Open Access Article
Open Access ArticleMultimodal high-resolution nano-DESI MSI and immunofluorescence imaging reveal molecular signatures of skeletal muscle fiber types†
Daisy
Unsihuay
 ad,
Hang
Hu
ad,
Hang
Hu
 a,
Jiamin
Qiu
b,
Alessandra
Latorre-Palomino
a,
Jiamin
Qiu
b,
Alessandra
Latorre-Palomino
 c,
Manxi
Yang
a,
Feng
Yue
b,
Ruichuan
Yin
a,
Shihuan
Kuang
b and
Julia
Laskin
c,
Manxi
Yang
a,
Feng
Yue
b,
Ruichuan
Yin
a,
Shihuan
Kuang
b and
Julia
Laskin
 *a
*a
aDepartment of Chemistry, Purdue University, West Lafayette, IN 47907, USA. E-mail: jlaskin@purdue.edu
bDepartment of Animal Sciences, Purdue University, West Lafayette, IN 47907, USA
cDepartment of Chemistry, Universidad Nacional Mayor de San Marcos, Lima, Peru
dDepartment of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA
First published on 23rd March 2023
Abstract
The skeletal muscle is a highly heterogeneous tissue comprised of different fiber types with varying contractile and metabolic properties. The complexity in the analysis of skeletal muscle fibers associated with their small size (30–50 μm) and mosaic-like distribution across the tissue tnecessitates the use of high-resolution imaging to differentiate between fiber types. Herein, we use a multimodal approach to characterize the chemical composition of skeletal fibers in a limb muscle, the gastrocnemius. Specifically, we combine high-resolution nanospray desorption electrospray ionization (nano-DESI) mass spectrometry imaging (MSI) with immunofluorescence (IF)-based fiber type identification. Computational image registration and segmentation approaches are used to integrate the information obtained with both techniques. Our results indicate that the transition between oxidative and glycolytic fibers is associated with shallow chemical gradients (<2.5 fold change in signals). Interestingly, we did not find any fiber type-specific molecule. We hypothesize that these findings might be linked to muscle plasticity thereby facilitating a switch in the metabolic properties of fibers in response to different conditions such as exercise and diet, among others. Despite the shallow chemical gradients, cardiolipins (CLs), acylcarnitines (CAR), monoglycerides (MGs), fatty acids, highly polyunsaturated phospholipids, and oxidized phospholipids, were identified as molecular signatures of oxidative metabolism. In contrast, histidine-related compounds were found as molecular signatures of glycolytic fibers. Additionally, the presence of highly polyunsaturated acyl chains in phospholipids was found in oxidative fibers whereas more saturated acyl chains in phospholipids were found in glycolytic fibers which suggests an effect of the membrane fluidity on the metabolic properties of skeletal myofibers.
Introduction
Skeletal muscle plays a primary role in human physiology enabling locomotion, maintenance of posture, metabolic homeostasis, and respiration.1 The contractile muscle cells also known as muscle fibers, are responsible for the wide array of capabilities that a muscle displays. Muscle fibers are generally classified according to their speed of contraction into two major groups: slow-twitch (type I) and fast-twitch (type II) fibers.2 Type I fibers are least powerful and most fatigue-resistant, and therefore are involved in endurance activities. In contrast, type II fibers include three subtypes (IIa, IIx, IIb) that are sequentially more powerful but fatigue more quickly, and therefore are mainly used to deliver rapid bursts of force. This classification of fiber types (I, IIa, IIx, IIb) are based on myosin heavy chain (MyHC) isoforms as the common molecular motor present in a fiber type.3 Another classification of fibers into oxidative/aerobic (I, IIa) or glycolytic/anaerobic (IIx, IIb) types is based on the metabolic properties through which they generate ATP.4 The diversity of myofibers is a result of the combination of different metabolic, contractile, and physiological properties. Based on these properties, myofibers are classified as slow-oxidative (type I), fast-oxidative (type IIa), fast-intermediate (type IIx), and fast-glycolytic (type IIb).2,5 Understanding of the chemical composition of muscle fibers and how it is linked to their biochemical and contractile properties provides the basis for the design of physical therapies, intervention, and treatments targeting a wide variety of muscle-related pathologies including sarcopenia,6 metabolic syndrome7 among others.A skeletal muscle typically contains heterogeneous myofibers that are arranged in a mosaic-like pattern. This feature makes it difficult to separate the individual myofiber types to examine their molecular signatures using traditional bulk characterization approaches. Metabolic profiling of skeletal myofibers typically involves liquid chromatography mass spectrometry (LC-MS) analysis of tissue extracts from a muscle enriched in a specific fiber type.8,9 However, this approach does not provide information on the spatial localization of molecules enhanced in a specific fiber type. Other strategies such as histochemical staining provide the localization of some lipid classes but cannot identify individual lipid or metabolite species.
Mass spectrometry imaging (MSI) is a powerful label-free technique that enables the visualization of molecular distributions across biological samples.10 In contrast to traditional staining approaches, MSI provides spatial maps of hundreds of compounds in a single experiment which can be used to track organ development, the progression of disease and the effectiveness of treatment.11 Several studies used matrix assisted laser desorption ionization (MALDI) and secondary ion mass spectrometry (SIMS) for imaging of bundles of different types of muscle fibers thereby providing important insights into key biochemical processes in these systems.7,12–16 Most of MALDI MSI studies were carried out using moderate spatial resolution of ∼100 μm and therefore could not differentiate fibers that are present in the same bundle. Tsai et al., demonstrated that it is possible to distinguish individual muscle fibers using MALDI MSI performed with a spatial resolution of 10 μm.17 They noted that both the crystal size and uniformity of the matrix influence the spatial resolution of these experiments. Meanwhile, Song et al. used SIMS with a spatial resolution of 0.2 μm to examine the distribution of a several intracellular lipids across different myofibers.16 Despite the impressive resolution achieved in these two studies, the molecular coverage was limited either because of the reduced sensitivity of MALDI MSI performed with high spatial resolution or fragmentation of molecules in SIMS, which limits metabolite identification.
Ambient ionization techniques do not require matrix application and provide a softer ionization thereby expanding lipid and metabolite coverage, which is advantageous for high-resolution MSI applications. Nanospray desorption electrospray ionization (nano-DESI) used in this study is an ambient ionization technique, in which analytes are extracted from tissues into a dynamic liquid bridge formed between two glass capillaries.18 The spatial resolution of nano-DESI MSI experiments is mainly determined by the size of the liquid bridge formed between the nano-DESI probe and sample surface. A spatial resolution of 10 μm has been achieved using a nano-DESI probe composed of two finely pulled capillaries.19 High spatial resolution nano-DESI MSI experiments have been used for mapping lipids and metabolites in small tissue samples including pancreatic islets (∼100 μm diameter),20 lung airways (∼50 μm diameter),21 and mouse uterine tissues (∼1000 μm diameter),19,22 which cannot be performed with moderate spatial resolution.
Herein, we used high resolution nano-DESI MSI for imaging of individual skeletal muscle fibers that have a diameter of ∼30–50 μm. This tissue represents one of the most challenging systems for MSI experiments as the fibers are distributed in a mosaic-like pattern. We used immunofluorescence (IF) imaging of serial tissue sections to distinguish between different types of fibers in mouse muscle tissues. IF images were subsequently used as roadmaps for identifying molecular signatures of different fiber types in adjacent sections analyzed using nano-DESI MSI. Image registration between IF and nano-DESI MSI is difficult for this system given the lack of major anatomical markers which have been traditionally used to train a variety of computational models for feature detection,23 co-localization24 and segmentation.25 Furthermore, high spatial resolution imposes additional constraints on the accuracy of image registration. To address these challenges, we have developed a robust computational method for image registration and extraction of ion abundances from well-defined regions of interest (ROIs) and performed relative quantification across different fiber types guided by IF imaging data. Using this approach, we have identified groups of molecules that are enhanced in specific fibers. Our results indicate that myofibers display shallow variations in their chemical gradients in a progression from more oxidative (type I) to the most glycolytic fibers (type IIb). The lack of drastic changes in the chemical composition may be attributed to metabolic flexibility of muscle fibers allowing them to switch from one type to another in response to different stimuli.
Results
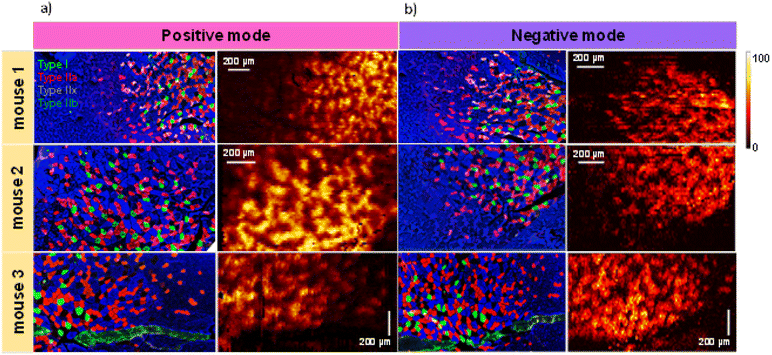
In this study, we performed multimodal imaging of mouse gastrocnemius muscle (GAS) tissues using a combination of IF with nano-DESI MSI on adjacent serial cross-sections to elucidate differences in the chemical composition of different fiber types. Table S1† provides a summary of all the annotated species whose identity was confirmed by matching their exact mass with open databases and manual analysis of their MS/MS spectra. The gastrocnemius muscle contains all four fiber types (type I, type IIa, type IIx and type IIb) that are intermixed in the tissue making their molecular imaging particularly challenging.Fig. 1 shows the results of IF and nano-DESI imaging obtained for three biological replicates. For each replicate, we used three adjacent 12 μm-thick sections: one for IF imaging, one for positive mode and one for negative mode nano-DESI MSI. Representative positive and negative mode nano-DESI MSI data are displayed along with the corresponding IF images in Fig. 1a and b, respectively. Additional ion images from replicate 1 can be found in Fig. S1.† A complete view of the optical image and IF images are provided in Fig. S2.† We use IF images as roadmaps of fiber localization. A mosaic pattern of fibers is observed in all the IF images, in which type I fibers are labelled in green, type IIa are labelled in red, type IIx are labelled in black, and type IIb are labelled in blue. As observed in Fig. 1 and S2,† GAS tissue displays fiber regionalization with the largest proportion of oxidative fibers present in the deep region closer to the blood supply. Meanwhile, glycolytic fibers are mainly found on the edges of the muscle.26 Despite that, little or no fiber segregation is observed in IF images. The IF data were used to guide nano-DESI experiments to areas where a transition from the deep region to the surface region of GAS tissue was observed. Representative ion images normalized to the total ion current (TIC) of m/z 856.5830 corresponding to phosphatidylcholine (PC) 40:6 and m/z 622.3716 corresponding to the oxidized PC 22:1; O2 observed in positive and negative mode, respectively, are also shown in Fig. 1. These ions correspond to two phospholipids enriched in the deep region of the tissue. There is a good qualitative correspondence between the patterns observed in the ion images and IF images. Fig. S3† shows the calculation of the spatial resolution using the 20/80 rule.19,27 In this approach, the spatial resolution is estimated by examining the distance over which the signal along the steepest chemical gradient in the sample changes between 20 and 80% of its maximum value. Our measurements indicate that the spatial resolution of our experiments is about 6 μm, which is the best value reported for nano-DESI MSI so far. We achieved this resolution by using a slightly smaller diameter of the nano-DESI capillaries of 20 μm instead of 25 μm used in previous work.22 We also used a slower scanning rate of 10 μm s−1, which reduces the pixel size in the direction of the scan. High spatial resolution is critical to the visualization of chemical gradients generated by individual muscle fibers.
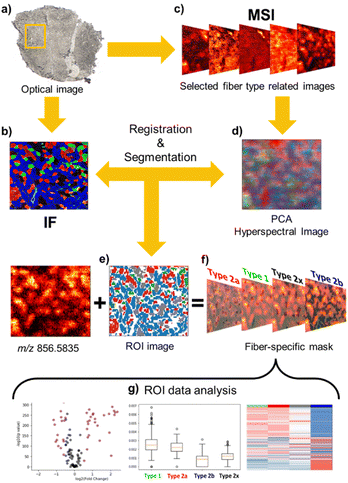
Despite the high quality of high-resolution nano-DESI ion images, the complex distribution of muscle fibers observed in Fig. 1 presents a challenge for feature extraction and image registration to specific fiber types. Although the chemical gradient between the deep region and outer part of the tissue is evident in ion images, the absence of anatomical markers and small dimensions of individual fibers make it difficult to discern the individual fibers and perform image registration with an accuracy comparable to the spatial resolution of nano-DESI MSI experiments. To address this problem, we have developed a robust image registration and segmentation approach, which is summarized in Fig. 2. A yellow box in the optical image of the tissue (Fig. 2a) marks the region analyzed using nano-DESI MSI. First, the IF image (Fig. 2b) is cropped to roughly cover the same region. Next, we generate MSI images (Fig. 2c) using a mass list of peaks. We use principal component analysis (PCA) to reduce data dimensionality of ion images and generate an RGB representation of the chemical gradients in the sample (Fig. 2d). A grey representation of the IF image is used for the affine registration to the PCA image as shown in Fig. S4.† This step resizes and reorients the IF image to align it with the pattern displayed by the PCA image. Next, we perform segmentation of the IF image to obtain fiber type-specific ROIs (Fig. 2e) using an image segmentation approach developed by our group.28 Finally, the segmented ROI image is registered to the RGB representation image and hence to all the ion images using a stochastic gradient descent algorithm thereby yielding well-defined ROIs specific to each fiber type (Fig. 2f). A closer view of this step is depicted in Fig. S5.† The segmented ROI image is subjected to an erosion step to avoid the extraction of pixels from fibers located in the vicinity of the fiber of interest (Fig. S5b†). As a result, fiber-specific masks for type I fibers (Fig. S5c†), type IIa fibers (Fig. S5d†), type IIx fibers (Fig. S5e†), and type IIb fibers (Fig. S5f†) are generated. Although the cellular outlines for type IIb are difficult to visualize in the glycolytic region, in which type IIb is the dominant fiber, individual type IIb fibers are clearly visible in the oxidative region where all fibers coexist. Fig. S5f† shows that our registration approach successfully extracts pixels corresponding to type IIb fibers generating a mask specific for type IIb fibers across the entire scanned area. In order to validate our approach of identifying fiber-specific pixels in ion images, we performed additional correlation analysis of the patterns observed in ion images with ROI-generated masks. This process is described in detail in Fig. S6† using type IIa fibers as an example. Briefly, a dilation morphology operation is used to define contour ROIs surrounding each fiber type at different distances away from the fiber ROI. An overlay of the fiber and contour ROIs onto ion images confirms that fiber ROI is colocalized with an ion image of an abundant species at m/z 856.5835. We also performed a quantitative analysis of the correlation between ion signals in the fiber ROI and contour ROI as a function of the spatial distance between them. This analysis found a statistically significant linear relationship (r = −0.86, p value < 0.001), which indicates the decrease of ion signal in tissue locations with an increase in the distance from fiber ROI. This change is in agreement with the chemical gradient extending from a specific fiber type into the surrounding tissue. Subsequent ROI analysis generates ion abundances for each fiber type. This workflow allowed us to evaluate differences between molecular markers of different myofiber types based on high-resolution nano-DESI MSI data (Fig. 2g).
In this work, we only report molecular markers that were detected in all three biological replicates and identified using MS/MS. Based on these criteria, we analyzed a total of 140 unique species observed in both positive and negative ionization mode.
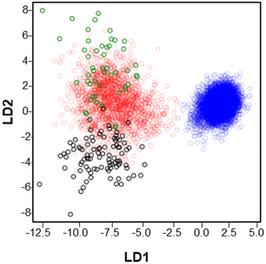
Fig. 3 shows the results of a linear discriminant analysis (LDA) performed on muscle fibers which provided a separation of all the observed m/z features into two large groups. The blue circles corresponding to type IIb fibers are well separated from the rest of the fibers across the LD1 and LD2 axis. In contrast, type I (green circles), type IIa (red circles) and type IIx (black circles) display overlapping regions indicating a greater overlap between the chemical composition of these fibers. These results provide a general overview of the distribution of the analyzed m/z features and their association with specific fiber types.
For each feature, we calculated its mean abundance in different fiber types as a percent of its maximum abundance across the fibers. The relative abundances obtained in this analysis are summarized in Table S1.† Following an approach described in a recent proteomics study where a series of criteria were used to discriminate specific fiber patterns,29 we established several criteria for the identification of myofibers based on the high-resolution spatial lipidomics experiments performed in this study. We then used these criteria to discriminate between myofibers.
First, we used a stringent criterion for identifying unique fiber-specific markers. We define a unique marker as a species that shows more than a 2.5-fold change in abundance in one fiber type in comparison with other fibers. This criterion provided only one unique marker of type IIb fibers at m/z 175.0245 corresponding to ascorbate (Fig. S7a†). Confirmation of the identity of this molecule was performed by comparison with MS/MS reported in the literature.30,31 Ion images of m/z 175.0245 in all three replicates show a gradual decrease of the abundance of ascorbate when moving from the more glycolytic region to the more oxidative region (Fig. S7b†). No other unique markers of fiber types have been identified, which is consistent with the results of the LDA analysis discussed earlier.
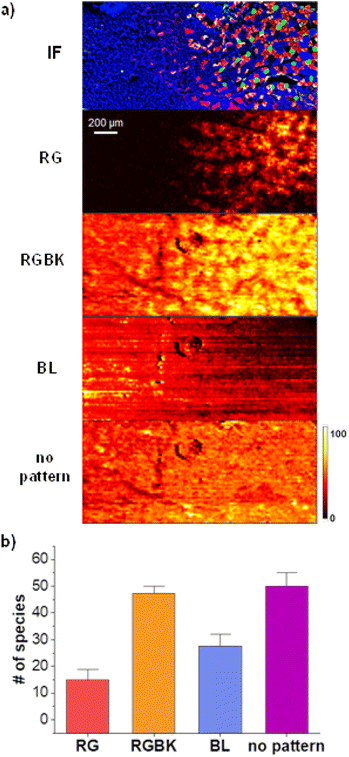
Next, we use less stringent criteria to identify groups of molecular markers that display distinct spatial patterns. Fig. 4a shows representative ion images of the four patterns that we observed which we refer to as red-green (RG) displayed by PC O-44:11, red–green–black (RGBK) displayed by PC 38:4, blue (BL) displayed by carnosine, and “no pattern” displayed by PC 32:0. The conditions used to define each pattern are summarized in Table 1. The RG pattern corresponds to species enhanced in type I and type IIa fibers by at least 1.2-fold in comparison to type IIx and type IIb, and with a ratio between type I and type IIa of less than 1.5. The RGBK pattern corresponds to molecules enhanced in type I, type IIa and type IIx fibers by at least 1.2-fold relative to type IIb, and with ratios between type I, IIa and IIx of less than 1.5. The BL pattern corresponds to molecules enhanced in type IIb fibers. Because chemical gradients in the glycolytic region are very shallow, we set a criterion that any molecule, for which the abundance in type IIb is by at least 1.05 higher than in other types of fibers is assigned to this pattern. Finally, molecules that do not meet any of these criteria are classified as species with “no pattern” indicating that they are uniformly distributed across the tissue. Fig. 4b shows a bar graph summarizing the results of this analysis. A detailed summary obtained for all the replicates is provided in Table S1.† The RGBK pattern (orange bar) is the most dominant pattern observed for 47 species, followed by the BL (blue bar) with 28 species, and RG (red bar) pattern with 15 species. Interestingly, 50 species (magenta bar) do not display a specific localization across the tissue. None of the molecules observed in nano-DESI MSI data displayed a distinct localization to either type I (green), type IIa (red), or type IIx (black) fibers making it difficult to differentiate between these three types of fibers.
| Conditions | |
|---|---|
| R–G | R/BL > 1.2, R/BK > 1.2, G/BL > 1.2, G/BK > 1.2, R/G < 1.5 |
| R–G–BK | R/BL > 1.2, G/BL > 1.2, BK/BL > 1.2, G/BK < 1.5, R/BK < 1.5 |
| BL | BL/R > 1.05, BL/G > 1.05, BL/BK > 1.05 |
| No pattern | Does not meet any conditions |
In order to uncover statistically significant differences between these fibers, we use volcano plots that compare p-values and fold-change (FC) of different species identified in nano-DESI MSI experiments. The results are shown in Fig. S8† for type I vs. type IIa, Fig. S9† for type I vs. type IIx and Fig. S10† for type IIa vs. type IIx for all replicates. Using log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) of less than −0.5 or greater than 0.5 and p-value <0.05, we observe the decreased abundance (left side) of SM(d36:1), PE(36:1), PC(36:4), PC(32:2), PC (30:0) and PE(36:1) and enhanced abundance (right side) of PC(20:2/12-HETE), CAR(14:2), CAR(18:2), PC(e44:11) and PC(36:1) when comparing type I vs. type IIa, type I vs. type IIx and type IIa vs. type IIx. These molecules are highlighted in blue to indicate their reproducibility across all replicates. The fact that similar molecules are suppressed or enhanced across these fibers suggests a gradual change in their abundance from more oxidative fibers to more glycolytic fiber (type I- > type IIa- > type IIx- > type IIb).
2(FC) of less than −0.5 or greater than 0.5 and p-value <0.05, we observe the decreased abundance (left side) of SM(d36:1), PE(36:1), PC(36:4), PC(32:2), PC (30:0) and PE(36:1) and enhanced abundance (right side) of PC(20:2/12-HETE), CAR(14:2), CAR(18:2), PC(e44:11) and PC(36:1) when comparing type I vs. type IIa, type I vs. type IIx and type IIa vs. type IIx. These molecules are highlighted in blue to indicate their reproducibility across all replicates. The fact that similar molecules are suppressed or enhanced across these fibers suggests a gradual change in their abundance from more oxidative fibers to more glycolytic fiber (type I- > type IIa- > type IIx- > type IIb).
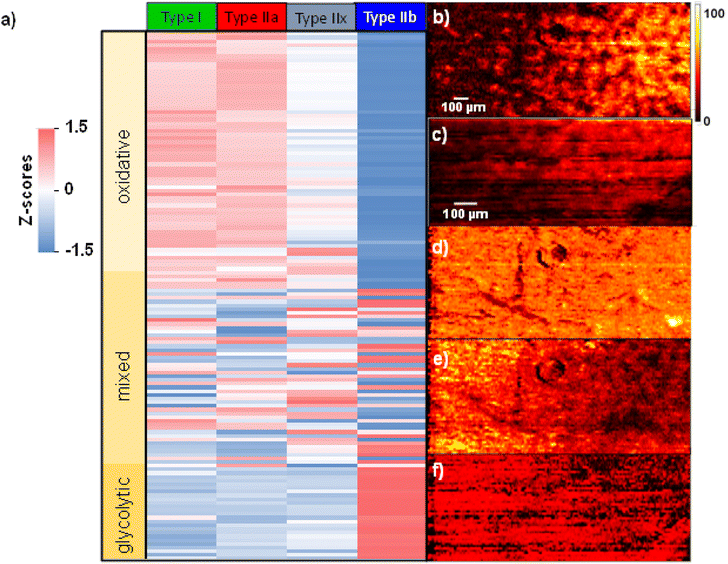
Fig. 5 shows a heatmap summarizing the variations in the abundance of species in each fiber type. A z-score is calculated indicating the number of standard deviations from the mean abundance. In this plot, the more positive z-score highlighted in red indicates that the abundance of a molecule in a specific fiber type is higher than the mean abundance across all fibers. Meanwhile, a more negative z-score highlighted in blue indicates an abundance lower than the mean value across the fibers. Finally, a z-score closer to zero shown in white indicates an abundance close to the mean value. The calculated z-scores across all replicates are provided in Table S2.† Molecules were sorted out based on the spatial localization they showed across the three replicates. If a molecule was assigned to either RG or RGBK pattern in more than 2 two replicates, it is considered to correspond to oxidative metabolism. The RG pattern clusters type I and type IIa fibers which are known to rely on oxidative phosphorylation for energy supply. The RGBK pattern adds type IIx into the group of oxidative fibers given the overlap in the chemical composition as displayed in Fig. 3. Meanwhile, molecules assigned to the BL pattern in more than two replicates are part of glycolytic metabolism. The BL pattern only clusters type IIb fibers which rely on glycolysis for energy supply. When extending this classification to the heatmap (Fig. 5), we observe three distinct blocks depicting different trends: the top block contains all the molecules associated with oxidative metabolism (light yellow), the middle block contains all the molecules associated with mixed metabolism (intermediate yellow), and the bottom block contains the molecules associated with glycolytic metabolism (dark yellow).
A complete list of molecules and their classification are provided in Table S2.† Overall, there is a larger number of molecules depicting oxidative metabolism than glycolytic metabolism. Specifically, species like cardiolipins (CL), monoacylglycerols (MG), oxidized lipids and fatty acids (FA), acylcarnitines and highly polyunsaturated phospholipids are detected as fingerprints of oxidative metabolism as shown in Fig. S11 and S12.† In contrast, saturated and monounsaturated phospholipids and histidine-related compounds including carnosine, anserine, and histidine are mainly identified as fingerprints of glycolytic metabolism Fig. S13.† Regardless of the type of metabolism, it is evident that there is always a gradual transition in molecular abundances from type I/type IIa → type IIx → type IIb across all the replicates (Table S2†). These observations are also confirmed by ion images shown in Fig. 5. For example, the abundances of PC 36:2 (Fig. 5b) and PE 40:6 (Fig. 5c) increase from left (glycolytic) to right (oxidative). Meanwhile, PC 34:0 (Fig. 5d) does not show a specific localization. Lastly, SM d36:1 and PC 16:0/5:0(COOH) shown in Fig. 5e and f, respectively, display a decreasing chemical gradient from left to right. In summary, these results confirm that the chemical composition across fiber types does not change dramatically but rather undergoes relatively small changes especially in the region where all the fiber types coexist such as the deep region of GAS tissue.
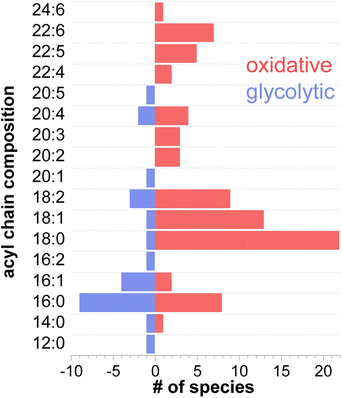
A deeper look at the structural information of membrane components such as phospholipids, revealed different acyl chain composition in fibers with oxidative or glycolytic metabolism. Fig. 6 displays the distribution of all the constituent fatty acids from 12C to 24C found in phospholipid species. A detailed list of all acyl chains can be found in Table S3.† Among the 91 identified phospholipids (including isomers with different acyl chain compositions), 52 are more abundant in oxidative fibers, 16 are enhanced in glycolytic fibers, and 23 are uniformly distributed across the tissue. Interestingly, a higher number of polyunsaturated fatty acids (PUFAs) including docosahexaenoic acid (DHA) FA 22:6, docosapentaenoic acid (DPA) FA 22:5, FA 22:4, and arachidonic acid (AA), FA 20:4 were identified in phospholipids associated with oxidative metabolism in comparison with phospholipids localized to glycolytic fibers, which do not contain these acyl chains. This trend in the acyl chain composition in phospholipids suggests an important relationship of their structures to the function of the fiber.
Discussion
The approach developed in this study uses high-precision registration of IF and high-resolution nano-DESI MSI data to provide unique insights into lipid signatures of different fiber types in muscle tissues. This approach is particularly advantageous for muscle tissues, in which heterogeneous fibers are intermingled making it difficult to separate individual fiber types for traditional bulk lipidomics and metabolomics analyses. Indeed, omics analyses are typically performed using muscle tissues enriched in a specific fiber type.7,12,32 In contrast, chemical gradients observed using high-resolution nano-DESI MSI enable a direct comparison of the chemical composition of different types of myofibers in the same experiment.The results reported herein provide a global overview of the chemical composition of the four main myofibers identified in mammals: type I, type IIa, type IIx, and type IIb. Our results indicate that although myofibers are classified into discrete categories expressing distinct MyHC isoforms, their chemical signatures are not drastically different. Instead, we observe a gradual change in the relative abundance of lipids and metabolites between oxidative and glycolytic fibers. The lack of fiber-specific patterns and prevalence of species that are uniformly distributed across the muscle tissue (50 species in Fig. 4b) supports this claim thereby suggesting that a substantial fraction of molecules is conserved across different myofiber types for a proper function of the muscle. The shallow chemical gradients observed in muscle fibers are likely linked to their inherent ‘muscle plasticity’, a term used to describe the ability of muscles to modify their fiber structure or fine-tune their aerobic capacity in response to changes in their contractile activity, loading conditions, substrate supply among others.2,33 For example, endurance training increases the oxidative capacity of all fibers types,34 which we propose is facilitated by the relatively small chemical differences between myofibers. Another example is found in our previous study in which the influence of defective fatty acid oxidation due to the loss of carnitine palmitoyltransferase 2 (CPT2) was tested in different muscles.35 The lack of CPT2 caused a shift in mitochondrial energy metabolism to non-lipid substrates. As a result, oxidative fibers underwent metabolic change towards a more glycolytic metabolic profile without necessarily changing their myosin heavy chain isoform which is more connected to myofiber contractile properties. Hence, it is reasonable to assume that from the metabolic point of view, the lack of substantial chemical gradients across myofibers makes it possible for the muscle to adapt to meet the needs that the new activity/condition demands and, in some cases, undergo metabolic transformation.
Among the four myofiber types analyzed in this study, type IIb fibers are the most chemically distinct (Fig. 3). Indeed, type IIb is the only fiber type for which we detected a specific molecular marker: ascorbate. Aside of being a powerful antioxidant, it has been suggested that ascorbate breakdown through the pentose phosphate pathway can be a potential substrate for glycolysis,36,37 the dominant metabolism of type IIb fibers. In contrast, type I, type IIa, and type IIx do not have a specific molecular marker and may be distinguished using an in-depth statistical analysis of the relative abundances of lipids and metabolites (Fig. S8–S10†). One key finding of this study is that although type IIx fibers are more similar to type IIb in their contractile properties and are traditionally considered to present a glycolytic metabolism,38,39 their chemical composition is more comparable to oxidative fibers: type I and type IIa as shown in Fig. 3 and 4. Indeed, 47 species are present in comparable abundances in fibers type I, type IIa and type IIx (RGBK pattern in Fig. 4b) in contrast to only 15 species present with similar abundances in fibers type I and type IIa (RG pattern in Fig. 4b). These results are consistent with recent studies acknowledging the oxidative properties of type IIx fibers which may allow to sustain high running speeds for long periods of time in sprinting wild animals.40,41 Of note is that this is the first MSI study that reports the chemical profile of the type IIx fiber, which could only be obtained using our accurate image registration approach.
Increased breakdown of ATP triggers the glycolytic pathway in an effort to keep up with the energy demand.4 In anaerobic glycolysis, glycogen is broken down to produce three ATP molecules and lactic acid. Accumulation of lactic acid causes a decrease in the pH thereby promoting muscle fatigue, disrupting phosphocreatine recycling, and causing muscle injury during high-intensity exercise.44 Therefore, it is not surprising that fast-glycolytic fibers have some mechanisms to protect themselves against oxidatively-induced tissue damage. Our results indicate that histidine-related compounds (Fig. S13†) including histidine, methyl histidine, carnosine, and anserine are enhanced in type IIb fibers. These species act as antioxidants thereby preventing tissue damage due to the oxidative stress.45 The antioxidant capacity of these compounds derive from the imidazole ring, which efficiently scavenges reactive oxygen species (ROS) and harmful alpha–beta-unsaturated aldehydes.45,46 Moreover, the presence of the imidazole moiety results in a high muscle buffering capacity of histidine-related compounds that helps regulate the acidosis conditions during glycolysis. For example, carnosine with a pKa = 6.72 is an efficient antioxidant that also helps maintaining the pH around physiological values thereby delaying the onset of fatigue following short bouts of exercise.47 The acidosis environment promoted by glycolysis also favors the synthesis of creatinine through the breakdown of phosphocreatine that consumes H+ for the phosphorylation of ADP.48,49
Oxidative phosphorylation becomes the main source of energy in events lasting from minutes to hours and is mainly manifested in slow-oxidative (type I) and fast-oxidative-intermediate (type IIa and type IIx) fibers. In this pathway, ATP is generated through the oxidation of fats and carbohydrates.4 The enhanced abundance of acylcarnitines including CAR 18:2, CAR 18:1, CAR 16:0 and CAR 14:2 in oxidative fibers is a fingerprint of this dynamic metabolism (Fig. S12†). The conversion of fatty acyl-coAs into acylcarnitines enables the transport of fatty acids across the mitochondrial membrane to support energy production through β-oxidation. Because of the higher content of mitochondria in oxidative fibers, it is reasonable to observe greater β-oxidation activity in these fibers. Our findings are consistent with a study by Yu-Hsuan et al. that reported a higher abundance of acylcarnitines in fibers with an oxidative metabolism in a GAS tissue.12 Interestingly, free carnitine and acetylcarnitine, CAR 2:0, do not show a specific localization and are uniformly distributed across all the tissue. Because of the efficient enzymatic conversion between carnitine and acetylcarnitine, it is reasonable that these two molecules have a similar spatial distribution. These results may be attributed to another important role of carnitine aside from translocating long-chain fatty acids from cytosol to the mitochondrial matrix discussed earlier. In particular, carnitine is also involved in the formation of acetylcarnitine from acetyl-CoA thereby preventing extreme fluctuations in acetyl-CoA and free CoA that otherwise can be deleterious to cellular function.50,51 Our results indicate that acetylcarnitine maintains this metabolic flexibility in all the fiber types.
Another molecular marker of oxidative metabolism is observed in the distributions of MG 18:2, MG 18:1, and MG 16:0 which are enhanced in oxidative fibers (Fig. S12†). We believe that the presence of MGs might reflect the lipolysis state of triglycerides (TGs) contained in these fibers which serve as energy reservoirs. The enzymes adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL) and monoacylglycerol lipase (MGL) sequentially hydrolyze TG, DG, and MG, respectively, thereby releasing fatty acids at every step.52 This lipolysis activity is known to be enhanced in oxidative fibers.53 Moreover, the high expression of ATGL and HSL enzymes in type I fibers reported in the literature further supports our findings.52,54
The acyl chain composition of most abundant phospholipids shown in Fig. 6 indicates that palmitic and stearic acids (PA and SA) are the most abundant acyl chains found in the total pool of phospholipids. This result is consistent with the known prevalence of PA and SA in cell membranes.60 Furthermore, highly polyunsaturated acyl tails such as DHA and DPA are exclusively found in oxidative fibers. Considering that all FAs can serve as energy source via β-oxidation, the observed preference towards specific acyl chains in different fiber types is likely attributed to their role as membrane building blocks rather than their involvement in energy production.61,62 The degree of unsaturation of acyl tails in phospholipids directly affects membrane fluidity. In particular, the presence of unsaturated acyl chains decreases the packing efficiency of membrane phospholipids thereby keeping the membrane more fluid at physiological temperatures.63,64 The fluidity of skeletal muscle membranes plays an important role in regulating the local lipid environment and protein conformations involved in multiple signaling processes. These may include Ca2+ signaling, which regulates the contractile properties and fatigue resistance,65 activation of carnitine palmitoyltransferases, which regulates fatty acid oxidation,66 and insulin-mediated glucose metabolism which regulates glucose disposal and hence blood sugar levels,56 among others. Given the prevalence of PUFAs in oxidative fibers, it is reasonable to assume that the high content of polyunsaturated acyl chains in oxidative fibers, especially DPA and DHA, results in a more fluid and permeable membrane thus facilitating many protein/receptor interactions associated with an endurance phenotype.67 In contrast, the prevalence of PA and relatively small number of PUFA in lipids observed in glycolytic fibers increases the rigidity and thereby constrains molecular diffusion in the lateral plane of the membrane, which may inhibit the activation of oxidative metabolism. Indeed, it has been reported that a more rigid membrane can impair insulin's ability to buffer plasma glucose concentration by altering the insulin receptor binding affinity or the ability to translocate glucose transporters.68–70 As a result, rigid membranes likely present greater insulin resistance, a feature displayed mainly by glycolytic fibers.56 In contrast, the incorporation of DHA and DPA into membrane phospholipids through dietary supplementation has been reported to enhance insulin action,71–73 improve fatigue resistance and oxidative capacity. Moreover, EPA and DHA are natural ligands of the peroxisome proliferator-activated receptors (PPARs) that regulate the expression of genes associated with lipid metabolism.67 All these properties are found in an endurance/oxidative fiber profile.
We have also observed a series of truncated oxidized phospholipids that display the same enhancement in oxidative fibers as phospholipids with highly unsaturated acyl tails Fig. S11 and S12.† Oxidized phospholipids are known to be formed under oxidative stress conditions.74 ROS formed during aerobic respiration attack C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds in PUFAs thereby promoting lipid radical peroxidation. In this study, we observed multiple oxidized species including PC 18:0_5:0(COOH) and PC 18:0_4:1(COOH) produced by oxidation of PC 18:0_20:4; PC 18:0_9:0(COOH) and PC 18:0_12:1;O; COOH produced by oxidation of PC 18:0_18:2; and PC 18:0_22:6; O produced by oxidation of PC 18:0_22:6.75 These assignments were manually confirmed using MS/MS as shown in Fig. S14† and are also listed in Table S3.† The enhanced abundance of PUFAs in oxidative fibers can also lead to the formation of oxylipins, another important group of lipid mediators.76 Because precursor FAs are enhanced in oxidative fibers, it is likely that this fiber type is also enriched in oxylipins as reported in a previous study.77 In this study, we identified 9-HODE and its derivative 9-oxoODE, which are produced from the oxidation of linoleic acid, LA (Fig. S11†). In summary, these results indicate that the membrane acyl chain composition is strongly linked to the regulation of metabolic properties of oxidative and glycolytic skeletal muscles.
C bonds in PUFAs thereby promoting lipid radical peroxidation. In this study, we observed multiple oxidized species including PC 18:0_5:0(COOH) and PC 18:0_4:1(COOH) produced by oxidation of PC 18:0_20:4; PC 18:0_9:0(COOH) and PC 18:0_12:1;O; COOH produced by oxidation of PC 18:0_18:2; and PC 18:0_22:6; O produced by oxidation of PC 18:0_22:6.75 These assignments were manually confirmed using MS/MS as shown in Fig. S14† and are also listed in Table S3.† The enhanced abundance of PUFAs in oxidative fibers can also lead to the formation of oxylipins, another important group of lipid mediators.76 Because precursor FAs are enhanced in oxidative fibers, it is likely that this fiber type is also enriched in oxylipins as reported in a previous study.77 In this study, we identified 9-HODE and its derivative 9-oxoODE, which are produced from the oxidation of linoleic acid, LA (Fig. S11†). In summary, these results indicate that the membrane acyl chain composition is strongly linked to the regulation of metabolic properties of oxidative and glycolytic skeletal muscles.
Conclusions
In this work, we demonstrate the power of high-resolution molecular imaging using nano-DESI MSI in combination with immunohistochemical fiber typing to examining molecular signatures of skeletal muscle fibers. The high specificity towards each fiber type allows us to accurately measure chemical gradients across muscle tissue, especially in complex regions where multiple myofiber types coexist. Our results indicate that moderate chemical gradients are present between oxidative and glycolytic fibers. We found that only type IIb myofiber may be differentiated from other fiber types based on the LDI analysis. The molecular signatures of type IIb myofibers are consistent with glycolytic metabolism. Interestingly, molecular signatures of type IIx fibers, which are commonly categorized as glycolytic fibers, are indicative of oxidative metabolism. Finally, we provide insights into the biological role of the species detected in this study either as membrane components, energy-related compounds, or antioxidants. Our results provide important insights into the skeletal muscle physiology by linking the biochemical pathways with the known contractile properties of different types of myofibers. The multimodal workflow established in this study may be extended to the analysis of other highly heterogenous and complex tissues such as pancreatic islets.Experimental
Materials
LC-MS grade methanol (MeOH) and LC-MS water were purchased from Sigma-Aldrich (St Louis, MO). Lipid standards lyso-phosphatidylcholine (LPC) 19:0, lyso-phosphatidylethalonamine (LPE) 17:1, lyso-phosphatidylglycerol (LPG) 17:1, lyso-phosphatidylserine (LPS) 17:1 and lyso-phosphatidylinositol (LPI) 17:1 were purchased from Avanti Polar Lipids (Alabaster, AL). Arachidonic acid (AA)-d8 standard was purchased from Cayman (Ann Arbor, MI).Primary antibodies used for fiber types staining were purchased from Developmental Studies Hybridoma Bank (DSHB) (Iowa City, IA): Myh7 MyHC-1 (BA-F8) for type I fiber, Myh2 MyHC-2A (2F7) for type IIA fiber, and Myh4 MyHC-2B (10F5) for type IIB fiber. Secondary antibodies used in the staining were purchased from Thermo Fisher Scientific (Waltham, MA): goat anti-mouse IgG1, Alexa Fluor 568 (A-21124); goat anti-mouse IgG2b, Alexa Fluor 647 (A-21242); goat anti-mouse IgM, Alexa Fluor 488 (A-210420).
Tissue collection
Three C57BL/6 mice (3-4 month old) used in this study were originally obtained from Jackson Laboratory (Bar Harbor, ME) and maintained in the animal facility with free access to standard rodent chow and water. All the procedures involving mice were approved by the Purdue University Animal Care and Use Committee (Protocol# 1112000440). Whole gastrocnemius muscle was dissected and frozen immediately in liquid nitrogen. Frozen muscle was fixed on optimal cutting temperature compound (OCT compound) with 60% of the tissue intact from OCT compound. The muscle was cross-sectioned with a thickness of 12 μm using a Leica CM1850 cryostat.Immunofluorescence staining
Gastrocnemius sections were directly incubated in blocking buffer (5% goat serum, 2% bovine serum albumin, 0.1% Triton X-100 and 0.1% sodium azide in PBS) for 1 h at room temperature. Samples were then incubated with primary antibodies diluted in blocking buffer overnight at 4 °C. After washing with PBS, the samples were incubated with secondary antibodies for 1 h at room temperature. Images were captured using a Leica DM 6000B microscope with a 20× objective and merged by Photoshop software.Nano-DESI MSI
Nano-DESI MSI experiments were carried out on a Q-Exactive HF-X Orbitrap mass spectrometer from Thermo Fisher Scientific (Waltham, MA) using a custom-designed nano-DESI source.21,78 The high-resolution nano-DESI probe was assembled by forming a ∼90° angle between two finely pulled fused silica capillaries with o.d. ∼20 μm: the primary and nanospray capillaries. A third finely pulled capillary was placed next to the nano-DESI probe to serve as a shear force probe. Detailed explanation of the preparation of the primary, nanospray and shear force capillaries and their assembling can be found in our previous work.19 A solution of MeOH:H2O (9:1) (v/v) containing 200 nM of lipid standards was used as the extraction solvent and was infused using a syringe pump at 0.5 μL min−1. Ionization was achieved by applying a 4 kV potential to the syringe needle. The heated capillary inlet was held at 30 V and 250 °C.For all the data reported in this study, we used a scan rate of 10 μm s−1, a step between the lines of 15 μm and an acquisition rate of 7 Hz resulting in an average pixel size of 0.7 μm × 15 μm. The spatial resolution of 6 μm was estimated using the 20/80 rule as shown in Fig. S2.† Both positive and negative mode data were collected for each biological replicate (n = 3) using a mass resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m/Δm at m/z 200 with a mass range of m/z 133–2000. MS/MS data for the endogenous species observed in nano-DESI MSI experiments were acquired directly from tissue sections using data dependent MS/MS mode at a normalized collision energy of 25 V and mass isolation window of 0.5 m/z.
000 m/Δm at m/z 200 with a mass range of m/z 133–2000. MS/MS data for the endogenous species observed in nano-DESI MSI experiments were acquired directly from tissue sections using data dependent MS/MS mode at a normalized collision energy of 25 V and mass isolation window of 0.5 m/z.
MSI data processing
MS data from nano-DESI experiments were acquired as RAW files using Xcalibur software (Thermo Electron, Bremen, Germany). Ion images were generated from. RAW files for a list of m/z values using a custom Python script (https://github.com/LabLaskin/MSI-image-generator). We used a mass window of 10 ppm to extract peak intensities and aligned MS signals with respect to the acquisition time. In each pixel, peak intensities were normalized to the TIC to compensate for signal fluctuations during imaging experiments. Ions in the m/z list were annotated by searching against METLIN database (https://metlin.scripps.edu) using accurate mass and MS/MS data.Image registration
Computational image registration was used to correlate the molecular information from both MSI and IF imaging experiments. We used a strategy reported in previous studies.79,80 Herein, we regarded the MSI image as the fixed image and immunofluorescence image as the moving image, respectively. A linear transformation model was adopted, in which translation and affine transformations were performed in tandem. To optimize the parameters in the transformation model, we selected mutual information as a criterion and adaptive stochastic gradient descent as an optimizer in this computation. After the optimal parameters were determined, the linear model can transform the moving IF image to spatially align it to the fixed MSI image. Since the mutual information metric only evaluates intensity similarity between two images with the same dimensions, we converted both MSI and immunofluorescence images into grayscale images before the registration. For MSI data, we used PCA to compress the hyperspectral data into a low-dimensional space while preserving molecular distribution information in the original high-dimensional data.80 Additional details of the PCA analysis of nano-DESI MSI data can be found in our previous report.28We validated the fiber ROI generated from IF images for ion imaging analysis using quantitative correlation analysis. Surrounding contour ROIs with varying spatial distance away from fiber locations were obtained using the dilation morphology algorithm with registered IF imaging data. This enables the extraction of MS signals in locations surrounding fiber ROI. Finally, ion signals were correlated with their spatial distances away from fiber locations using a linear regression analysis.
Statistical analysis
Both multivariate and univariate statistics were exploited for the exploratory analysis of muscle biomarkers. To create a heatmap of metabolite abundance in different muscle fibers, MS signals were scaled by a z-score transformation, representing the number of standard deviations from the mean value. Linear discriminant analysis (LDA) was used to map mass spectra from muscle fibers into a 2D space of the co-registered immunofluorescence images. In univariate analysis, the difference between metabolite MS signals in muscle fibers was analyzed using the volcano plot by plotting a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 fold change versus p-value. Herein, the p-value of one metabolite observed in mass spectra of two types of muscle fibers was calculated using the t-test.
2 fold change versus p-value. Herein, the p-value of one metabolite observed in mass spectra of two types of muscle fibers was calculated using the t-test.
Abbreviations
| AA | Arachidonic acid |
| ATGL | Adipose triglyceride lipase |
| CAR | Carnitine |
| CID | Collision-induced dissociation |
| CL | Cardiolipin |
| DG | Diglyceride |
| DHA | Docosahexaenoic acid |
| DPA | Docosapentaenoic acid |
| FA | Fatty acid |
| FC | Fold-change |
| GAS | Gastrocnemius muscle |
| HSL | Hormone sensitive lipase |
| IF | Immunofluorescence |
| LA | Linoleic acid |
| LC-MS | Liquid chromatography-mass spectrometry |
| LDA | Linear discriminant analysis |
| LDs | Lipid droplets |
| MALDI | Matrix assisted laser desorption ionization |
| MG | Monoglyceride |
| MGL | Monoacylglycerol lipase |
| MS/MS | Tandem mass spectrometry |
| MSI | Mass spectrometry imaging |
| MUFA | Monounsaturated fatty acid |
| MyHC | Myosin heavy chain |
| Nano-DESI | Nanospray desorption electrospray ionization |
| OCT | Optimal cutting temperature |
| PA | Palmitic acid |
| PC | Phosphatidylcholine |
| PCA | Principal component analysis |
| PE | Phosphatidylethanolamine |
| PPARs | Peroxisome proliferator-activated receptors |
| PUFA | Polyunsaturated fatty acid |
| ROI | Region of interest |
| ROS | Reactive oxygen species |
| SA | Stearic acid |
| SFA | Saturated fatty acid |
| TG | Triglyceride |
| TIC | Total ion current |
Data availability
The custom Python script developed for data visualization and registration is available at https://github.com/LabLaskin/MSI-image-generator.Author contributions
DU and JL designed the study and co-wrote the manuscript. DU performed the imaging experiments and processed the data. HH developed approaches for processing and visualization of the imaging data and for image registration. RY, ALP and MY contributed to data analysis. JQ, FY and SK provided the gastrocnemius tissue sections along with the immunofluorescence information and were involved in the interpretation of the results. All authors contributed to the reviewing and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is supported by the grants from the National Science Foundation (NSF-1808136 and NSF-2108729, JL) and National Institute of Health (R01 DK132819, SK).References
- K. Mukund and S. Subramaniam, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2020, 12, e1462 Search PubMed.
- S. Schiaffino and C. Reggiani, Physiol. Rev., 2011, 91, 1447–1531 CrossRef CAS PubMed.
- S. Schiaffino, FEBS J., 2018, 285, 3688–3694 CrossRef CAS PubMed.
- M. Hargreaves and L. L. Spriet, Nat. Metab., 2020, 2, 817–828 CrossRef CAS PubMed.
- W. Scott, J. Stevens and S. A. Binder-Macleod, Phys. Ther., 2001, 81, 1810–1816 CrossRef CAS PubMed.
- C. Mo, Y. Du and T. M. O'Connell, Curr. Osteoporosis Rep., 2021, 19, 151–157 CrossRef PubMed.
- M. Gueugneau, C. Coudy-Gandilhon, L. Théron, B. Meunier, C. Barboiron, L. Combaret, D. Taillandier, C. Polge, D. Attaix, B. Picard, J. Verney, F. Roche, L. Féasson, J. C. Barthélémy and D. Béchet, J. Gerontol., Ser. A, 2015, 70, 566–576 CrossRef CAS PubMed.
- R. Uchitomi, Y. Hatazawa, N. Senoo, K. Yoshioka, M. Fujita, T. Shimizu, S. Miura, Y. Ono and Y. Kamei, Sci. Rep., 2019, 9, 1–11 CrossRef CAS PubMed.
- N. Goto-Inoue, K. Yamada, A. Inagaki, Y. Furuichi, S. Ogino, Y. Manabe, M. Setou and N. L. Fujii, Sci. Rep., 2013, 3, 1–9 Search PubMed.
- B. Spengler, Anal. Chem., 2015, 87, 64–82 CrossRef CAS PubMed.
- Y. Hu, Z. Wang, L. Liu, J. Zhu, D. Zhang, M. Xu, Y. Zhang, F. Xu and Y. Chen, Chem. Sci., 2021, 12, 7993 RSC.
- Y. H. Tsai, T. J. Garrett, C. S. Carter and R. A. Yost, J. Am. Soc. Mass Spectrom., 2015, 26, 915–923 CrossRef CAS PubMed.
- N. Goto-Inoue, M. Morisasa, K. Machida, Y. Furuichi, N. L. Fujii, S. Miura and T. Mori, Rapid Commun. Mass Spectrom., 2019, 33, 185–192 CrossRef CAS PubMed.
- Y. Furuichi, N. Goto-Inoue, Y. Manabe, M. Setou, K. Masuda and N. L. Fujii, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 1699–1706 CrossRef CAS PubMed.
- N. Goto-Inoue, Y. Manabe, S. Miyatake, S. Ogino, A. Morishita, T. Hayasaka, N. Masaki, M. Setou and N. L. Fujii, Analytical and Bioanalytical Chemistry, Anal. Bioanal. Chem., 2012, 403, 1863–1871 CrossRef CAS PubMed.
- Z. Song, Z. Wang, H. Zhao, L. Cai, Z. Li, S. Zhang and X. Zhang, Analyst, 2020, 145, 6901–6909 RSC.
- Y. H. Tsai, D. R. Bhandari, T. J. Garrett, C. S. Carter, B. Spengler and R. A. Yost, Proteomics, 2016, 16, 1822–1824 CrossRef CAS PubMed.
- P. J. Roach, J. Laskin and A. Laskin, Analyst, 2010, 135, 2233–2236 RSC.
- R. Yin, K. E. Burnum-Johnson, X. Sun, S. K. Dey and J. Laskin, Nat. Protoc., 2019, 14, 3445–3470 CrossRef CAS PubMed.
- R. Yin, J. Kyle, K. Burnum-Johnson, K. J. Bloodsworth, L. Sussel, C. Ansong and J. Laskin, Anal. Chem., 2018, 90, 6548–6555 CrossRef CAS PubMed.
- S. N. Nguyen, R. L. Sontag, J. P. Carson, R. A. Corley, C. Ansong and J. Laskin, J. Am. Soc. Mass Spectrom., 2018, 29, 316–322 CrossRef CAS PubMed.
- D. Unsihuay, R. Yin, D. M. Sanchez, M. Yang, Y. Li, X. Sun, S. K. Dey and J. Laskin, Anal. Chim. Acta, 2021, 1186, 339085 CrossRef CAS PubMed.
- B. Paul, K. Kysenius, J. B. Hilton, M. W. M. Jones, R. W. Hutchinson, D. D. Buchanan, C. Rosty, F. Fryer, A. I. Bush, J. M. Hergt, J. D. Woodhead, D. P. Bishop, P. A. Doble, M. M. Hill, P. J. Crouch and D. J. Hare, Chem. Sci., 2021, 12, 10321–10333 RSC.
- H. Hu, J. P. Bindu and J. Laskin, Chem. Sci., 2022, 13, 90–98 RSC.
- P. Inglese, J. S. McKenzie, A. Mroz, J. Kinross, K. Veselkov, E. Holmes, Z. Takats, J. K. Nicholson and R. C. Glen, Chem. Sci., 2017, 8, 3500–3511 RSC.
- J. Sher and C. Cardasis, Acta Neurol. Scand., 1976, 54, 45–56 CrossRef CAS PubMed.
- S. L. Luxembourg, T. H. Mize, L. A. McDonnell and R. M. A. Heeren, Anal. Chem., 2004, 76, 5339–5344 CrossRef CAS PubMed.
- H. Hu, R. Yin, H. M. Brown and J. Laskin, Anal. Chem., 2021, 93, 3477–3485 CrossRef CAS PubMed.
- M. Murgia, L. Nogara, M. Baraldo, C. Reggiani, M. Mann and S. Schiaffino, Skeletal Muscle, 2021, 11, 1–19 CrossRef PubMed.
- J. C. Lavoie, P. Chessex, T. Rouleau, D. Migneault and B. Comte, Clin. Chem., 2004, 50, 135–140 CrossRef CAS PubMed.
- J. Fenoll, A. Martínez, P. Hellín and P. Flores, Food Chem., 2011, 127, 340–344 CrossRef CAS.
- N. Goto-Inoue, M. Morisasa, K. Kimura, T. Mori, Y. Furuichi, Y. Manabe and N. L. Fujii, Biosci., Biotechnol., Biochem., 2022, 86, 730–738 CrossRef PubMed.
- M. Flück and H. Hoppeler, Rev. Physiol., Biochem. Pharmacol., 2003, 146, 159–216 Search PubMed.
- J. Henriksson, Diabetes Care, 1992, 15, 1701–1711 CrossRef CAS PubMed.
- A. S. Pereyra, C. Te Lin, D. M. Sanchez, J. Laskin, E. E. Spangenburg, P. D. Neufer, K. Fisher-Wellman and J. M. Ellis, Mol. Metab., 2022, 59, 101456 CrossRef CAS PubMed.
- L. Braun, F. Puskás, M. Csala, G. Mészáros, J. Mandl and G. Bánhegyi, Free Radical Biol. Med., 1997, 23, 804–808 CrossRef CAS PubMed.
- A. Y. Cheng, E. Y. Tsui, A. J. G. Hanley and B. Zinman, Diabetes Care, 1999, 22, 1218–1219 CrossRef PubMed.
- L. Toniolo, L. Maccatrozzo, M. Patruno, F. Caliaro, F. Mascarello and C. Reggiani, J. Exp. Biol., 2005, 208, 4243–4253 CrossRef CAS PubMed.
- P. Hettige, U. Tahir, K. C. Nishikawa and M. J. Gage, BMC Genomics, 2020, 21, 1–16 CrossRef PubMed.
- J. C. Correia, Y. Kelahmetoglu, P. R. Jannig, C. Schweingruber, D. Shvaikovskaya, L. Zhengye, I. Cervenka, N. Khan, M. Stec, M. Oliveira, J. Nijssen, V. Martínez-Redondo, S. Ducommun, M. Azzolini, J. T. Lanner, S. Kleiner, E. Hedlund and J. L. Ruas, Cell Metab., 2021, 33, 2215–2230 CrossRef CAS PubMed.
- J. W. Curry, R. Hohl, T. D. Noakes and T. A. Kohn, J. Exp. Biol., 2012, 215, 3997–4005 CAS.
- M. Kohlmeier, in Nutrient Metabolism, 2015, pp. 265–477 Search PubMed.
- S. Rehunen, P. Karli and M. Härkönen, J. Neurol. Sci., 1985, 67, 299–306 CrossRef CAS PubMed.
- J. J. Wan, Z. Qin, P. Y. Wang, Y. Sun and X. Liu, Exp. Mol. Med., 2017, 49, e384 CrossRef CAS PubMed.
- R. Kohen, Y. Yamamoto, K. C. Cundy and B. N. Ames, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 3175–3179 CrossRef CAS PubMed.
- S. K. Powers, R. Deminice, M. Ozdemir, T. Yoshihara, M. P. Bomkamp and H. Hyatt, J. Sport Health Sci., 2020, 9, 415–425 CrossRef PubMed.
- S. Doğru-Abbasoğlu, N. Koçak-Toker and M. Uysal, in The Liver: Oxidative Stress and Dietary Antioxidants, 2018, pp. 295–304 Search PubMed.
- M. Wyss and R. Kaddurah-Daouk, Physiol. Rev., 2000, 80, 1107–1213 CrossRef CAS PubMed.
- A. Casey and P. L. Greenhaff, Am. J. Clin. Nutr., 2000, 72(2), 607S–617S CrossRef CAS PubMed.
- Y. M. Bruls, M. de Ligt, L. Lindeboom, E. Phielix, B. Havekes, G. Schaart, E. Kornips, J. E. Wildberger, M. K. Hesselink, D. Muoio, P. Schrauwen and V. B. Schrauwen-Hinderling, EBioMedicine, 2019, 49, 318–330 CrossRef CAS PubMed.
- M. R. McCann, M. V. G. De la Rosa, G. R. Rosania and K. A. Stringer, Metabolites, 2021, 11, 1–21 CrossRef PubMed.
- M. J. Watt, B. J. W. Van Denderen, L. A. Castelli, C. R. Bruce, A. J. Hoy, E. W. Kraegen, L. Macaulay and B. E. Kemp, Mol. Endocrinol., 2008, 22, 1200–1212 CrossRef CAS PubMed.
- M. J. Watt and A. J. Hoy, Am. J. Physiol., 2012, 302, 1315–1328 Search PubMed.
- W. W. T. Phua, M. X. Y. Wong, Z. Liao and N. S. Tan, Int. J. Mol. Sci., 2018, 19, 1425 CrossRef PubMed.
- A. Andersson, A. Sjödin, A. Hedman, R. Olsson and B. Vessby, Am. J. Physiol., 2000, 279, 42–44 Search PubMed.
- A. D. Kriketos, D. A. Pan, J. R. Sutton, J. F. Y. Hoh, L. A. Baur, G. J. Cooney, A. B. Jenkins and L. H. Storlien, Am. J. Physiol., 1995, 269, 38–45 Search PubMed.
- S. De Smet, K. Raes and D. Demeyer, Anim. Res., 2004, 53, 81–98 CrossRef CAS.
- J. E. Hergenreder, J. F. Legako, T. T. N. Dinh, P. R. Broadway, K. S. Spivey, J. O. Baggerman, J. P. Hutcheson, M. E. Corrigan and B. J. Johnson, Meat Muscle Biol., 2017, 1, 192–206 Search PubMed.
- L. E. Stefanyk, N. Coverdale, B. D. Roy, S. J. Peters and P. J. Leblanc, J. Membr. Biol., 2010, 234, 207–215 CrossRef CAS PubMed.
- F. Gao, E. J. Helm and D. Skowronska-Krawczyk, in Encyclopedia of Biological Chemistry, Elsevier, 3rd edn, 2021, vol. 2, pp. 695–706 Search PubMed.
- A. J. Hulbert, M. A. Kelly and S. K. Abbott, J. Comp. Physiol., B, 2014, 184, 149–166 CrossRef CAS PubMed.
- J. Leyton, P. J. Drury and M. A. Crawford, Br. J. Nutr., 1987, 51, 383–393 CrossRef PubMed.
- S. Ballweg, E. Sezgin, M. Doktorova, R. Covino, J. Reinhard, D. Wunnicke, I. Hänelt, I. Levental, G. Hummer and R. Ernst, Nat. Commun., 2020, 11, 756 CrossRef CAS PubMed.
- J. R. Silvius, in Lipid–Protein Interactions, ed. P. C. Jost and O. H. Griffith, John Wiley and Sons Inc., New York, 1982, vol. 2, pp. 239–281 Search PubMed.
- E. R. Chin, Exercise Sport Sci. Rev., 2010, 38, 76–85 CrossRef PubMed.
- K. Kashfi, R. L. Mynatt, E. A. Park and G. A. Cook, in Biochemical Society Transactions, Portland Press, 2011, vol. 39, pp. 833–837 Search PubMed.
- J.-M. Weber, J. Exp. Biol., 2009, 212, 593–597 CrossRef CAS PubMed.
- C. Grunfeld, K. L. Baird and C. R. Kahn, Biochem. Biophys. Res. Commun., 1981, 103, 219–226 CrossRef CAS PubMed.
- L. H. Storlien, D. A. Pan, A. D. Kriketos, J. O'Connor, I. D. Caterson, G. J. Cooney, A. B. Jenkins and L. A. Baur, Lipids, 1996, 31, S261–S265 CrossRef CAS PubMed.
- C. Moro and F. Capel, in Nutrition and Skeletal Muscle, Academic Press, 2018, pp. 367–378 Search PubMed.
- H. E. Kunz, S. Dasari and I. R. Lanza, Am. J. Physiol., 2019, 317, E460–E472 CAS.
- L. A. Baur, J. O'Connor, D. A. Pan, A. D. Kriketos and L. H. Storlien, Metab., Clin. Exp., 1998, 47, 106–112 CrossRef CAS PubMed.
- L. H. Storlien, A. B. Jenkins, D. J. Chisholm, W. S. Pascoe, S. Khouri and E. W. Kraegen, Diabetes, 1991, 40, 280–289 CrossRef CAS PubMed.
- G. O. Fruhwirth, A. Loidl and A. Hermetter, Biochim. Biophys. Acta, Mol. Basis Dis., 2007, 1772, 718–736 CrossRef CAS PubMed.
- Y. Matsuoka, M. Takahashi, Y. Sugiura, Y. Izumi, K. Nishiyama, M. Nishida, M. Suematsu, T. Bamba and K. ichi Yamada, Nat. Commun., 2021, 121(12), 1–12 Search PubMed.
- M. Gabbs, S. Leng, J. G. Devassy, M. Monirujjaman and H. M. Aukema, Adv. Nutr., 2015, 6, 513–540 CrossRef CAS PubMed.
- A. L. Penner, V. Waytt, T. Winter, S. Leng, T. A. Duhamel and H. M. Aukema, Appl. Physiol., Nutr., Metab., 2021, 46, 1378–1388 CrossRef CAS PubMed.
- I. Lanekoff, B. S. Heath, A. Liyu, M. Thomas, J. P. Carson and J. Laskin, Anal. Chem., 2012, 84, 8351–8356 CrossRef CAS PubMed.
- W. M. Abdelmoula, K. Škrášková, B. Balluff, R. J. Carreira, E. A. Tolner, B. P. F. Lelieveldt, L. Van Der Maaten, H. Morreau, A. M. J. M. Van Den Maagdenberg, R. M. A. Heeren, L. A. McDonnell and J. Dijkstra, Anal. Chem., 2014, 86, 9204–9211 CrossRef CAS PubMed.
- N. Verbeeck, J. M. Spraggins, M. J. M. Murphy, H. dong Wang, A. Y. Deutch, R. M. Caprioli and R. Van de Plas, Biochim. Biophys. Acta, Proteins Proteomics, 2017, 1865, 967–977 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Methods and data analysis, MSI images and complete clustering results. See DOI: https://doi.org/10.1039/d2sc06020e |
| This journal is © The Royal Society of Chemistry 2023 |