Open Access Article
Open Access ArticleSmart down/upconversion nanomachines integrated with “AND” logic computation and enzyme-free amplification for NIR-II fluorescence-assisted precise and enhanced photodynamic therapy†
Lifang
Pang‡
,
Xiaolan
Tang‡
,
Lijia
Yao
,
Liuyan
Zhou
,
Shengqiang
Hu
*,
Shulin
Zhao
 and
Liangliang
Zhang
and
Liangliang
Zhang
 *
*
State Key Laboratory for the Chemistry and Molecular Engineering of Medicinal Resources, School of Chemistry and Pharmaceutical Sciences, Guangxi Normal University, Guilin 541004, P. R. China. E-mail: zn_sqhu@163.com; liangzhang319@163.com
First published on 21st February 2023
Abstract
Upconversion nanoparticles enable indirect activation of photodynamic therapy (PDT) using near-infrared (NIR) light, providing an excellent alternative for treating deep tumors. However, conventional NIR light-triggered PDT systems suffered from low spatiotemporal accuracy and restricted therapeutic efficiency in vivo. In this work, DNA logic circuits were functionally modified on down/upconversion nanoparticles (D/UCNPs) to construct smart down/upconversion nanomachines (D/UCNMs) for NIR light-triggered PDT toward target tumors. Upon dual inputs of tumor-associated GSH and TK1 mRNA, DNA logic circuits perform “AND” logic computation and initiate the toehold-mediated strand displacement reaction. Meanwhile, the quenched upconversion fluorescence was recovered and then the approaching photosensitizers were activated, leading to in situ output of singlet oxygen (1O2) for precise and enhanced PDT. Importantly, the biodistribution of the D/UCNMs in vivo could be visualized by second near-infrared (NIR-II) fluorescence imaging via the downconversion luminance of D/UCNPs, which further contributed to performing precise PDT. This work provides new insights into the development of precise and highly efficient PDT systems.
Introduction
Upon specific light irradiation, photosensitizers are capable of producing toxic reactive oxygen species (ROS) such as singlet oxygen (1O2), which have been widely employed to kill cancer cells.1 Since most available photosensitizers simply work under the condition of visible light,2–4 upconversion nanoparticles (UCNPs) were always introduced to implement near-infrared (NIR) light-triggered indirect activation of photodynamic therapy (PDT) for treating deep tumors taking advantage of the property of converting NIR light into ultraviolet-visible light.5–7 Although much progress has been made, such a NIR light-triggered PDT system still suffers from some challenges in treating tumors in vivo. For example, nontargeted photosensitizers were mainly activated by a single tumor-associated stimulus, which is difficult to precisely control the ROS production in desired tumor sites due to nonspecific spatial distribution of the stimulus.8,9 In addition, the activation process of photosensitizers was generally carried out at the expense of consuming a large amount of low-level stimuli,10 leading to restricted therapeutic efficiency. Therefore, developing smart photosensitive systems, which could be selectively activated by multiple tumor-associated stimuli and show an amplified signal response, holds great promise for precise and highly efficient PDT of target tumors.Inspired by signal amplifiers in electronic circuits, programmable DNA circuits could be rationally designed as artificial molecular catalysts to obtain a specific amplification response.11,12 For example, the toehold-mediated strand displacement reaction (TMSD) utilizes an invader strand to recognize the toehold of a partially hybridized double-stranded (ds) DNA to direct branch migration (i.e., the first TMSD), followed by the release of the incumbent strand due to the additional DNA-mediated second TMSD.13,14 Such TMSD-based DNA circuits have been widely applied for enzyme-free signal amplification. Since the activation of TMSD requires at least two kinds of stimuli, the enzyme-free TMSD provided a promising alternative for DNA circuits to construct multiple stimuli-dependent logic gates.15,16 Consequently, the construction of TMSD-based DNA logic circuits, allowing logic computation and signal amplification, holds great potential in precise and efficient PDT.
Herein, smart down/upconversion nanomachines (D/UCNMs) were constructed by modifying photosensitizer-engineered DNA logic circuits on down/upconversion nanoparticles (D/UCNPs) to achieve conditionally activated and enhanced PDT for treating target tumors. By rationally designing DNA logical circuits, tumor-associated GSH and TK1 mRNA could serve as endogenous dual-gated inputs to perform “AND” logic computation and to initiate the enzyme-free TMSD. Meanwhile, the quenched upconversion luminance of D/UCNPs was recovered and then NIR-triggered PDT systems were activated with high spatiotemporal accuracy and enhanced efficiency. Additionally, the downconversion fluorescence of D/UCNPs in the second near-infrared (NIR-II) window was applied for visually positioning smart D/UCNMs in vivo.
Results and discussion
Principle of smart D/UCNMs for precise and enhanced PDT
Considering that both GSH and TK1 mRNA are over-expressed in some specific tumors,17,18 a smart DNA circuit is rationally designed using a redox-cleavage dsDNA probe to activate a TMSD-related dsDNA probe with the aid of TK1 mRNA, which integrated the functions of “AND” logic computation and enzyme-free amplification (Fig. 1). To achieve conditional activation and enhanced efficiency of NIR light-triggered PDT, the designed dsDNA probes in DNA circuits were selectively labeled with photochlorin e6 (Ce6) and black hole quencher 3 (BHQ3), respectively, followed by assembling on D/UCNPs to prepare smart D/UCNMs. In order to obtain low background PDT efficiency, the original modification site of BHQ3 was in close proximity to the D/UCNP surface, while the modified Ce6 photosensitizer agent was far away from D/UCNP. | ||
| Fig. 1 Schematic illustration of the preparation and application of smart D/UCNMs for precise and enhanced PDT of target tumors. | ||
Upon internalization of smart D/UCNMs into target cancer cells, the endogenous TK1 mRNA could recognize the first toehold of the anchoring strand (TP) of a dsDNA probe and displace the assistant strand (AP), leading to the exposure of another toehold in the middle domain. Meanwhile, the Ce6-labeled strand (Ce6-DNA) was released from another partially complementary dsDNA probe since the hybridized SS-DNA containing one disulfide bond (S–S) could be cleaved by endogenous GSH via the thiol–disulfide exchange reaction.19 Subsequently, the released Ce6-DNA recognized the generated toehold in the middle domain and initiated a branch migration process. In this case, the Ce6-DNA approached the surface of D/UCNPs, along with the liberation of both the BHQ3-labeled protection strand (BHQ3-PP) and the initial TK1 mRNA. Accordingly, the quenched upconversion emission of D/UCNPs at 660 nm by BHQ3 via Förster resonance energy transfer (FRET) was recovered and then activated the photosensitizer Ce6, giving rise to logically activatable generation of 1O2. Besides, the released TK1 mRNA triggered the next TMSD with recycling cascade amplification. The TMSD process was conducted at the expense of high concentration of GSH (1.0–10.0 mM) instead of low-level TK1 mRNA,20 which guaranteed an enhanced PDT efficiency. Therefore, the integration of “AND” logic computation and TMSD endowed smart D/UCNMs with high accuracy and enhanced efficiency for PDT of target tumors. Furthermore, the biodistribution of smart D/UCNMs could be visually positioned by NIR-II fluorescence imaging using the downconversion luminescence of D/UCNPs at 1550 nm under irradiation with a 980 nm laser, which further assisted to perform precise PDT.
Construction of smart D/UCNMs
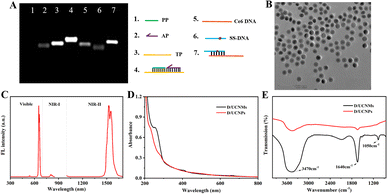
First, the preparation of two kinds of double-stranded (ds) DNA probes was demonstrated using agarose gel electrophoresis (AGE) analysis. As shown in Fig. 2A, only one band in lanes 1–3 verified a pure single-stranded structure for all the PP, AP, and TP strands (see Table S1† for the sequence). A new band appeared with a much lower migration rate in lane 4, suggesting the formation of a stable dsDNA probe from the hybridization of AP, TP, and PP strands. The hybridization of SS-DNA (lane 5) with Ce6-DNA (lane 6) formed another stable dsDNA probe, evidenced by a much brighter band with a lower migration rate (lane 7). Consequently, DNA circuits could be acquired simply by mixing the obtained two kinds of dsDNA probes. However, it is difficult to verify the logic response of DNA circuits toward TK1 mRNA and GSH using the AGE assay, because of a comparable migration rate before and after the activation of DNA circuits with dual inputs of TK1 mRNA and GSH (data not shown).Then, carboxylated D/UCNPs were introduced to prepare smart D/UCNMs. As observed from the transmission electron microscopy (TEM) image (Fig. 2B), the used NaErF4@NaYF4 D/UCNPs possess a spherical core–shell structure with an average diameter of about 28.7 nm. Upon irradiation with a 980 nm laser, the D/UCNPs emitted dual fluorescence (Fig. 2C), in which the upconversion emission wavelength was at 660 nm and the downconversion emission wavelength was located in the NIR-II region (1550 nm). The carboxyl group in the surface of D/UCNPs allowed further covalent binding of amino-substituted dsDNA probes. With the aid of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), smart D/UCNMs were successfully constructed using a cross-linking reaction, as judged by the appearance of a typical absorbance peak of DNA at 260 nm (black curve, Fig. 2D). Since dsDNA probes in DNA circuits were negatively charged, a lower potential was observed for smart D/UCNMs relative to that of D/UCNPs (Fig. S1†). In addition, the newly generated in-plane bending vibration of the carbonamide bond confirmed the successful construction of smart D/UCNMs, since two new bands at around 1640 cm−1 and 1050 cm−1 appeared except for the characteristic stretching vibrations of the –OH bond at 3470 cm−1 in Fourier transform infrared (FTIR) characterization (Fig. 2E).21
“AND” logically gated and enhanced PDT efficiency
The feasibility of a TMSD-based “AND” logic gate on smart D/UCNMs was first investigated using upconversion fluorescence at 660 nm (Fig. 3A). To this end, dsDNA probes in DNA circuits were labelled with BHQ3 but without the Ce6 photosensitizer, and the significantly enhanced upconversion fluorescence of D/UCNMs was defined as the computation output of the “1” state and the unchanged fluorescence intensity as the “0” state. After exposure to a 980 nm laser, low upconversion luminance of D/UCNMs was attained due to the FRET between D/UCNPs and BHQ3 (black curve, Fig. S2†). Single input of TK1 mRNA (1, 0) or GSH (0, 1) could not trigger TMSD, leading to logical “0” output (pale brown and pale blue columns, Fig. 3B). With the dual inputs of both TK1 mRNA and GSH (1, 1), logical “1” output was attained (red column, Fig. 3B), indicating that the TMSD was initiated and a large number of BHQ3-PP strands detached from the surface of D/UCNPs. Furthermore, the increased fluorescence intensity was positively proportional to the added amount of TK1 mRNA in the presence of 7 mM GSH (Fig. S3A†), which further verified the possibility of the TMSD-based “AND” logic gate on smart D/UCNMs using TK1 mRNA and GSH dual-responsive DNA circuits. Importantly, the limit of detection for TK1 mRNA analysis was calculated to be 0.5 nM (Fig. S3B†), which proved the advantage of TMSD-dependent signal amplification. Since the fluorescence output was determined by the simultaneous inputs of GSH and TK1 mRNA, the proposed “AND” logic gate strategy showed an excellent selectivity (Fig. 3C), as evidenced by the comparable F/F0 value for the samples with single or dual inputs of other common biomolecules to that of the blank group (F0 and F represent the upconversion fluorescence intensity of D/UCNPs before and after different treatments).Since the upconversion emission spectrum of D/UCNPs matched well with the absorption peaks of both the photosensitizer Ce6 and BHQ3, respectively (Fig. 3D), the upconversion emission of D/UCNPs could be quenched by BHQ3 and could activate the photosensitizer Ce6, which served as an excellent candidate to tune PDT efficiency through the TMSD-based DNA computation strategy. Using 2,2,6,6-tetramethylpiperidine (TEMP) as the spin trap of 1O2, the generated 1O2 could be quantified by measuring the formed paramagnetic products using electron paramagnetic resonance (EPR) spectroscopy.22 As expected, the quenched upconversion fluorescence of D/UCNPs by BHQ3 was unfavourable for photocontrollable generation of 1O2 (black curve, Fig. 3E). Only in the presence of both TK1 mRNA and GSH, indirect activation of the photosensitizer Ce6 by 980 nm light was able to produce 1O2 (red curve, Fig. 3E). Furthermore, the absorbance intensity of 1,3-diphenylisobenzofuran (DPBF) decreased with the irradiation time (Figure 3F),23 which demonstrated that such a TMSD-based “AND” logically gated PDT possesses high spatiotemporal resolutions. Consequently, dual inputs of GSH and TK1 mRNA into smart D/UCNMs yielded toxic 1O2 and the corresponding truth table for “AND” logically gated PDT could be summarized in Fig. S4.†
Photodynamic therapeutic effect in living cells
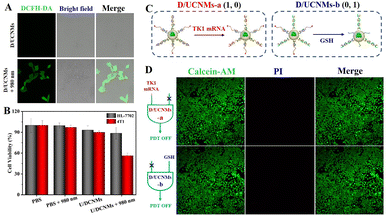
The possibility of TMSD-based “AND” logically gated therapeutic efficiency in vitro was evaluated by incubating smart D/UCNMs with 4T1 breast tumor cells overnight. Since the photogenerated 1O2 was one of reactive oxygen species, intracellular 1O2 generation was verified using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) staining assay.24–26 Bright green fluorescence of 2′,7′-dichlorofluorescein (DCF) was observed for the sample treated with D/UCNMs and 980 nm laser irradiation, implying the generation of 1O2 by the uptake nanomachines (bottom panel, Fig. 4A). In contrast, the sample treated with smart D/UCNMs (without photoirradiation) exerted a negligible generation efficiency (upper panel, Fig. 4A). Then, quantitative analysis of therapeutic efficiency was performed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 4B).27 The cell viability remained over 90% for both 4T1 cells and HL-7702 normal liver cells after treatment with smart D/UCNMs, demonstrating the excellent biocompatibility of smart D/UCNMs. After exposure to a 980 nm laser, the cell viability of smart D/UCNM-treated 4T1 cells decreased to be 56%, whereas almost all the HL-7702 cells were visually alive in the dead/live staining test (Fig. S5†), which verified the great potential of smart D/UCNMs for tumor PDT with high specificity. The different therapeutic efficiency might be ascribed to the difference in the operation of the “AND” logical gate in 4T1 and HL-7702 cells because of their different expression levels in GSH and TK1 mRNA.To further confirm that the intracellular TMSD-based “AND” logical computation was responsible for the therapeutic efficiency, the cellular uptake efficiency of smart D/UCNMs was compared between 4T1 and HL-7702 cells. Taking advantage of the luminance at 660 nm of smart D/UCNMs (without BHQ3 and Ce6 modification), their cellular delivery could be visually confirmed with an almost comparable uptake level for both two cell lines (Fig. S6†). Considering that the introduction of additional agents to regulate the intracellular content of TK1 mRNA and GSH may cause an unpredictable damage to cancer cells, two kinds of control D/UCNMs-a and D/UCNMs-b were then prepared to further validate the logically gated PDT in cancer cells. Since the used dsDNA probes in DNA circuits for preparing D/UCNMs were designed without a disulfide bond or with a blocked recognition toehold for TK1 mRNA, the prepared D/UCNMs-a and D/UCNMs-b could selectively respond to TK1 mRNA (1, 0) and GSH (0, 1), respectively (Fig. 4C). After treatment with D/UCNMs-a, all 4T1 cells remained alive, since Ce6-DNA could not be effectively released to activate the subsequent detachment of BHQ3-DNA (the upper panel, Fig. 4D). For the case treated with D/UCNMs-b, the TMSD process could not be activated in spite of the presence of free Ce6-DNA (the bottom panel, Fig. 4D), leading to negligible cytotoxicity toward 4T1 cells. These results suggested that intracellular 1O2 could not be effectively generated for the 4T1 cells after treatment with D/UCNMs-a and D/UCNMs-b, which further confirmed that GSH and TK1 mRNA dual-input activatable D/UCNMs allowed specific and enhanced tumor treatment via the TMSD-based “AND” logically gated PDT.
NIR-II fluorescence imaging-assisted PDT in vivo
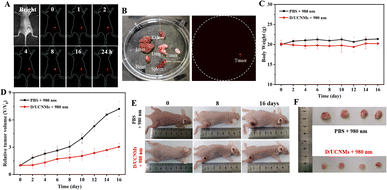
Inspired by the excellent PDT efficiency in vitro, smart D/UCNMs were attempted for in vivo anticancer treatment using 4T1 tumor bearing BALB/c mice as an animal model. Prior to in vivo experiments, a hemolysis test was carried out to examine the blood compatibility of smart D/UCNMs with a concentration ranging from 20 μM to 80 μM. As indicated in Fig. S7,† a low hemolysis level was maintained for that treated with 80 μM D/UCNMs, which reflected an excellent biocompatibility of smart D/UCNMs.Since both GSH and TK1 mRNA displayed little effect on the downconversion fluorescence of D/UCNPs (Fig. S8†), smart D/UCNMs were then intratumorally administered to 4T1 tumor bearing mice to assess the biodistribution of smart D/UCNMs in vivo, taking advantage of low background autofluorescence and light scattering of the NIR-II downconversion fluorescence.28–30 At 24 h post-injection, smart D/UCNMs still kept a high accumulation rate in the tumor region via monitoring the time-dependent downconversion fluorescence of D/UCNPs at 1550 nm, which might be associated with the enhanced permeability and retention (EPR) effect (Fig. 5A). In order to further explore the biodistribution of smart D/UCNMs, the major organs (heart, liver, spleen, lung, and kidney) and tumor extracted from sacrificed mice after 24 h treatments were collected to conduct ex vivo NIR-II fluorescence imaging (Fig. 5B). Only the tumor tissue exhibited obvious fluorescence, revealing good retention properties of the smart D/UCNMs at tumor sites.
Taking advantage of NIR-II fluorescence imaging-assisted positioning of smart D/UCNMs, in vivo antitumor treatment using smart D/UCNMs was executed with two groups of 4T1 tumor bearing mice, in which a saline-treated mouse served as the control group and the other mice were injected with smart D/UCNMs. For each mouse, the tumor was irradiated with a 980 nm laser for 20 min at 2 h post-injection every other day. Afterwards, the body weight and tumor volume were documented. During 16 days of treatment, the body weight in both groups remained in a normal fluctuated range (Fig. 5C). Furthermore, the hematoxylin & eosin (H&E) staining assay proved that 16 day treatment caused no distinct damage to the normal tissues (Fig. S9†). The little side effect of smart D/UCNMs could be explained by high accumulation of smart D/UCNMs at tumor sites, as well as “AND” logically gated PDT efficiency. As shown in Fig. 5D, a gradual increase in the tumor volume was anticipated for PBS-treated mice (the black profile), while the tumor growth in the smart D/UCNMs-treated mouse was substantially suppressed (the red profile). After visually comparing the tumor size both in vivo (Fig. 5E) and ex vivo (Fig. 5F), smart D/UCNMs were further demonstrated with outstanding phototherapeutic effects.
Conclusions
In summary, smart D/UCNMs were developed for activatable PDT of target tumors in response to multiple tumor-associated stimuli. Based on the TMSD, DNA circuits effectively integrated the functions of logic computation and signal amplification, which enabled high spatiotemporal accuracy and high therapeutic efficiency. Taking advantage of the down/upconversion fluorescence of D/UCNPs, the constructed nanomachines have been successfully employed for NIR-triggered PDT of target tumors in vivo. This study not only provides a novel design of DNA circuits for cellular logic operation and enzyme-free signal amplification, but also inspires smart integration of different functions for biological and biomedical applications.Data availability
Data are available from the corresponding author upon request.Author contributions
S. Hu and L. Zhang conceptualized the project. L. Pang and X. Tang performed the main experiments and analyzed the results. L. Yao assisted with cell imaging. L. Zhou assisted with animal experiments and discussion. S. Zhao contributed to the design of the study and the analysis of the results.Ethical statement
All related experiments were performed in compliance with the relevant laws or guidelines of China and approved by the Animal Ethical Committee of Guangxi Normal University.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21964005), Natural Science Foundation of Guangxi Province (No. 2019GXNSFFA245006 and 2019GXNSFBA245035), and Guangxi Provincial Science and Technology Bases and Special Fund for Talented Persons (No. GUIKEAD20159072).References
- T. C. Pham, V.-N. Nguyen, Y. Choi, S. Lee and J. Yoon, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS.
- J. Xie, Y. Wang, W. Choi, P. Jangili, Y. Ge, Y. Xu, J. Kang, L. Liu, B. Zhang, Z. Xie, J. He, N. Xie, G. Nie, H. Zhang and J. S. Kim, Chem. Soc. Rev., 2021, 50, 9152–9201 RSC.
- T. Luo, G. T. Nash, Z. Xu, X. Jiang, J. Liu and W. Lin, J. Am. Chem. Soc., 2021, 143, 13519–13524 CrossRef CAS PubMed.
- N. M. Idris, M. K. Gnanasammandhan, J. Zhang, P. C. Ho, R. Mahendran and Y. Zhang, Nat. Med., 2012, 18, 1580–1585 CrossRef CAS PubMed.
- C. Liu, B. Liu, J. Zhao, Z. Di, D. Chen, Z. Gu, L. Li and Y. Zhao, Angew. Chem., Int. Ed., 2020, 59, 2634–2638 CrossRef CAS PubMed.
- D. B. L. Teh, A. Bansal, C. Chai, T. B. Toh, R. A. J. Tucker, G. G. L. Gammad, Y. Yeo, Z. Lei, X. Zheng, F. Yang, J. S. Ho, N. Bolem, B. C. Wu, M. K. Gnanasammandhan, L. Hooi, G. S. Dawe, C. Libedinsky, W.-Y. Ong, B. Halliwell, E. K.-H. Chow, K.-L. Lim, Y. Zhang and B. K. Kennedy, Adv. Mater., 2020, 32, 2001459 CrossRef CAS PubMed.
- F. Yu, Y. Shao, X. Chai, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2022, 61, e202203238 CAS.
- F. Li, Y. Du, J. Liu, H. Sun, J. Wang, R. Li, D. Kim, T. Hyeon and D. Ling, Adv. Mater., 2018, 30, 1802808 CrossRef.
- Z. Yu, Y. Ge, Q. Sun, W. Pan, X. Wan, N. Li and B. Tang, Chem. Sci., 2018, 9, 3563–3569 RSC.
- M. Zou, Y. Zhao, B. Ding, F. Jiang, Y. Chen, P. A. Ma and J. Lin, Inorg. Chem. Front., 2021, 8, 2624–2633 RSC.
- J. Chen, S. Fu, C. Zhang, H. Liu and X. Su, Small, 2022, 18, 2108008 CrossRef CAS PubMed.
- W. E. Arter, Y. Yusim, Q. Peter, C. G. Taylor, D. Klenerman, U. F. Keyser and T. P. J. Knowles, ACS Nano, 2020, 14, 5763–5771 CrossRef CAS PubMed.
- H. Zhou, J. Zhang, B. Li, J. Liu, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2021, 93, 6120–6127 CrossRef CAS PubMed.
- W. Liu, A. Chen, S. Li, K. Peng, Y. Chai and R. Yuan, Anal. Chem., 2019, 91, 1516–1523 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- W. Li, Y. Yang, H. Yan and Y. Liu, Nano Lett., 2013, 13, 2980–2988 CrossRef CAS PubMed.
- S. Wang, L. Zhang, J. Zhao, M. He, Y. Huang and S. Zhao, Sci. Adv., 2021, 7, eabe3588 CrossRef CAS PubMed.
- S. Yu, Y. Zhou, Y. Sun, S. Wu, T. Xu, Y.-C. Chang, S. Bi, L.-P. Jiang and J.-J. Zhu, Angew. Chem., Int. Ed., 2021, 60, 5948–5958 CrossRef CAS PubMed.
- F. Kong, Z. Liang, D. Luan, X. Liu, K. Xu and B. Tang, Anal. Chem., 2016, 88, 6450–6456 CrossRef CAS.
- S. Hu, L. Huang, L. Zhou, T. Wu, S. Zhao and L. Zhang, Anal. Chem., 2023, 95, 3830–3839 CrossRef CAS PubMed.
- Y. Liu, X. Shen, H. Zhou, Y. Wang and L. Deng, Appl. Surf. Sci., 2016, 370, 270–278 CrossRef CAS.
- L. Gao, R. Liu, F. Gao, Y. Wang, X. Jiang and X. Gao, ACS Nano, 2014, 8, 7260–7271 CrossRef CAS PubMed.
- H. J. Halpern, S. Pou, M. Peric, C. Yu, E. Barth and G. M. Rosen, J. Am. Chem. Soc., 1993, 115, 218–223 CrossRef CAS.
- S. Hu, B. Ye, H. Tang, F. Wu, X. Yi, T. Yi, D. Wu, L. Wu and J. Wang, J. Mater. Chem. B, 2018, 6, 1187–1194 RSC.
- X. Liu, Y. Liu, X. Li, J. Huang, X. Guo, J. Zhang, Z. Luo, Y. Shi, M. Jiang, B. Qin, Y. Du, L. Luo and J. You, ACS Nano, 2022, 16, 9240–9253 CrossRef CAS PubMed.
- B. Sun, X. Guo, M. Feng, S. Cao, H. Yang, H. Wu, M. H. M. E. van Stevendaal, R. A. J. F. Oerlemans, J. Liang, Y. Ouyang and J. C. M. van Hest, Angew. Chem., Int. Ed., 2022, 61, e202208732 CAS.
- Z. Mao, Z. Liu, L. Chen, J. Yang, B. Zhao, Y. M. Jung, X. Wang and C. Zhao, Anal. Chem., 2013, 85, 7361–7368 CrossRef CAS PubMed.
- P. Pei, Y. Chen, C. Sun, Y. Fan, Y. Yang, X. Liu, L. Lu, M. Zhao, H. Zhang, D. Zhao, X. Liu and F. Zhang, Nat. Nanotechnol., 2021, 16, 1011–1018 CrossRef CAS PubMed.
- Y. Fan, P. Wang, Y. Lu, R. Wang, L. Zhou, X. Zheng, X. Li, J. A. Piper and F. Zhang, Nat. Nanotechnol., 2018, 13, 941–946 CrossRef CAS PubMed.
- M. Zhao, B. Li, Y. Wu, H. He, X. Zhu, H. Zhang, C. Dou, L. Feng, Y. Fan and F. Zhang, Adv. Mater., 2020, 32, 2001172 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc06601g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |