Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biosynthesis of pleuromutilin congeners using an Aspergillus oryzae expression platform†
Fabrizio
Alberti‡§
 *a,
Khairunisa
Khairudin‡
a,
Jonathan A.
Davies
*a,
Khairunisa
Khairudin‡
a,
Jonathan A.
Davies
 b,
Suphattra
Sangmalee
a,
Christine L.
Willis
b,
Suphattra
Sangmalee
a,
Christine L.
Willis
 b,
Gary D.
Foster
a and
Andy M.
Bailey
b,
Gary D.
Foster
a and
Andy M.
Bailey
 *a
*a
aSchool of Biological Sciences, University of Bristol, 24 Tyndall Avenue, Bristol BS8 1TQ, UK. E-mail: F.Alberti@warwick.ac.uk; Andy.Bailey@bristol.ac.uk
bSchool of Chemistry, University of Bristol, Cantock's Close, Bristol BS8 1TS, UK
First published on 15th March 2023
Abstract
Pleuromutilin is an antibiotic diterpenoid made by Clitopilus passeckerianus and related fungi, and it is the progenitor of a growing class of semi-synthetic antibiotics used in veterinary and human medicine. To harness the biotechnological potential of this natural product class, a full understanding of its biosynthetic pathway is essential. Previously, a linear pathway for pleuromutilin biosynthesis was established. Here we report two shunt pathways involving Pl-sdr and Pl-atf that were identified through the rational heterologous expression of combinations of pleuromutilin biosynthetic genes in Aspergillus oryzae. Three novel pleuromutilin congeners were isolated, and their antimicrobial activity was investigated, alongside that of an additional derivative produced through a semi-synthetic approach. It was observed that the absence of various functional groups – 3 ketone, 11 hydroxyl group or 21 ketone – from the pleuromutilin framework affected the antibacterial activity of pleuromutilin congeners. This study expands our knowledge on the biosynthesis of pleuromutilin and provides avenues for the development of novel pleuromutilin analogues by combining synthetic biology and synthetic chemistry.
Introduction
Pleuromutilin is a diterpenoid antimicrobial isolated from cultures of Clitopilus passeckerianus and related basidiomycete fungi.1,2 Pleuromutilin's antibacterial properties rely on the inhibition of protein synthesis by interfering with the peptidyl transferase centre (PTC) of the bacterial ribosome and subsequently preventing the formation of peptide bonds between amino acids.3Numerous efforts have been made to modify the structure of pleuromutilin with the aim to improve its bioactivity and pharmacokinetic properties. Functionalisation of the C-14 side chain has led to the development of two semisynthetic derivatives used in veterinary formulations, tiamulin and valnemulin,4 as well as of two more derivatives, retapamulin and lefamulin, which are used in human medicine to treat skin infections and community-acquired bacterial pneumonia, respectively.5,6 Efforts to expand this class of antibiotics are still ongoing, including reports of new semisynthetic derivatives of pleuromutilin generated through photoinduced addition reactions at the alkene position C19–C20, which resulted in improved antimicrobial activity against Gram-positive bacteria upon introduction of an N-acetyl-L-cysteine side chain.7
Studies on the biosynthesis of pleuromutilin have been aided by the identification of the corresponding biosynthetic gene cluster in C. passeckerianus,8 which includes the seven enzyme-coding genes Pl-ggs, Pl-cyc, Pl-p450-1, Pl-p450-2, Pl-p450-3, Pl-sdr and Pl-atf (Fig. 1a). Independent work from Yamane et al.9 and Alberti et al.10 led to the elucidation of the biosynthetic pathway to pleuromutilin via heterologous gene expression in Aspergillus oryzae (Fig. 1b). The pathway to pleuromutilin 1 begins with Pl-ggs producing geranylgeranyl diphosphate (GGPP), while Pl-cyc is a bifunctional (di)terpene synthase11 that catalyses the cyclisation of GGPP to the first tricyclic intermediate 3-deoxo-11-dehydroxymutilin (also called premutilin) 2. Two cytochrome P450s – Pl-p450-1 and Pl-p450-2 – add hydroxy groups at C-11 (producing intermediate 3) and C-3 (producing intermediate 4), respectively. The hydroxy group at C-3 is then oxidised to a keto by the short-chain dehydrogenase/reductase Pl-sdr, producing mutilin 5. The acetyltransferase Pl-atf adds an acetate group to C-14, giving 14-O-acetylmutilin 6. Lastly Pl-p450-3 oxidises C-22 on the acetate side chain to give pleuromutilin 1. Interestingly, Yamane et al.9 reported that Pl-p450-2 can act independently from Pl-p450-1 on the substrate 2 and leads to the accumulation of 7, which can then be converted to 4 by Pl-p450-1 and re-join the rest of the pathway. This suggests that both Pl-p450-1 and Pl-p450-2 have substrate flexibility. We therefore set out to investigate in the present study, if the other enzymes involved in pleuromutilin biosynthesis are also able to accept diverse substrates. Instances of alternative biosynthetic paths for secondary metabolites have been reported for natural products, such as for actinorhodin in Streptomyces coelicolor, which shows two alternative routes for the quinone formation by C-6 oxygenation.12
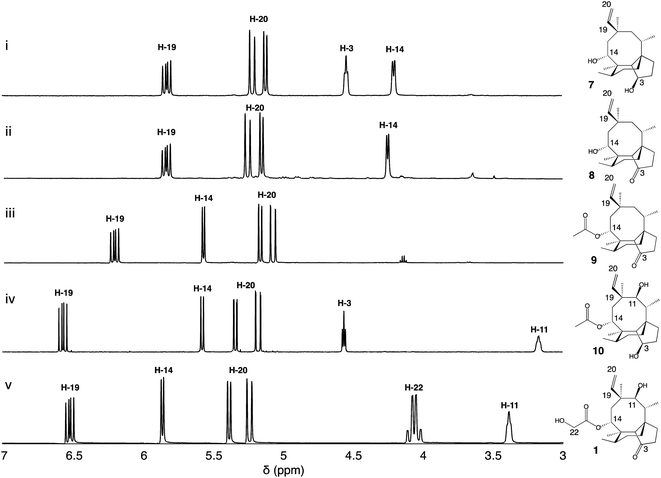
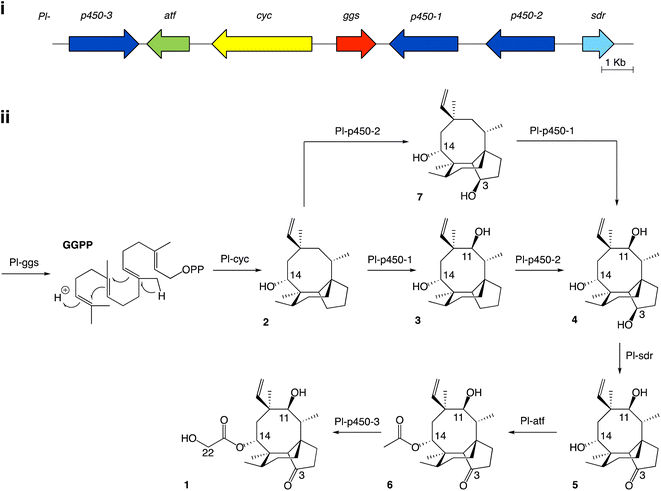 | ||
| Fig. 1 (i) Pleuromutilin biosynthetic gene cluster. The core biosynthetic genes Pl-ggs and Pl-cyc are represented in red and yellow, respectively. The acetyltransferase Pl-atf is represented in green. Genes coding for oxidizing enzymes Pl-p450-1, 2 and 3 are represented in dark blue, and Pl-sdr in light blue. (ii) Proposed biosynthetic route to pleuromutilin.9,10 | ||
In this study, we report the rational heterologous expression of combinations of pleuromutilin biosynthesis genes in A. oryzae, which led to the isolation of new pleuromutilin congeners. Synthetic chemistry routes were also adopted to introduce further functionalisation, and all pleuromutilin congeners were assessed for antimicrobial activity. Results from this study aid understanding of pleuromutilin biosynthetic reactions and provide the basis for further engineering of the pleuromutilin pathway to generate new unnatural analogues.
Results and discussion
Biosynthesis and isolation of novel pleuromutilin congeners
In this study, we set out to investigate the substrate tolerance of pleuromutilin biosynthetic enzymes and expand our knowledge of pleuromutilin biosynthesis. For this purpose, we used as the recipient host organism the premutilin-producing strain A. oryzae GC, which carries the core biosynthetic genes Pl-ggs and Pl-cyc,10 to further express downstream biosynthetic genes from the pleuromutilin cluster and isolate the corresponding biosynthetic products (Table 1).| A. oryzae strain | Heterologous genes from C. passeckerianus | Compounds produced de novo |
|---|---|---|
| GCP2 | Pl-ggs, Pl-cyc, Pl-p450-2 | 7 |
| GCP2S | Pl-ggs, Pl-cyc, Pl-p450-2, Pl-sdr | 8 |
| GCP2P3SA | Pl-ggs, Pl-cyc, Pl-p450-2, Pl-p450-3, Pl-sdr, Pl-atf | 9 |
| GCP1P2P3A | Pl-ggs, Pl-cyc, Pl-p450-1, Pl-p450-2, Pl-p450-3, Pl-atf | 10 |
Firstly, we introduced Pl-p450-2 in A. oryzae GC, producing strain A. oryzae GCP2. One major compound was produced de novo (see ESI Fig. 1† for mass spectra in positive and negative ion mode) and then purified using preparative HPLC. Approximately 17 mg of white precipitate was isolated from a one-litre culture of A. oryzae GCP2. The compound was identified as the 3,14-diol 7 through NMR spectroscopy experiments (Fig. 2, ESI Fig. 2–6†), in accordance with previous reports from Yamane et al.9
We next co-expressed Pl-p450-2 and Pl-sdr in A. oryzae GC, producing strain GCP2S, and successfully purified 8.5 mg of a new product (8, see ESI Fig. 7† for mass spectra in positive and negative ion mode) from a one-litre culture of the organism. The 1H-NMR of the product revealed similar chemical shifts as 7 except for the broad signal at δ 4.6 correlated to 3-OH, which was missing in 8. Further extensive NMR spectroscopy studies (Fig. 2, ESI Fig. 8–12†) confirmed the presence of a 3-keto as shown by the signal at δ 218.7 in the 13C-NMR spectrum, hence the structure of 8 was assigned as 11-dehydroxy mutilin. The isolation of 8 from A. oryzae GCP2S suggested that oxidation of the C-3 hydroxy group to a keto can take place independently of the hydroxylation of C-11.
The next steps in the production of mature pleuromutilin analogues involve the addition of the acetyl group on C-14 and further hydroxylation of C-22 catalysed by Pl-atf and Pl-p450-3, respectively. Thus, the expression cassettes for Pl-p450-2 and Pl-p450-3 alongside those for Pl-sdr and Pl-atf were transformed into A. oryzae GC, obtaining strain A. oryzae GCP2P3SA. It should be noted that this strain includes all but one – Pl-p450-1 – pleuromutilin biosynthetic genes. HPLC analysis of crude extracts from this strain revealed a new product (9, see ESI Fig. 13† for mass spectra in positive and negative ion mode). The isolation of the new product was carried out through column chromatography. The crude fungal extract from a one-litre culture of GCP2P3SA was eluted with petroleum ether/ethyl acetate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) and the fractions containing the targeted compound were pooled and concentrated in vacuo, yielding 18 mg of white precipitate. Metabolite 9 showed similar 1H-NMR shift patterns as 8, except for the signal at δ 4.25 being deshielded to δ 5.57 (Fig. 2). 2D-NMR spectroscopy experiments confirmed that this signal was correlated to H-14. Two additional carbon signals having similar chemical shifts as those observed in 6 were also observed in the 13C-NMR spectrum of 8, suggesting that acetylation took place on C-14. The isolation of 9 from A. oryzae GCP2P3SA shows that Pl-atf can catalyse acetylation of mutilin without needing pre-hydroxylation at C-11 (ESI Fig. 14–18†). Despite the presence of Pl-p450-3 in A. oryzae GCP2P3SA, no detectable hydroxylation of the acetate (at C-22) took place. It should be noted that Pl-p450-3 shows poor catalytic activity when heterologously expressed in A. oryzae, which has been observed both on the native substrate 14-O-acetylmutilin,8 and on its congeners – as seen from the incomplete functionalisation of C22 on a C1–C2 unsaturated 14-O-acetylmutilin analogue.10 Nevertheless, based on these results, we propose that intermediate 7 can be derivatised at positions C-3 and C-14 leading to two previously unreported pleuromutilin congeners, 8 and 9, through the catalytic activity of Pl-sdr and Pl-atf, respectively.
9) and the fractions containing the targeted compound were pooled and concentrated in vacuo, yielding 18 mg of white precipitate. Metabolite 9 showed similar 1H-NMR shift patterns as 8, except for the signal at δ 4.25 being deshielded to δ 5.57 (Fig. 2). 2D-NMR spectroscopy experiments confirmed that this signal was correlated to H-14. Two additional carbon signals having similar chemical shifts as those observed in 6 were also observed in the 13C-NMR spectrum of 8, suggesting that acetylation took place on C-14. The isolation of 9 from A. oryzae GCP2P3SA shows that Pl-atf can catalyse acetylation of mutilin without needing pre-hydroxylation at C-11 (ESI Fig. 14–18†). Despite the presence of Pl-p450-3 in A. oryzae GCP2P3SA, no detectable hydroxylation of the acetate (at C-22) took place. It should be noted that Pl-p450-3 shows poor catalytic activity when heterologously expressed in A. oryzae, which has been observed both on the native substrate 14-O-acetylmutilin,8 and on its congeners – as seen from the incomplete functionalisation of C22 on a C1–C2 unsaturated 14-O-acetylmutilin analogue.10 Nevertheless, based on these results, we propose that intermediate 7 can be derivatised at positions C-3 and C-14 leading to two previously unreported pleuromutilin congeners, 8 and 9, through the catalytic activity of Pl-sdr and Pl-atf, respectively.
Next the impact of omitting Pl-sdr from the gene cluster was investigated. We therefore generated strain A. oryzae GCP1P2P3A, which included the three pathway cytochrome P450s and Pl-atf in the A. oryzae GC background. The A. oryzae GCP1P2P3A strain accumulated a novel compound (10, see ESI Fig. 19† for mass spectra in positive and negative ion mode), which was purified through preparative HPLC yielding 20 mg of white precipitate. Analysis of the NMR spectroscopy data for 10 revealed the presence of two hydroxy groups. From the 13C-NMR spectrum (ESI Fig. 20–24†), it appeared that this molecule would be related to the previously characterized 4, though bearing two additional carbons. The chemical shifts of these two carbons matched with the shift pattern of C-21 and C-22 from 6 and the 1H-NMR data also demonstrated that the C-14 had incorporated an acetyl group, giving 10. The isolation of 10 from A. oryzae GCP1P2P3A proved that the C-14 acetylation reaction catalysed by Pl-atf can occur not only when there is a keto group at C-3 – as it is the case for mutilin 5 in the main pathway – but also in the presence of a hydroxy group at the same position – as it is the case for 4. As with metabolite 9, the acetate group on C-14 of 10 was not oxidized despite Pl-p450-3 being expressed in A. oryzae GCP1P2P3A. Isolation of this shunt product suggested that the acetylation catalysed by Pl-atf could take place independently from the oxidation of the C-3 hydroxy to keto. Overall, it appeared that when the activity of Pl-p450-1 and Pl-sdr were omitted, alternative pathways could be followed bypassing the catalytic reactions of the missing enzymes and leading to shunt products. These results suggest that some of the pleuromutilin biosynthetic enzymes possess relaxed substrate tolerance, which makes them useful as potential biocatalysts to generate novel pleuromutilin analogues. Using a similar approach to the one we adopted here, analogues of the final biosynthetic products from other pathways have been obtained in other studies. For instance, the biosynthetic enzymes involved in the production of the communesins show a good degree of substrate tolerance to make analogues of the final pathway products, as Lin et al.13 were able to generate communesin C, E, F and J, analogous to communesin A and B, from blocked mutants lacking the methyltransferase and epoxidase enzymes CnsE and CnsJ.
Production of metabolites 9 and 10 through feeding experiments
The rational heterologous expression of all genes of the pleuromutilin cluster minus Pl-p450-1 or Pl-sdr showed production of new metabolites, 9 and 10. However, the final hydroxylation of C-22 catalysed by Pl-p450-3 did not take place on either of these two shunt products. To investigate this reaction further, we performed feeding studies with 11-dehydroxy mutilin 8 and triol 4 on A. oryzae AP3, which harbours Pl-p450-3 and Pl-atf and was previously shown to promote conversion of 5 into 1.10 Metabolites 8 and 4 were isolated from strains A. oryzae GCP2S and GCP1P2, respectively. A. oryzae AP3 was cultured in the presence of 100 mg l−1 of 8 and 4 in separate experiments. Metabolite analyses of the fed cultures confirmed the conversion of 8 to 9 (ESI Fig. 25†) and of 4 to 10 (ESI Fig. 26†), however no congener with hydroxylated C-22 could be detected in either assay, in accordance with our previous observations of metabolites production in strains GCP2P3SA and GCP1P2P3A.Antimicrobial activity testing of novel pleuromutilin congeners
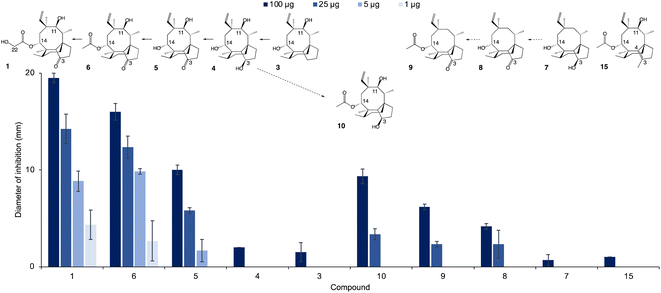
The new shunt products isolated in this work were assayed for antimicrobial activity on Bacillus subtilis ATCC 6633 and compared with the intermediates known to be part of the main pathway, following the same procedure reported by Alberti et al.10 Pleuromutilin 1 showed the highest antimicrobial activity, followed by 14-O-acetylmutilin 6 (Fig. 3). The small difference in bioactivity between 1 and 6 is in accordance with observations that the C-14 extension of pleuromutilin antibiotics is responsible for only minor hydrophobic interactions with bacterial ribosomal nucleotides.3 Conversely, the loss of the keto group at C-21 had a considerable impact on bioactivity, as can be seen in the reduced inhibitory activity of 3, 4, 7 and 10. This is in line with observations of the interaction of pleuromutilins with the ribosomal nucleotides, in which the C-21 keto group is known to form a network of hydrogen bonds with G2061 of the 23S RNA domain V.3 Mutilin 5 shows approximately half the activity of pleuromutilin while earlier intermediates with reduction of keto to hydroxy group or absence of the hydroxy group from C-3, 4 and 3 respectively, resulted in almost complete loss of activity. This is an agreement with previous studies showing that mutilin and other derivatives with a free C-14 hydroxy group were inactive.14 Absence of the 11-hydroxy group – known to form hydrogen bonding with the G2505 phosphate of the 23S RNA domain V3 – also resulted in a considerable decline in bioactivity for 7 and 8, although 8 showed a slightly stronger inhibition putatively due to the presence of a keto group at C-3. Similar to pleuromutilin intermediates, a decline in antimicrobial activity was also observed in the shunt products 10 and 9 when these are compared to 14-O-acetylmutilin 6, highlighting the importance of the C-3 keto and the C-11 hydroxy groups, respectively, for bioactivity (Fig. 4).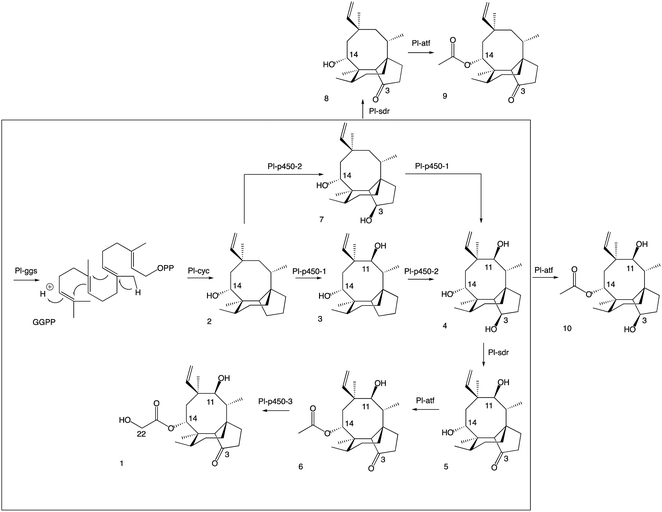 | ||
| Fig. 4 Revised pleuromutilin biosynthetic pathway. The main pathway is in the box, the shunt products are outside. | ||
Given the importance of the C-3 keto group in bioactivity, we decided to investigate whether substitution of this group with other functional groups would lead to an altered antimicrobial activity. Furthermore, the five-membered ring of pleuromutilin antibiotics is prone to attack by liver cytochrome P450s, which leads to their rapid metabolism.15 Therefore, their functionalisation may result in pleuromutilin derivatives with longer half-life or even altered bioactivity, as shown recently by the development of an anti-idiopathic pulmonary fibrosis lead compound through the functionalisation of the five-membered ring of pleuromutilin.16 In this study, we endeavoured to functionalise the five-membered ring by replacing the C-3 keto of pleuromutilin with a methyl group coupled with C3–C4 unsaturation. Hydrolysis of commercially available tiamulin hydrogen fumarate was performed to produce mutilin 5 (see ESI Fig. 27 and 28† for 1H-NMR and 13C-NMR spectra, and ESI Methods† for reaction conditions). Mutilin was used to prepare TMS-mutilin 12 (see ESI Fig. 29 and 30† for 1H-NMR and 13C-NMR spectra, and ESI Methods† for reaction conditions), which was then used to make a C-3 methyl, C3–C4 unsaturated analogue 14 (see ESI Fig. 31 and 32† for 1H-NMR and 13C-NMR spectra, and ESI Methods† for reaction conditions). Analogue 14 was fed to A. oryzae AP3 at a concentration of 100 mg l−1 for further functionalisation of the C-14 side chain. Successful acetylation by Pl-atf was observed, leading to the new semi-synthetic analogue 15, which was purified using a combination of column chromatography (10–100% ethyl acetate in petrol) and preparative HPLC, yielding 23 mg of pure compound (see ESI Fig. 33 and 34† for 1H-NMR and 13C-NMR spectra). Once again, no congener with hydroxylated C-22 could be detected, as previously observed when attempting to produce pleuromutilin analogues starting from 4 and 8. The semi-synthetic compound 15 was assayed for antimicrobial activity against B. subtilis. Compared to the closest naturally occurring intermediate 6, a considerable decrease in activity was observed when the five-membered ring was functionalised with a C-3 methyl and a C3–C4 double bond (Fig. 3). Thus, from our study, it can be concluded that retaining the three naturally occurring pleuromutilin functional groups – C-3 keto, C-11 hydroxy and C-21 keto – is important to maintain the bioactivity of the pleuromutilin analogues.
Conclusions
A more detailed picture of the biosynthetic pathway for pleuromutilin was obtained in this work, showing that biosynthetic genes can be expressed in different combinations to produce pleuromutilin congeners that have not been reported in C. passeckerianus. By omitting selected biosynthetic genes, we were able to isolate three novel pleuromutilin congeners, 8, 9 and 10. Overall, it appeared that when the activity of Pl-p450-1 and Pl-sdr were omitted, alternative pathways could be followed bypassing the catalytic reactions of the missing enzymes and leading to shunt products. These results suggest that some of the pleuromutilin biosynthetic enzymes possess relaxed substrate tolerance, which makes them useful as potential biocatalysts to generate novel pleuromutilin analogues. Heterologous gene expression in A. oryzae proved to be a flexible approach for developing modified pleuromutilin analogues. This strategy, when combined with synthetic endeavours, opens the possibility of generating new semi-synthetic pleuromutilins.Materials and methods
Reagents, strains and conditions for growth of microorganisms
Chemicals and media ingredients used in this study were obtained from Sigma, Fisher, Oxoid, ForMedium, Melford, Bioline, Thermo Scientific or VWR unless otherwise stated. Media were prepared and sterilized by autoclaving using a standard programme at 121 °C for 15 minutes. Deionized water was used for all solutions unless stated.Growth condition for fungal and bacterial strains
The host strain used for heterologous expression, A. oryzae NSAR1 (genotype niaD-, sC-, ΔargB, adeA-)17 was maintained on MEA plates with appropriate supplements (15 g l−1 malt extract, 1.5 g l−1 arginine, 1.5 g l−1 methionine, 0.1 g l−1 adenine, 2 g l−1 ammonium sulphate, 15 g l−1 agar) at 28 °C. The yeast strain used for homologous recombination-based construction of plasmids, S. cerevisiae BY4742 (genotype MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0), was maintained on YPAD plates (10 g l−1 yeast extract, 20 g l−1 bactopeptone, 20 g l−1D-glucose, 40 mg l−1 adenine hemisulfate 15 g l−1 agar). Propagation of plasmids was performed in E. coli One Shot® ccdB SurvivalTM 2 T1R competent cells (Life Technologies), which was grown in LB agar plates (10 g l−1 NaCl, 10 g l−1 tryptone, 5 g l−1 yeast extract, pH 7) at 37 °C.Construction of expression vectors
To construct fungal expression vector containing intron-free genes of the pleuromutilin cluster, three pTYGS18 plasmids with either arginine, adenine or basta selectable marker were used as backbones (plasmid maps are shown in ESI Methods 1–4†). The assembly of expression vectors pTYGSargGC, pTYGSadeP1P2P3, pTYGSadeP3, pTYGSbarAS, pTYGSbarA and pTYGSbarS was reported in Alberti et al.10 The construction of expression vectors pTYGSadeP2P3 and pTYGSadeP2 was carried out through yeast-based homologous recombinant as described by Ma et al.19 using PCR-amplified genes of interest with Phusion high-fidelity DNA polymerase according to the manufacturer's instructions (details on all plasmid components are reported in ESI Table 1,† plasmid maps are represented in ESI Fig. 35†). Primers used for amplification of the genes of interest are listed in ESI Table 2.†Transformation of A. oryzae NSAR1
Protoplast–polyethylene glycol method adapted from Halo et al.20 was used to transform A. oryzae NSAR1 (see ESI Table 3† for a list of strains generated in this study). The strain was firstly transformed with vector pTYGSargGC (which included Pl-ggs and Pl-cyc) to generate strain GC (producer of 2). Further transformation of GC with pTYGSadeP2 (containing Pl-p450-2) achieved production of strain GCP2 (producer of 7). Likewise, co-transformation of GC with pTYGSadeP2 (containing Pl-p450-2) and pTYGSbarS produced strain GCP2S (producer of 8). To construct GCP2P3SA (producer of 9), a combination of pTYGSadeP2P3 (containing Pl-p450-2 and Pl-p450-3) and pTYGSbarSA (containing Pl-sdr and Pl-atf) was introduced into GC, whereas a combination of pTYGSadeP1P2P3 (containing the three Pl-p450s) and pTYGSbarA (containing Pl-atf) was used to transform strain GC to obtain strain GCP1P2P3A (producer of 10). Strain AP3, used for feeding experiments with 4 and 8, was generated by transforming A. oryzae NSAR1 with a combination of pTYGSadeP3 (containing Pl-p450-3) and pTYGSbarA (containing Pl-atf), as previously reported.10A. oryzae transformant strains were screened by PCR for integration of the genes of interest using DreamTaq Green PCR Master Mix (Thermo Scientific) in combination with the screening primers reported in ESI Table 2.†Metabolite analysis of A. oryzae transformants
A. oryzae transformant strains were analysed for production of metabolites through HPLC-MS. Each strain was grown in 100 ml of CMP medium (35 g l−1 Czapek-Dox liquid, 20 g l−1 maltose, 10 g l−1 peptone) at 28 °C for 10 days prior to performing metabolite extractions with ethyl acetate, followed by concentration of the crude extract in vacuo methanol. Crude extracts were dissolved in methanol and analysed by HPLC-MS including a Waters 2767 HPLC system with a Waters 2545 pump, a Phenomenex LUNA column (2.6 μ, C18, 100 Å, 4.6 × 100 mm) and a Phenomenex Security Guard precolumn (Luna C5 300 Å) for reverse-phase chromatography. UV absorbance was detected between 200 and 400 nm through a Waters 2998 diode array detector while a mass range between 150 and 800 Da in positive and negative ion mode was scanned by the Waters Quattro Micro spectrometer. Chromatography was achieved using a gradient of solvents ((A) HPLC grade H2O containing 0.05% formic acid; (B) HPLC grade CH3CN containing 0.045% formic acid) with the following programme: 0 minutes, 20% B; 15 minutes, 90% B; 16 minutes 95% B; 17 minutes 95% B; 18 minutes 10% B, 20 minutes 10% B. The flow rate was set at 1 ml min−1.Isolation and purification of metabolites of interest
Large scale metabolite extractions from 1 litre cultures of chosen fungal strains in CMP medium were performed with the same conditions as described above. Purification of compound 9 was carried out through column chromatography by passing the concentrated crude extract through a packed silica gel 60 (Merck) in a column. An appropriate gradient solvent system consisting of a mixture of ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether (9
petroleum ether (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 7
1 to 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) was used to elute the metabolites into fractions. Fractions with the targeted compound were pooled in a round bottom flask and dried in vacuo. Isolation of other shunt products was performed through preparative HPLC with a reverse-phase Phenomenex LUNA column (5 μ, C18, 100 Å, 10 × 250 mm) using a flow rate at 16 ml min−1. The mobile phase was a gradient mixture of acetonitrile and formic acid in water following the elution programme: 0 minutes, 5% B; 1 minute, 5% B; 2 minutes, 40% B; 15 minutes 90% B; 17 minutes 95% B; 18 minutes 5% B, 20 minutes 5% B ((A) HPLC grade H2O containing 0.05% formic acid; (B) HPLC grade CH3CN containing 0.045% formic acid). The eluent was monitored by UV detection at 200–400 nm and mass range 100–600 Da. Targeted compounds were isolated and collected in glass vials prior to being pooled and concentrated in vacuo.
3, v/v) was used to elute the metabolites into fractions. Fractions with the targeted compound were pooled in a round bottom flask and dried in vacuo. Isolation of other shunt products was performed through preparative HPLC with a reverse-phase Phenomenex LUNA column (5 μ, C18, 100 Å, 10 × 250 mm) using a flow rate at 16 ml min−1. The mobile phase was a gradient mixture of acetonitrile and formic acid in water following the elution programme: 0 minutes, 5% B; 1 minute, 5% B; 2 minutes, 40% B; 15 minutes 90% B; 17 minutes 95% B; 18 minutes 5% B, 20 minutes 5% B ((A) HPLC grade H2O containing 0.05% formic acid; (B) HPLC grade CH3CN containing 0.045% formic acid). The eluent was monitored by UV detection at 200–400 nm and mass range 100–600 Da. Targeted compounds were isolated and collected in glass vials prior to being pooled and concentrated in vacuo.
NMR spectroscopy analysis of purified metabolites
Nuclear magnetic resonance (NMR) spectroscopy was used routinely to characterise purified metabolites. Dried compounds were dissolved in CDCl3 and characterised by NMR spectroscopy, which was conducted either on an Agilent VNMRS500 spectrometer (1H NMR at 500 MHz and 13C NMR at 125 MHz) or on an Agilent V400-MR spectrometer (1H NMR at 400 MHz and 13C NMR at 100 MHz). Chemical shifts were recorded in parts per million (ppm) and coupling constant (J) in Hz.Assessing antimicrobial activity of purified metabolites
The purified compounds were tested against Bacillus subtilis to study antimicrobial activity. A 1 × 109 ml−1 spore suspension of B. subtilis with 4% 2,3,5-triphenyl-2H-tetrazolium chloride (TTC) was overlaid onto TSA agar (30 g l−1 tryptic soy, 15 g l−1 agar). Purified compounds in different amounts of 100, 25, 5 and 1 μg were each released onto individual sterile 6 mm paper discs and placed on the bacterial lawn. All plates were incubated overnight at 28 °C, following which the growth inhibition zones were measured.Author contributions
GDF and AMB coordinated the project. FA, KK and SS performed the heterologous expression and characterisation of metabolites produced de novo, under the supervision of AMB and GDF. JAD performed chemical synthesis under the supervision of CLW. KK and FA drafted the manuscript with edits and contributions from AMB and GDF.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Dr Colin Lazarus is thanked for providing expression vectors for the transformation of A. oryzae; the NMR spectroscopy and MS facilities and teams of the University of Bristol are thanked for data collection and helpful discussion. KK was supported by a scholarship from Majlis Amanah Rakyat. JAD was supported by a scholarship from the EPSRC, Bristol Chemical Sciences Centre for Doctoral Training (EP/L015366/1). FA would like to acknowledge current funding from UKRI through a Future Leaders Fellowship (MR/V022334/1).References
- F. Kavanagh, A. Hervey and W. J. Robbins, Antibiotic Substances from Basidiomycetes: VIII. Pleurotus Multilus (Fr.) Sacc. and Pleurotus Passeckerianus Pilat, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 570–574 CrossRef CAS PubMed.
- A. J. Hartley, et al., Investigating pleuromutilin-producing Clitopilus species and related basidiomycetes, FEMS Microbiol. Lett., 2009, 297, 24–30 CrossRef CAS PubMed.
- C. Davidovich, et al., Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 4291–4296 CrossRef CAS.
- O. Goethe, A. Heuer, X. Ma, Z. Wang and S. B. Herzon, Antibacterial properties and clinical potential of pleuromutilins, Nat. Prod. Rep., 2019, 36, 220–247 RSC.
- S. Rittenhouse, et al., Selection of retapamulin, a novel pleuromutilin for topical use, Antimicrob. Agents Chemother., 2006, 50, 3882–3885 CrossRef CAS PubMed.
- G. G. Zhanel, et al., Lefamulin: A Novel Oral and Intravenous Pleuromutilin for the Treatment of Community-Acquired Bacterial Pneumonia, Drugs, 2021, 81, 233–256 CrossRef CAS PubMed.
- S. Thai Le, et al., The Very First Modification of Pleuromutilin and Lefamulin by Photoinitiated Radical Addition Reactions - Synthesis and Antibacterial Studies, Pharmaceutics, 2021, 13, 2028 CrossRef CAS PubMed.
- A. M. Bailey, et al., Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production, Sci. Rep., 2016, 6, 25202 CrossRef CAS.
- M. Yamane, et al., Biosynthetic Machinery of Diterpene Pleuromutilin Isolated from Basidiomycete Fungi, ChemBioChem, 2017, 18, 2317–2322 CrossRef CAS PubMed.
- F. Alberti, et al., Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives, Nat. Commun., 2017, 8, 1831 CrossRef PubMed.
- M. Xu, et al., Premutilin Synthase: Ring Rearrangement by a Class II Diterpene Cyclase, Org. Lett., 2018, 20, 1200–1202 CrossRef CAS PubMed.
- S. Okamoto, T. Taguchi, K. Ochi and K. Ichinose, Biosynthesis of Actinorhodin and Related Antibiotics: Discovery of Alternative Routes for Quinone Formation Encoded in the Act Gene Cluster, Chem. Biol., 2009, 16, 226–236 CrossRef CAS PubMed.
- H. C. Lin, et al., Elucidation of the concise biosynthetic pathway of the communesin indole alkaloids, Angew. Chem., Int. Ed., 2015, 54, 3004–3007 CrossRef CAS PubMed.
- H. Egger and H. Reinshagen, New pleuromutilin derivatives with enhanced antimicrobial activity. II. Structure-activity correlations, J. Antibiot., 1976, 29, 923–927 CrossRef CAS PubMed.
- F. Sun, et al., Unraveling the metabolic routes of retapamulin: insights into drug development of pleuromutilins, Antimicrob. Agents Chemother., 2018, 62, e02388 CAS.
- K. Zhang, et al., Discovery of a novel Pleuromutilin derivative as anti-IPF lead compound via high-throughput assay, Eur. J. Med. Chem., 2022, 241, 114643 CrossRef CAS PubMed.
- F. J. Jin, J. I. Maruyama, P. R. Juvvadi, M. Arioka and K. Kitamoto, Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae, FEMS Microbiol. Lett., 2004, 239, 79–85 CrossRef CAS.
- K. A. K. Pahirulzaman, K. Williams and C. M. Lazarus, A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae, Methods Enzymol., 2012, 517, 241–260 CAS.
- H. Ma, S. Kunes, P. J. Schatz and D. Botstein, Plasmid construction by homologous recombination in yeast, Gene, 1987, 58, 253–3623 CrossRef PubMed.
- L. M. Halo, et al., Late stage oxidations during the biosynthesis of the 2-pyridone tenellin in the entomopathogenic fungus Beauveria bassiana, J. Am. Chem. Soc., 2008, 130, 17988–17996 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc06638f |
| ‡ Authors contributed equally. |
| § Present address: School of Life Sciences, University of Warwick, Gibbet Hill Road, Coventry CV4 7AL, UK. |
| This journal is © The Royal Society of Chemistry 2023 |