Open Access Article
Open Access ArticleRationale design and synthesis of new apoptotic thiadiazole derivatives targeting VEGFR-2: computational and in vitro studies†
Walid E. Elgammal a,
Hazem Elkady
a,
Hazem Elkady b,
Hazem A. Mahdy
b,
Hazem A. Mahdy b,
Dalal Z. Huseinc,
Aisha A. Alsfoukd,
Bshra A. Alsfoukd,
Ibrahim M. Ibrahime,
Eslam B. Elkaeed*f,
Ahmed M. Metwaly
b,
Dalal Z. Huseinc,
Aisha A. Alsfoukd,
Bshra A. Alsfoukd,
Ibrahim M. Ibrahime,
Eslam B. Elkaeed*f,
Ahmed M. Metwaly *gh and
Ibrahim H. Eissa
*gh and
Ibrahim H. Eissa *b
*b
aDepartment of Chemistry, Faculty of Science, Al-Azhar University, Nasr City, Cairo, Egypt
bPharmaceutical Medicinal Chemistry & Drug Design Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo 11884, Egypt
cChemistry Department, Faculty of Science, New Valley University, El-Kharja 72511, Egypt
dDepartment of Pharmaceutical Sciences, College of Pharmacy, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh 11671, Saudi Arabia
eBiophysics Department, Faculty of Science, Cairo University, Giza 12613, Egypt. E-mail: Ibrahim_mohamed@cu.edu.eg
fDepartment of Pharmaceutical Sciences, College of Pharmacy, AlMaarefa University, Riyadh 13713, Saudi Arabia. E-mail: ekaeed@um.edu.sa
gPharmacognosy and Medicinal Plants Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo 11884, Egypt. E-mail: ametwaly@azhar.edu.eg
hBiopharmaceutical Products Research Department, Genetic Engineering and Biotechnology Research Institute, City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt
First published on 19th December 2023
Abstract
This work presents the synthesis and in vitro, and in silico analyses of new thiadiazole derivatives that are designed to mimic the pharmacophoric characteristics of vascular endothelial growth factor receptor-2 (VEGFR-2) inhibitors. A comprehensive evaluation of the inhibitory properties of the synthesized thiadiazole derivatives against the cancer cell lines MCF-7 and HepG2 identified several auspicious candidates. Among them, compound 14 showed remarkably low IC50 values of 0.04 μM and 0.18 μM against MCF-7 and HepG2, respectively. VEGFR-2 inhibitory evaluation of compound 14 revealed a promising IC50 value in the nanomolar range (103 nM). Further examination of the cell cycle revealed that compound 14 has the ability to stop the progression of the cell cycle in MCF-7 cells via G0–G1 phase arrest. Interestingly, compound 14 also demonstrated a noteworthy pro-apoptotic effect in MCF-7 cells, with notable increases in early apoptosis (16.53%) and late apoptosis (29.57%), along with a slight increase in the population of necrotic cells (5.95%). Furthermore, compound 14 showed a significant drop in MCF-7 cells' ability to migrate and heal wounds. Additionally, compound 14 promoted apoptosis by boosting BAX (6-fold) while lowering Bcl-2 (6.2-fold). The binding affinities of the synthesized candidates to their target (VEGFR-2) were confirmed by computational investigations, including molecular docking, principal component analysis of trajectories (PCAT), and molecular dynamics (MD) simulations. Additionally, compound 14's stability and reactivity were investigated using density functional theory (DFT). These thorough results highlight compound 14's potential as a lead contender for additional research in the creation of anticancer drugs that target VEGFR-2. This work establishes a foundation for promising thiadiazole derivatives for future therapeutic developments in anticancer- and angiogenesis-related scientific fields.
1. Introduction
In 2021, the International Agency for Research on Cancer (IARC), a division of the World Health Organization (WHO), reported that there were 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 292
292![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 789 new cases of cancer worldwide in 2020, resulting in 9
789 new cases of cancer worldwide in 2020, resulting in 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958
958![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 133 fatalities.1 These numbers underscore how serious and widespread cancer is, underlining the crucial need for ongoing, thorough scientific exploration to find new ways to treat it. This effort is vital for advancing our understanding and developing more effective methods in our fight against this tough disease.
133 fatalities.1 These numbers underscore how serious and widespread cancer is, underlining the crucial need for ongoing, thorough scientific exploration to find new ways to treat it. This effort is vital for advancing our understanding and developing more effective methods in our fight against this tough disease.
Vascular endothelial growth factor receptor 2 (VEGFR-2) is a transmembrane receptor primarily found on the surface of endothelial cells, which form the inner lining of blood vessels. In the context of cancer, VEGFR-2 is of particular interest because it is overexpressed in many types of tumor.2,3 This overexpression leads to increased signaling through the VEGF pathway, which promotes the growth of new blood vessels around the tumor, a phenomenon known as angiogenesis,4 which is a crucial process for tumor survival, growth, and metastasis.5,6 Furthermore, recent research has observed a correlation between heightened expression of VEGFR-2 and heightened resistance to cancer drugs, amplified angiogenesis,7 and diminished apoptosis.8 Consequently, targeting VEGFR-2 has emerged as a promising therapeutic strategy.
VEGFR-2 inhibitors are a class of drugs designed to block the activity of this receptor. The inhibition of VEGFR-2 will disrupt the angiogenic process and starve the tumor of vital resources, impeding its ability to grow and spread.9 Unfortunately, inhibitors targeting VEGFR-2 have been linked to severe adverse effects, such as elevated blood pressure,10 proteinuria,11 instances of bleeding,12 germline polymorphisms,13 and reversible posterior leukoencephalopathy syndrome.10,14 These adverse effects underscore the pressing need for research into new safer alternatives. The development of novel VEGFR-2 inhibitors holds the potential to enhance treatment effectiveness and overall well-being for patients, thereby advancing the quality of cancer care.
Computational chemistry revolutionized anticancer research by enabling the precise design of targeted inhibitors.15 This technology employs computational power to design molecules tailored to interact specifically with specific receptors in the cancer cells.16,17 It also predicts the stability of these designed molecules, streamlining drug development and the optimization of lead compounds.18 Additionally, it anticipates the binding affinity of these molecules with the targeted receptors, expediting screening and enhancing efficacy against cancer cells.19 Furthermore, this approach shows promise in minimizing side effects associated with traditional chemotherapy through the precise targeting of cancer-specific pathways.20,21 This selectivity could revolutionize cancer treatment, providing therapies that are not only more potent against the disease but also safer for patients.22,23
Computational chemistry encompasses several approaches, such as design,24 docking,25 and prediction of ADMET (absorption, distribution, metabolism, excretion, and toxicity),26 as well as DFT (density functional theory).27 These tools have been utilized to identify compounds that show potential for effective anticancer activity in numerous research studies.28–31 Our laboratory has employed computational chemistry to present new VEGFR-2 inhibitors, including quinolones,32 isatins,33 theobromines,34–38 benzoxazoles,39,40 nicotinamides,41,42 thiazolidines,43 pyridines,44 naphthalenes,45 and indoles.46 This manuscript introduces novel apoptotic thiadiazole analogues, which were synthesized for the first time and exhibited promising anticancer properties in silico and in vitro.
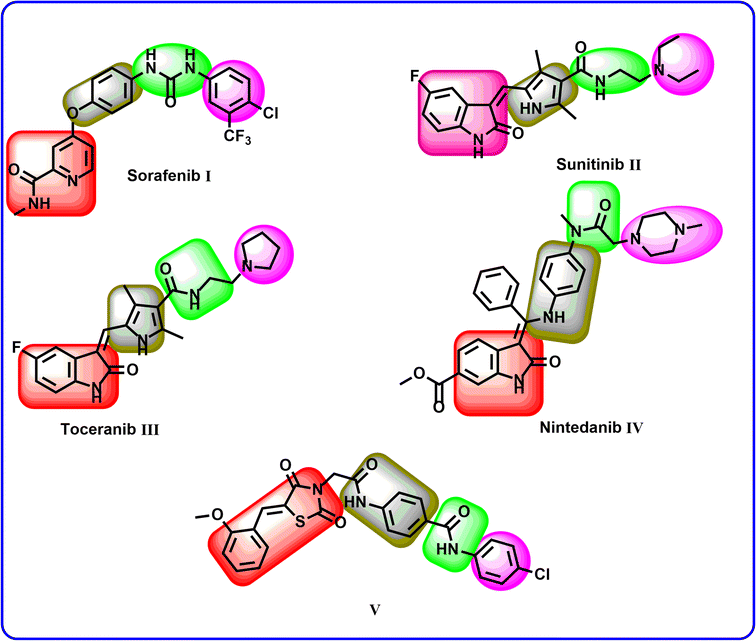
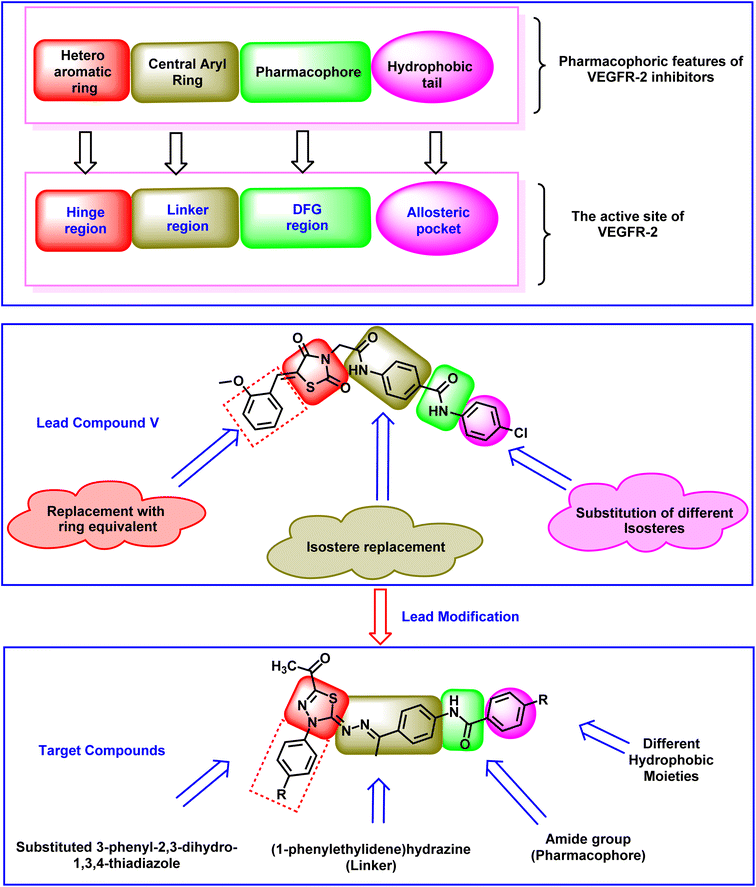
2. Rationale
The clinically used VEGFR-2 inhibitors and those under investigation are reported to have four pharmacophoric features for good binding with their biological target. The reported features are a hetero-aromatic head, a linker group, a pharmacophore group, and a hydrophobic tail47–49 (Fig. 1). The different regions of the active site are the hinge, the gatekeeper, the DFG motif, and the allosteric region. The pharmacophore group is the most important one. It should have at least one hydrogen bond (H-bond) donor and one H-bond acceptor atom to form essential hydrogen bonds with Asp1044 and Glu883 in the DFG motif region. Drug discovery efforts have provided many FDA-approved VEGFR-2 inhibitors, such as sorafenib I,50 sunitinib II,51 toceranib III,52 and nintedanib IV.53 As shown in Fig. 1, the reported VEGFR-2 inhibitors share the same pharmacophoric features.Recently, our team introduced compound V (ref. 54) as a potent VEGFR-2 inhibitor. This compound is a (Z)-5-benzylidenethiazolidine-2,4-dione derivative and it exhibited a high ability to inhibit VEGFR-2 (IC50 = 0.081 μM) and stop the growth of three different cancer cell types (HT-29, A-549, and HCT-116). In addition, compound V exhibited a remarkable apoptotic effect in HT-29 cancer cells. Furthermore, it increased the levels of BAX, caspase-8, and caspase-9 and decreased the level of Bcl-2 in the same cells. Computational studies revealed the high potential of compound V to bind with its prospective target (VEGFR-2) with extended stability. The obtained results suggested compound V as a lead compound to guide future attempts to develop new drugs.
In the current work, compound V was used as a lead compound for further modifications, hoping to obtain more efficient VEGFR-2 inhibitors. First, the (Z)-5-(2-methoxy benzylidene)thiazolidine-2,4-dione moiety (hetero cyclic head) of compound V was replaced by substituted 3-phenyl-2,3-dihydro-1,3,4-thiadiazole moieties. In this modification, the five-membered ring (thiazolidine-2,4-dione) was replaced by an equivalent five-membered ring (2,3-dihydro-1,3,4-thiadiazole). In addition, the attached phenyl ring was unchanged while the one-carbon bridge was removed. The second modification involved the replacement of the N-phenylacetamide (linker) moiety with (1-phenylethylidene)hydrazine. In this modification, the length of the linker moiety was not changed. Regarding the pharmacophore (amide) moiety of the lead compound, it was kept as it is to match the pharmacophore moiety of sunitinib II,51 toceranib III,52 and nintedanib IV.53 Finally, the 4-chlorophenyl (hydrophobic tail) moiety of the lead compound was replaced by methyl and different substituted phenyl rings (Fig. 2).
3. Results and discussion
3.1. Chemistry
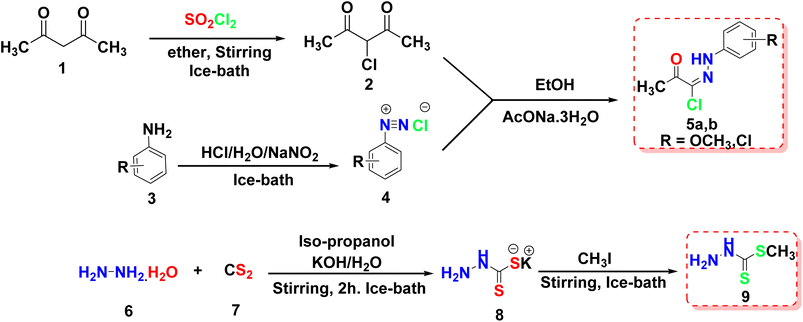
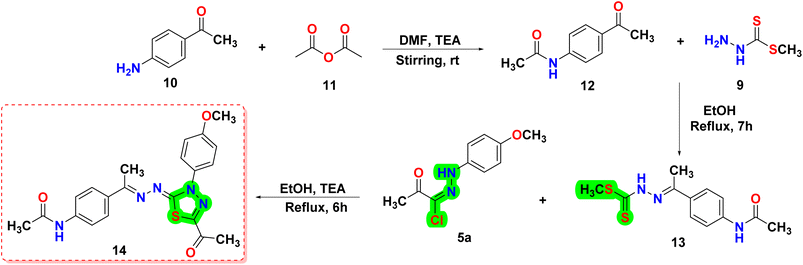
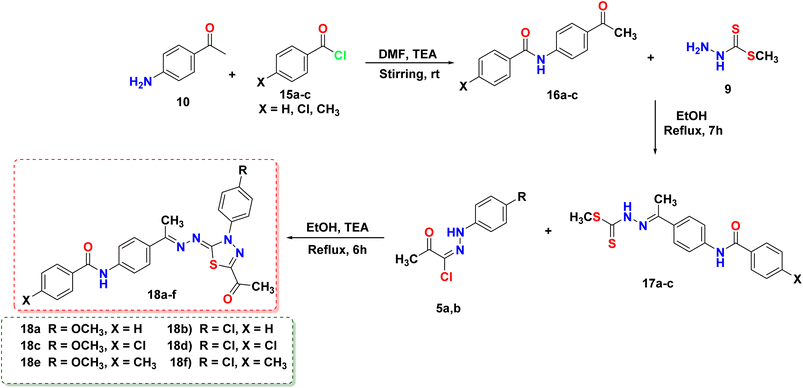
This article focuses on the synthesis of a series of innovative thiadiazole derivatives. We obtained the novel targeted compounds through the synthetic pathways illustrated in Schemes 1–3. Briefly, precursor 2 (ref. 55) was synthesized by chlorination of acetylacetone with sulfuryl chloride in anhydrous diethyl ether in an ice/NaCl bath with stirring. In the next step, compound 2 was coupled with appropriate benzene diazonium chloride 4 [prepared from substituted aniline derivatives 3a, 3b and a solution of sodium nitrite in the presence of diluted hydrochloric acid in an ice bath] to afford corresponding hydrazonoyl chlorides 5a and 5b.56 The key intermediate 9 was obtained by first reacting hydrazine hydrate with carbon disulfide in isopropyl alcohol and potassium hydroxide, leading to the formation of intermediate 8. This intermediate was then subjected to methylation using iodomethane, resulting in the formation of reagent 9,57 as presented in Scheme 1.As shown in Scheme 2, compound 12 (ref. 58) was prepared via acetylation of 4-aminoacetophenone 10 with acetic anhydride under stirring at ambient room temperature. Then, compound 12 was heated under reflux with methyl hydrazinecarbodithioate 9 in absolute ethanol to provide methyl (E)-2-(1-(4-acetamidophenyl)ethylidene)hydrazine-1-carbodithioate 13. Treatment of compound 13 with a hydrazonoyl chloride derivative 5a in ethanol in the presence of triethylamine as a catalyst under refluxing temperature afforded the target thiadiazole derivative 14.
In a similar vein, the reaction of 4-aminoacetophenone 10 with benzoyl chloride derivatives 15a–c was carried out in the presence of triethylamine as a catalyst and DMF as a solvent. This reaction produced the derivatives 16a and 16b, which then reacted with hydrazinecarbodithioate 9 in boiling ethanol to form compounds 17a–c. Afterward, compounds 17a–c were allowed to react with hydrazonoyl chloride derivatives 5a or 5b in the presence of ethanol and TEA under heating, resulting in the formation of the respective 1,3,4-thiadiazole derivatives (18a–f) in good purity/yield, as demonstrated in Scheme 3.
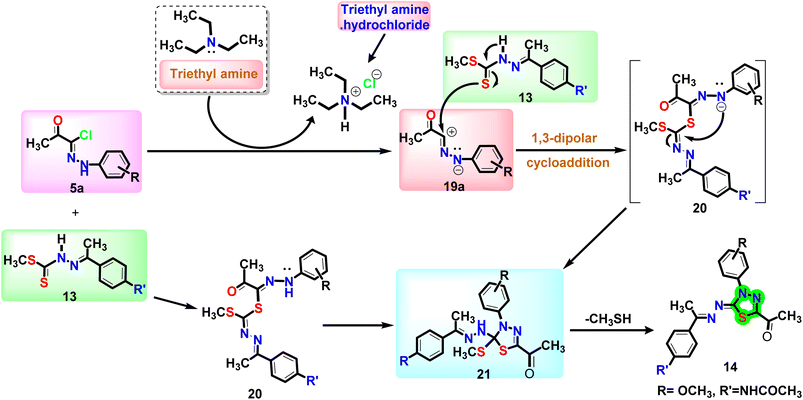
The mechanism outlined in Fig. 3 seems likely to be responsible for the formation of compound 14, via the reaction of 13 with 5a. As soon as thiohydrazonate 20 is formed, it undergoes intermolecular cyclization to form intermediate 21, or through 1,3-dipolar cycloaddition of nitrileimine 19a or 19b [prepared in situ from 5a and 5b, respectively, with triethylamine] to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S double bond of 13. During the elimination of the alkanethiol (CH3SH↥), compound 21 was converted to compound 14. In a similar way, combining 17a–c with 5a and 5b resulted in the formation of 1,3,4-thiadiazoles 18a–f with satisfactory yields (Fig. 1).
S double bond of 13. During the elimination of the alkanethiol (CH3SH↥), compound 21 was converted to compound 14. In a similar way, combining 17a–c with 5a and 5b resulted in the formation of 1,3,4-thiadiazoles 18a–f with satisfactory yields (Fig. 1).
3.2. Biological evaluation
| Compound | In vitro cytotoxicity IC50a (μM) | |
|---|---|---|
| MCF-7 | HepG2 | |
| a Data are presented as mean of the IC50 values from three different experiments. | ||
| 14 | 0.04 ± 0.009 | 0.18 ± 0.006 |
| 18a | 0.16 ± 0.017 | 0.80 ± 0.012 |
| 18b | 0.22 ± 0.098 | 0.20 ± 0.011 |
| 18c | 0.19 ± 0.014 | 0.16 ± 0.032 |
| 18d | 5.00 ± 0.192 | 6.17 ± 0.019 |
| 18e | 0.19 ± 0.045 | 0.21 ± 0.045 |
| 18f | 0.44 ± 0.065 | 0.18 ± 0.018 |
| Sorafenib | 0.15 ± 0.013 | 0.14 ± 0.052 |
In the context of the cytotoxicity of the benzamide counterparts 18a, 18c, and 18e, it was found that unsubstituted benzamide analogue 18a showed more significant cytotoxicity compared to the 4-chlorobenzamide 18c or 4-methybenzamide 18e ones.
Indeed, the incorporation of the 3-(4-chlorophenyl)-2,3-dihydro-1,3,4-thiadiazole moiety in 18b, 18d, and 10g decreased the anti-proliferative action. Additionally, appending of unsubstituted benzamide 18b is the most beneficial pattern within these counterparts 18b, 18d, and 10f; replacement of the benzamide moiety with a 4-methybenzamide moiety (as in compound 18f) led to a slightly decrease in the activity, while the incorporation of the 4-chlorobenzamide moiety (as in compound 18d) led to a marked decrease in the activity.
3.2.4.1. Cell cycle analysis. Compound 14 showed potent cytotoxic and VEGFR-2 inhibition activities and was selected to explore its activity on the cell cycle distribution of MCF-7 cells. The MCF-7 cells were exposed to compound 14 (0.041 μM, equal to its anti-proliferative IC50) for 72 hours, and cell cycle progression was monitored by flow cytometry; the results are reported in Table 3.
| Sample | Cell cycle distribution (%) | ||
|---|---|---|---|
| G0–G1 | S | G2/M | |
| Compound 14-treated MCF-7 cells | 68.43 | 22.27 | 9.3 |
| MCF-7 cells | 62.37 | 23.91 | 13.72 |
The obtained data revealed that compound 14 decreased the distribution in the S phase (22.27%) and the G2/M phase (9.3%) compared with the control (23.91 and 13.72%, respectively). In addition, the percentage of cell population increased at the G0–G1 phase by 1.09-fold more than the control. These findings revealed that compound 14 induced arrest of the cell cycle of the MCF-7 cells at the G0–G1 phase (Fig. 4).
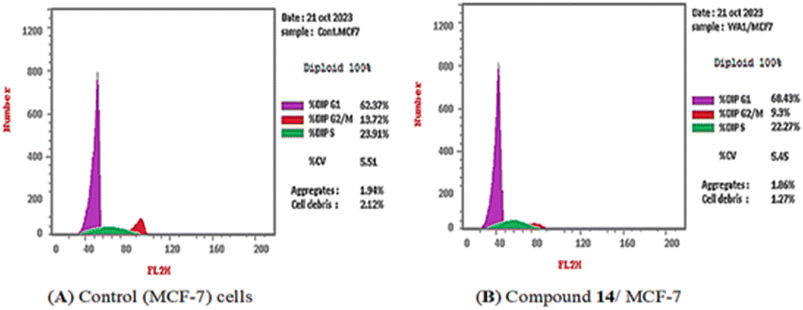 | ||
| Fig. 4 Cell cycle distribution of MCF-7 cells as determined by flow cytometry. (A) Control and (B) compound 14. | ||
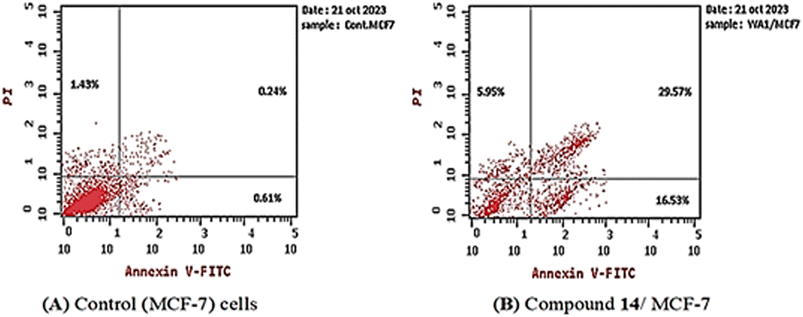
3.2.4.2. Apoptosis assay. The MCF-7 cells were treated with compound 14 (0.04 μM for 72 h), and the apoptotic effect was assessed using the annexin V-FITC/PI assay. The results (Table 4) demonstrated that, compound 14 enhanced total apoptosis by 22-fold compared to the control (52.05% and 2.28%, respectively). Additionally, 14 increased the percentage of early apoptosis compared with the control MCF-7 cells (16.53% and 0.61%, respectively). Moreover, it increased the percentage of late apoptotic cells by 132-fold more than the control cells (from 0.24% to 29.57%). In addition, compound 14 enhanced the necrosis percentage four-fold more than the control. These details suggest that compound 14 can induce the apoptotic mechanism of programed cell death in the MCF-7 cell line (Fig. 5).
| Sample | Total apoptosis (%) | Early apoptosis (%) | Late apoptosis (%) | Necrosis (%) |
|---|---|---|---|---|
| Compound 14-treated MCF-7 cells | 52.05 | 16.53 | 29.57 | 5.95 |
| MCF-7 cells | 2.28 | 0.61 | 0.24 | 1.43 |
3.2.4.3. BAX and BCl-2 analysis. This investigation examined the effects of compound 14 at a concentration of 0.04 μM on the expression levels of well-known apoptotic markers, BAX and Bcl-2, in MCF-7 cells. The findings of this examination showed that compound 14 significantly boosted the level of the pro-apoptotic protein (BAX) by six-fold when compared to the control MCF-7 cells. Furthermore, compound 14 dramatically lowered levels of the anti-apoptotic protein Bcl-2 by 6.2-fold compared to the control MCF-7, as presented in Table 5.
| Sample | RT-PCR fold change | |
|---|---|---|
| Bcl2 | BAX | |
| Compound 14 | 0.159 | 6 |
| Control MCF-7 cells | 1 | 1 |
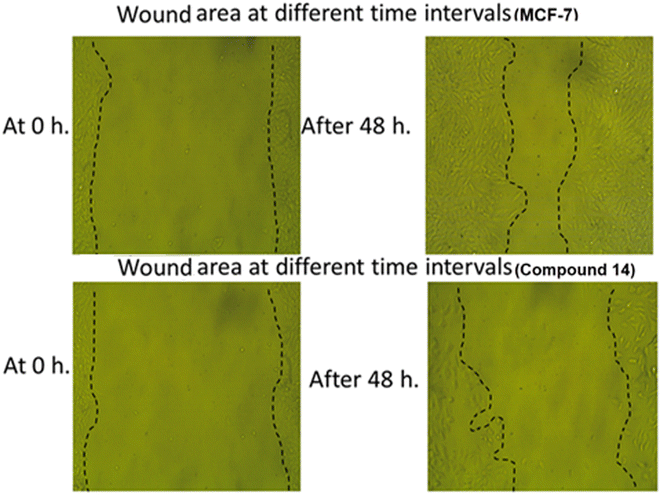
3.2.4.4. Cell migration and wound healing assay. Cell migration is a pivotal step in cancer development and progression. Assessment of in vitro migration processes in cancer cell lines can be a crucial tool for studying new potential therapeutic anticancer drugs in order to understand cellular mechanistic pathways. The impact of compound 14 on the cell migration of MCF-7 cells was evaluated by using the in vitro scratch wound healing assay protocol.60,61 This protocol involves the generation of a scratch on a monolayer of MCF-7 cancer cells, measuring the initial diameter, and checking the scratch's closure for both treated and untreated cells after 0 and 48 hours.
As shown in Table 6 and Fig. 6, the scratch width of the control MCF-7 cells was reduced by about 67.55% after 48 hours and appeared to be approximately closed, while the scratch width of the MCF-7 cells treated with compound 14 was only reduced by about 38.76% after 48 hours. This finding showed the ability of compound 14 to inhibit the healing and migration of the treated cells.
| Item | At 0 h | At 24 h | At 48 h | RM (μm) | Wound closure (%) | Area difference (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Area (μm2) | Width (μm) | Area (μm2) | Width (μm) | Area (μm2) | Width (μm) | ||||
| Control MCF-7 cells | 876.333 | 875.422 | 673.000 | 672.059 | 284.333 | 283.351 | 12.335 | 67.554 | 592.000 |
| MCF-7 cells treated with 14 | 933.000 | 932.006 | 836.333 | 835.443 | 571.333 | 570.472 | 7.532 | 38.764 | 361.667 |
3.3. Computational studies
| IP | EA | μ (eV) | χ (eV) | η (eV) | σ (eV) | ω (eV) | Dm (Debye) | TE (eV) | ΔNmax | ΔE (eV) |
|---|---|---|---|---|---|---|---|---|---|---|
| 5.653 | 2.472 | −4.063 | 4.063 | 1.591 | 0.629 | 13.126 | 8.516 | −46546.1 | 2.554 | −13.126 |
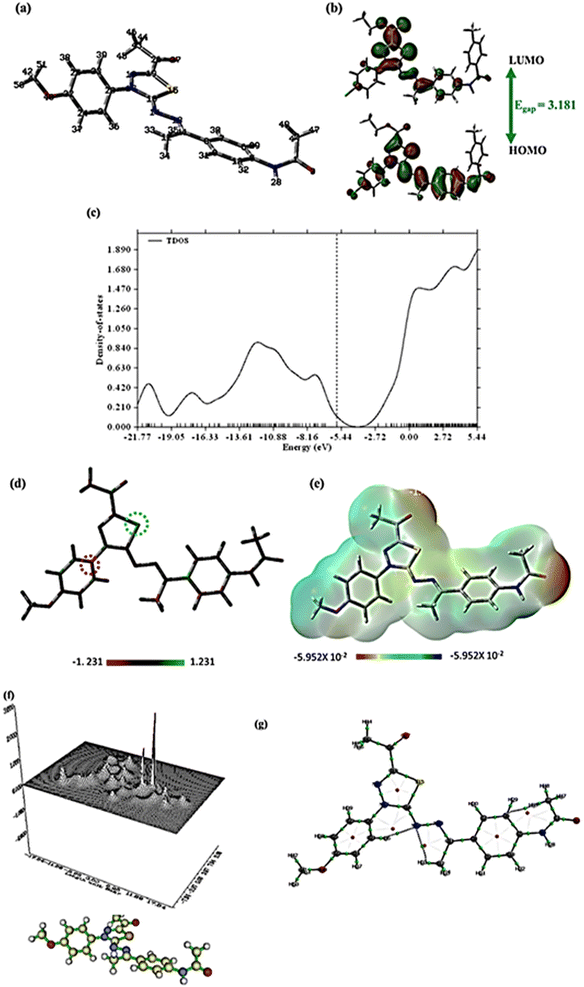
The title molecule's non-planarity may be seen in the optimized geometric structure, which has 222 electrons and 51 atoms. The two terminal benzene rings are slightly angled towards the average plane of the molecule, according to geometry optimization, and the optimized geometrical parameter TE, or total ground energy, is −46546.1 eV. The same level of theory was used to estimate the electronic dipole moment (Dm), which is large (8.516 Debye), indicating considerable charge separation. The HOMO–LUMO gap (Egap), a crucial indicator of kinetic stability, was calculated after the quantum evaluation of HOMO and LUMO energies, as shown in Fig. 7b. Chemical reactivity and the HOMO/LUMO border orbitals have a strong relationship as low reactivity is indicated by high molecular orbital gap values (Egap). The small Egap indicates that the produced drug has high chemical reactivity because of charge transfer interactions. Subsequently, the Gauss Sum 3.0 program calculated the gas phase's density of states (TDOS). The TDOS diagram for compound 14 is shown in Fig. 7c.
Koopman's' hypothesis was used to study the global reactivity descriptors; the values in Table 7 of the ESI† are used. Due to its high HOMO energy and low HOMO–LUMO Egap, which indicate significant inhibitory reactivity, the molecule under examination exhibits a high softness (σ) and a low hardness (μ). A further property indicated in Table 7 that is used to assess the inhibitory reactivity and quantify a compound's capacity to absorb electrons from the target is the electrophilicity index (ω). The results showed that the title compound has significant inhibitory reactivity.62
Mulliken atomic charge distribution is a common technique for calculating inhibitory sites for adsorption. The investigated compound has good ability to bond with the target surface using the donor–acceptor method, as shown in Fig. 7d. The highest positive charge was found at C15 while C20 acquired the highest negative atomic charge.
For predicting drug–receptor connections, surface molecular electrostatic potential (ESP) can be mapped using DFT. The ESP increases recognition of electrophilic interactions, substituent impacts, inhibitory reactivity, and both inter- and intra-bonding forces after being connected to the active sites of the prepared compound. Red (electron-rich), yellow (slightly electron-rich), blue (electron deficient), and green (neutral zone) are the colors in which the value of ESP increases. Fig. 7e shows that the blue zone of the drug is localized over the hydrogen atoms, whereas the red and yellow portions are generated by the lone pair of oxygen atoms. The ring system is encircled by neutral green. As a result, the drug's electrophilic active sites are anticipated to be located around the hydrogen atoms, while the negative electrostatic potential is encapsulated over the oxygen and nitrogen atoms, making this the preferred location for electrophilic assault by the target sites.63
Microelectronic and structural investigations might not be sufficient to completely comprehend a drug's intermolecular interactions. Consequently, charge density parameters were evaluated using the Multiwfn and AIMALL programs. These programs enable the computation of topological features such as electronic charge density (ρ), total electron energy density (H(r)), Laplacian of electronic charge density (∇2ρ), and the two components of H(r); kinetic electron energy density (G(r) and potential electron energy density (V(r)) at the bond critical points (BCP). These computations are possible via the Quantum Theory of Atoms in Molecules (QTAIM).
Table S1† makes it clear that the values of ∇2ρ and H(r) are negative, suggesting that all BCP connections are covalent except for N14–H33 and N14–H36, which have weak electrostatic attractions. The relief map in Fig. 7f depicts the electron density (ρ) in the XY plane, whereas Fig. 7g shows all bond paths within the compound.
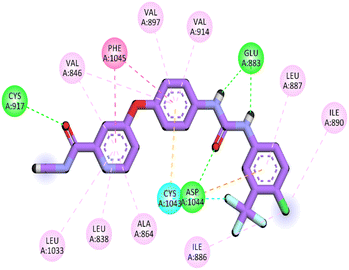
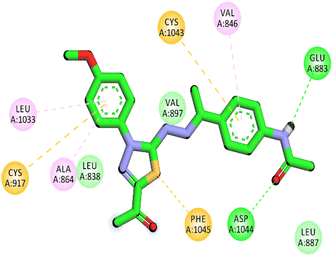
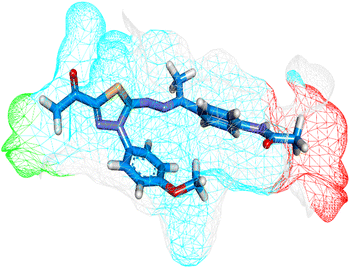
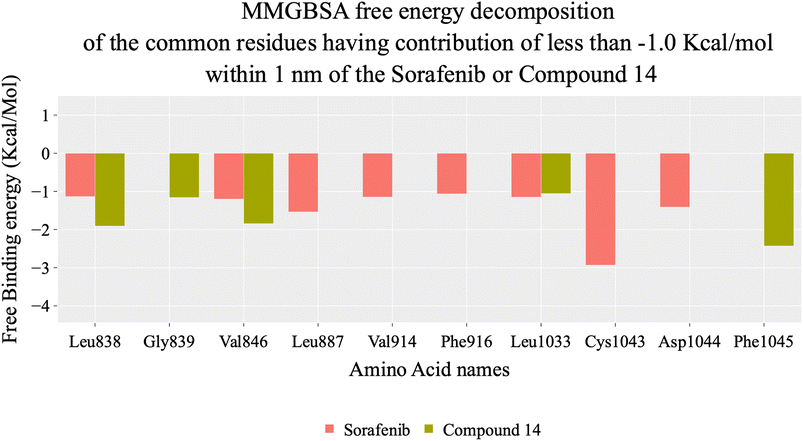
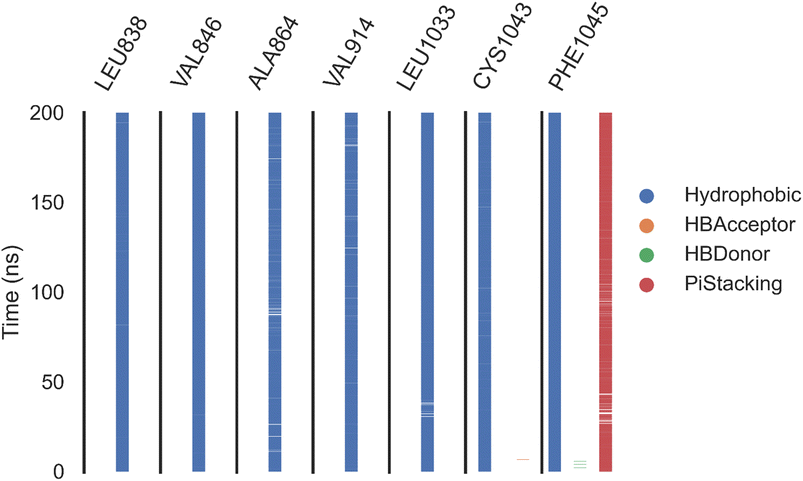
Herein, the examined molecule (14) kept the crucial hydrogen bonds to Glu883 and Asp1044 through its amide group verifying its positioning with the identical orientation inside the VEGFR-2 active pocket (Table 8). In addition, it formed two electrostatic interactions with Cys917 and Phe1045 in the hinge region through its 5-acetyl-3-(4-methoxyphenyl)-1,3,4-thiadiazole moiety. Furthermore, additional electrostatic and pi–pi interactions were observed in the linker area between the central phenyl moiety and Cys1043 and Val846, respectively. Thus, the results discussed above strongly support candidate 14's extremely promising inhibitory capability towards the VEGFR-2 target receptor. This may be why it produced better biological outcomes as a prospective VEGFR-2 antagonist when compared to the reference drug sorafenib.
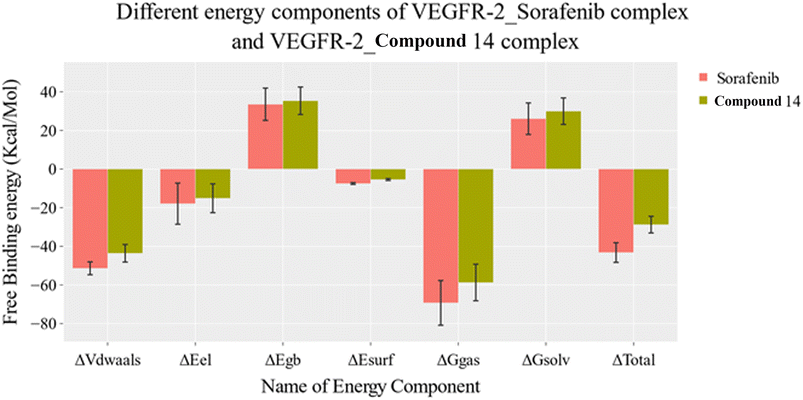 | ||
| Fig. 9 MM-GBSA analysis as well as the detailed values for both systems. Bars represent the standard deviations. | ||
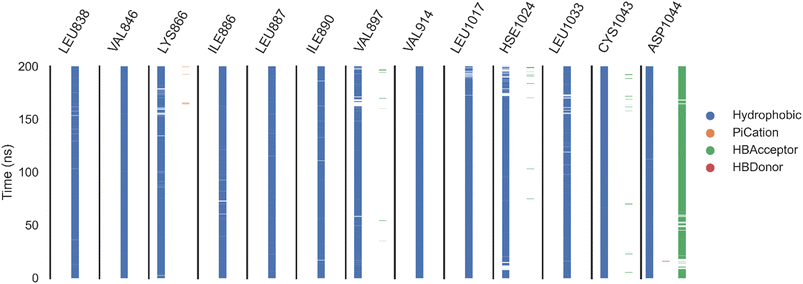 | ||
| Fig. 11 The amino acids, the types of interactions with sorafenib, and their occurrence during the whole simulation time using the ProLIF python library. | ||
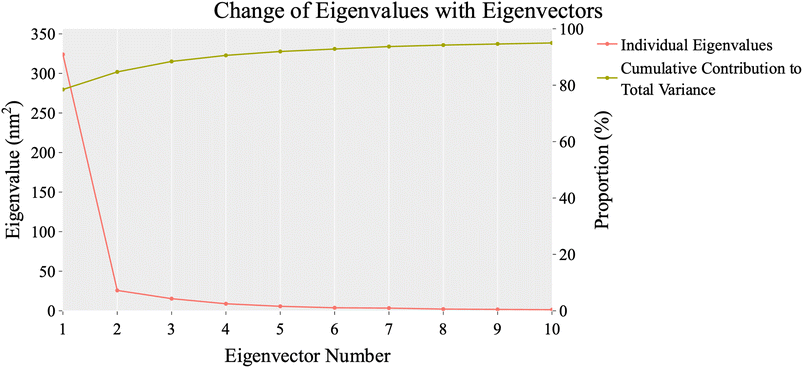 | ||
| Fig. 13 The change in eigenvalues with increasing eigenvectors (blue line). In addition, the cumulative variance retained in the eigenvectors is shown (red line). | ||
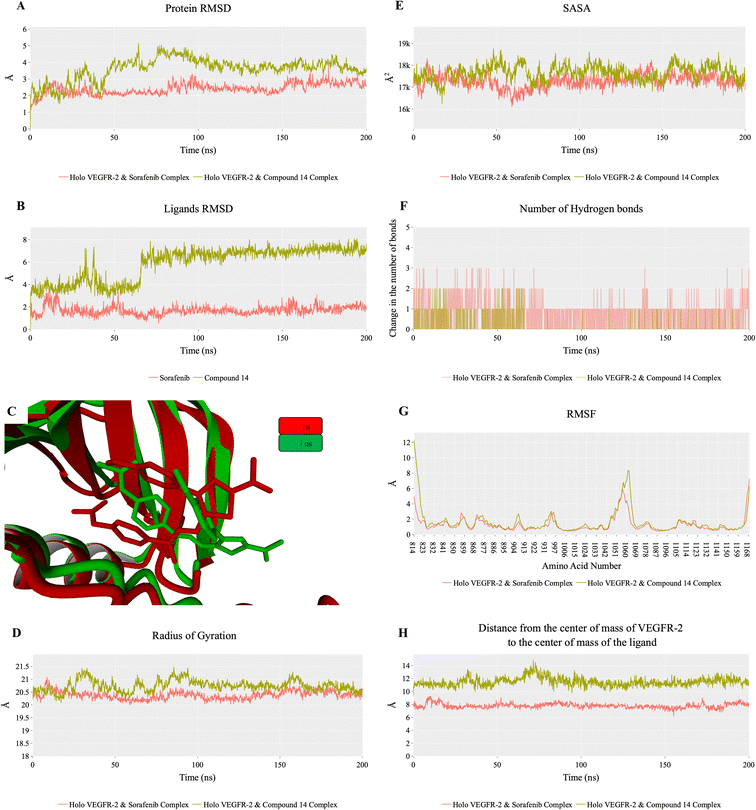
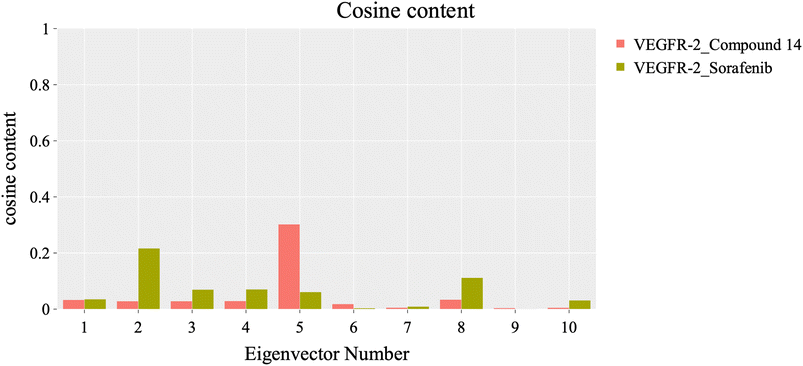
To evaluate the degree of randomness in the behavior of the first ten eigenvectors, we calculated the cosine content for both systems (Fig. 15). Our analysis revealed that, with the exception of the second principal component of the VEGFR-2–sorafenib system (0.21) and the fifth PC value of the VEGFR-2–compound 14 system, the cosine content of the first 10 eigenvectors remained below 0.2 in both systems. Furthermore, the root-mean-square inner product (RMSIP) indicated a limited overlap between the two subspaces, especially for the first three eigenvectors, which had an overlap of approximately 16%. Additionally, the RMSIP analysis showed that the C matrices of the two systems were only 27.9% similar, indicating distinct sampling characteristics for each system.
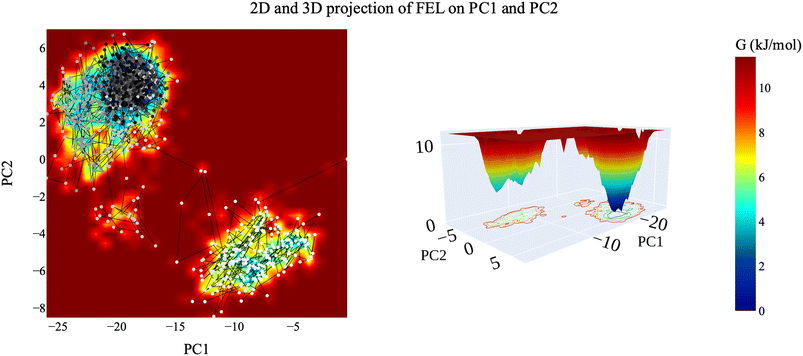 | ||
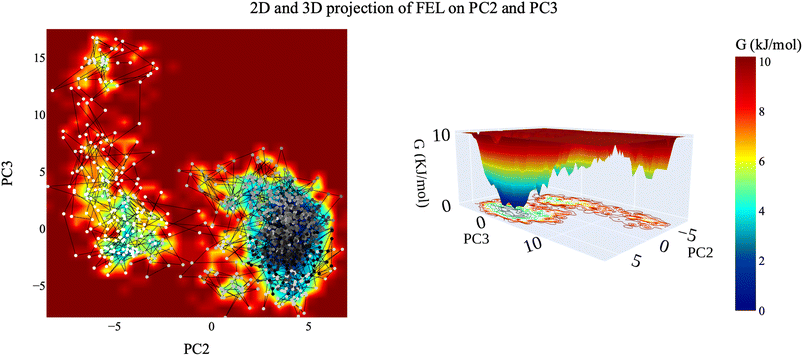
| Fig. 16 The 2D and 3D projection of the VEGFR-2–compound 14 trajectory's FEL on the first two eigenvectors. | ||
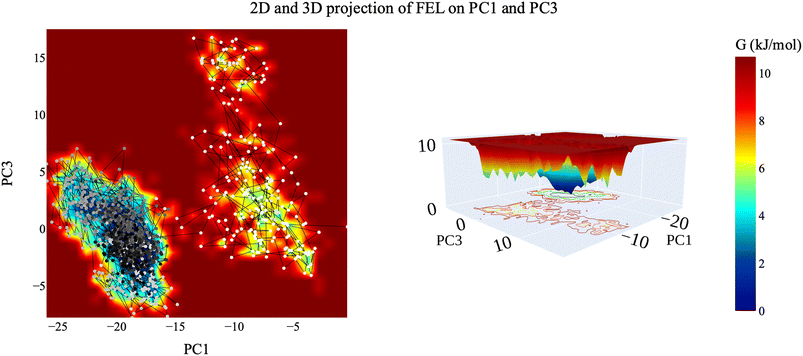 | ||
| Fig. 17 2D and 3D projection of the VEGFR-2–compound 14 trajectory's FEL on the first and third eigenvectors. | ||
| Compound | BBB level | Solubility level | Absorption level | CYP2D6 prediction | PPB prediction |
|---|---|---|---|---|---|
| 14 | Low | Low | Good | Non-inhibitors | Less than 90% |
| 18a | Very low | Very low | Moderate | More than 90% | |
| 18b | |||||
| 18c | |||||
| 18d | Low | Poor | |||
| 18e | Moderate | ||||
| 18f | Very low | Poor | |||
| Sorafenib | Good |
| Compound | Carcinogenic potency TD50 (mouse) a | Ames prediction | FDA rodent carcinogenicity (mouse–female) | Rat oral LD50b | Rat chronic LOAELb | Skin irritancy | Ocular irritancy |
|---|---|---|---|---|---|---|---|
| a Unit: mg kg−1 day−1.b Unit: g kg−1. | |||||||
| 14 | 25.1941 | Non-mutagen | Non-carcinogen | 0.307791 | 0.0340816 | Non-irritant | Mild |
| 18a | 19.1051 | 0.211142 | 0.0407246 | ||||
| 18b | 10.1397 | 0.342938 | 0.0653748 | ||||
| 18c | 5.86494 | 0.147569 | 0.012965 | ||||
| 18d | 3.51292 | 0.292276 | 0.0298888 | ||||
| 18e | 10.9809 | 0.357591 | 0.0180164 | ||||
| 18f | 5.82645 | 0.580656 | 0.0289143 | ||||
| Sorafenib | 19.2359 | Single-carcinogen | 0.822583 | 0.00482816 | |||
4. Conclusion
In conclusion, in this study we successfully synthesized and comprehensively evaluated a series of thiadiazole derivatives designed to mimic VEGFR-2 inhibitors. Among the synthesized compounds, compound 14 emerged as a highly promising candidate, displaying exceptional inhibitory properties against cancer cell lines MCF-7 and HepG2, as well as a remarkable affinity for VEGFR-2. Furthermore, compound 14 exhibited a unique ability to induce cell cycle arrest and apoptosis in MCF-7 cells, while also inhibiting cell migration and wound healing. Furthermore, compound 14 increased apoptosis by 6-fold while decreasing Bcl-2 levels (6.2-fold). The computational investigations confirmed the binding affinities, stability, and reactivity of compound 14. Collectively, these findings strongly support compound 14 as a lead contender for further research in the development of anticancer drugs targeting VEGFR-2. This work lays a solid foundation for future advancements in the fields of anticancer- and angiogenesis-related research, driven by the potential of these thiadiazole derivatives.5. Experimental
5.1. Chemistry
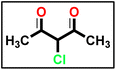 The synthesis of α-chloro-2,4-pentanedione 2 was carried out in accordance with the methodology described in ref. 55 with minor adjustments. A 100 mL Erlenmeyer flask equipped with a magnetic stirring bar was filled with acetylacetone (1 eq.) in anhydrous diethyl ether and chilled in an ice/NaCl bath to 0 °C. The sulfuryl chloride (1 eq.) was added gradually to the conical flask using the dropping funnel such that the internal temperature was kept below 5 °C. In the next step, the reaction mixture was stirred for 2 hours, the solvent was extracted under reduced pressure, and the mixture was washed with sodium bicarbonate and dried over magnesium sulfate, yielding a crude product suitable for further processing without further purification.
The synthesis of α-chloro-2,4-pentanedione 2 was carried out in accordance with the methodology described in ref. 55 with minor adjustments. A 100 mL Erlenmeyer flask equipped with a magnetic stirring bar was filled with acetylacetone (1 eq.) in anhydrous diethyl ether and chilled in an ice/NaCl bath to 0 °C. The sulfuryl chloride (1 eq.) was added gradually to the conical flask using the dropping funnel such that the internal temperature was kept below 5 °C. In the next step, the reaction mixture was stirred for 2 hours, the solvent was extracted under reduced pressure, and the mixture was washed with sodium bicarbonate and dried over magnesium sulfate, yielding a crude product suitable for further processing without further purification.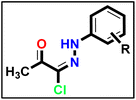 A solution of appropriate aromatic amine (1 eq.), namely (4-methoxyaniline and/or 4-chloroaniline) in concentrated hydrochloric acid (12 M) plus water (200 mL) was stirred in a 500 mL round-bottomed flask for 30 min. Then, the reaction flask was cooled by immersion in ice and salt, until the temperature of the acid reached 0 °C. A solution of sodium nitrite (1.1 eq.) in water (10 mL) was gradually added to the reaction mixture so that the reaction temperature was maintained below 5 °C. This required about 30 minutes under stirring conditions. A separate solution containing sodium acetate (1 eq.) and α-chloro-2,4-pentanedione (2) (1 eq.) in commercially available ethyl alcohol was added in portions. The mixture was then warmed to 25 °C and stirred continuously for 16 h, and the formed precipitate was separated by filtration, rinsed with water, and subsequently dried, resulting in the formation of 5a and 5b.
A solution of appropriate aromatic amine (1 eq.), namely (4-methoxyaniline and/or 4-chloroaniline) in concentrated hydrochloric acid (12 M) plus water (200 mL) was stirred in a 500 mL round-bottomed flask for 30 min. Then, the reaction flask was cooled by immersion in ice and salt, until the temperature of the acid reached 0 °C. A solution of sodium nitrite (1.1 eq.) in water (10 mL) was gradually added to the reaction mixture so that the reaction temperature was maintained below 5 °C. This required about 30 minutes under stirring conditions. A separate solution containing sodium acetate (1 eq.) and α-chloro-2,4-pentanedione (2) (1 eq.) in commercially available ethyl alcohol was added in portions. The mixture was then warmed to 25 °C and stirred continuously for 16 h, and the formed precipitate was separated by filtration, rinsed with water, and subsequently dried, resulting in the formation of 5a and 5b.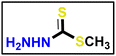 Methyl hydrazinecarbodithioate 9 was synthesized using hydrazine hydrate, carbon disulfide, and methyl iodide, following the same methodology described in a previous publication.57
Methyl hydrazinecarbodithioate 9 was synthesized using hydrazine hydrate, carbon disulfide, and methyl iodide, following the same methodology described in a previous publication.57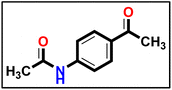 The compound was synthesized using a method described in the literature58 with a few minor adjustments. An equal amount of triethylamine (1 eq.) was added to a solution of 4-aminoacetophenone (1 eq.) and acetic anhydride (2 eq.) in dimethyl formamide (10 mL), as a reaction solvent. Afterward, the mixture was stirred for 1 hour at ambient temperature. The precipitate which formed was separated and collected on a suction funnel, washed with ice-cold, and dried and purified by recrystallization from ethanol to afford the desired compound (the product was white with yield of 90%).
The compound was synthesized using a method described in the literature58 with a few minor adjustments. An equal amount of triethylamine (1 eq.) was added to a solution of 4-aminoacetophenone (1 eq.) and acetic anhydride (2 eq.) in dimethyl formamide (10 mL), as a reaction solvent. Afterward, the mixture was stirred for 1 hour at ambient temperature. The precipitate which formed was separated and collected on a suction funnel, washed with ice-cold, and dried and purified by recrystallization from ethanol to afford the desired compound (the product was white with yield of 90%).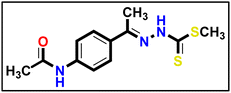 Compound 12 (1 eq.) was solubilized in absolute ethanol (10 mL) and 1 eq. of methyl hydrazinecarbodithioate 9 was added. The reaction mixture was heated under reflux for 7 h (TLC analysis). The obtained solid was filtered while hot, washed with hot ethanol, dried, and recrystallized from dioxane to produce the corresponding compound 13 in 80% yield.
Compound 12 (1 eq.) was solubilized in absolute ethanol (10 mL) and 1 eq. of methyl hydrazinecarbodithioate 9 was added. The reaction mixture was heated under reflux for 7 h (TLC analysis). The obtained solid was filtered while hot, washed with hot ethanol, dried, and recrystallized from dioxane to produce the corresponding compound 13 in 80% yield.Yellow powder (yield, 73%); m.p. = 255–257 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.35 (s, 1H, NH), 10.10 (s, 1H, NH), 7.85–7.73 (m, 2H, Ar–H), 7.66–7.53 (m, 2H, Ar–H), 2.47 (s, 3H, CH3), 2.32 (s, 3H, CH3), 2.03 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 199.44, 151.45, 141.00, 140.57, 131.58, 127.24, 127.08, 118.53, 24.12, 17.03, 14.43 for C12H15N3OS2 (281.39).
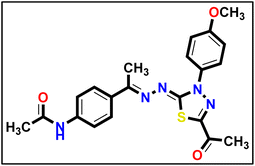 To prepare the derivative 14, applicable quantities of hydrazonoyl chloride 5a (1 eq.) and intermediate 13 (1 eq.) were mixed in absolute ethanol (20 mL). An equivalent amount of triethylamine (1 eq.) was added to the mixture, which was then heated under reflux for 6 hours. The resulting solid was filtered, washed with hot ethanol, and then purified with hexane to obtain the desired final product.
To prepare the derivative 14, applicable quantities of hydrazonoyl chloride 5a (1 eq.) and intermediate 13 (1 eq.) were mixed in absolute ethanol (20 mL). An equivalent amount of triethylamine (1 eq.) was added to the mixture, which was then heated under reflux for 6 hours. The resulting solid was filtered, washed with hot ethanol, and then purified with hexane to obtain the desired final product.Yellow powder (yield, 70%); m.p. = 283–285 °C. FT-IR (vmax, cm−1): 3387, 3305 (NH), 3084, 3007 (C–H aromatic), 2952, 2929 (C–H aliphatic), 1688, 1665 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 10.14 (s, 1H, NH), 7.96–7.88 (m, 2H, Ar–H), 7.82 (d, J = 8.5 Hz, 2H, Ar–H), 7.66 (d, J = 8.5 Hz, 2H, Ar–H), 7.17–7.09 (m, 2H, Ar–H), 3.84 (s, 3H, OCH3), 2.57 (s, 3H, CH3), 2.36 (s, 3H, CH3), 2.08 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.13, 169.02, 164.68, 160.01, 158.63, 150.52, 141.32, 132.27, 132.23, 127.52, 124.47, 119.01, 114.66, 55.96, 25.46, 24.56, 15.55; mass (m/z): 423 (M+, 5%), 85 (100%, base peak); anal. calcd. for C21H21N5O3S (423.49): C, 59.56; H, 5.00; N, 16.54; found: C, 59.84; H, 5.13; N, 16.78%.
O); 1H NMR (400 MHz, DMSO-d6) δ 10.14 (s, 1H, NH), 7.96–7.88 (m, 2H, Ar–H), 7.82 (d, J = 8.5 Hz, 2H, Ar–H), 7.66 (d, J = 8.5 Hz, 2H, Ar–H), 7.17–7.09 (m, 2H, Ar–H), 3.84 (s, 3H, OCH3), 2.57 (s, 3H, CH3), 2.36 (s, 3H, CH3), 2.08 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.13, 169.02, 164.68, 160.01, 158.63, 150.52, 141.32, 132.27, 132.23, 127.52, 124.47, 119.01, 114.66, 55.96, 25.46, 24.56, 15.55; mass (m/z): 423 (M+, 5%), 85 (100%, base peak); anal. calcd. for C21H21N5O3S (423.49): C, 59.56; H, 5.00; N, 16.54; found: C, 59.84; H, 5.13; N, 16.78%.
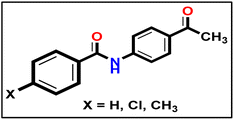 To synthesize intermediates 16a–c, we followed a process analogous to that used for 12. We combined derivative 10 (1 eq.) with diverse (un)substituted benzoyl chlorides 15a–c (1 eq.).
To synthesize intermediates 16a–c, we followed a process analogous to that used for 12. We combined derivative 10 (1 eq.) with diverse (un)substituted benzoyl chlorides 15a–c (1 eq.).5.1.9.1. Methyl (E)-2-(1-(4-benzamidophenyl)ethylidene)hydrazine-1-carbodithioate 17a.
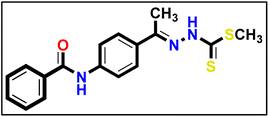 Yellow powder (yield, 80%); m.p. = 250–252 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H, NH), 10.41 (s, 1H, NH), 8.00–7.92 (m, 3H, Ar–H), 7.84 (m, 3H, Ar–H), 7.58 (m, 1H, Ar–H), 7.52 (d, J = 7.5 Hz, 2H, Ar–H), 2.49 (s, 3H, CH3), 2.35 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 199.51, 165.99, 151.43, 140.90, 132.18, 131.74, 129.30, 128.42, 127.80, 127.72, 127.10, 119.83, 119.42, 17.01, 14.45 for C17H17N3OS2 (343.46).
Yellow powder (yield, 80%); m.p. = 250–252 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H, NH), 10.41 (s, 1H, NH), 8.00–7.92 (m, 3H, Ar–H), 7.84 (m, 3H, Ar–H), 7.58 (m, 1H, Ar–H), 7.52 (d, J = 7.5 Hz, 2H, Ar–H), 2.49 (s, 3H, CH3), 2.35 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 199.51, 165.99, 151.43, 140.90, 132.18, 131.74, 129.30, 128.42, 127.80, 127.72, 127.10, 119.83, 119.42, 17.01, 14.45 for C17H17N3OS2 (343.46).
5.1.9.2. Methyl (E)-2-(1-(4-(4-chlorobenzamido)phenyl)ethylidene)hydrazine-1-carbodi thioate 17b.
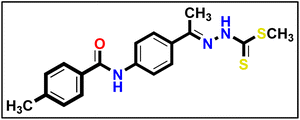 Yellowish white powder (yield, 85%); m.p. = 265–267 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H, NH), 10.46 (s, 1H, NH), 8.00–7.95 (m, 3H, Ar–H), 7.84 (t, J = 6.2 Hz, 3H, Ar–H), 7.59 (dd, J = 8.3, 4.5 Hz, 2H, Ar–H), 2.52 (s, 3H, CH3), 2.35 (s, 3H, CH3); for C17H16ClN3OS2 (377.91).
Yellowish white powder (yield, 85%); m.p. = 265–267 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H, NH), 10.46 (s, 1H, NH), 8.00–7.95 (m, 3H, Ar–H), 7.84 (t, J = 6.2 Hz, 3H, Ar–H), 7.59 (dd, J = 8.3, 4.5 Hz, 2H, Ar–H), 2.52 (s, 3H, CH3), 2.35 (s, 3H, CH3); for C17H16ClN3OS2 (377.91).
5.1.9.3. Methyl (E)-2-(1-(4-(4-methylbenzamido)phenyl)ethylidene)hydrazine-1-carbodi thioate 17c.
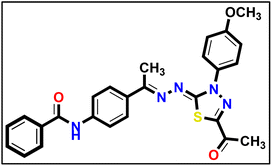 Orange powder (yield, 82%); m.p. = 270–272 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H), 10.31 (s, 1H), 7.93 (d, J = 5.9 Hz, 1H), 7.87–7.83 (m, 5H), 7.32 (d, J = 7.6 Hz, 2H), 2.52 (s, 3H), 2.36 (s, 3H), 2.35 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ 199.58, 165.54, 151.47, 141.85, 141.02, 129.33, 128.98, 127.81, 127.12, 119.84, 119.42, 21.08, 17.05, 14.48 for C18H19N3OS2 (357.49).
Orange powder (yield, 82%); m.p. = 270–272 °C. 1H NMR (500 MHz, DMSO-d6) δ 12.39 (s, 1H), 10.31 (s, 1H), 7.93 (d, J = 5.9 Hz, 1H), 7.87–7.83 (m, 5H), 7.32 (d, J = 7.6 Hz, 2H), 2.52 (s, 3H), 2.36 (s, 3H), 2.35 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ 199.58, 165.54, 151.47, 141.85, 141.02, 129.33, 128.98, 127.81, 127.12, 119.84, 119.42, 21.08, 17.05, 14.48 for C18H19N3OS2 (357.49).
5.1.10.1. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-methoxyphenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono)ethyl)phenyl) benzamide 18a.
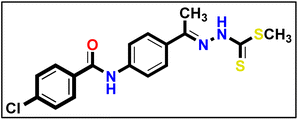 Canary yellow powder (yield, 78%); m.p. = 258–260 °C. FT-IR (vmax, cm−1): 3285 (NH), 3058, 3010 (C–H aromatic), 2923, 2834 (C–H aliphatic), 1686, 1654 (C
Canary yellow powder (yield, 78%); m.p. = 258–260 °C. FT-IR (vmax, cm−1): 3285 (NH), 3058, 3010 (C–H aromatic), 2923, 2834 (C–H aliphatic), 1686, 1654 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 10.46 (s, 1H, NH), 8.02–7.98 (m, 3H, Ar–H), 7.91 (m, 5H, Ar–H), 7.63–7.60 (m, 1H, Ar–H), 7.58–7.55 (m, 2H, Ar–H), 7.18–7.12 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.59 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.14, 166.16, 164.80, 160.04, 158.65, 150.56, 141.24, 135.25, 132.88, 132.23, 132.20, 128.91, 128.25, 128.22, 128.02, 127.40, 124.52, 120.32, 114.68, 55.96, 25.48, 15.61; mass (m/z): 485 (M+, 6.72%), 66 (100%, base peak); anal. calcd. for C26H23N5O3S (485.56): C, 64.31; H, 4.77; N, 14.42; found: C, 64.19; H, 4.89; N, 14.70%.
O); 1H NMR (400 MHz, DMSO-d6) δ 10.46 (s, 1H, NH), 8.02–7.98 (m, 3H, Ar–H), 7.91 (m, 5H, Ar–H), 7.63–7.60 (m, 1H, Ar–H), 7.58–7.55 (m, 2H, Ar–H), 7.18–7.12 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.59 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.14, 166.16, 164.80, 160.04, 158.65, 150.56, 141.24, 135.25, 132.88, 132.23, 132.20, 128.91, 128.25, 128.22, 128.02, 127.40, 124.52, 120.32, 114.68, 55.96, 25.48, 15.61; mass (m/z): 485 (M+, 6.72%), 66 (100%, base peak); anal. calcd. for C26H23N5O3S (485.56): C, 64.31; H, 4.77; N, 14.42; found: C, 64.19; H, 4.89; N, 14.70%.
5.1.10.2. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-chlorophenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono)ethyl)phenyl) benzamide 18b.
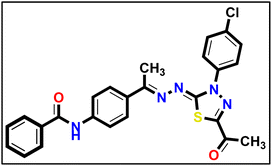 Yellow powder (yield, 72%); m.p. = 267–269 °C. FT-IR (vmax, cm−1): 3331, 3103 (NH), 3067, 3014 (C–H aromatic), 1682, 1654 (C
Yellow powder (yield, 72%); m.p. = 267–269 °C. FT-IR (vmax, cm−1): 3331, 3103 (NH), 3067, 3014 (C–H aromatic), 1682, 1654 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O);1H NMR (400 MHz, DMSO-d6) δ 10.46 (s, 1H), 8.14 (d, J = 8.5 Hz, 2H, Ar–H), 7.96 (m, 6H, Ar–H), 7.73–7.50 (m, 5H, Ar–H), 2.61 (s, 3H, CH3), 2.44 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.24, 166.17, 164.32, 160.79, 151.46, 141.40, 138.14, 135.25, 132.68, 132.21, 131.48, 129.59, 128.90, 128.23, 127.52, 123.82, 120.30, 25.53, 15.81; mass (m/z): 489 (M+, 27.03%), 77 (100%, base peak); anal. calcd. for C25H20ClN5O2S (489.98): C, 61.28; H, 4.11; N, 14.29; found: C, 61.44; H, 4.29; N, 14.57%.
O);1H NMR (400 MHz, DMSO-d6) δ 10.46 (s, 1H), 8.14 (d, J = 8.5 Hz, 2H, Ar–H), 7.96 (m, 6H, Ar–H), 7.73–7.50 (m, 5H, Ar–H), 2.61 (s, 3H, CH3), 2.44 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.24, 166.17, 164.32, 160.79, 151.46, 141.40, 138.14, 135.25, 132.68, 132.21, 131.48, 129.59, 128.90, 128.23, 127.52, 123.82, 120.30, 25.53, 15.81; mass (m/z): 489 (M+, 27.03%), 77 (100%, base peak); anal. calcd. for C25H20ClN5O2S (489.98): C, 61.28; H, 4.11; N, 14.29; found: C, 61.44; H, 4.29; N, 14.57%.
5.1.10.3. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-methoxyphenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono)ethyl)phenyl)-4-chlorobenzamide 18c.
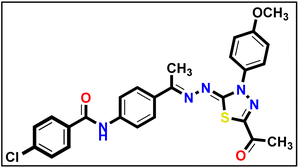 Yellow powder (yield, 80%); m.p. = 245–247 °C. FT-IR (vmax, cm−1): 3248, 3105 (NH), 3003 (C–H aromatic), 2931, 2838 (C–H aliphatic), 1691, 1643 (C
Yellow powder (yield, 80%); m.p. = 245–247 °C. FT-IR (vmax, cm−1): 3248, 3105 (NH), 3003 (C–H aromatic), 2931, 2838 (C–H aliphatic), 1691, 1643 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 10.50 (s, 1H, NH), 8.06–7.99 (m, 2H, Ar–H), 8.00–7.84 (m, 6H, Ar–H), 7.68–7.60 (m, 2H, Ar–H), 7.19–7.10 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.58 (s, 3H, CH3), 2.40 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.13, 165.02, 164.83, 160.00, 158.65, 150.57, 141.02, 137.03, 133.93, 133.04, 132.23, 130.19, 128.97, 127.41, 124.50, 120.38, 114.68, 55.96, 25.47, 15.60; mass (m/z): 520 (M+, 34.8%), 294 (100%, base peak); anal. calcd. for C26H22ClN5O3S (520.00): C, 60.05; H, 4.26; N, 13.47; found: C, 60.26; H, 4.41; N, 13.73%.
O); 1H NMR (400 MHz, DMSO-d6) δ 10.50 (s, 1H, NH), 8.06–7.99 (m, 2H, Ar–H), 8.00–7.84 (m, 6H, Ar–H), 7.68–7.60 (m, 2H, Ar–H), 7.19–7.10 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.58 (s, 3H, CH3), 2.40 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.13, 165.02, 164.83, 160.00, 158.65, 150.57, 141.02, 137.03, 133.93, 133.04, 132.23, 130.19, 128.97, 127.41, 124.50, 120.38, 114.68, 55.96, 25.47, 15.60; mass (m/z): 520 (M+, 34.8%), 294 (100%, base peak); anal. calcd. for C26H22ClN5O3S (520.00): C, 60.05; H, 4.26; N, 13.47; found: C, 60.26; H, 4.41; N, 13.73%.
5.1.10.4. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-chlorophenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono) ethyl)phenyl)-4-chlorobenzamide 18d.
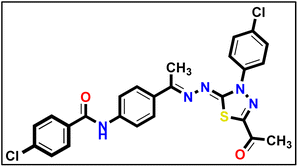 Yellow powder (yield, 75%); m.p. = 275–277 °C. FT-IR (vmax, cm−1): 3411 (NH), 3098 (C–H aromatic), 2923 (C–H aliphatic), 1689, 1659 (C
Yellow powder (yield, 75%); m.p. = 275–277 °C. FT-IR (vmax, cm−1): 3411 (NH), 3098 (C–H aromatic), 2923 (C–H aliphatic), 1689, 1659 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 10.52 (s, 1H, NH), 8.21–8.12 (m, 2H, Ar–H), 8.07–8.00 (m, 2H, Ar–H), 7.95–7.86 (m, 4H, Ar–H), 7.73–7.60 (m, 4H, Ar–H), 2.62 (s, 3H, CH3), 2.46 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.28, 165.05, 160.81, 151.50, 141.19, 138.14, 137.05, 131.54, 130.21, 129.62, 128.99, 127.56, 123.94, 120.41, 15.83. Mass (m/z): 524 (M+, 25.53%), 264 (100%, base peak); anal. calcd. for C25H19Cl2N5O2S (524.42): C, 57.26; H, 3.65; N, 13.35; found: C, 57.53; H, 3.91; N, 13.61%.
O); 1H NMR (400 MHz, DMSO-d6) δ 10.52 (s, 1H, NH), 8.21–8.12 (m, 2H, Ar–H), 8.07–8.00 (m, 2H, Ar–H), 7.95–7.86 (m, 4H, Ar–H), 7.73–7.60 (m, 4H, Ar–H), 2.62 (s, 3H, CH3), 2.46 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.28, 165.05, 160.81, 151.50, 141.19, 138.14, 137.05, 131.54, 130.21, 129.62, 128.99, 127.56, 123.94, 120.41, 15.83. Mass (m/z): 524 (M+, 25.53%), 264 (100%, base peak); anal. calcd. for C25H19Cl2N5O2S (524.42): C, 57.26; H, 3.65; N, 13.35; found: C, 57.53; H, 3.91; N, 13.61%.
5.1.10.5. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-methoxyphenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono) ethyl)phenyl)-4-methylbenzamide 18e.
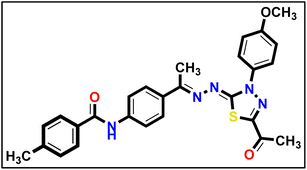 Mustard yellow powder (yield, 80%); m.p. = 240–242 °C. FT-IR (vmax, cm−1): 3290 (NH), 3086, 3063, 3006 (C–H aromatic), 2962, 2930, 2835 (C–H aliphatic), 1684, 1643 (C
Mustard yellow powder (yield, 80%); m.p. = 240–242 °C. FT-IR (vmax, cm−1): 3290 (NH), 3086, 3063, 3006 (C–H aromatic), 2962, 2930, 2835 (C–H aliphatic), 1684, 1643 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 10.36 (s, 1H, NH), 7.95–7.87 (m, 8H, Ar–H), 7.37 (d, J = 7.9 Hz, 2H, Ar–H), 7.17–7.12 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.59 (s, 3H, CH3), 2.40 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.14, 165.94, 164.75, 160.07, 158.66, 150.56, 142.25, 141.33, 132.76, 132.35, 132.24, 129.42, 128.26, 127.38, 124.52, 120.30, 114.68, 55.97, 25.48, 21.52, 15.61; mass (m/z): 499 (M+, 16.1%), 158 (100%, base peak); anal. calcd. for C27H25N5O3S (499.59): C, 64.91; H, 5.04; N, 14.02; found: C, 64.79; H, 5.23; N, 14.29%.
O); 1H NMR (400 MHz, DMSO-d6) δ 10.36 (s, 1H, NH), 7.95–7.87 (m, 8H, Ar–H), 7.37 (d, J = 7.9 Hz, 2H, Ar–H), 7.17–7.12 (m, 2H, Ar–H), 3.85 (s, 3H, CH3), 2.59 (s, 3H, CH3), 2.40 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (101 MHz, DMSO-d6) δ 190.14, 165.94, 164.75, 160.07, 158.66, 150.56, 142.25, 141.33, 132.76, 132.35, 132.24, 129.42, 128.26, 127.38, 124.52, 120.30, 114.68, 55.97, 25.48, 21.52, 15.61; mass (m/z): 499 (M+, 16.1%), 158 (100%, base peak); anal. calcd. for C27H25N5O3S (499.59): C, 64.91; H, 5.04; N, 14.02; found: C, 64.79; H, 5.23; N, 14.29%.
5.1.10.6. N-(4-((E)-1-(((Z)-5-Acetyl-3-(4-chlorophenyl)-1,3,4-thiadiazol-2(3H)-ylidene) hydrazono) ethyl)phenyl)-4-methylbenzamide 18f.
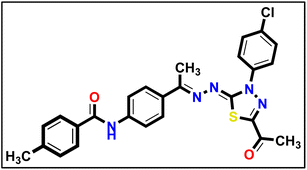 Pale yellow powder (yield, 77%); m.p. = 235–237 °C. FT-IR (vmax, cm−1): 3328, 3100 (NH), 3066, 3012 (C–H aromatic), 2917 (C–H aliphatic), 1692, 1651 (C
Pale yellow powder (yield, 77%); m.p. = 235–237 °C. FT-IR (vmax, cm−1): 3328, 3100 (NH), 3066, 3012 (C–H aromatic), 2917 (C–H aliphatic), 1692, 1651 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ 8.20–8.12 (m, 2H), 7.92 (m, 7H, Ar–H), 7.73–7.64 (m, 2H, Ar–H), 7.37 (d, J = 8.0 Hz, 2H, Ar–H), 2.63 (s, 3H, CH3), 2.46 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (126 MHz, DMSO-d6) δ 166.03, 164.16, 161.00, 151.49, 142.23, 141.53, 138.27, 132.51, 131.64, 129.59, 129.41, 128.26, 127.51, 123.99, 120.47, 25.50, 21.49, 15.82; mass (m/z): 504 (M+, 7.55%), 301 (100%, base peak); anal. calcd. for C26H22ClN5O2S (504.01): C, 61.96; H, 4.40; N, 13.90. found: C, 61.82; H, 4.62; N, 14.17%.
O); 1H NMR (400 MHz, DMSO-d6) δ 8.20–8.12 (m, 2H), 7.92 (m, 7H, Ar–H), 7.73–7.64 (m, 2H, Ar–H), 7.37 (d, J = 8.0 Hz, 2H, Ar–H), 2.63 (s, 3H, CH3), 2.46 (s, 3H, CH3), 2.41 (s, 3H, CH3); 13C NMR (126 MHz, DMSO-d6) δ 166.03, 164.16, 161.00, 151.49, 142.23, 141.53, 138.27, 132.51, 131.64, 129.59, 129.41, 128.26, 127.51, 123.99, 120.47, 25.50, 21.49, 15.82; mass (m/z): 504 (M+, 7.55%), 301 (100%, base peak); anal. calcd. for C26H22ClN5O2S (504.01): C, 61.96; H, 4.40; N, 13.90. found: C, 61.82; H, 4.62; N, 14.17%.
5.2. Biological evaluation
5.3. In silico studies
Conflicts of interest
None.Acknowledgements
This research was funded by Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R116), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors extend their appreciation to the Research Center at AlMaarefa University for funding this work.References
- International Agency for Research on Cancer, Cancer-Population Fact Sheets, https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf, (accessed 10-10-2023, 2023) Search PubMed.
- J.-D. Yan, Y. Liu, Z.-Y. Zhang, G.-Y. Liu, J.-H. Xu, L.-Y. Liu and Y.-M. Hu, Pathol., Res. Pract., 2015, 211, 539–543 CrossRef CAS PubMed.
- J. Bai, J. Wu, R. Tang, C. Sun, J. Ji, Z. Yin, G. Ma and W. Yang, Invest. New Drugs, 2020, 38, 229–245 CrossRef CAS.
- X. Wang, A. M. Bove, G. Simone and B. Ma, Front. Cell Dev. Biol., 2020, 8, 599281 CrossRef.
- R. Lugano, M. Ramachandran and A. Dimberg, Cell. Mol. Life Sci., 2020, 77, 1745–1770 CrossRef CAS PubMed.
- N. Fathi Maroufi, S. Taefehshokr, M.-R. Rashidi, N. Taefehshokr, M. Khoshakhlagh, A. Isazadeh, N. Mokarizadeh, B. Baradaran and M. Nouri, Mol. Biol. Rep., 2020, 47, 4749–4765 CrossRef CAS PubMed.
- A. A. Shah, M. A. Kamal and S. Akhtar, Curr. Drug Metab., 2021, 22, 50–59 CAS.
- W. A. Spannuth, A. M. Nick, N. B. Jennings, G. N. Armaiz-Pena, L. S. Mangala, C. G. Danes, Y. G. Lin, W. M. Merritt, P. H. Thaker and A. A. Kamat, Int. J. Cancer, 2009, 124, 1045–1053 CrossRef CAS.
- F. W. Peng, D. K. Liu, Q. W. Zhang, Y. G. Xu and L. Shi, Expert Opin. Ther. Pat., 2017, 27, 987–1004 CrossRef CAS PubMed.
- X. Li, J. Chai, Z. Wang, L. Lu, Q. Zhao, J. Zhou and F. Ju, OncoTargets Ther., 2018, 11, 4407–4411 CrossRef.
- P. Tesařová and V. Tesař, Folia Biol., 2013, 59, 15–25 Search PubMed.
- C. Fontanella, E. Ongaro, S. Bolzonello, M. Guardascione, G. Fasola and G. Aprile, Ann. Transl. Med., 2014, 2, 123 Search PubMed.
- L. Erdem, E. Giovannetti, L. G. Leon, R. Honeywell and G. J. Peters, Curr. Top. Med. Chem., 2012, 12, 1649–1659 CrossRef CAS.
- A. Levy, L. Benmoussa, S. Ammari, L. Albiges and B. Escudier, Clin. Genitourin. Cancer, 2014, 12, e33–e34 CrossRef PubMed.
- S. Grimme and P. R. Schreiner, Angew. Chem., Int. Ed. Engl., 2018, 57, 4170–4176 CrossRef CAS PubMed.
- D. Reker and G. J. D. d. t, Schneider, 2015, 20, 458–465 Search PubMed.
- X. Yang, Y. Wang, R. Byrne, G. Schneider and S. Yang, Chem. Rev., 2019, 119, 10520–10594 CrossRef CAS.
- H. González-Díaz, Curr. Top. Med. Chem., 2021, 21, 789 CrossRef.
- H. Willems, S. De Cesco and F. Svensson, J. Med. Chem., 2020, 63, 10158–10169 CrossRef CAS.
- B. De, K. Bhandari, F. J. B. Mendonça, M. T. Scotti and L. Scotti, Anti-Cancer Agents Med. Chem., 2019, 19, 587–591 CrossRef CAS.
- G. D. Geromichalos, J. B.U.ON., 2007, 12(Suppl 1), S101–S118 Search PubMed.
- R. Hameed, A. Khan, S. Khan and S. Perveen, Anti-Cancer Agents Med. Chem., 2019, 19, 592–598 CrossRef CAS PubMed.
- G. D. Geromichalos, C. E. Alifieris, E. G. Geromichalou and D. T. Trafalis, J. B.U.ON., 2016, 21, 1337–1358 Search PubMed.
- D. C. Elton, Z. Boukouvalas, M. D. Fuge and P. W. Chung, Mol. Syst. Des. Eng., 2019, 4, 828–849 RSC.
- J. Fan, A. Fu and L. Zhang, Quant. Biol., 2019, 7, 83–89 CrossRef CAS.
- L. L. Ferreira and A. D. Andricopulo, Drug discovery today, 2019, 24, 1157–1165 CrossRef CAS PubMed.
- I. Obot, D. Macdonald and Z. Gasem, Corros. Sci., 2015, 99, 1–30 CrossRef CAS.
- N. del Carmen Quintal Bojórquez and M. R. Campos, Curr. Cancer Drug Targets, 2023, 23, 333–345 CrossRef.
- Q. B. NDC and M. Campos, Curr. Cancer Drug Targets, 2022, 23(5), 333–345 Search PubMed.
- O. Daoui, S. N. Mali, K. Elkhattabi, S. Elkhattabi and S. Chtita, Heliyon, 2023, 9, e15545 CrossRef CAS.
- I. J. d. S. Nascimento, T. M. de Aquino and E. F. da Silva-Júnior, Lett. Drug Des. Discovery, 2022, 19, 951–955 CrossRef CAS.
- M. S. Taghour, H. Elkady, W. M. Eldehna, N. El-Deeb, A. M. Kenawy, A. E. Abd El-Wahab, E. B. Elkaeed, B. A. Alsfouk, A. M. Metwaly and I. H. Eissa, J. Biomol. Struct. Dyn., 2022, 1–16 Search PubMed.
- E. B. Elkaeed, M. S. Taghour, H. A. Mahdy, W. M. Eldehna, N. M. El-Deeb, A. M. Kenawy, B. A. Alsfouk, M. A. Dahab, A. M. Metwaly and I. H. Eissa, J. Enzyme Inhib. Med. Chem., 2022, 37, 2191–2205 CrossRef CAS PubMed.
- I. H. Eissa, R. G. Yousef, H. Elkady, E. B. Elkaeed, A. A. Alsfouk, D. Z. Husein, I. M. Ibrahim, M. A. Elhendawy, M. Godfrey and A. M. Metwaly, Comput. Biol. Chem., 2023, 107, 107953 CrossRef CAS PubMed.
- I. H. Eissa, R. G. Yousef, H. Elkady, E. B. Elkaeed, A. A. Alsfouk, D. Z. Husein, I. M. Ibrahim, M. M. Radwan and A. M. Metwaly, ChemistryOpen, 2023, 12, e202300066 CrossRef CAS PubMed.
- H. A. Mahdy, H. Elkady, M. S. Taghour, A. Elwan, M. A. Dahab, M. A. Elkady, E. G. Elsakka, E. B. Elkaeed, B. A. Alsfouk, I. M. Ibrahim, I. H. Eissa and A. M. Metwaly, Future Med. Chem., 2023, 15, 1233–1250 CrossRef CAS.
- M. A. Dahab, H. A. Mahdy, H. Elkady, M. S. Taghour, A. Elwan, M. A. Elkady, E. G. E. Elsakka, E. B. Elkaeed, A. A. Alsfouk, I. M. Ibrahim, A. M. Metwaly and I. H. Eissa, J. Biomol. Struct. Dyn., 2023, 1–20, DOI:10.1080/07391102.2023.2219333.
- I. H. Eissa, R. G. Yousef, H. Elkady, E. B. Elkaeed, B. A. Alsfouk, D. Z. Husein, M. A. Asmaey, I. M. Ibrahim and A. M. Metwaly, Pathol., Res. Pract., 2023, 251, 154894 CrossRef CAS PubMed.
- A. Elwan, A. E. Abdallah, H. A. Mahdy, M. A. Dahab, M. S. Taghour, E. B. Elkaeed, A. B. Mehany, A. Nabeeh, M. Adel and A. A. Alsfouk, Molecules, 2022, 27, 5047 CrossRef CAS PubMed.
- M. S. Taghour, H. A. Mahdy, M. H. Gomaa, A. Aglan, M. G. Eldeib, A. Elwan, M. A. Dahab, E. B. Elkaeed, A. A. Alsfouk and M. M. Khalifa, J. Enzyme Inhib. Med. Chem., 2022, 37, 2063–2077 CrossRef CAS.
- E. B. Elkaeed, R. G. Yousef, H. Elkady, I. M. Gobaara, B. A. Alsfouk, D. Z. Husein, I. M. Ibrahim, A. M. Metwaly and I. H. Eissa, Molecules, 2022, 27, 4606 CrossRef CAS PubMed.
- R. G. Yousef, A. Elwan, I. M. Gobaara, A. B. Mehany, W. M. Eldehna, S. A. El-Metwally, B. A. Alsfouk, E. B. Elkaeed, A. M. Metwaly and I. H. Eissa, J. Enzyme Inhib. Med. Chem., 2022, 37, 2206–2222 CrossRef CAS.
- H. Elkady, A. A. Abuelkhir, M. Rashed, M. S. Taghour, M. A. Dahab, H. A. Mahdy, A. Elwan, H. A. Al-Ghulikah, E. B. Elkaeed, I. M. Ibrahim, D. Z. Husein, A. Metwaly and I. H. Eissa, Comput. Biol. Chem., 2023, 107, 107958 CrossRef CAS.
- R. G. Yousef, H. Elkady, E. B. Elkaeed, I. M. Gobaara, H. A. Al-Ghulikah, D. Z. Husein, I. M. Ibrahim, A. M. Metwaly and I. H. Eissa, Molecules, 2022, 27, 7719 CrossRef CAS PubMed.
- E. B. Elkaeed, R. G. Yousef, H. Elkady, A. B. Mehany, B. A. Alsfouk, D. Z. Husein, I. M. Ibrahim, A. M. Metwaly and I. H. Eissa, J. Biomol. Struct. Dyn., 2022, 1–16 Search PubMed.
- M. A. El-Zahabi, H. Elkady, H. Sakr, A. S. Abdelraheem, S. I. Eissa and K. El-Adl, J. Biomol. Struct. Dyn., 2023, 1–18 Search PubMed.
- M. M. Alanazi, H. A. Mahdy, N. A. Alsaif, A. J. Obaidullah, H. M. Alkahtani, A. A. Al-Mehizia, S. M. Alsubaie, M. A. Dahab and I. H. Eissa, Bioorg. Chem., 2021, 112, 104949 CrossRef CAS PubMed.
- M. M. Alanazi, H. Elkady, N. A. Alsaif, A. J. Obaidullah, H. M. Alkahtani, M. M. Alanazi, M. A. Alharbi, I. H. Eissa and M. A. Dahab, RSC Adv., 2021, 11, 30315–30328 RSC.
- R. G. Yousef, H. M. Sakr, I. H. Eissa, A. B. Mehany, A. M. Metwaly, M. A. Elhendawy, M. M. Radwan, M. A. ElSohly, H. S. Abulkhair and K. El-Adl, New J. Chem., 2021, 45, 16949–16964 RSC.
- L. Adnane, P. A. Trail, I. Taylor and S. M. Wilhelm, Methods Enzymol., 2006, 407, 597–612 CAS.
- F.-W. Peng, D.-K. Liu, Q.-W. Zhang, Y.-G. Xu and L. Shi, Expert Opin. Ther. Pat., 2017, 27, 987–1004 CrossRef CAS.
- M. G. Papich, Papich Handbook of Veterinary Drugs-E-Book, Elsevier Health Sciences, 2020 Search PubMed.
- G. J. Roth, R. Binder, F. Colbatzky, C. Dallinger, R. Schlenker-Herceg, F. Hilberg, S.-L. Wollin and R. Kaiser, J. Med. Chem., 2015, 58, 1053–1063 CrossRef CAS PubMed.
- H. Elkady, O. A. El-Dardir, A. Elwan, M. S. Taghour, H. A. Mahdy, M. A. Dahab, E. B. Elkaeed, B. A. Alsfouk, I. M. Ibrahim and D. Z. Husein, RSC Adv., 2023, 13, 27801–27827 RSC.
- R. Verhé, L. De Buyck, N. De Kimpe, A. De Rooze and N. Schamp, Bull. Soc. Chim. Belg., 1978, 87, 143–152 CrossRef.
- S. R. Donohue, R. F. Dannals, C. Halldin and V. W. Pike, J. Med. Chem., 2011, 54, 2961–2970 CrossRef CAS.
- D. L. Klayman, J. F. Bartosevich, T. S. Griffin, C. J. Mason and J. P. Scovill, J. Med. Chem., 1979, 22, 855–862 CrossRef CAS.
- L. Yurttaş, Y. Özkay, G. Akalın-Çiftçi and Ş. Ulusoylar-Yıldırım, J. Enzyme Inhib. Med. Chem., 2014, 29, 175–184 CrossRef.
- J. S. Chu, F. J. Ge, B. Zhang, Y. Wang, N. Silvestris, L. J. Liu, C. H. Zhao, L. Lin, A. E. Brunetti and Y. L. Fu, J. Exp. Clin. Cancer Res., 2013, 32, 1–8 CrossRef.
- J. Pijuan, C. Barceló, D. F. Moreno, O. Maiques, P. Sisó, R. M. Marti, A. Macià and A. Panosa, Front. Cell Dev. Biol., 2019, 7, 107 CrossRef PubMed.
- I. Arranz-Valsero, L. Soriano-Romaní, L. García-Posadas, A. López-García and Y. Diebold, Exp. Eye Res., 2014, 125, 183–192 CrossRef CAS.
- D. Z. Husein, R. Hassanien and M. Khamis, RSC Adv., 2021, 11, 27027–27041 RSC.
- T. Wang and D. Z. Husein, Environ. Sci. Pollut. Res., 2023, 30, 8928–8955 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- F. Denizot and R. Lang, J. Immunol. Methods, 1986, 89, 271–277 CrossRef CAS PubMed.
- M. Thabrew, R. D. HUGHES and I. G. MCFARLANE, J. Pharm. Pharmacol., 1997, 49, 1132–1135 CrossRef CAS.
- N. A. Alsaif, M. S. Taghour, M. M. Alanazi, A. J. Obaidullah, W. A. Alanazi, A. Alasmari, H. Albassam, M. A. Dahab and H. A. Mahdy, Bioorg. Med. Chem., 2021, 46, 116384 CrossRef CAS.
- H. A. Mahdy, H. Elkady, M. S. Taghour, A. Elwan, M. A. Dahab, M. A. Elkady, E. G. Elsakka, E. B. Elkaeed, B. A. Alsfouk and I. M. Ibrahim, Future Med. Chem., 2023, 15, 1233–1250 CrossRef CAS.
- A. A. Nasser, I. H. Eissa, M. R. Oun, M. A. El-Zahabi, M. S. Taghour, A. Belal, A. M. Saleh, A. B. Mehany, H. Luesch and A. E. Mostafa, Org. Biomol. Chem., 2020, 18, 7608–7634 RSC.
- D. Z. Husein, R. Hassanien and M. J. R. A. Khamis, RSC adv., 2021, 11, 27027–27041 RSC.
- Y. M. Suleimen, R. A. Jose, G. K. Mamytbekova, R. N. Suleimen, M. Y. Ishmuratova, W. Dehaen, B. A. Alsfouk, E. B. Elkaeed, I. H. Eissa and A. M. Metwaly, Plants, 2022, 11, 2072 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1, 19–25 CrossRef.
- M. S. Valdés-Tresanco, M. E. Valdés-Tresanco, P. A. Valiente and E. Moreno, J. Chem. Theory Comput., 2021, 17, 6281–6291 CrossRef.
- A. Amadei, A. B. Linssen and H. J. J. P. S. Berendsen, Funct., Bioinf., 1993, 17, 412–425 CrossRef CAS.
- E. Papaleo, P. Mereghetti, P. Fantucci, R. Grandori and L. De Gioia, J. Mol. Graphics Modell., 2009, 27, 889–899 CrossRef CAS.
- D. S. Biovia, Release, 2017 Search PubMed.
- A. M. Metwaly, E. B. Elkaeed, B. A. Alsfouk, A. M. Saleh, A. E. Mostafa and I. H. Eissa, Plants, 2022, 11, 1886–1905 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07562a |
| This journal is © The Royal Society of Chemistry 2023 |