Open Access Article
Open Access ArticleLong-term spatiotemporal and highly specific imaging of the plasma membrane of diverse plant cells using a near-infrared AIE probe†
Jiaqi
Zuo‡
a,
Engao
Zhu‡
b,
Wenjing
Yin
b,
Chuangye
Yao
a,
Jiajia
Liao
a,
Xinni
Ping
a,
Yuqing
Zhu
a,
Xuting
Cai
a,
Yuchun
Rao
b,
Hui
Feng
 a,
Kewei
Zhang
a,
Kewei
Zhang
 *b and
Zhaosheng
Qian
*b and
Zhaosheng
Qian
 *a
*a
aKey Laboratory of the Ministry of Education for Advanced Catalysis Materials, College of Chemistry and Material Sciences, Zhejiang Normal University, Yingbin Road 688, Jinhua 321004, China. E-mail: qianzhaosheng@zjnu.cn
bCollege of Life Sciences, Zhejiang Normal University, Yingbin Road 688, Jinhua 321004, China. E-mail: kwzhang@zjnu.edu.cn
First published on 19th January 2023
Abstract
Fluorescent probes are valuable tools to visualize plasma membranes intuitively and clearly and their related physiological processes in a spatiotemporal manner. However, most existing probes have only realized the specific staining of the plasma membranes of animal/human cells within a very short time period, while almost no fluorescent probes have been developed for the long-term imaging of the plasma membranes of plant cells. Herein, we designed an AIE-active probe with NIR emission to achieve four-dimensional spatiotemporal imaging of the plasma membranes of plant cells based on a collaboration approach involving multiple strategies, demonstrated long-term real-time monitoring of morphological changes of plasma membranes for the first time, and further proved its wide applicability to plant cells of different types and diverse plant species. In the design concept, three effective strategies including the similarity and intermiscibility principle, antipermeability strategy and strong electrostatic interactions were combined to allow the probe to specifically target and anchor the plasma membrane for an ultralong amount of time on the premise of guaranteeing its sufficiently high aqueous solubility. The designed APMem-1 can quickly penetrate cell walls to specifically stain the plasma membranes of all plant cells in a very short time with advanced features (ultrafast staining, wash-free, and desirable biocompatibility) and the probe shows excellent plasma membrane specificity without staining other areas of the cell in comparison to commercial FM dyes. The longest imaging time of APMem-1 can be up to 10 h with comparable performance in both imaging contrast and imaging integrity. The validation experiments on different types of plant cells and diverse plants convincingly proved the universality of APMem-1. The development of plasma membrane probes with four-dimensional spatial and ultralong-term imaging ability provides a valuable tool to monitor the dynamic processes of plasma membrane-related events in an intuitive and real-time manner.
Introduction
Cells are at the central position of biology because they are the basic units of animal and plant structures and the activities of life. As an indispensable component of cells, the plasma membrane is an important barrier to isolate the internal and external environments of cells and plays a key role in cellular physiological processes such as cell migration, spreading, endocytosis, exocytosis and material transport across the membrane.1 Moreover, plasma membrane integrity is directly related to the integrity of biological cells, and thus an abnormality of the plasma membrane structure is closely related to cellular senescence, apoptosis and some physiological diseases.2 Therefore, it is of great significance to realize long-time in situ tracking imaging of plasma membranes. Fluorescence imaging technology is a powerful tool to help us conveniently and intuitively display the structure of living cells and monitor cellular processes within in a complex and large system.3 Currently, plasma membrane imaging mainly depends on fluorescent proteins and molecular fluorescent probes. Biologists generally track and monitor the endocytosis process in living cells using specific fluorescent proteins on the plasma membrane, but fluorescent proteins have also shown several disadvantages such as cumbersome operation and low photostability. The fluorescent proteins currently known are nearly always accompanied by a significant decrease in brightness during long-term fluorescence imaging,4,5 incomplete fluorophore maturation of fluorescent proteins frequently impairs signal readouts and their large size also restricts functionality for site-specific labeling.6 In addition to fluorescent proteins, scientists working in chemistry and biology and have developed a variety of small molecular dyes which can be used as markers of diverse subcellular organelles and track various biochemical and biophysical processes of cells.7–10 A wide choice of fluorophore and controlled structural design make molecular fluorescent probes promising tools to realize long-term in situ dynamic plasma membrane labelling for live-cell imaging.The currently-known molecular fluorescence probes for plasma membranes are mainly built on traditional fluorescent dyes with planar structures, and thus they inevitably have strict requirements of low-dose use and repetitive washing before imaging due to aggregation-caused quenching (ACQ).11 In contrast to those fluorescent dyes, luminogens with aggregation-induced emission (AIEgens) intensely fluoresce in the aggregate state but exhibit no fluorescence in solution, and a variety of AIEgens have been developed.12–16 It has been demonstrated that AIEgens have good potential for biological applications because they largely avoid the fluorescence quenching effect caused by aggregation.17–21 Diverse AIEgen-based molecular probes have been designed and used in subcellular localization,22 photodynamic therapy23,24 and cancer therapy,25,26 which provides an opportunity to design new plasma membrane probes based on AIEgens without the requirements of low-dose use and washing before imaging. The first set of AIEgen-based probes for plasma membranes showed good photostability and wash-free ability, but their poor solubility in water makes them lack imaging integrity and plasma membrane specificity.27,28 To solve this problem, charged groups like pyridinium and quaternary ammonium units were introduced into AIE-active probes to simultaneously increase the water solubility of the probes and enhance the interaction between the probes and the plasma membrane.29–33 These modified probes showed significant improvement in the imaging of plasma membranes including signal brightness and targeting integrity, but they still lack sufficient long-term tracking ability for plasma membranes because only half an hour of specific imaging of the plasma membrane can be achieved, and other parts of the cell such as the endoplasmic reticulum or mitochondria membrane can also be stained simultaneously in a short time.34,35 Therefore, it is extremely challenging and complex to achieve long-term and highly specific imaging of plasma membranes based on AIE-active molecular probes due to the high permeability of molecular probes to plasma membranes.36
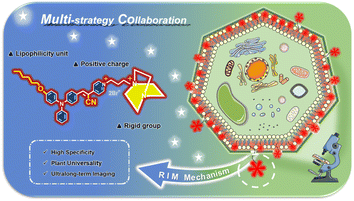
In contrast to the plasma membranes of animals, the complex structures of plant cells and the presence of diverse natural fluorescent compounds like chlorophylls in plant cells make the design of molecular probes for the plasma membranes of plants more challenging.37,38 Fluorescent staining of plant plasma membranes is generally indispensable to observe plasma-membrane-related processes, and FM dyes are widely used in selectively staining plant plasma membranes among the existing probes because of their good imaging performance.39,40 However, all probes including FM dyes penetrate fast through the plasma membrane to stain almost the whole plasma area within less than 30 min, and thus there is currently no fluorescent probe that can achieve long-term fluorescent staining of plant plasma membranes. Preliminary results of our previous work showed that the introduction of a rigid group might effectively enhance the retention ability of the probe inside the plasma membrane, but only a short-term imaging (1.5 h) experiment in animal cells was demonstrated with red-emitting probes. This provides a promising pathway to realize the ultralong-term imaging of plasma membranes of plant cells.41 For this purpose, several strategies including the similarity and intermiscibility principle, antipermeability strategy based on the steric hindrance effect, and strong electrostatic interactions with multiple charges were combined into an AIE-active molecular skeleton with near-infrared (NIR) emission in this work. α,β-Diaryl-acrylonitrile with a D–A pattern was chosen as the molecular skeleton since polar diarylethene derivatives generally possess longer emission wavelengths and the cyano group as a typical auxochrome can cause dramatic red-shift of the emission of the whole luminogen, which guarantees that the designed probe will possess near-infrared or red emission favouring bioimaging. Based on the similarity and intermiscibility principle, the target probe was designed as an amphiphilic molecule resembling phospholipid molecules (a major component of plasma membranes) by introducing a long alkyl chain at one end and a charged group at the other end. The charged group was endowed with two positive charges to enhance electrostatic interactions between the probe and plasma membrane, and a rigid group with large steric hindrance was introduced to effectively deter the diffusion-driven permeation of the molecular probe. Three-dimensional spatial imaging experiments and imaging experiments using various times demonstrate that the designed probe is capable of imaging plasma membranes of plant cells with high contrast and high specificity, and the probe still shows excellent imaging performance after 10 h of imaging time. It is also proved that this probe is generally applicable to plasma membranes of different cell types and diverse plants with comparable imaging performance, which convincingly verifies the validity of the multi-strategy collaboration for the plasma membrane probe and general universality of the probe.
Results and discussion
Design and synthesis of the NIR-emitting fluorescent probe via multi-strategy collaboration
A series of reported probes are based on AIEgens to improve their solubility and fluorescence properties for plasma membrane imaging.29,32 However, it is not difficult to find that these existing probes perform unsatisfactorily in imaging, such as regarding the imaging time and imaging integrity. Most physiological studies need a long imaging time to specifically track biological processes or functions. In addition, most currently developed plasma membrane probes based on AIEgens are applied to animal and human cells without cell walls.42–44 In contrast, the existence of a cell wall makes the plasma membrane imaging of plant cells more challenging. Regrettably, to date there have been few reports on fluorescent probes with AIE activity for the long-term specific tracking of plant plasma membranes. To overcome these problems, we propose a multi-strategy collaboration design principle for a plasma membrane probe by introducing different functional groups to synergistically achieve ultralong-term and specific imaging of plasma membranes. In the probe APMem-1 as shown in Scheme 1, AIE-active α,β-diaryl-acrylonitrile with a D–A pattern is exploited as the molecular skeleton, and a cyano group is introduced onto the central ethene unit to ensure the NIR emission of the whole molecule.45 A lipophilic unit with a suitable length is introduced to ensure a sufficiently strong interaction between the probe and the plasma membrane, while guaranteeing the penetration of the probe from the small cell wall pores to the plasma membrane. At the same time, two positive charges are included in another tail of the probe to ensure a sufficiently large water solubility while the rigid group 1,4-diazabicyclo[2.2.2]octane with large steric hindrance is introduced to effectively block the permeability of the probe through the plasma membrane, achieving ultralong-term imaging. Under the guidelines of the multiple-strategy collaboration, it is predicted that APMem-1 can show NIR emission, and the dynamic rotations of the aromatic rotors can guarantee its emission enhancement behavior when anchoring on the plasma membrane via restriction of the intramolecular motion mechanism. In actual cell tracking applications, emission in the NIR region is beneficial to deep tissue penetration and imaging, and can effectively avoid spectral overlap with biosubstrate autofluorescence. The introduction of a cyanide unit not only can contribute to the NIR emission but also change the linear structure of the central ethene of the molecular structure to be more rigid. The synthesis routes of APMem-1 are shown in Scheme S1.† All of the compounds were purified and characterized using 1H NMR, 13C NMR, and high-resolution mass spectrometry (HRMS).Photophysical properties and emission enhancement behaviour caused by restriction of the intramolecular motion of APMem-1
As shown in Fig. 1A, APMem-1 exists as a dark purple solid in daylight, which means that it has a wide absorption band in the visible light region. Its UV-visible spectrum in Fig. S1† confirms that APMem-1 in DMSO has a maximum absorption peak located at 525 nm with a broad absorption band from 420 nm to 660 nm, plus two major absorption peaks at 300 nm and 350 nm in the UV region. This feature is perfectly suited for fluorescent bioimaging via excitation by visible light. Amphiphilic APMem-1 can dissolve in highly polar solvents such as ethanol, H2O and DMSO, and its solution is almost non-emissive under UV irradiation. However, its solid can intensely emit red light located at 675 nm covering the NIR region upon UV irradiation, suggesting that it has significant aggregation-induced emission behaviour. Compared to the aggregation-induced emission enhancement, the emission enhancement induced by the restriction of intramolecular motion (RIM) of the probe in the environmental matrix is more practically valuable for plasma membrane probes, and thus the restriction of intramolecular motion behaviour was examined using adamantane as the solid matrix. The emission peak of APMem-1 is located at 688 nm when it is dispersed in adamantane (nAPMem-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nAdamantane = 1
nAdamantane = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in molar), and thus the dispersed state of the probe is very similar to its solid in terms of its fluorescence. The slight difference in the emission maximum can be attributed to a small difference in the conformation of the two states. They also have similarly short lifetimes of 1.8 ns and 2.6 ns in the solid and dispersed states, respectively (Fig. S2†). The dramatic emission enhancement of APMem-1 dispersed in adamantane where the APMem-1 molecules are restricted by surrounding adamantane molecules in comparison to its solution where the APMem-1 molecules are free without restriction clearly verifies the restriction of intramolecular motion behaviour of APMem-1. Due to the characteristic D–A structure of the probe, it is deduced that it might exhibit solvatochromic behaviour. Fig. S4† shows that the probe has polarity-dependent emission behaviour, but the determined values for the emission efficiency in most solvents are generally lower than 0.01, which excludes polarity change of the surroundings as the major contribution to emission enhancement in lighting up the plasma membrane. The photostability of APMem-1 was evaluated using UV-visible spectroscopy and PL spectroscopy under irradiation at 561 nm, and the results showed that no apparent changes in both absorption and emission were observed after 60 min of irradiation (Fig. S5 and S6†), indicating its good photostability. To further evaluate its fluorescence imaging performance for the plasma membrane, we used a stable single-layer micelle formed by sodium dodecylbenzene sulfonate (SDBS) in water to mimic the structure of the plasma membrane. The probe exhibits almost no emission in aqueous solution upon irradiation, and thus its red emission cannot be observed by the naked eye. However, its red emission can be gradually increased when APMem-1 is added into an aqueous solution containing SDBS as shown in Fig. 1B. It is noted that the fluorescence intensity progressively increases as the amount of APMem-1 added is increased from 0.0 to 20.0 μM, while this sharp emission enhancement is easily observed by the naked eye under UV light. It is easily understood that a large number of single-layer micelles are formed in solution when a large amount of SDBS is dissolved in water because SDBS is composed of a charged head and a long alkyl chain tail. Due to the similarity and intermiscibility principle, APMem-1 molecules tend to insert into the single-layer micelles formed by SDBS after it is added, and thus these APMem-1 molecules can be restricted inside the micelles leading to the dramatic emission enhancement as shown in Fig. 1C. As the concentration of APMem-1 increases, more and more APMem-1 molecules are inserted into the SDBS micelle, which makes the red emission brighter and brighter in solution. To exclude the possibility of autofluorescence of the probe, different amounts of SDBS were added into a solution of APMem-1, as shown in Fig. S3,† and the results indicate that the autofluorescence of the probe in solution is very weak, but its fluorescence is sharply increased upon the addition of SDBS. These observations provide direct and convincing evidence that APMem-1 can be used to image the plasma membrane-like micelles based on the working mechanism of restriction of intramolecular motion. In addition, intracellular pH frequently varies over a small range in many cellular events including endocytosis, senescence, transmembrane transport and cellular homeostasis,46,47 and thus resistance to small variations of pH is also very important for plasma membrane probes. Therefore, the influence of pH on APMem-1 was further evaluated using a micelle consisting of SDBS to mimic the plasma membrane as shown in Fig. S7.† It was found that the bright fluorescence of APMem-1 inserted in the SDBS micelle is hardly affected by the surrounding pH variation when the pH is changed from 5.7 to 8.0, indicating that the small pH variation frequently occurring inside live cells will not apparently influence the fluorescence stability of APMem-1. Such a feature of APMem-1 favours specific imaging of the plasma membrane without the interference of other factors.
50 in molar), and thus the dispersed state of the probe is very similar to its solid in terms of its fluorescence. The slight difference in the emission maximum can be attributed to a small difference in the conformation of the two states. They also have similarly short lifetimes of 1.8 ns and 2.6 ns in the solid and dispersed states, respectively (Fig. S2†). The dramatic emission enhancement of APMem-1 dispersed in adamantane where the APMem-1 molecules are restricted by surrounding adamantane molecules in comparison to its solution where the APMem-1 molecules are free without restriction clearly verifies the restriction of intramolecular motion behaviour of APMem-1. Due to the characteristic D–A structure of the probe, it is deduced that it might exhibit solvatochromic behaviour. Fig. S4† shows that the probe has polarity-dependent emission behaviour, but the determined values for the emission efficiency in most solvents are generally lower than 0.01, which excludes polarity change of the surroundings as the major contribution to emission enhancement in lighting up the plasma membrane. The photostability of APMem-1 was evaluated using UV-visible spectroscopy and PL spectroscopy under irradiation at 561 nm, and the results showed that no apparent changes in both absorption and emission were observed after 60 min of irradiation (Fig. S5 and S6†), indicating its good photostability. To further evaluate its fluorescence imaging performance for the plasma membrane, we used a stable single-layer micelle formed by sodium dodecylbenzene sulfonate (SDBS) in water to mimic the structure of the plasma membrane. The probe exhibits almost no emission in aqueous solution upon irradiation, and thus its red emission cannot be observed by the naked eye. However, its red emission can be gradually increased when APMem-1 is added into an aqueous solution containing SDBS as shown in Fig. 1B. It is noted that the fluorescence intensity progressively increases as the amount of APMem-1 added is increased from 0.0 to 20.0 μM, while this sharp emission enhancement is easily observed by the naked eye under UV light. It is easily understood that a large number of single-layer micelles are formed in solution when a large amount of SDBS is dissolved in water because SDBS is composed of a charged head and a long alkyl chain tail. Due to the similarity and intermiscibility principle, APMem-1 molecules tend to insert into the single-layer micelles formed by SDBS after it is added, and thus these APMem-1 molecules can be restricted inside the micelles leading to the dramatic emission enhancement as shown in Fig. 1C. As the concentration of APMem-1 increases, more and more APMem-1 molecules are inserted into the SDBS micelle, which makes the red emission brighter and brighter in solution. To exclude the possibility of autofluorescence of the probe, different amounts of SDBS were added into a solution of APMem-1, as shown in Fig. S3,† and the results indicate that the autofluorescence of the probe in solution is very weak, but its fluorescence is sharply increased upon the addition of SDBS. These observations provide direct and convincing evidence that APMem-1 can be used to image the plasma membrane-like micelles based on the working mechanism of restriction of intramolecular motion. In addition, intracellular pH frequently varies over a small range in many cellular events including endocytosis, senescence, transmembrane transport and cellular homeostasis,46,47 and thus resistance to small variations of pH is also very important for plasma membrane probes. Therefore, the influence of pH on APMem-1 was further evaluated using a micelle consisting of SDBS to mimic the plasma membrane as shown in Fig. S7.† It was found that the bright fluorescence of APMem-1 inserted in the SDBS micelle is hardly affected by the surrounding pH variation when the pH is changed from 5.7 to 8.0, indicating that the small pH variation frequently occurring inside live cells will not apparently influence the fluorescence stability of APMem-1. Such a feature of APMem-1 favours specific imaging of the plasma membrane without the interference of other factors.
In vivo three-dimensional spatial imaging of plasma membranes of plant cells
In contrast to plasma membrane probes for animal and human cells, the design of molecular probes for plant plasma membranes is challenging because plant cells have complex cell walls around their cells in addition to plasma membranes. The small size of the pores of the plant cell wall range between 3.5 nm and 6 nm, which requires that the designed probe has the right molecular size and sufficiently high solubility, enabling it to easily diffuse into the plant plasma membrane through the cell wall. Thus, we introduced an intermediate length of alkyl chain with six carbons into the APMem-1 to meet the size requirement and achieve the rapid staining of the plasma membrane of all the cells in a plant. Before evaluating its performance in three-dimensional spatial imaging of plasma membranes, the toxicity of APMem-1 was first tested using the whole morphology changes of Arabidopsis thaliana. The toxicity tests in Fig. S8† show that the whole morphologies of the seedlings treated with APMem-1 are very similar to those of the mock group, and the average root lengths of the seedlings incubated with APMem-1 are also comparable to those of the mock group when the incubation concentrations are lower than 20.0 μM, indicating that a concentration of APMem-1 lower than 20.0 μM can be used to stain plant cells without toxicity to the whole plant. To evaluate the three-dimensional spatial imaging performance of APMem-1, Arabidopsis thaliana was selected as the model plant, and various concentrations of APMem-1 from 5.0 μM to 20.0 μM were used to treat the its seedlings for condition optimization (Fig. S10†). A control experiment was also performed to confirm the negligible influence of autofluorescence from plant cells under the same recording conditions (Fig. S9†). It was found that the plasma membranes of all the cells in the root tip could be stained after the seedlings had been treated with a solution of APMem-1 at 5.0 μM with incubation for 5 min, but the signal from the cells at the central transection was very weak so that the plasma membrane in those zones could not be clearly observed. When the concentration of APMem-1 is increased to 10.0 μM, all of the plasma membrane can be clearly stained, and bright red light is observed. The imaging performance at a higher concentration of 20.0 μM is comparable to that at 10.0 μM under the same incubation conditions, indicating that such a concentration is suitable for precise imaging of the plasma membranes of all the cells. Thus, the three-dimensional spatial staining images of the seedling root tip were recorded with a Leica TCS SP5 model confocal laser scanning microscope under the optimized conditions. Fig. 2A shows three-dimensional spatial staining images and three transection staining images of the seedling root tips along the X, Y and Z axes. It can be clearly observed that the plasma membranes of all the cells of the whole root tip are clearly stained with bright red signal, and the signal gradually intensifies from the central part to the surface which is consistent with the diffusion-controlled distribution for APMem-1. The three-axis transection staining images also verify the successful staining of plasma membranes of all the cells of the root tip, and the signal from the surface cells is much brighter than that from the central parts. However, a clear signal from the cells in the central transection can still be recorded, suggesting that APMem-1 is able to stain plasma membranes of all the target plant cells in a very short incubation time. To further quantify the performance of APMem-1 in spatial imaging, high-resolution image analysis of a transection of the root tip was performed, as shown in Fig. 2B. It can be directly observed from the high-resolution image that all the cells are clearly marked with bright red emission from the region of the plasma membranes. The ratio of the imaging signal area to the whole-cell area (SImaging) is only 10.2%, and the imaging integrity of all the cells is up to 100%, which clearly proves that APMem-1 is able to specifically stain complete plasma membranes of all the cells without staining the other areas of the cell apart from the cell walls. The ratio of the signal intensity from the plasma membrane to that from the cell interior (fSignal/fBackground) was also determined for the central cells and peripheral epidermal cells with different brightnesses due to the different concentrations of APMem-1 caused by diffusion. The average values of fSignal/fBackground for plasma membranes from the central cells and peripheral epidermal cells are 106.3 and 144.4 respectively, which is consistent with the direct observation by the naked eye. These large values clearly indicate that the signal-to-noise ratio is sufficiently high to distinguish between plasma membranes and the other regions of the cells. Finally, plasmolysis experiments were performed using onion epidermal cells as an example to verify whether the plasma membrane, cell wall, or both can show an emission signal due to APMem-1. The onion epidermal cells were first treated with APMem-1 (10.0 μM) for 5 min, and their laser scanning confocal microscopy images show that both the plasma membranes and cell walls of the observed cells show a clear red emission signal under such conditions (Fig. S11†). After 20 min of treatment with a sucrose solution, it is clearly observed that all the cells have dramatically dehydrated and shrunk to similarly oval shapes, and the cell walls and plasma membranes are remarkably separated so that they can be easily distinguished by the naked eye. It is noted that all the plasma membranes separated from the surrounding cell walls exhibit a continuous bright red signal whereas the cell walls show discontinuous signals, which convincingly verifies that APMem-1 is capable of effectively penetrating the cell walls to stain the plasma membranes with high specificity and high brightness. For comparision, the same experiments were conducted using commercial FM 1-43 and FM 4-46. Fig. S12† shows that both can clearly stain the plasma membrane and cell wall, but FM 1-43 is mainly located at cell walls in a discontinuous manner, and only a small amount of it is distributed at plasma membranes after plasmolysis, whereas FM 4-64 performs well in both staining of cell walls and plasma membranes, comparable to the performance of our APMeme-1. To avoid the emission enhancement effect of the probe caused by viscous sucrose in the cell walls, sodium chloride was used to replace sucrose, and the results in Fig. S13† clearly show that the probe mainly stains the plasma membrane rather than both the plasma membrane and cell wall. To further verify the targeting capability of APMem-1 for plasma membranes, the plasma membranes of root cells of Arabidopsis thaliana were first stained with APMem-1 and then destroyed by treatment with DMSO. As shown in Fig. S14,† only the red signal from APMem-1 located at the plasma membranes can be clearly recorded before treatment of the root cells with DMSO. After the treatment with DMSO, the other areas of the plant cells also exhibited a red signal, which convincingly proves that APMem-1 can only specifically stain plasma membranes when the membrane structure is complete.Ultralong-term imaging of the plasma membranes of plant cells
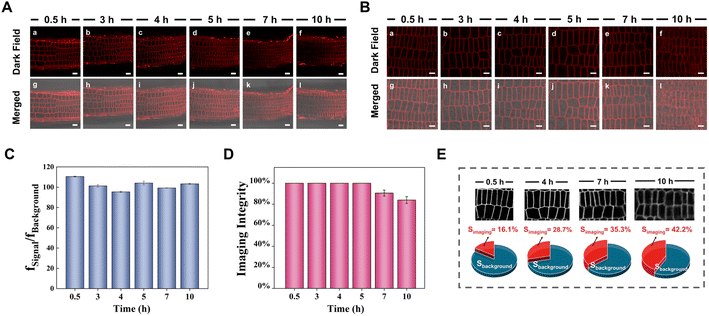
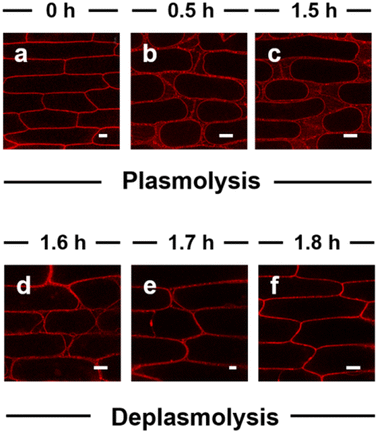
Long-term tracking of plasma membranes is generally required for understanding some specific biological processes relating to the plasma membrane, but few plasma membrane probes can achieve long-term imaging with high specificity without staining the other areas of the cells due to the diffusion-driven homogenous distribution. To evaluate the ultralong-term imaging performance of APMem-1 for plasma membranes of seedling root cells of Arabidopsis thaliana, the photostability of APMem-1 was first investigated through continuous irradiation at 561 nm with the excitation source of the laser scanning confocal microscope (Fig. S15†). It is noted that no detectable changes in the emission signals from the plasma membranes of APMem-1-stained cells are observed after continuous irradiation for 90 min, and the bright red signal from the plasma membrane is well retained under such continuous irradiation, which suggests that APMem-1 has good photostability suited to long-term imaging of plasma membranes. Imaging experiments with various times of plasma membranes stained with APMem-1 were carried out to explore the longest imaging time using seedling root cells of Arabidopsis thaliana as the example, as shown in Fig. 3 and S16.† All the seedling roots were first treated with APMem-1 at a concentration of 10.0 μM for 5 min, and then the corresponding staining images of the central transections of the root tips were recorded at various time ranging from 0.5 to 10 h, during which the seedlings were continuously incubated with the culture medium to maintain their biological activity. Fig. 3A shows the staining images of the overall sections of the root tips at various times, and it can be seen that the brightness of the entire red signal from the plasma membranes is maintained, and all the plasma membranes are marked with a red signal with nearly 100% imaging integrity at various times from 0.5 h to 10 h. However, the signal from the plasma membranes of a small number of cells has a lower intensity than that from the others for the seedlings recorded at 7 h and 10 h, indicating that a remarkably heterogenous distribution of APMem-1 among different cells caused by molecular diffusion takes place after more than 7 h. To more clearly observe the change in imaging performance of APMem-1 at various imaging times, enlarged high-resolution images of a small number of well-stained cells of the seedlings with the probe are shown in Fig. 3B. It is clear that there is no detectable change in signal brightness for these stained cells at different recording times, and the plasma membranes of all the cells in the selected regions recorded at different times are clearly stained with the red signal, but the red signals from the plasma membranes start becoming blurred after 7 h and become obviously obscured after 10 h, indicating that permeation of APMem-1 through the plasma membranes takes place after 7 h. The same imaging experiments at various times were also conducted using FM 1-43 and FM 4-64 for comparison. Fig. S17† shows that FM 1-43 stained most of the plasma area of the stained cells after only 5 min while almost the entirety of the cells exhibited emission after 20 min of incubation with FM 4-64 except the cell nuclei, which convincingly proves that both FM 1-43 and 4-64 are not suitable for long-term imaging of plasma membranes. In order to more intuitively quantify the imaging performance of APMem-1 for plasma membranes at different imaging times, we quantitatively analyzed these enlarged high-resolution images considering three aspects: average fluorescence contrast ratio (fSignal/fBackground), imaging integrity of the plasma membranes, and the ratio of the signal area to the whole area (SImaging). It is found that the values of fSignal/fBackground remain stable with relatively high values of around 100 for various imaging times from 0.5 to 10 h, and the value of fSignal/fBackground still remains at 103 even if the imaging time is up to 10 h, which indicates that plasma membranes can be clearly stained with high contrast ratios during different imaging times due to the high specificity and brightness of APMem-1 (Fig. 3C and S18†). The imaging integrity of the plasma membrane represents the ability of the probe to homogenously target plasma membranes by penetrating cell walls. Fig. 3D shows that the values of the imaging integrity reach 100% for imaging times of less than 5 h, but the values are slightly decreased to 91% for an imaging time of 7 h and to 81% for an imaging time of 10 h, which also suggests that APMem-1 can specifically stain the whole plasma membranes of all the cells after 5 h of free diffusion. These observations prove that APMem-1 has excellent resistance to diffusion-driven signal weakening within 10 h. The specificity of APMem-1 to the plasma membrane was further quantified using the ratio of the signal area to the whole area of the cell (SImaging). The staining of other cell areas caused by time-dependent permeation is generally enhanced as the imaging time is increased as shown in Fig. 3E. The value of SImaging remains at a relatively low level of less than 36% for imaging times of less than 7 h, and its value is gradually increased to 42.2% after 10 h (Fig. S19†), indicating that less than a half of the whole cell area is stained with APMem-1 after such a long imaging time. These facts provide clear and convincing evidence that APMem-1 is capable of anchoring plasma membranes with high contrast and brightness without remarkable staining of other areas of the cells at a long imaging time of 10 h, which further validates the ultralong-term imaging ability of the molecular probes for plant plasma membranes based on the multi-strategy collaboration principle.The ultralong-term imaging ability of APMem-1 enables us to monitor in real time the morphological changes of the plasma membranes of plant cells caused by the external environment over a long timescale. Herein, we demonstrate real-time imaging of the morphological changes of the plasma membrane of onion epidermal cells during plasmolysis and deplasmolysis processes using the probe (Fig. 4 and S20†). Different concentrations of sodium chloride were used for plasmolysis, and it is found that when its concentration is lower than 0.2 M, no apparent plasmolysis is observed within 3 h. When the concentration of NaCl is up to 0.5–1.0 M, clear plasmolysis takes place within 1.5 h, in which the plasma membrane is gradually detached from its neighboring cell wall, and the plasma membrane progressively shrinks to an elliptical shape. It is clearly observed that the plasma membrane is stained with a bright red signal, and a discontinuous signal is distributed among the plasma membranes, while no signal can be observed inside the plasma area. During the following deplasmolysis, the plasma membrane quickly recovers to its initial shape, and its morphological shape is clearly marked by the probe within 0.3 h. It is noted that the plasma membrane was clearly marked in different shapes during the 1.8 h of plasmolysis and deplasmolysis. A comparison experiment with FM 4-64 showed that FM 4-64 is not able to always clearly stain the plasma membrane during plasmolysis because a large part of its signal is lost after 1.5 h during the plasmolysis process (Fig. S21†). This demonstration confirms that APMem-1 has real-time tracking ability for plasma membranes with high specificity in a biological process over a long period of time, which currently existing probes do not possess.
Universality of APMem-1 to stain the plasma membranes of plant cells of different types and diverse plants
In the previous section we demonstrated that APMem-1 is able to stain plant plasma membranes with extremely high specificity and ultralong imaging time using the model plant Arabidopsis thaliana as an example. To evaluate the universality of APMem-1 to stain plant plasma membranes of diverse plants, the imaging of plasma membranes with APMem-1 was further expanded to an additional three distinct plants including one vegetable (onion), one main grain crop (rice) and one economic crop (tobacco). The epidermis of onion, and seedling roots of rice and tobacco were incubated with APMem-1 under the same conditions as the seedling roots of Arabidopsis thaliana, and the images were recorded and analysed. Fig. 5 shows high-resolution fluorescence images of the epidermal cells of onion, and seedling root cells of Arabidopsis thaliana, rice and tobacco stained with APMem-1. It is worth noting that the plasma membranes of all four plants can be clearly and completely marked by bright red by APMem-1 without apparent differences, although they have very distinct cell shapes. The close values of fSignal/fBackground and the imaging integrity for the four plants suggest that comparable imaging performance of APMem-1 for onion, rice and tobacco is achieved to that for Arabidopsis thaliana, which further confirms the universality of the probe for plasma membranes of diverse plants. Compared to the transparent root cells of plants, leaf cells consist of more diverse cell types with distinct plant functions and more complex structural components, and thus specific staining of plasma membranes of leaf cells is more challenging due to their non-transparent and complex structures. Generally, a plant leaf is composed of a waxy cuticle, an upper epidermis, a lower epidermis and mesophyll cells abundant in chloroplasts, and guard cells are scattered among the epidermal cells to control the opening and closing of the stomates. Herein we used tobacco leaves as an example to demonstrate the specific staining of leaf plasma membranes with APMem-1. In contrast to the short treatment of root cells with the probe (5 min), the incubation of tobacco leaves with APMem-1 requires a relatively long time (60 min) to achieve effective staining performance because of the existence of the waxy cuticles on the leaf surfaces. Fig. 6 shows laser scanning confocal microscopy images of epidermal cells and guard cells on the upper surfaces of tobacco leaves treated with APMem-1 at different scales. It is worth noting that plasma membranes of both irregularly corrugated epidermal cells and semicircular coupled guard cells are clearly and completely marked with the red signal. The enlarged images clearly show that the whole plasma membranes of both larger epidermal cells and smaller guard cells are specifically stained, and no apparent signals are observed from the other areas in either case, indicating that APMem-1 can stain complex and nontransparent leaf cells with comparable imaging performance to that of the transparent root cells of a plant. Interestingly, it is found that the opening and closing states of the stomates are clearly tracked by imaging two distinct shapes of guard cells stained with APMem-1, which is valuable for monitoring the respiration and transpiration of plants involving stomata. Despite the complex and nontransparent nature of the leaves, the average value of fSignal/fBackground for leaf plasma membranes stained with APMem-1 (100) is very close to that for root plasma membranes (113), indicating that comparable imaging performance to that for more complex leaf plasma membranes can be achieved by APMem-1 by slightly changing the staining conditions. Compared to an imaging integrity of almost 100% for the root plasma membranes of tobacco, the imaging integrity for the leaf plasma membranes is slightly lower at 85%, as indicated by statistical analysis, which can be attributed to the protection effect of the waxy cuticles of the leaf surface. Therefore, it is convincingly concluded that APMem-1 is generally applicable to the highly specific imaging of plasma membranes of different cell types and diverse plant species.Conclusions
In summary, we proposed an effective design principle based on multiple-strategy collaboration for the AIE-active probe APMem-1 for the plasma membranes of live cells, and demonstrated its highly specific and ultralong-term imaging performance for plasma membranes of different cell types and diverse plant species. In the proposed design concept, the similarity and intermiscibility principle was used to target plasma membranes, while an antipermeability strategy based on the steric hindrance effect and strong electrostatic interactions through multiple charges guaranteed retention of the probe molecules inside the plasma membranes for a long time, and sufficiently high aqueous solubility of the probe, avoiding self-precipitation. In the APMem-1 probe, an AIE-active molecular skeleton with NIR emission was constructed to avoid or reduce the interference from the autofluorescence of plant cells, and its suitable molecular size allows the probe to quickly penetrate cell walls with small pores. Compared to the previous AIE probes and commercial dyes for plasma membranes, APMem-1 exhibits excellent features such as ultrafast staining, wash-free, and desirable biocompatibility, which may contribute to its suitability for the high-quality imaging of the plasma membranes of plant cells, as opposed to animal and human plasma membranes. The imaging time of APMem-1 can be up to 10 h to guarantee high-quality imaging performance. Long-term real-time monitoring of the morphological changes of plasma membranes during plasmolysis and deplasmolysis was achieved with the fluorescence probe for the first time, which provides the possibility to continuously monitor plasma membrane-related physiological processes on a long timescale. The validation of the imaging performance of the probe for plasma membranes of various plant cells convincingly proved that this probe is generally applicable to different cell types and diverse plant species.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
Jiaqi Zuo, Engao Zhu — conceive study, live-cell imaging and write original draft (contribute equally); Wenjing Yin, Chuangye Yao, Jiajia Liao, Xinni Ping, Yuqing Zhu, Xuting Cai, Yuchun Rao — discuss the results and refining of the ideas; Hui Feng — review & editing; Kewei Zhang — review editing, funding acquisition, supervision; Zhaosheng Qian — design of central ideas, carried out additional analyses, finalized this article and funding acquisition, supervision.Conflicts of interest
Z. Q, H. F. and J. Z. have submitted a patent application based on the APMem-1 dye described in this work.Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation of China (Grant No. 21775139 and 21675143) and the Natural Science Foundation of Zhejiang Province (Grant No. LR18B050001).Notes and references
- (a) E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS PubMed; (b) D. A. Ammendolia, W. M. Bement and J. H. Brumell, BMC Biol., 2021, 19, 71–99 CrossRef CAS PubMed; (c) A. J. Desai and L. J. Miller, Br. J. Pharmacol., 2018, 175, 4009–4025 CrossRef CAS PubMed; (d) S. M. Mylvaganam, S. Grinstein and S. A. Freeman, Semin. Immunopathol., 2018, 40, 605–615 CrossRef CAS PubMed.
- (a) B. Fadeel and D. Xue, Crit. Rev. Biochem. Mol. Biol., 2009, 44, 264–277 CrossRef CAS PubMed; (b) Z. Shi, Z. T. Graber, T. Baumgart, H. A. Stone and A. E. Cohen, Cell, 2018, 175, 1769–1779 CrossRef CAS PubMed; (c) J. A. Vance and N. K. Devaraj, J. Am. Chem. Soc., 2021, 143, 8223–8231 CrossRef CAS PubMed; (d) Y. Zhang, X. Chen, C. Gueydan and J. Han, Cell Res., 2018, 28, 9–21 CrossRef CAS PubMed.
- (a) P. Gao, W. Pan, N. Li and B. Tang, Chem. Sci., 2019, 10, 6035–6071 RSC; (b) S. I. Reja, M. Minoshima, Y. Hori and K. Kikuchi, Chem. Sci., 2021, 12, 3437–3447 RSC; (c) N. Niu, Y. Yu, Z. Zhang, M. Kang, L. Wang, Z. Zhao, D. Wang and B. Z. Tang, Chem. Sci., 2022, 13, 5929–5937 RSC.
- N. Yagi, A. Yoshinari, R. J. Iwatate, R. Isoda, W. B. Frommer and M. Nakamura, Plant Cell Physiol., 2021, 62, 1259–1268 CrossRef CAS PubMed.
- M. Hirano, R. Ando, S. Shimozono, M. Sugiyama, N. Takeda, H. Kurokawa, R. Deguchi, K. Endo, K. Haga, R. Takai-Todaka, S. Inaura, Y. Matsumura, H. Hama, Y. Okada, T. Fujiwara, T. Morimoto, K. Katayama and A. Miyawaki, Nat. Biotechnol., 2022, 40, 1132–1142 CrossRef CAS PubMed.
- (a) J. Liu and Z. Cui, Bioconjugate Chem., 2020, 31, 1587–1595 CrossRef CAS PubMed; (b) T. Taniyama, N. Tsuda and S. Sueda, ACS Chem. Biol., 2018, 13, 1463–1469 CrossRef CAS PubMed; (c) X. Zhang, Y. Deng, H. Chang, C. Ji, M. Zhang, J. Peng, T. Xu and P. Xu, Protein Cell, 2014, 5, 800–803 CrossRef CAS PubMed; (d) Y. Yano and K. Matsuzaki, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 2124–2131 CrossRef CAS PubMed.
- Y. Wang, S. Xu, L. Shi, C. Teh, G. Qi and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 14945–14953 CrossRef CAS PubMed.
- H. Park, G. Niu, C. Wu, C. Park, H. Liu, H. Park, R. T. K. Kwok, J. Zhang, B. He and B. Z. Tang, Chem. Sci., 2022, 13, 2965–2970 RSC.
- X. Wu, X. Wang, Y. Li, F. Kong, K. Xu, L. Li and B. Tang, Anal. Chem., 2022, 94, 4881–4888 CrossRef CAS PubMed.
- M. Jiang, X. Gu, J. W. Y. Lam, Y. Zhang, R. T. K. Kwok, K. S. Wong and B. Z. Tang, Chem. Sci., 2017, 8, 5440–5446 RSC.
- C. Liu, X. Gao, J. Yuan and R. Zhang, Trends Anal. Chem., 2020, 133, 116092–116127 CrossRef CAS.
- J. Luo, Z. Xie, J. W. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Chem. Commun., 2001, 18, 1740–1741 RSC.
- B. Tang, X. Zhan, G. Yu, P. P. Sze Lee, Y. Liu and D. Zhu, J. Mater. Chem., 2001, 11, 2974–2978 RSC.
- H. Tong, Y. Hong, Y. Dong, Y. Ren, M. Häussler, J. W. Y. Lam, K. S. Wong and B. Z. Tang, J. Phys. Chem. B, 2007, 111, 2000–2007 CrossRef CAS PubMed.
- J. Wang, J. Mei, R. Hu, J. Z. Sun, A. Qin and B. Z. Tang, J. Am. Chem. Soc., 2012, 134, 9956–9966 CrossRef CAS PubMed.
- J. Mei, Y. Hong, J. W. Lam, A. Qin, Y. Tang and B. Z. Tang, Adv. Mater., 2014, 26, 5429–5479 CrossRef CAS PubMed.
- J. Lin, X. Zeng, Y. Xiao, L. Tang, J. Nong, Y. Liu, H. Zhou, B. Ding, F. Xu, H. Tong, Z. Deng and X. Hong, Chem. Sci., 2019, 10, 1219–1226 RSC.
- C. Zhao, Y. Liu, W. Wang, Z. Wang and W. Lin, J. Photochem. Photobiol., A, 2021, 412, 113245–113252 CrossRef CAS.
- M.-Y. Wu, J.-K. Leung, C. Kam, T. Y. Chou, J.-L. Wang, X. Zhao, S. Feng and S. Chen, Sci. China: Chem., 2022, 65, 979–988 CrossRef CAS.
- B. He, J. Huang, J. Zhang, H. H. Y. Sung, J. W. Y. Lam, Z. Zhang, S. Yan, D. Wang, J. Zhang and B. Z. Tang, Angew. Chem., Int. Ed., 2022, 61, 202117709–202117719 Search PubMed.
- G. Niu, R. Zhang, X. Shi, H. Park, S. Xie, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Trends Anal. Chem., 2020, 123, 115769–115797 CrossRef CAS.
- F. Hu and B. Liu, Org. Biomol. Chem., 2016, 14, 9931–9944 RSC.
- M. Y. Wu, M. Gu, J. K. Leung, X. Li, Y. Yuan, C. Shen, L. Wang, E. Zhao and S. Chen, Small, 2021, 17, 2101770–2101779 CrossRef CAS PubMed.
- F. Hu, S. Xu and B. Liu, Adv. Mater., 2018, 30, 1801350–1801378 CrossRef PubMed.
- H. Liu, L. H. Xiong, R. T. K. Kwok, X. He, J. W. Y. Lam and B. Z. Tang, Adv. Opt. Mater., 2020, 8, 2000162–2000189 CrossRef CAS.
- D. Wang, M. M. S. Lee, G. Shan, R. T. K. Kwok, J. W. Y. Lam, H. Su, Y. Cai and B. Z. Tang, Adv. Mater., 2018, 30, 1802105–1802113 CrossRef PubMed.
- Y. Li, Y. Wu, J. Chang, M. Chen, R. Liu and F. Li, Chem. Commun., 2013, 49, 11335–11337 RSC.
- L. Shi, Y. H. Liu, K. Li, A. Sharma, K. K. Yu, M. S. Ji, L. L. Li, Q. Zhou, H. Zhang, J. S. Kim and X. Q. Yu, Angew. Chem., Int. Ed., 2020, 59, 9962–9966 CrossRef CAS PubMed.
- L. Hu, B. Xu, H. Chen and H. Wang, Sens. Actuators, B, 2021, 340, 129950–129957 CrossRef CAS.
- F. Meng, Y. Liu, J. Niu and W. Lin, RSC Adv., 2017, 7, 16087–16091 RSC.
- L. Shi, K. Li, Y. H. Liu, X. Liu, Q. Zhou, Q. Xu, S. Y. Chen and X. Q. Yu, Chem. Commun., 2020, 56, 3661–3664 RSC.
- D. Wang, H. Su, R. T. K. Kwok, X. Hu, H. Zou, Q. Luo, M. M. S. Lee, W. Xu, J. W. Y. Lam and B. Z. Tang, Chem. Sci., 2018, 9, 3685–3693 RSC.
- X. Zheng, D. Wang, W. Xu, S. Cao, Q. Peng and B. Z. Tang, Mater. Horiz., 2019, 6, 2016–2023 RSC.
- Y. Zheng, Y. Ding, J. Ren, Y. Xiang, Z. Shuai and A. Tong, Anal. Chem., 2020, 92, 14494–14500 CrossRef CAS PubMed.
- P. Alam, W. He, N. L. C. Leung, C. Ma, R. T. K. Kwok, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, K. S. Wong and B. Z. Tang, Adv. Funct. Mater., 2020, 30, 1909268 CrossRef CAS.
- (a) K. Li, Y. Lyu, Y. Huang, S. Xu, H. W. Liu, L. Chen, T. B. Ren, M. Xiong, S. Huan, L. Yuan, X. B. Zhang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2018033118 CrossRef CAS PubMed; (b) K. Hac Wydro, P. Wydro, A. Jagoda and J. Kapusta, Chem. Phys. Lipids, 2007, 150, 22–34 CrossRef CAS PubMed.
- M. Collot, E. Boutant, K. T. Fam, L. Danglot and A. S. Klymchenko, Bioconjugate Chem., 2020, 31, 875–883 CrossRef CAS PubMed.
- Z. Liu, J. Gao, Y. Cui, S. Klumpe, Y. Xiang, P. S. Erdmann and L. Jiang, Plant Physiol., 2021, 185, 562–576 CrossRef CAS PubMed.
- M. Kohari, N. Shibuya and H. Kaku, Plant Cell Rep., 2020, 39, 1517–1523 CrossRef CAS PubMed.
- A. Jelinkova, K. Malinska, S. Simon, J. Kleine-Vehn, M. Parezova, P. Pejchar, M. Kubes, J. Martinec, J. Friml, E. Zazimalova and J. Petrasek, Plant J., 2010, 61, 883–892 CrossRef CAS PubMed.
- X. Zhang, Z. Wang, H. Chu, Z. Xiong, Y. Li, Y. Chen, Q. Zhu, H. Feng, E. Zhu, J. Zhou, P. Huang and Z. Qian, Anal. Chem., 2022, 94, 4048–4058 CrossRef CAS PubMed.
- I. Lopez-Duarte, P. Chairatana, Y. Wu, J. Perez-Moreno, P. M. Bennett, J. E. Reeve, I. Boczarow, W. Kaluza, N. A. Hosny, S. D. Stranks, R. J. Nicholas, K. Clays, M. K. Kuimova and H. L. Anderson, Org. Biomol. Chem., 2015, 13, 3792–3802 RSC.
- M. R. Dent, I. Lopez-Duarte, C. J. Dickson, P. Chairatana, H. L. Anderson, I. R. Gould, D. Wylie, A. Vysniauskas, N. J. Brooks and M. K. Kuimova, Chem. Commun., 2016, 52, 13269–13272 RSC.
- L. Guo, R. Zhang, Y. Sun, M. Tian, G. Zhang, R. Feng, X. Li, X. Yu and X. He, Analyst, 2016, 141, 3228–3232 RSC.
- L. Fan, J. Ge, Q. Zan, X. Wang, S. Wang, Y. Zhang, W. Dong, S. Shuang and C. Dong, Sens. Actuators, B, 2021, 327, 128929–128937 CrossRef CAS.
- B. Tang, F. Yu, P. Li, L. Tong, X. Duan, T. Xie and X. Wang, J. Am. Chem. Soc., 2009, 131, 3016–3023 CrossRef CAS PubMed.
- Q. Ma, X. Cao, Y. Xie, Y. Gu, Y. Feng, W. Mi, X. Yang and L. Wu, Environ. Exp. Bot., 2017, 133, 139–150 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: UV-visible and PL spectra, NMR and MS spectra, fluorescence images and experimental details. See DOI: https://doi.org/10.1039/d2sc05727a |
| ‡ These authors contributed to this work equally. |
| This journal is © The Royal Society of Chemistry 2023 |