Open Access Article
Open Access ArticleThe compact integration of a cascaded HCR circuit for highly reliable cancer cell discrimination†
Pei
Dong‡
a,
Ruomeng
Li‡
a,
Shizhen
He‡
a,
Qingqing
Zhang
a,
Jinhua
Shang
a,
Yuqian
Jiang
a,
Xiaoqing
Liu
 ab and
Fuan
Wang
ab and
Fuan
Wang
 *ab
*ab
aCollege of Chemistry and Molecular Sciences, Wuhan University, 430072 Wuhan, P. R. China
bResearch Institute of Shenzhen, Wuhan University, Shenzhen, 518057, P. R. China. E-mail: fuanwang@whu.edu.cn
First published on 24th January 2023
Abstract
The accurate identification of multiple biomarkers involved in disease plays a vital role in effectively distinguishing cancer cells from normal cells, facilitating reliable cancer diagnosis. Motivated by this knowledge, we have engineered a compact and clamped cascaded DNA circuit for specifically discriminating cancer cells from normal cells via the amplified multi-microRNA imaging strategy. The proposed DNA circuit combines the traditional cascaded DNA circuit with multiply localized responsive character through the elaboration of two super-hairpin reactants, thus concurrently streamlining the circuit components and realizing localization-intensified cascaded signal amplification. In parallel, the multiple microRNA-stimulated sequential activations of the compact circuit, combined with a handy logic operation, significantly elevated the cell-discriminating reliability. Applications of the present DNA circuit in vitro and in cellular imaging experiments were executed with expected results, therefore illustrating that our DNA circuit is useful for precise cell discrimination and further clinical diagnosis.
Introduction
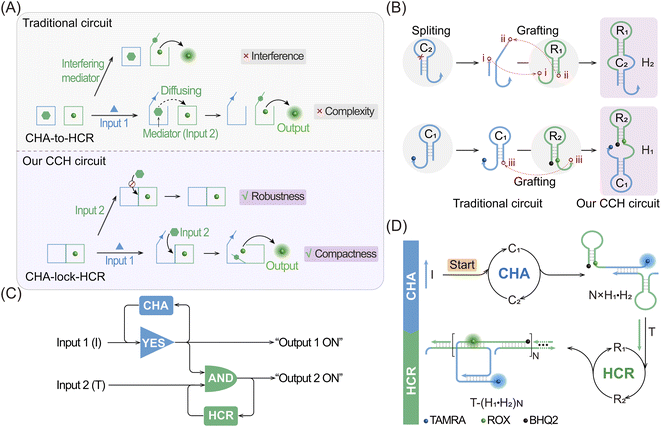
Live-cell imaging techniques play a significant role in biomedical research, such as cytology, hematology, histopathology and immunology.1–3 It can provide critical information on the basic properties of different cellular functions, and has been developed as a powerful tool for monitoring biomarkers of various key diseases.4–7 Numerous approaches have thus been developed for the intracellular imaging of biomarkers,8–12 including tools involving DNA, which are helpful for clinical diagnosis and therapeutics.13,14 Among these DNA-based methods, DNA circuits have attracted much attention due to their unique features, such as high programmability, high biocompatibility and inherent molecular recognition features.15–19 DNA circuits, based on the toehold-mediated strand-displacement reaction (SDR), have been widely used for signal amplification,20 logic operation21 and drug delivery applications.22 As one of the classic amplification circuits, the hybridization chain reaction (HCR) involves the analyte-driven isothermal cross-opening of two DNA hairpin substrates into long double-stranded DNA (dsDNA) nanowires.23–27 However, the intrinsic low reaction efficiency of the HCR prevents their extensive application in detecting biomarkers with low abundance,28,29 and needs to be urgently solved before their extensive utilization can be achieved.To address this issue, effort has been devoted to improve the sensing performance of the HCR circuit, including localized HCR,30–33 cascaded HCR34–39 and so on. By confining HCR substrates on DNA scaffolds (e.g. DNA origami) or inorganic nanoparticles, the local concentration of HCR reactants could be dramatically increased (10-fold) and the speed of the HCR assembly process could be increased to achieve sensitive microRNA (miRNA) imaging in live cells. This faster reaction kinetics of localized HCR are attributed to the non-diffusion-controlled DNA assembly process which is known to be the main reaction-limiting parameter of conventional HCR. However, most of the localized HCR systems are based on a non-responsive scaffold that might harm the specificity of the HCR-sensing system. Apart from localized HCR systems, various cascaded HCR methods have also been developed to accelerate the overall reaction rate with improved signal transduction. In a cascaded HCR circuit, the product of the upstream circuit system acts as a versatile mediator for triggering the downstream HCR procedure. Compared with the single-stage HCR circuit, such cascade circuits could achieve an improved sensing performance. However, they introduce new reliability concerns originating from the poor uniformity of cell delivery of the multiple circuitry components and the complex reaction procedures that might introduce undesired crosstalk between the multiple components. Additionally, the cell-specificity of the HCR process is still unsolved considering that some cancer cells are integrated with the same biomarkers, and, even worse, conventional single-stimuli-based sensing procedures could not distinguish cancer cells from normal cells. Thus, it is highly desirable to construct a compact and cascaded HCR circuit with multiply localized responsive features that could achieve the cell-specific amplified imaging of multiple biomarkers involved in disease.
Herein, we constructed a compact and clamped HCR (CHA-control-HCR, CCH) circuit for the more reliable intracellular imaging of miRNAs. As shown in Fig. 1A and B, to decrease the complexity of the traditional cascade circuitry system, the two circuit constituents (CHA and HCR) are integrated and simplified into only two hairpin reactants. Additionally, benefiting from the multiple miRNA-stimulated logic design (Fig. 1C), the current CCH circuit can substantially improve the reliability and specificity of live cell imaging. Here the CHA (catalytic hairpin assembly) represents another efficient amplification circuit that mediates the target-catalyzed assembly of numerous dsDNA products.40–43 In upstream CHA, the reaction of these two compact hairpins was catalyzed by microRNA-155 (miR-155, input 1) to generate a high number of dsDNA intermediates. The toehold region of downstream HCR was exposed and activated by microRNA-21 (miR-21, input 2) to generate numerous co-polymeric dsDNA nanowires with a remarkably enhanced fluorescence signal (Fig. 1D). The compact CCH circuit was capable of accurately discriminating targets from their mutants, which is ascribed to the high selectivity of the multiple target-recognition and the sequential activation of this cascaded circuit. Our CCH circuit achieved highly selective and sensitive detection for miRNAs, and realized the discrimination of different cells by monitoring the variation of miRNA expression in living cells. Our compact CCH circuit provides a sensitive and reliable approach towards the intracellular imaging of miRNA and therefore holds great potential for clinical diagnosis.
Results and discussion
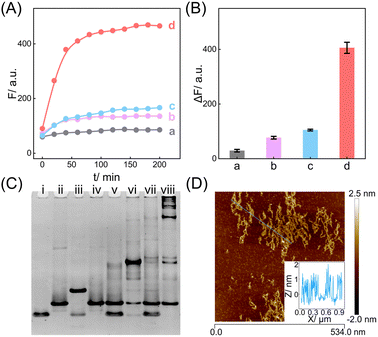
The compact CCH circuit has two integrated signal amplification circuits (Fig. 1B), the upstream CHA circuit and the downstream HCR circuit, in which all the reactants are integrated and simplified into two super-hairpin reactants (H1 and H2). In contrast to the traditional hairpin reactants of a single DNA circuit, the coexistence of two DNA circuits could allow elaborate encoding within the super-hairpins, whereby the clamped upstream CHA hairpins (C1 and C2) could thus control the downstream HCR hairpins (R1 and R2). To monitor the reaction procedure and the amplification performance of the CCH circuit, a set of fluorophores was introduced with a variable fluorescence signal readout. H1 was labeled with a Black Hole Quencher (BHQ2) at the 5′-end, a carboxytetramethylrhodamine (TAMRA) fluorophore at the 3′-end, and a carboxy-X-Rhodamine (ROX) fluorophore in the stem (domain g*). In the initial state of H1, the fluorescence of both the TAMRA and ROX fluorophores is simultaneously quenched by BHQ2, relying on the close proximity-based Förster resonance energy transfer (FRET), thus effectively restraining the background leakage signal. As shown in Fig. 1D and S1,† when input 1 (initiator I) is introduced, part of H1 (C1) is opened via a toehold-mediated strand-displacement reaction, accompanied by the generation of an intermediate I-H1 with the fluorescence recovery of TAMRA and a newly exposed toehold (b–c*). Then, the toehold b–c* hybridizes with H2, and C2 is opened, forming the intermediate I-H1·H2 and further generating the CHA product H1·H2 with the concomitant release of I for further participation in the upstream CHA circuit. Following the generation of H1·H2, the toehold e of R1 is released with the unfolded downstream HCR reactants. In the presence of input 2 (target T), the unfolded R1 is opened and hybridizes with T to form an intermediate T-H1·H2 (Fig. S2†). Concurrently, the exposed domain g-f* of R1 opens R2via CHA-localized molecular hybridization, generating the monomer CHA-HCR product T-H1·H2 with the second fluorescence recovery of ROX. The accompanying liberated toehold f*–e* contains the same T sequence that triggers the cyclic cross-opening of R1 and R2 from the upstream unfolded H1·H2, consequently generating HCR-assembled dsDNA nanowires decorated with CHA product [T-(H1·H2)N]. Meanwhile, the varying fluorescence response of the integrated upstream-to-downstream circuit is instantly captured, originating from the stepwise fluorescence recovery of the two fluorophore pairs.The CCH circuit was verified by monitoring the stepwise fluorescence variation via fluorescence assay, in which the fluorescence changes of TAMRA and ROX were used to trace the upstream CHA circuit and downstream HCR circuit, respectively. As shown in Fig. S3A and B,† a negligible fluorescence change of TAMRA was observed without initiator I and target T (curve a) or with just T (curve c). Once I was introduced, a remarkable fluorescence enhancement of TAMRA was observed (curves b and d, Fig. S3A and B†), demonstrating that the I-stimulated activation of the upstream CHA circuit is indispensable for the fluorescence recovery of TAMRA. In parallel, a slight fluorescence variation of TAMRA was observed with the introduction of T, which might be due to the possible fluorescence interference between TAMRA and ROX. Next, the fluorescence change of ROX was recorded to visualize the downstream HCR circuit (Fig. 2A and B). In the presence of I and T, significantly enhanced fluorescence of ROX was observed with increasing duration, and the fluorescence intensity reached a plateau after 2.5 h (group d, Fig. 2A and B). However, no obvious fluorescence change was observed with no input (group a, Fig. 2A and B), just I (group b, Fig. 2A and B), or just T (group c, Fig. 2A and B). Notably, in contrast to the fluorescence of TAMRA, simultaneous activations of the CHA and HCR circuits were required for the fluorescence recovery of ROX by pulling ROX away from BHQ2. These results indicated that the integrated CCH circuit could be executed by the full exposure of these two compact DNA hairpins. The upstream CHA circuit was initiated by I to liberate the downstream HCR circuit and generate the CCH product [T-(H1·H2)N] via the T trigger. Accordingly, based on the TAMRA-to-ROX fluorescence verification, the feasibility of our CCH circuit was confirmed, and the fluorescence change of ROX was considered as a versatile readout signal for monitoring CCH assembly.
From the working principle of the compact CCH circuit, it is clear that the super-hairpin-simplified reactant integration could reduce the number of reactants and thus increase the local concentration of HCR reactants, aiding their stimulated close proximity, via upstream CHA. Thus, a multiply localization-accelerated reaction format was realized to facilitate the amplified detection of the analyte. Here a non-localization-based CHA-HCR circuit, consisting of three DNA hairpins (H1′, H2′, and H3′), was introduced to comprehensively verify the multiply localized CCH circuit. As shown in Fig. S4,† H2′ retains the same sequence as H2 of the CCH circuit, and H1 of the CCH circuit is split into H1′ and H3′. A fluorophore/quencher pair (ROX/BHQ2) is labeled in H3′ to monitor the formation of the product T-(H1′·H2′·H3′)N. In the presence of I, the CHA circuit was activated, and H1′ was opened to generate I-H1’ with an exposed toehold b–c*. Then, the exposed toehold b-c* hybridized with C2′ of H2′ and I was released to participate again in the CHA circuit. Following the generation of the CHA product H1′·H2′, R1′ was uncovered with a newly exposed toehold e. With the additional introduction of T, R1′ was opened to cross-hybridize with H3′, thus generating the ultimate product T-(H1′·H2′·H3′)N with the fluorescence recovery of ROX. Then, the feasibility of the three-hairpin CHA-HCR circuit was evaluated (Fig. S5†). A low fluorescence recovery was observed with I and T (group d, Fig. S5A and B†). Note that a remarkable background fluorescence was observed with just T (group c, Fig. S5A and B†), compared to the groups without inputs and with I (groups a and b, respectively, Fig. S5A and B†), which was due to signal leakage of the always-exposed toehold g* in the free H3′. Accordingly, the localization-based CCH circuit can provide an enhanced amplification ability for target detection while removing the signal leakage via CHA-controlled HCR responsive exposure.
In addition, native polyacrylamide gel electrophoresis (PAGE) experiments were used to validate the proposed CCH reaction mechanism (Fig. 2C; for the detailed DNA structures of the corresponding bands, see Fig. S6 and S7†). In accordance with the above fluorescence results, I opened H1 to generate an intermediate product I-H1 (lane iii). With the introduction of H2, I-H1 hybridized with H2 to generate the CHA product H1·H2 with the unfolded HCR reactants (lane vi), which were triggered by T to further generate the higher-molecular-weight CCH product (lane viii). However, neither the clamped R1 (lane iv) nor the clamped R1 and R2 (lane vii), could be triggered by T. In parallel, the unfolded R1 of H2, as a result of the I-activated CHA process, was unable to activate the HCR reaction without T (lane vi). Additionally, the bands of H1 and H2 were unaffected in the mixture sample of H1 and H2 (lane v). Overall, these phenomena indicated that the integrated CCH circuit could guarantee highly effective amplification while retaining low signal leakage. Furthermore, the morphology of the produced CCH nanostructures was determined by atomic force microscopy (AFM). Plenty of long dsDNA nanowires are shown in Fig. 2D, and the height of the dsDNA nanowires is about 1 nm, which is consistent with the typical height of dsDNA. Consequently, we can conclude that the compact CCH can be successfully activated via the CHA-controlled HCR circuitry mechanism to generate localization-intensified amplification products.
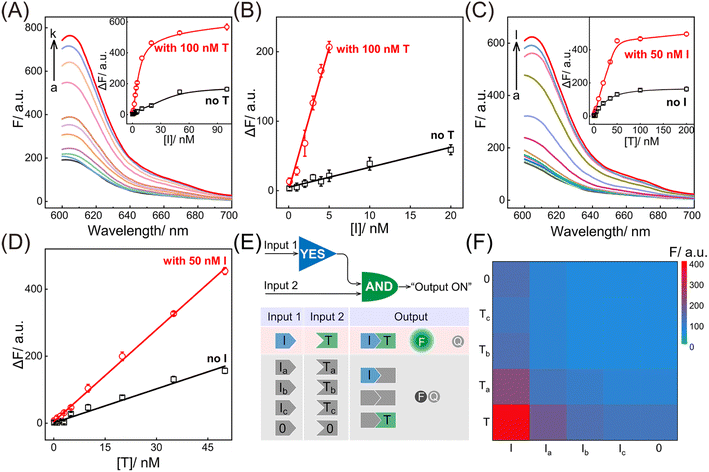
Having verified the feasibility of the compact CCH circuit, the sensing performance was investigated for detecting DNA targets. After optimization of the CCH reaction conditions (for details, see Fig. S8†), the corresponding sensing performances of the DNA targets were interrogated. For the upstream analysis of I involving CHA, a concentration-dependent fluorescence increase of the CCH circuit was observed at a fixed concentration of T (100 nM). However, a negligible increase of the T-absent CCH circuit was observed (Fig. 3A and 3A inset). From Fig. 3B, the detection limit (LOD) of the T-activated CCH circuit was determined to be 15 pM based on a traditional 3-σ method, which was about 15-fold lower than in the HCR-inactivated CCH circuit (224 pM). Similarly, with a fixed concentration of I (50 nM), the downstream detection of T involving HCR was investigated. As shown in Fig. 3C and its inset, the fluorescence of the I-incubated CCH circuit showed an increasing trend with increasing concentration of T. However, a smaller increase of the fluorescence was observed for the I-absent CCH circuit, indicating that the I-activated CHA circuit is indispensable to the downstream HCR circuit for liberating and localizing the HCR reactants. After calculations using Fig. 3D, the LOD of the I-activated CCH circuit was determined to be 80 pM, which is lower than that in the CHA-inactivated CCH circuit (190 pM). When compared with the detection of T, a better sensing performance was observed for the detection of I, which was attributed to the upstream-to-downstream cascaded signal amplification. Consequently, the integrated CCH circuit simultaneously enables the assay of I and T, relying on the localization-intensified cascaded amplification of the well-coordinated CCH circuit. In addition to the high amplification capacity, a satisfactory selectivity of the compact CCH circuit was required for targets I and T. Thus, the CCH circuit was applied to the analysis of three groups of mutant DNAs with different mismatches, including one-, two-, and three-base mismatches in each group (labeled as Ia and Ta, Ib and Tb, and Ic and Tc, respectively) (Fig. 3E). As expected, a distinctly high fluorescence was observed only when I and T were matched, meaning they could be clearly distinguished from any other mismatched group (Fig. 3F). These results proved that the compact CCH circuit possesses high sensitivity and selectivity for concurrently detecting DNA targets.
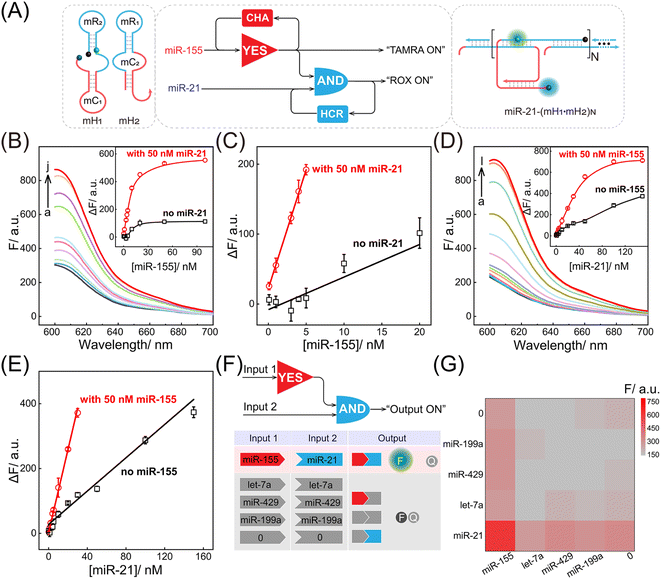
Motivated by the above successful exploration of the dual-stimuli CCH system, the practical availability of the current system was considered. The CCH sensing system was updated by engineering multiple miRNA-recognizing super-hairpin reactants to detect the miRNAs involved in disease. MiR-155 and miR-21 with synchronous abnormal expressions can induce multiple human cancers, e.g. by regulating DNA damage and repair, and are regarded as a vital clue in early diagnosis and clinical treatment.44–48 Hence miR-155 and miR-21 were chosen as CCH targets to investigate its practicability. As shown in Fig. 4A, the sequences of H1 and H2 were replaced by mH1 and mH2, respectively. The upstream CHA circuit could be initiated by miR-155, and then the unfolded downstream HCR circuit was triggered by miR-21, further generating the CCH product [miR-21-(mH1·mH2)N] with a high-gain readout signal. Similar to the above DNA-triggered system, the feasibility of the miRNA-targeted CCH system was assessed by monitoring the stepwise fluorescence variation. As expected, the upstream CHA-amplified fluorescence recovery of TAMRA was achieved by the introduction of miR-155 (curves b and d, Fig. S9A and B†). However, no fluorescence recovery of TAMRA was observed in the miR-155-absent CCH circuits (curves a and c, Fig. S9A and B†). For the downstream HCR-propelled fluorescence transduction, only the CCH system was fully activated by miR-155 and miR-21, as revealed by the remarkable fluorescence recovery of ROX (curve d, Fig. S9C and D†). In the absence of any miRNA, the downstream HCR circuit maintains a passive state without ROX fluorescence recovery (curves a, b, and c, Fig. S9C and D†). Consequently, the construction of a multiple miRNA-stimulated CCH system is viable for exclusively dual-biomarker analysis, and the fluorescence transduction of ROX can facilitate the monitoring of the CCH circuit in a convenient way. Additionally, a tailored three-hairpin CHA-HCR circuit was established to validate the localization-accelerated reaction profiles as mentioned above (for details, see Fig. S10†). The result demonstrated that the localization-based CCH circuit could indeed significantly improve the signal amplification efficiency relative to traditional diffusion-controlled DNA assembly (three-hairpin CHA-HCR circuit).
After optimization of the reaction conditions of the CCH circuit (for details, see Fig. S11†), the miRNA-sensing performance was explored. For the miR-155 detection involving upstream CHA, gradually increasing fluorescence was observed in the miR-21-incubated CCH circuit, as the concentration of miR-155 increased to 50 nM. However, the miR-21-absent CCH system enabled a weak fluorescence fluctuation (Fig. 4B and 4B inset). From the calibration curves in Fig. 4C, the LOD of the miR-21-incubated CCH circuit was determined to be 21 pM, which was approximately 11-fold lower than in the miR-21-absent CCH circuit (153 pM). Then, the miR-21 detection involving downstream HCR was investigated with a fixed concentration of miR-155 (50 nM). As shown in Fig. 4D and its inset, with increasing concentration of miR-21, a clear distinction between the fluorescence for the miR-155-activated CCH circuit and the miR-155-absent CCH circuit was observed. The LOD of the miR-155-activated CCH circuit was determined to be 56 pM (Fig. 4E), which was 4-fold lower than in the miR-155-absent CCH circuit (279 pM). Therefore, the miRNA-targeting CCH system can sensitively and simultaneously detect miR-155 and miR-21. In addition, the selectivity of the CCH system for miR-155 and miR-21 was investigated (Fig. 4F). The CCH circuit was used to analyze 16 different combinations of interfering RNAs. As expected, a satisfactory specificity was obtained (Fig. 4G), and a remarkable fluorescence was observed only with miR-155 and miR-21. Additionally, the robustness of the CCH system was verified by monitoring fluorescence fluctuations in different serum samples. No obvious fluorescence changes were observed in 5% and 10% fetal bovine serums (Fig. S12†), indicating good stability of the compact CCH circuit that could facilate in-depth biological applications. A 3-(4,5-dimethylthialzol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) experiment was carried out to evaluate the cytotoxicity of the CCH system. The cell viability remained high even upon treatment with 400 nM of CCH probe for 24 h (Fig. S13†), indicating low cytotoxicity of the CCH system that further favours its application in complex intracellular environments.
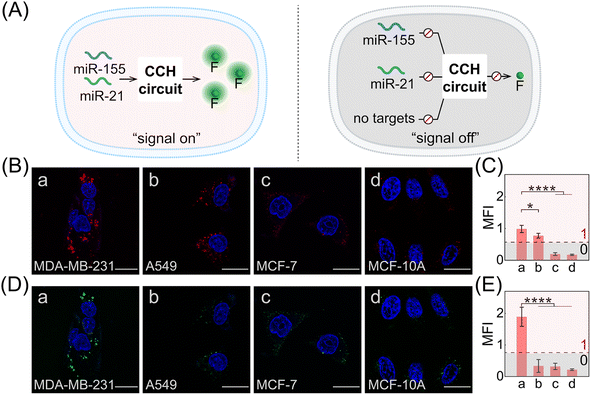
Based on the above systematic verifications, the compact CCH system was applied for imaging miRNAs in living cells. According to previous reports,49,50 the representative cells used exhibited different expressions of miR-155 and miR-21, and included MDA-MB-231 cells (human breast cancer cells) with overexpressed miR-155 and miR-21 profiles, A549 cells (human lung cancer cells) with high miR-155 and negligible miR-21 expression profiles, MCF-7 cells (human breast cancer cells) with overexpressed miR-21 and negligible miR-155 expression profiles, and MCF-10A cells (human normal breast epithelial cells) with negligible miR-155 and miR-21 expression profiles. These cells were used to evaluate the miRNA-imaging competence of our CCH system. The fluorescence of TAMRA and ROX was recorded using confocal laser scanning microscopy (CLSM). TAMRA fluorescence was employed for imaging miR-155, and ROX fluorescence was employed for simultaneously imaging miR-155 and miR-21. First, the cellular uptake of the compact DNA probes was explored. ROX fluorescence of MDA-MB-231 cells was monitored by flow cytometry, and an increasing fluorescence signal was detected with increasing incubation duration up to 4 h (Fig. S14†). Evidently, the robust CCH system can be delivered into living cells for fluorescence transduction. Our cascaded CCH system was compared with the single-staged HCR system to evaluate the imaging performance for biomarkers of low abundance (Fig. S15†). Clear fluorescence was observed for CCH-treated MDA-MB-231 cells, yet only faint fluorescence was observed for HCR-treated MDA-MB-231 cells, demonstrating that the cascaded CCH system can achieve more sensitive and accurate intracellular imaging in living cells than the single-staged HCR system. Meanwhile, our homogeneous CCH system was also compared with the heterogeneous three-hairpin CHA-HCR system to further assess the delivery efficiency of the DNA probes (Fig. S16†). Remarkably enhanced fluorescence of the compact CCH system was observed, demonstrating that the high cell-delivering uniformity of the CCH system could eliminate the undesired crosstalk between the heterogeneous multiple-components, and thus could assure a reliable site-specific signal response.
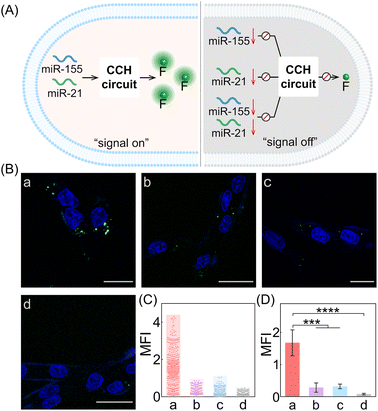
Next, the CCH system was utilized for discriminating different cells with diverse miR-155 and miR-21 expressions (Fig. 5A; for imaging details, see Fig. S17†). As shown in Fig. 5B, for the miR-155-stimulated TAMRA fluorescence, clear red fluorescence was observed in the MDA-MB-231 cells with overexpressed miR-155 (group a, Fig. 5B and C), moderate red fluorescence of TAMRA was observed in A549 cells with high miR-155 expression (group b, Fig. 5B and C), and faint red fluorescence of TAMRA was observed in MCF-7 cells and MCF-10A cells, both with extremely low miR-155 expression (groups c and d, respectively, Fig. 5B and C). Thus, the compact CCH system can distinguish different cells based on their miR-155 expression. In parallel, for ROX fluorescence stimulated by miR-155 and miR-21, only strong green fluorescence was observed for MDA-MB-231 cells with overexpressed miR-155 and miR-21 (group a, Fig. 5D and E), yet negligible green fluorescence was observed for A549 cells with high miR-155 and negligible miR-21 expression, MCF-7 cells with overexpressed miR-21 and negligible miR-155 expression, as well as MCF-10A cells with insignificant miR-155 and miR-21 expression (groups b, c, and d, respectively, Fig. 5D and E). Flow cytometry analysis was executed (Fig. S18 and S19†) and the results were consistent with those of the CLSM imaging. The miRNA expression of different cells, as reflected by fluorescence variations, are consistent with the above reports. Note that MDA-MB-231 cells and A549 cells could not be clearly distinguished by miR-155 (as revealed by the fluorescence of TAMRA), yet could be accurately identified by both miR-155 and miR-21 (as revealed by the fluorescence of ROX). Therefore, the CCH system can accomplish cell-specific amplified imaging of multiple miRNAs without false-positive signals, as a result of the pervasive distribution of the same miRNA in different cells. To further explore the imaging performance of the compact CCH system, MDA-MB-231 cells with regulated expressions of miRNAs were analyzed (Fig. 6A; for details, see Fig. S20†). Compared to the intact cells with a distinct fluorescence signal (group a, Fig. 6B–D), a weak fluorescence signal was discerned for the anti-miR-155-pretreated cells and the anti-miR-21-pretreated cells (groups b and c, Fig. 6B–D), and very low fluorescence was observed in cells that were pretreated with the anti-miR-155 and anti-miR-21 (group d, Fig. 6B–D). These results provided further support for the miRNA activation principle, showing that the CCH circuit can accurately sense multiple miRNAs with diverse expressions in living cells.
Conclusions
In summary, we have developed a multiple biomarker-stimulated and compact CHA-control-HCR (CCH) circuitry system for precisely distinguishing cancer cells from normal cells. The compact CCH circuitry system simplified the multiple components of a traditional cascaded circuit into only two-hairpin reactants. This guaranteed high cell-delivering uniformity and reaction kinetics determined by the CHA-localized HCR system. Combined with the logic-screened accurate identification of multiple biomarkers, the current CCH circuitry system can be used to implement robust and high-performance cell imaging with varied and amplified signal transductions. By virtue of the multi-targeted cell-selectivity and the localization-intensified cascaded signal amplification, a highly reliable and sensitive discrimination method for different cells has been achieved with simpler circuit constituents (two-hairpin reactants), further promoting the operability and universality of the proposed method. We have verified the feasibility and practicality of the compact CCH circuitry system through systematic in vitro demonstrations and cellular imaging experiments. We are encouraged by these satisfactory results, and the compact CCH circuitry system is expected to have considerable potential for early cancer diagnosis and therapeutic intervention applications.Data availability
All relevant data is presented in the paper and ESI.†Author contributions
F. W. and X. L. conceived and designed the experiment. P. D., R. L., and S. H. performed the experiment and analysed the data. P. D. and R. L. collected fluorescence spectra. J. S. and Y. J. cultured cells and took CLSM images. R. L., S. H., X. L., and F. W. wrote the manuscript. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (22274121, 22274123 and 22074112), the Fundamental Research Funds for the Central Universities (2042022kf1175), and Shenzhen Science and Technology Program (JCYJ20220530140609021).Notes and references
- A. Farhadi, G. H. Ho, D. P. Sawyer, R. W. Bourdeau and M. G. Shapiro, Science, 2019, 365, 1469–1475 CrossRef CAS PubMed.
- M. A. Miller and R. Weissleder, Nat. Rev. Cancer, 2017, 17, 399–414 CrossRef CAS PubMed.
- M. Yasui, M. Hiroshima, J. Kozuka, Y. Sako and M. Ueda, Nat. Commun., 2018, 9, 1–11 CrossRef CAS PubMed.
- S. H. Shim, Nature, 2017, 546, 39–40 CrossRef CAS PubMed.
- K. Zhang, R. Deng, X. Teng, Y. Li, Y. Sun, X. Ren and J. Li, J. Am. Chem. Soc., 2018, 140, 11293–11301 CrossRef CAS PubMed.
- S. McArdle, Z. Mikulski and K. Ley, J. Exp. Med., 2016, 213, 1117–1131 CrossRef CAS PubMed.
- X. Luo, B. Xue, G. Feng, J. Zhang, B. Lin, P. Zeng, H. Li, H. Yi, X. L. Zhang, H. Zhu and Z. Nie, J. Am. Chem. Soc., 2019, 141, 5182–5191 CrossRef CAS PubMed.
- C. Wiraja, D. C. Yeo, K. C. Tham, S. W. T. Chew, X. Lim and C. Xu, Small, 2018, 14, 1703440 CrossRef PubMed.
- Q. Wang, K. Tan, H. Wang, J. Shang, Y. Wan, X. Liu, X. Weng and F. Wang, J. Am. Chem. Soc., 2021, 143, 6895–6904 CrossRef CAS PubMed.
- S. Kumar, N. Harrison, R. Richards-Kortum and K. Sokolov, Nano Lett., 2007, 7, 1338–1343 CrossRef CAS PubMed.
- X. Deng, Z. Yin, J. Lu, X. Li, L. Shao, C. Zhao, Y. Yang, Q. Hu, Y. Wu and W. Sheng, Adv. Sci., 2018, 5, 1700542 CrossRef PubMed.
- R. C. Qian, Y. Cao, L. J. Zhao, Z. Gu and Y. T. Long, Angew. Chem., Int. Ed., 2017, 129, 4880–4883 CrossRef.
- Y. Wu, Z. Li, M. Shi, K. Yuan, H. M. Meng, L. Qu and Z. Li, Anal. Chem., 2021, 93, 12456–12463 CrossRef CAS PubMed.
- H. Zhang, X. Huang, J. Liu and B. Liu, Chem. Sci., 2020, 11, 3812–3819 RSC.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- C. Yuan, J. Fang, M. L. de la Chapelle, Y. Zhang, X. Zeng, G. Huang, X. Yang and W. Fu, Trends Anal. Chem., 2021, 143, 116401 CrossRef CAS.
- H. Zhou, J. Liu, J. J. Xu, S. S. Zhang and H. Y. Chen, Chem. Soc. Rev., 2018, 47, 1996–2019 RSC.
- C. Zhang, T. Belwal, Z. Luo, B. Su and X. Lin, Small, 2022, 18, 2102711 CrossRef CAS PubMed.
- D. Li, T. Zhang, F. Yang, R. Yuan and Y. Xiang, Anal. Chem., 2019, 92, 2074–2079 CrossRef PubMed.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed.
- T. Song, A. Eshra, S. Shah, H. Bui, D. Fu, M. Yang, R. Mokhtar and J. Reif, Nat. Nanotechnol., 2019, 14, 1075–1081 CrossRef CAS PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2018, 119, 6459–6506 CrossRef PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- X. Gong, H. Wang, R. Li, K. Tan, J. Wei, J. Wang, C. Hong, J. Shang, X. Liu, J. Liu and F. Wang, Nat. Commun., 2021, 12, 3953 CrossRef CAS PubMed.
- D. J. Huang, Z. M. Huang, H. Y. Xiao, Z. K Wu, L. J. Tang and J. H. Jiang, Chem. Sci., 2018, 9, 4892–4897 RSC.
- D. Ye, M. Li, T. Zhai, P. Song, L. Song, H. Wang, X. Mao, F. Wang, X. Zhang, Z. Ge, J. Shi, L. Wang, C. Fan, Q. Li and X. Zuo, Nat. Protoc., 2020, 15, 2163–2185 CrossRef CAS PubMed.
- H. Chu, J. Zhao, Y. Mi, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2019, 58, 14877–14881 CrossRef CAS PubMed.
- J. Wang, D. X. Wang, J. Y. Ma, Y. X. Wang and D. M. Kong, Chem. Sci., 2019, 10, 9758–9767 RSC.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- H. Bui, S. Shah, R. Mokhtar, T. Song, S. Garg and J. Reif, ACS Nano, 2018, 12, 1146–1155 CrossRef CAS PubMed.
- Z. Wu, G. Q. Liu, X. L. Yang and J. H Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS PubMed.
- C. Song, J. Zhang, Y. Liu, X. Guo, Y. Guo, X. Jiang and L. Wang, Sens. Actuators, B, 2020, 325, 128970 CrossRef CAS PubMed.
- Y. Zhang, J. Wang, S. Chen and R. Yuan, Sens. Actuators, B, 2021, 342, 130040 CrossRef CAS.
- H. Wang, C. Li, X. Liu, X. Zhou and F. Wang, Chem. Sci., 2018, 9, 5842–5849 RSC.
- K. Quan, J. Li, J. Wang, N. Xie, Q. Wei, J. Tang, X. Yang, K. Wang and J. Huang, Chem. Sci., 2019, 10, 1442–1449 RSC.
- J. Wei, X. Gong, Q. Wang, M. Pan, X. Liu, J. Liu, F. Xia and F. Wang, Chem. Sci., 2018, 9, 52–61 RSC.
- J. Wei, J. Shang, S. He, Y. Ouyang, I. Willner and F. Wang, CCS Chem., 2022, 4, 3549–3562 CrossRef CAS.
- W. Y. Lv, C. H. Li, Y. F. Li, S. J. Zhen and C. Z. Huang, Anal. Chem., 2021, 93, 3411–3417 CrossRef CAS PubMed.
- S. He, S. Yu, R. Li, Y. Chen, Q. Wang, Y. He, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2022, 61, e202206529 CAS.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- X. Gong, S. He, R. Li, Y. Chen, K. Tan, Y. Wan, X. Liu and F. Wang, Chem. Sci., 2022, 13, 10428–10436 RSC.
- X. Chen, N. Briggs, J. R. McLain and A. D. Ellington, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5386–5391 CrossRef CAS PubMed.
- H. Wang, Y. He, J. Wei, H. Wang, K. Ma, Y. Zhou, X. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2022, 61, e202115489 CAS.
- L. Mulrane, S. F. McGee, W. M. Gallagher and D. P. O'Connor, Cancer Res., 2013, 73, 6554–6562 CrossRef CAS PubMed.
- J. Chen, B. C. Wang and J. H. Tang, J. Surg. Oncol., 2012, 106, 260–266 CrossRef CAS PubMed.
- S. Jiang, H. W. Zhang, M. H. Lu, X. H. He, Y. Li, H. Gu, M. F. Liu and E. D. Wang, Cancer Res., 2010, 70, 3119–3127 CrossRef CAS PubMed.
- L. Y. W. Bourguignon, C. C. Spevak, G. Wong, W. Xia and E. Gilad, J. Biol. Chem., 2009, 284, 26533–26546 CrossRef CAS PubMed.
- J. Shen, N. W. Todd, H. Zhang, L. Yu, X. Lingxiao, Y. Mei, M. Guarnera, J. Liao, A. Chou, C. L. Lu, Z. Jiang, H. B. Fang, R. L. Katz and F. Jiang, Lab. Invest., 2011, 91, 579–587 CrossRef CAS PubMed.
- X. Gong, J. Wei, J. Liu, R. Li, X. Liu and F. Wang, Chem. Sci., 2019, 10, 2989–2997 RSC.
- S. Hajalirezay Yazdi, M. Paryan and S. Mohammadi-Yeganeh, Cell. Mol. Biol. Lett., 2018, 23, 1–13 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, DNA sequences, fluorescence data, cell imaging, flow cytometric analysis, and additional figures, schemes and tables. See DOI: https://doi.org/10.1039/d2sc05568f |
| ‡ P. Dong, R. Li and S. He contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |