Open Access Article
Open Access ArticleFunctional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4′-di-O-glycosyltransferase from Ziziphus jujuba var. spinosa†
Zi-Long
Wang‡
 a,
Wanqing
Wei‡
b,
Hai-Dong
Wang‡
a,
Jia-Jing
Zhou
a,
Hao-Tian
Wang
a,
Kuan
Chen
a,
Wanqing
Wei‡
b,
Hai-Dong
Wang‡
a,
Jia-Jing
Zhou
a,
Hao-Tian
Wang
a,
Kuan
Chen
 a,
Rong-Shen
Wang
a,
Fu-Dong
Li
c,
Xue
Qiao
a,
Rong-Shen
Wang
a,
Fu-Dong
Li
c,
Xue
Qiao
 a,
Huan
Zhou
*d,
Yong
Liang
a,
Huan
Zhou
*d,
Yong
Liang
 *e and
Min
Ye
*e and
Min
Ye
 *a
*a
aState Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, 38 Xueyuan Road, Beijing 100191, China. E-mail: yemin@bjmu.edu.cn
bState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi 214122, China
cHefei National Laboratory for Physical Science at Microscale, School of Life Sciences, University of Science and Technology of China, Hefei 230036, China
dShanghai Synchrotron Radiation Facility, Shanghai Advanced Research Institute, Chinese Academy of Sciences, 239 Zhangheng Road, Pudong District, Shanghai 201204, China. E-mail: zhouhuan@sari.ac.cn
eState Key Laboratory of Coordination Chemistry, Jiangsu Key Laboratory of Advanced Organic Materials, Chemistry and Biomedicine Innovation Centre (ChemBIC), School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: yongliang@nju.edu.cn
First published on 29th March 2023
Abstract
A highly efficient and promiscuous 7,4′-di-O-glycosyltransferase ZjOGT3 was discovered from the medicinal plant Ziziphus jujuba var. spinosa. ZjOGT3 could sequentially catalyse 4′- and 7-O-glycosylation of flavones to produce 7,4′-di-O-glycosides with obvious regio-selectivity. For 7,4′-dihydroxyl flavanones and 3-O-glycosylated 7,4′-dihydroxyl flavones, ZjOGT3 selectively catalyses 7-O-glycosylation. The crystal structure of ZjOGT3 was solved. Structural analysis, DFT calculations, MD simulations, and site-directed mutagenesis reveal that the regio-selectivity is mainly controlled by the enzyme microenvironment for 7,4′-dihydroxyl flavones and 3-O-glycosylated 7,4′-dihydroxyl flavones. For 7,4′-dihydroxyl flavanones, the selectivity is mainly controlled by intrinsic reactivity. ZjOGT3 is the first plant flavonoid 7,4′-di-O-glycosyltransferase with a crystal structure. This work could help understand the catalytic mechanisms of multi-site glycosyltransferases and provides an efficient approach to synthesise O-glycosides with medicinal potential.
Introduction
Glycosylation is an important post-modification biosynthetic reaction in plant secondary metabolism.1 This type of reaction is generally catalyzed by uridine diphosphate (UDP)-dependent glycosyltransferases (UGTs).2 Thus far, a huge family of glycosyltransferases (GTs) has been discovered from plants.3 They provide powerful tools to prepare important bioactive glycosides, including schaftoside,4 stevioside,5 glycyrrhizin,6 and ginsenosides.7 Glycosylation of polyhydroxyphenolics by chemical synthesis is usually limited by poor regio- and stereoselectivity and protection and deprotection of unstable hydroxyl groups under severe conditions.8 In contrast, GT-mediated glycosylation reactions take only one step and exhibit high catalytic efficiency.9 However, many GTs suffer from poor selectivity, particularly regio-selectivity when multiple glycosylation sites are present.3c,d,10Flavonoids represent a large class of polyphenols and are widely distributed in plants.11 They possess a variety of bioactivities, including anti-inflammatory, antioxidative, anti-viral, antitumor, hepatoprotective, and cardio-cerebrovascular protective activities.12 For example, diosmin and scutellarin are used as clinical drugs to treat lymphatic insufficiency and brain stroke in China, respectively. Flavonoids also contribute to plant tastes13 and flower pigments.14
Thus far, more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 flavonoids have been reported.11,15 Due to the common biosynthetic pathway, a large proportion of flavonoids possess two conserved hydroxyl groups at 7-OH and 4′-OH.16 Although many GTs have been reported to catalyse 7- and 4′-O-glycosylation reactions, very few of them show high catalytic efficiency and high regio-selectivity.17 F4′GT and F4′G7GT from Nemophila menziesii could catalyse stepwise 7,4′-O-glycosylation to form flavonoid 4′-O-glycosides and 7,4′-di-O-glycosides, respectively.18 However, they could only accept a limited range of substrates.
000 flavonoids have been reported.11,15 Due to the common biosynthetic pathway, a large proportion of flavonoids possess two conserved hydroxyl groups at 7-OH and 4′-OH.16 Although many GTs have been reported to catalyse 7- and 4′-O-glycosylation reactions, very few of them show high catalytic efficiency and high regio-selectivity.17 F4′GT and F4′G7GT from Nemophila menziesii could catalyse stepwise 7,4′-O-glycosylation to form flavonoid 4′-O-glycosides and 7,4′-di-O-glycosides, respectively.18 However, they could only accept a limited range of substrates.
Wild jujube (Ziziphus jujuba var. spinosa, Rhamnaceae family) is a medicinal plant native to China. Its seeds have been used as the traditional Chinese medicine Suan-Zao-Ren for a long time to treat anxiety and depression.19 This plant contains abundant flavonoid 4′-O-glycosides.
In this work, we report a regio-selective flavonoid 7,4′-di-O-glycosyltransferase ZjOGT3 (UGT84A68) from Z. Jujuba var. spinosa. Mechanisms for its regio-selectivity were dissected by crystal structure analysis, density functional theory (DFT) calculations, molecular docking, molecular dynamics (MD) simulation, binding free energy calculations, and site-directed mutagenesis.
Results and discussion
Molecular cloning and functional characterization of ZjOGT3
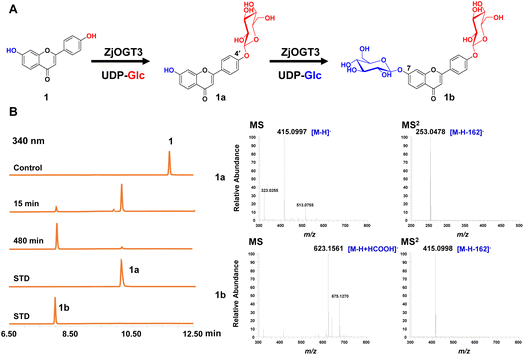
Six transcriptome data sets of Ziziphus jujuba var. spinosa (SRR9721936, SRR9721937, SRR9721938, SRR9721939, SRR9721941, and SRR9721944) were downloaded from the Sequence Read Archive (SRA) database of NCBI, and clean reads were de novo assembled using the Trinity program (K-mer = 31) with default parameters. Several reported UDP-glycosyltransferase genes were selected as query sequences (Table S1†). By using local BLAST search, 24 putative full-length GT genes (ZjOGT1-24) were discovered.20 After heterologous expression and functional characterization, ZjOGT3 was identified as a flavone 7,4′-di-O-glycosyltransferase. The cDNA sequence of ZjOGT3 contains an open reading frame (ORF) of 1440 bp encoding 479 amino acids (Table S2 and Fig. S1†). It was named UGT84A68 (accession number: OQ603459) by the UGT Nomenclature Committee.The catalytic function of ZjOGT3 was characterized using kumatakenin B (1, 7,4′-dihydroxy flavone) as a sugar acceptor and uridine 5′-diphosphate glucose (UDP-Glc) as a sugar donor. The enzymatic reaction system contained 25 μg purified protein, 0.1 mM substrate (1), and 0.5 mM UDP-Glc in 100 μL of 50 mM NaH2PO4–Na2HPO4 buffer (pH 8.0). After co-incubation at 37 °C for 8 h, the products were analyzed by liquid chromatography coupled with mass spectrometry (LC/MS) (Table S3†). ZjOGT3 could completely convert 1 into a more polar product 1b (Fig. 1A and B). The mass spectrum of 1b showed an [M-H+HCOOH]− ion at m/z 623, which was 370 amu greater (2Glc + HCOOH) than 1, indicating 1b as a di-O-glucoside. The structure of 1b was fully identified as 7,4′-di-O-glycoside flavone by comparing with an authentic reference standard.10a However, after 15 min of co-incubation, 1 was converted into 1a as the main product, which was identified as kumatakenin B 4′-O-glucoside (Fig. 1B and S2†). These results indicated that ZjOGT3 could catalyse glucosylation of 1 at 4′- and 7-OH consecutively. Kumatakenin B 7-O-glucoside was never observed as a main product.
The biochemical properties of recombinant ZjOGT3 were further investigated using 1 and UDP-Glc as substrates. ZjOGT3 showed its maximum activity at pH 8.0 (50 mM NaH2PO4–Na2HPO4 buffer) and 45 °C and was independent of divalent cations (Fig. S3†). The apparent kinetic parameters for 1 and 1a with saturated UDP-Glc were measured (Fig. S4†). The Km value for 1 and 1a was 15.57 μM and 197.70 μM, respectively. The catalytic efficiency (kcat/Km) for the 4′-O-glycosylation of 1 (0.010462 μM−1 s−1) was around 53 times higher than that for 7-O-glycosylation of 1a (0.0002014 μM−1 s−1). Thus, the second step was rate-limiting to form 1b.
Substrate promiscuity and synthetic applicability of ZjOGT3
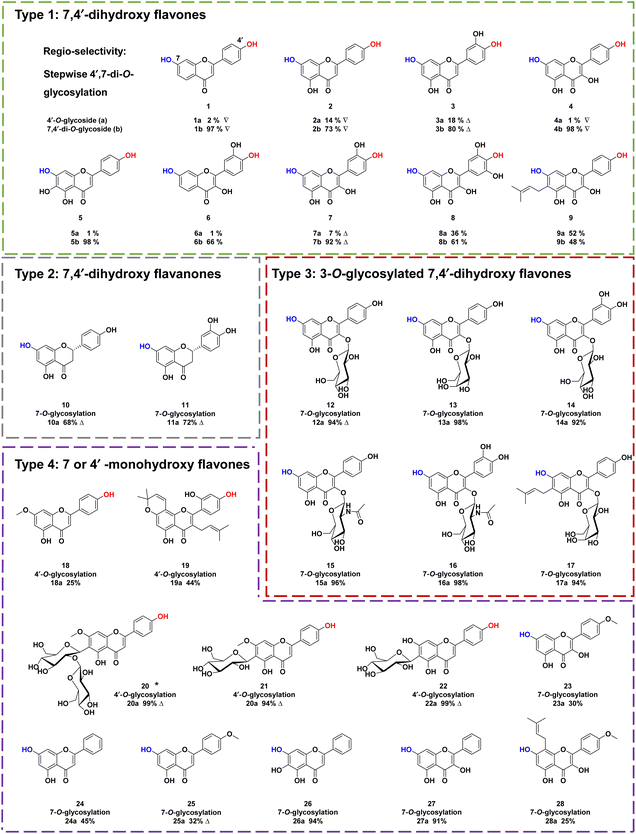
To explore the substrate specificity of ZjOGT3, 34 flavonoids were screened by enzymatic assay using UDP-Glc as the sugar donor (Fig. 2). These substrates included 7,4′-dihydroxy flavones (1–9), 7,4′-dihydroxy flavanones (10–11), 3-O-glycosylated 7,4′-dihydroxy flavones (12–17), 7 or 4′ monohydroxy flavones (18–28), isoflavones (29–31), isoflavans (32), isoflavone glycoside (33), and chalone (34). LC/MS analysis revealed that ZjOGT3 could catalyse glucosylation of all 34 substrates. The glucosylated products were characterized by the diagnostic fragment ions [M-H-162]− in the MS/MS spectra (Fig. S5–S38†). Some of the products were purified and fully identified by NMR spectroscopy or by comparing with reference standards.Interestingly, ZjOGT3 could efficiently glycosylate 1–9 (Type 1) to generate one mono-O-glycoside and one di-O-glycoside. The di-O-glycoside was usually the major product (Fig. S5–S13†). Furthermore, we fully identified 1a, 2a, 3a, 4a and 7a as 4′-mono-O-glycosides and 1b, 2b, 3b, 4b and 7b as 7,4′-di-O-glycosides by NMR spectroscopic analyses and standards (Fig. S39–S53†). For 10–11 which are flavanones with 7- and 4′-OH (Type 2), ZjOGT3 could only produce 7-mono-O-glycosides (10a and 11a) (Fig. S14–S21 and S54–S60†). Similarly, for 3-O-glycosylated 7,4′-dihydroxy flavones with 7- and 4′-OH (12–17, Type 3), ZjOGT3 also could only produce 7-O-glycosides, and the structure of 12a was fully identified (Fig. S61–S64†). For Type 4 flavonoids which contain either one 7-OH or one 4′-OH (18–28), ZjOGT3 could produce the corresponding 7 or 4′-O-glucosides (Fig. S22–S32 and S65–S79†). Moreover, ZjOGT3 could also accept isoflavones, isoflavans, isoflavone glycosides and chalcones (29–34) as substrates (Fig. S33–S38†). The above results indicate that the C2–C3 double bond and 3-O-glucosyl substitution could remarkably alter the regio-selectivity of ZjOGT3. A 6-C-glucosyl substituent could also inhibit 7-O-glycosylation, as ZjOGT3 only catalysed 4′-O-glycosylation of 22.
To explore the sugar donor promiscuity of ZjOGT3, we tested six other sugar donors, including UDP-galactose (UDP-Gal), UDP-xylose (UDP-Xyl), UDP-arabinose (UDP-Ara), UDP-glucuronic acid (UDP-GlcA), UDP-N-acetylglucosamine (UDP-GlcNAc), and UDP-rhamnose (UDP-Rha). Compound 1 was used as the substrate. The results demonstrated that ZjOGT3 could accept UDP-Glc, UDP-Xyl, UDP-Ara, and UDP-GlcNAc. Among these four positive sugar donors, ZjOGT3 produced di-O-glycoside as the major product for UDP-Glc. While the conversion rate for UDP-Xyl was almost 100%, mono-O-glycoside was the major product. The conversion rates for UDP-Ara and UDP-GlcNAc were relatively low (Fig. S80†).
Crystal structure of the ZjOGT3/UDP complex
Although the crystal structures of many plant glycosyltransferases have been reported, little is known about catalytic mechanisms for stepwise di-O-glycosylation and regio-selectivity.5,21 In this work, we obtained the crystal structure of ZjOGT3 in complex with UDP at 2.50 Å resolution (PDB: 8INH) (Fig. 3, S81 and Table S4†). To our knowledge, ZjOGT3 is the first plant flavonoid di-O-glycosyltransferase with a crystal structure. ZjOGT3 adopts a canonical GT-B fold consisting of two Rossmann-like β/α/β domains that face each other and are separated by a deep cleft.9 The N-terminal domain (NTD, residues 1–246 and 443–470) and the C-terminal domain (CTD, residues 247–442) are responsible primarily for sugar acceptor and sugar donor binding, respectively (Fig. 3B and C). We made a lot of efforts to obtain the crystal structure in complex with the sugar acceptor by optimizing protein/ligand concentrations, pH values, temperatures, and precipitant concentrations, but we were not successful.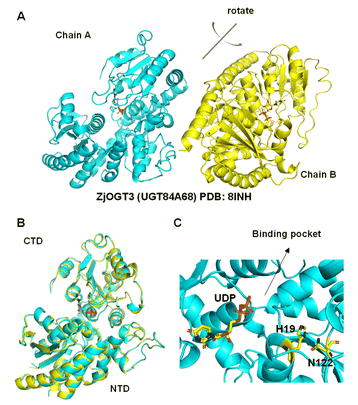 | ||
| Fig. 3 Structural basis for the catalytic mechanisms of ZjOGT3. (A) The crystal structure of ZjOGT3 (PDB: 8INH); (B) superimposition of chain A and chain B of ZjOGT3; (C) a close view of the binding pocket. | ||
Catalytic mechanisms for the regio-selectivity
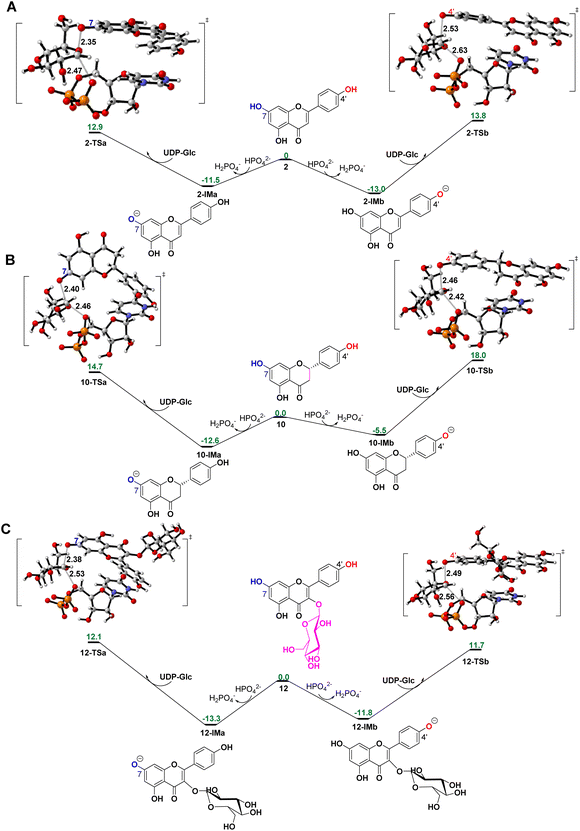
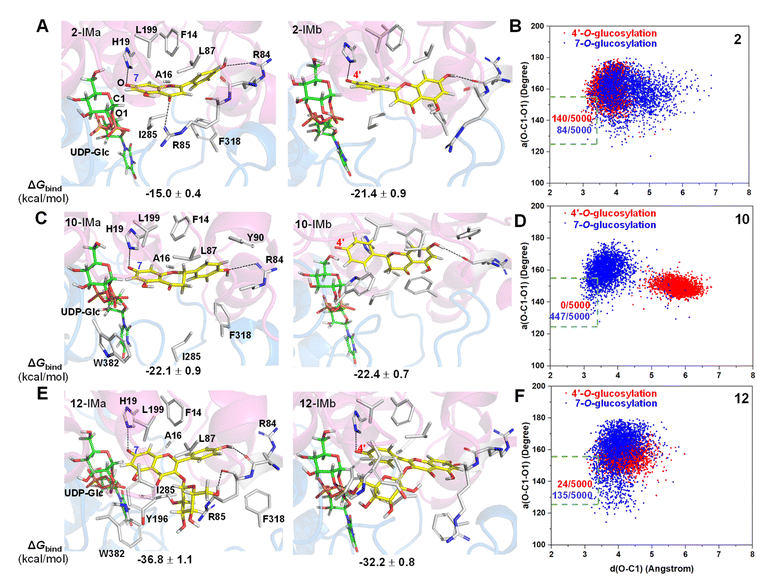
To reveal the catalytic mechanisms for regio-selectivity of ZjOGT3, we conducted DFT calculations on the glycosylation of substrate 2 (Fig. 4A and Table S5†). The 7-OH of 2 is in a deprotonated form in the pH 8.0 buffer, since its pKa was estimated to be around 7.22 In the presence of HPO42− ions,23 deprotonation of 2 is spontaneous and computed to be exergonic by 13.0 and 11.5 kcal mol−1 in water at 4′-OH and 7-OH, respectively (Fig. 4A), suggesting that 2 is deprotonated without enzyme catalysis under the experimental conditions. This is remarkably different from reported glycosyltransferase mechanisms, where deprotonation of a hydroxyl group activated by a histidine (His) residue is critical to initiate GT-mediated glycosylation.3,9 Subsequently, the anionic intermediate performs nucleophilic attack via a SN2-like transition state and generates the corresponding O-glucoside. The overall glycosylation barrier for 4′-OH in 2 (viaTS2-b, 26.8 kcal mol−1) is 0.9 kcal mol−1 higher than that for 7-OH (viaTS2-a, 25.9 kcal mol−1) (Fig. 4A), indicating that the innate nucleophilicity of the oxyanion at the 4′-position is not more favored than that of 7-OH. This result is not, however, consistent with the observed 4′-O-glucosylation catalysed by ZjOGT3. Therefore, the enzyme microenvironment is also a critical factor affecting the regio-selectivity of ZjOGT3.Subsequently, we employed the combination of molecular docking,24 MD simulations,25 and binding free energy calculations26 to reveal the binding mechanisms of 2 in its two phenolate forms (2-IMa, the oxyanion at the 7-position; and 2-IMb, the oxyanion at the 4′-position) into ZjOGT3. First, we constructed the binary complex configuration of ZjOGT3 in complex with UDP-Glc based on the crystal structure and selected a representative MD-equilibrated snapshot as the target receptor for molecular docking. Then, we docked 2-IMa and 2-IMb into the active site and identified catalytically active binding modes as initial configurations to perform MD simulations of the ternary complexes (Fig. S82†). 2-IMa and 2-IMb locate above UDP-Glc in a hydrophobic pocket constituted by F14, A16, L87, L199, I285 and F318 residues, defining a cavity in which acceptors with an aromatic group can be easily accommodated (Fig. 5A). Additionally, H19 stabilizes these two anionic intermediates by forming a hydrogen bond with the nucleophilic attacking phenolic anion. According to the DFT-optimized structures, we defined the active conformation (shown in the light green dotted rectangle) that would lead to the formation of O-glucoside as O–C1 distance ≤3.4 Å (the sum of van der Waals atomic radii of carbon and oxygen atoms27) and angle O–C1–O1 in the range of 140 ± 15°. Our calculations showed that the oxyanion at the 4′-position in 2-IMb has a greater probability of being situated in a catalytically competent position and is more poised for nucleophilic attack than 2-IMa (140/5000 for 2-IMbvs. 84/5000 for 2-IMa, Fig. 5B). To qualitatively rank the binding modes, we evaluated the binding free energies by the molecular mechanics generalized born surface area (MM/GBSA) method. The binding free energy of 2-IMb is about 6.4 kcal mol−1 (Fig. 5A) lower than that of 2-IMa, which is large enough to reverse the intrinsic reactivity. Taken together, 4′-O-glucoside has around 5.5 kcal mol−1 (6.4–0.9 kcal mol−1) priority over 7-O-glucoside for substrate 2, which is consistent with the experimental data. Overall, the regio-selectivity of 2-O-glycosylation is mainly controlled by the enzymatic environment. Since H19 is closer to the nucleophilic attacking phenolic anion in 2-IMb than 2-IMa, there may be a stronger stabilizing effect of H19 in 2-IMb (Fig. S83†). Per-residue contribution of binding energies of 2 in the two phenolate forms suggested that F14, A16, H19, H44, R84, R85, L87, F124, Q145, Y196, L199, I285, F318 and W382 may be key amino acids for substrate binding (Fig. S84†). As expected, alanine scanning mutagenesis for the above residues reduced the conversion rates to different extents (Fig. S85†). Interestingly, W382A and L199A mutants changed the catalytic sequence. They first catalyzed 2 to generate 7-O-glucoside and then 4′-O-glucosylation to generate di-O-glycoside. In these two mutants, we found that the oxyanion at the 4′-position in 2-IMb has a lower probability of being situated in a catalytically competent position and is less poised for nucleophilic attack than 2-IMa by using molecular docking and MD simulations (Fig. S86 and S87†). In addition, the probability that the distance between the phenolic anion and the imidazole ring hydrogen atom of the H19 side chain is less than 2.0 Å in 2-IMa is significantly higher than that in 2-IMb. Overall, 2 should preferentially undergo 7-O-glycosylation in W382A and L199A mutants.
To further test our hypothesis, we studied the glycosylation of substrates 10 and 12. Similarly, 10 and 12 would be deprotonated spontaneously under buffer solution conditions (Fig. 4B and C). For 10, where the C2–C3 double bond is reduced, the conjugation effect of the oxyanion is significantly decreased, resulting in a remarkable increase in the total energy barrier, especially at 4′-OH (30.6 kcal mol−1 for 10-TSbvs. 26.8 kcal mol−1 for 2-TSb; 27.3 kcal mol−1 for 10-TSavs. 25.9 kcal mol−1 for 2-TSa; Fig. 4B). Consistent with these calculations, only 10 underwent the 7-O-glucosylation reaction. Given that the binding free energies of the two anionic intermediates for compound 10 are almost the same (−22.1 vs. −22.4 kcal mol−1 in Fig. 5C), the site selectivity depends primarily on the innate reactivity. In addition, the enzyme microenvironment could further enhance the regio-selectivity (Fig. 5D).
For 12, the introduction of a 3-O-glucosyl substituent has little effect on the intrinsic nucleophilicity because it is far from the reactive sites. The similar energy barriers for 7-O- and 4′-O-glycosylation also support our deduction (25.4 kcal mol−1 for 12-TSavs. 25.0 kcal mol−1 for 12-TSb; Fig. 4C). In fact, ZjOGT3 exhibited high regio-selectivity toward 7-O-glycosylation for 12. It is possible that the size and shape of the enzyme cavity make it easier to place 12-IMa in a catalytically active conformation (135/5000 for 12-IMavs. 24/5000 for 12-IMb; Fig. 5F). The favorable binding free energy (Δ 4.6 kcal mol−1; Fig. 5E) makes 7-O-glucosylation the dominant reaction. Moreover, R84 and R85 could form two extra hydrogen bonds in 12-IMa.
To interpret why only the free flavones or flavonols (1–9) could be catalysed to generate di-O-glycosides, we conducted DFT calculations and molecular docking (Fig. S88†). The energy barrier for 7-O-glycosylation of 2a is 23.9 kcal mol−1, and molecular docking and molecular dynamics simulations show that the conformation of 2a in the active pocket is reasonable. Thus, 7-O-glycosylation of 2a could occur. However, the docking results show that 10a, 12a and 22a have obvious steric hindrance (Fig. S89†). Thus, the second glycosylation is difficult to take place for 10, 12, and 22.
Conclusions
In summary, we report an efficient flavonoid 7,4′-di-O-glycosyltransferase ZjOGT3 from Z. jujuba var. spinosa. ZjOGT3 has broad substrate promiscuity and could catalyse at least 34 flavonoids. The structures of 19 products were fully identified. For free 7,4′-hydroxy flavones, ZjOGT3 could catalyse 4′-O-glycosylation and 7-O-glycosylation sequentially to produce 7,4′-di-O-glycosides. For 7,4′-hydroxy flavanones and 3-O-glycosylated 7,4′-dihydroxyl flavones, ZjOGT3 selectively catalyses 7-O-glycosylation. For flavone-6-C-glycosides, however, ZjOGT3 only catalyses 4′-O-glycosylation. Furthermore, we solved the crystal structure of ZjOGT3 and interpreted mechanisms for the regio-selectivity by structural analysis, DFT calculations, molecular docking, MD simulation, MM/GBSA binding free energy calculations, and site-directed mutagenesis. For 7,4′-hydroxy flavones and 3-O-glycosylated 7,4′-dihydroxyl flavones such as 2 and 12, the regio-selectivity is mainly controlled by the enzyme microenvironment, especially the binding orientation of substrates. For 7,4′-hydroxy flavanones such as 10, the regio-selectivity is mainly controlled by the intrinsic reactivity. This work provides a highly efficient and regio-selective biocatalyst to prepare flavonoid 7,4′-O-glycosides and highlights regio-selectivity mechanisms based on structural analysis and theoretical calculations.Data availability
Sequences for all the genes described in this paper and the crystal structure data have been submitted to the NCBI database and PDB, respectively. Accession number: ZjOGT3 (UGT84A68), OQ603459; ZjOGT2 (UGT84A67), OQ603460; ZjOGT6, OQ603461; ZjOGT10 (UGT71BJ1), OQ603462; ZjOGT13 (UGT72A17), OQ603463; ZjOGT15, OQ603464. PDB ID: ZjOGT3, 8INH. The other data needed to evaluate the conclusions are presented in the paper and/or the ESI.† Any additional data related to this paper may be requested from Min Ye (yemin@bjmu.edu.cn).Author contributions
Min Ye and Zi-Long Wang conceived this study; Zi-Long Wang and Hai-Dong Wang conducted major investigations; Wanqing Wei and Yong Liang contributed to theoretical calculations; Huan Zhou and Fu-Dong Li supervised the crystallography experiments. Jia-Jing Zhou, Hao-Tian Wang, Kuan Chen, Rong-Shen Wang and Xue Qiao assisted with data acquisition; Min Ye, Zi-Long Wang and Wanqing Wei acquired funding; Zi-Long Wang and Wanqing Wei wrote the original draft; Min Ye and Yong Liang revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the staff of the National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility, Shanghai, China, for assistance during data collection. We thank Dr Hong-Li Jia and Dr Fen Liu at State Key Laboratory of Natural and Biomimetic Drugs of Peking University for technical help in crystal and NMR experiments. This work was supported by the National Natural Science Foundation of China (Grant No. 81725023 to M. Y.), China National Postdoctoral Program for Innovation Talents (Grant No. BX20220022 to Z. L. W.), and Jiangsu Provincial Research Foundation for Basic Research (Grant No. BK20200335 to W. W.). We thank the High Performance Computing Center (HPCC) of Nanjing University for supporting the numerical calculations on its blade cluster system.Notes and references
- (a) D. Yi, T. Bayer, C. P. S. Badenhorst, S. Wu, M. Doerr, M. Hohne and U. T. Bornscheuer, Chem. Soc. Rev., 2021, 50, 8003–8049 RSC; (b) M. C. Tang, Y. Zou, K. Watanabe, C. T. Walsh and Y. Tang, Chem. Rev., 2017, 117, 5226–5333 CrossRef CAS PubMed; (c) W. J. C. de Bruijn, M. Levisson, J. Beekwilder, W. J. H. van Berkel and J. P. Vincken, Trends Biotechnol., 2020, 38, 917–934 CrossRef CAS PubMed.
- Y. Q. Liu, Q. Wang, X. N. Liu, J. Cheng, L. Zhang, H. Y. Chu, R. Y. Wang, H. R. Li, H. Chang, N. Ahmed, Z. H. Wang, X. P. Liao and H. F. Jiang, Mol. Plant, 2023 DOI:10.1016/j.molp.2023.01.003.
- (a) N. Putkaradze, D. Teze, F. Fredslund and D. H. Welner, Nat. Prod. Rep., 2021, 38, 432–443 RSC; (b) Y. Q. Zhang, M. Zhang, Z. L. Wang, X. Qiao and M. Ye, Biotechnol. Adv., 2022, 60, 108030 CrossRef CAS PubMed; (c) E. Kurze, M. Wust, J. R. Liao, K. McGraphery, T. Hoffmann, C. K. Song and W. Schwab, Nat. Prod. Rep., 2022, 39, 389–409 RSC; (d) U. M. Vasudevan and E. Y. Lee, Biotechnol. Adv., 2020, 41, 107550 CrossRef PubMed.
- Z. L. Wang, H. M. Gao, S. Wang, M. Zhang, K. Chen, Y. Q. Zhang, H. D. Wang, B. Y. Han, L. L. Xu, T. Q. Song, C. H. Yun, X. Qiao and M. Ye, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 30816–30823 CrossRef CAS PubMed.
- (a) S. G. Lee, E. Salomon, O. Yu and J. M. Jez, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 13131–13136 CrossRef CAS PubMed; (b) J. Zhang, M. Tang, Y. Chen, D. Ke, J. Zhou, X. Xu, W. Yang, J. He, H. Dong, Y. Wei, J. H. Naismith, Y. Lin, X. Zhu and W. Cheng, Nat. Commun., 2021, 12, 7030 CrossRef CAS PubMed.
- (a) A. Jozwiak, P. D. Sonawane, S. Panda, C. Garagounis, K. K. Papadopoulou, B. Abebie, H. Massalha, E. Almekias-Siegl, T. Scherf and A. Aharoni, Nat. Chem. Biol., 2020, 16, 740–748 CrossRef CAS PubMed; (b) S. Y. Chung, H. Seki, Y. Fujisawa, Y. Shimoda, S. Hiraga, Y. Nomura, K. Saito, M. Ishimoto and T. Muranaka, Nat. Commun., 2020, 11, 5664 CrossRef CAS PubMed.
- (a) X. Yan, Y. Fan, W. Wei, P. Wang, Q. Liu, Y. Wei, L. Zhang, G. Zhao, J. Yue and Z. Zhou, Cell Res., 2014, 24, 770–773 CrossRef CAS PubMed; (b) X. Li, Y. Wang, Z. Fan, Y. Wang, P. Wang, X. Yan and Z. Zhou, Metab. Eng., 2021, 66, 87–97 CrossRef CAS PubMed.
- M. Jager and A. J. Minnaard, Chem. Commun., 2016, 52, 656–664 RSC.
- D. M. Liang, J. H. Liu, H. Wu, B. B. Wang, H. J. Zhu and J. J. Qiao, Chem. Soc. Rev., 2015, 44, 8350–8374 RSC.
- (a) K. Chen, Z. M. Hu, W. Song, Z. L. Wang, J. B. He, X. M. Shi, Q. H. Cui, X. Qiao and M. Ye, ACS Synth. Biol., 2019, 8, 1858–1866 CrossRef CAS PubMed; (b) K. B. Xie, R. D. Chen, J. H. Li, R. S. Wang, D. W. Chen, X. X. Dou and J. G. Dai, Org. Lett., 2014, 16, 4874–4877 CrossRef CAS PubMed.
- N. C. Veitch and R. J. Grayer, Nat. Prod. Rep., 2011, 28, 1626–1695 RSC.
- J. B. Johnson, J. S. Mani, D. Broszczak, S. S. Prasad, C. P. Ekanayake, P. Strappe, P. Valeris and M. Naiker, Phytother. Res., 2021, 35, 3484–3508 CrossRef CAS PubMed.
- A. Frydman, O. Weisshaus, M. Bar-Peled, D. V. Huhman, L. W. Sumner, F. R. Marin, E. Lewinsohn, R. Fluhr, J. Gressel and Y. Eyal, Plant J., 2004, 40, 88–100 CrossRef CAS PubMed.
- (a) J. Ogata, Y. Kanno, Y. Itoh, H. Tsugawa and M. Suzuki, Nature, 2005, 435, 757–758 CrossRef CAS PubMed; (b) N. Noda, S. Yoshioka, S. Kishimoto, M. Nakayama, M. Douzono, Y. Tanaka and R. Aida, Sci. Adv., 2017, 3, e1602785 CrossRef PubMed.
- (a) C. A. Williams and R. J. Grayer, Nat. Prod. Rep., 2004, 21, 539–573 RSC; (b) N. C. Veitch and R. J. Grayer, Nat. Prod. Rep., 2008, 25, 555–611 RSC.
- Q. Zhao, Y. Zhang, G. Wang, L. Hill, J. K. Weng, X. Y. Chen, H. W. Xue and C. Martin, Sci. Adv., 2016, 2, e1501780 CrossRef PubMed.
- (a) X. Liu, J. Cheng, G. Zhang, W. Ding, L. Duan, J. Yang, L. Kui, X. Cheng, J. Ruan, W. Fan, J. Chen, G. Long, Y. Zhao, J. Cai, W. Wang, Y. Ma, Y. Dong, S. Yang and H. Jiang, Nat. Commun., 2018, 9, 448 CrossRef PubMed; (b) Z. L. Wang, S. Wang, Z. Xu, M. W. Li, K. Chen, Y. Q. Zhang, Z. M. Hu, M. Zhang, Z. Y. Zhang, X. Qiao and M. Ye, Org. Lett., 2019, 21, 2241–2245 CrossRef CAS PubMed; (c) Z. Wen, Z. M. Zhang, L. Zhong, J. Fan, M. Li, Y. Ma, Y. Zhou, W. Zhang, B. Guo, B. Chen and J. B. Wang, ACS Catal., 2021, 11, 14781–14790 CrossRef CAS.
- N. Okitsu, K. Matsui, M. Horikawa, K. Sugahara and Y. Tanaka, Plant Cell Physiol., 2018, 59, 2075–2085 CrossRef CAS PubMed.
- Y. Hua, X. X. Xu, S. Guo, H. Xie, H. Yan, X. F. Ma, Y. Niu and J. A. Duan, J. Agric. Food Chem., 2022, 70, 7871–7886 CrossRef CAS PubMed.
- Y. Q. Zhang, Z. L. Wang, Z. Chen, Z. T. Jin, A. Hasan, H. D. Wang, Y. W. Sun, X. Qiao, Y. Wang and M. Ye, Chem. Commun., 2022, 58, 2472–2475 RSC.
- W. Huang, Y. He, R. Jiang, Z. Deng and F. Long, ACS Catal., 2022, 12, 2927–2937 CrossRef CAS.
- J. M. Cabot, E. Fuguet and M. Roses, ACS Comb. Sci., 2014, 16, 518–525 CrossRef CAS PubMed.
- (a) J. Shi, Y. Shi, J. C. Li, W. Wei, Y. Chen, P. Cheng, C. L. Liu, H. Zhang, R. Wu, B. Zhang, R. H. Jiao, S. Yu, Y. Liang, R. X. Tan and H. M. Ge, J. Am. Chem. Soc., 2022, 144, 7939–7948 CrossRef CAS PubMed; (b) Y. Zhang, W. Zhao, Y. Chen, H. Yuan, H. Fang, S. Yao, C. Zhang, H. Xu, N. Li, Z. Guo, Q. Zhao, Y. Liang and W. He, Nat. Commun., 2021, 12, 2772 CrossRef CAS PubMed.
- S. Forli, R. Huey, M. E. Pique, M. F. Sanner, D. S. Goodsell and A. J. Olson, Nat. Protoc., 2016, 11, 905–919 CrossRef CAS PubMed.
- J. L. Klepeis, K. Lindorff-Larsen, R. O. Dror and D. E. Shaw, Curr. Opin. Struct. Biol., 2009, 19, 120–127 CrossRef CAS PubMed.
- P. A. Kollman, I. Massova, C. Reyes, B. Kuhn, S. H. Huo, L. Chong, M. Lee, T. Lee, Y. Duan, W. Wang, O. Donini, P. Cieplak, J. Srinivasan, D. A. Case and T. E. Cheatham, Acc. Chem. Res., 2000, 33, 889–897 CrossRef CAS PubMed.
- R. S. Rowland and R. Taylor, J. Phys. Chem., 1996, 100, 7384–7739 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of experimental procedures, including molecular cloning, protein expression and purification, catalytic reaction analysis, product purification, structural characterisation, crystal structure and DFT calculations, LC/MS analysis, and NMR and ESI-MS spectra. See DOI: https://doi.org/10.1039/d2sc06504e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |