Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vivo singlet state filtered nuclear magnetic resonance: towards monitoring toxic responses inside living organisms†
Daniel H.
Lysak‡
 a,
Flavio V. C.
Kock‡
ab,
Salvatore
Mamone‡
c,
Ronald
Soong
a,
Stefan
Glöggler
*c and
Andre J.
Simpson
a,
Flavio V. C.
Kock‡
ab,
Salvatore
Mamone‡
c,
Ronald
Soong
a,
Stefan
Glöggler
*c and
Andre J.
Simpson
 *c
*c
aEnvironmental NMR Centre, University of Toronto Scarborough, 1265 Military Trail, Scarborough, Ontario, Canada
bDepartment of Chemistry, Federal University of São Carlos (UFSCar), Rod. Washington Luís, Monjolinho, São Carlos–SP, 13565-905, Brazil
cNMR Signal Enhancement Group, Max Planck Institute for Multidisciplinary Sciences, Am Fassberg, 11 37077, Göttingen, Germany. E-mail: andre.simpson@utoronto.ca; stefan.gloeggler@mpinat.mpg.de
First published on 9th January 2023
Abstract
In line with recent paradigm shifts in toxicity testing, in vivo nuclear magnetic resonance (NMR) is a powerful tool for studying the biological impacts and perturbations caused by toxicants in living organisms. However, despite the excellent molecular insights that can be obtained through this technique, in vivo NMR applications are hampered by considerable experimental challenges such as poor line shape and spectral overlap. Here, we demonstrate the application of singlet-filtered NMR to target specific metabolites and facilitate the study of metabolite fluxes in living Daphnia magna, an aquatic keystone species and model organism. Informed by mathematical simulations and experiments on ex vivo organisms, singlet state NMR is used to monitor the flux of metabolites such as D-glucose and serine in living D. magna, during the environmentally relevant processes of anoxic stress and reduced food availability. Overall, singlet state NMR is shown to have significant future potential for studying metabolic processes in vivo.
Introduction
Whether used to determine the structure of an unknown compound, examine the carbon distribution of a soil or study non-covalent interactions in macromolecules, nuclear magnetic resonance (NMR) spectroscopy is a powerful analytical technique with wide ranging applications. Numerous advantages, including molecular insights allowing de novo structural determination, facile quantification, unrivalled reproducibility and inherently non-destructive analysis have made NMR a pillar in the field of metabolomics.1–3 Of particular note, the non-destructive nature of NMR opens the door towards the study of organisms in vivo.4 This ability to study living organisms allows for the monitoring of metabolic changes in vivo, for example in response to changing environmental conditions such as decreased oxygen or food, presence of pollutants or varying pH, has resulted in increasing interest in this technique for the purposes of studying toxicity.5The landmark report Toxicity testing in the 21st century: a vision and a strategy, commissioned by the U.S. National Academy of Sciences, noted that traditional toxicity testing “relies primarily on apical endpoints (e.g. death, loss of movement) upon exposure to high doses of a test chemical”.6 However, these tests “provide little to no information on the toxic modes of action and sublethal toxicity – and a paradigm shift towards examining biological perturbations as opposed to apical endpoints is required”.6 When one considers that, in the environment, pollutants are rarely found in the concentrations used in acute toxicity tests,5 it becomes clear that such sub-lethal insights are invaluable for understanding toxicity in the real world, as well as developing effective environmental regulations.4
Nuclear magnetic resonance is uniquely poised to help address these existing knowledge gaps. Specifically, in vivo NMR has the ability to examine the metabolic profile of a living organism upon exposure to sublethal toxicant concentrations in real-time and even the potential to examine recovery in the same organisms, after the stressor is removed.5 However, despite the exceptional potential and considerable previous success of this technique, there are significant experimental challenges faced in vivo. Of note, the line shapes resulting from an in vivo spectrum are typically much broader than for true solutions, due to the differences in magnetic susceptibility caused by the different “compartments” of the organism.5 Thus, when combined with the inherent natural complexity of a living organism, along with the fact that lipids often dominate the 1H spectral envelope, it becomes difficult to isolate metabolite signals directly from 1H NMR.7
One solution, that has been applied for the study of small aquatic organisms in an environmental context, has been to culture organisms on a purely 13C diet and then use the increased signal to obtain heteronuclear 2D 1H–13C spectra that provide the additional spectral dispersion required to assign and monitor metabolites in vivo.7 While an elegant solution, the approach limits studies to organisms raised in the lab and is prohibitively expensive over the long term. On the other hand, highly selective NMR approaches have been introduced, that allow multiple targets inside organisms to be isolated and monitored.8 However, such approaches are challenging to implement and involve the generation of tailored waveforms that must be changed for every metabolite or metabolite combination. On the other hand, singlet state NMR provides the potential to isolate signals without the need for any selective excitation, offering a simple and robust approach for targeted in vivo monitoring.
Singlet states are effective spin 0 states that can be created between spin pairs. They received increased attention as soon as it was realized that they can persist for longer times (up to hours) compared to longitudinal magnetization states in favourable situations, depending on the molecular structure and spin network.9–11 Over time, they have been proposed as a tool for studying slow diffusion,12–14 drug binding and protein folding,15 self-assembling and stimuli-response phenomena,16–18 and storage for signal in hyperpolarization.19,20 More recently, singlet states have been proposed as quantum filters to increase the contrast of certain resonances from undesired background signals.21–24
In this work the gc-M2S2M sequence was used to bring longitudinal magnetization into the singlet state and back24 (see the ESI† for experimental details and simulations). The gc-M2S2M sequence can generate singlet states in spin pairs in any coupling regime by appropriate settings of the sequence parameters. It is insensitive to B0 inhomogeneities and pulse offsets (within the bandwidth of the pulses). The signal selectivity depends strongly on chemical shift differences and spin J-couplings, and it was observed that the sequence is very effective in suppressing signals that do not pass through the singlet state (as selected by the sequence parameters).
To illustrate the potential of singlet filtered NMR for improving contrast in vivo, we demonstrate here, to our knowledge, the first reported use of singlet filtered nuclear magnetic resonance to study metabolic changes in vivo. The gc-M2S2M sequence24 allows for selective identification of metabolites in vivo, and thus for monitoring of individual or small groups of metabolites during stress responses. Daphnia magna (water fleas) are studied, which are among the most common species for aquatic toxicity testing and are highly responsive to environmental stresses.25,26 Further, D. magna have recently been shown to be important in the transport of pollutants such as polystyrene nanoparticles through upper trophic levels, including fish that are consumed by humans.27Daphnia also provide an important link between aquatic producers such as algae and aquatic consumers such as fish27,28 and, as such, are considered an ecological keystone species.7 Studies have shown that Daphnia are effective model organisms for human health and disease,29 and have recently been listed as a National Institute of Health model organism,26 making their study of particular interest.
Results and discussion
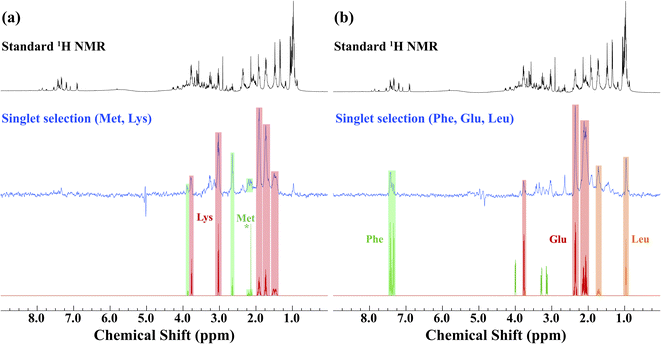
Prior to the application of the gc-M2S2M sequence to living D. magna, mathematical simulations (see ESI Section 2.1†) were used to help identify starting experimental conditions for the selection of individual metabolites, and the conditions were tested and optimized on ex vivo organisms (Fig. 1). The singlet filtering part of the sequence is defined by the following 4 parameters: the number of echoes in the single-quantum and zero-quantum block n1 and n2, the echo delay τ and the zero-quantum delay Δ. For the selected proton pairs, the efficiency f was determined by spanning the corresponding 4-dimensional parameter space, see Section 2.2 of the ESI† for details. In practice, signal losses may arise when the sequence duration becomes long compared to the relaxation mechanisms, that are more effective in vivo. Therefore, the parameters were chosen by trying to balance the theoretical transfer efficiency and the total sequence time T = 4n1τ + 2(Δ + n2τ).We identified sets of metabolites that can be selected concurrently. This can be a considerable advantage simply for the fact that it can result in significant time savings, allowing for improved throughput, and greater information content from a single experiment. If, for example, one wishes to track a process that is known to involve changes of two metabolites, it would increase confidence if both trends can be seen in the same experiment. This concept is demonstrated first on ex vivo D. magna (see Fig. 1) before moving on to living Daphnia. In this case, the sequence was modified to contain a total correlation spectroscopy (TOCSY) mixing block and a zero-spoil block. The TOCSY block allows magnetization transfer through the selected 1H–1H spin system30 which aids in identification of the selected metabolite/metabolites, while the zero quantum spoil suppresses zero quantum components, improving phase and lineshape.31 However, application of the TOCSY block comes at the cost of sensitivity and therefore it was used only for metabolite identification, but not for monitoring. Though the loss in sensitivity is dependant on the individual spin system and mixing times chosen, a sensitivity decrease on the order of 40–60% occurs using a 120 ms mixing time for D-glucose (See Fig. S4† for more details). The full pulse sequence code is provided in the ESI,† Section 3.
Starting with lysine, the singlet state filter with TOCSY pulls out all the signals. Indeed, all the protons belonging to the spin system which are coupled to the selected singlet state sub-unit become observable. Methionine, on the other hand, contains an isolated CH3 group (see * in Fig. 1). This group is not spin coupled to the remaining spins that are selected by the singlet state TOCSY filter and thus is not detected. In the case of phenylalanine, the singlet state target is in the aromatic ring which is not strongly coupled to the aliphatic side chain and thus not detected. Conversely, both leucine and glutamic acid contain fully coupled spin systems and all spins in these molecules are detected. Here, we note how singlet filtration simplifies the spectrum and allows a trustworthy metabolite identification. The standard 1H NMR spectrum shows a wide range of overlapping peaks, while the selective spectrum is heavily biased towards the desired compounds, and signals from unwanted compounds are strongly (although not completely) suppressed.
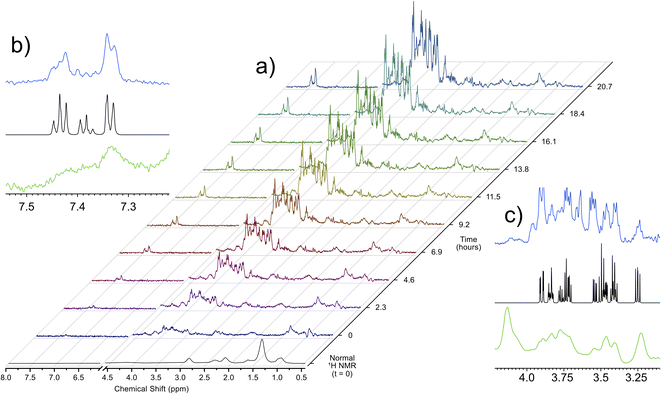
Utilizing the parameters that were optimized through simulations and validated on ex vivo samples, the singlet state sequence was applied to trace the process of anoxic stress in vivo. For a typical in vivo experiment, the organisms are sustained in a 5 mm flow system, which provides food, oxygenated water, and removes waste products. This system, described in a previous work,32 allows for D. magna to be sustained under low stress inside the spectrometer indefinitely. Turning off the flow induces anoxic stress in the organisms and Fig. 2 shows a time lapse of a singlet filtered experiment throughout this process. Fig. S3† shows expanded spectral regions demonstrating that without the singlet-filter the metabolites of interest cannot be identified. Here, the target compounds were: glucose, which has a central role in energy metabolism, and phenylalanine, an amino acid that is important as a protein building block as well as a precursor to numerous signalling molecules such as dopamine.33 As can be seen in the insets in Fig. 2, the singlet filtered spectrum shows an excellent match to the target metabolite spectrum, despite the considerable matrix effects that can influence in vivo data.5 Neither compound can be discerned in the standard 1H NMR in vivo data (first spectrum in Fig. 2a). The increase in both phenylalanine and glucose concentrations as time progresses is a result of the biological response to increasingly severe anoxia. As the oxygen concentration in the water decreases, the potential for aerobic respiration is reduced, and the organism breaks down energy storage molecules (i.e., glycogen) into glucose in order to perform anaerobic glycolysis.34 The increase in phenylalanine can be attributed to broad-scale protein breakdown, which occurs in order to provide free amino acids, that are in turn used for energy production.35,36
By comparing the intensities of the selected peaks across spectra acquired under the same conditions, it is possible to quantify metabolite changes on a relative basis. Although relative quantification can be performed quite easily, due to the variations in singlet transition efficiency for different spin systems, absolute quantification is more challenging. For absolute quantification a standard addition of the target compound would be required. This approach has been previously described in the literature for other selective NMR experiments.8Fig. 3 shows plots of the relative concentrations of glucose and serine in vivo, throughout two different environmental conditions: anoxic stress (red data points) and decreased availability of nutrients (blue data points).
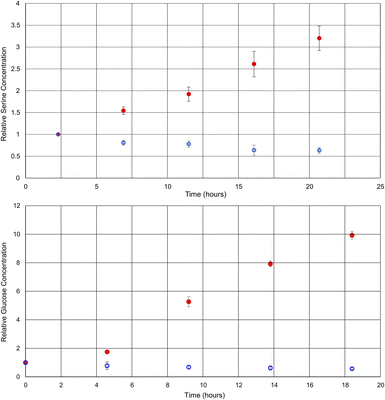 | ||
| Fig. 3 Graphs of the relative concentrations of serine and glucose through the process of anoxic stress (red) and halved food availability (blue). Each experiment was repeated in triplicate. | ||
Here, experiments focusing on serine and glucose were interleaved such that both could be monitored and repeated in triplicate. As such, the time axes for the metabolites are staggered and the temporal resolution reduced in comparison to Fig. 2. Phenylalanine was not quantified, as the initial concentration was below the limits of quantification at time zero in some of the D. magna samples. In the case of the anoxic stress experiment, the first data point is gathered with the flow system on, and it is turned off for subsequent data points. As expected, anoxia has a large impact on the metabolite concentrations.7 Glucose increases due to anaerobic glycolysis34 and serine increases due to protein breakdown to provide free amino acids for energy production35,36
In contrast, the reduction of food over a relatively short period (24 h) has been documented to have a much smaller impact,37 making this a test of more nuanced metabolite tracking. In this case, the flow system was kept on for the duration of the experiment, but the tank water was diluted in half with dechlorinated tap water. The decreased food availability caused by the halving of the algae concentration can be expected to cause a decrease in short term energy stores such as glucose, as metabolism continues, but the opportunity for replenishment is decreased. Similarly, the decrease in serine concentration may be attributed to its role as a precursor to pyruvate,38 which feeds into the Krebs cycle.36
Conclusions
In conclusion, singlet-filtered NMR has been shown, for the first time, to be able to track a metabolic response to stressors in vivo. Specifically, singlet filtered NMR allowed the monitoring of selected metabolites in vivo in both an acute (anoxic stress) and chronic (reduced food) scenario and holds promise for future studies to better understand various biological processes including growth, toxic responses and recovery inside living organisms. The selection and detection of multiple metabolites has been demonstrated as well. This technique yields significantly simplified NMR spectra with reduced overlap and has considerable potential for non-destructive in vivo metabolomics. In the future, this technique could be used to examine the effects of and recovery from exposure to environmentally relevant toxicants such as polyfluorinated compounds, pesticides or microplastics. Further, the analytes studied here are of considerable importance to human health: glucose imbalance is, of course, integral in diabetes and can be a biomarker for various types of cancer,39 phenylketonuria is caused by the inability to breakdown phenylalanine,40 and serine is a metabolic precursor to glutathione, which is responsible for defense against reactive oxygen species.41 As such, using these compounds, there are numerous additional metabolic pathways that are impactful for human health that could be studied, demonstrating considerable potential for this approach.Data availability
The pulse program and associated code are provided in the ESI.† Example data sets are available on request.Author contributions
AJS and SG supervised the project and were responsible for funding acquisition. DHL and FVCK prepared, optimized and performed the NMR experiments. SM performed the singlet state simulations. DHL, SM and FVCK contributed to formal analysis, data curation and validation. DHL contributed to data visualization and wrote the original draft. All authors contributed to conceptualization of the project and editing/review of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
AJS would like to thank the Natural Sciences and Engineering Research Council of Canada (NSERC) [Alliance (ALLRP 549399), Alliance (ALLRP 555452) and Discovery Programs (RGPIN-2019-04165)], the Canada Foundation for Innovation (CFI), the Ontario Ministry of Research and Innovation (MRI), and the Krembil Foundation for providing funding. SG would like to thank the German Research Foundation (DFG) project number 450146057 for funding.Notes and references
- A. H. Emwas, R. Roy, R. T. McKay, L. Tenori, E. Saccenti, G. A. Nagana Gowda, D. Raftery, F. Alahmari, L. Jaremko, M. Jaremko and D. S. Wishart, Metabolites, 2019, 9, 1–39 CrossRef PubMed.
- A. Smolinska, L. Blanchet, L. M. C. Buydens and S. S. Wijmenga, Anal. Chim. Acta, 2012, 750, 82–97 CrossRef CAS PubMed.
- P. G. Takis, V. Ghini, L. Tenori, P. Turano and C. Luchinat, TrAC, Trends Anal. Chem., 2019, 120, 115300 CrossRef.
- M. T. Anaraki, D. H. Lysak, K. Downey, F. V. C. Kock, X. You, R. D. Majumdar, A. Barison, L. M. Lião, A. G. Ferreira, V. Decker, B. Goerling, M. Spraul, M. Godejohann, P. A. Helm, S. Kleywegt, K. Jobst, R. Soong, M. J. Simpson and A. J. Simpson, Prog. Nucl. Magn. Reson. Spectrosc., 2021, 126–127, 121–180 CrossRef CAS PubMed.
- M. Bastawrous, A. Jenne, M. Tabatabaei Anaraki and A. Simpson, Metabolites, 2018, 8, 35 CrossRef PubMed.
- D. Krewski, D. Acosta, M. Andersen, H. Anderson, J. C. Bailar, K. Boekelheide, R. Brent, G. Charnley, V. G. Cheung, S. Green, K. T. Kelsey, N. I. Kerkvliet, A. A. Li, L. McCray, O. Meyer, R. D. Patterson, W. Pennie, R. A. Scala, G. M. Solomon, M. Stephens, J. Yager and L. Zeise, Staff of Committee on Toxicity Test, J. Toxicol. Environ. Health, Part B, 2010, 13, 51–138 CAS.
- M. T. Anaraki, D. H. Lysak, R. Soong, M. J. Simpson, M. Spraul, W. Bermel, H. Heumann, M. Gundy, H. Boenisch and A. J. Simpson, Analyst, 2020, 145, 5787–5800 RSC.
- A. Jenne, W. Bermel, C. A. Michal, O. Gruschke, R. Soong, R. Ghosh Biswas, M. Bastawrous and A. J. Simpson, Angew. Chem., Int. Ed., 2022, 61, e202110044 CrossRef CAS PubMed.
- M. Carravetta and M. H. Levitt, J. Am. Chem. Soc., 2004, 126, 6228–6229 CrossRef CAS PubMed.
- M. Carravetta, O. G. Johannessen and M. H. Levitt, Phys. Rev. Lett., 2004, 92, 1–4 CrossRef PubMed.
- M. Carravetta and M. H. Levitt, J. Chem. Phys., 2005, 122, 214505 CrossRef PubMed.
- P. Ahuja, R. Sarkar, P. R. Vasos and G. Bodenhausen, J. Am. Chem. Soc., 2009, 131, 7498–7499 CrossRef CAS PubMed.
- R. Sarkar, P. R. Vasos and G. Bodenhausen, J. Am. Chem. Soc., 2007, 129, 328–334 CrossRef CAS PubMed.
- G. Pileio, J. N. Dumez, I. A. Pop, J. T. Hill-Cousins and R. C. D. Brown, J. Magn. Reson., 2015, 252, 130–134 CrossRef CAS PubMed.
- A. Bornet, P. Ahuja, R. Sarkar, L. Fernandes, S. Hadji, S. Y. Lee, A. Haririnia, D. Fushman, G. Bodenhausen and P. R. Vasos, ChemPhysChem, 2011, 12, 2729–2734 CrossRef CAS PubMed.
- S. Mamone and S. Glöggler, Phys. Chem. Chem. Phys., 2018, 20, 22463–22467 RSC.
- P. Saul, S. Mamone and S. Glöggler, Chem. Sci., 2019, 10, 413–417 RSC.
- P. Saul, S. Yang, S. Mamone, F. Opazo, A. Meyer, S. O. Rizzoli and S. Glöggler, Phys. Chem. Chem. Phys., 2021, 23, 26349–26355 RSC.
- P. Ahuja, R. Sarkar, S. Jannin, P. R. Vasos and G. Bodenhausen, Chem. Commun., 2010, 46, 8192–8194 RSC.
- G. Pileio, S. Bowen, C. Laustsen, M. C. D. Tayler, J. T. Hill-Cousins, L. J. Brown, R. C. D. Brown, J. H. Ardenkjaer-Larsen and M. H. Levitt, J. Am. Chem. Soc., 2013, 135, 5084–5088 CrossRef CAS PubMed.
- S. J. Devience, R. L. Walsworth and M. S. Rosen, NMR Biomed., 2013, 26, 1204–1212 CrossRef CAS PubMed.
- A. N. Pravdivtsev, A. S. Kiryutin, A. V. Yurkovskaya, H. M. Vieth and K. L. Ivanov, J. Magn. Reson., 2016, 273, 56–64 CrossRef CAS PubMed.
- A. S. Kiryutin, A. N. Pravdivtsev, A. V. Yurkovskaya, H. M. Vieth and K. L. Ivanov, J. Phys. Chem. B, 2016, 120, 11978–11986 CrossRef CAS PubMed.
- S. Mamone, N. Rezaei-Ghaleh, F. Opazo, C. Griesinger and S. Glöggler, Sci. Adv., 2020, 6, 1–7 Search PubMed.
- A. Martins and L. Guilhermino, Sci. Total Environ., 2018, 631–632, 421–428 CrossRef CAS PubMed.
- J. K. Colbourne, M. E. Pfrender, D. Gilbert, W. K. Thomas, A. Tucker, T. H. Oakley, S. Tokishita, A. Aerts, G. J. Arnold, M. K. Basu, D. J. Bauer, C. E. Cáceres, L. Carmel, C. Casola, J. H. Choi, J. C. Detter, Q. Dong, S. Dusheyko, B. D. Eads, T. Fröhlich, K. A. Geiler-Samerotte, D. Gerlach, P. Hatcher, S. Jogdeo, J. Krijgsveld, E. V. Kriventseva, D. Kültz, C. Laforsch, E. Lindquist, J. Lopez, J. R. Manak, J. Muller, J. Pangilinan, R. P. Patwardhan, S. Pitluck, E. J. Pritham, A. Rechtsteiner, M. Rho, I. B. Rogozin, O. Sakarya, A. Salamov, S. Schaack, H. Shapiro, Y. Shiga, C. Skalitzky, Z. Smith, A. Souvorov, W. Sung, Z. Tang, D. Tsuchiya, H. Tu, H. Vos, M. Wang, Y. I. Wolf, H. Yamagata, T. Yamada, Y. Ye, J. R. Shaw, J. Andrews, T. J. Crease, H. Tang, S. M. Lucas, H. M. Robertson, P. Bork, E. V. Koonin, E. M. Zdobnov, I. V. Grigoriev, M. Lynch and J. L. Boore, Science, 2011, 331, 555–561 CrossRef CAS PubMed.
- A. Elizalde-Velázquez, A. M. Carcano, J. Crago, M. J. Green, S. A. Shah and J. E. Cañas-Carrell, Environ. Pollut., 2020, 259, 1–8 CrossRef PubMed.
- J. Xu, C. S. Guo, Y. Zhang and W. Meng, Environ. Pollut., 2014, 184, 254–261 CrossRef CAS PubMed.
- A. S. Edison, R. D. Hall, C. Junot, P. D. Karp, I. J. Kurland, R. Mistrik, L. K. Reed, K. Saito, R. M. Salek, C. Steinbeck, L. W. Sumner and M. R. Viant, Metabolites, 2016, 6, 1–7 CrossRef PubMed.
- J. Cavanagh and M. Rance, J. Magn. Reson., 1990, 88, 72–85 CAS.
- M. J. Thrippleton and J. Keeler, Angew. Chem., Int. Ed., 2003, 42, 3938–3941 CrossRef CAS PubMed.
- M. Tabatabaei Anaraki, R. Dutta Majumdar, N. Wagner, R. Soong, V. Kovacevic, E. J. Reiner, S. P. Bhavsar, X. Ortiz Almirall, D. Lane, M. J. Simpson, H. Heumann, S. Schmidt and A. J. Simpson, Anal. Chem., 2018, 90, 7912–7921 CrossRef CAS PubMed.
- M. D. McCoole, N. J. Atkinson, D. I. Graham, E. B. Grasser, A. L. Joselow, N. M. McCall, A. M. Welker, E. J. Wilsterman, K. N. Baer, A. R. Tilden and A. E. Christie, Comp. Biochem. Physiol., Part D: Genomics Proteomics, 2012, 7, 35–58 CAS.
- R. J. Paul, M. Colmorgen, R. Pirow, Y. H. Chen and M. C. Tsai, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 1998, 120, 519–530 CrossRef.
- A. Van Waarde, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1988, 91, 207–228 CrossRef.
- N. D. Wagner, Z. Yang, A. B. Scott and P. C. Frost, Aquat. Sci., 2017, 79, 127–137 CrossRef CAS.
- T. U. B. Filho, A. M. V. M. Soares and S. Loureiro, Environ. Sci. Pollut. Res., 2011, 18, 655–662 CrossRef.
- V. Kovacevic, A. J. Simpson and M. J. Simpson, Metabolites, 2018, 8, 34 CrossRef PubMed.
- E. A. Richter, Compr. Physiol., 1996, 912–951 Search PubMed.
- N. Blau, F. J. Van Spronsen and H. L. Levy, Lancet, 2010, 376, 1417–1427 CrossRef CAS.
- T. J. Koning, K. Snell, M. Duran, R. Berger, B.-T. Poll-The and R. Surtees, Biochem. J., 2003, 371, 653–661 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, further discussion and pulse sequence. See DOI: https://doi.org/10.1039/d2sc06624f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |