Open Access Article
Open Access ArticleA review of lipase immobilization on hydrophobic supports incorporating systematic mapping principles†
José Renato
Guimarães
ab,
Kaíque Souza Gonçalves Cordeiro
Oliveira
b,
Maria Carolina Pereira
Gonçalves
 b,
João Paulo
Romanelli
c,
Laiane Antunes
Lopes
b,
Ángel
Berenguer-Murcia
b,
João Paulo
Romanelli
c,
Laiane Antunes
Lopes
b,
Ángel
Berenguer-Murcia
 d,
Roberto
Fernandez-Lafuente
d,
Roberto
Fernandez-Lafuente
 *e and
Paulo Waldir
Tardioli
*e and
Paulo Waldir
Tardioli
 *b
*b
aInstitute of Natural Resources, Graduate in Bioprocess Engineering, Laboratory of Fermentative and Enzymatic Processes (LPFEnz), Federal University of Itajubá, Av. Benedito Pereira dos Santos, 1303, 37500-903, Itajubá, MG, Brazil. E-mail: jrenatoguimaraes@unifei.edu.br
bGraduate Program in Chemical Engineering (PPGEQ), Laboratory of Enzyme Technologies (LabEnz), Department of Chemical Engineering, Federal University of São Carlos (DEQ/UFSCar), Rod. Washington Luís, km 235, 13565-905 São Carlos, SP, Brazil. E-mail: kaiquesg.eq@gmail.com; mariacarolinapgoncalves@gmail.com; laiane_antunes@hotmail.com; pwtardioli@ufscar.br; Tel: +55 16 3351 9362
cLaboratory of Ecology and Forest Restoration, Department of Forest Sciences, “Luiz de Queiroz” College of Agriculture, University of São Paulo, 11 Av. Pádua Dias, 13418-900 Piracicaba, SP, Brazil. E-mail: joaopromanelli@usp.br
dDepartamento de Química Inorgánica e Instituto Universitario de Materiales, Universidad de Alicante, 03080 Alicante, Spain. E-mail: a.berenguer@ua.es
eDepartamento de Biocatálisis, ICP-CSIC, Campus UAM-CSIC, 28049 Madrid, Spain. E-mail: rfl@icp.csic.es; Tel: +34 91594804
First published on 8th September 2023
Abstract
A review of the literature covering research on the immobilization of lipases on hydrophobic supports was performed using systematic mapping (SM) concepts. This approach consists of a rigorous review of the methodology used to catalog evidence, to identify gaps at the frontier of knowledge, to identify unknown trends, and to list research groups. Our results show a wide variety of available lipases, including commercial, wild-type and recombinant strains. However, the most commonly used lipases are lipases from Thermomyces lanuginosus (TLL), Candida rugosa (CRL) or Rhizomucor miehei (RML) and lipase B from Candida antarctica (CALB). A wide variety of supports with different degrees of hydrophobicity were identified and the supports activated with a layer of octyl or octadecyl groups were the most commonly used. The advantages of lipase immobilization on these supports are discussed. Among them, the immobilization, purification, stabilization and hyperactivation of lipases in a single step stand out. Moreover, problems related to lipase immobilization by interfacial activation are highlighted (mainly enzyme release). Strategies to overcome these problems include immobilization on heterofunctional supports or intermolecular crosslinking of enzymes immobilized by physical and/or chemical agents. The possibility of increasing the capacity of supports by lipase multilayer immobilization is also discussed. Finally, the structure, distribution of the network and the frequency of co-occurrence between lipases and supports are elucidated to determine the possible hotspots and hitherto unexplored advances in knowledge.
Introduction
Biocatalytic processes have been expanding due to their promising role in the development and optimization of industrial technologies in different areas. Among the most commonly used biocatalysts, lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) are gaining a privileged position due to their numerous advantages over the use of traditional chemical catalysts, especially those concerning the high selectivity and specificity of these enzymes.1–5 However, the use of such enzymes in their soluble form for large-scale industrial processes is not very attractive, because of their high production cost, moderate operational stability and difficulties in the downstream product raised by lipases.6 To overcome these drawbacks, the immobilization of these biocatalysts on a solid support has been widely exploited. If properly designed, this approach can provide several advantages from an industrial point of view, such as an increase in the operational stability of the biocatalyst, a widening of the operational window, easy recovery from the product stream, and the possibility of lipase reuse.6–11Lipases have been extensively immobilized by adsorption on supports bearing hydrophobic surfaces12–16 due to their peculiar catalytic mechanism, which is called interfacial activation (Fig. 1). This mechanism relies on the existence of two forms of the lipase in equilibrium: 1) a closed form (hydrophobic regions of the lid interact with hydrophobic regions around the active center, isolating it from the reaction medium); 2) an open form (the lid moves and allows lipase adsorption on a hydrophobic surfaces (e.g., drops of oils or air bubbles),17–20 exposing the active center to the medium).20
 | ||
| Fig. 1 Interfacial activation of lipases versus drops of insoluble substrates or the surfaces of hydrophobic supports. | ||
This immobilization protocol is a simple technique and presents several advantages: 1) pH is not a limiting factor as long as the enzyme is soluble and stable at the utilized pH;21 2) the use of low ionic strength can lead to the simultaneous immobilization and purification of lipases;21–25 3) the immobilization method is reversible; 4) the open and adsorbed form of this enzyme is very stable,26–28 even regarding covalently immobilized multipoint lipases;29,30 5) lipases immobilized on hydrophobic supports have the active center more exposed to the medium and the lid does not move in the presence of a medium of high ionic strength;31 6) reduction in the accumulation of hydrophilic compounds on the porous biocatalyst surface;32–34 7) higher enzymatic activities can be achieved regarding the free enzyme, due to stabilization of open-form lipase.22
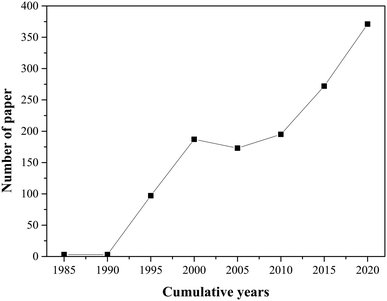
Moreover, the relevance of the research addressing the immobilization of lipases on hydrophobic supports is indisputable, since this theme has led to an increased number of published scientific research papers over the years (Fig. 2). Accordingly, in an effort to advance rigorous methods for conducting evidence reviews in this topic, we incorporated some principles of systematic mapping (SM) to evaluate research involving the immobilization of lipases on hydrophobic supports. SM methodologies investigate and reveal trends, knowledge gaps, and existing heterogeneity in studies conducted on benches. This is because SM retrieves comprehensive research from various bibliographic sources, transparently screens articles, and critically assesses the quality of these studies.35–39
Guidelines to standardize SM methods are formulated by formal coordinating review bodies from various disciplines, such as Cochrane in healthcare, the Campbell Collaboration in social welfare, and the Collaboration for Environmental Evidence (CEE) in conservation and environmental management.40 Nonetheless, there is no specialized organization devoted to guiding the conduct of evidence synthesis in the chemical engineering sector, which suggests that the majority of reviews in this area are classified as nonsystematic. Consequently, using SM methods to conduct a literature review in this field may allow the extraction of relevant information from our research subject.
In this context, the purpose of this review is to present trends and gaps across the literature on the immobilization of lipases on hydrophobic supports in order to support future studies on this topic. Our evaluation is restricted to a specific subset of scientific articles retrieved from four bibliographic sources: Web of Science (Core Collection: SCI-E and ESCI), Scopus, PubMed (Central; PMC), and SciELO.
Methodology
Literature search and database building
This review was designed by following the Collaboration for Environmental Evidence guidelines,41 in close collaboration with environmentalists and specialists on the subject. There is no dedicated body for retrieving evidence in the area of chemical engineering. The following search string was used to retrieve titles, abstracts, and keywords of related publications: “interfacial activation*” OR “activation immobilization*” OR “interfacial immobilization*” OR “interfacial activation immobilization*” OR “interfacial adsorption*” OR “adsorption immobilization*” OR “hydrophobic adsorption*” OR “hydrophobic immobilization*” OR “hydrophobic adsorption immobilization*” OR “hydrophobic support*” OR “hydrophobic interaction*” AND “lipase*”. Searches were restricted to articles published between 2011 and 2020 in order to obtain a sample of recent publications. The database was updated on April 26, 2021 (see the complete search procedures in Fig. S1 – ESI†).Bibliographic searches were performed in four different databases to minimize possible bias in the review process, returning a total of 1855 articles. We retrieved 368 publications from Scopus, 644 from Web of Science, 843 from PubMed, and none from SciELO (Table S1 – ESI†). The SCI-E database present within the Web of Science platform was used because it covers most significant scientific results and other online databases that also contain citation information, such as Science Direct and Google Scholar. The ESCI was also used because it contains complete records of articles indexed by journals not yet covered by the SCI-E which are nevertheless under evaluation for indexing in the SCI-E. Thus, relevant scientific results that can be found in this database may have an influence on bibliometric metrics. After an analysis of coverage and overlap to remove replicates, the database was reduced to 1433 publications. Eligibility criteria were used to eliminate unrelated publications inadvertently included in the reference list after this search. Initially, the screening process was conducted by analyzing titles and abstracts of primary research publications (i.e., original data for a specific research study), considering the following inclusion criteria: (i) lipase immobilization; (ii) lipase immobilization on solid supports; (iii) lipase immobilization on hydrophobic supports. A total of 370 records were then submitted to a full-text analysis and 264 primary studies were selected to be included in this study. A summary of the methodology employed is presented in Fig. 3 and details of the papers eliminated after title/abstract screening and full-text analysis are shown in Table S2 and Fig. S2 (ESI†), respectively.
 | ||
| Fig. 3 Flow diagram for the selection of studies (adapted from ROSES Flow Diagram for Systematic Maps. Version 1.0). | ||
A sample of 30 random articles was double screened by two different assessors (J. R. G. and L. A. L.) to account for subjective decisions in the inclusion/exclusion of eligible studies.41 Decisions were compared using the kappa test of agreement to ensure the repeatability of the process. Kappa values range from 0 to 1: high values indicate greater agreement and values lower than 0.6 indicate inconsistency between raters when the inclusion criteria should be redefined.42 In this analysis, a kappa score of 0.91 (0.95 lower/upper confidence limit) was obtained, which indicates almost perfect agreement between the reviewers, as well as decisions being sufficiently repeatable (Table S3 – ESI†).
Data synthesis
Articles meeting our inclusion criteria (n = 264) were subjected to data coding and synthesis of results. Two categories of keyword (types of lipase and hydrophobic support) were created and assigned to each paper to build network maps. Despite the extensive literature that exists on immobilization of lipases on hydrophobic supports, important aspects of this immobilization protocol remain to be clarified. Considering this, here we elucidate some of the relevant aspects, including the main enzymes and supports used, besides problems and solutions that are related to this research topic.Data handling
For data processing, Origin® (version 9.0) and MS Excel (v. 2016) were used to perform the calculations and prepare the graphs. R software was used to perform the screening process – ‘revtools package’.43 This analysis allows the removal of duplicate publications that were retrieved from different academic bibliographic sources (a necessary step in systematic mapping).Furthermore, the set of keywords created was applied to VOSviewer software (version 1.16.15) through the criterion of co-occurrence of terms to build network maps containing information related to the important topics of immobilization of lipases on hydrophobic supports. The purpose of co-occurrence analysis is to examine the degree of association between the elements of a “collection unit”. If pairing is performed multiple times, then the association is strengthened with every additional match. However, if only one match is made as part of the support, the association is tenuous or spurious.
Results and discussion
The different lipases and solid supports present in our database are available in Table S4 (ESI†).Enzymes and supports used in hydrophobic immobilization
The immobilization of a lipase on a hydrophobic support can allow the synthesis of a library of biocatalysts with modulated catalytic properties for application in industrial biotransformation processes. This is possible due to the structure and properties of the different existing supports44–47 and the affinity of the lipases with these carriers,48,49 as well as the immobilization conditions.50–54 These factors, alone or collectively, can alter the selectivity, specificity and final activity of lipases and in some cases lead to an immobilized biocatalyst design that is much more suitable for the production process than the free-enzyme formulation.51,55–58 Thus, there is no way to say whether a specific lipase is adequate or inadequate for a specific process as its properties may be strongly tailored upon immobilization.57As shown in Fig. 4, the most frequently immobilized enzymes on hydrophobic supports are lipases from Candida antarctica (form B) (CALB), Thermomyces lanuginosus (TLL) and Candida rugosa (CRL). These lipases have probably been used due to their stability and activity after immobilization. CALB and TLL are enzymes that have high stability and can be used in various industrial conditions, and are already commercialized as immobilized formulations in resins.59,60 On the other hand, CRL presents moderate stability61 and an immobilized formulation is not available on the market. The use of stability is a delicate factor to infer the sovereignty of these lipases in this bibliographic survey, considering that lipase A from Candida antarctica (CALA) has high stability and is in the seventh position of the most used lipases in the survey. In addition, Eversa® Transform 2.0 (EV2.0) is a commercial evolution of TLL and has high stability, but is not in the top 10 due to its recent appearance in the market.62
The cost of acquiring enzymes is a factor that reduces the technical and economic viability of biocatalytic processes, but it is probably not the main variable that determined the ranking of lipases in our study. Lipases such as CRL and porcine pancreas lipase (PPL) are formulations that have a low cost compared to other lipases,61,63 but PPL is the tenth most used lipase in the bibliographic survey. Probably, the specificity of these lipases is the main factor that led to their classification in this survey. Non-specific enzymes can be applied in numerous industrial processes, but enzymes that present strict specificity are limited to restricted reactions, but are no less important than non-specific lipases. Some lipases, such as TLL and Eversa® Transform, are 1,3-specific enzymes whose specificities are affected when immobilized mainly on hydrophobic supports. Thus, this increases the range of application of these enzymes.15,51
Fig. 5 shows a compilation of the most commonly used hydrophobic supports from 2011 to 2020. It is possible to observe a wide variety of matrices with different degrees of hydrophobicity used for lipase immobilization. Among them, there are commercial supports, such as octyl-agarose, (and some derivatives such as octyl-glyoxyl agarose), EC-octadecyl Sepabeads™, Accurel MP 1000, octyl and phenyl silica and Lewatit VP OC 1600. This last support is used in the industrial production of Novozym® 435.59 Moreover, the use of supports prepared from agro-industrial residues is also observed, such as rice husk silica (RHS) with triethoxy(phenyl)silane, chitin-polyhedral oligomeric silsesquioxanes support, and cellulose/Fe2O3 hydrogel microspheres. Given the variety of available supports, a library of lipase biocatalysts with different properties for industrial application can be obtained. This is only possible because the structure and properties of the supports condition the catalytic properties of the lipases. This effect can produce changes in the specificity and selectivity of the enzymes, modulating the biocatalyst to the desired biotechnological process.52–54,64–66
The main acyl groups present on supports are the octyl and octadecyl groups. In general, the use of these acyl groups can lead to the formation of a lipase biocatalyst with different properties, even using the same conditions during hydrophobic immobilization. TLL was immobilized on octyl-agarose (OC) and Purolite® C18 (Lifetech ECR8806M) and the results demonstrated that the biocatalyst immobilized on resin with octadecyl groups led to better catalytic properties in certain processes and higher stabilization compared to the biocatalyst immobilized on OC.67 Furthermore, the greater degree of hydrophobicity of octadecyl groups can improve the lipase adsorption strength and in that way reduce enzyme leakage.24,68
Problems of lipase immobilization on hydrophobic supports
The main problem with lipase immobilization on a hydrophobic support is the possibility of enzyme desorption when the biocatalyst is subjected to drastic conditions, such as high temperature or the presence of an organic solvent.69 Furthermore, when applied in biosurfactant synthesis, reactions where a biosurfactant is an intermediate product of the process or in a heterogeneous medium containing substances with detergent properties can also facilitate enzyme release.70 A strategy used to minimize this problem is lipase immobilization on a heterofunctional support. This type of support has a unique surface exhibiting various physicochemical capabilities, e.g., a layer of acyl groups to obtain lipase interfacial activation or a layer of groups able to give other physical interactions (e.g., ionic exchange71) or a covalent bond that will make immobilization irreversible.72–76 These new biocatalysts are generally even more stable than standard biocatalysts immobilized only by hydrophobic interaction.77The use of heterofunctional supports can provide different modulating effects on the catalytic properties of lipases due to changes in the interactions between enzyme and support,78 and it can also provide new systems for the co-immobilization of lipases by different mechanisms,94 increasing the possibilities for application of the biocatalyst in industry.77,79,92 On heterofunctional supports, the enzyme is usually initially immobilized by interfacial activation under conditions of low ionic strength and pH in the range of 5.0–7.0, and later, the conditions are modified to favor covalent bonds, such as incubation in alkaline solution. The limitation of the use of these supports is related to the sensitivity of some lipases to alkaline pH when incubated to produce covalent immobilization or the lack of nucleophiles near the active center.72 Some heterofunctional supports and their functional groups are presented in this review, as shown in Table 1.
| Lipase source | Support | Functional groups | Reference |
|---|---|---|---|
| CRL; RML; TLL | OCDVS | Octyl and vinyl sulfone | 79 |
| CALB | Lifetech™ ECR 8285F | Butyl and epoxy | 80 |
| CALB | SMMP-octyl-glu | Octyl and aldehyde | 81 |
| CRL; LipC12 | Aga-C8-GLU | Octyl and aldehyde | 82 |
| BaL | GO-NH2-PMAO | Hydrophobic groups present on the support and aldehyde | 83 |
| PLL | PBA–PAD | Inner hydrophobic PBA domain and aldehyde | 84 |
| CRL | Lifetech™ ECR1030M; Lifetech™ ECR8285 | Butyl and epoxy | 85 |
| CRL; CALB; RAL; ASL; HPL; MML; PCL; RNL | P(SAN-DVB)-GMA | Hydrophobic groups of the matrix poly(styrene-co-acrylonitrile-co-divinylbenzene) and epoxide | 86 |
| LipC12 | OCA; ODA | Octyl and aldehyde; octadecyl and aldehyde | 87 |
| SHL | PMA-co-DVB | Inner hydrophobic PBA domain and aldehyde | 88 |
| CALB; CCL; PCL; PFL; HPL | ChiS-G | Hydrophobic portion of chitosan and aldehyde | 89 |
| RML | OCEPX | Octyl and epoxide | 90 |
| CALB | UndGLX, OCEPX; UndGLXS; OCEPXS | Undecanol and glyoxyl; octyl and epoxide | 75 |
| BTL2 | UndGLXS; OCEPX | Undecanol and glyoxyl; octyl and epoxide | 91 |
| PsL; AsL | OGS | Octyl and glyoxyl | 74 |
| CALA; CALB; CRL; TLL; RML; LU | OCDVS; OCGLX | Octyl and vinyl sulfone; octyl and glyoxyl | 77 |
| CALB | OCGLX | Octyl and glyoxyl | 92 |
| CALB; TLL | OCGLX | Octyl and glyoxyl | 93 |
| LU; RML; PFL | OCGLX | Octyl and glyoxyl | 94 |
| CALB; TLL; RML | OCGLX | Octyl and glyoxyl | 95 |
| PFL | OCGLX | Octyl and glyoxyl | 96 |
| CRL; CALA | OCGLX | Octyl and glyoxyl | 73 |
| RML; CRL | OCGLX | Octyl and glyoxyl | 97 |
| CALB; TLL; RML | OCGLX | Octyl and glyoxyl | 72 |
| CALB; RML; LEU | OCGLX; OCGLXR | Octyl and glyoxyl | 98 |
| PsL; AsL | OCGLXS | Octyl and glyoxyl | 99 |
| CALA; CALB; TLL; RML; CRL; LU | OCEDA; OCHDA | Octyl and glyoxyl | 71 |
Table 1 demonstrates the predominance of heterofunctional supports activated with octyl and glyoxyl groups for the immobilization of different lipases,71,73–75,77,91,92,94,95,97–99 followed by the use of supports activated with octyl and epoxy groups.75,90,91 The other heterofunctional supports present distinct groups for immobilization by interfacial and covalent activation. A support activated with octyl and vinyl sulfone groups has not been used frequently, very likely because it has only recently been proposed. However, it should be highlighted as a support that can lead to the construction of a library of biocatalysts.77,79 This is possible because at the end of the covalent immobilization, there is a need for a final step that requires the blocking of the other reactive groups of the support with a nucleophile.77,79 This step allows the adaptation of enzyme–support interactions using reagents with very different physical properties.77,79 Thus, the use of different blocking reagents can lead to the formation of biocatalysts with different structures and functional properties, even starting from a collection of immobilized enzymes with exactly the same distribution of enzymes present on the support, orientation in relation to the support surface and number of bindings of enzymes to the support,30,100,101 as shown in Fig. 6.
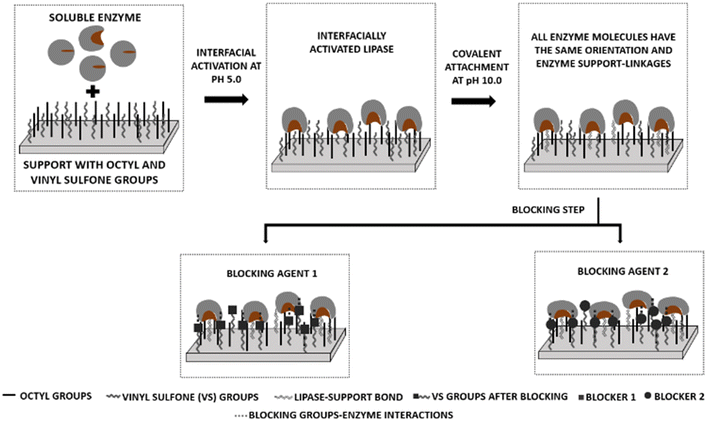 | ||
| Fig. 6 Schematic representation of enzyme immobilization on octyl and vinyl sulfone heterofunctionalized support. | ||
The most commonly used acyl group in the supports is octyl, but the octadecyl group can lead to higher lipase stabilization.67 No paper showing a combination of octadecyl and vinyl sulfone groups in a heterofunctional support was retrieved in the systemic analysis, although one was found in 2023.102 Also, no record was found when performing a brief analysis of data from the last years extracted from the Web of Science using “octadecyl” AND “vinyl sulfone” as a search criterion. This is a gap in the advancement of knowledge to be explored for the study of the synthesis of heterofunctional supports with these characteristics. A possible problem is the size mismatch of the spacer spleens of vinyl sulfone and octadecyl groups. In this case, strategies must be sought to minimize this problem.102
Another strategy that has been used to minimize the desorption problems of hydrophobic supports is intermolecular crosslinking of immobilized enzymes with physical and/or chemical agents. The association of these agents and the immobilization of the lipase on the support provides the formation of a biocatalyst with large enzyme planar aggregates simultaneously interacting with the support at multiple sites.103–106 Therefore, to achieve desorption of the lipase, it will be necessary to release all the enzyme molecules forming each planar physical or chemical aggregate, which is much more difficult than the release of a single lipase protein. Another advantage of crosslinking with physical or chemical agents is the modulation of catalytic properties and, in some cases, improvements in the stability of the biocatalyst.66,107–109 However, recovery of the support after enzyme inactivation becomes complicated if intermolecular chemical crosslinking of all enzyme molecules is achieved, physical crosslinking being better in this respect. Table 2 shows the main physical and/or chemical agents used in the papers obtained in the bibliographic survey.
| Lipase source | Support | Chemical or physical agent | Reference |
|---|---|---|---|
| TLL | Lifetech™ ECR8806M; EC-OD; Lewatit VP OC 1600 | PEG | 15 |
| CALB; TLL; RML | OCGLX | EDA | 95 |
| TLL | MNPs | EDA, GA and DA | 120 |
| CALA; CALB; RML; TLL; LU | OC | PEI and GA | 121 |
| RML | OC | DA | 69 |
| CALB; RML | OC | PEI and DS | 103 |
| CALB | OC | PEI | 122 |
| CALB | OC | DA | 123 |
| AFE | EC-OD | GA | 111 |
| RML | OC | Poly-allylamine (PAA) and DA | 104 |
| CALB | OC | EDA and TNBS | 64 |
| CALB; TLL; RML | OC | EDA, amino groups with succinic anhydride and PEI | 66 |
| CALA; CALB; TLL; RML; LU | OC; OVS | PEI | 124 |
| LipC12 | OCA; ODA | PEG | 87 |
| CALB | Lewatit VP OC1600 | TNBS, EDA, PEI | 125 |
| CALB | OC | PEI and DS | 119 |
| CALB | NKA | PEG | 16 |
| RML | OC | PEI and DS | 118 |
| CALB; TLL; RML | EC-OD | PEI and DS | 65 |
| CALB | OC | PEI | 105 |
| TLL; RML; CALB | OC | EDA and DA | 106 |
| CALB; TLL; RML | Lewatit VP OC1600 | PEI | 126 |
| TLL | EC-OD | PEG | 127 |
| LU | OC | EDA and TNBS | 109 |
| CALB | OS | PEG | 128 |
| CALA; CALB; RML; TLL; LU | OC | PEI and GA | 129 |
| CALB | PTMOS | GA | 76 |
| CRL | PHB | GA | 112 |
| CRL | MSU-H | GA | 113 |
| Eversa2.0; CALB | OC | PEI | 130 |
| CALA; CALB | OC | PEI | 131 |
Physical or chemical crosslinking agents have different characteristics. Glutaraldehyde (GA) is a small bifunctional crosslinking molecule110 that has been used in situations where the enzymes are immobilized at high load on the support76,111–113 or when the surface of the immobilized enzyme is aminated with ethylenediamine (EDA),109 since close proximity between molecules is required so that the amine groups located in different enzyme molecules are clustered together for intermolecular crosslinking to be efficient.114 On the other hand, dextran aldehyde (DA) is a large multifunctional molecule115 that does not demonstrate the limitations involved with crosslinking with glutaraldehyde. Due to its being a multifunctional reagent, it provides very intense crosslinking that makes immobilization of the enzyme on the support irreversible. This makes it impossible to reuse the support after enzyme inactivation.116 Poly-ionic polymers, such as polyethyleneimine (PEI)117 or dextran sulfate (DS), are alternatives to dextran aldehydes, as they provide highly efficient crosslinking, but do not preclude the reuse of the support after enzyme inactivation.103,118,119
The crosslinking of the enzyme immobilized on a hydrophobic support has been carried out with physical and chemical agents of carbonic origin; however, a possibility that should be studied is the crosslinking of immobilized lipases with metal phosphate. This strategy arises from the frontier of knowledge related to the use of metal phosphates for free-enzyme mineralization by an immobilization technique called nanoflower self-assembly.132 In this case, the metal phosphate interacts with a protein nucleation center and starts the self-assembly of an organic–inorganic flower, leading to the formation of an insoluble biocatalyst. Expanding this approach to an immobilized enzyme, an immobilized biocatalyst with intermolecular crosslinking using an inorganic agent can be obtained. In this case, the metal phosphate binds to the enzyme nucleation point and begins the growth of the inorganic structure with the formation of an inorganic film on the immobilized enzyme. The nanoflower self-assembly step is probably not reached because the enzyme is immobilized in a plane.132 The combination of immobilization and inorganic crosslinking can combine the benefits of both protocols and contribute to the formation of a biocatalyst with catalytic properties and greater stability than the biocatalyst without crosslinking.67,102
Another problem that is related to all preexisting supports, not restricted to hydrophobic supports, is their limited load capacity: that is, the amount of enzyme that can be immobilized per gram of support did not permit the filling of the particle volume with enzyme when using preexisting supports. If the loading capacity of the support is increased, the impact of the cost of the support on the total expenses of biocatalyst production can be reduced.133 An alternative is the construction of biocatalysts with a three-dimensional design composed of multilayers of enzymes (an enzyme layer immobilized over the previous one to multiply the final load capacity of the support). This allows the use of a smaller amount of support by increasing the volumetric or mass activity of the biocatalyst.124,129 In addition, this strategy allows the synthesis of combilipases that can be used in a cascade reaction.121,124
PEI is a crosslinking agent that can be used to build a multilayer biocatalyst starting from a first layer of immobilized enzyme.117 This polymer is able to adsorb onto immobilized enzyme molecules through strong ionic exchange. In addition, the PEI-coated enzyme is able to immobilize other enzyme molecules also via ion exchange121,122,124 involving the enzyme in a three-dimensional structure instead of a flat surface.134 After each overlapping enzyme layer, crosslinking with glutaraldehyde of the structural network formed by PEI and enzyme can be carried out; this strategy of immobilization by covalent bonding is adopted to prevent the release of the enzyme during the PEI incubation121 (Fig. 7). Dextran sulfate is an alternative polymer to PEI in the construction of multilayer biocatalysts.135
Co-occurrence analysis of keywords
We performed a co-occurrence analysis between the main lipases and supports in our database. Based on the retrieved publications, it is possible to elucidate the structure, distribution of the network and the frequency of co-occurrence of these keywords to determine the possible hotspots and frontiers of knowledge not yet explored.Bibliometric maps revealed that 75 lipases and 45 hydrophobic supports were cited by the studies we investigated (Fig. 8). Fig. 8a shows the use of more than one lipase in ∼23% of the studies, which may indicate that these studies are looking for an enzyme with catalytic properties suitable for the productive process being evaluated, since there is no universal biocatalyst for application in all processes. CALB showed the highest number of co-occurrences with different lipases (N = 34), the main ones being TLL, CRL, RML, PFL, CALA and Lecitase Ultra (LU). Next, RML (N = 32), TLL (N = 28) and CRL (N = 17) were the lipases that showed the highest co-occurrence. The use of different lipases in the same publication may be related to the development of new methods of immobilization and/or stabilization of the final biocatalyst, making it necessary to validate these strategies with different lipases that have different characteristics. In addition, this may demonstrate the need to develop a library of biocatalysts with suitable catalytic properties for the industrial process to be studied.120,122
As can be seen in this systemic analysis, several studies are focused on the immobilization of CALB, TLL, CRL, RML, PFL and CALA for application in industrial processes or studies of new stabilization strategies for these enzymes. However, Eversa Transform is a new lipase on the market that has been gaining pace in several applications in aqueous and organic media but only presented 3 co-occurrences. This is an enzyme that emerged as an improved version of TLL and has low cost compared to the mentioned enzymes, high thermal stability and greater resistance to the presence of methanol.62 Another enzyme worth mentioning is PPL, which presented only one co-occurrence. This enzyme has a very low cost, so this factor associated with the immobilization technique by interfacial adsorption can minimize the costs of the process and the simultaneous purification and immobilization of this enzyme may be carried out. The main limitation is its specificity, since it is a 1,3-specific lipase.63
In Fig. 8b, a co-occurrence network between hydrophobic supports occurs in ∼30% of publications. This result suggests that the use of supports with different degrees of hydrophobicity served as priorities in studies to obtain biocatalysts with different catalytic properties. We found a high number of co-occurrences between commercial and non-commercial supports (N = 22). This demonstrates two possible issues: the first would be the use of commercial supports to validate the advantages of non-commercial supports; the second would be the use of commercially pre-existing supports, aiming at easing their acquisition and, possibly, reducing process costs. There is also the co-occurrence of heterofunctional supports (N = 5) and the occurrence of many supports prepared using materials that are agro-industrial residues, although there is no co-occurrence between them. Even though supports prepared from agro-industrial residues are evident, there is a gap and there is a need to explore other materials, such as glycerol, lignin, and soybean husk, among others. These residues can be used for the synthesis of activated carbon, are rich in oxygen and nitrogen surface groups, and can be used in natura or activated with desired reactive groups.136,137
Limitations
Our strategy was mapped using two Web of Science databases (SCI-E and ESCI), Scopus, PubMed and SciELO. We chose not to use scientific evidence, such as theses and dissertations, committee or government reports, documents from congresses, conferences and seminars, or ongoing research. This limits it to a set of primary data published in scientific journals; that is, the information extracted from publications obtained in the bibliographic survey may have been underestimated. Thus, our conclusions must be evaluated under these circumstances. However, the contribution of this review is very important to show the direction of research related to the immobilization of lipases on hydrophobic supports.Conclusions
The results presented in this research demonstrate a systematic review of the immobilization of lipases on hydrophobic supports. Here, a wide variety of lipases with different applicability are presented. It is not possible to obtain an optimal universal biocatalyst for use in different applications. However, the choice of support and enzyme immobilization and stabilization strategies are factors that can influence the catalytic properties and final stability of the immobilized biocatalyst. Various supports with different degrees of hydrophobicity are presented. Furthermore, problems related to immobilization by interfacial adsorption and the strategies used to overcome them are described. Immobilization on a heterofunctional support and crosslinking with physical or chemical reagents are the main solutions to this problem. A co-occurrence analysis between the main lipases and supports in our database was performed. Furthermore, the structure, network distribution and frequency of co-occurrence between lipases and supports to determine possible hotspots and as yet unexplored frontiers of knowledge were elucidated. Therefore, the contribution of this review is very important for showing the direction of research related to the immobilization of lipases on hydrophobic supports, their bottlenecks and solutions to problems.Author contributions
J. R. G.: conceptualization, methodology, investigation, visualization, formal analysis, writing – original draft, review and editing. K. S. G. C. O.: visualization, formal analysis, review and editing. M. C. P. G.: visualization, formal analysis. J. P. R.: methodology, visualization, formal analysis; L. A. L.: visualization, formal analysis. A. B.-M.: visualization, formal analysis, review and editing. R. F.-L.: resources, conceptualization, methodology, writing – review and editing, supervision. P. W. T.: resources, conceptualization, methodology, writing – review and editing, supervision. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was funded by São Paulo Research Foundation (FAPESP, grant number 2016/10636-8), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES, Finance Code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, project numbers 405889/2016-0 and 308212/2017-7), Ministerio de Ciencia e Innovación and Agencia Estatal de Investigación (Spanish Government) (project PID2022-136535OB-I00 and MCIN/AEI/10.13039/501100011033/) and by FEDER A Way of Making Europe funds (project PID2021-123079OB-I00), and Generalitat Valenciana (CIPROM/2021/70).References
- N. Sarmah, D. Revathi, G. Sheelu, K. Yamuna Rani, S. Sridhar, V. Mehtab and C. Sumana, Biotechnol. Prog., 2018, 34, 5–28 CrossRef CAS PubMed.
- E. C. G. Aguieiras, D. S. N. de Barros, H. Sousa, R. Fernandez-Lafuente and D. M. G. Freire, Renewable Energy, 2017, 113, 112–118 CrossRef CAS.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS.
- J. J. Virgen-Ortíz, J. C. S. dos Santos, C. Ortiz, Á. Berenguer-Murcia, O. Barbosa, R. C. Rodrigues and R. Fernandez-Lafuente, Mol. Catal., 2019, 473, 110405 CrossRef.
- R. R. C. Monteiro, J. C. S. dos Santos, A. R. Alcántara and R. Fernandez-Lafuente, Catalysts, 2020, 10, 891 CrossRef CAS.
- R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Chem. Soc. Rev., 2013, 42, 6437 RSC.
- P. Adlercreutz, Chem. Soc. Rev., 2013, 42, 6406 RSC.
- O. Barbosa, R. Torres, C. Ortiz, Á. Berenguer-Murcia, R. C. Rodrigues and R. Fernandez-Lafuente, Biomacromolecules, 2013, 14, 2433–2462 CrossRef CAS PubMed.
- C. Garcia-Galan, Á. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Adv. Synth. Catal., 2011, 353, 2885–2904 CrossRef CAS.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS.
- F. L. C. Almeida, M. P. J. Castro, B. M. Travália and M. B. S. Forte, Process Biochem., 2021, 110, 37–51 CrossRef CAS.
- T. D. Nalder, I. Kurtovic, C. J. Barrow and S. N. Marshall, Enzyme Microb. Technol., 2018, 113, 18–23 CrossRef CAS PubMed.
- E. A. Manoel, M. F. P. Ribeiro, J. C. S. Dos Santos, M. A. Z. Coelho, A. B. C. Simas, R. Fernandez-Lafuente and D. M. G. Freire, Process Biochem., 2015, 50, 1557–1564 CrossRef CAS.
- W. Li, H. Shen, Y. Tao, B. Chen and T. Tan, Process Biochem., 2014, 49, 1488–1496 CrossRef CAS.
- E. A. Silveira, S. Moreno-Perez, A. Basso, S. Serban, R. Pestana-Mamede, P. W. Tardioli, C. Sanchez-Farinas, N. Castejon, G. Fernandez-Lorente, J. Rocha-Martin and J. M. Guisan, J. Biotechnol., 2019, 289, 126–134 CrossRef PubMed.
- H. Wang, B. Duan, H. Li, Y. Lu, Z. Liu, W. Su, H. Wang, B. Duan, H. Li, Y. Lu, Z. Liu, W. Su and S. Li, New J. Chem., 2020, 44, 3463–3470 RSC.
- L. Brady, A. M. Brzozowski, Z. S. Derewenda, E. Dodson, G. Dodson, S. Tolley, J. P. Turkenburg, L. Christiansen, B. Huge-Jensen, L. Norskov, L. Thim and U. Menge, Nature, 1990, 343, 767 CrossRef CAS PubMed.
- N. Miled, F. Beisson, J. de Caro, A. de Caro, V. Arondel and R. Verger, J. Mol. Catal. B: Enzym., 2001, 11, 165–171 CrossRef CAS.
- R. D. Schmid and R. Verger, Angew. Chem., 1998, 110, 1694–1720 CrossRef.
- R. Verger, Trends Biotechnol., 1997, 15, 32–38 CrossRef CAS.
- R. Fernandez-Lafuente, P. Armisén, P. Sabuquillo, G. Fernández-Lorente and J. M. Guisán, Chem. Phys. Lipids, 1998, 93, 185–197 CrossRef CAS PubMed.
- A. Bastida, P. Sabuquillo, P. Armisen, R. Fernández-Lafuente, J. Huguet and J. M. Guisán, Biotechnol. Bioeng., 1998, 58, 486–493 CrossRef CAS PubMed.
- L. Abreu, R. Fernandez-Lafuente, R. C. Rodrigues, G. Volpato and M. A. Z. Ayub, J. Mol. Catal. B: Enzym., 2014, 99, 51–55 CrossRef.
- J. Brabcová, Z. Demianová, J. Vondrášek, M. Jágr, M. Zarevúcka and J. M. Palomo, J. Mol. Catal. B: Enzym., 2013, 98, 62–72 CrossRef.
- N. Ghattas, M. Filice, F. Abidi, J. M. Guisan and A. Ben Salah, J. Mol. Catal. B: Enzym., 2014, 110, 111–116 CrossRef CAS.
- M. Cygler and J. D. Schrag, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 1999, 1441, 205–214 CrossRef CAS PubMed.
- K.-E. Jaeger, S. Ransac, H. B. Koch, F. Ferrato and B. W. Dijkstra, FEBS Lett., 1993, 332, 143–149 CrossRef CAS PubMed.
- K. K. Kim, H. K. Song, D. H. Shin, K. Y. Hwang and S. W. Suh, Structure, 1997, 5, 173–185 CrossRef CAS PubMed.
- J. M. Santos, E. S. de Moura, A. S. da Silva, J. M. B. Cavalcanti, R. D. S. Torres and M. L. A. Vidal, Pattern Recognit. Lett., 2015, 65, 1–7 CrossRef.
- J. C. S. Santos, N. Rueda, R. Torres, O. Barbosa, L. R. B. Gonçalves and R. Fernandez-Lafuente, Process Biochem., 2015, 50, 918–927 CrossRef.
- E. A. Manoel, J. C. S. dos Santos, D. M. G. Freire, N. Rueda and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2015, 71, 53–57 CrossRef CAS PubMed.
- V. Dossat, D. Combes and A. Marty, Enzyme Microb. Technol., 1999, 25, 194–200 CrossRef CAS.
- A. Marty, V. Dossat and J.-S. Condoret, Biotechnol. Bioeng., 1997, 56, 232–237 CrossRef CAS PubMed.
- E. Séverac, O. Galy, F. Turon, C. A. Pantel, J.-S. Condoret, P. Monsan and A. Marty, Enzyme Microb. Technol., 2011, 48, 61–70 CrossRef PubMed.
- J. P. Romanelli, P. Meli, R. P. Naves, M. C. Alves and R. R. Rodrigues, Conserv. Biol., 2021, 35, 142–154 CrossRef PubMed.
- K. Dwan, C. Gamble, P. R. Williamson and J. J. Kirkham, PLoS One, 2013, 8, e66844 CrossRef CAS PubMed.
- K. Petersen, S. Vakkalanka and L. Kuzniarz, Inf. Softw. Technol., 2015, 64, 1–18 CrossRef.
- K. L. James, N. P. Randall and N. R. Haddaway, Environ. Evid., 2016, 5, 7 CrossRef.
- M. C. P. Gonçalves, J. P. Romanelli, J. R. Guimarães, A. C. Vieira, B. P. de Azevedo and P. W. Tardioli, Crit. Rev. Biotechnol., 2021, 41, 865–878 CrossRef PubMed.
- N. R. Haddaway, A. Feierman, M. J. Grainger, C. T. Gray, E. Tanriver-Ayder, S. Dhaubanjar and M. J. Westgate, Environ. Evid., 2019, 8, 22 CrossRef.
- Collaboration for Environmental Evidence, Guidelines and Standards for Evidence synthesis in Environmental Management, Version 5.0, ed. A. S. Pullin, G. K. Frampton, B. Livoreil and G. Petrokofsky, The Environmental Evidence Library, Bangor (UK), 2018, https://www.environmentalevidence.org/information-for-authors, [18/09/2021] Search PubMed.
- J. Cohen, Educ. Psychol. Meas., 1960, 20, 37–46 CrossRef.
- R. Core, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna (Austria), 2019 Search PubMed.
- Z. Cabrera, G. Fernandez-Lorente, R. Fernandez-Lafuente, J. M. Palomo and J. M. Guisan, J. Mol. Catal. B: Enzym., 2009, 57, 171–176 CrossRef CAS.
- G. Fernández-Lorente, Z. Cabrera, C. Godoy, R. Fernandez-Lafuente, J. M. Palomo and J. M. Guisan, Process Biochem., 2008, 43, 1061–1067 CrossRef.
- V. G. Tacias-Pascacio, S. Peirce, B. Torrestiana-Sanchez, M. Yates, A. Rosales-Quintero, J. J. Virgen-Ortíz and R. Fernandez-Lafuente, RSC Adv., 2016, 6, 100281–100294 RSC.
- M. C. C. Pinto, S. S. Everton, L. C. M. Cirilo, T. N. de Melo, E. P. Cipolatti, E. A. Manoel, J. C. Pinto and D. M. G. Freire, Biocatal. Biotransform., 2020, 38, 304–314 CAS.
- G. Fernández-Lorente, C. Ortiz, R. L. Segura, R. Fernández-Lafuente, J. M. Guisán and J. M. Palomo, Biotechnol. Bioeng., 2005, 92, 773–779 CrossRef PubMed.
- R. L. Segura, L. Betancor, J. M. Palomo, A. Hidalgo, G. Fernández-Lorente, M. Terreni, C. Mateo, A. Cortés, R. Fernández-Lafuente and J. M. Guisán, Enzyme Microb. Technol., 2006, 39, 817–823 CrossRef CAS.
- J. L. R. Friedrich, F. P. Peña, C. Garcia-Galan, R. Fernandez-Lafuente, M. A. Z. Ayub and R. C. Rodrigues, J. Chem. Technol. Biotechnol., 2013, 88, 1089–1095 CrossRef CAS.
- E. A. Silveira, S. Moreno-Perez, A. Basso, S. Serban, R. P. Mamede, P. W. Tardioli, C. S. Farinas, J. Rocha-Martin, G. Fernandez-Lorente and J. M. Guisan, BMC Biotechnol., 2017, 17, 88 CrossRef CAS PubMed.
- S. Arana-Peña, N. S. Rios, D. Carballares, L. R. B. Gonçalves and R. Fernandez-Lafuente, Catal. Today, 2021, 362, 130–140 CrossRef.
- S. Arana-Peña, N. S. Rios, D. Carballares, C. Mendez-Sanchez, Y. Lokha, L. R. B. Gonçalves and R. Fernandez-Lafuente, Front. Bioeng. Biotechnol., 2020, 8, 1–13 Search PubMed.
- Y. Lokha, S. Arana-Peña, N. S. Rios, C. Mendez-Sanchez, L. R. B. Gonçalves, F. Lopez-Gallego and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2020, 133, 109461 CrossRef CAS PubMed.
- G. Fernández-Lorente, M. Terreni, C. Mateo, A. Bastida, R. Fernández-Lafuente, P. Dalmases, J. Huguet and J. M. Guisán, Enzyme Microb. Technol., 2001, 28, 389–396 CrossRef PubMed.
- V. G. Tacias-Pascacio, J. J. Virgen-Ortíz, M. Jiménez-Pérez, M. Yates, B. Torrestiana-Sanchez, A. Rosales-Quintero and R. Fernandez-Lafuente, Fuel, 2017, 200, 1–10 CrossRef CAS.
- R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- G. Fernández-Lorente, J. M. Palomo, Z. Cabrera, J. M. Guisán and R. Fernández-Lafuente, Enzyme Microb. Technol., 2007, 41, 565–569 CrossRef.
- C. Ortiz, M. L. Ferreira, O. Barbosa, J. C. S. dos Santos, R. C. Rodrigues, Á. Berenguer-Murcia, L. E. Briand and R. Fernandez-Lafuente, Catal. Sci. Technol., 2019, 9, 2380–2420 RSC.
- R. Fernandez-Lafuente, J. Mol. Catal. B: Enzym., 2010, 62, 197–212 CrossRef CAS.
- P. Domínguez De María, J. M. Sánchez-Montero, J. V. Sinisterra and A. R. Alcántara, Biotechnol. Adv., 2006, 24, 180–196 CrossRef PubMed.
- R. R. C. Monteiro, S. Arana-Peña, T. N. da Rocha, L. P. Miranda, Á. Berenguer-Murcia, P. W. Tardioli, J. C. S. dos Santos and R. Fernandez-Lafuente, Renewable Energy, 2021, 164, 1566–1587 CrossRef CAS.
- A. A. Mendes, P. C. Oliveira and H. F. Castro, J. Mol. Catal. B: Enzym., 2012, 78, 119–134 CrossRef CAS.
- O. Barbosa, M. Ruiz, C. Ortiz, M. Fernández, R. Torres and R. Fernandez-Lafuente, Process Biochem., 2012, 47, 867–876 CrossRef CAS.
- S. Moreno-Pérez, J. M. Guisan and G. Fernandez-Lorente, J. Am. Oil Chem. Soc., 2014, 91, 63–69 CrossRef.
- G. Fernández-Lorente, L. Betancor, A. V. Carrascosa, J. M. Palomo and J. M. Guisan, J. Am. Oil Chem. Soc., 2012, 89, 97–102 CrossRef.
- J. R. Guimarães, D. Carballares, J. Rocha-Martin, P. W. Tardioli and R. Fernandez-Lafuente, Catalysts, 2022, 12, 1552 CrossRef.
- M. V. Tacin, T. A. Costa-Silva, A. V. de Paula, J. M. Palomo and V. de C. Santos-Ebinuma, Prep. Biochem. Biotechnol., 2021, 51, 749–760 CrossRef CAS PubMed.
- G. Fernandez-Lorente, M. Filice, D. Lopez-Vela, C. Pizarro, L. Wilson, L. Betancor, Y. Avila and J. M. Guisan, J. Am. Oil Chem. Soc., 2011, 88, 801–807 CrossRef CAS.
- M. C. P. Gonçalves, J. C. Amaral, L. A. Lopes, R. Fernandez-Lafuente and P. W. Tardioli, Int. J. Biol. Macromol., 2021, 192, 665–674 CrossRef PubMed.
- N. Rueda, T. L. Albuquerque, R. Bartolome-Cabrero, L. Fernandez-Lopez, R. Torres, C. Ortiz, J. C. S. Dos Santos, O. Barbosa and R. Fernandez-Lafuente, Molecules, 2016, 21, 646 CrossRef PubMed.
- N. Rueda, J. C. S. Dos Santos, R. Torres, C. Ortiz, O. Barbosa and R. Fernandez-Lafuente, RSC Adv., 2015, 5, 11212–11222 RSC.
- A. Suescun, N. Rueda, J. C. S. Dos Santos, J. J. Castillo, C. Ortiz, R. Torres, O. Barbosa and R. Fernandez-Lafuente, Process Biochem., 2015, 50, 1211–1217 CrossRef CAS.
- C. Bernal, A. Illanes and L. Wilson, Langmuir, 2014, 30, 3557–3566 CrossRef CAS PubMed.
- N. Guajardo, C. Bernal, L. Wilson and Z. Cabrera, Process Biochem., 2015, 50, 1870–1877 CrossRef CAS.
- Z. Boros, D. Weiser, M. Márkus, E. Abaháziová, Á. Magyar, A. Tomin, B. Koczka, P. Kovács and L. Poppe, Process Biochem., 2013, 48, 1039–1047 CrossRef CAS.
- D. B. Hirata, T. L. Albuquerque, N. Rueda, J. J. Virgen-Ortíz, V. G. Tacias-Pascacio and R. Fernandez-Lafuente, J. Mol. Catal. B: Enzym., 2016, 133, 117–123 CrossRef CAS.
- J. R. Guimarães, D. Carballares, J. Rocha-Martin, P. W. Tardioli and R. Fernandez-Lafuente, Int. J. Biol. Macromol., 2022, 222, 2452–2466 CrossRef PubMed.
- T. L. D. Albuquerque, N. Rueda, J. C. S. Dos Santos, O. Barbosa, C. Ortiz, B. Binay, E. Özdemir, L. R. B. Gonçalves and R. Fernandez-Lafuente, Process Biochem., 2016, 51, 865–874 CrossRef.
- A. Milivojević, M. Ćorović, M. Simović, K. Banjanac and D. Bezbradica, Biochem. Eng. J., 2020, 163, 107748 CrossRef.
- L. N. Lima, G. N. A. Vieira, W. Kopp, P. W. Tardioli and R. L. C. Giordano, J. Mol. Catal. B: Enzym., 2016, 133, S491–S499 CrossRef.
- R. R. de Melo, R. C. Alnoch, A. F. L. Vilela, E. M. de Souza, N. Krieger, R. Ruller, H. H. Sato and C. Mateo, Molecules, 2017, 22, 1088 CrossRef PubMed.
- A. Ameri, M. Shakibaie, M. Khoobi, M. A. Faramarzi, E. Gholibegloo, A. Ameri and H. Forootanfar, Int. J. Biol. Macromol., 2020, 162, 1790–1806 CrossRef CAS PubMed.
- X. Sun, W. Zhu and K. Matyjaszewski, Macromolecules, 2018, 51, 289–296 CrossRef CAS PubMed.
- A. Anand, P. Gnanasekaran, A. M. Allgeier and L. R. Weatherley, Food Bioprod. Process., 2020, 123, 164–176 CrossRef CAS.
- H. Yang, L. Wang, S. Chen, M. Wang and Z. Feng, Polym. Int., 2015, 64, 915–923 CrossRef CAS.
- R. Alnoch, R. R. Melo, J. Palomo, E. M. Souza, N. Krieger and C. Mateo, Catalysts, 2016, 6, 191 CrossRef.
- S. H. Kim, S.-J. Kim, S. Park and H. K. Kim, J. Mol. Catal. B: Enzym., 2013, 85–86, 10–16 CrossRef CAS.
- M. D. Mello, D. Cordeiro, L. T. Costa and C. Follmer, Biotechnol. Bioprocess Eng., 2013, 18, 1090–1100 CrossRef.
- M. Garmroodi, M. Mohammadi, A. Ramazani, M. Ashjari, J. Mohammadi, B. Sabour and M. Yousefi, Int. J. Biol. Macromol., 2016, 86, 208–215 CrossRef CAS PubMed.
- N. Guajardo, C. Bernal, L. Wilson and Z. Cabrera, Catal. Today, 2015, 255, 21–26 CrossRef CAS.
- D. B. Hirata, T. L. Albuquerque, N. Rueda, J. M. Sánchez-Montero, E. Garcia-Verdugo, R. Porcar and R. Fernandez-Lafuente, ChemistrySelect, 2016, 1, 3259–3270 CrossRef CAS.
- N. Rueda, J. C. S. Dos Santos, R. Torres, O. Barbosa, C. Ortiz and R. Fernandez-Lafuente, RSC Adv., 2015, 5, 55588–55594 RSC.
- N. Rios, S. Arana-Peña, C. Mendez-Sanchez, C. Ortiz, L. Gonçalves and R. Fernandez-Lafuente, Catalysts, 2019, 9, 487 CrossRef CAS.
- N. Rueda, J. C. S. Dos Santos, C. Ortiz, O. Barbosa, R. Fernandez-Lafuente and R. Torres, Catal. Today, 2016, 259, 107–118 CrossRef CAS.
- N. S. Rios, C. Mendez-Sanchez, S. Arana-Peña, N. Rueda, C. Ortiz, L. R. B. Gonçalves and R. Fernandez-Lafuente, Biochim. Biophys. Acta, Proteins Proteomics, 2019, 1867, 741–747 CrossRef CAS PubMed.
- L. Fernandez-Lopez, N. Rueda, R. Bartolome-Cabrero, M. D. Rodriguez, T. L. Albuquerque, J. C. S. Dos Santos, O. Barbosa and R. Fernandez-Lafuente, Process Biochem., 2016, 51, 48–52 CrossRef CAS.
- S. Arana-Peña, C. Mendez-Sanchez, N. S. Rios, C. Ortiz, L. R. B. Gonçalves and R. Fernandez-Lafuente, Int. J. Biol. Macromol., 2019, 131, 989–997 CrossRef PubMed.
- C. Bernal, A. Illanes and L. Wilson, J. Agric. Food Chem., 2015, 63, 3716–3724 CrossRef CAS PubMed.
- J. C. S. Santos, N. Rueda, A. Sanchez, R. Villalonga, L. R. B. Gonçalves and R. Fernandez-Lafuente, RSC Adv., 2015, 5, 35801–35810 RSC.
- J. C. S. Santos, N. Rueda, L. R. B. Gonçalves and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2015, 77, 1–7 CrossRef CAS PubMed.
- J. R. Guimarães, D. Carballares, J. Rocha-Martin, A. R. Alcántara, P. W. Tardioli and R. Fernandez-Lafuente, Catalysts, 2023, 13, 108 CrossRef.
- J. Virgen-Ortíz, S. Pedrero, L. Fernandez-Lopez, N. Lopez-Carrobles, B. Gorines, C. Otero and R. Fernandez-Lafuente, Molecules, 2017, 22, 91 CrossRef PubMed.
- J. Cejudo-Sanches, A. H. Orrego, A. Jaime-Mendoza, R. Ghobadi, S. Moreno-Perez, G. Fernandez-Lorente, J. Rocha-Martin and J. M. Guisan, Process Biochem., 2020, 92, 156–163 CrossRef CAS.
- S. Peirce, V. Tacias-Pascacio, M. Russo, A. Marzocchella, J. Virgen-Ortíz and R. Fernandez-Lafuente, Molecules, 2016, 21, 751 CrossRef PubMed.
- A. Orrego, R. Ghobadi, S. Moreno-Perez, A. Mendoza, G. Fernandez-Lorente, J. Guisan and J. Rocha-Martin, Int. J. Mol. Sci., 2018, 19, 553 CrossRef PubMed.
- O. Barbosa, M. Ruiz, C. Ortiz, M. Fernández, R. Torres and R. Fernandez-Lafuente, Process Biochem., 2012, 47, 867–876 CrossRef CAS.
- J. C. S. dos Santos, C. Garcia-Galan, R. C. Rodrigues, H. B. de Sant'Ana, L. R. B. Gonçalves and R. Fernandez-Lafuente, Process Biochem., 2014, 49, 1511–1515 CrossRef CAS.
- C. Garcia-Galan, J. C. S. dos Santos, O. Barbosa, R. Torres, E. B. Pereira, V. C. Corberan, L. R. B. Gonçalves and R. Fernandez-Lafuente, Process Biochem., 2014, 49, 604–616 CrossRef CAS.
- O. Barbosa, C. Ortiz, Á. Berenguer-Murcia, R. Torres, R. C. Rodrigues and R. Fernandez-Lafuente, RSC Adv., 2014, 4, 1583–1600 RSC.
- M. Wang, H. Shi, D. Wu, H. Han, J. Zhang, Z. Xing, S. Wang and Q. Li, Molecules, 2014, 19, 9838–9849 CrossRef PubMed.
- N. Binhayeeding, T. Yunu, N. Pichid, S. Klomklao and K. Sangkharak, Process Biochem., 2020, 95, 174–185 CrossRef CAS.
- W. H. Yu, M. Fang, D. S. Tong, P. Shao, T. N. Xu and C. H. Zhou, Biochem. Eng. J., 2013, 70, 97–105 CrossRef CAS.
- R. Fernandez-Lafuente, C. M. Rosell, V. Rodriguez and J. M. Guisan, Enzyme Microb. Technol., 1995, 17, 517–523 CrossRef CAS.
- V. G. Tacias-Pascacio, C. Ortiz, N. Rueda, A. Berenguer-Murcia, N. Acosta, I. Aranaz, C. Civera, R. Fernandez-Lafuente and A. R. Alcántara, Catalysts, 2019, 9, 622 CrossRef CAS.
- R. C. Rodrigues, J. J. Virgen-Ortíz, J. C. S. dos Santos, Á. Berenguer-Murcia, A. R. Alcantara, O. Barbosa, C. Ortiz and R. Fernandez-Lafuente, Biotechnol. Adv., 2019, 37, 746–770 CrossRef CAS PubMed.
- J. J. Virgen-Ortíz, J. C. S. dos Santos, Á. Berenguer-Murcia, O. Barbosa, R. C. Rodrigues and R. Fernandez-Lafuente, J. Mater. Chem. B, 2017, 5, 7461–7490 RSC.
- L. Fernandez-Lopez, S. G. Pedrero, N. Lopez-Carrobles, J. J. Virgen-Ortíz, B. C. Gorines, C. Otero and R. Fernandez-Lafuente, Process Biochem., 2017, 54, 81–88 CrossRef CAS.
- L. Fernandez-Lopez, J. J. Virgen-OrtÍz, S. G. Pedrero, N. Lopez-Carrobles, B. C. Gorines, C. Otero and R. Fernandez-Lafuente, Biocatal. Biotransform., 2018, 36, 47–56 CrossRef CAS.
- R. O. Henriques, J. A. Bork, G. Fernandez-Lorente, J. M. Guisan, A. Furigo, D. de Oliveira and B. C. Pessela, Mol. Catal., 2018, 453, 12–21 CrossRef CAS.
- S. Arana-Peña, N. S. Rios, C. Mendez-Sanchez, Y. Lokha, D. Carballares, L. R. B. Gonçalves and R. Fernandez-Lafuente, Int. J. Biol. Macromol., 2020, 145, 856–864 CrossRef PubMed.
- S. Peirce, J. J. Virgen-Ortíz, V. G. Tacias-Pascacio, N. Rueda, R. Bartolome-Cabrero, L. Fernandez-Lopez, M. E. Russo, A. Marzocchella and R. Fernandez-Lafuente, RSC Adv., 2016, 6, 61707–61715 RSC.
- M. L. E. Gutarra, O. Romero, O. Abian, F. A. G. Torres, D. M. G. Freire, A. M. Castro, J. M. Guisan and J. M. Palomo, ChemCatChem, 2011, 3, 1902–1910 CrossRef CAS.
- S. Arana-Peña, D. Carballares, V. Cortés Corberan and R. Fernandez-Lafuente, Catalysts, 2020, 10, 1207 CrossRef.
- M. Villalba, C. M. Verdasco-Martín, J. C. S. dos Santos, R. Fernandez-Lafuente and C. Otero, Enzyme Microb. Technol., 2016, 90, 35–44 CrossRef CAS PubMed.
- S. Moreno-Perez, M. Filice, J. M. Guisan and G. Fernandez-Lorente, Chem. Phys. Lipids, 2013, 174, 48–54 CrossRef CAS PubMed.
- N. Castejón, S. Moreno-Pérez, E. A. Silveira, G. Fernández-Lorente, J. M. Guisán and F. J. Señoráns, Food Chem., 2019, 271, 433–439 CrossRef PubMed.
- R. M. Blanco and I. Roldán, Biocatal. Biotransform., 2018, 36, 224–232 CrossRef CAS.
- S. Arana-Peña, N. S. Rios, C. Mendez-Sanchez, Y. Lokha, L. R. B. Gonçalves and R. Fernández-Lafuente, Enzyme Microb. Technol., 2020, 137, 109535 CrossRef PubMed.
- S. Arana-Peña, Y. Lokha and R. Fernández-Lafuente, Catalysts, 2018, 8, 511 CrossRef.
- S. Arana-Peña, Y. Lokha and R. Fernández-Lafuente, Biotechnol. Prog., 2019, 35, 1–8 CrossRef PubMed.
- J. R. Guimarães, D. Carballares, J. Rocha-Martin, P. W. Tardioli and R. Fernandez-Lafuente, Int. J. Biol. Macromol., 2022, 213, 43–54 CrossRef PubMed.
- N. S. Rios, S. Arana-Peña, C. Mendez-Sanchez, Y. Lokha, V. Cortes-Corberan, L. R. B. Gonçalves and R. Fernandez-Lafuente, Catalysts, 2019, 9, 576 CrossRef CAS.
- R. Torres, B. C. C. Pessela, M. Fuentes, C. Mateo, R. Munilla, R. Fernandez-Lafuente and J. M. Guisán, Enzyme Microb. Technol., 2006, 39, 711–716 CrossRef CAS.
- D. Carballares, J. Rocha-Martin and R. Fernandez-Lafuente, ACS Sustainable Chem. Eng., 2022, 10, 3920–3934 CrossRef CAS.
- P. T. Juchen, K. M. Barcelos, K. S. G. C. Oliveira and L. A. M. Ruotolo, Chem. Eng. J., 2022, 429, 132209 CrossRef CAS.
- L. F. Costa, L. A. M. Ruotolo, L. S. Ribeiro, M. C. Pereira, E. R. Camargo and F. G. E. Nogueira, J. Environ. Chem. Eng., 2021, 9, 105061 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3re00420a |
| This journal is © The Royal Society of Chemistry 2023 |