectiveness. Meanwhile, the detection of neutralizing antibodies also plays a crucial role in assessing the level of human immune response after vaccination. Antibody assays, such as SARS-CoV-2 IgG II Quant, Actim ELISA SARS-CoV-2 IgG and AMP ELISA Test SARS-CoV-2 IgG, that use spiked protein as the detection antigen will not distinguish the antibody response to vaccination from the reaction to natural viral infection. In contrast, the assays that use the N protein as a detection antigen, such as Platelia SARS-CoV-2 Total Ab, Biohit SARS-CoV-2 IgM/IgG Antibody Test Kit, and New York SARS-CoV microsphere immunoassay, may only detect the response to viral infection, but not for response to vaccination. The main constraint of all the above-mentioned antibody tests is that they all detect binding antibodies against the SARS-CoV-2 antigen, which serves only for population-based serosurveillance to understand the epidemiology of COVID-19, such as seroprevalence; however, the analysis results will not directly indicate whether an individual is immune to SARS-CoV-2 infection. The neutralization potential of antibodies in serum can only be ascertained using neutralization tests. Table 5 presents a summary of the characteristics of current neutralizing antibody (nAb) assays.| nAb test method | Test time | Biosafety Level | Pros | Cons |
|---|---|---|---|---|
| Live virus neutralization test | 3–4 days | 3 | Accurate, gold-standard testing conditions close to the reality | Prolonged, low security, expensive and time-consuming, complex analysis leading to high variability |
| Pseudovirus neutralization test | 3–4 days | 2 | More accessible, safer, and more sensitive than PRNT | Still very slow and complicated |
| Surrogate virus neutralization test | 2–3 h | 1 | Fast and easy, simple system, high throughput, high sensitivity, no virus required | Only detecting partial nAb, unable to measure fusion-blocking antibodies |
| Lateral flow assay for neutralization test | 10–15 min | 0–1 | Most accessible, fastest, and easiest to use outside the lab | Similar to the surrogate virus neutralization test |
4.1 Live virus neutralization test
The gold standard for detecting nAb is the plaque reduction neutralization test (PRNT), which requires serial dilution of patient serum and incubation with authentic virus, followed by infection of cells (including animals, chicken embryos, and cells). After several proliferation cycles, it forms a restricted cytopathic cell area known as “plaque” (Fig. 10).243–245 For assays, the nAb titer was evaluated regarding 50% (PRNT50) or 90% (PRNT90) plaque reduction. PRNT seldom requires particular reagents and offers excellent sensitivity. However, it has several limitations, including high technical requirements, cumbersome operation, low throughput and difficulty in automation. Thus, to address the shortcomings of PRNT, an alternative analytical method called the focus reduction neutralization test (FRNT) was developed. FRNT is carried out in a 96-well plate and utilizes immunostaining to display infected lesions that can be counted using a computer-controlled imager, significantly increasing the analytical throughput compared to manual counting performed in PRNT.246 FRNT has been frequently employed to longitudinally assess the antibody response in SARS-CoV-2 infection owing to its high sensitivity and capability to quantify neutralizing antibody titers in serum.228,247–249 In addition, many researchers verified their experimental results using this method as a control experiment.250–252 | ||
Fig. 10 Cartoon illustrating the detection of nAb. (A) For the analysis, the sera were 2-fold serially diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 1 20 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640) and incubated with SARS-CoV-2. The PRNT titer was calculated based on a 50% reduction in plaque counts (PRNT50). The PRNT50 titer was chosen according to the WHO guideline. Reproduced with permission from Viruses, 2022, 14, 410, 10.3390/v14020410.253 (B) When nAb is absent, spike proteins of SARS-CoV-2 interact with ACE2 receptors (ACE2) to allow viral entry, replication, and subsequent plaque formation. The presence of nAb inhibits viral entry and HEK293T is commonly expressed ACE2 together with TMPRSS2. Reproduced with permission from J. Mol. Sci., 2021, 22, 2268, 10.3390/ijms22052268.254 640) and incubated with SARS-CoV-2. The PRNT titer was calculated based on a 50% reduction in plaque counts (PRNT50). The PRNT50 titer was chosen according to the WHO guideline. Reproduced with permission from Viruses, 2022, 14, 410, 10.3390/v14020410.253 (B) When nAb is absent, spike proteins of SARS-CoV-2 interact with ACE2 receptors (ACE2) to allow viral entry, replication, and subsequent plaque formation. The presence of nAb inhibits viral entry and HEK293T is commonly expressed ACE2 together with TMPRSS2. Reproduced with permission from J. Mol. Sci., 2021, 22, 2268, 10.3390/ijms22052268.254 | ||
In summary, for the detection of nAbs against SARS-CoV-2, live virus nAb assays are highly effective methods. However, the turnaround time required for the virus to form visible plaques is too long, and all the experimental procedures must be processed in a biosafety level 3 (BSL-3) containment facility. Hence, this approach is not feasible for large-scale serological diagnosis and vaccine evaluation.
4.2 Pseudovirus neutralization test
To overcome the containment of BSL-3, pseudotyped virus engineering has been developed. Pseudovirus (or pseudotyped virus) is a recombinant viral particle that expresses the SARS-CoV-2 S protein on its surface. Pseudovirus binds to ACE2, which is over-expressed by target cells and highly mimics the invasion process of SARS-CoV-2.255–257 To facilitate readout when designing, pseudotyped viruses typically harbor reporter genes encoding NanoLuc luciferase or green fluorescent protein (GFP).258,259 The selection of cell lines and viral models will influence the neutralization activity of the antibody. In most pseudovirus neutralization assays, the Vero, Huh7 and HEK293T cell lines were frequently utilized.260–263 Nie et al. infected six diverse cells (human- and animal-derived cell lines) using the developed vesicular stomatitis virus (VSV) pseudovirus and found that all the cells exhibited high susceptibility, where Huh7 cell signaling was the highest,264 which is consistent with the previously reported result.265 However, it is worth noting that SARS-CoV-2 replicates weakly in the Huh7 cell lines, as shown in some live virus neutralization assays.266 To enhance the infectivity of cells in pseudovirus neutralization assays, HEK293T cells are frequently modified to express both ACE2 and TMPRSS2, making them more susceptible to infection.267,268 VSV, human immunodeficiency virus-1 (HIV-1) and murine leukemia virus (MLV) are often chosen as vectors in the construction of pseudoviruses. Table 6 presents a summary of the SARS-CoV-2 neutralization assays that used these vectors. Among them, VSV is the most frequently utilized vector because it can be engineered into two formats, i.e., a replication-deficient VSV that lacks the G protein (VSVΔG) and a replication-capable VSV/SARS-CoV-2 chimeric virus.269–272 Case et al. produced a high-titer, replication-competent chimeric VSV, which expressed the SARS-CoV-2 S protein and performed similarly to the SARS-CoV-2 clinical isolate in multiple neutralization assays.273 In another study, Schmidt et al. developed a series of SARS-CoV-2 S-pseudotype, single-cycle, HIV-1 and VSV-based methods, as well as replication-capable VSV/SARS-CoV-2 chimeric viruses for the detection of nAb. They simultaneously assessed the differences among these pseudoviruses and live viruses and found that the HIV-1 and VSV-based pseudotyped viruses were slightly less sensitive to neutralization compared to the live SARS-CoV-2, especially by weakly neutralizing plasma. In contrast, VSV/SARS-CoV-2 chimeric viruses were similarly susceptible to neutralization by monoclonal antibodies to authentic SARS-CoV-2. More interestingly, VSV/SARS-CoV-2 chimeric viruses have been shown to be more sensitive to plasma neutralization than SARS-CoV-2.274 Notably, the irreproducible HIV-1 and VSV-based pseudoviruses used by other researchers displayed the identical reliable performance as live SARS-CoV-2 in detecting SARS-CoV-2 nAbs275,276 In addition, many studies have constructed pseudoviruses by truncating the C-terminus of the S protein and introducing a D614G mutant spike to increase the infectivity titer of SARS-CoV-2 pseudotypes in neutralization experiments.277–280 In the assay, single-cycle pseudovirus neutralization assays allow direct readout of the percentage of virus blocked from entering in a single round of infection, and replicating chimeric viruses can be employed to assess the capability of nAb to reduce virus particle growth or eliminate the virus. Thus, the selection of cell and pseudovirus models is warranted for the assay. A reagent public repository is provided for our selection (https://www.beiresources.org/).| Sample type | Pseudoviral vector | Fluorescent gene | Target cell | TMPRSS2 expression | Type of SP | Effect compared with SARS-CoV-2 virus | Turnaround time | Biosafety level | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Healthy adults immunized with the mRNA vaccine | VSVΔG | Firefly Luciferase | A549 | Engineered | Full-length SP, D614G mutation | Similar | ∼2 days | 2 | 281 |
| COVID-19 seropositive samples | VSVΔG | Luciferase | Vero E6 | No | Full-length SP | — | ∼3 days | 2 | 282 |
| Convalescent patient sera | VSVΔG | Firefly luciferase | Huh7 | No | Full-length SP | — | ∼3 days | 2 | 283 |
| Serum samples from COVID-19 and health donors | VSVΔG | Luciferase | Vero E6 | No | Spike-Δ19 | — | ∼1 day | 2 | 284 |
| Healthy adults immunized with the vaccine | VSVΔG | GFP and luciferase | Vero 76 | No | — | Consistent | ∼3 days | 2 | 269 |
| Germline-encoded interferon | VSV chimeric virus | GFP | HEK293 | Engineered | Spike-Δ19 | — | ∼3 days | 2 | 285 |
| Human plasma samples COV-47, COV-72 and COV-107 and monoclonal antibodies C144, C135 and C121 | VSV chimeric virus | GFP | HEK293T | No | Spike-Δ19 | — | ∼2 days | 2 | 286 |
| mAbs against the RBD and convalescent sera | VSV chimeric virus | eGFP | Vero E6 | Engineered | Spike-Δ21 | — | ∼2 days | 2 | 256 |
| mAbs, animal immune sera, human convalescent sera, and human sera immunized by mRNA vaccine | VSV chimeric virus | Luciferase | Vero E6 | Engineered | Spike-Δ19 | Consistent | ∼2 days | 2 | 287 |
| Convalescent plasma and human monoclonal antibodies | VSV chimeric virus | GFP | Vero E6 | No | Spike-Δ18 | Better than SARS-CoV-2 | ∼3 days | 2 | 288 |
| Rhesus macaque anti-SARS-CoV-2 serum | HIV-1 | Firefly luciferase | HEK293T | No | Spike-ΔCT13 | — | ∼3 days | 2 | 289 |
| Human monoclonal antibodies | HIV-1 | Firefly luciferase | HEK293T | Engineered | Full-length SP | — | ∼3 days | 2 | 290 |
| Animal immune sera, human convalescent sera, and human sera immunized by mRNA vaccine | HIV-1 | Luciferase | Vero E6 | Engineered | Spike-Δ19 | Inferior to SARS-CoV-2 | ∼3 days | 2 | 291 |
| Convalescent plasma and human mAbs | HIV-1 | NanoLuc luciferase and GFP | Vero E6 | No | Spike-Δ19 | Inferior to SARS-CoV-2 | ∼3 days | 2 | 288 |
| Convalescent COVID-19 patient sera | HIV-1 | Luciferase | HEK293T | No | Full-length SP | — | ∼3 days | 2 | 292 |
| S2X259 mAb | MLV | Luciferase | Vero E6 | No | Spike-Δ19 | Inferior to SARS-CoV-2 | ∼4 days | 2 | 293 |
| Blood samples | MLV | Firefly luciferase | HEK293T | No | Spike-Δ19 | — | ∼3 days | 2 | 294 |
| Mouse mAb 10G6H5 against SARS-COV2 S protein, human plasma from COVID-19 patients | MLV | Luciferase | HEK293T | Engineered | Full-length SP | — | ∼3 days | 267 |
Overall, pseudoviruses are relatively safe and reliable, with similar testing results to live SARS-CoV-2 viruses, which can be used for high-throughput assays. Nevertheless, the pseudovirus has different kinetics in comparison with live SARS-CoV-2 when expressing SP, which may cause testing bias.
4.3 Surrogate virus neutralization test (sVNT)
Although the pseudovirus-based nAb test overcomes the limitation of BSL-3, it still requires the use of live viruses and cells. In addition, the analytical results are heterogeneous between laboratories due to the different culture conditions, virus strains and cell lines. In this case, sVNT can address the aforementioned issues.The majority of sVNTs are based on the mechanism of blocking the interaction between RBD and ACE2 as well as the high immunogenicity of the RBD. Typically, 96-well plates are coated with the human angiotensin-converting enzyme 2 (hACE2) receptor, and then test sera are co-incubated with horseradish peroxidase (HRP)-conjugated recombinant SARS-CoV-2 RBD fragments (HRP-RBD). The binding of HRP-RBD and hACE2 is blocked by nAbs against SARS-CoV-2, and this blocking effect can be detected by reducing the HRP luminescence signal.295–298 Alternatively, plates can be encapsulated with RBD, soluble hACE2 for competition with nAbs.299,300 After comparing these two immobilization assays, Abe et al. found that the immobilized RBD and soluble hACE2 were more sensitive.301
At present, most sVNTs are ELISA-like methods. To date, many commercial kits based on this method have been developed. Krüttgen et al. evaluated two ELISA-based sVNT kits and discovered that they both displayed high sensitivity and specificity.302 Michiels et al. compared the sVNT commercial kit (GenScript cPass™) with a live virus neutralization assay and Luminex multiplex immunoassay (MIA), and studied two different cohorts using these three methods.303 The sVNT obtained a sensitivity of 94% (CI 90–96%) and 89% (CI 81–93%) compared with the other two methods. They also found that the different methods had a strong antibody titer correlation (r2 > 0.8). Interestingly, Tan et al. found that sVNT was as specific as the live virus neutralization assay and more sensitive than it.304 Besides, Bošnjak et al. compared sVNT with the pseudovirus neutralization test (PVNT) and found a strong positive correlation between sVNT and PVNT50 (r2 = 0.7135, p < 0.0001) as well as PVNT90 (r2 = 0.5042, p < 0.0001) inhibitory titers.305 Embregts et al. also studied 298 PCR-confirmed patient sera using sVNT and PVNT and found that sVNT had moderate to high sensitivity (91.3%) and specificity (100%), but it was not sensitive to detect low titers.306 However, when using a cut-off value of 50% inhibition, highly neutralized samples could be identified, which is similar to a previous report.295
In addition to the above-mentioned ELISA-based sVNTs, many other forms of sVNTs have been developed. Wang et al. developed a track-etched microporous membrane filtration microplate (TEM) and optical fibers transmitted immunosensing smartphone platform (TEMFIS)-based surrogate virus neutralization test (TEMFIS-sVNT) for rapid one-step testing of nAb to SARS-CoV-2.307 The entire assay process only took 45 min. The pore size of the TEM is 3 μm, allowing softer red blood cells, smaller platelets and fresh plasma blood components to pass through or be washed away, and thus this device can be used to assay serum, plasma and whole blood samples. In addition, TEMFIS-sVNT has a strong statistical correlation (R2 > 0.8) with ELISA-sVNT and pVNT. However, it requires HRP-tagged RBD. Thus, to address this issue, Luo et al. established an alternative virus neutralization test on a label-free immunoassay platform (LF-sVNT).308 The LF-sVNT mimics the SARS-CoV-2 surface using a sensing probe coated with RBD and simulates host cells using ACE2, which models virus-host cell interactions. Compared to other sVNTs, LF-sVNT eliminates the effects of attachment of secondary antibody-labeled enzymes,309 extension of streptavidin-labeled enzymes or fluorophores,301,310 and performing color-generating enzyme reactions.301,304,309,311 In addition, this platform allows real-time monitoring of RBD-ACE2 interactions to reduce the experimental error rates (Fig. 11).
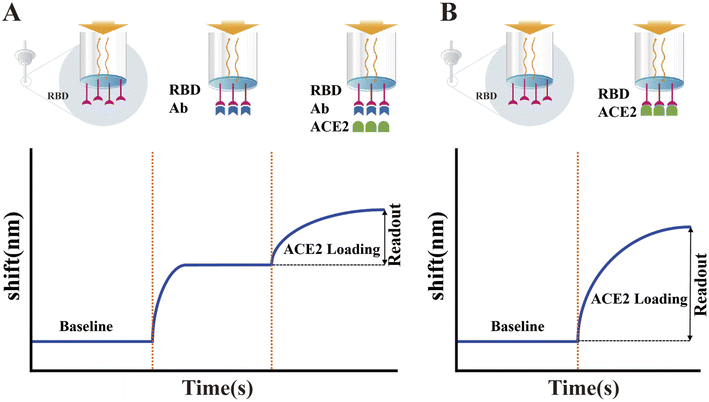 | ||
| Fig. 11 Illustration of the LF-sVNT protocol and example sensorgrams. (A) First cycle measuring the binding ability of RBD to ACE2 after neutralization. (B) Second cycle measuring the full binding ability of RBD without neutralization. Reproduced with permission from J. Clin. Microbiol., 2021, 59, e0019321, 10.1128/JCM.00193-21.308 | ||
Overall, sVNT is more straightforward, cheaper and faster (typically 1–2 h) than other nAb assays, making it more suitable for rapidly screening many samples. In addition, because the test is not species antibody-dependent, it can detect nAbs in any animal species used in preclinical testing of the SARS-CoV-2 vaccine. However, the drawback of sVNT is that it is unable to detect nAbs other than that bound to SARS-CoV-2 S-RBD. However, these non-RBD-targeting antibodies play only a minor role in SARS-CoV-2 neutralization.312,313 Nevertheless, although sVNT analysis may never completely replace conventional virus neutralization test (cVNT), the performance of sVNT is closely related to that of cVNT and pVNT.
4.4 Lateral flow assay (LFA)-based neutralization test
Currently, the majority of LFAs are based on the principle of antibody–antigen interaction. In general, the higher the affinity of the antibody, the higher the sensitivity of the assay. Thus, antibodies with high affinity for the antigen can also be developed with modern antibody technologies, making this type of assay relatively easy to implement. Protein–protein interaction without antibodies can also be applied in lateral flow assays. However, it is very rare, given that the binding affinity between proteins is commonly lower than that between antibodies and antigens and cannot be artificially increased. A lower binding affinity or lower analytical sensitivity often results in a longer analysis time, which requires high-sensitivity detection systems such as fluorescent labeling and fluorescent detection instruments.27Fortunately, the SARS-CoV-2 S-RBD exhibits high affinity to ACE2. The lateral flow competitive protein binding assay was developed based on the interaction between RBD and ACE2 or between the S1 protein and ACE2.312,314,315 One of the recombinant proteins is labeled with a colored material (usually colloidal gold), and another is dispensed on a nitrocellulose membrane to form the test line. During the assay, the presence of nAb in the sample will block the interaction between the labeled-RBD and ACE2, by which the intensity of the detection line on the nitrocellulose membrane can be measured. Alternatively, the colored material labeled S1 or RBD may also influence the detection sensitivity. Zhang et al. compared the analytical performance of AuNPs-tagged RBD and AuNPs-tagged S1 and found that LFA using AuNPs-tagged S1 was more sensitive.316 However, although this finding is predictable, given that there are many reports that RBD is the main part of the S1 protein, which plays a neutralizing role, some nAbs also bind to the NTD of the S1 protein, playing a neutralizing role.317–320 Although the sensitivity is different using RBD or S1, nAb-LFA is as effective as the pseudovirus neutralization assay for detection.316
Considering the convenience, speed and simplicity of the LFA-based neutralization test, it can be used both in professional laboratories and by individuals at home. Therefore, the development of LFA-based SARS-CoV-2 nAb assays can complement the available methods for detecting nAb. Table 7 presents a summary of the commercially available LFAs for the detection of SARS-CoV-2 nAb.
| Kit name | Regulatory status | Time to result (min) | Self-testing/self-collection | Validated sample types | Impact of SARS-CoV-2 variant | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 nAb Assay (colloidal gold) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 97.62% | 100.00% |
| Serum | Beta – unknown | ||||||
| Whole blood | Gamma – unknown | ||||||
| 2019-nCoV nAbs Test (fluorescence immunochromatographic assay) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 98.40% | 98.40% |
| Serum | Beta – unknown | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Rapid Test Device | CE-IVD | 15 | Intended for self-collection (kit available) | Plasma | Alpha – unknown | 93.86% | 99.89% |
| Serum | Beta – unknown | ||||||
| Gamma – unknown | |||||||
| reOpenTest™ SARS-CoV-2 nAb Rapid Test | CE-IVD | 10 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 98.82% | 99.17% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAbs Test | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 98.80% | 99.14% |
| Serum | Beta – unknown | ||||||
| Whole blood | Gamma – unknown | ||||||
| Coronavirus (SARS-CoV-2) nAb Rapid Test | CE-IVD | 10 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 95.70% | 100.00% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Rapid Test | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 98.51% | 99.50% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Test Kit (fluorescence immunoassay) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 95.06% | 99.00% |
| Serum | Beta – unknown | ||||||
| Gamma – unknown | |||||||
| SARS-CoV-2 nAb TestKit (colloidal gold chromatographic immunoassay) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 96.50% | 100.00% |
| Serum | Beta – unknown | ||||||
| Venous blood | Gamma – unknown | ||||||
| V-Pass SARS-CoV-2 nAb Rapid Test (colloidal gold) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 96.19% | 95.00% |
| Serum | Beta – unknown | ||||||
| Venous blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Test Kit (colloidal gold) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 90.70% | 98.63% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – unknown | ||||||
| COVID-19 nAb Rapid Test Kit (whole blood/serum/plasma) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 99.29% | 99.76% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – unknown | ||||||
| 2019-nCoV nAb Test Cassette (whole blood/serum/plasma) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 94.17% | 98.18% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – unknown | ||||||
| PRIMACOVID™ Covid-19 nAb Rapid Test | CE-IVD | 10 | Intended for self-testing (in development) | Plasma | Alpha – no expected impact (in silico analyses) | 92.60% | 97.40% |
| Serum | Beta – unknown | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAbs Test Kit (dry fluorescence Immunoassay) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 95.00% | 98.00% |
| Serum | Beta – unknown | ||||||
| Venous blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Rapid Test (fluorescence dry quantitative immunoassay) | CE-IVD | 10 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 97.40% | 100.00% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – no expected impact (in silico analyses) | ||||||
| SARS-CoV-2 nAb Rapid Test (colloidal gold) | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 94.70% | 100.00% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – no expected impact (in silico analyses) | ||||||
| Corona Virus (COVID-19) Combined (IgM/IgG/nAb) Rapid Test | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – unknown | 99.00% | 97.00% |
| Serum | Beta – unknown | ||||||
| Venous blood | Gamma – unknown | ||||||
| COVID-19 nAb Test Kit | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 95.40% | 98.40% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – unknow | ||||||
| Wesail COVID-19 nAb Test Kit | CE-IVD | 15 | Intended for professional use only | Plasma | Alpha – no expected impact (in silico analyses) | 95.40% | 98.40% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Venous blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Test Kit (colloidal gold) | CE-IVD | 15 | Intended for self-testing (version available) | Plasma | Alpha – no expected impact (in silico analyses) | 98.13% | 99.26% |
| Serum | Beta – no expected impact (in silico analyses) | ||||||
| Whole blood | Gamma – unknown | ||||||
| SARS-CoV-2 nAb Rapid Test (colloidal gold) | CE-IVD | 10 | Intended for professional use only | Plasma | Alpha – unknown | 98.56% | 99.65% |
| Serum | Beta – unknown | ||||||
| Whole blood | Gamma – unknown |
nAb-LFA can be performed similar to other neutralization tests by diluting the sample multiple times in triplicate to calculate the inhibitory concentration 50 (IC50), effective concentration 50 (EC50) or PRNT50. However, for nAb-LFA, which is intended for individual home users, it is impossible to prepare blood samples at home at different dilutions accurately. Therefore, a single dilution must be selected that will cover the protective titer of nAb. Currently, the FDA recommends an nAb titer at least 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160; however, the titer of 1
160; however, the titer of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 may be acceptable if no alternative matching units are available. For nAb-LFA, the optimal cut-off value should be the IC50 of the selected single dilution. Zhang et al. tested 80 COVID-19 plasma samples using nAb-LFA with an artificially set 50% inhibition at 1
80 may be acceptable if no alternative matching units are available. For nAb-LFA, the optimal cut-off value should be the IC50 of the selected single dilution. Zhang et al. tested 80 COVID-19 plasma samples using nAb-LFA with an artificially set 50% inhibition at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 dilution as the cut-off value and found that the final result was similar to the ELISA-based neutralization assay (R2 = 0.79).321 Wang et al. speculated that the nAb-LFA using a single dilution of the test sample could be designed to cover the range of color changes equivalent to 1
12 dilution as the cut-off value and found that the final result was similar to the ELISA-based neutralization assay (R2 = 0.79).321 Wang et al. speculated that the nAb-LFA using a single dilution of the test sample could be designed to cover the range of color changes equivalent to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 1
20 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PRNT.322 However, the cut-off value should be determined by large-scale clinical trials, such as the IC50 diluted sera for 1
100 in PRNT.322 However, the cut-off value should be determined by large-scale clinical trials, such as the IC50 diluted sera for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 for Japanese Encephalitis323 and 1
10 for Japanese Encephalitis323 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 for Mumps.324
22 for Mumps.324
Overall, nAb-LFA is inexpensive and highly portable, which can be used to assist in the detection and longitudinal evaluation of nAbs in resource-poor areas where traditional virus neutralization tests or ELISA-based neutralization assays are not available. Considering that the design and manufacture of nAb-LFA are comparatively easy, even if the virus mutates, this can be addressed by modifying the labeled recombinant protein. However, the significant limitation of nAb-LFA is that it cannot facilitate the detection of nAb without high affinity for the S1 protein and a large number of nAbs with low affinity.
5. SARS-CoV-2 antigen detection
Since the beginning of the COVID-19 pandemic, nucleic acid amplification assays (e.g., RT-PCR assays) have dominated to detect the SARS-CoV-2 virus. Thus far, clinical laboratories worldwide have performed over 3 billion molecular diagnostic tests for SARS-CoV-2. More than 850 million tests were performed in the United States (an average of 2.5 tests per person),305 although China has not yet reported complete testing data, but the absolute number should be greater than any other countries. The execution of mass nucleic acid amplification assays is technically challenging, labor-intensive, and dependent on efficient sample transport and reporting systems, all of which contribute to detection bias. Thus, to facilitate the diagnosis and treatment of COVID-19, antigen rapid diagnostic tests (Ag-RDTs) for SARS-CoV-2 have evolved rapidly, with at least 600 antigen testing kits now available worldwide and widely used in healthcare settings as well as high- and low-resource settings.306 An analysis on the performance of several mainstream test kits currently available in the market is shown in Table 8.| Method | Developer | Test | Sensitivity | Specificity | Antigen | Time (min) | Specimen |
|---|---|---|---|---|---|---|---|
| LFIA | SD Biosensor, Inc. | COVID-19 At-Home Test | 95.3% | 100.0% | SARS-CoV-2 NP | 15–30 | Human nasal sample |
| InBios International Inc. | SCoV-2 Ag Detect Rapid Self-Test | 85.7% | 100.0% | SARS-CoV-2 NP | ∼20 | Human nasal sample | |
| Oceanit Foundry LLC | ASSURE-100 Rapid COVID-19 Test | 89.00% | 100.0% | SARS-CoV-2 NP | ∼20 | Human nasal sample | |
| PHASE Scientific International, Ltd | INDICAID COVID-19 Rapid Antigen Test | 84.4% | 96.3% | SARS-CoV-2 NP | ∼20 | Human nasal sample | |
| Nano-Ditech Corp. | Nano-Check COVID-19 Antigen Test | 90.32% | 100.0% | SARS-CoV-2 NP | 15–20 | Human nasal sample | |
| Siemens Healthineers | CLINITEST Rapid COVID-19 Antigen Self-Test | 86.5% | 99.30% | SARS-CoV-2 NP | ∼15 | Human nasal sample | |
| Abbott Diagnostics Scarborough, Inc. | BinaxNOW COVID-19 Ag Card Home Test | 91.7% | 100.00% | SARS-CoV-2 NP | 15–30 | Human nasal sample | |
| iHealth Labs, Inc. | iHealth COVID-19 Antigen Rapid Test Pro | 88.2% | 100.00% | SARS-CoV-2 NP | ∼15 | Human nasal sample | |
| iHealth Labs, Inc. | iHealth COVID-19 Antigen Rapid Test | 94.3% | 98.10% | SARS-CoV-2 NP | ∼15 | Human nasal sample | |
| Celltrion USA, Inc. | Celltrion DiaTrust COVID-19 Ag Home Test | 86.7% | 99.80% | SARS-CoV-2 NP and S-RBD antigens | ∼15 | Human nasal sample | |
| Xtrava Health | SPERA COVID-19 Ag Test | 91.8% | 96.90% | SARS-CoV-2 NP | ∼15 | Human nasal sample | |
| ANP Technologies, Inc. | NIDS COVID-19 Antigen Rapid Test Kit | 95.1% | 97.00% | SARS-CoV-2 NP | ∼15 | Human nasal sample | |
| CLIA | Siemens Healthcare Diagnostics, Inc. | ADVIA Centaur SARS-CoV-2 Antigen (CoV2Ag) | 85.1% | 100.0% | SARS-CoV-2 NP | 10 | Human nasal sample |
| Siemens Healthcare Diagnostics, Inc. | Atellica IM SARS-CoV-2 Antigen (CoV2Ag) | 85.1% | 100.0% | SARS-CoV-2 NP | 26 | Human nasal sample | |
| LIAISON SARS-CoV-2 Ag | DiaSorin, Inc. | 97.00% | 100% | SARS-CoV-2 NP | ∼30 | Human nasal sample |
Antigen-based diagnostic tests are based on the principle of detecting viral proteins by antigen capture methods (e.g., using antibodies and aptamers), and in clinical testing, are mainly used to detect protein fragments on or within the virus from NP swabs or nasal swabs. To date, most antigen tests use portable devices (e.g., LFIA). Compared to RT-PCR, this type of test can detect active infection within 15 min, and it minimizes the risk of virus transmission by avoiding crowd gathering. Therefore, this method offers significant benefits in preventing early infection transmission. Besides, enzyme immunoassay techniques such as ELISA or CLIA on semi-automated or automated instruments also enable the detection of antigens with high-throughput. To date, many other antigen detection technologies have been developed for SARS-CoV-2 such as biosensors using nanotechnology, field-effect transistors (FETs), microfluidic platforms and electrochemical methods.
Given the current widespread use of Ag-RDT in clinical, community and home settings, the progress of antigen detection is summarized and its clinical application together with existing challenges is evaluated in this section.
5.1 SARS-CoV-2 NP detection
The most common protein in the structure of SARS-CoV-2 is NP, which is an evolutionarily conserved, highly immunogenic protein. NP is essential for the early replication of the virus in the host cell, which plays a major role in packaging viral RNA into a ribonucleoprotein (RNP) complex called nucleocapsid.325,326 When the virus enters the host cell, NP supports the replication of the viral RNA and releases the virus particles into the host cell.327,328 However, because NP is highly conserved among all coronaviruses, the specificity of its detection is low.329 NP is released in large quantities in the serum, nasopharyngeal aspirate, throat wash samples, feces and urine in the early stages of infection.330 Therefore, the detection of NP antigen may be an effective strategy for the early screening of patients with suspected SARS-CoV-2 infection.331Currently, most Ag-RDTs are quantitative immunoassays that use SARS-CoV-2-specific antibodies to bind viral proteins and generate visual or fluorescent signals. This strategy is primarily performed in the LFA on a nitrocellulose membrane and provides assay results within 10 to 30 min.332–335 In LFIA, the probe that can specifically bind to NP is the key factor in determining the assay performance. To obtain high-performance immunoprobes, Yamaoka et al. used a wheat germ cell-free protein production system yielding a monoclonal antibody (mAb) that specifically binds to the SARS-CoV-2 NP.336 The modified antibodies not only had no cross-reactivity with other coronaviruses, but this mAb-based LFIA could also be used to detect variants 501Y.V1-V3. Kim's team also obtained four SARS-CoV-2 NP-specific single-chain variable fragment-crystallizable fragment (scFv-Fc) fusion antibodies by phage display.337 They screened the specific scFv-Fc antibody pair as the LFIA detection probe, and the developed cellulose nanobead LFIA platform could detect 2 ng of antigenic protein and 2.5 × 104 pfu of cultured virus (Fig. 12A). In addition to probes, the color-labeling of LFIA can greatly affect the detection performance.338,339 Oh et al. synthesized a plasmonic exciton color-preserving (PLASCOP) gold nanocluster to detect NPs with an LOD 5.9 to 23.8-times lower than that of GNPs (Fig. 12B).340 Wang et al. developed a triple quantum dot shell (MagTQD) nanotag and integrated it in the LFIA system, enabling the simultaneous detection of the SARS-CoV-2 SP and NP antigens on one strip. The LOD for the two antigens in direct and enrichment modes was 1 and 0.5 pg mL−1, respectively.341 Novel nanomaterials can be used in the LFIA platform to design other detection formats. Guo et al. synthesized a versatile magnetic all-solid Z-scheme heterojunction (Fe3 O4@SiO2@TiO2@CdS/Au, FSTCA) nanocomposite, which has various advantages such as simplified separation and washing process, to improve the reproducibility and stability. Then, the authors developed a photoelectrochemical immunosensor to detect NP with a wide linear range of 10 pg mL−1 to 100 ng mL−1 and low detection limit down to 2.9 pg mL−1.342 The Si-FITC NPs prepared by Mao et al. not only solved the problem of reduced quantum yield during the coupling reaction but also achieved the sensitive detection of NP (LOD of 3 pg mL−1) (Fig. 12C).343 Besides antibody probes, aptamer probes were used in the NP assay.344 Zhang et al. constructed a sensor using an aptamer capable of detecting pM-level NP.345 In addition, other sensors are also being actively expanded for NP detection, such as FET sensors,346,347 microfluidic platforms348,349 and electrochemical sensors.350,351 Recently, Novodchuk et al. developed an FET sensor using boron/nitrogen co-doped graphene oxide gels (BN-GO gels), which could detect viral proteins with an LOD of 10 ag mL−1 (Fig. 12D).352
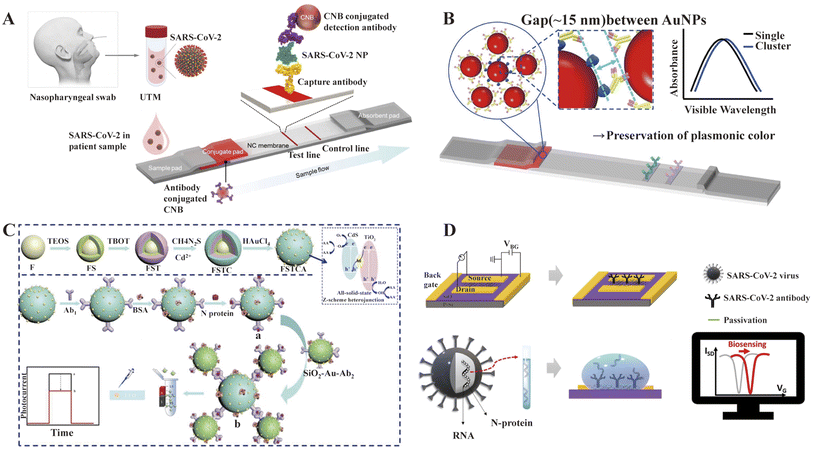 | ||
| Fig. 12 Schematic diagram of some strategies for SARS-CoV-2 NP assay. (A) Schematic illustration of the development processes of SARS-CoV-2-specific scFv-Fc fusion proteins based on phage display technology and LFIA-based biosensor using scFv-Fc fusion proteins. Reproduced with permission from Biosens. Bioelectron., 2021, 175, 112868, 10.1016/j.bios.2020.112868.337 (B) Structure of AuNP-based clusters LFIA. Reproduced with permission from Biosens. Bioelectron., 2022, 205, 114094, 10.1016/j.bios.2022.114094.340 (C) Preparation and detection process of signal-off photoelectrochemical immunosensor based on magnetic FSTCA. Reproduced with permission from Sens. Actuators, B, 2023, 374, 132800, 10.1016/j.snb.2022.132800.342 (D) The Schematic of the device configuration and biosensing mechanism of the BN-GO gel FET NP biosensor. Reproduced with permission from Biosens. Bioelectron., 2022, 210, 114331, 10.1016/10.1016/j.bios.2022.114331.352 | ||
5.2 SARS-CoV-2 SP detection
SP is a glycoprotein (∼140 kDa) that consists of two subunits (S1 and S2) and forms a trimer on the viral membrane.353 Compared with other SARS-related coronaviruses, the nucleotide sequence similarity of the S gene is less than 75%.71 Therefore, SP may be one of the most valuable antigenic biomarkers for diagnosing COVID-19.Similar to the NP assay, most LFIA assays for SP are based on the hybridization of antigen–antibody immunoreactivity.332,354–356 However, Baker et al. found that N-acetylneuraminic acid had affinity and specificity for SARS-COV-2 SP, based on which they developed a paper-based LFA and verified its feasibility using pseudovirus.357 Similarly, Kim et al. found that glycoproteins also can bind to the SARS-COV-2 S protein, based on which they developed an SP test platform called GlycoGrip.358 These studies broaden the idea of exploring new biological probes. More recently, several nanoscale integrated architectures based on optical and electronic systems have been introduced to detect SP with much higher sensitivity, such as terahertz plasmonic biosensor,359 gated graphene-enhanced FET-based biosensors,360 In2O3 nanoribbon transistors biosensor,361 and electrical-double-layer gated FET-based biosensor.347 Generally, most of these devices have favorable sensitivity and possess the advantages of low cost, simplicity, easy miniaturization and mass fabrication. However, there is still a continuous demand for fast, ultrasensitive biosensors to detect SP. SERS is known as an ultrasensitive molecular spectroscopy technique that is not interfered by water, giving it a distinct advantage in identifying biological samples.362,363 Compared to LFIA, photothermal methods and electrical biosensors, SERS-based immunoassay techniques do not require sample pretreatment and are highly sensitive for the detection of trace bioparticles.363,364 Currently, SERS technology has been well developed and applied in SP detection.365–367 Besides direct testing for unprocessed samples,368 SERS platforms combined with other techniques have also been developed. Huang et al. developed a deep learning-based SERS technique for the on-site detection of SP from human throat swabs and sputum with an accuracy of 87.7% in 20 min.369 Yang et al. introduced machine algorithms in SERS technology to further develop a sensor capable of detecting SARS-CoV-2 SP and differentiating among 13 respiratory viruses with >99% accuracy.370 Shim et al. proposed a sensing platform based on core–shell SERS with single nanoparticle (SNP)-based digital SERS analysis capability. This platform has a better detection range and lower detection limit than conventional ELISA, which can distinguish multiple mutants.371 Peptides have merits over antibodies including smaller size, stability and scaled-up preparation. Recently, our group screened a peptide probe Pn that can specifically recognize SP through phage display. By combining a specific peptide with the tyramine signal amplification (TSA) method, an ultra-sensitive peptide-ELISA (p-ELISA) detection method was developed, capable of detecting SARS-CoV-2 SP as low as 0.4 pg mL−1 and SARS-CoV-2 pseudoviruses with 3 TCID 50 per mL (Fig. 13).372
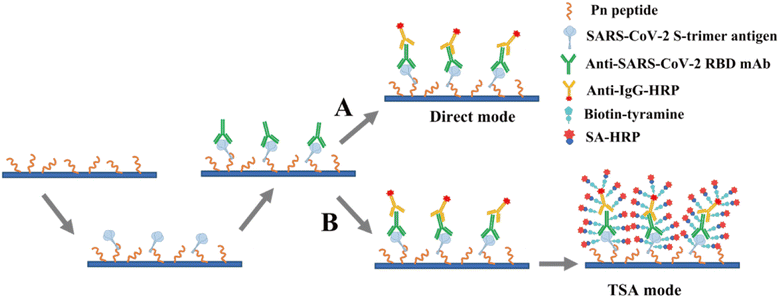 | ||
| Fig. 13 Schematic illustrating the p-ELISA to detect SARS-CoV-2 S antigen. (A) In the direct mode, the peptide Pn was used as the capture probe and an antibody (anti-SARS-CoV-2 RBD mAb) bound to the S-RBD was selected as the detection antibody, by which a “peptide-antigen-antibody” sandwich p-ELISA was constructed to detect SARS-CoV-2 SP antigen. (B) TSA mode. Upon catalysis by HRP, biotinylated tyramine is deposited at the site of signal amplification. Subsequently, SA-HRP is added to introduce more HRP in the positive microplate wells where the sample is tested, taking advantage of the high affinity between streptavidin and biotin. Reproduced with permission from Sens. Actuators, B, 2023, 387, 133746, 10.1016/j.snb.2023.133746.372 | ||
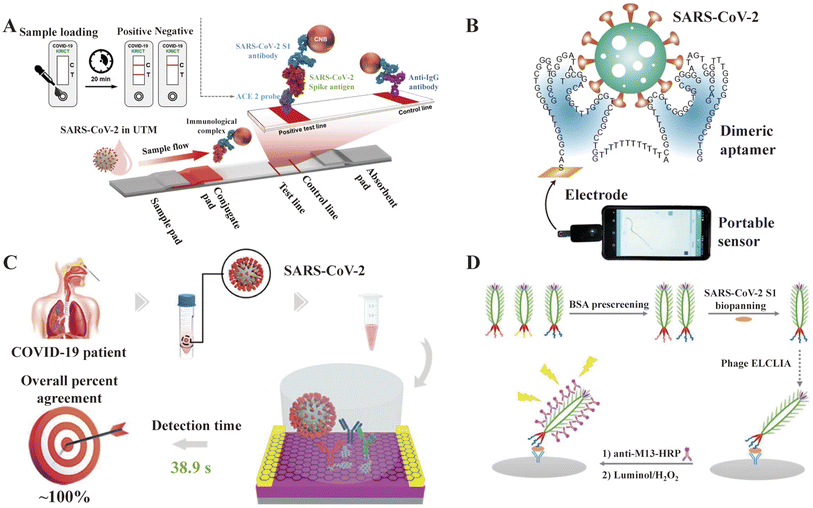 | ||
| Fig. 14 Schematic diagram of some strategies for S1 protein detection. (A) Schematic of an ACE2-based LFIA consisting of a sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad. The test line placed on the nitrocellulose membrane contains ACE2 to detect SARS-CoV-2 SP. Reproduced with permission from Biosens. Bioelectron., 2021, 171, 112715, 10.1016/j.bios.2020.112715.374 (B) Platform for the rapid electrochemical detection of S1 protein with high-affinity dimer aptamer. Reproduced with permission from Angew. Chem., 2021, 60, 24266–24274, 10.1002/anie.202110819.388 (C) Multiantibody FET sensor. Reproduced with permission from J. Am. Chem. Soc., 2021, 143, 19794–19801, 10.1021/jacs.1c08598.386 (D) Schematic diagram of antibody/S1/specific peptide-displayed phage/HRP-M13 antibody-based ELCLIA. Reproduced with permission from Anal. Chem., 2022, 94, 11591–11599, 10.1021/acs.analchem.2c01988.387 | ||
Functionally, the binding of RBD to ACE2 is crucial for the SARS-CoV-2 virus to enter human cells. SARS-CoV-2 S-RBD shares only 70% sequence identity with SARS-CoV S-RBD, and the latter has been evaluated for vaccine and therapeutic drug development.389 Therefore, the S-RBD of SARS-CoV-2 is an excellent target for diagnosis and therapeutic intervention. Many of the technologies described above were developed and designed to detect RBD to diagnose SARS-CoV-2. For example, Raman spectroscopy,390–393 electrochemical methods,394–396 localized surface plasmon resonance (LSPR)397 and lateral flow tomography.333,398 Recently, Yang et al. reported a high-affinity SARS-CoV-2 Omicron variant RBD-binding aptamer (SCORe) that binds to the Omicron BA.1 and BA.2 variant RBD with affinity at the nM level. Multiplex flow assays were developed to distinguish the Omicron variant from wild-type down to 100 pM.399 In addition, Ravalin et al. developed a biosensor that uses binding-activated tandem split-enzyme technology to detect RBD. The biosensor is a single-component, recombinant, luminescent sensor that can be expressed in laboratory strains of Escherichia coli and Saccharomyces cerevisiae, which was detected by a regular cell phone camera to record the chemiluminescent signal.400 Murtaza et al. obtained a new high-affinity aptamer for RBD by computer methods to fabricate a photonic crystal (PC)-decorated aptamer sensor, for which the aptamer is cross-linked in the polyacrylamide hydrogel network and selectively binds to SARS-CoV-2 in saliva samples. The binding reaction can be visually monitored by swelling of the hydrogel and color generated by light diffraction from the PC and can be quantified by the diffraction ring diameter or spectroscopy.401
Several studies suggest that in some coronaviruses, NTD may recognize specific glycosyl groups during initial attachment and play an essential role in the transition of S proteins from pre-fusion to post-fusion. Thus, it may be involved in the regulation of viral infection.402–404 Currently, most variants are mutated mainly in the RBD region.405 Therefore, the NTD assay may diagnose multiple variants more effectively.
Kacherovsky et al. obtained the SNAP1 aptamer with good affinity to NTD, which was applied in LFIA and ELISA to detect SARS-CoV-2 down to 5 × 105 copies per mL.406 Recently, this team obtained another aptamer SNAP4 binding NTD with 2-fold higher affinity than that of SNAP1. In addition, the constructed sandwich LFIA with both SNAP1 and SNAP4 could detect 106 copies per mL SARS-CoV-2.407
The analytical performance of some antigen detection methods are summarized in Table 9. Currently, many antigen detection kits based on these technologies have been approved for clinical testing to tackle SARS-CoV-2. The WHO suggests that all countries to use high-quality rapid antigen tests to effectively respond to the global COVID-19 pandemic.408
| Methods | Target antigen | Sample | Linear range | LOD | Detection time | Ref |
|---|---|---|---|---|---|---|
| LFIA | NP | Clinical isolate of SARS-CoV-2 | — | 2.5 × 104 pfu | 20 min | 337 |
| Double-antibody sandwich ELISA | NP | Recombinant protein | — | 0.78 ng mL−1 | About 1 day | 409 |
| Nanomechanical sensor | S1, NP | Nasopharyngeal swab | — | 0.71 ng mL−1 | ≤5 min | 410 |
| Microfluidic magneto immunosensor | NP | Whole serum | 1–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1 000 pg mL−1 |
0.23 ng mL−1 | <1 h | 16 |
| Half-strip LFA | NP | Recombinant protein | — | 0.65 ng mL−1 | About 10 min | 411 |
| LFIA | NP | Human serum or urine | — | 1.0 ng mL−1 | <15 min | 345 |
| MagPlex fluid array assays | NP | Recombinant protein | — | 0.05 ng mL−1 | <1 h | 412 |
| Cotton-tipped electrochemical immunosensor | NP | Clinical samples | 1–1000 ng mL−1 | 0.8 pg mL−1 | About 25 min | 350 |
| Electrochemical biosensor | NP | Recombinant protein | 1 pg mL−1–100 ng mL−1 | 0.4 pg mL−1 | About 20 min | 413 |
| Bright fluorescent probe assay | NP | Nasopharyngeal swab samples | — | 30 ng mL−1 | — | 414 |
| Electrochemical sensor | NP | Clinical samples | 2.22–111 fM | 15 fM | About 20 min | 415 |
| Chemiluminescent functionalized magnetic nanomaterial | NP | Recombinant protein | 0.1 pg mL−1–10 ng mL−1 | 69 fg mL−1 | — | 416 |
| Immunochromatographic assay augmented with silver amplification | NP | Nasopharyngeal swab | — | 6.3 ×104 copies per mL | About 15 min | 336 |
| Nanozyme-linked immunochromatographic sensor | NP | Clinical samples | 0.05–1.6 ng mL−1 | 0.026 ng mL−1 | ≤25 min | 417 |
| Smartphone-based nanozyme-linked immunosorbent assay | NP | Serum samples | 30 pg mL−1–3 ng mL−1 | 10 pg mL−1 | <1 h | 418 |
| AuNP-based plasmonic sensor | NP | Recombinant protein | 150–550 ng mL−1 | 150 ng mL−1 | <5 min | 419 |
| Colorimetric LFIA | NP | Human saliva | 0–1000 ng mL−1 | 54 TCID50 per mL | 15 min | 340 |
| MagTQD-LFIA | SP, NP | Saliva and nasal swab samples | 1 pg mL−1–1000 ng mL−1 | 0.5 pg mL−1 | 10 min | 341 |
| Photoelectrochemical immunosensor | NP | Recombinant protein | 10 pg mL−1–100 ng mL−1 | 2.9 pg mL−1 | — | 342 |
| Ratiometric fluorescent Si-FITC nanoprobe for immunoassay | NP | Human serum | 0.02–50 ng mL−1 | 0.003 ng mL−1 | — | 343 |
| Electrical-double-layer gated FET-based biosensing | NP | Artificial saliva | 0.4 ng mL−1–400 ng mL−1 | 0.14 ng mL−1 (2.96 pM) | 65 min | 347 |
| FET | NP | Calibration samples | 16 fg mL−1–16 pg mL−1 | 0.016 fg mL−1 | 5 min | 346 |
| Handheld microfluidic filtration platform | NP | Nasal swab samples | 1 × 103–1 × 106 particles per mL | 100 copies per mL | 15–30 min | 348 |
| Microelectrode array chip | NP | Throat swabs | 10−5–10−2 ng mL−1 | 3.16 × 10 −6 ng mL−1 | 15 s | 349 |
| Electrochemical immuno-biosensing | NP | Spiked sample | 1–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1 000 pg mL−1 |
21 fg mL−1 | 15 min | 351 |
| Paper-based electrochemical platform | NP | Serum sample | 1–100 ng mL−1 | 0.11 ng mL−1 | — | 420 |
| FET biosensor | SP | Cultured SARS-CoV-2 | — | 1.6 × 101 pfu mL−1 | — | 360 |
| Microfluidic biochip | SP | Serum | — | 0.3 pg mL−1 | 40 min | 421 |
| Electrochemical detection | SP | Recombinant protein | 1 pg mL−1–10 ng mL−1 | 1 pg mL−1 | About 10 min | 422 |
| Electrochemical biosensor | SP | Saliva, artificial nasal, UTM | 0.25 fg mL−1–1 μg per mL | 0.04 fg mL−1 | 20 min | 423 |
| G-quadruplex aptamer-based sensor | SP | Nasopharyngeal swab specimens | 0.5–125 nM | 2 nM | — | 424 |
| Graphite electrode functionalized with AuNPs | SP | Recombinant protein | 1 pg mL−1–1 ng mL−1 | 229 fg mL−1 | 6.5 min | 425 |
| WSe2 based-FET | SP | Recombinant protein | 25 fg μL−1–10 ng μL−1 | 25 fg μL−1 | — | 426 |
| SERS (Au/Si with labels) | SP | Untreated saliva | 1 fg mL−1–1ng mL−1 | 0.77 fg mL−1 | About 5 h | 427 |
| Multi-antibody transistor assay | S1 | Saliva | — | 3.5 × 10−17 g per mL | 38.9 s | 386 |
| Nanozyme chemiluminescence paper test | SP | Nasopharyngeal swab sample | 0.1–100 ng mL−1 | 0.1 ng mL−1 | 30 min | 428 |
| Lateral flow device | SP | Pseudotyped lentivirus | — | 1.5 × 104 TU per mL | <30 min | 357 |
| Cell surface-inspired universal sensor | SP | Recombinant protein | 3.13–50 μg mL−1 | 3.13 μg mL−1 | < 30 min | 358 |
| Functionalized terahertz plasmonic metasensor | SP | Recombinant protein | 4–12 fM | ∼4.2 fM | — | 359 |
| In2O3 nanoribbon transistor | SP | Recombinant protein in UTM | — | 100 fg mL−1 | — | 361 |
| Single-particle SERS immunoassay | SP | Saliva | 105–107 genomes per mL | 1.94 × 103 genomes per mL or 4.7 fg per mL | — | 365 |
| SERS | S1 | Recombinant protein | 0.01 ng mL−1–100 ng mL−1 | 10 pg mL−1 | 2 h | 366 |
| Plasmonic nanostructure-enhanced Raman scattering | SP | Recombinant protein | 1.0 pg mL−1–1.0 mg mL−1 | 1 fg mL−1 | — | 367 |
| SERS-based biosensor | SP | Saliva | 10 fg mL−1–10 ng mL−1 | 6.07 fg mL−1 | — | 368 |
| Deep learning-based SERS | SP | Throat swabs or sputum | — | 10 fg mL−1 | <20 min | 369 |
| SERS | SP | Saliva | 400 PFU mL−1–105 PFU mL−1 | 391 PFU mL−1 | — | 370 |
| Bumpy core–shell SERS | SP | Recombinant protein and clinical sample | 0.1 fg mL−1 to 10 ng mL−1 | 0.1 fg mL−1 | < 30 min | 371 |
| Peptide-ELISA | SP | Recombinant protein and pseudoviruses | — | 1.4 fM, 3TCID50 per mL | 2 days | 372 |
| Fluorescence LFIA | SP and NP | Saliva and nasal swab | 1 pg mL−1–100 ng mL−1 | 0.5 pg mL−1 | 10 min | 374 |
| LFIA | S1 | Nasal swab | 1.86 × 105 copies per mL | — | 20 min | 375 |
| LFIA | S1 | Nasopharyngeal swabs | 1 × 106 pfu mL−1–1 × 105 pfu mL−1 | 500 pg mL−1 | 20 min | 376 |
| Colorimetric and fluorescent dual-functional LFIA | S1 | Saliva samples | 33 pg mL−1–1 ng mL−1 | 37 pg mL−1 | 30 min | 378 |
| Impedimetric sensor | S1 | — | 0.0059 fg mL−1–0.020 fg mL−1 | 7.2 fg mL−1 | — | 380 |
| Electrochemical biosensor | S1 | Saliva swab | 0.5–5 ng mL−1 | 0.15 ng mL−1 | — | 388 |
| Electrochemical biosensor | S1 | Saliva swab | — | 1000 copies per mL | 10 min | 382 |
| SPR biosensor | S1 | Artificial saliva and human serum samples | 0.1 pg mL−1–1000 ng mL−1 | 12 fg mL−1 | — | 429 |
| Electrochemical sensor | S1 | Saliva swab | 0.1 fg mL−1 to 5.0 pg mL−1 | 4.12 fg mL−1 | 2–3 min | 383 |
| Immunoassay technique | S1 | Mid-turbinate swabs and exhaled breath aerosol samples | 1 ag mL−1–1 μg mL−1 | 60 copies per mL | 0.6 s | 384 |
| Functionalization of graphene oxide-glazed double-interdigitated capacitive biosensing platform | S1 | Recombinant protein | 1.0 mg mL−1–1.0 fg mL−1 | 1 fg mL−1 | 3 s | 385 |
| Phage-based enzyme-linked CLIA | S1 | Saliva swab | — | 60 TU mL−1 | — | 387 |
| LFIA | S1 | Nasal swabs | — | 1.86 × 105 copies per mL | About 10 min | 430 |
| Cell-based biosensor | S1 | Recombinant protein | 10 fg–1 μg mL−1 | 1 fg mL−1 | 3 min | 431 |
| Electrochemical biosensor | S1 | Recombinant SARS-CoV-2 | — | 20 μg mL−1 | ∼45 min | 432 |
| AuNP-based immunochromatographic strip | S-RBD | Recombinant protein | 62.5–4000 ng mL−1 | 62.5 ng mL−1 | 15–30 min | 433 |
| Electrochemical sensor | SP and NP | Untreated saliva | — | 19 ng mL−1 | About 2 h | 434 |
| CNT-FET | S1 | Recombinant protein | 0.1 fg mL−1–5.0 pg mL−1 | 4.12 fg mL−1 | 2–3 min | 429 |
| Electrochemical sensor | S1 | Nasopharyngeal samples | — | 64 fM | 15 min | 435 |
| Impedimetric biosensor | S-RBD | Nasal secretions | 0.0012–120 pg mL−1 | 0.58 fg mL−1 | About 2 h | 436 |
| ACE2-SWCNT nanosensors | S-RBD | Virus-like particles | — | 12.6 nM | About 90 min | 437 |
| Nanozyme chemiluminescence paper | S-RBD | Pseudovirus | 0.2–100 ng mL−1 | 0.1 ng mL−1 | 15 min | 428 |
| SERS | S-RBD | Saliva | — | 1 nM | — | 390 |
| SERS | S-RBD | Recombinant protein | — | 6.4 × 104 molecules | — | 391 |
| SERS | S-RBD | Recombinant protein | — | 1 fM | — | 392 |
| SERS | S-RBD | Recombinant protein | 1.25 to 20 ng mL−1 | 0.625 ng mL−1 | ≤15 min | 393 |
| SERS | S-RBD | Recombinant protein | 0.0012–120 pg mL−1 | 0.58 fg mL−1 | — | 394 |
| Photonic crystals | S-RBD | Saliva swab | 12.15 ng mL−1–13.25 ng mL−1 | 12.15 ng mL−1 | 5 min | 401 |
| Luminescent biosensor | S-RBD | Both recombinant form and cultured virus | 0.1 nM and 10 nM | 0.1 nM | — | 400 |
| Electrochemical immunosensor | S-RBD | Nasopharyngeal samples | 1 fM–1 μM | 0.73 fM | ≤ 30 s | 395 |
| Lateral flow assays | S-NTD | UV-inactivated SARS-CoV-2 virus | — | 5 × 105 copies per mL | — | 406 |
| Aptamer sandwich lateral flow assay | S-NTD | Inactivated SARS-CoV-2 virus | — | 106 copies per mL | 1 h | 407 |
5.3 Clinical applications and challenges
In clinical testing, antigen detection is mainly used in the acute infection phase of SARS-CoV-2, i.e., testing of samples within 5–7 days of the onset of symptoms in the suspected population (Omicron variant infection can be considered within 5 days). According to the WHO, the incubation period is an average of 3 days for the Omicron variant and 5–6 days for other SARS-CoV-2 variants.438 During incubation, virus shedding occurs in individuals who were infected with SARS-CoV-2. Clinical studies by Harvard Medical School showed that virus shedding was the highest in those infected with the Omicron variant within 3–6 days after onset.439 It is also revealed that the virus was undetected 10 days after onset in COVID-19 vaccine recipients infected with the Omicron variant. These studies suggest that antigen detection faces a limited window of optimal detection time, and therefore early infection and latency may be missed. However, according to viral culture research, SARS-CoV-2 may only replicate 10 to 14 days after the onset of symptoms. Antigen-based assays remain positive for 5 to 12 days after symptom onset and perform better in individuals with high viral loads.440 Thus, antigen-based assays correlate better with replication-competent SARS-CoV-2, which can provide information about potential transmissibility. Although hundreds of antigen-based tests are available for clinical testing worldwide, they are generally less sensitive than molecular tests compared to the reference standard of laboratory-based RT-PCR tests, especially in populations with low or no replication of viral load-capable viruses. Dinnes et al. summarized data from five studies involving 943 samples, with an average sensitivity of 56.2% (95% CI, 29.5–79.8%) and an average specificity of 99.5% (95% CI, 98.1–99.9%) for antigen detection compared to an average sensitivity of 95.2% (95% CI 86.7% to 98.3%) and an average specificity of 98.9% (95% CI, 97.3% to 99.5%) for molecular assays.441 After comparing Ag-RDT with viral culture and RT-PCR methods, Mak et al. concluded that the LOD of Ag-RDT was approximately 103-fold lower than the culture-based SARS-CoV-2 assay and 105-fold lower than the molecular assay.442 Mertens et al. stated that the sensitivity of Ag-RDT increased to 74.8% when samples with high viral loads (samples with RT-PCR CT values of ≤25) were evaluated, while the overall sensitivity was 57.6% when all the samples were considered.443 Due to the false negatives caused by antigen testing sensitivity, antigen testing is currently only used as an adjunct to RT-PCR testing. However, the potential application of antigen testing cannot be ignored. According to the recommendations of the WHO and CDC,444 anyone with symptoms of COVID-19 should be tested for SARS-CoV-2. Asymptomatic individuals in close contact with someone known or likely to be infected with SARS-CoV-2 should undergo diagnostic testing. In addition, asymptomatic individuals in a high-transmission risk setting should also be tested.Presently, the global manufacturing, delivery, and implementation of antigen tests remain challenging. Firstly, antigen testing may not yield accuracy comparable to laboratory-based molecular testing, and thus diagnostic tests should be available to underserved and affordable populations. Secondly, although home-based Ag-RDTs have increased antigen testing, Ag-RDTs performed by trained healthcare providers have proven to be more accurate than that performed by untrained individuals. Therefore, test kit instructions should be carefully followed by those performing testing at home.
6. Tackling SARS-CoV-2 variants
6.1 Evolution of SARS-CoV-2
New variants caused by viral mutations are emerging and spreading rapidly worldwide. Coronaviruses contain a special nucleic acid exonuclease (ExoN), which increases the replication fidelity of their genetic material by approximately 15-fold in vitro;445 however, when variants with different mutations infect the same host, recombination between them probably results in new SARS-CoV-2 variants. The accumulation of mutations will then gradually lead to SARS-CoV-2 diversity.446 In addition, human cytidine deaminases (APOBECs/ADARs) are also involved in the editing of SARS-CoV-2 transcriptome RNA, which also accelerates the SARS-CoV-2 diversity.447 The evolutionary rate of SARS-CoV-2 was estimated to accumulate 1–2 nucleotide mutations per month.448 Thus far, many variant strains have emerged, which contain constellations of mutations rather than a single site. Most mutations occur at similar locations and exhibit comparable properties, suggesting the recombination and evolution of SARS-CoV-2 in individuals. Although most mutations are harmless or neutral, some may result in increased disease severity, evasion of immune responses, reduced effectiveness of antiviral therapy, or enhanced ability of the virus to infect new animal hosts.449 Due to the growing emergence of variant strains, WHO designates variants that cause altered virus behavior as variants of interest (VOI) or VOC to prioritize global surveillance and research (Fig. 15A).450Beginning in April 2020, the original SARS-CoV-2 strain was replaced by a variant called D614G (aspartate to glycine at position 614 of the S protein).451 By June 2020, the virus with the D614G mutation rapidly became the dominant variant worldwide, which was justified to be more infectious than the original strain by subsequent studies.452 Currently, both VOC and VOI have this mutation site. In October 2020, sequencing analysis in the UK detected an emerging variant, later called B.1.1.7 or 20I/501Y.V1. This variant contains ten mutations in the S protein.453 Among them, N501Y appeared to further increase the interaction of the S protein with ACE2, making this variant approximately 70% more transmissible than the original strain.454,455 Although the impact of this variant on disease severity remains uncertain, the B.1.1.7 variant may still lead to a dramatic increase in hospitalizations and deaths due to its high transmissibility. This was also confirmed in a retrospective observational study by Stirrup et al.456 The B.1.351 variant (20H/510Y.V2) was initially detected in South Africa in the end of 2020,457 while the P.1 variant (20J/501Y.V3) emerged in Brazil in November 2020.458 These variants possess three mutations (K417N/T, E484K, and N501Y) located on the RBD of the S protein, are associated with increased transmission potential. In addition, these two variants can mediate immune escape against protective antibodies produced by humoral immunity and vaccination.459,460 The Delta variant was first detected in India in December 2020, exhibiting a greater ability to evade the host's immune system than the original strain, which resulted in its rapid global spread and emergence as the dominant strain in numerous countries within just a few months after May 2021.461 There are multiple mutation sites in the S protein of the Delta variant (T19R, L452R, T478K, D614G, P681R and d960N and deletions at positions 157 and 158), with L452R and P681R being the two most notable mutations. The mutation at position 452 significantly increases the affinity of B.1.617.2 for ACE2, potentially allowing it to evade the vaccine-induced production of protective antibodies that bind to the S protein.462 Other studies showed that the mutation allows the B.1.617.2 variant to evade CD8 T cells. In addition, the mutation at position 681 helps cleave the precursor S protein into an activated form, permitting better fusion and integration of the virus into the host cell.463 The reasons for this may be that the B.1.617.2 variant has strong propagation and immune escape ability. The B.1.1.529 variant, later named Omicron, was first reported to the WHO on November 24, 2021, and quickly classified as a VOC just 2 days later. This variant contains an unusually high number of mutations, with over 30 identified in its S protein genes, including many located in the RBD and NTD regions.464 Among them, the deletion of 69/70 led to false negative results for S detection in TaqPath test.465 K417N and E484A enhanced the immune escape ability.466 G496S, Q498R, N501Y and Y505H significantly increased the binding affinity for ACE2,464 resulting in loss or reduction of neutralization, while H655Y, N679K and P681H enhanced their propagation properties.467 Currently, increasing evidence supports that this variant is more transmissible than B.1.617.2 and has surpassed its prevalence within one month of discovery (Fig. 15B).
Since the emergence of the Omicron variant, SARS-CoV-2 has continued to mutate. Currently, over 500 sub-lineages of the Omicron variant are in circulation, with the BA.1, BA.2, and BA.3 strains being the most common.468 The BA.1 strain has 37 mutations in its spike protein, while the BA.2 and BA.3 strains have 31 and 33 mutations, respectively. Among these mutations, 21 mutations are commonly shared by all three lineages, including N501Y and Q498R, to enhance the virus's binding affinity with the ACE2 receptor, and H655Y, N679K and P681H, which are thought to increase its transmission capability.469 Recently, the XBB.1.5 variant, recombining the BA.2.10.1 and BA.2.75 sub-variants, has been widely circulating in the United States.470 According to the WHO, XBB has been identified in at least 70 countries/regions, leading to a surge in infections in some parts of Asia, including India and Singapore.471 However, overall, the pathogenicity of the Omicron variant is not as severe as that of the Delta variant, which may be due to various factors such as the virus replicating more efficiently in the upper respiratory tract and the steady improvement of population immunity worldwide.
6.2 Sequencing for identification of SARS-CoV-2 variants
Genomics is the only method to identify and characterize new variants and clarify existing types. Whole genome sequencing (WGS) is an essential method for the genetic identification of viruses, allowing untargeted, unbiased sequencing of nucleic acids in a sample, and thus viral RNA or DNA can be identified if a sufficiently high copy number of DNA or RNA is present relative to other sources.472,473 The tiled amplicon method or shotgun sequencing is usually used in sequencing, which can also be compared with other circulating strains.474 Three groups used this method to identify the causative pathogen of COVID-19 at the beginning of the outbreak.475 With the emergence of various variants, this approach has also been employed to detect the variations and analyze the patterns and factors that influence the spread of these variants.16,476 The implementation of WGS can be found in WHO's Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. However, although sequencing costs have decreased significantly, WGS is still relatively expensive compared to other molecular diagnostic techniques (e.g., RT-PCR and isothermal amplification). In addition, WGS is a resource-intensive method that can generate results in several days. Therefore, it cannot be routinely used to diagnose various variants. Sanger-based or amplicon-based NGS can sequence all or part of the genes of a variant and is an effective alternative method for identifying and characterizing multiple variants. HiSpike, developed by Fass and colleagues, can identify variants within the S gene for the 20I/501Y.V1 and 20H/501Y.V2 variants with the help of a small Illumina MiSeq instrument.477 Compared with the conventional sequencing method, HiSpike improves the detection throughput and significantly reduces the detection costs and time. In another study, Castañeda developed ADSSpike similar to HiSpike. Compared to HiSpike, ADSSpike also sequences the S gene, but it uses deep sequencing (700 read depth) to identify SARS-CoV-2 mutants, capable of identifying variant-specific single nucleotide polymorphism (SNP) with a Ct value of up to 30.17 for the variant characterized.478 However, both strategies require tens of primer pairs to detect variants to cover the entire S gene. In fact, a dedicated SARS-CoV-2 variant diagnostic and assay monitoring strategy deployed globally will require cheap instruments and a reduced number of reagents to allow long-term storage. This problem was well addressed by Stüder et al., who used long DNA fragments compatible with nanopore DNA sequencing technology.479 The solution requires only four primers targeting the Spike region, one random primer for the cDNA step, and a common Gibson sequence for real-time diagnosis and variant tracking on a portable MinION DNA sequencer. To further improve the detection throughput of sequencing, the pooling of samples has been attempted using primer barcodes. Cohen-Aharonov et al. used primers with barcodes to reverse transcribe and amplify RNA extracted from samples in one step, pooled and sequenced the resulting amplicons and identified infected individuals from all pooled test samples by subject-specific barcodes.480 The feasibility of this strategy for variant detection was verified by pooled testing of 960 samples. Chappleboim et al. also developed ApharSeq based on a pooled sample sequencing strategy, which combined samples as early as possible by hybridizing barcode primers.481 This allows multiplex reverse transcription, PCR and sequencing of hundreds of mixed samples to detect and classify variant sequences with high sensitivity and negligible contamination. Validation of the method on hundreds of clinical samples demonstrated excellent results (detection limit of Ct33, specificity > 99.5%) and a significant reduction in detection cost. The WGS and NGS methods for sequencing SARS-CoV-2 variants face the same challenges of expensive equipment and the need for specialized bioinformatics analysis, but these methods provide the basis for other detection technologies.The SARS-CoV-2 genomic sequence data obtained by sequencing can help identify viral proteins that may be strongly antigenic and indicate how to generate these antigens for serological analysis. When SARS-CoV-2 acquires genomic substitutions, a spectrum with altered antigenic properties may emerge, indicating that serological assays cannot detect an infected individuals because the antigen used in the assay differs from the antigen the individual was exposed to. In addition, the rapid release of mutant sequences is important for designing primers and probes for molecular assays. These genomes are necessary for the design of primers and probes, and mismatches between primers or probes and corresponding binding sites within the SARS-CoV-2 genome may reduce the sensitivity of molecular diagnostic methods or lead to false negatives. As SARS-CoV-2 continues to acquire genetic changes, the continued generation and sharing of the viral genome are critical for monitoring the expected sensitivity of various diagnostic assays at different sites.
6.3 Other molecular assays for the detection of SARS-CoV-2 variants
The PCR-based diagnostic method is an effective alternative to WGS and NGS, with a flexible testing strategy and the ability for detection needs. This method can be used to assess the spread of different SARS-CoV-2 variants in the community and for genetic characterization to monitor virus evolution, while providing information for outbreak analysis.482,483 Several countries/area have released their RT-PCR protocols. A compilation of primer sequences was developed in China, Hong Kong, Germany, France, Thailand and the United States.484 The average mismatches and significant mismatches obtained from PrimerScan comparing the primers from each region with all SARS-COV-2 whole genome sequences were published on GISAID143 (Table 10). Since the S protein is the main target of SARS-CoV-2 nAbs, it will be more meaningful to develop RT-qPCR assays for some key mutations in the S gene, such as E484K, N501Y and 69-70del. Many manufacturers have developed kits for different sites of the S gene, such as ID SARS-CoV-2/UK/SA Variant Triplex®, PKampVariantDetect SARS-CoV-2 RT-PCR combination 1 and 3® PerkinElmer, and cobas® SARS-CoV-2 Variant Set 1 Test.144,146 Many recent studies have evaluated different screening kits using NGS methods, demonstrating a high level of agreement between methods.485–487 In addition, many scientific papers report RT-qPCR methods for S mutation detection with promising results. The detection of S-gene mutations mainly involves two strategies, including a diagnostic screening approach that utilizes negative or weakly positive S gene results (caused by deletions at nt 207–212) from multiplex RT-PCR analysis, together with positive results for other targets, to identify specific variants with deletions at this site (Fig. 16A).488,489 This method was extensively used when B.1.1.7 variant was prevalent in the UK and was also used for diagnosing this variant after the appearance of the B.1.1.529 variant.486 It is important to note that S target failure is not restricted to these two variants, given that reports indicate that it occurred in 1–5% of samples sequenced before the advent of B.1.17 VOC.426 Besides, target failure can also detect ORF1aΔ3675-3677 in variants (B.1.1.7, B.1.351, P.1, and C.37). Vogels et al. designed and validated an open-source PCR assay to detect B.1.1.7, B.1.351, and P.1 using ORF1aΔ3675-3677 as the primary target and using spikes Δ69-70 for differentiation.490 In addition, given that ORF1aΔ3674-3676 is present in B.1.1.529, this combined target failure strategy also has potential to be developed to differentiate between B.1.1.7 and B.1.1.529. This target failure strategy works well when the prevalence of VOC in the environment is already high, but in low prevalence settings, it is preferable to confirm the presence of both deletions by sequencing, which will be necessary to increase the confidence in the results. An alternative approach is to use S gene SNP-based analysis, which involves the design of allelic primers or specific primer probes incorporating lock nucleic acids (LNA) to target the mutation site (Fig. 16B).491 For example, the RT-qPCR analysis for the N501Y, 69-70del, K417N and E484K mutations developed by Vega-Magaña et al. and the one-step multiplex allele-specific RT-qPCR assay developed by Lee et al. facilitated the detection of all common and uncommon VOCs/VOIs containing L452R, E484K or N501Y mutations.492,493 One advantage of these methods is that they allow the rapid (<1 h) estimation of the prevalence of specific mutation-positive variants in the community, and considering that SNPs at certain sites are not only present in VOCs merely in VOIs, we should also sequence a portion of the samples for validation at the time of detection. Moreover, some RT-PCR platforms allow melting curve analysis, and based on this many genotyping methods have been developed to identify specific amino acids substitutions, such as HV69-70del, K417N, N439K, Y453F, E484K, N501Y, A570D, D614G, P681H or V1176F.494| Region | Primer/Probe | Target gene | Sequence (5′-3′) | Primer length | Genomic regiona | Average mismatchesb | Significant mismatchesc |
|---|---|---|---|---|---|---|---|
| a Genomic Region, Site numbering using access MN908947.3 as the reference. b Average mismatches per sequence per assay. This value represents the overall similarity between the primers/probes and the target sequences in the database. (The data was obtained by blasting primers and probes with the database in the GISAID). c Significant mismatches. Significant mismatches are those in the probe or the last 5 bases of a primer. These mismatches can significantly affect the specificity and sensitivity of the detection. (The data was obtained by blasting primers and probes with the database in the GISAID.) | |||||||
| Charité Germany | E-Sarbeco-F | E gene | ACAGGTACGTTA ATAGTTAATAGC GT | 26 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 269–26 269–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294 294 |
0.0031 | — |
| Charité Germany | E-Sarbeco-R | E gene | ATATTGCAGCAG TACGCACACA | 22 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360–26 360–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 381 381 |
||
| Charité Germany | E-Sarbeco-P | E gene | ACACTAGCCATC CTTACTGCGCTT CG | 26 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332–26 332–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357 357 |
||
| Charité Germany | N-Sarbeco-F | N gene | CACATTGGCACC CGCAATC | 19 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 706–28 706–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 724 |
0.0411 | 0.0125 |
| Charité Germany | N -Sarbeco-R | N gene | GAGGAACGA GAA GAGGCT TG | 20 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814–28 814–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 833 |
||
| Charité Germany | N-Sarbeco-P | N gene | ACTTCCTCAAGG AACAACATTGCC A | 25 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753–28 753–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 777 777 |
||
| China CDC | China-CDC-N-F | N gene | GGG GAA CTT CTC CTG CTA GAA T | 22 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881–28 881–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 902 902 |
1.1619 | 0.0081 |
| China CDC | China-CDC-N-R | N gene | CAGACA TTT TGC TCTCAA GCTG | 22 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958–28 958–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 979 979 |
||
| China CDC | China-CDC-N-P | N gene | TTGCTGCTGCTT GACAGATT | 20 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 934–28 934–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 953 953 |
||
| China CDC | China-CDC-ORF1-F | ORF1ab | CCCTGTGGG TTT TACACTTAA | 21 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342–13 342–13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 362 362 |
0.0069 | 0.0030 |
| China CDC | China-CDC-ORF1-R | ORF1ab | ACGATT GTGCAT CAGCTGA | 19 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 442–13 442–13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 460 |
||
| China CDC | China-CDC-ORF1-P | ORF1ab | CCGTCT GCGGTA TGTGGA AAGGTT ATGG | 28 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 377–13 377–13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 404 404 |
||
| Hong Kong University | HKU-NF | N gene | TAATCAGACAAG GAACTG ATTA | 22 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 145–29 145–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 166 166 |
0.0225 | 0.0117 |
| Hong Kong University | HKU-NR | N gene | CGAAGGTGT GACTTCCATG | 19 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236–29 236–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 254 254 |
||
| Hong Kong University | HKU-NP | N gene | GCAAATTGTGCA ATTTGCGG | 20 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 177–29 177–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 196 |
||
| Hong Kong University | HKU-ORF1b-nsp141F | ORF1b | TGGGGYTTT ACRGGTAACCT | 20 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 778–18 778–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 797 |
0.0097 | 0.0037 |
| Hong Kong University | HKU-ORF1b-nsp141R | ORF1b | AACRCGCTT AACAAAGCA CTC | 21 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 889–18 889–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 909 909 |
||
| Hong Kong University | HKU-ORF1b-nsp141P | ORF1b | TAGTTGTGATGC WATCATGACTAG | 24 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849–18 849–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 872 872 |
||
| Pasteur | NCoV-IP2-12669Fw | RdRp IP2 | ATGAGCTTAGTC CTGTTG | 18 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690–12 690–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 707 707 |
0.0129 | 0.0056 |
| Pasteur | NCoV-IP2-12759Rv | RdRp IP2 | CTCCCTTTGTTG TGTTGT | 18 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780–12 780–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 797 |
||
| Pasteur | nCoV_IP2-12696 a crucial probe | RdRp IP2 | AGATGTCTTGTG CTGCCGGTA | 21 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 719–12 719–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 737 |
||
| Pasteur | NCoV-IP4-14059Fw | RdRp IP4 | GGTAACTGGTAT GATTTCG | 19 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080–14 080–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 098 098 |
0.0131 | 0.0085 |
| Pasteur | NCoV-IP4-14146Rv | RdRp IP4 | CTGGTCAAG GTT AATATAGG | 20 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 167–14 167–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 186 186 |
||
| Pasteur | NCoV-IP4-14084 probe | RdRp IP4 | TCATACAAACCA CGCCAGG | 19 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105–14 105–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 123 123 |
||
| Public Health Thailand | WH-NIC-N-F | N gene | CGTTTGGTGGAC CCTCAGAT | 20 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320–28 320–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339 339 |
0.0214 | 0.0090 |
| Public Health Thailand | WH-NIC-N-R | N gene | CCCCACTGCGTT CTCCATT | 19 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 358–28 358–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 376 |
||
| Public Health Thailand | WH-NIC-N-P | N gene | CAACTGGCAGTA ACCA | 16 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 341–38 341–38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 356 356 |
||
| US CDC | 2019-nCoV-N1-F | N gene | GACCCCAAAATC AGCGAAAT | 20 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287–28 287–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 306 306 |
0.0345 | 0.0214 |
| US CDC | 2019-nCoV-N1-R | N gene | TCTGGTTACTGC CAGTTGAATCTG | 24 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335–28 335–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 358 358 |
||
| US CDC | 2019-nCoV-N1-P | N gene | ACCCCGCATTAC GTTTGGTGGACC | 24 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309–28 309–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332 332 |
||
| US CDC | 2019-nCoV-N2-F | N gene | TTACAAACATTG GCCGCAAA | 20 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 164–29 164–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 183 183 |
0.0383 | 0.0149 |
| US CDC | 2019-nCoV-N2-R | N gene | GCGCGACATTCC GAAGAA | 18 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213–29 213–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 230 |
||
| US CDC | 2019-nCoV-N2-P | N gene | ACAATTTGCCCC CAGCGCTTCAG | 23 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 188–29 188–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 210 210 |
||
| US CDC | 2019-nCoV-N3-F | N gene | GGGAGCCTT GAATACACCAAA A | 22 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681–28 681–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 702 702 |
0.0200 | 0.0113 |
| US CDC | 2019-nCoV-N3-R | N gene | TGTAGCACGATT GCAGCATTG | 21 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732–28 732–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 752 752 |
||
| US CDC | 2019-nCoV-N3-P | N gene | AYCACATTGGCA CCCGCAATCCTG | 24 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704–28 704–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 727 727 |
||
With the increasing number of sequences becoming publicly available, many other molecular diagnostic techniques have been developed for variant detection. RT-LAMP for variant detection can also be performed directly by visual color change,128 fluorescence,495 sequence-specific probes such as Detection of Amplification by Releasing of Quenching (DARQ) and molecular beacons,496 or coupled to secondary molecular analysis platforms such as CRISPR and NGS (e.g., LamPORE and LAMP-Seq).497 Currently the available protocols cannot discriminate between specific VOCs/VOIs; however, RT-LAMP technology offers certain advantages such as faster test results and requirement of less resources, while maintaining high sensitivity and specificity. CRISPR-based variant detection techniques focus on designing crRNAs with mismatches for mutation positions in known sequences498,499 or developing chimeric crRNAs to distinguish mutations in different positions specifically.500 Most of these technologies use fluorescence501 or LFAs502 for the readout of the detection results, and some have integrated technology into micro devices to develop POCT devices that integrate extraction, purification, and concentration of viral RNA, amplification and detection.503 CRISPR technology has the merits of easy-to-use and easy-to-set-up, which can be rapidly deployed in response to outbreaks. By combination with RT-PCR, these techniques rely on oligonucleotide primers and mutations in the target region, which may have an impact on amplification. Therefore, appropriate clinical validation studies of the protocols are also needed to assess their potential role in different settings.
7. Conclusions and prospect
The global epidemic is still spreading, and the SARS-CoV-2 strains are mutating. Recently, a study by Harvard University revealed that 94% of Americans were infected with COVID-19 at least once, and 97.8% had immunity to SARS-CoV-2.504 Although protectiveness in preventing COVID-19 has increased from 22% to 63% among the U.S. population, the rate of reinfection is increasing because the Omicron variant is extremely contagious and a large portion of Americans has stopped wearing masks. Additionally, this study also warned that as the virus evolves and the immune antibodies continuously decline, new waves of COVID-19 infections need to be cautioned. A timely and accurate identification of patients infected with SARS-CoV-2 is still the cornerstone of public health containment and mitigation of the pandemic. Presently, significant improvements have been achieved in techniques for detecting SARS-CoV-2, such as molecular assay for viral RNA and immunoassay for viral antigens or virus-specific antibodies produced by the immune system. Among them, RT-PCR is the gold standard for COVID-19 diagnosis. Mini RT-PCR devices that combine RT-PCR with different detection modalities have been deployed to address the current low sensitivity of rapid POC assays, and simultaneously analyze samples in a high-throughput format beyond the centralized laboratory.505 RT-dPCR has become an efficient high-throughput detection system.506 CRISPR/Cas-based systems have further overcome the problem of pre-test nucleic acid extraction and amplification.507 Moreover, diverse isothermal amplification technologies (RT-LAMP, RT-RPA, TMA) with robust detection capabilities have been implemented for rapid detection laboratories and POC applications, screening large patient populations, or rapid deployment to assess target regions508 Sequencing-based assays have enabled the monitoring of SARS-CoV-2 genomic sequences, especially for variant-associated detection.509 However, we must acknowledge that all molecular diagnostic methods are subject to a constant limitation, which is the sequence mismatch between the detection probe and the target region due to the mutations in SARS-CoV-2. Currently, false negatives caused by mutants are minimized by detecting multiple genes simultaneously or by increasing the binding to the mutant target using abbreviated primers and probes.510,511 During the pandemic, SARS-CoV-2 detection mainly relied on molecular diagnostic methods, especially RT-PCR, which brought many challenges globally. For example, laboratories need to procure large quantities of supplies quickly, acquire instruments, and train additional personnel. For testing, they are also faced with many challenges such as the shortage of human resources for sample collection and testing, and the unsustainable and rapid supply of swab collection resources and testing reagents. Sampling often results in crowd gathering, which poses the risk of causing virus transmission.Antigen-based assays play an invaluable role in the rapid identification of highly infectious cases, which can generally provide rapid results without the need for complex instrumentation. However, antigen-based assays exhibit low sensitivity compared to molecular diagnostics (mean sensitivity: 56.2%, 95% CI (29.5–79.8%) for rapid antigen-based assays vs. 95.2%, 95% CI (86.7–98.3%) for molecular diagnostics).512 A possible strategy to mitigate the relatively low sensitivity of antigen-based assays is to increase the frequency of assays in the patient population over time, and to increase the chance of identifying individuals at high viral shedding stages. Serological tests are valuable for seroepidemiological studies, which contribute to ongoing outbreak investigations and the detection of suspected cases where molecular diagnostics are consistently negative or molecular diagnostics are not available. However, the performance of serological assays varies with techniques, time of onset and comparative methods. Recent meta-analyses and systematic evaluations show high levels of heterogeneity in these methods, with risks of bias and applicability.513 The key area of research in serology is the relevance, magnitude, and duration of protective antibody responses, especially in the case of widespread vaccination. Longitudinal studies are needed for the protective relevance of nAbs, although various neutralization assays can assess and quantify SARS-CoV-2 nAbs in serum or plasma.
Overall, RNA-, antigen-, or antibody-based assays have their respective place in diagnosing SARS-CoV-2, but ongoing research is critical to further improve them for practical applications. Eventually, the understanding of the strengths and limitations of the available methods and the development of new methods will help us unravel the unknowns of disease pathogenesis, epidemiology and transmissibility, which can restrain the SARS-CoV-2 virus.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.Author contributions
Aihua Liu: conceptualization, funding acquisition, project administration, resources, visualization, writing – review & editing; Tao Dong: conceptualization, visualization, writing – original draft, writing – review & editing; Mingyang Wang: visualization, writing – review & editing; Junchong Liu: visualization, writing – review & editing; Pengxin Ma: visualization, writing – review & editing; Shuang Pang: visualization, writing – review & editing. Wanjian Liu: visualization, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
National Natural Science Foundation of China (22174081, 81673172) and the National Key Research and Development Program of China (2021YFA0910400) are acknowledged.References
- O. T. Idoko, E. Usuf, U. Okomo, C. Wonodi, K. Jambo, B. Kampmann, S. Madhi and I. Adetifa, Clin. Infect. Dis., 2022, 75, S136–S140 CrossRef PubMed.
- S. Pecetta, S. Kratochvil, Y. Kato, K. Vadivelu and R. Rappuoli, Pharmacol. Rev., 2022, 74, 313–339 CrossRef CAS PubMed.
- R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang and N. Zhu, Lancet, 2020, 395, 565–574 CrossRef CAS PubMed.
- K. Dhama, S. Khan, R. Tiwari, S. Sircar, S. Bhat, S. Malik Yashpal, P. Singh Karam, W. Chaicumpa, D. K. Bonilla-Aldana and J. Rodriguez-Morales Alfonso, Clin. Microbiol. Rev., 2020, 33, e00028–e00020 CrossRef CAS PubMed.
- T. Chaibun, J. Puenpa, T. Ngamdee, N. Boonapatcharoen, P. Athamanolap, A. P. O'Mullane, S. Vongpunsawad, Y. Poovorawan, S. Y. Lee and B. Lertanantawong, Nat. Commun., 2021, 12, 802 CrossRef CAS PubMed.
- N. Sethuraman, S. S. Jeremiah and A. Ryo, JAMA, 2020, 323, 2249–2251 CrossRef CAS PubMed.
- B. Bošnjak, S. C. Stein, S. Willenzon, A. K. Cordes, W. Puppe, G. Bernhardt, I. Ravens, C. Ritter, C. R. Schultze-Florey and N. Gödecke, MedRxiv, 2020, preprint, DOI:10.1038/s41423-020-00573-9.
- M. Letko, A. Marzi and V. Munster, Nat. Microbiol., 2020, 5, 562–569 CrossRef CAS PubMed.
- T. N. Starr, S. K. Zepeda, A. C. Walls, A. J. Greaney, S. Alkhovsky, D. Veesler and J. D. Bloom, Nature, 2022, 603, 913–918 CrossRef CAS PubMed.
- C. B. Jackson, M. Farzan, B. Chen and H. Choe, Nat. Rev. Mol. Cell Biol., 2022, 23, 3–20 CrossRef CAS PubMed.
- M. Scudellari, Nature, 2021, 595, 640–644 CrossRef CAS PubMed.
- A. R. Fehr and S. Perlman, Coronaviruses, 2015, 1–23 CAS.
- E. Callaway, Nature, 2020, 585, 174–177 CrossRef CAS PubMed.
- C. A. Ascoli, Nat. Biotechnol., 2021, 39, 274–275 CrossRef CAS PubMed.
- Q. Wang, S.-B. Ye, Z.-J. Zhou, J.-Y. Li, J.-Z. Lv, B. Hu, S. Yuan, Y. Qiu and X.-Y. Ge, bioRxiv, 2022, preprint, DOI:10.1002/jmv.28116.
- O. T. Ng, V. Koh, C. J. Chiew, K. Marimuthu, N. M. Thevasagayam, T. M. Mak, J. K. Chua, S. S. H. Ong, Y. K. Lim, Z. Ferdous, A. K. B. Johari, M. I. Chen, S. Maurer-Stroh, L. Cui, R. T. P. Lin, K. B. Tan, A. R. Cook, P. Y. Leo and P. V. J. Lee, Lancet Reg. Health West. Pac., 2021, 17, 100299 CrossRef PubMed.
- A. Escalera, A. S. Gonzalez-Reiche, S. Aslam, I. Mena, M. Laporte, R. L. Pearl, A. Fossati, R. Rathnasinghe, H. Alshammary and A. van de Guchte, Cell Host Microbe, 2022, 30, 373–387 CrossRef CAS PubMed.
- T. Mourier, M. Shuaib, S. Hala, S. Mfarrej, F. Alofi, R. Naeem, A. Alsomali, D. Jorgensen, A. K. Subudhi and F. Ben Rached, Nat. Commun., 2022, 13, 1–14 Search PubMed.
- H. Guo, Q. Fan, S. Song, S. Shen, B. Zhou, H. Wang, L. Cheng, X. Ge, B. Ju and Z. Zhang, J. Clin. Virol., 2022, 150, 105162 CrossRef PubMed.
- E. H. Lau, O. T. Tsang, D. S. Hui, M. Y. Kwan, W.-h. Chan, S. S. Chiu, R. L. Ko, K. H. Chan, S. Cheng and R. A. Perera, Nat. Commun., 2021, 12, 1–7 CrossRef PubMed.
- Y. Zhang, H. Zhang and W. Zhang, Front. Med., 2022, 16, 1–12 CrossRef PubMed.
- C. Chakraborty, A. R. Sharma, M. Bhattacharya and S.-S. Lee, Front. Immunol., 2022, 13, 801522 CrossRef CAS PubMed.
- W. J. Wiersinga, A. Rhodes, A. C. Cheng, S. J. Peacock and H. C. Prescott, JAMA, 2020, 324, 782–793 CrossRef CAS PubMed.
- C. Elias, A. Sekri, P. Leblanc, M. Cucherat and P. Vanhems, Int. J. Infect. Dis., 2021, 104, 708–710 CrossRef CAS PubMed.
- P. Lia, J.-B. Fub and K.-F. Lia, Int. J. Infect. Dis., 2020, 96, 452–453 CrossRef PubMed.
- M. F. Osuchowski, M. S. Winkler, T. Skirecki, S. Cajander, M. Shankar-Hari, G. Lachmann, G. Monneret, F. Venet, M. Bauer, F. M. Brunkhorst, S. Weis, A. Garcia-Salido, M. Kox, J.-M. Cavaillon, F. Uhle, M. A. Weigand, S. B. Flohé, W. J. Wiersinga, R. Almansa, A. de la Fuente, I. Martin-Loeches, C. Meisel, T. Spinetti, J. C. Schefold, C. Cilloniz, A. Torres, E. J. Giamarellos-Bourboulis, R. Ferrer, M. Girardis, A. Cossarizza, M. G. Netea, T. van der Poll, J. F. Bermejo-Martín and I. Rubio, Lancet Respir. Med., 2021, 9, 622–642 CrossRef CAS PubMed.
- A. Carfì, R. Bernabei and F. Landi, JAMA, 2020, 324, 603–605 CrossRef PubMed.
- Y. Zhao, Y. Cao, S. Wang, K. Cai and K. Xu, Br. J. Surg., 2021, 108, E319 CrossRef CAS PubMed.
- A. H. Attaway, R. G. Scheraga, A. Bhimraj, M. Biehl and U. Hatipoğlu, BMJ, 2021, 372, n436 CrossRef PubMed.
- N. Shental, S. Levy, V. Wuvshet, S. Skorniakov, B. Shalem, A. Ottolenghi, Y. Greenshpan, R. Steinberg, A. Edri, R. Gillis, M. Goldhirsh, K. Moscovici, S. Sachren, L. M. Friedman, L. Nesher, Y. Shemer-Avni, A. Porgador and T. Hertz, Sci. Adv., 2020, 6, eabc5961 CrossRef CAS PubMed.
- T. Konishi, PLoS One, 2021, 16, e0247626 CrossRef CAS PubMed.
- Z. Al-Aly, B. Bowe and Y. Xie, Nat. Med., 2022, 28, 1461–1467 CrossRef CAS PubMed.
- C. Serviente, S. T. Decker and G. Layec, J. Appl. Physiol., 2022,(132), 581–592 CrossRef CAS PubMed.
- K. L. Newell and A. T. Waickman, Curr. Opin. Immunol., 2022, 77, 102228 CrossRef CAS PubMed.
- S. Sauleda, L. Palacios, V. Bres, M. Pinana, L. Alonso-Hernandez, M. Bes, M. Piron, E. Entrena, A. M. Minguez-Micolau, J. M. Marimon, A. Gurrola, G. Soria, L. Puig, A. Anton, T. Pumarola and J. M. Linnen, Diagn. Microbiol. Infect. Dis., 2022, 102, 115560 CrossRef CAS PubMed.
- M. R. Gitman, M. V. Shaban, A. E. Paniz-Mondolfi and E. M. Sordillo, Diagnostics, 2021, 11, 2075–4418 CrossRef PubMed.
- E. Degli-Angeli, J. Dragavon, M. L. Huang, D. Lucic, G. Cloherty, K. R. Jerome, A. L. Greninger and R. W. Coombs, J. Clin. Virol., 2020, 129, 104474 CrossRef CAS PubMed.
- H. Ceylan Koydemir and A. Ray, Diagnostics, 2020, 10, 2075–4418 CrossRef PubMed.
- H. H. Mostafa, K. C. Carroll, R. Hicken, G. J. Berry, R. Manji, E. Smith, J. L. Rakeman, R. C. Fowler, M. Leelawong, S. M. Butler-Wu, D. Quintero, M. Umali-Wilcox, R. W. Kwiatkowski, D. H. Persing, F. Weir and M. J. Loeffelholz, J. Clin. Microbiol., 2021, 59, 1098–1660X CrossRef PubMed.
- L. J. Carter, L. V. Garner, J. W. Smoot, Y. Li, Q. Zhou, C. J. Saveson, J. M. Sasso, A. C. Gregg, D. J. Soares, T. R. Beskid, S. R. Jervey and C. Liu, ACS Cent. Sci., 2020, 6, 591–605 CrossRef CAS PubMed.
- M. J. Mina, R. Parker and D. B. Larremore, N. Engl. J. Med., 2020, 383, e120 CrossRef CAS PubMed.
- Fact Sheet for Healthcare Providers: AMPIPROBE SARS-CoV-2 Test System, Enzo Life Sciences, Inc, https://pesquisa.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/resource/pt/grc-740851, accessed July 25, 2022 Search PubMed.
- C. B. F. Vogels, A. E. Watkins, C. A. Harden, D. E. Brackney, J. Shafer, J. Wang, C. Caraballo, C. C. Kalinich, I. M. Ott, J. R. Fauver, E. Kudo, P. Lu, A. Venkataraman, M. Tokuyama, A. J. Moore, M. C. Muenker, A. Casanovas-Massana, J. Fournier, S. Bermejo, M. Campbell, R. Datta, A. Nelson, C. S. Dela Cruz, A. I. Ko, A. Iwasaki, H. M. Krumholz, J. D. Matheus, P. Hui, C. Liu, S. F. Farhadian, R. Sikka, A. L. Wyllie and N. D. Grubaugh, MedRxiv, 2020, preprint, DOI:10.1101/2020.08.03.20167791.
- M. Ghofrani, M. T. Casas, R. K. Pelz, C. Kroll, N. Blum and S. D. Foster, MedRxiv, 2020, preprint, DOI:10.1101/2020.06.03.20116327.
- J. J. Forde, D. Goldberg, D. Sussman, F. Soriano, J. A. Barkin and S. Amin, Gastroenterology, 2020, 159, 1538–1540 CrossRef CAS PubMed.
- S. J. Mascuch, S. Fakhretaha-Aval, J. C. Bowman, M. T. H. Ma, G. Thomas, B. Bommarius, C. Ito, L. Zhao, G. P. Newnam, K. R. Matange, H. R. Thapa, B. Barlow, R. K. Donegan, N. A. Nguyen, E. G. Saccuzzo, C. T. Obianyor, S. C. Karunakaran, P. Pollet, B. Rothschild-Mancinelli, S. Mestre-Fos, R. Guth-Metzler, A. V. Bryksin, A. S. Petrov, M. Hazell, C. B. Ibberson, P. I. Penev, R. G. Mannino, W. A. Lam, A. J. Garcia, J. Kubanek, V. Agarwal, N. V. Hud, J. B. Glass, L. D. Williams and R. L. Lieberman, J. Biol. Chem., 2020, 295, 15438–15453 CrossRef CAS PubMed.
- T. Shaffaf and E. Ghafar-Zadeh, Bioengineering, 2021, 8, 2306–5354 Search PubMed.
- E. T. Beck, W. Paar, L. Fojut, J. Serwe and R. R. Jahnke, J. Clin. Microbiol., 2021, 59, e02727–e02720 CrossRef CAS PubMed.
- B. S. Karon, L. J. Donato, A. R. Bridgeman, J. H. Blommel, B. Kipp, A. Maus, S. Renuse, J. Kemp, A. K. Madugundu, P. M. Vanderboom, S. Chavan, S. Dasari, R. J. Singh, S. K. Grebe and A. Pandey, Clin. Chem., 2021, 67, 1545–1553 CrossRef PubMed.
- E. Valera, A. Jankelow, J. Lim, V. Kindratenko, A. Ganguli, K. White, J. Kumar and R. Bashir, ACS Nano, 2021, 15, 7899–7906 CrossRef CAS PubMed.
- M. Rybicka, E. Milosz and K. P. Bielawski, Viruses, 2021, 13, 730 CrossRef CAS PubMed.
- D. D. Rhoads, D. Plunkett, J. Nakitandwe, A. Dempsey, Z. J. Tu, G. W. Procop, D. Bosler, B. P. Rubin, M. J. Loeffelholz and J. E. Brock, J. Clin. Microbiol., 2021, 59, e00913–e00921 CAS.
- M. W. McCarthy, Expert Rev. Mol. Diagn., 2021, 21, 1–2 CrossRef CAS PubMed.
- S. A. van Asten, S. A. Boers, J. D. de Groot, R. Schuurman and E. C. Claas, BMC Microbiol., 2021, 21, 1–5 CrossRef PubMed.
- S. A. Mahmoud, S. Ganesan, E. Ibrahim, B. Thakre, J. G. Teddy, P. Raheja and W. Abbas Zaher, J. Med. Virol., 2021, 93, 5538–5543 CrossRef CAS PubMed.
- C. Y. Yu, K. G. Chan, C. Y. Yean and G. Y. Ang, Diagnostics, 2021, 11, 53 CrossRef CAS PubMed.
- W. Stokes, B. M. Berenger, T. Singh, I. Adeghe, A. Schneider, D. Portnoy, T. King, B. Scott, K. Pabbaraju and S. Shokoples, J. Med. Microbiol., 2021, 70, 001372 CrossRef CAS PubMed.
- J. Kerr, C. Panagopoulos and S. van der Linden, Pers. Individ. Differ., 2021, 179, 110892 CrossRef PubMed.
- R. Rahbari, N. Moradi and M. Abdi, Clin. Chim. Acta, 2021, 516, 1–7 CrossRef CAS PubMed.
- A. Eftekhari, M. Alipour, L. Chodari, S. Maleki Dizaj, M. Ardalan, M. Samiei, S. Sharifi, S. Zununi Vahed, I. Huseynova, R. Khalilov, E. Ahmadian and M. Cucchiarini, Microorganisms, 2021, 9, 232 CrossRef CAS PubMed.
- B. A. McGrath, M. J. Brenner, S. J. Warrillow, V. Pandian, A. Arora, T. S. Cameron, J. M. Añon, G. H. Martínez, R. D. Truog and S. D. Block, Lancet Respir. Med., 2020, 8, 717–725 CrossRef CAS PubMed.
- P. B. van Kasteren, B. van der Veer, S. van den Brink, L. Wijsman, J. de Jonge, A. van den Brandt, R. Molenkamp, C. Reusken and A. Meijer, J. Clin. Virol., 2020, 128, 104412 CrossRef CAS PubMed.
- S. Klein, T. G. Muller, D. Khalid, V. Sonntag-Buck, A. M. Heuser, B. Glass, M. Meurer, I. Morales, A. Schillak, A. Freistaedter, I. Ambiel, S. L. Winter, L. Zimmermann, T. Naumoska, F. Bubeck, D. Kirrmaier, S. Ullrich, I. Barreto Miranda, S. Anders, D. Grimm, P. Schnitzler, M. Knop, H. G. Krausslich, V. L. Dao Thi, K. Borner and P. Chlanda, Viruses, 2020, 12, 863 CrossRef CAS PubMed.
- S. Lee, J. Song and S. Kim, Analyst, 2021, 146, 6917–6923 RSC.
- Z. Zhao, H. Cui, W. Song, X. Ru, W. Zhou and X. Yu, bioRxiv, 2020, preprint, DOI:10.1101/2020.02.22.961268.
- C. Hwang, N. Park, E. S. Kim, M. Kim, S. D. Kim, S. Park, N. Y. Kim and J. H. Kim, Biosens. Bioelectron., 2021, 185, 113177 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- B. Dharavath, N. Yadav, S. Desai, R. Sunder, R. Mishra, M. Ketkar, P. Bhanshe, A. Gupta, A. K. Redhu, N. Patkar, S. Dutt, S. Gupta and A. Dutt, Heliyon, 2020, 6, e04405 CrossRef PubMed.
- T. Phan, S. Boes, M. McCullough, J. Gribschaw, J. Marsh, L. H. Harrison and A. Wells, J. Clin. Microbiol., 2022, 60, e00024–e00022 CrossRef CAS PubMed.
- T. Li, H. K. Chung, P. K. Pireku, B. F. Beitzel, M. A. Sanborn, C. Y. Tang, R. D. Hammer, D. Ritter, X.-F. Wan and I. Maljkovic Berry, J. Clin. Microbiol., 2021, 59, e02784–e02720 CAS.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- T. Alsuliman, R. Sulaiman, S. Ismail, M. Srour and A. Alrstom, Curr. Res. Transl. Med., 2020, 68, 83–91 CrossRef PubMed.
- C. B. F. Vogels, A. F. Brito, A. L. Wyllie, J. R. Fauver, I. M. Ott, C. C. Kalinich, M. E. Petrone, A. Casanovas-Massana, M. Catherine Muenker, A. J. Moore, J. Klein, P. Lu, A. Lu-Culligan, X. Jiang, D. J. Kim, E. Kudo, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takahashi, M. Taura, M. Tokuyama, A. Venkataraman, O. E. Weizman, P. Wong, Y. Yang, N. R. Cheemarla, E. B. White, S. Lapidus, R. Earnest, B. Geng, P. Vijayakumar, C. Odio, J. Fournier, S. Bermejo, S. Farhadian, C. S. Dela Cruz, A. Iwasaki, A. I. Ko, M. L. Landry, E. F. Foxman and N. D. Grubaugh, Nat. Microbiol., 2020, 5, 1299–1305 CrossRef CAS PubMed.
- C. B. F. Vogels, A. F. Brito, A. L. Wyllie, J. R. Fauver, I. M. Ott, C. C. Kalinich, M. E. Petrone, A. Casanovas-Massana, M. Catherine Muenker, A. J. Moore, J. Klein, P. Lu, A. Lu-Culligan, X. Jiang, D. J. Kim, E. Kudo, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takahashi, M. Taura, M. Tokuyama, A. Venkataraman, O. E. Weizman, P. Wong, Y. Yang, N. R. Cheemarla, E. B. White, S. Lapidus, R. Earnest, B. Geng, P. Vijayakumar, C. Odio, J. Fournier, S. Bermejo, S. Farhadian, C. S. Dela Cruz, A. Iwasaki, A. I. Ko, M. L. Landry, E. F. Foxman and N. D. Grubaugh, Nat. Microbiol., 2020, 5, 1299–1305 CrossRef CAS PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brunink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J. L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 2000045 Search PubMed.
- Y. Matsumura, T. Shimizu, T. Noguchi, S. Nakano, M. Yamamoto and M. Nagao, J. Mol. Diagn., 2021, 23, 164–170 CrossRef CAS PubMed.
- A. K. Nalla, A. M. Casto, M.-L. W. Huang, G. A. Perchetti, R. Sampoleo, L. Shrestha, Y. Wei, H. Zhu, K. R. Jerome, A. L. Greninger and A. J. McAdam, J. Clin. Microbiol., 2020, 58, e00557–e00520 CrossRef PubMed.
- P. B. van Kasteren, B. van der Veer, S. van den Brink, L. Wijsman, J. de Jonge, A. van den Brandt, R. Molenkamp, C. Reusken and A. Meijer, J. Clin. Virol., 2020, 128, 104412 CrossRef CAS PubMed.
- P. Asrani, M. S. Eapen, C. Chia, G. Haug, H. C. Weber, M. I. Hassan and S. S. Sohal, Expert Rev. Respir. Med., 2021, 15, 197–212 CrossRef CAS PubMed.
- Z. Gong, J.-W. Zhu, C.-P. Li, S. Jiang, L.-N. Ma, B.-X. Tang, D. Zou, M.-L. Chen, Y.-B. Sun, S.-H. Song, Z. Zhang, J.-F. Xiao, Y.-B. Xue, Y.-M. Bao, Z.-L. Du and W.-M. Zhao, Zool. Res., 2020, 41, 705 Search PubMed.
- M. Artesi, S. Bontems, P. Göbbels, M. Franckh, P. Maes, R. Boreux, C. Meex, P. Melin, M.-P. Hayette, V. Bours and K. Durkin, J. Clin. Microbiol., 2020, 58, e01598–e01520 CrossRef CAS PubMed.
- M. R. Hasan, S. Sundararaju, C. Manickam, F. Mirza, H. Al-Hail, S. Lorenz and P. Tang, J. Clin. Microbiol., 2021, 59, e03278–e03220 CrossRef CAS PubMed.
- Y. Shu and J. McCauley, Eurosurveillance, 2017, 22, 30494 CrossRef PubMed.
- Y. Pan, D. Zhang, P. Yang, L. L. M. Poon and Q. Wang, Lancet Infect. Dis., 2020, 20, 411–412 CrossRef CAS PubMed.
- Y. Yang, M. Yang, J. Yuan, F. Wang, Z. Wang, J. Li, M. Zhang, L. Xing, J. Wei, L. Peng, G. Wong, H. Zheng, W. Wu, C. Shen, M. Liao, K. Feng, J. Li, Q. Yang, J. Zhao, L. Liu and Y. Liu, Innovation, 2020, 1, 100061 CAS.
- G. Ye, Z. Pan, Y. Pan, Q. Deng, L. Chen, J. Li, Y. Li and X. Wang, J. Infect., 2020, 80, e14–e17 CrossRef CAS PubMed.
- C. Sentis, G. Billaud, A. Bal, E. Frobert, M. Bouscambert, G. Destras, L. Josset, B. Lina, F. Morfin and A. Gaymard, Viruses, 2022, 14, 919 CrossRef CAS PubMed.
- X. Wang, J. Zheng, L. Guo, H. Yao, L. Wang, X. Xia and W. Zhang, Virus Res., 2020, 289, 198147 CrossRef CAS PubMed.
- S. Woloshin, N. Patel and A. S. Kesselheim, N. Engl. J. Med., 2020, 383, e38 CrossRef CAS PubMed.
- T. Kageyama, S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda and K. Katayama, J. Clin. Microbiol., 2003, 41, 1548–1557 CrossRef CAS PubMed.
- T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun and L. Xia, Radiology, 2020, 296, E32–E40 CrossRef PubMed.
- A. Stang, J. Robers, B. Schonert, K.-H. Jöckel, A. Spelsberg, U. Keil and P. Cullen, J. Infect., 2021, 83, 237–279 CrossRef PubMed.
- Y. Liu, Z. Ning, Y. Chen, M. Guo, Y. Liu, N. K. Gali, L. Sun, Y. Duan, J. Cai, D. Westerdahl, X. Liu, K. Xu, K. F. Ho, H. Kan, Q. Fu and K. Lan, Nature, 2020, 582, 557–560 CrossRef CAS PubMed.
- O. Erster, O. Shkedi, G. Benedek, E. Zilber, I. Varkovitzky, R. Shirazi, D. Oriya Shorka, Y. Cohen, T. Bar, R. Yechieli, M. Tepperberg Oikawa, D. Venkert, M. Linial, E. Oiknine-Djian, M. Mandelboim, Z. Livneh, G. Shenhav-Saltzman, E. Mendelson, D. Wolf, M. Szwarcwort-Cohen, O. Mor, Y. Lewis and D. Zeevi, PLoS One, 2021, 16, e0249149 CrossRef CAS PubMed.
- N. W. West, A. A. Vasquez, A. Bahmani, M. F. Khan, J. Hartrick, C. L. Turner, W. Shuster and J. L. Ram, Sci. Total Environ., 2022, 847, 157547 CrossRef CAS PubMed.
- Y. Jiang, H. Wang, S. Hao, Y. Chen, J. He, Y. Liu, L. Chen, Y. Yu and S. Hua, Clin. Chim. Acta, 2020, 511, 346–351 CrossRef CAS PubMed.
- A. S. Whale, C. A. Bushell, P. R. Grant, S. Cowen, I. Gutierrez-Aguirre, D. M. O'Sullivan, J. Zel, M. Milavec, C. A. Foy, E. Nastouli, J. A. Garson and J. F. Huggett, J. Clin. Microbiol., 2016, 54, 392–400 CrossRef CAS PubMed.
- J. F. Huggett, S. Cowen and C. A. Foy, Clin. Chem., 2015, 61, 79–88 CrossRef CAS PubMed.
- M. Vynck, W. Trypsteen, O. Thas, L. Vandekerckhove and W. De Spiegelaere, Mol. Diagn. Ther., 2016, 20, 437–447 CrossRef CAS PubMed.
- P. L. Quan, M. Sauzade and E. Brouzes, Sensors, 2018, 18, 1271 CrossRef PubMed.
- L. Cao, X. Cui, J. Hu, Z. Li, J. R. Choi, Q. Yang, M. Lin, L. Ying Hui and F. Xu, Biosens. Bioelectron., 2017, 90, 459–474 CrossRef CAS PubMed.
- C. Tan, D. Fan, N. Wang, F. Wang, B. Wang, L. Zhu and Y. Guo, View, 2021, 20200082, DOI:10.1002/VIW.20200082.
- F. Scollo, L. A. Egea, A. Gentile, S. La Malfa, G. Dorado and P. Hernandez, Food Chem., 2016, 213, 388–394 CrossRef CAS PubMed.
- P. Wang, F. Jing, G. Li, Z. Wu, Z. Cheng, J. Zhang, H. Zhang, C. Jia, Q. Jin, H. Mao and J. Zhao, Biosens. Bioelectron., 2015, 74, 836–842 CrossRef CAS PubMed.
- L. Zhang and H. Guo, Adv. Biomarker Sci. Technol., 2020, 2, 1–23 CrossRef PubMed.
- H. G. Barreto, F. A. de Padua Milagres, G. C. de Araujo, M. M. Daude and V. A. Benedito, J. Mol. Med., 2020, 98, 1727–1736 CrossRef CAS PubMed.
- J. F. Zhang, K. Yan, H. H. Ye, J. Lin, J. J. Zheng and T. Cai, Int. J. Infect. Dis., 2020, 97, 212–214 CrossRef CAS PubMed.
- K. Narang, E. A. L. Enninga, M. Gunaratne, E. R. Ibirogba, A. T. A. Trad, A. Elrefaei, R. N. Theiler, R. Ruano, L. M. Szymanski, R. Chakraborty and V. D. Garovic, Mayo Clin. Proc., 2020, 95, 1750–1765 CrossRef CAS PubMed.
- L. Lan, D. Xu, G. Ye, C. Xia, S. Wang, Y. Li and H. Xu, JAMA, 2020, 323, 1502–1503 CrossRef CAS PubMed.
- J.-f. Zhang, K. Yan, H.-h. Ye, J. Lin, J.-j. Zheng and T. Cai, Int. J. Infect. Dis., 2020, 97, 212–214 CrossRef CAS PubMed.
- R. Lu, J. Wang, M. Li, Y. Wang, J. Dong and W. J. M. Cai, 2020.
- C. Alteri, V. Cento, M. Antonello, L. Colagrossi, M. Merli, N. Ughi, S. Renica, E. Matarazzo, F. Di Ruscio, L. Tartaglione, J. Colombo, C. Grimaldi, S. Carta, A. Nava, V. Costabile, C. Baiguera, D. Campisi, D. Fanti, C. Vismara, R. Fumagalli, F. Scaglione, O. M. Epis, M. Puoti and C. F. Perno, PLoS One, 2020, 15, e0236311 CrossRef CAS PubMed.
- D. Chirizzi, M. Conte, M. Feltracco, A. Dinoi, E. Gregoris, E. Barbaro, G. La Bella, G. Ciccarese, G. La Salandra, A. Gambaro and D. Contini, Environ. Int., 2021, 146, 106255 CrossRef CAS PubMed.
- I. D. Amoah, L. Pillay, N. Deepnarian, O. Awolusi, K. Pillay, P. Ramlal, S. Kumari and F. Bux, Int. J. Hyg. Environ. Health, 2020, 236, 113807 CrossRef PubMed.
- A. Zali, M. R. Sohrabi, A. Mahdavi, N. Khalili, M. S. Taheri, A. Maher, M. Sadoughi, A. Zarghi, S. A. Ziai, A. A. Shabestari, M. Bakhshayeshkaram, H. Haghighatkhah, B. Salevatipour, A. Abrishami, M. Raoufi, P. Dehghan, A. K. Bagheri, R. J. Khoshnoud and K. Hanani, Acad. Radiol., 2021, 28, 1654–1661 CrossRef PubMed.
- B. Perez-Lopez and M. Mir, Talanta, 2021, 225, 121898 CrossRef CAS PubMed.
- M. Ikeda, K. Imai, S. Tabata, K. Miyoshi, N. Murahara, T. Mizuno, M. Horiuchi, K. Kato, Y. Imoto and M. Iwata, Infect. Dis., 2020, 58, e01438–20 Search PubMed.
- B. Schermer, F. Fabretti, M. Damagnez, V. Di Cristanziano, E. Heger, S. Arjune, N. A. Tanner, T. Imhof, M. Koch, A. Ladha, J. Joung, J. S. Gootenberg, O. O. Abudayyeh, V. Burst, F. Zhang, F. Klein, T. Benzing and R. U. Muller, PLoS One, 2020, 15, e0238612 CrossRef CAS PubMed.
- W. E. Huang, B. Lim, C. C. Hsu, D. Xiong, W. Wu, Y. Yu, H. Jia, Y. Wang, Y. Zeng, M. Ji, H. Chang, X. Zhang, H. Wang and Z. Cui, Microb. Biotechnol., 2020, 13, 950–961 CrossRef CAS PubMed.
- C. Yan, J. Cui, L. Huang, B. Du, L. Chen, G. Xue, S. Li, W. Zhang, L. Zhao, Y. Sun, H. Yao, N. Li, H. Zhao, Y. Feng, S. Liu, Q. Zhang, D. Liu and J. Yuan, Clin. Microbiol. Infect., 2020, 26, 773–779 CrossRef CAS PubMed.
- Y. Lin, S. Wu, X. Hao, X. Dong, L. Mao, V. Pelechano, C. Wei-Hua and X. Yin, Clin. Chem., 2020, 66, 975–977 CrossRef PubMed.
- L. E. Lamb, S. N. Bartolone, E. Ward and M. B. Chancellor, PLoS One, 2020, 15, e0234682 CrossRef CAS PubMed.
- X. Hu, Q. Deng, J. Li, J. Chen, Z. Wang, X. Zhang, Z. Fang, H. Li, Y. Zhao, P. Yu, W. Li, X. Wang, S. Li, L. Zhang and T. Hou, mSphere, 2020, 5, e00808–e00820 CrossRef CAS PubMed.
- A. Premraj, A. G. Aleyas, B. Nautiyal and T. J. Rasool, Diagnostics, 2020, 10, 866 CrossRef CAS PubMed.
- K. Minami, R. Masutani, Y. Suzuki, M. Kubota, N. Osaka, T. Nakanishi, T. Nakano and A. Ukimura, J. Infect. Chemother., 2021, 27, 1068–1071 CrossRef CAS PubMed.
- Y. Zhang, M. Chen, C. Liu, J. Chen, X. Luo, Y. Xue, Q. Liang, L. Zhou, Y. Tao, M. Li, D. Wang, J. Zhou and J. Wang, Sens. Actuators, B, 2021, 345, 130411 CrossRef CAS PubMed.
- Y. Zhang, M. Chen, C. Liu, J. Chen, X. Luo, Y. Xue, Q. Liang, L. Zhou, Y. Tao and M. Li, Sens. Actuators, B, 2021, 345, 130411 CrossRef CAS PubMed.
- V. L. D. Thi, K. Herbst, K. Boerner, M. Meurer, L. P. Kremer, D. Kirrmaier, A. Freistaedter, D. Papagiannidis, C. Galmozzi and M. L. Stanifer, Sci. Transl. Med., 2020, 12, 7075 CrossRef PubMed.
- M. A. Rohaim, E. Clayton, I. Sahin, J. Vilela, M. E. Khalifa, M. Q. Al-Natour, M. Bayoumi, A. C. Poirier, M. Branavan, M. Tharmakulasingam, N. S. Chaudhry, R. Sodi, A. Brown, P. Burkhart, W. Hacking, J. Botham, J. Boyce, H. Wilkinson, C. Williams, J. Whittingham-Dowd, E. Shaw, M. Hodges, L. Butler, M. D. Bates, R. La Ragione, W. Balachandran, A. Fernando and M. Munir, Viruses, 2020, 12, 972 CrossRef CAS PubMed.
- P. Gill and A. Ghaemi, Nucleosides, Nucleotides Nucleic Acids, 2008, 27, 224–243 CrossRef CAS PubMed.
- M. V. Neguembor, L. Martin, Á. Castells-García, P. A. Gómez-García, C. Vicario, D. Carnevali, J. A. Abed, A. Granados, R. Sebastian-Perez and F. Sottile, Mol. Cell, 2021, 81, 3065–3081 CrossRef CAS PubMed.
- Y. P. Liu and C. Y. Yao, World J. Gastroenterol., 2015, 21, 11954–11963 CrossRef CAS PubMed.
- H. H. Mostafa, J. Hardick, E. Morehead, J. A. Miller, C. A. Gaydos and Y. C. Manabe, J. Clin. Virol., 2020, 130, 104578 CrossRef CAS PubMed.
- J. Pham, S. Meyer, C. Nguyen, A. Williams, M. Hunsicker, I. McHardy, I. Gendlina, D. Y. Goldstein, A. S. Fox and A. Hudson, J. Clin. Microbiol., 2020, 58, e01669–e01620 CrossRef CAS PubMed.
- S. L. Mitchell and S. E. Ventura, J. Clin. Microbiol., 2020, 58, e02241–e02220 CAS.
- H. Wang, C. A. Hogan, J. A. Miller, M. K. Sahoo, C. Huang, K. O. Mfuh, M. Sibai, J. Zehnder, B. Hickey, N. Sinnott-Armstrong and B. A. Pinsky, Emerging Infect. Dis., 2021, 27, 92 CrossRef CAS PubMed.
- K. Newsom, Y. Zhang, S. Chamala, K. Martinez, M. Clare-Salzler and P. Starostik, J. Clin. Lab. Anal., 2021, 35, e23888 CrossRef CAS PubMed.
- J. L. Cantera, H. N. White, M. S. Forrest, O. W. Stringer, V. Y. Belizario Jr, H. L. Storey, E. L. de Hostos and T. de Los Santos, PLoS Neglected Trop. Dis., 2021, 15, e0009782 CrossRef CAS PubMed.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry, H. M. Sallum, E. M. Zhao, J. B. Niemi and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- O. Faye, O. Faye, B. Soropogui, P. Patel, A. A. El Wahed, C. Loucoubar, G. Fall, D. Kiory, N. Magassouba, S. Keita, M. K. Konde, A. A. Diallo, L. Koivogui, H. Karlberg, A. Mirazimi, O. Nentwich, O. Piepenburg, M. Niedrig, M. Weidmann and A. A. Sall, Eurosurveillance, 2015, 20, 30053 CrossRef PubMed.
- N. I. Vasileva Wand, L. C. Bonney, R. J. Watson, V. Graham and R. Hewson, J. Gen. Virol., 2018, 99, 1012–1026 CrossRef CAS PubMed.
- P. Huang, H. Jin, Y. Zhao, E. Li, F. Yan, H. Chi, Q. Wang, Q. Han, R. Mo, Y. Song, J. Bi, C. Jiao, W. Li, H. He, H. Wang, A. Ma, N. Feng, J. Wang, T. Wang, S. Yang, Y. Gao, X. Xia and H. Wang, PLoS Neglected Trop. Dis., 2021, 15, e0009227 CrossRef CAS PubMed.
- S. Xia and X. Chen, Cell Discovery, 2020, 6, 37 CrossRef CAS PubMed.
- W. Guo, G. Xu, Y. Wang, Y. Nan, C. Wu and E. M. Zhou, J. Virol. Methods, 2019, 272, 113710 CrossRef CAS PubMed.
- R. Aman, A. Mahas, T. Marsic, N. Hassan and M. M. Mahfouz, Front. Microb., 2020, 11, 610872 CrossRef PubMed.
- F. E. Chen, P. W. Lee, A. Y. Trick, J. S. Park, L. Chen, K. Shah, H. Mostafa, K. C. Carroll, K. Hsieh and T. H. Wang, Biosens. Bioelectron., 2021, 190, 113390 CrossRef CAS PubMed.
- A. A. El Wahed, P. Patel, M. Maier, C. Pietsch, D. Ruster, S. Bohlken-Fascher, J. Kissenkotter, O. Behrmann, M. Frimpong, M. M. Diagne, M. Faye, N. Dia, M. A. Shalaby, H. Amer, M. Elgamal, A. Zaki, G. Ismail, M. Kaiser, V. M. Corman, M. Niedrig, O. Landt, O. Faye, A. A. Sall, F. T. Hufert, U. Truyen, U. G. Liebert and M. Weidmann, Anal. Chem., 2021, 93, 2627–2634 CrossRef CAS PubMed.
- Y. Sun, P. Qin, J. He, W. Li, Y. Shi, J. Xu, Q. Wu, Q. Chen, W. Li, X. Wang, G. Liu and W. Chen, Biosens. Bioelectron., 2022, 197, 113771 CrossRef CAS PubMed.
- P. Khan, L. M. Aufdembrink and A. E. Engelhart, ACS Synth. Biol., 2020, 9, 2861–2880 CrossRef CAS PubMed.
- Y. Ju, J. Kim, Y. Park, C. Y. Lee, K. Kim, K. H. Hong, H. Lee, D. Yong and H. G. Park, Biosens. Bioelectron., 2022, 196, 113689 CrossRef CAS PubMed.
- M. Mohammadniaei, M. Zhang, J. Ashley, U. B. Christensen, L. J. Friis-Hansen, R. Gregersen, J. G. Lisby, T. L. Benfield, F. E. Nielsen and J. Henning Rasmussen, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- S. Al-Attar, E. R. Westra, J. van der Oost and S. J. Brouns, Biol. Chem., 2011, 392, 277–289 CAS.
- R. Barrangou, Curr. Opin. Immunol., 2015, 32, 36–41 CrossRef CAS PubMed.
- Y. Dai, Y. Wu, G. Liu and J. J. Gooding, Angew. Chem., 2020, 59, 20754–20766 CrossRef CAS PubMed.
- W. Feng, A. M. Newbigging, J. Tao, Y. Cao, H. Peng, C. Le, J. Wu, B. Pang, J. Li, D. L. Tyrrell, H. Zhang and X. C. Le, Chem. Sci., 2021, 12, 4683–4698 RSC.
- S. Jolany Vangah, C. Katalani, H. A. Booneh, A. Hajizade, A. Sijercic and G. Ahmadian, Biol. Proced. Online, 2020, 22, 22 CrossRef CAS PubMed.
- R. Aman, A. Mahas and M. Mahfouz, ACS Synth. Biol., 2020, 9, 1226–1233 CrossRef CAS PubMed.
- Y. Li, S. Li, J. Wang and G. Liu, Trends Biotechnol., 2019, 37, 730–743 CrossRef CAS PubMed.
- D. G. Sashital, Genome Med., 2018, 10, 32 CrossRef PubMed.
- Z. Li, J. Wei, D. Di, X. Wang, C. Li, B. Li, Y. Qiu, K. Liu, F. Gu, M. Tong, S. Wang, X. Wu and Z. Ma, Acta Biochim. Biophys., 2020, 52, 1413–1419 CAS.
- T. Yuan, O. Mukama, Z. Li, W. Chen, Y. Zhang, J. de Dieu Habimana, Y. Zhang, R. Zeng, C. Nie, Z. He and L. Zeng, Analyst, 2020, 145, 6388–6394 RSC.
- C. M. LaManna, B. Pyhtila and R. Barrangou, CRISPR J., 2020, 3, 248–252 CrossRef PubMed.
- F. Zhang, O. O. Abudayyeh and J. S. Gootenberg, Nat. Protoc., 2020, 14, 2986–3012 Search PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam, T. Wongsatit, N. Athipanyasilp, B. Eiamthong, B. Lakkanasirorat, T. Phoodokmai, N. Niljianskul, D. Pakotiprapha, S. Chanarat, A. Homchan, R. Tinikul, P. Kamutira, K. Phiwkaow, S. Soithongcharoen, C. Kantiwiriyawanitch, V. Pongsupasa, D. Trisrivirat, J. Jaroensuk, T. Wongnate, S. Maenpuen, P. Chaiyen, S. Kamnerdnakta, J. Swangsri, S. Chuthapisith, Y. Sirivatanauksorn, C. Chaimayo, R. Sutthent, W. Kantakamalakul, J. Joung, A. Ladha, X. Jin, J. S. Gootenberg, O. O. Abudayyeh, F. Zhang, N. Horthongkham and C. Uttamapinant, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- Z. Ali, R. Aman, A. Mahas, G. S. Rao, M. Tehseen, T. Marsic, R. Salunke, A. K. Subudhi, S. M. Hala, S. M. Hamdan, A. Pain, F. S. Alofi, A. Alsomali, A. M. Hashem, A. Khogeer, N. A. M. Almontashiri, M. Abedalthagafi, N. Hassan and M. M. Mahfouz, Virus Res., 2020, 288, 198129 CrossRef CAS PubMed.
- J. Joung, A. Ladha, M. Saito, N.-G. Kim, A. E. Woolley, M. Segel, R. P. Barretto, A. Ranu, R. K. Macrae and G. Faure, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- J. Joung, A. Ladha, M. Saito, N.-G. Kim, A. E. Woolley, M. Segel, R. P. Barretto, A. Ranu, R. K. Macrae and G. Faure, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li and C. Liu, bioRxiv, 2020, preprint, DOI:10.1101/2020.03.19.998724.
- Z. Huang, D. Tian, Y. Liu, Z. Lin, C. J. Lyon, W. Lai, D. Fusco, A. Drouin, X. Yin, T. Hu and B. Ning, Biosens. Bioelectron., 2020, 164, 112316 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, C. K. Boehm, B. A. Petros, S. Siddiqui, B. M. Shaw, G. Adams, T. F. Kosoko-Thoroddsen, M. E. Kemball, R. Gross, L. Wronka, K. Caviness, L. E. Hensley, N. H. Bergman, B. L. MacInnis, J. E. Lemieux, P. C. Sabeti and C. Myhrvold, bioRxiv, 2020, preprint, DOI:10.1101/2020.05.28.119131.
- J. Arizti-Sanz, A. D. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T.-S. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai and S. Mantena, Nat. Biomed. Eng., 2022, 1–12, DOI:10.1038/s41551-022-00889-z.
- F. Wu, S. Zhao, B. Yu, Y. M. Chen, W. Wang, Z. G. Song, Y. Hu, Z. W. Tao, J. H. Tian, Y. Y. Pei, M. L. Yuan, Y. L. Zhang, F. H. Dai, Y. Liu, Q. M. Wang, J. J. Zheng, L. Xu, E. C. Holmes and Y. Z. Zhang, Nature, 2020, 579, 265–269 CrossRef CAS PubMed.
- P. Khan, L. M. Aufdembrink and A. E. Engelhart, ACS Synth. Biol., 2020, 9, 2861–2880 CrossRef CAS PubMed.
- A. Diekstra, E. Bosgoed, A. Rikken, B. van Lier, E. J. Kamsteeg, M. Tychon, R. C. Derks, R. A. van Soest, A. R. Mensenkamp, H. Scheffer, K. Neveling and M. R. Nelen, Clin. Chem., 2015, 61, 154–162 CrossRef CAS PubMed.
- B. L. Brown, M. Watson, S. S. Minot, M. C. Rivera and R. B. Franklin, Gigascience, 2017, 6, 1–10 CrossRef PubMed.
- A. Payne, N. Holmes, T. Clarke, R. Munro, B. J. Debebe and M. Loose, Nat. Biotechnol., 2021, 39, 442–450 CrossRef CAS PubMed.
- F. Bayat and H. Tajalli, Heliyon, 2020, 6, e03382 CrossRef PubMed.
- D. Muzzey, E. A. Evans and C. Lieber, Curr. Genet. Med. Rep., 2015, 3, 158–165 CrossRef PubMed.
- M. L. Metzker, Nat. Rev. Genet., 2010, 11, 31–46 CrossRef CAS PubMed.
- K. R. Kumar, M. J. Cowley and R. L. Davis, Semin. Thromb. Hemostasis, 2019, 45, 661–673 CrossRef CAS PubMed.
- J. F. Hess, T. A. Kohl, M. Kotrova, K. Ronsch, T. Paprotka, V. Mohr, T. Hutzenlaub, M. Bruggemann, R. Zengerle, S. Niemann and N. Paust, Biotechnol. Adv., 2020, 41, 107537 CrossRef CAS PubMed.
- D. Chandler-Brown, A. M. Bueno, O. Atay and D. S. Tsao, bioRxiv, 2020, preprint, DOI:10.1101/2020.04.07.029199.
- B. Furlani, K. Kouter, D. Rozman and A. Videtič Paska, Acta Chim. Slov., 2021, 68, 268–278 CrossRef CAS PubMed.
- A. Raza and M. S. Shahid, Appl. Plant Virol., 2020, 131–140, DOI:10.1016/b978-0-12-818654-1.00010-4.
- J. A. Nasir, R. A. Kozak, P. Aftanas, A. R. Raphenya, K. M. Smith, F. Maguire, H. Maan, M. Alruwaili, A. Banerjee, H. Mbareche, B. P. Alcock, N. C. Knox, K. Mossman, B. Wang, J. A. Hiscox, A. G. McArthur and S. Mubareka, Viruses, 2020, 12, 895 CrossRef CAS PubMed.
- B. G. St Hilaire, N. C. Durand, N. Mitra, S. G. Pulido, R. Mahajan, A. Blackburn, Z. L. Colaric, J. W. Theisen, D. Weisz and O. Dudchenko, bioRxiv, 2020, preprint, DOI:10.1101/2020.04.25.061499.
- R. C. Bhoyar, A. Jain, P. Sehgal, M. K. Divakar, D. Sharma, M. Imran, B. Jolly, G. Ranjan, M. Rophina, S. Sharma, S. Siwach, K. Pandhare, S. Sahoo, M. Sahoo, A. Nayak, J. N. Mohanty, J. Das, S. Bhandari, S. K. Mathur, A. Kumar, R. Sahlot, P. Rojarani, J. V. Lakshmi, A. Surekha, P. C. Sekhar, S. Mahajan, S. Masih, P. Singh, V. Kumar, B. Jose, V. Mahajan, V. Gupta, R. Gupta, P. Arumugam, A. Singh, A. Nandy, P. Ragavendran, R. M. Jha, A. Kumari, S. Gandotra, V. Rao, M. Faruq, S. Kumar, G. B. Reshma, G. N. Varma, S. S. Roy, A. Sengupta, S. Chattopadhyay, K. Singhal, S. Pradhan, D. Jha, S. Naushin, S. Wadhwa, N. Tyagi, M. Poojary, V. Scaria and S. Sivasubbu, PLoS One, 2021, 16, e0247115 CrossRef CAS PubMed.
- L. W. Meredith, W. L. Hamilton, B. Warne, C. J. Houldcroft, M. Hosmillo, A. S. Jahun, M. D. Curran, S. Parmar, L. G. Caller, S. L. Caddy, F. A. Khokhar, A. Yakovleva, G. Hall, T. Feltwell, S. Forrest, S. Sridhar, M. P. Weekes, S. Baker, N. Brown, E. Moore, A. Popay, I. Roddick, M. Reacher, T. Gouliouris, S. J. Peacock, G. Dougan, M. E. Török and I. Goodfellow, Lancet Infect. Dis., 2020, 20, 1263–1271 CrossRef CAS PubMed.
- J. S. Bloom, L. Sathe, C. Munugala, E. M. Jones, M. Gasperini, N. B. Lubock, F. Yarza, E. M. Thompson, K. M. Kovary, J. Park, D. Marquette, S. Kay, M. Lucas, T. Love, A. Sina Booeshaghi, O. F. Brandenberg, L. Guo, J. Boocock, M. Hochman, S. W. Simpkins, I. Lin, N. LaPierre, D. Hong, Y. Zhang, G. Oland, B. J. Choe, S. Chandrasekaran, E. E. Hilt, M. J. Butte, R. Damoiseaux, C. Kravit, A. R. Cooper, Y. Yin, L. Pachter, O. B. Garner, J. Flint, E. Eskin, C. Luo, S. Kosuri, L. Kruglyak and V. A. Arboleda, Nat. Biomed. Eng., 2021, 5, 657–665 CrossRef CAS PubMed.
- M. Krause, A. M. Niazi, K. Labun, Y. N. T. Cleuren, F. S. Müller and E. Valen, RNA, 2019, 25, 1229–1241 CrossRef CAS PubMed.
- R. Bowden, R. W. Davies, A. Heger, A. T. Pagnamenta, M. de Cesare, L. E. Oikkonen, D. Parkes, C. Freeman, F. Dhalla, S. Y. Patel, N. Popitsch, C. L. C. Ip, H. E. Roberts, S. Salatino, H. Lockstone, G. Lunter, J. C. Taylor, D. Buck, M. A. Simpson and P. Donnelly, Nat. Commun., 2019, 10, 1869 CrossRef PubMed.
- M. Wang, A. Fu, B. Hu, Y. Tong, R. Liu, Z. Liu, J. Gu, B. Xiang, J. Liu, W. Jiang, G. Shen, W. Zhao, D. Men, Z. Deng, L. Yu, W. Wei, Y. Li and T. Liu, Small, 2020, 16, e2002169 CrossRef PubMed.
- M. Infantino, V. Grossi, B. Lari, R. Bambi, A. Perri, M. Manneschi, G. Terenzi, I. Liotti, G. Ciotta and C. J. Taddei, J. Med. Virol., 2020, 92, 1671–1675 CrossRef CAS PubMed.
- G. Alderton, Science, 2020, 368, 1076–1078 CrossRef.
- D. Jacofsky, E. M. Jacofsky and M. Jacofsky, J. Arthroplasty, 2020, 35, S74–S81 CrossRef PubMed.
- M. A. Black, G. Shen, X. Feng, W. F. G. Beltran, Y. Feng, V. Vasudevaraja, D. Allison, L. H. Lin, T. Gindin and M. Astudillo, J. Immunol. Methods, 2021, 489, 112909 CrossRef CAS PubMed.
- A. Saghazadeh and N. Rezaei, Int. Immunopharmacol., 2020, 84, 106560 CrossRef CAS PubMed.
- M. A. Black, G. Shen, X. Feng, W. F. G. Beltran, Y. Feng, V. Vasudevaraja, D. Allison, L. H. Lin, T. Gindin and M. Astudillo, J. Immunol. Methods, 2021, 489, 112909 CrossRef CAS PubMed.
- A. Saghazadeh and N. Rezaei, Int. Immunopharmacol., 2020, 84, 106560 CrossRef CAS PubMed.
- E. O Murchu, P. Byrne, K. A. Walsh, P. G. Carty, M. Connolly, C. De Gascun, K. Jordan, M. Keoghan, K. K. O'Brien and M. O'Neill, Rev. Med. Virol., 2021, 31, e2162 CrossRef CAS PubMed.
- B. Lou, T.-D. Li, S.-F. Zheng, Y.-Y. Su, Z.-Y. Li, W. Liu, F. Yu, S.-X. Ge, Q.-D. Zou and Q. Yuan, Eur. Respir. J., 2020, 56, 2000763 CrossRef CAS PubMed.
- H.-w. Jiang, Y. Li, H.-n. Zhang, W. Wang, X. Yang, H. Qi, H. Li, D. Men, J. Zhou and S.-c. Tao, Nat. Commun., 2020, 11, 1–11 CrossRef PubMed.
- K. Harley and I. L. Gunsolus, J. Clin. Microbiol., 2020, 59, e02104–e02120 CrossRef PubMed.
- Z. Wang, Z. Zheng, H. Hu, Q. Zhou, W. Liu, X. Li, Z. Liu, Y. Wang and Y. Ma, Lab Chip, 2020, 20, 4255–4261 RSC.
- K. Harley and I. L. Gunsolus, J. Clin. Microbiol., 2020, 59, e02104–e02120 CrossRef PubMed.
- Q. X. Long, B. Z. Liu, H. J. Deng, G. C. Wu, K. Deng, Y. K. Chen, P. Liao, J. F. Qiu, Y. Lin, X. F. Cai, D. Q. Wang, Y. Hu, J. H. Ren, N. Tang, Y. Y. Xu, L. H. Yu, Z. Mo, F. Gong, X. L. Zhang, W. G. Tian, L. Hu, X. X. Zhang, J. L. Xiang, H. X. Du, H. W. Liu, C. H. Lang, X. H. Luo, S. B. Wu, X. P. Cui, Z. Zhou, M. M. Zhu, J. Wang, C. J. Xue, X. F. Li, L. Wang, Z. J. Li, K. Wang, C. C. Niu, Q. J. Yang, X. J. Tang, Y. Zhang, X. M. Liu, J. J. Li, D. C. Zhang, F. Zhang, P. Liu, J. Yuan, Q. Li, J. L. Hu, J. Chen and A. L. Huang, Nat. Med., 2020, 26, 845–848 CrossRef CAS PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- N. M. Okba, M. A. Müller, W. Li, C. Wang, C. H. GeurtsvanKessel, V. M. Corman, M. M. Lamers, R. S. Sikkema, E. De Bruin and F. D. Chandler, Emerging Infect. Dis., 2020, 26, 1478 CrossRef CAS PubMed.
- B. A. McGrath, M. J. Brenner, S. J. Warrillow, V. Pandian, A. Arora, T. S. Cameron, J. M. Añon, G. Hernández Martínez, R. D. Truog, S. D. Block, G. C. Y. Lui, C. McDonald, C. H. Rassekh, J. Atkins, L. Qiang, S. Vergez, P. Dulguerov, J. Zenk, M. Antonelli, P. Pelosi, B. K. Walsh, E. Ward, Y. Shang, S. Gasparini, A. Donati, M. Singer, P. J. M. Openshaw, N. Tolley, H. Markel and D. J. Feller-Kopman, Lancet Respir. Med., 2020, 8, 717–725 CrossRef CAS PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. Luong, Trends Biotechnol., 2015, 33, 692–705 CrossRef CAS PubMed.
- S. Sharma, J. Zapatero-Rodríguez, P. Estrela and R. O'Kennedy, Biosensors, 2015, 5, 577–601 CrossRef PubMed.
- C. Zheng, X. Wang, Y. Lu and Y. Liu, Food Control, 2012, 26, 446–452 CrossRef CAS.
- C. Zheng, X. Wang, Y. Lu and Y. Liu, Food Control, 2012, 26, 446–452 CrossRef CAS.
- J.-L. Wu, W.-P. Tseng, C.-H. Lin, T.-F. Lee, M.-Y. Chung, C.-H. Huang, S.-Y. Chen, P.-R. Hsueh and S.-C. Chen, J. Infect., 2020, 81, 435–442 CrossRef CAS PubMed.
- G. V. Guedez-López, M. Alguacil-Guillén, P. González-Donapetry, I. Bloise, C. Tornero-Marin, J. González-García, J. Mingorance, J. García-Rodríguez and I. Diseases, Eur. J. Clin. Microbiol. Infect. Dis., 2020, 39, 2289–2297 CrossRef PubMed.
- M. Lisboa Bastos, G. Tavaziva, S. K. Abidi, J. R. Campbell, L.-P. Haraoui, J. C. Johnston, Z. Lan, S. Law, E. MacLean, A. Trajman, D. Menzies, A. Benedetti and F. Ahmad Khan, BMJ, 2020, 370, 32611558–32611558 Search PubMed.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu and W. Chen, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS PubMed.
- S. Cavalera, B. Colitti, S. Rosati, G. Ferrara, L. Bertolotti, C. Nogarol, C. Guiotto, C. Cagnazzo, M. Denina, F. Fagioli, F. Di Nardo, M. Chiarello, C. Baggiani and L. Anfossi, Talanta, 2021, 223, 121737 CrossRef CAS PubMed.
- C. Wang, D. Shi, N. Wan, X. Yang, H. Liu, H. Gao, M. Zhang, Z. Bai, D. Li, E. Dai, Z. Rong and S. Wang, Analyst, 2021, 146, 3908–3917 RSC.
- C. Zhang, L. Zhou, H. Liu, S. Zhang, Y. Tian, J. Huo, F. Li, Y. Zhang, B. Wei, D. Xu, J. Hu, J. Wang, Y. Cheng, W. Shi, X. Xu, J. Zhou, P. Sang, X. Tan, W. Wang, M. Zhang, B. Wang, Y. Zhou, K. Zhang and K. He, Emerging Microbes Infect., 2020, 9, 2020–2029 CrossRef CAS PubMed.
- R. Chen, C. Ren, M. Liu, X. Ge, M. Qu, X. Zhou, M. Liang, Y. Liu and F. Li, ACS Nano, 2021, 15(5), 8996–9004 CrossRef CAS PubMed , 33928784.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu, W. Chen, Y. Zhang, J. Wang, B. Huang, Y. Lin, J. Yang, W. Cai, X. Wang, J. Cheng, Z. Chen, K. Sun, W. Pan, Z. Zhan, L. Chen and F. Ye, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS PubMed.
- G. E. Kenny and C. L. Dunsmoor, J. Clin. Microbiol., 1983, 17, 655–665 CrossRef CAS PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS PubMed.
- S. D. Gan and K. R. Patel, J. Invest. Dermatol., 2013, 133, e12 CrossRef CAS PubMed.
- E. H. Y. Lau, O. T. Y. Tsang, D. S. C. Hui, M. Y. W. Kwan, W. H. Chan, S. S. Chiu, R. L. W. Ko, K. H. Chan, S. M. S. Cheng, R. Perera, B. J. Cowling, L. L. M. Poon and M. Peiris, Nat. Commun., 2021, 12, 63 CrossRef CAS PubMed.
- Z. Lopandić, I. Protić-Rosić, A. Todorović, S. Glamočlija, M. Gnjatović, D. Ćujic and M. Gavrović-Jankulović, Int. J. Mol. Sci., 2021, 22, 4951 CrossRef PubMed.
- X.-f. Cai, J. Chen, J.-. li Hu, Q.-x. Long, H.-j. Deng, P. Liu, K. Fan, P. Liao, B.-z. Liu and G.-c. Wu, J. Infect. Dis., 2020, 222, 189–193 CrossRef CAS PubMed.
- L. F. Huergo, M. S. Conzentino, E. C. Marques Gerhardt, A. R. Schenberger Santos, F. O. Pedrosa, E. M. Souza, M. B. Nogueira, K. Forchhammer, F. G. Moraes Rego, S. M. Raboni and R. A. Reis, MedRxiv, 2020, preprint, DOI:10.1101/2020.07.26.20162255.
- F. Gong, H. X. Wei, J. Qi, H. Ma, L. Liu, J. Weng, X. Zheng, Q. Li, D. Zhao, H. Fang, L. Liu, H. He, C. Ma, J. Han, A. Sun, B. Wang, T. Jin, B. Li and B. Li, ACS Sens., 2021, 6, 2709–2719 CrossRef CAS PubMed.
- Y. Liu, Y. Tan, Q. Fu, M. Lin, J. He, S. He, M. Yang, S. Chen and J. Zhou, Biosens. Bioelectron., 2021, 176, 112920 CrossRef CAS PubMed.
- E. S. Theel, J. Harring, H. Hilgart and D. Granger, J. Clin. Microbiol., 2020, 58, e01243–e01220 CAS.
- D. Lin, L. Liu, M. Zhang, Y. Hu, Q. Yang, J. Guo, Y. Dai, Y. Xu, Y. Cai and X. Chen, Eur. J. Clin. Microbiol. Infect. Dis., 2020, 39, 2271–2277 CrossRef CAS PubMed.
- A. N. Grossberg, L. A. Koza, A. Ledreux, C. Prusmack, H. K. Krishnamurthy, V. Jayaraman, A. C. Granholm and D. A. Linseman, Nat. Commun., 2021, 12, 740 CrossRef CAS PubMed.
- Q. X. Long, B. Z. Liu, H. J. Deng, G. C. Wu, K. Deng, Y. K. Chen, P. Liao, J. F. Qiu, Y. Lin, X. F. Cai, D. Q. Wang, Y. Hu, J. H. Ren, N. Tang, Y. Y. Xu, L. H. Yu, Z. Mo, F. Gong, X. L. Zhang, W. G. Tian, L. Hu, X. X. Zhang, J. L. Xiang, H. X. Du, H. W. Liu, C. H. Lang, X. H. Luo, S. B. Wu, X. P. Cui, Z. Zhou, M. M. Zhu, J. Wang, C. J. Xue, X. F. Li, L. Wang, Z. J. Li, K. Wang, C. C. Niu, Q. J. Yang, X. J. Tang, Y. Zhang, X. M. Liu, J. J. Li, D. C. Zhang, F. Zhang, P. Liu, J. Yuan, Q. Li, J. L. Hu, J. Chen and A. L. Huang, Nat. Med., 2020, 26, 845–848 CrossRef CAS PubMed.
- W. H. Kong, R. Zhao, J. B. Zhou, F. Wang, D. G. Kong, J. B. Sun, Q. F. Ruan and M. Q. Liu, Virol. Sin., 2020, 35, 752–757 CrossRef CAS PubMed.
- A. Cameron, C. A. Porterfield, L. D. Byron, J. Wang, Z. Pearson, J. L. Bohrhunter, A. B. Cardillo, L. Ryan-Muntz, R. A. Sorensen and M. T. Caserta, J. Clin. Microbiol., 2021, 59, e02489–e02420 CAS.
- M. Miyara, D. Sterlin, F. Anna, S. Marot, A. Mathian, M. Atif, P. Quentric, A. Mohr, L. Claer and K. Dorgham, MedRxiv, 2020, preprint, DOI:10.1101/2020.08.14.20173393.
- M. Möckel, V. M. Corman, M. S. Stegemann, J. Hofmann, A. Stein, T. C. Jones, P. Gastmeier, J. Seybold, R. Offermann and U. J. B. Bachmann, Biomarkers, 2021, 26, 213–220 Search PubMed.
- FDA In Brief: FDA Advises Against Use of SARS-CoV-2 Antibody Test Results to Evaluate Immunity or Protection From COVID-19, Including After Vaccination, https://www.fda.gov/news-events/press-announcements/fda-brief-fda-advises-against-use-sars-cov-2-antibody-test-results-evaluate-immunity-or-protection, accessed May 25, 2022 Search PubMed.
- J. T. Roehrig, J. Hombach and A. D. Barrett, Viral Immunol., 2008, 21, 123–132 CrossRef CAS PubMed.
- S. J. Thomas, A. Nisalak, K. B. Anderson, D. H. Libraty, S. Kalayanarooj, D. W. Vaughn, R. Putnak, R. V. Gibbons, R. Jarman and T. P. Endy, Am. J. Trop. Med. Hyg., 2009, 81, 825–833 CrossRef PubMed.
- S. Ratnam, V. Gadag, R. West, J. Burris, E. Oates, F. Stead and N. Bouilianne, J. Clin. Microbiol., 1995, 33, 811–815 CrossRef CAS PubMed.
- K. R. Bewley, N. S. Coombes, L. Gagnon, L. McInroy, N. Baker, I. Shaik, J. R. St-Jean, N. St-Amant, K. R. Buttigieg, H. E. Humphries, K. J. Godwin, E. Brunt, L. Allen, S. Leung, P. J. Brown, E. J. Penn, K. Thomas, G. Kulnis, B. Hallis, M. Carroll, S. Funnell and S. Charlton, Nat. Protoc., 2021, 16, 3114–3140 CrossRef CAS PubMed.
- L. Liu, K. K. To, K. H. Chan, Y. C. Wong, R. Zhou, K. Y. Kwan, C. H. Fong, L. L. Chen, C. Y. Choi, L. Lu, O. T. Tsang, W. S. Leung, W. K. To, I. F. Hung, K. Y. Yuen and Z. Chen, Emerging Microbes Infect., 2020, 9, 1664–1670 CrossRef CAS PubMed.
- E. T. Stone, E. Geerling, T. L. Steffen, M. Hassert, A. Dickson, J. F. Spencer, K. Toth, R. J. DiPaolo, J. D. Brien and A. K. Pinto, bioRxiv, 2020, preprint, DOI:10.1101/2020.08.20.259838.
- J. Seow, C. Graham, B. Merrick, S. Acors, S. Pickering, K. J. A. Steel, O. Hemmings, A. O'Byrne, N. Kouphou, R. P. Galao, G. Betancor, H. D. Wilson, A. W. Signell, H. Winstone, C. Kerridge, I. Huettner, J. M. Jimenez-Guardeno, M. J. Lista, N. Temperton, L. B. Snell, K. Bisnauthsing, A. Moore, A. Green, L. Martinez, B. Stokes, J. Honey, A. Izquierdo-Barras, G. Arbane, A. Patel, M. K. I. Tan, L. O'Connell, G. O'Hara, E. MacMahon, S. Douthwaite, G. Nebbia, R. Batra, R. Martinez-Nunez, M. Shankar-Hari, J. D. Edgeworth, S. J. D. Neil, M. H. Malim and K. J. Doores, Nat. Microbiol., 2020, 5, 1598–1607 CrossRef CAS PubMed.
- J. B. Case, P. W. Rothlauf, R. E. Chen, Z. Liu, H. Zhao, A. S. Kim, L. M. Bloyet, Q. Zeng, S. Tahan, L. Droit, M. X. G. Ilagan, M. A. Tartell, G. Amarasinghe, J. P. Henderson, S. Miersch, M. Ustav, S. Sidhu, H. W. Virgin, D. Wang, S. Ding, D. Corti, E. S. Theel, D. H. Fremont, M. S. Diamond and S. P. J. Whelan, Cell Host Microbe, 2020, 28, 475–485 CrossRef CAS PubMed.
- J. S. Schultz, M. K. McCarthy, C. Rester, K. R. Sabourin, K. Annen, M. DomBourian, E. Eisenmesser, A. Frazer-Abel, V. Knight, T. Jaenisch, T. E. Morrison, R. Rochford and R. M. Kedl, J. Clin. Microbiol., 2021, 59, e00290–e00221 CrossRef CAS PubMed.
- X. Xie, M. C. Nielsen, A. E. Muruato, C. R. Fontes-Garfias and P. Ren, Diagn. Microbiol. Infect. Dis., 2021, 99, 115248 CrossRef CAS PubMed.
- M.-L. Herrlein, S. Hein, T. Zahn, I. Mhedhbi, J. Raupach, Y. Husria, N. I. Benz, J. Eisert, D. Bender, V. Haberger, F. D. Hastert, L. Henss, B. S. Schnierle, J. C. Stingl, M. Dreher and E. Hildt, Viruses, 2022, 14, 410 CrossRef CAS PubMed.
- E. E. F. Brown, R. Rezaei, T. R. Jamieson, J. Dave, N. T. Martin, R. Singaravelu, M. J. F. Crupi, S. Boulton, S. Tucker, J. Duong, J. Poutou, A. Pelin, H. Yasavoli-Sharahi, Z. Taha, R. Arulanandam, A. Surendran, M. Ghahremani, B. Austin, C. Matar, J.-S. Diallo, J. C. Bell, C. S. Ilkow and T. Azad, Int. J. Mol. Sci., 2021, 22, 2268 CrossRef CAS PubMed.
- G. Donofrio, V. Franceschi, F. Macchi, L. Russo, A. Rocci, V. Marchica, F. Costa, N. Giuliani, C. Ferrari and G. Missale, Vaccines, 2021, 9, 389 CrossRef CAS PubMed.
- J. Nie, Q. Li, J. Wu, C. Zhao, H. Hao, H. Liu, L. Zhang, L. Nie, H. Qin and M. Wang, Emerging Microbes Infect., 2020, 9, 680–686 CrossRef CAS PubMed.
- C. Wu, Q. Xu, H. Wang, B. Tu, J. Zeng, P. Zhao, M. Shi, H. Qiu and Y. Huang, Acta Pharm. Sin. B, 2022, 12, 1523–1533 CrossRef CAS PubMed.
- C. Gaebler, Z. Wang, J. C. Lorenzi, F. Muecksch, S. Finkin, M. Tokuyama, A. Cho, M. Jankovic, D. Schaefer-Babajew and T. Y. Oliveira, Nature, 2021, 591, 639–644 CrossRef CAS PubMed.
- S. L. Sarkar, A. R. U. Alam, P. K. Das, M. H. A. Pramanik, H. M. Al-Emran, I. K. Jahid and M. A. Hossain, Sci. Rep., 2021, 2, 1–13 Search PubMed.
- S. A. V. van Asten, S. A. Boers, J. D. F. de Groot, R. Schuurman and E. C. J. Claas, BMC Microbiol., 2021, 21, 236 CrossRef CAS PubMed.
- L. L. Luchsinger, B. P. Ransegnola, D. K. Jin, F. Muecksch, Y. Weisblum, W. Bao, P. J. George, M. Rodriguez, N. Tricoche, F. Schmidt, C. Gao, S. Jawahar, M. Pal, E. Schnall, H. Zhang, D. Strauss, K. Yazdanbakhsh, C. D. Hillyer, P. D. Bieniasz and T. Hatziioannou, J. Clin. Microbiol., 2020, 58, e02005–e02020 CrossRef CAS PubMed.
- J. M. Condor Capcha, G. Lambert, D. M. Dykxhoorn, A. G. Salerno, J. M. Hare, M. A. Whitt, S. Pahwa, D. T. Jayaweera and L. A. Shehadeh, Front. Cardiovasc. Med., 2020, 7, 618651 CrossRef PubMed.
- R. Yang, B. Huang, R. H. A, W. Li, W. Wang, Y. Deng and W. Tan, Biosaf. Heal., 2020, 2, 226–231 CrossRef PubMed.
- J. Nie, Q. Li, J. Wu, C. Zhao, H. Hao, H. Liu, L. Zhang, L. Nie, H. Qin, M. Wang, Q. Lu, X. Li, Q. Sun, J. Liu, C. Fan, W. Huang, M. Xu and Y. Wang, Emerging Microbes Infect., 2020, 9, 680–686 CrossRef CAS PubMed.
- H. Hofmann, K. Hattermann, A. Marzi, T. Gramberg, M. Geier, M. Krumbiegel, S. Kuate, K. Uberla, M. Niedrig and S. Pohlmann, J. Virol., 2004, 78, 6134–6142 CrossRef CAS PubMed.
- A. Manenti, M. Maggetti, E. Casa, D. Martinuzzi, A. Torelli, C. M. Trombetta, S. Marchi and E. Montomoli, J. Med. Virol., 2020, 92, 2096–2104 CrossRef CAS PubMed.
- S. N. Neerukonda, R. Vassell, R. Herrup, S. Liu, T. Wang, K. Takeda, Y. Yang, T. L. Lin, W. Wang and C. D. Weiss, PLoS One, 2021, 16, e0248348 CrossRef CAS PubMed.
- A. T. Sampson, J. Heeney, D. Cantoni, M. Ferrari, M. S. Sans, C. George, C. Di Genova, M. Mayora Neto, S. Einhauser, B. Asbach, R. Wagner, H. Baxendale, N. Temperton and G. Carnell, Viruses, 2021, 13, 1579 CrossRef CAS PubMed.
- A. Muik, A.-K. Wallisch, B. Sänger, K. A. Swanson, J. Mühl, W. Chen, H. Cai, D. Maurus, R. Sarkar and Ö. Türeci, Science, 2021, 371, 1152–1153 CrossRef CAS PubMed.
- R. Zang, J. B. Case, E. Yutuc, X. Ma, S. Shen, M. F. Gomez Castro, Z. Liu, Q. Zeng, H. Zhao, J. Son, P. W. Rothlauf, A. J. B. Kreutzberger, G. Hou, H. Zhang, S. Bose, X. Wang, M. D. Vahey, K. Mani, W. J. Griffiths, T. Kirchhausen, D. H. Fremont, H. Guo, A. Diwan, Y. Wang, M. S. Diamond, S. P. J. Whelan and S. Ding, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 32105–32113 CrossRef CAS PubMed.
- D. C. Malherbe, D. Kurup, C. Wirblich, A. J. Ronk, C. Mire, N. Kuzmina, N. Shaik, S. Periasamy, M. A. Hyde, J. M. Williams, P. Y. Shi, M. J. Schnell and A. Bukreyev, npj Vaccines, 2021, 6, 91 CrossRef CAS PubMed.
- A. T. Ostadgavahi, R. Booth, G. Sisson, N. McMullen, M. Warhuus, P. Robertson, M. Miller, W. C. Allen, M. El Sherif, R. Brownlie, D. Falzarano and C. D. Richardson, J. Infect. Dev. Countries, 2021, 15, 653–656 CrossRef CAS PubMed.
- J. B. Case, P. W. Rothlauf, R. E. Chen, Z. Liu, H. Zhao, A. S. Kim, L. M. Bloyet, Q. Zeng, S. Tahan, L. Droit, M. X. G. Ilagan, M. A. Tartell, G. Amarasinghe, J. P. Henderson, S. Miersch, M. Ustav, S. Sidhu, H. W. Virgin, D. Wang, S. Ding, D. Corti, E. S. Theel, D. H. Fremont, M. S. Diamond and S. P. J. Whelan, Cell Host Microbe, 2020, 28, 475–485 CrossRef CAS PubMed.
- F. Schmidt, Y. Weisblum, F. Muecksch, H. H. Hoffmann, E. Michailidis, J. C. C. Lorenzi, P. Mendoza, M. Rutkowska, E. Bednarski, C. Gaebler, M. Agudelo, A. Cho, Z. Wang, A. Gazumyan, M. Cipolla, M. Caskey, D. F. Robbiani, M. C. Nussenzweig, C. M. Rice, T. Hatziioannou and P. D. Bieniasz, J. Exp. Med., 2020, 217, e20201181 CrossRef PubMed.
- A. M. K. Tolah, S. S. Sohrab, K. M. K. Tolah, A. M. Hassan, S. A. El-Kafrawy and E. I. Azhar, Diagnostics, 2021, 11, 994 CrossRef CAS PubMed.
- M. C. Johnson, T. D. Lyddon, R. Suarez, B. Salcedo, M. LePique, M. Graham, C. Ricana, C. Robinson and D. G. Ritter, J. Virol., 2020, 94, e01062–e01020 CAS.
- X. Fu, L. Tao and X. Zhang, Mol. Ther.–Methods Clin. Dev., 2021, 20, 350–356 CrossRef CAS PubMed.
- H. T. Nguyen, S. Zhang, Q. Wang, S. Anang, J. Wang, H. Ding, J. C. Kappes and J. Sodroski, J. Virol., 2020, 95, e02304–e02320 Search PubMed.
- M. C. Johnson, T. D. Lyddon, R. Suarez, B. Salcedo, M. LePique, M. Graham, C. Ricana, C. Robinson and D. G. Ritter, J. Virol., 2020, 94 Search PubMed.
- J. Yu, Z. Li, X. He, M. S. Gebre, E. A. Bondzie, H. Wan, C. Jacob-Dolan, D. R. Martinez, J. P. Nkolola, R. S. Baric and D. H. Barouch, J. Virol., 2021, 95, e00044–21 CAS.
- A. Choi, M. Koch, K. Wu, G. Dixon, J. Oestreicher, H. Legault, G. B. E. Stewart-Jones, T. Colpitts, R. Pajon, H. Bennett, A. Carfi and D. K. Edwards, J. Virol., 2021, 95, e0131321 CrossRef PubMed.
- S. A. Almahboub, A. Algaissi, M. A. Alfaleh, M. Z. ElAssouli and A. M. Hashem, Front. Microb., 2020, 11, 2020 CrossRef PubMed.
- J. Nie, Q. Li, J. Wu, C. Zhao, H. Hao, H. Liu, L. Zhang, L. Nie, H. Qin, M. Wang, Q. Lu, X. Li, Q. Sun, J. Liu, C. Fan, W. Huang, M. Xu and Y. Wang, Emerging Microbes Infect., 2020, 9, 680–686 CrossRef CAS PubMed.
- H. Tani, M. Kimura, L. Tan, Y. Yoshida, T. Ozawa, H. Kishi, S. Fukushi, M. Saijo, K. Sano, T. Suzuki, H. Kawasuji, A. Ueno, Y. Miyajima, Y. Fukui, I. Sakamaki, Y. Yamamoto and Y. Morinaga, Virol. J., 2021, 18, 16 CrossRef CAS PubMed.
- R. Zang, J. B. Case, E. Yutuc, X. Ma, S. Shen, M. F. Gomez Castro, Z. Liu, Q. Zeng, H. Zhao, J. Son, P. W. Rothlauf, A. J. B. Kreutzberger, G. Hou, H. Zhang, S. Bose, X. Wang, M. D. Vahey, K. Mani, W. J. Griffiths, T. Kirchhausen, D. H. Fremont, H. Guo, A. Diwan, Y. Wang, M. S. Diamond, S. P. J. Whelan and S. Ding, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 32105–32113 CrossRef CAS PubMed.
- Y. Weisblum, F. Schmidt, F. Zhang, J. DaSilva, D. Poston, J. C. Lorenzi, F. Muecksch, M. Rutkowska, H.-H. Hoffmann and E. Michailidis, eLife, 2020, 9, e61312 CrossRef CAS PubMed.
- M. Diamond, R. Chen, X. Xie, J. Case, X. Zhang, L. VanBlargan, Y. Liu, J. Liu, J. Errico, E. Winkler, N. Suryadevara, S. Tahan, J. Turner, W. Kim, A. Schmitz, M. Thapa, D. Wang, A. Boon, D. Pinto, R. Presti, J. O'Halloran, A. Kim, P. Deepak, D. Fremont, D. Corti, H. Virgin, J. Crowe, L. Droit, A. Ellebedy, P. Y. Shi and P. Gilchuk, Res. Sq., 2021, preprint, DOI:10.21203/rs.3.rs-228079/v1.
- F. Schmidt, Y. Weisblum, F. Muecksch, H. H. Hoffmann, E. Michailidis, J. C. C. Lorenzi, P. Mendoza, M. Rutkowska, E. Bednarski, C. Gaebler, M. Agudelo, A. Cho, Z. Wang, A. Gazumyan, M. Cipolla, M. Caskey, D. F. Robbiani, M. C. Nussenzweig, C. M. Rice, T. Hatziioannou and P. D. Bieniasz, J. Exp. Med., 2020, 217 Search PubMed.
- J. Yu, Z. Li, X. He, M. S. Gebre, E. A. Bondzie, H. Wan, C. Jacob-Dolan, D. R. Martinez, J. P. Nkolola, R. S. Baric and D. H. Barouch, J. Virol., 2021, 95, e00044–21 CAS.
- N. Mishra, S. Kumar, S. Singh, T. Bansal, N. Jain, S. Saluja, R. Kumar, S. Bhattacharyya, J. K. Palanichamy, R. A. Mir, S. Sinha and K. Luthra, PLoS Pathog., 2021, 17, e1009958 CrossRef CAS PubMed.
- Y. Ma, G. Mao, G. Wu, M. Chen, F. Qin, L. Zheng and X.-E. Zhang, ACS Appl. Mater., 2021, 13, 24477–24486 CrossRef CAS PubMed.
- Z. Gong, J. W. Zhu, C. P. Li, S. Jiang, L. N. Ma, B. X. Tang, D. Zou, M. L. Chen, Y. B. Sun, S. H. Song, Z. Zhang, J. F. Xiao, Y. B. Xue, Y. M. Bao, Z. L. Du and W. M. Zhao, Zool. Res., 2020, 41, 705–708 Search PubMed.
- M. A. Tortorici, N. Czudnochowski, T. N. Starr, R. Marzi, A. C. Walls, F. Zatta, J. E. Bowen, S. Jaconi, J. Di Iulio, Z. Wang, A. De Marco, S. K. Zepeda, D. Pinto, Z. Liu, M. Beltramello, I. Bartha, M. P. Housley, F. A. Lempp, L. E. Rosen, E. Dellota Jr, H. Kaiser, M. Montiel-Ruiz, J. Zhou, A. Addetia, B. Guarino, K. Culap, N. Sprugasci, C. Saliba, E. Vetti, I. Giacchetto-Sasselli, C. S. Fregni, R. Abdelnabi, S. C. Foo, C. Havenar-Daughton, M. A. Schmid, F. Benigni, E. Cameroni, J. Neyts, A. Telenti, H. W. Virgin, S. P. J. Whelan, G. Snell, J. D. Bloom, D. Corti, D. Veesler and M. S. Pizzuto, Nature, 2021, 597, 103–108 CrossRef CAS PubMed.
- Y. Zheng, E. T. Larragoite, E. Williams, J. Lama, I. Cisneros, J. C. Delgado, P. Slev, J. Rychert, E. A. Innis, M. Coiras, M. T. Rondina, A. M. Spivak and V. Planelles, Virol. J., 2021, 18, 1 CrossRef CAS PubMed.
- C. von Rhein, T. Scholz, L. Henss, R. Kronstein-Wiedemann, T. Schwarz, R. N. Rodionov, V. M. Corman, T. Tonn and B. S. Schnierle, J. Virol. Methods, 2021, 288, 114031 CrossRef CAS PubMed.
- B. Bosnjak, S. C. Stein, S. Willenzon, A. K. Cordes, W. Puppe, G. Bernhardt, I. Ravens, C. Ritter, C. R. Schultze-Florey, N. Godecke, J. Martens, H. Kleine-Weber, M. Hoffmann, A. Cossmann, M. Yilmaz, I. Pink, M. M. Hoeper, G. M. N. Behrens, S. Pohlmann, R. Blasczyk, T. F. Schulz and R. Forster, Cell. Mol. Immunol., 2021, 18, 936–944 CrossRef CAS PubMed.
- C. W. Tan, W. N. Chia, X. Qin, P. Liu, M. I. Chen, C. Tiu, Z. Hu, V. C. Chen, B. E. Young, W. R. Sia, Y. J. Tan, R. Foo, Y. Yi, D. C. Lye, D. E. Anderson and L. F. Wang, Nat. Biotechnol., 2020, 38, 1073–1078 CrossRef CAS PubMed.
- S. C. Taylor, B. Hurst, C. L. Charlton, A. Bailey, J. N. Kanji, M. K. McCarthy, T. E. Morrison, L. Huey, K. Annen and M. G. DomBourian, J. Clin. Microbiol., 2021, 59, e02438–e02420 CAS.
- J. R. Byrnes, X. X. Zhou, I. Lui, S. K. Elledge, J. E. Glasgow, S. A. Lim, R. P. Loudermilk, C. Y. Chiu, T. T. Wang, M. R. Wilson, K. K. Leung and J. A. Wells, mSphere, 2020, 5, e00802–e00820 CrossRef CAS PubMed.
- Y. Cao, B. Su, X. Guo, W. Sun, Y. Deng, L. Bao, Q. Zhu, X. Zhang, Y. Zheng, C. Geng, X. Chai, R. He, X. Li, Q. Lv, H. Zhu, W. Deng, Y. Xu, Y. Wang, L. Qiao, Y. Tan, L. Song, G. Wang, X. Du, N. Gao, J. Liu, J. Xiao, X. D. Su, Z. Du, Y. Feng, C. Qin, C. Qin, R. Jin and X. S. Xie, Cell, 2020, 182, 73–84 CrossRef CAS PubMed.
- K. T. Abe, Z. Li, R. Samson, P. Samavarchi-Tehrani, E. J. Valcourt, H. Wood, P. Budylowski, A. P. Dupuis 2nd, R. C. Girardin, B. Rathod, J. H. Wang, M. Barrios-Rodiles, K. Colwill, A. J. McGeer, S. Mubareka, J. L. Gommerman, Y. Durocher, M. Ostrowski, K. A. McDonough, M. A. Drebot, S. J. Drews, J. M. Rini and A. C. Gingras, JCI Insight, 2020, 5, e142362 CrossRef PubMed.
- A. Krüttgen, M. Lauen, H. Klingel, M. Imöhl and M. Kleines, J. Virol. Methods, 2022, 299, 114297 CrossRef PubMed.
- J. Michiels, L. Heyndrickx, A. Nkuba-Ndaye, A. Ceulemans, K. K. Bartholomeeusen, J. Madinga, P. Mbala-Kingebeni, V. Vanlerberghe, S. Ahuka-Mundeke and L. Wang, J. Virol. Methods, 2021, 294, 114228 Search PubMed.
- C. W. Tan, W. N. Chia, X. Qin, P. Liu, M. I. Chen, C. Tiu, Z. Hu, V. C. Chen, B. E. Young, W. R. Sia, Y. J. Tan, R. Foo, Y. Yi, D. C. Lye, D. E. Anderson and L. F. Wang, Nat. Biotechnol., 2020, 38, 1073–1078 CrossRef CAS PubMed.
- B. Bošnjak, S. C. Stein, S. Willenzon, A. K. Cordes, W. Puppe, G. Bernhardt, I. Ravens, C. Ritter, C. R. Schultze-Florey and N. J. C. Gödecke, Cell. Mol. Immunol., 2021, 18, 936–944 CrossRef PubMed.
- C. W. Embregts, B. Verstrepen, J. A. Langermans, K. P. Boszormenyi, R. S. Sikkema, R. D. de Vries, D. Hoffmann, K. Wernike, L. A. Smit and S. Zhao, One Health, 2021, 100313 CrossRef CAS PubMed.
- C. Wang, Z. Wu, B. Liu, P. Zhang, J. Lu, J. Li, P. Zou, T. Li, Y. Fu, R. Chen, L. Zhang, Q. Fu and C. Li, Biosens. Bioelectron., 2021, 192, 113550 CrossRef CAS PubMed.
- Y. R. Luo, C. Yun, I. Chakraborty, A. H. B. Wu and K. L. Lynch, J. Clin. Microbiol., 2021, 59, e0019321 CrossRef PubMed.
- B. Bošnjak, S. C. Stein, S. Willenzon, A. K. Cordes, W. Puppe, G. Bernhardt, I. Ravens, C. Ritter, C. R. Schultze-Florey and N. Gödecke, medRxiv, 2020, preprint, DOI:10.1101/2020.07.12.20151407.
- E. P. Gniffke, W. E. Harrington, N. Dambrauskas, Y. Jiang, O. Trakhimets, V. Vigdorovich, L. Frenkel, D. N. Sather and S. E. P. Smith, J. Infect. Dis., 2020, 222, 1965–1973 CrossRef CAS PubMed.
- R. Perera, R. Ko, O. T. Y. Tsang, D. S. C. Hui, M. Y. M. Kwan, C. J. Brackman, E. M. W. To, H. L. Yen, K. Leung, S. M. S. Cheng, K. H. Chan, K. C. K. Chan, K. C. Li, L. Saif, V. R. Barrs, J. T. Wu, T. H. C. Sit, L. L. M. Poon and M. Peiris, J. Clin. Microbiol., 2021, 59, e02504–e02520 CrossRef CAS PubMed.
- D. F. Robbiani, C. Gaebler, F. Muecksch, J. C. C. Lorenzi, Z. Wang, A. Cho, M. Agudelo, C. O. Barnes, A. Gazumyan, S. Finkin, T. Hagglof, T. Y. Oliveira, C. Viant, A. Hurley, H. H. Hoffmann, K. G. Millard, R. G. Kost, M. Cipolla, K. Gordon, F. Bianchini, S. T. Chen, V. Ramos, R. Patel, J. Dizon, I. Shimeliovich, P. Mendoza, H. Hartweger, L. Nogueira, M. Pack, J. Horowitz, F. Schmidt, Y. Weisblum, E. Michailidis, A. W. Ashbrook, E. Waltari, J. E. Pak, K. E. Huey-Tubman, N. Koranda, P. R. Hoffman, A. P. West Jr, C. M. Rice, T. Hatziioannou, P. J. Bjorkman, P. D. Bieniasz, M. Caskey and M. C. Nussenzweig, Nature, 2020, 584, 437–442 CrossRef CAS PubMed.
- L. Premkumar, B. Segovia-Chumbez, R. Jadi, D. R. Martinez, R. Raut, A. J. Markmann, C. Cornaby, L. Bartelt, S. Weiss and Y. Park, Sci. Immunol., 2020, 5, eabc8413 CrossRef CAS PubMed.
- T. S. Fulford, H. Van, N. A. Gherardin, S. Zheng, M. Ciula, H. E. Drummer, S. Redmond, H.-X. Tan, I. Boo and R. Center, EBioMedicine, 2021, 74, 103729 CrossRef CAS PubMed.
- D. F. Lake, A. J. Roeder, E. Kaleta, P. Jasbi, K. Pfeffer, C. Koelbela, S. Periasamy, N. Kuzmina, A. Bukreyev and T. E. Grys, J. Clin. Virol., 2021, 145, 105024 CrossRef CAS PubMed.
- H. T. Nguyen, S. Zhang, Q. Wang, S. Anang, J. Wang, H. Ding, J. C. Kappes and J. Sodroski, J. Virol., 2020, 95, e02304–e02320 Search PubMed.
- X. Chi, R. Yan, J. Zhang, G. Zhang, Y. Zhang, M. Hao, Z. Zhang, P. Fan, Y. Dong and Y. Yang, Science, 2020, 369, 650–655 CrossRef CAS PubMed.
- L. Liu, P. Wang, M. S. Nair, J. Yu, M. Rapp, Q. Wang, Y. Luo, J. F. Chan, V. Sahi, A. Figueroa, X. V. Guo, G. Cerutti, J. Bimela, J. Gorman, T. Zhou, Z. Chen, K. Y. Yuen, P. D. Kwong, J. G. Sodroski, M. T. Yin, Z. Sheng, Y. Huang, L. Shapiro and D. D. Ho, Nature, 2020, 584, 450–456 CrossRef CAS PubMed.
- M. McCallum, A. De Marco, F. A. Lempp, M. A. Tortorici, D. Pinto, A. C. Walls, M. Beltramello, A. Chen, Z. Liu and F. Zatta, Cell, 2021, 184, 2332–2347 CrossRef CAS PubMed.
- M. McCallum, A. De Marco, F. A. Lempp, M. A. Tortorici, D. Pinto, A. C. Walls, M. Beltramello, A. Chen, Z. Liu and F. Zatta, Cell, 2021, 184, 2332–2347 CrossRef CAS PubMed.
- N. Zhang, S. Chen, J. V. Wu, X. Yang and J. J. Wang, MedRxiv, 2020, preprint, DOI:10.1101/2020.11.05.20222596.
- R. E. Chen, X. Zhang, J. B. Case, E. S. Winkler, Y. Liu, L. A. VanBlargan, J. Liu, J. M. Errico, X. Xie, N. Suryadevara, P. Gilchuk, S. J. Zost, S. Tahan, L. Droit, J. S. Turner, W. Kim, A. J. Schmitz, M. Thapa, D. Wang, A. C. M. Boon, R. M. Presti, J. A. O'Halloran, A. H. J. Kim, P. Deepak, D. Pinto, D. H. Fremont, J. E. Crowe Jr, D. Corti, H. W. Virgin, A. H. Ellebedy, P. Y. Shi and M. S. Diamond, Nat. Med., 2021, 27, 717–726 CrossRef CAS PubMed.
- A. D. Barrett and L. R. Stanberry, Vaccines for biodefense and emerging and neglected diseases, Academic Press, 2009 Search PubMed.
- S. A. Rubin, S. A. Plotkin and O. Plotkin, Vaccines, 2012, 6, 419–446 Search PubMed.
- Q. Yang, A. Kelkar, A. Sriram, R. Hombu, T. A. Hughes and S. Neelamegham, Sci. Adv., 2022, 8, eabq8678 CrossRef CAS PubMed.
- P. Ma, J. Liu, S. Pang, W. Zhou, H. Yu, M. Wang, T. Dong, Y. Wang, Q. Wang and A. Liu, Anal. Chim. Acta, 2023, 1264, 341300 CrossRef CAS PubMed.
- W. H. Zeng, G. F. Liu, H. Ma, D. Zhao, Y. R. Yang, M. Y. Liu, A. Mohammed, C. C. Zhao, Y. Yang, J. J. Xie, C. C. Ding, X. L. Ma, J. P. Weng, Y. Gao, H. L. He and T. C. Jin, Biochem. Biophys. Res. Commun., 2020, 527, 618–623 CrossRef CAS PubMed.
- M. Surjit and S. K. Lal, Infect., Genet. Evol., 2008, 8, 397–405 CrossRef CAS PubMed.
- X. F. Cai, J. Chen, J. L. Hu, Q. X. Long, H. J. Deng, P. Liu, K. Fan, P. Liao, B. Z. Liu, G. C. Wu, Y. K. Chen, Z. J. Li, K. Wang, X. L. Zhang, W. G. Tian, J. L. Xiang, H. X. Du, J. Wang, Y. Hu, N. Tang, Y. Lin, J. H. Ren, L. Y. Huang, J. Wei, C. Y. Gan, Y. M. Chen, Q. Z. Gao, A. M. Chen, C. L. He, D. X. Wang, P. Hu, F. C. Zhou, A. L. Huang and D. Q. Wang, J. Infect. Dis., 2020, 222, 189–193 CrossRef CAS PubMed.
- D. Das, S. Kammila and M. R. Suresh, Clin. Vaccine Immunol., 2010, 17, 2033–2036 CrossRef CAS PubMed.
- P. C. Y. Woo, S. K. P. Lau, Y. X. Chen, E. Y. M. Wong, K. H. Chan, H. L. Chen, L. B. Zhang, N. S. Xia and K. Y. Yuen, Emerging Microbes Infect., 2018, 7, 18 Search PubMed.
- P. Srithong, S. Chaiyo, E. Pasomsub, S. Rengpipat, O. Chailapakul and N. Praphairaksit, Microchim. Acta, 2022, 189, 386 CrossRef CAS PubMed.
- Y. Liu, L. Zhan, J. W. Shen, B. Baro, A. Alemany, J. Sackrison, O. Mitjà and J. C. Bischof, ACS Appl. Nano Mater., 2021, 4, 13826–13837 CrossRef CAS PubMed.
- T. Peng, X. Jiao, Z. Liang, H. Zhao, Y. Zhao, J. Xie, Y. Jiang, X. Yu, X. Fang and X. Dai, Biosensors, 2021, 12, 13 CrossRef PubMed.
- G.-Q. Zhang, Z. Gao, J. Zhang, H. Ou, H. Gao, R. T. Kwok, D. Ding and B. Z. Tang, Cell Rep. Phys. Sci., 2022, 3, 100740 CrossRef CAS PubMed.
- Y. Yamaoka, K. Miyakawa, S. S. Jeremiah, R. Funabashi, K. Okudela, S. Kikuchi, J. Katada, A. Wada, T. Takei, M. Nishi, K. Shimizu, H. Ozawa, S. Usuku, C. Kawakami, N. Tanaka, T. Morita, H. Hayashi, H. Mitsui, K. Suzuki, D. Aizawa, Y. Yoshimura, T. Miyazaki, E. Yamazaki, T. Suzuki, H. Kimura, H. Shimizu, N. Okabe, H. Hasegawa and A. Ryo, Cell Rep. Med., 2021, 2, 100311 CrossRef CAS PubMed.
- H.-Y. Kim, J.-H. Lee, M. J. Kim, S. C. Park, M. Choi, W. Lee, K. B. Ku, B. T. Kim, E. Changkyun Park, H. G. Kim and S. I. Kim, Biosens. Bioelectron., 2021, 175, 112868 CrossRef CAS PubMed.
- D. Hong, E.-J. Jo, C. Jung and M.-G. Kim, ACS Appl. Mater., 2022, 14, 45189–45200 CrossRef CAS PubMed.
- Z. Xie, S. Feng, F. Pei, M. Xia, Q. Hao, B. Liu, Z. Tong, J. Wang, W. Lei and X. Mu, Anal. Chim. Acta, 2022, 1233, 340486 CrossRef CAS PubMed.
- H.-K. Oh, K. Kim, J. Park, H. Im, S. Maher and M.-G. Kim, Biosens. Bioelectron., 2022, 205, 114094 CrossRef CAS PubMed.
- C. Wang, X. Cheng, L. Liu, X. Zhang, X. Yang, S. Zheng, Z. Rong and S. Wang, ACS Appl. Mater., 2021, 13, 40342–40353 CrossRef CAS PubMed.
- A. Guo, F. Pei, S. Feng, W. Hu, P. Zhang, M. Xia, X. Mu, Z. Tong, F. Wang and B. Liu, Sens. Actuators, B, 2023, 374, 132800 CrossRef CAS PubMed.
- G. Mao, S. Ye, W. Yin, Y. Yang, X. Ji, J. He, Y. Liu, J. Dai, Z. He and Y. Ma, Nano Lett., 2022, 16, 2859–2865 Search PubMed.
- A. L. Chen and S. M. Yang, Biosens. Bioelectron., 2015, 71, 230–242 CrossRef CAS PubMed.
- L. Zhang, X. Fang, X. Liu, H. Ou, H. Zhang, J. Wang, Q. Li, H. Cheng, W. Zhang and Z. Luo, Chem. Commun., 2020, 56, 10235–10238 RSC.
- W. Shao, M. R. Shurin, S. E. Wheeler, X. He and A. Star, ACS Appl. Mater., 2021, 13, 10321–10327 CrossRef CAS PubMed.
- P.-H. Chen, C.-C. Huang, C.-C. Wu, P.-H. Chen, A. Tripathi and Y.-L. Wang, Sens. Actuators, B, 2022, 357, 131415 CrossRef CAS PubMed.
- J. Xu, W. Suo, Y. Goulev, L. Sun, L. Kerr, J. Paulsson, Y. Zhang and T. Lao, Small, 2021, 17, 2104009 CrossRef CAS PubMed.
- H. Qi, Z. Hu, Z. Yang, J. Zhang, J. J. Wu, C. Cheng, C. Wang and L. Zheng, Anal. Chem., 2022, 94, 2812–2819 CrossRef CAS PubMed.
- S. Eissa and M. Zourob, Anal. Chem., 2021, 93, 1826–1833 CrossRef CAS PubMed.
- F. Haghayegh, R. Salahandish, M. Hassani and A. Sanati-Nezhad, ACS Appl. Mater., 2022, 14, 10844–10855 CrossRef CAS PubMed.
- I. Novodchuk, M. Kayaharman, I. Prassas, A. Soosaipillai, R. Karimi, I. A. Goldthorpe, E. Abdel-Rahman, J. Sanderson, E. P. Diamandis, M. Bajcsy and M. Yavuz, Biosens. Bioelectron., 2022, 210, 114331 CrossRef CAS PubMed.
- J. K. Millet and G. R. Whittaker, Virology, 2018, 517, 3–8 CrossRef CAS PubMed.
- W. W.-W. Hsiao, N. Sharma, T.-N. Le, Y.-Y. Cheng, C.-C. Lee, D.-T. Vo, Y. Y. Hui, H.-C. Chang and W.-H. Chiang, Anal. Chim. Acta, 2022, 1230, 340389 CrossRef PubMed.
- J. L. Cantera, D. M. Cate, A. Golden, R. B. Peck, L. L. Lillis, G. J. Domingo, E. Murphy, B. C. Barnhart, C. A. Anderson, L. F. Alonzo, V. Glukhova, G. Hermansky, B. Barrios-Lopez, E. Spencer, S. Kuhn, Z. Islam, B. D. Grant, L. Kraft, K. Herve, V. de Puyraimond, Y. Hwang, P. K. Dewan, B. H. Weigl, K. P. Nichols and D. S. Boyle, ACS Omega, 2021, 6, 20139–20148 CrossRef CAS PubMed.
- D. Liu, C. Ju, C. Han, R. Shi, X. Chen, D. Duan, J. Yan and X. Yan, Biosens. Bioelectron., 2021, 173, 112817 CrossRef CAS PubMed.
- A. N. Baker, S.-J. Richards, C. S. Guy, T. R. Congdon, M. Hasan, A. J. Zwetsloot, A. Gallo, J. R. Lewandowski, P. J. Stansfeld, A. Straube, M. Walker, S. Chessa, G. Pergolizzi, S. Dedola, R. A. Field and M. I. Gibson, ACS Cent. Sci., 2020, 6, 2046–2052 CrossRef CAS PubMed.
- S. H. Kim, F. L. Kearns, M. A. Rosenfeld, L. Casalino, M. J. Papanikolas, C. Simmerling, R. E. Amaro and R. Freeman, ACS Cent. Sci., 2022, 8, 22–42 CrossRef CAS PubMed.
- A. Ahmadivand, B. Gerislioglu, Z. Ramezani, A. Kaushik, P. Manickam and S. A. Ghoreishi, Biosens. Bioelectron., 2021, 177, 112971 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- M. Chen, D. Cui, Z. Zhao, D. Kang, Z. Li, S. Albawardi, S. Alsageer, F. Alamri, A. Alhazmi, M. R. Amer and C. Zhou, Nano Lett., 2022, 15, 5510–5516 CAS.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef CAS PubMed.
- J. H. Granger, N. E. Schlotter, A. C. Crawford and M. D. Porter, Chem. Soc. Rev., 2016, 45, 3865–3882 RSC.
- N. Karimian, P. Hashemi, A. Khanmohammadi, A. Afkhami and H. Bagheri, Anal. Bioanal. Chem. Res., 2020, 7, 281–301 CAS.
- M. Mohammadi, D. Antoine, M. Vitt, J. M. Dickie, S. Sultana Jyoti, J. G. Wall, P. A. Johnson and K. E. Wawrousek, Anal. Chim. Acta, 2022, 1229, 340290 CrossRef CAS PubMed.
- M. J. Bistaffa, S. A. Camacho, W. M. Pazin, C. J. L. Constantino, O. N. Oliveira and P. H. B. Aoki, Talanta, 2022, 244, 123381 CrossRef CAS PubMed.
- Y.-J. Yeh, T.-N. Le, W. W.-W. Hsiao, K.-L. Tung, K. Ostrikov and W.-H. Chiang, Anal. Chim. Acta, 2022, 1239, 340651 CrossRef PubMed.
- M. Zhang, X. Li, J. Pan, Y. Zhang, L. Zhang, C. Wang, X. Yan, X. Liu and G. Lu, Biosens. Bioelectron., 2021, 190, 113421 CrossRef CAS PubMed.
- J. Huang, J. Wen, M. Zhou, S. Ni, W. Le, G. Chen, L. Wei, Y. Zeng, D. Qi, M. Pan, J. Xu, Y. Wu, Z. Li, Y. Feng, Z. Zhao, Z. He, B. Li, S. Zhao, B. Zhang, P. Xue, S. He, K. Fang, Y. Zhao and K. Du, Anal. Chem., 2021, 93, 9174–9182 CrossRef CAS PubMed.
- Y. Yang, B. Xu, J. Murray, J. Haverstick, X. Chen, R. A. Tripp and Y. Zhao, Biosens. Bioelectron., 2022, 217, 114721 CrossRef CAS PubMed.
- J.-E. Shim, Y. J. Kim, J.-H. Choe, T. G. Lee and E.-A. You, ACS Appl. Mater., 2022, 14, 38459–38470 CrossRef CAS PubMed.
- J. Liu, S. Pang, M. Wang, H. Yu, P. Ma, T. Dong, Z. Zheng, Y. Jiao, Y. Zhang and A. Liu, Sens. Actuators, B, 2023, 387, 133746 CrossRef CAS PubMed.
- D. Li and J. Li, J. Clin. Microbiol., 2021, 59, e02160–e02120 CAS.
- J.-H. Lee, M. Choi, Y. Jung, S. K. Lee, C.-S. Lee, J. Kim, J. Kim, N. H. Kim, B.-T. Kim and H. G. Kim, Biosens. Bioelectron., 2021, 171, 112715 CrossRef CAS PubMed.
- J.-H. Lee, Y. Lee, S. K. Lee, J. Kim, C.-S. Lee, N. H. Kim and H. G. Kim, Biosens. Bioelectron., 2022, 203, 114034 CrossRef CAS PubMed.
- H. Han, C. Wang, X. Yang, S. Zheng, X. Cheng, Z. Liu, B. Zhao and R. Xiao, Sens. Actuators, B, 2022, 351, 130897 CrossRef CAS PubMed.
- F. Curti, S. Fortunati, W. Knoll, M. Giannetto, R. Corradini, A. Bertucci and M. Careri, ACS Appl. Mater., 2022, 14, 19204–19211 CrossRef CAS PubMed.
- J. Li, Z. Zhang, J. Gu, H. D. Stacey, J. C. Ang, A. Capretta, C. D. M. Filipe, K. L. Mossman, C. Balion, B. J. Salena, D. Yamamura, L. Soleymani, M. S. Miller, J. D. Brennan and Y. Li, Nucleic Acids Res., 2021, 49, 7267–7279 CrossRef CAS PubMed , 34232998.
- S. Witt, A. Rogien, D. Werner, J. Siegenthaler, R. Lesiyon, N. Kurien, R. Rechenberg, N. Baule, A. Hardy and M. Becker, Diamond Relat. Mater., 2021, 118, 108542 CrossRef CAS PubMed.
- A. Erdem, H. Senturk, E. Yildiz and M. Maral, Talanta, 2022, 244, 123422 CrossRef CAS PubMed.
- Z. Zhang, R. Pandey, J. Li, J. Gu, D. White, H. D. Stacey, J. C. Ang, C. J. Steinberg, A. Capretta and C. D. Filipe, Angew. Chem., 2021, 60, 24266–24274 CrossRef CAS PubMed.
- Q. Wu, W. Wu, F. Chen and P. Ren, Analyst, 2022, 147, 2809–2818 RSC.
- M. A. Zamzami, G. Rabbani, A. Ahmad, A. A. Basalah, W. H. Al-Sabban, S. Nate Ahn and H. Choudhry, Bioelectrochemistry, 2022, 143, 107982 CrossRef CAS PubMed.
- S. Kim, H. Ryu, S. Tai, M. Pedowitz, J. R. Rzasa, D. J. Pennachio, J. R. Hajzus, D. K. Milton, R. Myers-Ward and K. M. Daniels, Biosens. Bioelectron., 2022, 197, 113803 CrossRef CAS PubMed.
- P. K. Sharma, E.-S. Kim, S. Mishra, E. Ganbold, R.-S. Seong, A. K. Kaushik and N.-Y. Kim, ACS Sens., 2021, 6, 3468–3476 CrossRef CAS PubMed.
- C. Dai, M. Guo, Y. Wu, B.-P. Cao, X. Wang, Y. Wu, H. Kang, D. Kong, Z. Zhu, T. Ying, Y. Liu and D. Wei, J. Am. Chem. Soc., 2021, 143, 19794–19801 CrossRef CAS PubMed.
- J. Liu, P. Ma, H. Yu, M. Wang, P. Yin, S. Pang, Y. Jiao, T. Dong and A. Liu, Anal. Chem., 2022, 94, 11591–11599 CrossRef CAS PubMed.
- Z. Zhang, R. Pandey, J. Li, J. Gu, D. White, H. D. Stacey, J. C. Ang, C. J. Steinberg, A. Capretta and C. D. Filipe, Angew. Chem., 2021, 133, 24468–24476 CrossRef.
- W. Tai, X. Zhang, Y. He, S. Jiang and L. Du, Antiviral Res., 2020, 179, 104820 CrossRef CAS PubMed.
- M. B. Abdullah, C. Dab, M. Almalki, A. Alnaim, A. Abuzir and C. Awada, Appl. Sci., 2022, 12, 5039 CrossRef CAS.
- A. K. Sarychev, A. Sukhanova, A. V. Ivanov, I. V. Bykov, N. V. Bakholdin, D. V. Vasina, V. A. Gushchin, A. P. Tkachuk, G. Nifontova, P. S. Samokhvalov, A. Karaulov and I. Nabiev, Biosensors, 2022, 12, 300 CrossRef CAS PubMed.
- C. Awada, M. M. B. Abdullah, H. Traboulsi, C. Dab and A. Alshoaibi, Sensors, 2021, 21, 4617 CrossRef CAS PubMed.
- G. Huang, H. Zhao, P. Li, J. Liu, S. Chen, M. Ge, M. Qin, G. Zhou, Y. Wang, S. Li, Y. Cheng, Q. Huang, J. Wang, H. Wang and L. Yang, Anal. Chem., 2021, 93, 16086–16095 CrossRef CAS PubMed.
- E. B. Aydın, M. Aydın and M. K. Sezginturk, ACS Biomater. Sci. Eng., 2021, 7, 3874–3885 CrossRef PubMed.
- D. Shahdeo, A. Roberts, G. J. Archana, N. S. Shrikrishna, S. Mahari, K. Nagamani and S. Gandhi, Biosens. Bioelectron., 2022, 212, 114406 CrossRef CAS PubMed.
- J. C. Abrego-Martinez, M. Jafari, S. Chergui, C. Pavel, D. Che and M. Siaj, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- Y. Yang, J. Murray, J. Haverstick, R. A. Tripp and Y. Zhao, Sens. Actuators, B, 2022, 359, 131604 CrossRef CAS PubMed.
- G.-Q. Zhang, Z. Gao, J. Zhang, H. Ou, H. Gao, R. T. K. Kwok, D. Ding and B. Z. Tang, Cell Rep. Phys. Sci., 2022, 3, 100740 CrossRef CAS PubMed.
- L. F. Yang, N. Kacherovsky, J. Liang, S. J. Salipante and S. H. Pun, Anal. Chem., 2022, 94, 12683–12690 CrossRef CAS PubMed.
- M. Ravalin, H. Roh, R. Suryawanshi, G. R. Kumar, J. E. Pak, M. Ott and A. Y. Ting, J. Am. Chem. Soc., 2022, 144, 13663–13672 CrossRef CAS PubMed.
- G. Murtaza, A. S. Rizvi, M. Xue, L. Qiu and Z. Meng, Anal. Chem., 2022, 94, 7391–7399 CrossRef CAS PubMed.
- C. Krempl, B. Schultze, H. Laude and G. Herrler, J. Virol., 1997, 71, 3285–3287 CrossRef CAS PubMed.
- F. Künkel and G. Herrler, Virology, 1993, 195, 195–202 CrossRef PubMed.
- G. Lu, Q. Wang and G. F. Gao, Trends Microbiol., 2015, 23, 468–478 CrossRef CAS PubMed.
- S. Di Gaetano, D. Capasso, P. Delre, L. Pirone, M. Saviano, E. Pedone and G. F. Mangiatordi, Int. J. Mol. Sci., 2021, 22, 6462 CrossRef CAS PubMed.
- N. Kacherovsky, L. F. Yang, H. V. Dang, E. L. Cheng, I. I. Cardle, A. C. Walls, M. McCallum, D. L. Sellers, F. DiMaio and S. J. Salipante, Angew. Chem., 2021, 133, 21381–21385 CrossRef.
- L. F. Yang, N. Kacherovsky, N. Panpradist, R. Wan, J. Liang, B. Zhang, S. J. Salipante, B. R. Lutz and S. H. Pun, Anal. Chem., 2022, 94, 7278–7285 CrossRef CAS PubMed.
- Statement on the tenth meeting of the International Health Regulations (2005) Emergency Committee regarding the coronavirus disease (COVID-19) pandemic, https://www.who.int/news/item/19-01-2022-statement-on-the-tenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic, accessed May 5, 2022 Search PubMed.
- W. Liu, Q. Zhang, J. Chen, R. Xiang, H. Song, S. Shu, L. Chen, L. Liang, J. Zhou and L. You, N. Engl. J. Med., 2020, 382, 1370–1371 CrossRef PubMed.
- D. K. Agarwal, V. Nandwana, S. E. Henrich, V. P. V. N. Josyula, C. S. Thaxton, C. Qi, L. M. Simons, J. F. Hultquist, E. A. Ozer, G. S. Shekhawat and V. P. Dravid, Biosens. Bioelectron., 2021, 195, 113647 CrossRef PubMed.
- B. D. Grant, C. E. Anderson, J. R. Williford, L. F. Alonzo, V. A. Glukhova, D. S. Boyle, B. H. Weigl and K. P. Nichols, Anal. Chem., 2020, 92, 11305–11309 CrossRef CAS PubMed.
- G. P. Anderson, J. L. Liu, T. J. Esparza, B. T. Voelker, E. R. Hofmann and E. R. Goldman, Anal. Chem., 2021, 93, 7283–7291 CrossRef CAS PubMed.
- S. Eissa, H. A. Alhadrami, M. Al-Mozaini, A. M. Hassan and M. Zourob, Mikrochim. Acta, 2021, 188, 199 CrossRef CAS PubMed.
- A. Stambaugh, J. W. Parks, M. A. Stott, G. G. Meena, A. R. Hawkins and H. Schmidt, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2103480118 CrossRef CAS PubMed.
- A. Raziq, A. Kidakova, R. Boroznjak, J. Reut, A. Öpik and V. Syritski, Biosens. Bioelectron., 2021, 178, 113029 CrossRef CAS PubMed.
- S. Wang, J. Shu, A. Lyu, X. Huang, W. Zeng, T. Jin and H. Cui, Anal. Chem., 2021, 93, 14238–14246 CrossRef CAS PubMed.
- C. Liang, B. Liu, J. Li, J. Lu, E. Zhang, Q. Deng, L. Zhang, R. Chen, Y. Fu, C. Li and T. Li, Sens. Actuators, B, 2021, 349, 130718 CrossRef CAS PubMed.
- B. Liu, Z. Wu, C. Liang, J. Lu, J. Li, L. Zhang, T. Li, W. Zhao, Y. Fu, S. Hou, X. Tang and C. Li, Front. Microb., 2021, 12, 692831 CrossRef PubMed.
- K. Behrouzi and L. Lin, Biosens. Bioelectron., 2022, 195, 113669 CrossRef CAS PubMed.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- C. Wang, C. Wang, J. Qiu, J. Gao, H. Liu, Y. Zhang and L. Han, Mikrochim. Acta, 2021, 188, 262 CrossRef CAS PubMed.
- E. Karakuş, E. Erdemir, N. Demirbilek and L. Liv, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef PubMed.
- Z. Rahmati, M. Roushani, H. Hosseini and H. Choobin, Microchim. Acta, 2021, 188, 105 CrossRef CAS PubMed.
- A. Gupta, A. Anand, N. Jain, S. Goswami, A. Anantharaj, S. Patil, R. Singh, A. Kumar, T. Shrivastava, S. Bhatnagar, G. R. Medigeshi and T. K. Sharma, Mol. Ther.–Nucleic Acids, 2021, 26, 321–332 CrossRef CAS PubMed.
- L. F. de Lima, A. L. Ferreira, M. D. T. Torres, W. R. de Araujo and C. de la Fuente-Nunez, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2106724118 CrossRef CAS PubMed.
- P. Fathi-Hafshejani, N. Azam, L. Wang, M. A. Kuroda, M. C. Hamilton, S. Hasim and M. Mahjouri-Samani, ACS Nano, 2021, 15, 11461–11469 CrossRef CAS PubMed.
- M. Zhang, X. Li, J. Pan, Y. Zhang, L. Zhang, C. Wang, X. Yan, X. Liu and G. Lu, Biosens. Bioelectron., 2021, 190, 113421 CrossRef CAS PubMed.
- D. Liu, C. Ju, C. Han, R. Shi, X. Chen, D. Duan, J. Yan and X. Yan, Biosens. Bioelectron., 2020, 173, 112817 CrossRef PubMed.
- M. A. Zamzami, G. Rabbani, A. Ahmad, A. A. Basalah, W. H. Al-Sabban, S. Nate Ahn and H. Choudhry, Bioelectrochemistry, 2021, 143, 107982 CrossRef PubMed.
- J.-H. Lee, M. Choi, Y. Jung, S. K. Lee, C.-S. Lee, J. Kim, J. Kim, N. H. Kim, B.-T. Kim and H. G. Kim, Biosens. Bioelectron., 2021, 171, 112715 CrossRef CAS PubMed.
- S. Mavrikou, G. Moschopoulou, V. Tsekouras and S. Kintzios, Sensors, 2020, 20, 3121 CrossRef CAS PubMed.
- B. Mojsoska, S. Larsen, D. A. Olsen, J. S. Madsen, I. Brandslund and F. A. a. Alatraktchi, Sensors, 2021, 21, 390 CrossRef CAS PubMed.
- G. Li, A. Wang, Y. Chen, Y. Sun, Y. Du, X. Wang, P. Ding, R. Jia, Y. Wang and G. Zhang, Front. Immunol., 2021, 12, 635677 CrossRef CAS PubMed.
- L. Fabiani, M. Saroglia, G. Galatà, R. De Santis, S. Fillo, V. Luca, G. Faggioni, N. D'Amore, E. Regalbuto, P. Salvatori, G. Terova, D. Moscone, F. Lista and F. Arduini, Biosens. Bioelectron., 2021, 171, 112686 CrossRef CAS PubMed.
- A. G. Ayankojo, R. Boroznjak, J. Reut, A. Öpik and V. Syritski, Sens. Actuators, B, 2022, 353, 131160 CrossRef CAS PubMed.
- E. B. Aydın, M. Aydın and M. K. Sezgintürk, ACS Biomater. Sci. Eng., 2021, 7, 3874–3885 CrossRef PubMed.
- R. L. Pinals, F. Ledesma, D. Yang, N. Navarro, S. Jeong, J. E. Pak, L. Kuo, Y.-C. Chuang, Y.-W. Cheng, H.-Y. Sun and M. P. Landry, Nano Lett., 2021, 21, 2272–2280 CrossRef CAS PubMed.
- W. S. Hart, E. Miller, N. J. Andrews, P. Waight, P. K. Maini, S. Funk and R. N. Thompson, Lancet Infect. Dis., 2022, 22, 603–610 CrossRef CAS PubMed.
- J. Boucau, C. Marino, J. Regan, R. Uddin, M. C. Choudhary, J. P. Flynn, G. Chen, A. M. Stuckwisch, J. Mathews, M. Y. Liew, A. Singh, T. Lipiner, A. Kittilson, M. Melberg, Y. Li, R. F. Gilbert, Z. Reynolds, S. L. Iyer, G. C. Chamberlin, T. D. Vyas, M. B. Goldberg, J. M. Vyas, J. Z. Li, J. E. Lemieux, M. J. Siedner and A. K. Barczak, MedRxiv, 2022, preprint, DOI:10.1101/2022.03.01.22271582.
- J. Bullard, K. Dust, D. Funk, J. E. Strong, D. Alexander, L. Garnett, C. Boodman, A. Bello, A. Hedley and Z. Schiffman, Clin. Infect. Dis., 2020, 71, 2663–2666 CrossRef CAS PubMed.
- J. Dinnes, J. J. Deeks, S. Berhane, M. Taylor, A. Adriano, C. Davenport, S. Dittrich, D. Emperador, Y. Takwoingi and J. Cunningham, Cochrane Database Syst. Rev., 2021, 21, 147–161 Search PubMed.
- G. C. Mak, P. K. Cheng, S. S. Lau, K. K. Wong, C. Lau, E. T. Lam, R. C. Chan and D. N. Tsang, J. Clin. Virol., 2020, 129, 104500 CrossRef CAS PubMed.
- P. Mertens, N. De Vos, D. Martiny, C. Jassoy, A. Mirazimi, L. Cuypers, S. Van den Wijngaert, V. Monteil, P. Melin and K. Stoffels, Front. Med., 2020, 7, 225 CrossRef PubMed.
- P. Banada, R. Green, S. Banik, A. Chopoorian, D. Streck, R. Jones, S. Chakravorty and D. Alland, J. Clin. Microbiol., 2021, 59, e0084521 CrossRef PubMed.
- E. Domingo, C. Garcia-Crespo, R. Lobo-Vega and C. Perales, Viruses, 2021, 13 Search PubMed.
- L. van Dorp, D. Richard, C. C. S. Tan, L. P. Shaw, M. Acman and F. Balloux, Nat. Commun., 2020, 11, 5986 CrossRef CAS PubMed.
- R. Pecori, S. Di Giorgio, J. Paulo Lorenzo and F. Nina Papavasiliou, Nat. Rev. Genet., 2022, 23, 505–518 CrossRef CAS PubMed.
- S. Portelli, M. Olshansky, C. H. M. Rodrigues, E. N. D'Souza, Y. Myung, M. Silk, A. Alavi, D. E. V. Pires and D. B. Ascher, Nat. Genet., 2020, 52, 999–1001 CrossRef CAS PubMed.
- L. Yurkovetskiy, X. Wang, K. E. Pascal, C. Tomkins-Tinch, T. Nyalile, Y. Wang, A. Baum, W. E. Diehl, A. Dauphin, C. Carbone, K. Veinotte, S. B. Egri, S. F. Schaffner, J. E. Lemieux, J. Munro, A. Rafique, A. Barve, P. C. Sabeti, C. A. Kyratsous, N. Dudkina, K. Shen and J. Luban, bioRxiv, 2020, preprint, DOI:10.1101/2020.07.04.187757.
- S. Ikegame, M. N. A. Siddiquey, C.-T. Hung, G. Haas, L. Brambilla, K. Y. Oguntuyo, S. Kowdle, H.-P. Chiu, C. S. Stevens, A. E. Vilardo, A. Edelstein, C. Perandones, J. P. Kamil and B. Lee, Nat. Commun., 2021, 12, 4598 CrossRef CAS PubMed.
- J. R. Mascola, B. S. Graham and A. S. Fauci, JAMA, 2021, 325, 1261–1262 CrossRef CAS PubMed.
- C. S. von Bartheld, M. M. Hagen and R. Butowt, ACS Chem. Neurosci., 2021, 12, 3535–3549 CrossRef CAS PubMed.
- H. Liu, Q. Zhang, P. Wei, Z. Chen, K. Aviszus, J. Yang, W. Downing, C. Jiang, B. Liang, L. Reynoso, G. P. Downey, S. K. Frankel, J. Kappler, P. Marrack and G. Zhang, Cell Res., 2021, 31, 720–722 CrossRef CAS PubMed.
- A. Khan, T. Zia, M. Suleman, T. Khan, S. S. Ali, A. A. Abbasi, A. Mohammad and D. Q. Wei, J. Cell. Physiol., 2021, 236, 7045–7057 CrossRef CAS PubMed.
- A. H. Williams and C. G. Zhan, J. Phys. Chem. B, 2021, 125, 4330–4336 CrossRef CAS PubMed.
- O. Stirrup, F. Boshier, C. Venturini, J. A. Guerra-Assuncao, A. Alcolea-Medina, A. Beckett, T. Charalampous, A. da Silva Filipe, S. Glaysher, T. Khan, R. Kulasegaran Shylini, B. Kele, I. Monahan, G. Mollett, M. Parker, E. Pelosi, P. Randell, S. Roy, J. Taylor, S. Weller, E. Wilson-Davies, P. Wade, R. Williams, C.-U.-H. V. s. consortium, C.-G. U. consortium, A. Copas, M. T. Cutino-Moguel, N. Freemantle, A. C. Hayward, A. Holmes, J. Hughes, T. Mahungu, G. Nebbia, D. Partridge, C. Pope, J. Price, S. Robson, K. Saeed, T. de Silva, L. Snell, E. Thomson, A. A. Witney and J. Breuer, BMJ Open Respir. Res., 2021, 8, e001029 CrossRef PubMed.
- D. Planas, T. Bruel, L. Grzelak, F. Guivel-Benhassine, I. Staropoli, F. Porrot, C. Planchais, J. Buchrieser, M. M. Rajah, E. Bishop, M. Albert, F. Donati, M. Prot, S. Behillil, V. Enouf, M. Maquart, M. Smati-Lafarge, E. Varon, F. Schortgen, L. Yahyaoui, M. Gonzalez, J. De Seze, H. Pere, D. Veyer, A. Seve, E. Simon-Loriere, S. Fafi-Kremer, K. Stefic, H. Mouquet, L. Hocqueloux, S. van der Werf, T. Prazuck and O. Schwartz, Nat. Med., 2021, 27, 917–924 CrossRef CAS PubMed.
- F. G. Naveca, V. Nascimento, V. C. de Souza, A. L. Corado, F. Nascimento, G. Silva, A. Costa, D. Duarte, K. Pessoa, M. Mejia, M. J. Brandao, M. Jesus, L. Goncalves, C. F. da Costa, V. Sampaio, D. Barros, M. Silva, T. Mattos, G. Pontes, L. Abdalla, J. H. Santos, I. Arantes, F. Z. Dezordi, M. M. Siqueira, G. L. Wallau, P. C. Resende, E. Delatorre, T. Graf and G. Bello, Nat. Med., 2021, 27, 1230–1238 CrossRef CAS PubMed.
- D. Geers, M. C. Shamier, S. Bogers, G. den Hartog, L. Gommers, N. N. Nieuwkoop, K. S. Schmitz, L. C. Rijsbergen, J. A. van Osch and E. Dijkhuizen, Sci. Immunol., 2021, 6, eabj1750 CrossRef PubMed.
- W. F. Garcia-Beltran, E. C. Lam, K. St Denis, A. D. Nitido, Z. H. Garcia, B. M. Hauser, J. Feldman, M. N. Pavlovic, D. J. Gregory, M. C. Poznansky, A. Sigal, A. G. Schmidt, A. J. Iafrate, V. Naranbhai and A. B. Balazs, Cell, 2021, 184, 2372–2383 CrossRef CAS PubMed.
- J. W. V. Thangaraj, P. Yadav, C. G. Kumar, A. Shete, D. A. Nyayanit, D. S. Rani, A. Kumar, M. S. Kumar, R. Sabarinathan and V. S. Kumar, J. Infect., 2021, 81, 94–118 Search PubMed.
- S. Shiehzadegan, N. Alaghemand, M. Fox and V. Venketaraman, Clin. Pract., 2021, 11, 778–784 CrossRef PubMed.
- J. Liu, Y. Liu, H. Xia, J. Zou, S. C. Weaver, K. A. Swanson, H. Cai, M. Cutler, D. Cooper, A. Muik, K. U. Jansen, U. Sahin, X. Xie, P. R. Dormitzer and P. Y. Shi, Nature, 2021, 596, 273–275 CrossRef CAS PubMed.
- J. Lan, X. He, Y. Ren, Z. Wang, H. Zhou, S. Fan, C. Zhu, D. Liu, B. Shao and T.-Y. Liu, Cell Res., 2022, 32, 593–595 CrossRef CAS PubMed.
- J. Chen, R. Wang, N. B. Gilby and G.-W. Wei, J. Chem. Inf. Model., 2022, 62, 412–422 CrossRef CAS PubMed.
- Y. Cao, J. Wang, F. Jian, T. Xiao, W. Song, A. Yisimayi, W. Huang, Q. Li, P. Wang and R. An, Nature, 2021, 602, 657–663 CrossRef PubMed.
- V. Thakur and R. Kanta Ratho, J. Med. Virol., 2021, 94, 1821–1824 CrossRef PubMed.
- J. Ai, X. Wang, X. He, X. Zhao, Y. Zhang, Y. Jiang, M. Li, Y. Cui, Y. Chen, R. Qiao, L. Li, L. Yang, Y. Li, Z. Hu, W. Zhang and P. Wang, Cell Host Microbe, 2022, 30, 1077–1083.e1074 CrossRef CAS PubMed.
- P. A. Desingu, K. Nagarajan and K. Dhama, J. Med. Virol., 2022, 94, 1808 CrossRef CAS PubMed.
- E. Mahase, BMJ, 2023, 153, DOI:10.1136/bmj.p153.
- TAG-VE statement on Omicron sublineages BQ.1 and XBB, https://www.who.int/news/item/27-10-2022-tag-ve-statement-on-omicron-sublineages-bq.1-and-xbb, accessed April 3, 2023 Search PubMed.
- F. Taylor, J. Bradford, P. J. Woll, D. Teare and A. Cox, in Circulating Nucleic Acids in Serum and Plasma–CNAPS IX, Springer, 2016, pp. 29–32 Search PubMed.
- K. Thompson, J. J. Collier, R. I. C. Glasgow, F. M. Robertson, A. Pyle, E. L. Blakely, C. L. Alston, M. Olahova, R. McFarland and R. W. Taylor, J. Inherited Metab. Dis., 2020, 43, 36–50 CrossRef PubMed.
- L. Mamanova, A. J. Coffey, C. E. Scott, I. Kozarewa, E. H. Turner, A. Kumar, E. Howard, J. Shendure and D. J. Turner, Nat. Methods, 2010, 7, 111–118 CrossRef CAS PubMed.
- S. Lu, J. Lin, Z. Zhang, L. Xiao, Z. Jiang, J. Chen, C. Hu and S. Luo, J. Med. Virol., 2021, 93, 518–521 CrossRef CAS PubMed.
- K. A. Lythgoe, M. Hall, L. Ferretti, M. de Cesare, G. MacIntyre-Cockett, A. Trebes, M. Andersson, N. Otecko, E. L. Wise, N. Moore, J. Lynch, S. Kidd, N. Cortes, M. Mori, R. Williams, G. Vernet, A. Justice, A. Green, S. M. Nicholls, M. A. Ansari, L. Abeler-Dorner, C. E. Moore, T. E. A. Peto, D. W. Eyre, R. Shaw, P. Simmonds, D. Buck, J. A. Todd, G. Oxford Virus Sequencing Analysis, T. R. Connor, S. Ashraf, A. da Silva Filipe, J. Shepherd, E. C. Thomson, C.-G. U. Consortium, D. Bonsall, C. Fraser and T. Golubchik, Science, 2021, 372, eabg0821 CrossRef CAS PubMed.
- E. Fass, G. Z. Valenci, M. Rubinstein, P. J. Freidlin, S. Rosencwaig, I. Kutikov, R. Werner, N. Ben-Tovim, E. Bucris and N. S. Zuckerman, Front. Med., 2021, 8, 798130 CrossRef PubMed.
- D. Castaneda-Mogollon, C. Kamaliddin, L. Fine, L. K. Oberding and D. R. Pillai, Diagn. Microbiol. Infect. Dis., 2021, 102, 115606 CrossRef PubMed.
- F. Stüder, J.-L. Petit, S. Engelen and M. A. Mendoza-Parra, Sci. Rep., 2021, 11, 1–11 CrossRef PubMed.
- L. A. Cohen-Aharonov, A. Rebibo-Sabbah, A. Yaacov, R. Z. Granit, M. Strauss, R. Colodner, O. Cheshin, S. Rosenberg and R. Eavri, PLoS One, 2022, 17, e0253404 CrossRef CAS PubMed.
- A. Chappleboim, D. Joseph-Strauss, A. Rahat, I. Sharkia, M. Adam, D. Kitsberg, G. Fialkoff, M. Lotem, O. Gershon and A.-K. Schmidtner, Sci. Transl. Med., 2021, 13, eabj2266 CrossRef CAS PubMed.
- J. Khateeb, Y. Li and H. Zhang, Crit. Care, 2021, 25, 244 CrossRef PubMed.
- P. Paul, A. M. France, Y. Aoki, D. Batra, M. Biggerstaff, V. Dugan, S. Galloway, A. J. Hall, M. A. Johansson and R. Kondor, Morb. Mortal. Wkly. Rep., 2021, 70, 846 CrossRef CAS PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brünink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J. L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 2000045 Search PubMed.
- M. Migueres, S. Lhomme, P. Tremeaux, C. Dimeglio, N. Ranger, J. Latour, M. Dubois, F. Nicot, M. Miedouge, J. M. Mansuy and J. Izopet, J. Clin. Virol., 2021, 143, 104969 CrossRef CAS PubMed.
- A. V. Robinson, G. Weaving, B. J. Philips, A. C. Eziefula, K. E. Shipman and T. Chevassut, Clin. Med., 2021, 21, e300–e305 CrossRef PubMed.
- Y. Wang, Y. Zhang, J. Chen, M. Wang, T. Zhang, W. Luo, Y. Li, Y. Wu, B. Zeng, K. Zhang, R. Deng and W. Li, Anal. Chem., 2021, 93, 3393–3402 CrossRef CAS PubMed.
- V. Borges, C. Sousa, L. Menezes, A. M. Goncalves, M. Picao, J. P. Almeida, M. Vieita, R. Santos, A. R. Silva, M. Costa, L. Carneiro, P. Casaca, P. Pinto-Leite, A. Peralta-Santos, J. Isidro, S. Duarte, L. Vieira, R. Guiomar, S. Silva, B. Nunes and J. P. Gomes, Eurosurveillance, 2021, 26, 2100131 CrossRef CAS PubMed.
- K. A. Brown, J. Gubbay, J. Hopkins, S. Patel, S. A. Buchan, N. Daneman and L. Goneau, MedRxiv, 2021, preprint, DOI:10.1101/2021.02.09.21251225.
- C. B. F. Vogels, M. I. Breban, I. M. Ott, T. Alpert, M. E. Petrone, A. E. Watkins, C. C. Kalinich, R. Earnest, J. E. Rothman, J. Goes de Jesus, I. Morales Claro, G. Magalhaes Ferreira, M. A. E. Crispim, U. K. C. G. N. Brazil, L. Singh, H. Tegally, U. J. Anyaneji, E. B. Hodcroft, C. E. Mason, G. Khullar, J. Metti, J. T. Dudley, M. J. MacKay, M. Nash, J. Wang, C. Liu, P. Hui, S. Murphy, C. Neal, E. Laszlo, M. L. Landry, A. Muyombwe, R. Downing, J. Razeq, T. de Oliveira, N. R. Faria, E. C. Sabino, R. A. Neher, J. R. Fauver and N. D. Grubaugh, PLoS Biol., 2021, 19, e3001236 CrossRef CAS PubMed.
- S. Hegde, Z. Tang, J. Zhao and J. Wang, Front. Chem., 2021, 9, 802766 CrossRef CAS PubMed.
- W. L. Lee, M. Imakaev, F. Armas, K. A. McElroy, X. Gu, C. Duvallet, F. Chandra, H. Chen, M. Leifels and S. Mendola, Environ. Sci. Technol. Lett., 2021, 8, 675–682 CrossRef CAS.
- N. Vega-Magaña, R. Sánchez-Sánchez, J. Hernández-Bello, A. A. Venancio-Landeros, M. Peña-Rodríguez, R. A. Vega-Zepeda, B. Galindo-Ornelas, M. Díaz-Sánchez, M. García-Chagollán and G. Macedo-Ojeda, Front. Cell. Infect. Microbiol., 2021, 11, 672562 CrossRef PubMed.
- S. M. Al-Jaf and S. S. Niranji, MedRxiv, 2021, preprint, DOI:10.1101/2021.04.17.21255656.
- L. Jones, A. Bakre, H. Naikare, R. Kolhe, S. Sanchez, Y. C. Mosley and R. A. Tripp, PLoS One, 2021, 16, e0257563 CrossRef CAS PubMed.
- E. Tamanaha, Y. Zhang and N. A. Tanner, PLoS One, 2021, 17, e0259610 CrossRef PubMed.
- P. James, D. Stoddart, E. D. Harrington, J. Beaulaurier, L. Ly, S. Reid, D. J. Turner and S. Juul, MedRxiv, 2020, preprint, DOI:10.1101/2020.08.07.20161737.
- M.-C. Marqués, R. Ruiz, R. Montagud-Martínez, R. Márquez-Costa, S. Albert, P. Domingo-Calap and G. Rodrigo, ACS Synth. Biol., 2021, 10, 3595–3599 CrossRef PubMed.
- Q. Meng, X. Wang, Y. Wang, L. Dang, X. Liu, X. Ma, T. Chi, X. Wang, Q. Zhao, G. Yang, M. Liu, X. Huang and P. Ma, Biotechnol. J., 2021, 16, e2100040 CrossRef PubMed.
- J. Yang, N. Barua, M. N. Rahman, N. Lo, T. F. Tsang, X. Yang, P. K. S. Chan, L. Zhang and M. Ip, PLoS One, 2021, 16, e0261778 CrossRef CAS PubMed.
- Y. Liang, H. Lin, L. Zou, J. Zhao, B. Li, H. Wang, J. Lu, J. Sun, X. Yang, X. Deng and S. Tang, Microbiol. Spectrum, 2021, 9, e0101721 CrossRef PubMed.
- M. Kumar, S. Gulati, A. H. Ansari, R. Phutela, S. Acharya, P. Kathpalia, A. Kanakan, R. Maurya, J. S. Vasudevan and A. Murali, MedRxiv, 2021, preprint, DOI:10.1101/2021.02.01.21250900.
- H. de Puig, R. Lee, D. Najjar, X. Tan, L. Soekensen, N. Angenent-Mari, N. Donghia, N. Weckman, A. Ory and C. Ng, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed.
- F. Klaassen, M. H. Chitwood, T. Cohen, V. E. Pitzer, M. Russi, N. A. Swartwood, J. A. Salomon and N. A. Menzies, MedRxiv, 2022, preprint, DOI:10.1101/2022.11.19.22282525.
- J. Cheong, H. Yu, C. Y. Lee, J. U. Lee, H. J. Choi, J. H. Lee, H. Lee and J. Cheon, Nat. Biomed. Eng., 2020, 4, 1159–1167 CrossRef CAS PubMed.
- W. Bu, W. Li, J. Li, T. Ao, Z. Li, B. Wu, S. Wu, W. Kong, T. Pan, Y. Ding, W. Tan, B. Li, Y. Chen and Y. Men, Sens. Actuators, B, 2021, 348, 130678 CrossRef CAS.
- P. Fozouni, S. Son, M. D. de León Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar and S. I. Stephens, MedRxiv, 2020, preprint, DOI:10.1101/2020.09.28.20201947.
- A. S. James and J. I. Alawneh, Diagnostics, 2020, 10, 399 CrossRef CAS PubMed.
- S. Srivastava, S. Banu, P. Singh, D. T. Sowpati and R. K. Mishra, J. Biosci., 2021, 46, 1–14 CrossRef.
- W. S. Chan, C. H. Au, H. Y. Lam, C. L. N. Wang, D. N. Ho, Y. M. Lam, D. K. W. Chu, L. L. M. Poon, T. L. Chan, J. S. Zee, E. S. K. Ma and B. S. F. Tang, Virol. J., 2020, 17, 183 CrossRef CAS PubMed.
- J. J. Leblanc, J. Pettipas, R. J. Davidson, G. A. Tipples, J. Hiebert and T. F. Hatchette, J. Clin. Microbiol., 2008, 46, 4049–4051 CrossRef CAS PubMed.
- J. Dinnes, J. J. Deeks, S. Berhane, M. Taylor, A. Adriano, C. Davenport, S. Dittrich, D. Emperador, Y. Takwoingi, J. Cunningham, S. Beese, J. Domen, J. Dretzke, L. Ferrante di Ruffano, I. M. Harris, M. J. Price, S. Taylor-Phillips, L. Hooft, M. M. Leeflang, M. D. McInnes, R. Spijker and A. Van den Bruel, Cochrane Database Syst. Rev., 2021, 3, CD013705 Search PubMed.
- X. Chen, Z. Chen, A. S. Azman, X. Deng, X. Chen, W. Lu, Z. Zhao, J. Yang, C. Viboud, M. Ajelli, D. T. Leung and H. Yu, Lancet Global Health, 2020, 9, e598–e609 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |