Open Access Article
Open Access ArticleGenome mining of actinomycin shunt products from Kitasatospora sp. YINM00002†
Zhou-Tian-Le Zhang‡
a,
Hui-Bing Sun‡a,
Zhen Ren‡b,
Tian-Peng Xiea,
Ying-Fang Wanga,
Yin Guoa,
Xiaoyu Sua,
Min Yin*a,
Hao Zhou *a and
Zhong-Tao Ding
*a and
Zhong-Tao Ding ac
ac
aKey Laboratory of Functional Molecules Analysis and Biotransformation of Universities in Yunnan Province, Yunnan Characteristic Plant Extraction Laboratory, School of Chemical Science and Technology, School of Medicine, Yunnan University, University Town East Outer Ring South Road, Kunming, Yunnan 650500, China. E-mail: yinmin@ynu.edu.cn; haozhou@ynu.edu.cn
bSchool of Agriculture and Life Sciences, Kunming University, 2 Pu Xin Road, Kunming, Yunnan 650214, China
cCollege of Traditional Chinese Medicine, Yunnan University of Chinese Medicine, 1076 Yu Hua Road, Kunming, Yunnan 650500, China
First published on 12th December 2023
Abstract
Actinomycins are known for their anti-tumor, antibacterial and antiviral activities, and in particular for the ability of actinomycin D as a clinical drug to treat a variety of cancers. In our ongoing work to obtain novel natural products from endophytic actinomycetes derived from traditional Chinese herbs, we identified the potential to produce actinomycins in YINM00002, a Kitasatospora strain derived from Polygonatum kingianum. According to genome mining, we isolated actinomycins D and V (1 and 2) and small amounts of 4-methyl-3-hydroxyanthranilic acid (4-MHA) derivates (3 and 4) from strain fermentation broth. The presence of actinrhater A (3) and actinrhater B (4) reveals a mysterious shunt pathway in the early stages of actinomycin D biosynthesis. Our study provides a fresh perspective for further discovery and modification of novel actinomycins.
Introduction
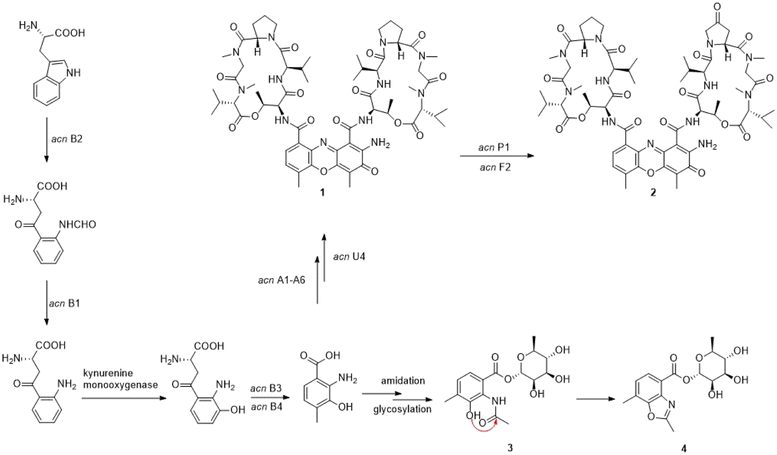
Actinomycins have strong anti-tumor, antibacterial and antiviral activities, among which actinomycin D has been used in the clinical treatment of a variety of tumors, such as sarcomas, choriocarcinoma and lymphomas, especially for gestational trophoblastic neoplasia in women1 and for pediatric tumors like Wilms tumor,2 childhood rhabdomyosarcoma3 and Ewing's sarcoma.4 Actinomycin D could intercalate in the guanine–cytosine-rich regions of the transcription initiation complex, and prevent the elongation of the RNA chain. The strong dipole moment of actinomycin D, which might be provided by the phenoxazinone chromophore moiety, was thought to be the cause of the intercalation.5,6 In addition, the crystal structure of actinomycin D bound to a specific DNA sequence, ATGCGGCAT, has been resolved.7Despite their strong biological activity, these compounds also have undesirable side-effects, limiting their clinical use.8,9 Therefore, it is necessary to investigate the biosynthetic mechanisms of these compounds and obtain novel actinomycins with high activity and low toxicity through synthetic biological modifications. At present, several actinomycin biosynthetic gene clusters have been cloned from different Streptomyces strains, and parts of their biosynthetic mechanisms have been elucidated.10–14 To date, more than 40 analogues have been reported.15–22 The skeleton of antinomycins is composed of a phenoxazinone chromophore core, an α-peptidolactone and a β-peptidolactone. The biosynthesis of actinomycins is initiated by using tryptophan as starter, then five enzymes, including a tryptophan oxygenase, a kynurenine formamidase, a kynurenine 3-monooxygenase, a kynureninase and a hydroxykynureninase, converted tryptophan into the key building block 4-MHA.14,23–27 After that, the non-ribosomal peptide synthetases (NRPS) assembly line loaded five amino acids to the 4-MHA to form the pentapeptide precursor. However, the details of how the two MHA-pentapeptide monomers come together to generate the intact actinomycin are still unknown.
In order to find more active natural products, we focused on the endogenous and rhizospheric soil actinomycetes of traditional Chinese herbs. Polygonatum kingianum Coll. et Hemsl, a medicinal plant that can be used to treat osteoporosis, feebleness, and fatigue, is widely cultivated in Southwest China.28 It is distinct in polysaccharides and possesses unique bioactivities, but the endogenous and rhizosphere actinomycetes live with it have not been studied. During the genome mining of the endophytic actinomycetes in this medicinal plant, we found that a strain of Kitasatospora possessed a putative actinomyctin biosynthetic gene cluster that also demonstrated strong inhibitory activity against several pathogens. In this study, we isolated actinomycins under the guidance of the genome mining study of strain YINM00002. In the meantime, we discovered two unexpected new compounds. The two compounds are the products of the amidation, glycosylation and cyclation of 4-MHA, a key building block in the actinomycin biosynthetic pathway. Based on these findings, we speculate that they are new shunt products in the actinomycin biosynthetic pathway.
Results and discussion
Taxonomy analysis of Kitasatospora sp. YINM00002
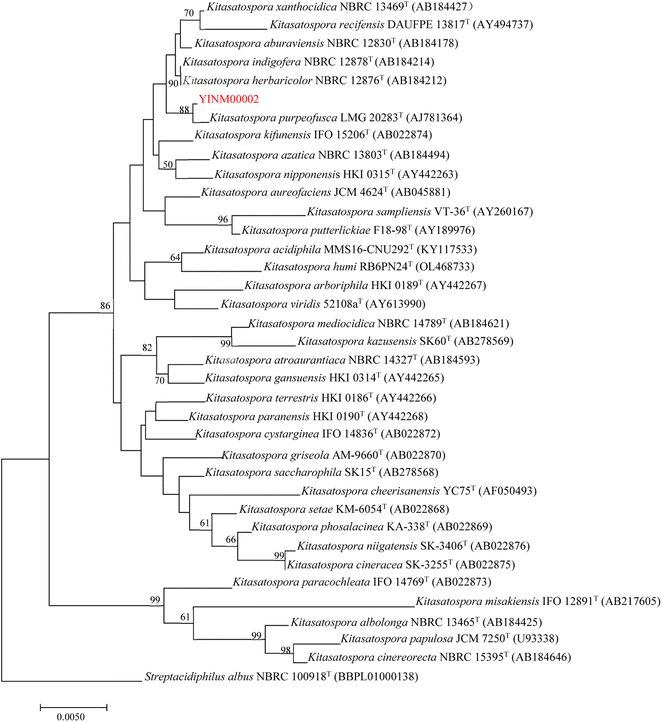
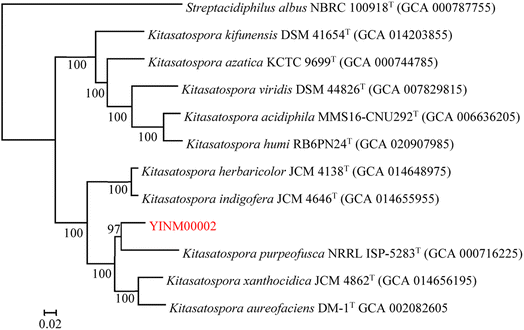
The neighbor-joining phylogenetic tree of the 16S rRNA gene sequences showed that strain YINM00002 formed a cluster with Kitasatospora purpeofusca LMG 20283T (99.79% similarity) (Fig. 1). The RAxML neighbor-joining phylogenomic tree demonstrated that YINM00002 formed a cluster with strain Kitasatospora purpeofusca NRRL ISP-5283T under the 97 bootstrap values (Fig. 2), the maximum-likelihood tree and maximum-parsimony tree also showed it clustered with strain Kitasatospora purpeofusca NRRL ISP-5283T under high bootstrap values (Fig. S1 and S2†). Phylogenetic analysis of strain YINM00002 indicates that it is a member of the genus Kitasatospora.Antimicrobial activity and antibiotic resistance of Kitasatospora sp. YINM00002
Strains of Kitasatospora genus are capable of producing various types of antibiotics. Kitasatospora sp. YINM00002 has shown strong inhibitory activity against several pathogens, including Escherichia coli, Staphylococcus aureus, Mycobacterium tuberculosis, and Bacillus subtilis. At the same time, it could grow well on ISP2 medium with bacitracin. Antimicrobial activity and antibiotic resistance screening results indicate that Kitasatospora sp. YINM00002 may be a good strain for bioactive secondary metabolites discovery.Genome mining of actinomycin gene cluster
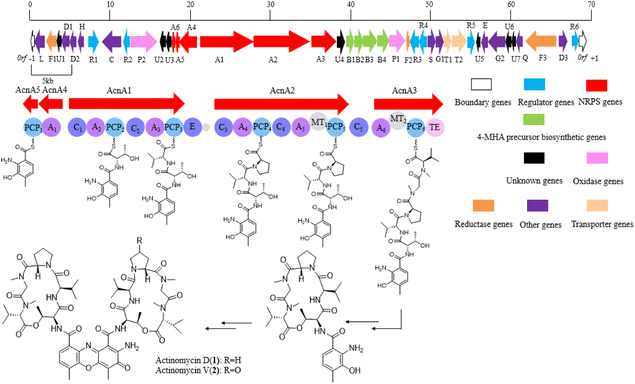
With the assistance of a genome mining software antiSMASH 6.0, a gene cluster which might be responsible for actinomycin biosynthesis was identified (Fig. 3). The putative gene cluster shows 89% similarity with the reported actinomycin gene cluster in a Streptomyces chrysomallus strain. Besides the start unit 4-MHA and the backbone non-ribosomal peptide (NRP) biosynthetic genes, additional genes, which coding regulators, oxidase, reductase, transporter and hypothetical proteins, are distributed on both sides of the entire gene cluster (Table 1). The high similarity of the core biosynthetic genes within the identified gene cluster and the reported actinomycin gene cluster showed strain YINM00002 is likely to produce actinomycins. Meanwhile, the inconsistency of the other genes between the two gene clusters indicated strain YINM00002 might have potential to generate novel members of actinomycin family (Fig. S3†).| Identifier | Sizea | Protein homolog and origin | ID/SM (%) | Origin (protein ID) |
|---|---|---|---|---|
| a Size in units of amino acids (aa). | ||||
| Orf (−1) | 195 | Hypothetical protein | 73/100 | WP_209415338.1 Kitasatospora sp. RG8 |
| acn L | 355 | SCO0930 family lipoprotein | 82/100 | WP_230210278.1 Streptomyces kaniharaensis |
| acn F1 | 480 | Ferredoxin | 97/100 | MBV2153232.1 Kitasatospora sp. SUK 42 |
| acn U1 | 325 | Hypothetical protein | 84/100 | WP_153464056.1 Streptomyces kaniharaensis |
| acn D1 | 464 | DUF1996 domain-containing protein | 91/100 | WP_209415335.1 Kitasatospora sp. RG8 |
| acn D2 | 244 | DUF4142 domain-containing protein | 88/100 | WP_148648111.1 Streptomyces sp. CB01881 |
| acn H | 268 | Helix-turn-helix domain-containing protein | 89/91 | WP_230210812.1 Streptomyces kaniharaensis |
| acn R1 | 309 | LysR family transcriptional regulator | 95/100 | MBV6703076.1 Kitasatospora aureofaciens |
| acn C | 856 | Collagenase | 76/100 | WP_221503753.1 Kitasatospora gansuensis |
| acn R2 | 224 | LuxR C-terminal-related transcriptional regulator | 81/100 | WP_188298505.1 Streptomyces sp. CBMA156 |
| acn P2 | 1049 | Cytochrome P450 | 83/98 | WP_218198784.1 Kitasatospora aureofaciens |
| acn U2 | 210 | Hypothetical protein | 86/100 | WP_051838397.1 Streptomyces sp. NRRL WC-3742 |
| acn U3 | 186 | Hypothetical protein | 86/100 | WP_057667199.1 Streptomyces anulatus |
| acn A6 | 66 | MbtH family protein | 97/100 | WP_199823413.1 Streptomyces sp. NRRL WC-3742 |
| acn A5 | 79 | 4-MHA carrier protein | 69/98 | WGD01484.1 Streptomyces sp. |
| acn A4 | 472 | 3-Hydroxy-4-methylanthranilate adenylyltransferase AcmA | 81/100 | WP_078911522.1 Streptomyces sp. NRRL WC-3742 |
| acn A1 | 2612 | Non-ribosomal peptide synthetase | 74/99 | QIT48436.1 Streptomyces antibioticus |
| acn A2 | 2964 | Non-ribosomal peptide synthetase | 79/94 | WP_104880054.1 Streptomyces dengpaensis |
| acn A3 | 1248 | Non-ribosomal peptide synthetase | 74/100 | WP_078636602.1 Streptomyces antibioticus |
| acn U4 | 210 | Hypothetical protein | 93/100 | WP_051838204.1 Streptomyces sp. NRRL WC-3742 |
| acn B1 | 292 | Arylformamidase | 53/96 | SBU91169.1 Streptomyces sp. Ncost-T6T-1 |
| acn B2 | 281 | Tryptophan 2,3-dioxygenase family protein | 87/100 | WP_031075029.1 Streptomyces sp. NRRL WC-3742 |
| acn B3 | 420 | Kynureninase | 86/100 | WP_096632687.1 Streptomyces sp. WZ.A104 |
| acn B4 | 346 | Methyltransferase | 90/100 | WP_275821042.1 Streptomyces ferralitis |
| acn P1 | 429 | Cytochrome P450 | 84/96 | WP_229893031.1 Streptomyces xanthochromogenes |
| acn F2 | 72 | Ferredoxin | 70/91 | WP_161359452.1 Streptomyces sp. SID3343 |
| acn R3 | 215 | LmbU family transcriptional regulator | 78/99 | WP_248634514.1 Streptomyces lichenis |
| acn R4 | 257 | TetR/AcrR family transcriptional regulator | 74/98 | WP_051781104.1 Streptomyces antibioticus |
| acn S | 295 | Siderophore-interacting protein | 87/100 | WP_031075040.1 Streptomyces sp. NRRL WC-3742 |
| acn G1 | 326 | ATP-binding cassette domain-containing protein | 87/100 | WP_255386436.1 Streptomyces parvus |
| acn T1 | 255 | ABC transporter permease | 96/100 | WP_057667158.1 Streptomyces anulatus |
| acn T2 | 753 | Excinuclease ABC subunit UvrA | 89/100 | WP_031075046.1 Streptomyces sp. NRRL WC-3742 |
| acn R5 | 288 | AfsR/SARP family transcriptional regulator | 89/100 | WP_238862110.1 Kitasatospora sp. A2-31 |
| acn U5 | 174 | Hypothetical protein | 79/99 | WP_238862109.1 Kitasatospora sp. A2-31 |
| acn E | 161 | S26 family signal peptidase | 87/96 | WP_238862108.1 Kitasatospora sp. A2-31 |
| acn G2 | 637 | ABC transporter ATP-binding protein | 92/95 | MCG6495646.1 Kitasatospora sp. A2-31 |
| acn U6 | 190 | Hypothetical protein | 85/100 | WP_238862106.1 Kitasatospora sp. A2-31 |
| acn U7 | 79 | Hypothetical protein | 96/100 | WP_238862105.1 Kitasatospora sp. A2-31 |
| acn Q | 191 | FAD binding domain-containing protein | 90/96 | WP_280701620.1 Kitasatospora sp. GP82 |
| acn F3 | 1423 | Bifunctional nitrate reductase | 80/100 | WP_280723810.1 Kitasatospora sp. MAA4 |
| acn D3 | 356 | DUF3048 domain-containing protein | 83/100 | WP_238862099.1 Kitasatospora sp. A2-31 |
| acn R6 | 245 | Crp/Fnr family transcriptional regulator | 82/99 | WP_117490432.1 Kitasatospora xanthocidica |
| Orf (−1) | 284 | Hypothetical protein | 66/99 | WP_074004766.1 Streptomyces sp. CB02056 |
Isolation of compounds guided by genome mining
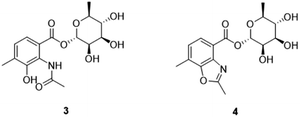
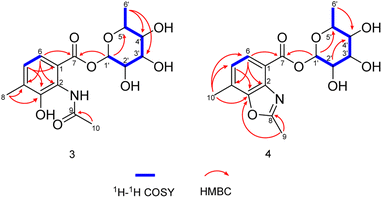
As expected, red amorphous powder actinomycin D (1) and actinomycin V (2) were isolated from the 9# medium ferments of strain YINM00002. From the same crude extracts, two compounds (3 and 4) (Fig. 4) were found and elucidated as new products.Compound 3 was obtained as faint yellow amorphous powder, the molecular formula of C16H21NO8 was deduced from its HRESIMS at m/z 354.1193 [M − H]− (calcd for C16H20NO8, 354.1189) and 13C NMR data, indicating seven degrees of unsaturation. The 13C NMR data of 3 exhibited 16 carbons including two carbonyls (δC 170.6, 164.9), six aromatic carbons (δC 121.3, 124.9, 128.0, 131.9, 139.8, 150.6), three methyls (δC 17.2, 18.4, 23.7), and five sugar-related methines (δC 69.8, 70.8, 71.7, 72.0, 95.2) (Table 2). Comparing the 1D and 2D NMR data of 3 to that of the known compound 2-acetylamino-3-hydroxyl-4-methyl-benzoic acid,29 the aromatic moiety of 3 was determined. This was confirmed by HMBC correlations from H-5 to C-1 and C-3, from H-6 to C-2, C-4 and C-7, from H-10 to C-3, C-4 and C-5, from H-9 to C-8, together with COSY correlation between H-5 and H-6 (Fig. 5). In addition, the 1H–1H COSY correlations of H-1′/H-2′/H-3′/H-4′/H-5′/H3-6′ revealed the spin-system corresponding to the C-1′ to C-6′ unit in 3. Combined with the HMBC correlation from H-1′ to C-5′, the moiety of rhamnose was identified. Comparison of the 1D NMR data of the sugar moiety in compound 3 with those of rhamnose30 also confirmed it. This was further verified by the hydrolysis experiment along with HRESIMS analysis of the sugar moiety (Fig. S28†). The specific rotation of the rhamnose moiety was [α]D25 −8.6 (c 0.1, MeOH) in accordance with that of authentic L-rhamnose ([α]D25 −4.9 (c 0.1, MeOH)). The HMBC correlation from H-1′ to C-7 indicated that the L-rhamnose moiety is connected to C-7 (Fig. 5). Thus, the structure of 3 was elucidated as a new benzoic acid glycosyl ester named as actinrhater A.
| No. | 3a | 4b | ||||||
|---|---|---|---|---|---|---|---|---|
| 13C, type | 1H (J in Hz) | HMBC | COSY | 13C, type | 1H (J in Hz) | HMBC | COSY | |
| a 3 was measured at 400 MHz.b 4 was measured at 600 MHz. | ||||||||
| 1 | 124.9, C | — | — | — | 118.2, C | — | — | — |
| 2 | 139.8, C | — | — | — | 140.3, C | — | — | — |
| 3 | 150.6, C | — | — | — | 150.2, C | — | — | — |
| 4 | 131.9, C | — | — | — | 126.3, C | — | — | — |
| 5 | 128.0, CH | 7.12, d (7.9) | C-1,3 | H-6 | 125.1, CH | 7.30, d (7.9) | C-1,3,4 | H-6 |
| 6 | 121.3, CH | 7.21, d (8.0) | C-2,4,7 | H-5 | 126.2, CH | 7.80, d (7.9) | C-2,4,7 | H-5 |
| 7 | 164.9, C | — | — | — | 162.7, C | — | — | — |
| 8 | 170.6, C | — | — | — | 165.4, C | — | — | — |
| 9 | 23.7, CH3 | 2.08, s | C-8 | — | 14.3, CH3 | 2.68, s | C-3,8 | — |
| 10 | 17.2, CH3 | 2.22, s | C-3,4,5 | — | 15.2, CH3 | 2.53, s | C-2,3,5,6 | — |
| 1′ | 95.2, CH | 5.91, d (1.8) | C-7,5′ | H-2′ | 94.5, CH | 6.07, d (1.7) | C-7,5′ | H-2′ |
| 2′ | 69.8, CH | 3.75, m | — | H-1′,3′ | 69.6, CH | 3.80, m | C-4′ | H-1′,3′ |
| 3′ | 70.8, CH | 3.53, m | — | H-2′,4′ | 70.2, CH | 3.83, m | — | H-2′,4′ |
| 4′ | 72.0, CH | 3.31, m | — | H-3′,5′ | 71.6, CH | 3.35, m | C-6′ | H-3′,5′ |
| 5′ | 71.7, CH | 3.57, m | — | H-6′,4′ | 71.2, CH | 3.78, m | — | H-4′,6′ |
| 6′ | 18.4, CH3 | 1.16, d (6.2) | C-3′,4′ | H-5′ | 17.9, CH3 | 1.17, d (6.2) | C-4′ | H-5′ |
| 2′-OH | 5.21, s | 5.22, s | ||||||
| 3′-OH | 4.75, s | 4.76, d (6.0) | ||||||
| 4′-OH | 4.97, s | 4.95, d (5.3) | ||||||
Actinrhater B (4) afforded a sodium adduct ion at m/z 360.1047 [M + Na]+ in agreement with a molecular formula of C16H19NO7. The signal systems of the NMR of 4 resembled those of compound 3, suggesting structural similarity. The most evident difference was that one more degree of unsaturation was required than compound 3. Further aided by 1H–1H COSY and HMBC experiments (Fig. 5), the planar structure of 4 was established as dimethylbenzo[d]oxazole with the C-7 rhamnosyl ester as in 3, which was supported by the HMBC correlation from H-9 to C-3 and C-8. Moreover, the moiety of dimethylbenzo[d]oxazole in 4 was also confirmed by comparing with a recently reported literature.18 Then, the rhamnose moiety of 4 was determined to be the same L-rhamnose as that of 3 by its negative specific rotation. Accordingly, the structure of 4 was defined.
Actinrhaters are shunt products of actinomycin biosynthesis
4-MHA is the key starting unit for actinomycin chromophore formation, and a cluster including acnB1–B4, which encodes an arylformamidase, a tryptophan-2, a 3-dioxygenase, and a methyltransferase, has been identified in the middle of the putative actinomycin gene cluster in YINM00002, showing high similarity to known 4-MHA biosynthetic enzymes.Previous in vitro studies have confirmed that the pathway of 4-MHA needed a tryptophan oxygenase, a kynurenine formamidase, a kynurenine 3-monooxygenase, a kynureninase and a hydroxykynureninase.14,23–27 Similar to the subcluster in an actinomycin producing strain of Streptomyces costaricanus SCSIO ZS0073, the putative kynurenine 3-monooxygenase is absent in YINM00002. After the biosynthesis of 4-MHA, it is loaded to PCP1 (AcnA5) and then attached a L-threonine, a D-valine, a L-proline, a L-sarcosine and a methyl-valine separately by a series of NRPS (AcnA5, AcnA1, AcnA2 and AcnA3) to form the 4-MHA-pentapeptide halves of actinomycin. A hypothetical protein (acnU4) located next to AcnA3 might be responsible for the formation of the mature actinomycin D from the 4-MHA-pentapeptide monomers. A cytochrome P450 (acnP1) gene and a ferredoxin (acnF2) gene are located downstream of the 4-MHA biosynthetic cassette, which might be involved in the formation of actinomycin V from actinomycin D (Fig. 6).
The presence of small amounts of compounds 3 and 4 reveals a mysterious shunt pathway in the early stages of actinomycin D biosynthesis. After the formation of 4-MHA, the main metabolites, catalyzed by the core enzymes described above, flow towards actinomycins biosynthesis, while a small amount of 4-MHA is converted to 3 by amidation and glycosylation, and then to 4 by cyclization. Whether the amidation, glycosylation and cyclization are catalyzed by enzymes or formed spontaneously remains to be discovered. No such enzymes were found in and around the acn gene cluster.
Experimental
General experimental procedures
UV-vis spectra were recorded using a Shimadzu UV-2550 PC spectrometer (Shimadzu Co., Ltd, Tokyo, Japan). The specific rotations were measured using a JASCO P-1020 digital polarimeter (Horiba, Tokyo, Japan). NMR spectra were recorded on a Bruker Avance-400 MHz instrument and a Bruker Avance-600 MHz instrument (Bruker, Karlsruhe, Germany) using tetramethylsilane as the internal standard. HRESIMS data were obtained by an Agilent 1200 Q-TOF mass instrument (Agilent, Santa Clara, CA, United States). The preparative HPLC was performed on an Agilent 1260 series equipped with a DAD detector and a Zorbax SB-C18 (250 × 9.4 mm, 5 mm) column. Sephadex LH-20 (GE Healthcare Bio-Science AB, Uppsala, Sweden), silica gel (200–300 mesh, Qingdao Marine Chemical Group Co., Qingdao, China) and Lichroprep RP-18 gel (40–63 mm, Merck, Darmstadt, Germany) were used for column chromatography (CC). Thinlayer chromatography (TLC) was performed on silica gel GF254 plates (Qingdao Haiyang Chemical Co., Ltd, Qingdao, China), which can be visualized by spraying with anisaldehyde–H2SO4 reagent. All solvents used were of analytical grade from Chengdu Titan Chron Chemical Co., Ltd (Chengdu, China).Bacterial isolation
Fresh rhizospheric soil samples of Polygonatum kingianum Coll. et Hemsl were randomly collected from the Chinese medicinal herb garden at Kunming University, Yunnan Province, China. All the samples were thoroughly mixed and then stored at 4 °C and processed within 12 h. Strain YINM00002 was isolated on M1 medium (asparagine 1 g, glycerol 10 g, K2HPO4·H2O 1 g, MgSO4·7H2O 0.05 g, CaCO3 0.3 g, vitamin complex and trace salt solution 1 mL, pH = 7.2) with nystatin and nalidixic acid (50 μg mL−1 each), which had been inoculated with the soil sample suspension and incubated at 28 °C for up to a month. The isolate was picked out and purified three times on ISP2 plates with nystatin and nalidixic acid. The purified culture was preserved in ISP2 slants and 20% (v/v) glycerol tubes, stored at 4 °C and −80 °C respectively for further using. Genomic DNA isolation, 16S rRNA gene amplification and sequencing were performed as described previously.31Antimicrobial activity and antibiotic resistance assay
Strain YINM00002 was inoculated on ISP2 agar disks and incubated at 28 °C for a week. The liquid cultures of activated pathogens, including Escherichia coli (CGMCC 1.2385), Pseudomonas aeruginosa (CGMCC 1.2387), Bacillus subtilis (CGMCC 1.1849), Staphylococcus aureus (CGMCC 1.2386), Mycobacterium tuberculosis (ATCC 25177), Fusarium oxysporum (MW149127.1), Candida albicans (CGMCC 2.2086) and Fusarium fulcrum (MW149128.1), were mixed well with Mueller–Hinton agar (beef extract powder 2 g, casein hydrolysate 17.5 g, starch 1.5 g, agar 12 g, pH = 7.3) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, then spread on the top of ISP2 agar. Once the top agar had solidified, about 5 mm diameter ISP2 agar with a single colony of each isolated strains were placed on the top agar, incubated at 28 °C for a week.
1 v/v, then spread on the top of ISP2 agar. Once the top agar had solidified, about 5 mm diameter ISP2 agar with a single colony of each isolated strains were placed on the top agar, incubated at 28 °C for a week.
Strain YINM00002 was inoculated on ISP2 agar supplemented with chloramphenicol (50 μg mL−1), kanamycin (200 μg mL−1), levofloxacin (50 μg mL−1), vancomycin (50 μg mL−1), rifampicin (50 μg mL−1), oxytetracycline (50 μg mL−1), apramycin (50 μg mL−1), bacitracin (50 μg mL−1), respectively, and incubated at 28 °C for 5 days.
Genome sequencing and actinomycin gene cluster identification
A single colony of strain YINM00002 was inoculated into 50 mL Tryptic Soy Broth (TSB) medium (casein tryptone 17 g, soy peptone 5 g, sodium chloride 5 g, D-glucose 2.5 g, dipotassium phosphate 2.5 g, H2O up to 1000 mL, pH = 7.3) for 36 h at 28 °C with 200 rpm vigorous shaking. Next the strain was stored with dry ice and sent to Majorbio (Shanghai, China) for complete genome sequencing. After sequencing by Pacbio and Illumina Hiseq Technologies, the biosynthetic gene cluster of actinomycin was identified using the antiSMASH 6.0,32 and then verified by manual inspection.Fermentation of strain YINM00002
For small-scale fermentation, the strain YINM00002 was activated in ISP2 medium at 28 °C for a week. The activated strain was inoculated into 500 mL Erlenmeyer flasks containing 100 mL of ISP2 medium and cultured for 2 days at 28 °C and 200 rpm. Then, 1.0 mL of the seed culture was transferred into 500 mL Erlenmeyer flasks containing 100 mL of 5 different types of fermentation media (2#, 3#, 9#, 10# and 15#, data not show), respectively. The crude exacts of 5, 10, 15 days ferments were detected with high performance liquid chromatograph (HPLC) and thin layer chromatography (TLC) to determine the optimal fermentation condition.For large-scale fermentation, seed cultures of strain YINM00002 were prepared as described above. Then, 1.0 mL of the seed cultures were transferred into 500 mL Erlenmeyer flasks containing 100 mL of 9# fermentation media (glucose 10 g, glycerol 10 mL, corn extract 2.5 g, peptone 5 g, soluble starch 10 g, yeast extract 2 g, calcium carbonate 3 g, sodium chloride 1 g, H2O up to 1000 mL, pH = 7.3), and cultured for 10 days at 28 °C and 200 rpm (13.5 L).
Isolation and identification of compounds
The fermentation broth of strain YINM00002 (13.5 L) was extracted with equal volume ethyl acetate three times to afford an EtOAc extract (10.5 g). The crude extract was separated into five fractions (Fr.1–Fr.5) by a silica gel column with a petroleum ether-acetone gradient system (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 30
0, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10
1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0
1, 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Fr.2 was fractionated using a Sephadex LH-20 column with MeOH to obtain Fr.2-1 to Fr.2-3. Fr.2-2 was fractionated by LiChroprep RP-18 column, and eluted stepwise with a MeOH–H2O gradient (40% MeOH, 60% MeOH, 80% MeOH and 100% MeOH) to afford Fr.2-2-1 to Fr.2-2-4. Fr.2-2-3 was separated by HPLC using a Zorbax SB-C18 column (250 × 9.4 mm, 5 μm; the mobile phase is 95% MeOH; the flow rate is 3.0 mL min−1; detection wavelength 440 nm) to give compounds 1 (20.7 mg, tR = 28.5 min) and 2 (13.2 mg, tR = 27.5 min). Fr.3 was divided into four parts (Fr.3-1 to Fr.3-4) by a Sephadex LH-20 column with MeOH. Compounds 3 (4.5 mg, tR = 10.0 min) and 4 (3.4 mg, tR = 14.8 min) were obtained from Fr.3-4 by HPLC purification. (Zorbax SB-C18 column: 250 × 9.4 mm, 5 μm; the mobile phase 55% MeOH; the flow rate 3.0 mL min−1; detection wavelength 300 nm).
1). Fr.2 was fractionated using a Sephadex LH-20 column with MeOH to obtain Fr.2-1 to Fr.2-3. Fr.2-2 was fractionated by LiChroprep RP-18 column, and eluted stepwise with a MeOH–H2O gradient (40% MeOH, 60% MeOH, 80% MeOH and 100% MeOH) to afford Fr.2-2-1 to Fr.2-2-4. Fr.2-2-3 was separated by HPLC using a Zorbax SB-C18 column (250 × 9.4 mm, 5 μm; the mobile phase is 95% MeOH; the flow rate is 3.0 mL min−1; detection wavelength 440 nm) to give compounds 1 (20.7 mg, tR = 28.5 min) and 2 (13.2 mg, tR = 27.5 min). Fr.3 was divided into four parts (Fr.3-1 to Fr.3-4) by a Sephadex LH-20 column with MeOH. Compounds 3 (4.5 mg, tR = 10.0 min) and 4 (3.4 mg, tR = 14.8 min) were obtained from Fr.3-4 by HPLC purification. (Zorbax SB-C18 column: 250 × 9.4 mm, 5 μm; the mobile phase 55% MeOH; the flow rate 3.0 mL min−1; detection wavelength 300 nm).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 220.0 (2.34), 304.0 (2.48); 1H and 13C NMR data, Table 2; HRESIMS m/z 354.1193 [M − H]− (calcd for C16H20NO8, 354.1194).
ε) 220.0 (2.34), 304.0 (2.48); 1H and 13C NMR data, Table 2; HRESIMS m/z 354.1193 [M − H]− (calcd for C16H20NO8, 354.1194).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 214.0 (2.33), 256.0 (2.40), 288.0 (2.45); 1H and 13C NMR data, Table 2; HRESIMS m/z 360.1047 [M + Na]+ (calcd for C16H19NO7Na, 360.1054).
ε) 214.0 (2.33), 256.0 (2.40), 288.0 (2.45); 1H and 13C NMR data, Table 2; HRESIMS m/z 360.1047 [M + Na]+ (calcd for C16H19NO7Na, 360.1054).Identification of the sugar moiety
Compounds 3 and 4 were hydrolyzed using 5% HCl (10 mL) at 60 °C for 6 h, respectively. Both dissociated sugar moieties were obtained and characterized by HRESIMS (m/z 163.0611 and 163.0608 [M − H]−, calcd for C6H11O5, 163.0612). The identification of L-rhamnose was then carried out by comparison of their specific rotations with that of an authentic sample under the same conditions. The specific rotation of rhamnose moieties from 3 and 4 were [α]D25 −8.6 (c 0.1, MeOH) and [α]D25 −6.9 (c 0.1, MeOH) respectively, in accordance with that of L-rhamnose, which was [α]D25 −4.9 (c 0.1, MeOH).33Conclusions
In this study, we identified a putative actinomycin biosynthetic gene cluster in Kitasatospora sp. YINM00002 through genome mining, and also found small amounts of two new compounds actinrhaters A (3) and B (4). The 4-MHA, a key node in the biosynthetic flow of actinomycins under NRPS catalysis, is also converted into actinrhaters A and B by amidation, glycosylation and cyclization. The discovery of a branch of actinomycin biosynthesis provides a fresh perspective for further discovery and modification of novel actinomycins.Data availability
This strain had been deposited at the Yunnan University under accession number YINM00002. The sequence of the actinomycin biosynthetic gene cluster in strain YINM00002 was deposited in GenBank under accession number OR217453.Author contributions
M. Y., H. Z. and Z. D. designed the study, carried out the data analysis and wrote the manuscript. Z. Z., H. S., and Z. R. carried out the experiments and participated in data analysis. T. X., Y. W., Y. G., X. S. participated in data analysis, all authors have read and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (82160674 and 31860017 for M. Y., 22267001 for Z. D.), Yunnan Revitalization Talent Support Program (YNWR-QNBJ-2019-031 for M. Y., YNWR-QNBJ-2020-096 for Z. R.), Natural Science Foundation of Yunnan Province (202201AS070007, 202001BB050069 and 202002AA100007 for M. Y., 202201AT070225 for H. Z. and 202101BA070001-035 for Z. R.), Yunnan Key Laboratory of Stomatology, The Affiliated Stomatology Hospital of Kunming Medical University (2022YNKQ005 for M. Y.) and Yunnan University Graduate Research Innovation Fund.References
- D. P. Goldstein and R. S. Berkowitz, Hematol./Oncol. Clin. North Am., 2012, 26, 111–131 CrossRef PubMed.
- M. Malogolowkin, C. A. Cotton, D. M. Green, N. E. Breslow, E. Perlman, J. Miser, M. L. Ritchey, P. R. M. Thomas, P. E. Grundy, G. J. D'Angio, J. B. Beckwith, R. C. Shamberger, G. M. Haase, M. Donaldson, R. Weetman, M. J. Coppes, P. Shearer, P. Coccia, M. Kletzel, R. Macklis, G. Tomlinson, V. Huff, R. Newbury and D. Weeks, Pediatr. Blood Cancer, 2008, 50, 236–241 CrossRef PubMed.
- S. X. Skapek, A. Ferrari, A. A. Gupta, P. J. Lupo, E. Butler, J. Shipley, F. G. Barr and D. S. Hawkins, Nature Reviews Disease Primers, 2019, 5, 1 CrossRef PubMed.
- N. Jaffe, D. Traggis, S. Salian and J. R. Cassady, Cancer, 1976, 38, 1925–1930 CrossRef CAS.
- J. Gallego, A. R. Ortiz, B. dePascualTeresa and F. Gago, J. Comput.-Aided Mol. Des., 1997, 11, 114–128 CrossRef CAS PubMed.
- N. R. Bachur, M. V. Gee and S. L. Gordon, Proceedings of the American Association for Cancer Research, 1978, 19, 75–83 Search PubMed.
- Y. S. Lo, W. H. Tseng, C. Y. Chuang and M. H. Hou, Nucleic Acids Res., 2013, 41, 4284–4294 CrossRef CAS PubMed.
- B. Langholz, J. M. Skolnik, J. S. Barrett, J. Renbarger, N. L. Seibel, A. Zajicek and C. A. S. Arndt, Pediatr. Blood Cancer, 2011, 57, 252–257 CrossRef PubMed.
- R. J. Osborne, V. Filiaci, J. C. Schink, R. S. Mannel, A. A. Secord, J. L. Kelley, D. Provencher, D. S. Miller and A. L. Covens, J. Clin. Oncol., 2011, 29, 825–831 CrossRef CAS PubMed.
- F. Pfennig, F. Schauwecker and U. Keller, J. Biol. Chem., 1999, 274, 12508–12516 CrossRef CAS PubMed.
- F. Fawaz and G. H. Jones, J. Biol. Chem., 1988, 263, 4602–4606 CrossRef CAS PubMed.
- M. C. Liu, Y. X. Jia, Y. C. Xie, C. Y. Zhang, J. Y. Ma, C. L. Sun and J. H. Ju, Mar. Drugs, 2019, 17, 240 CrossRef CAS PubMed.
- U. Keller, M. Lang, I. Crnovcic, F. Pfennig and F. Schauwecker, J. Bacteriol., 2010, 192, 2583–2595 CrossRef CAS PubMed.
- I. Crnovcic, C. Ruckert, S. Semsary, M. Lang, J. Kalinowski and U. Keller, Adv. Appl. Bioinf. Chem., 2017, 10, 29–46 Search PubMed.
- J. Bitzer, V. Gesheva and A. Zeeck, J. Nat. Prod., 2006, 69, 1153–1157 CrossRef CAS PubMed.
- W. L. Cai, X. C. Wang, S. I. Elshahawi, L. V. Ponomareva, X. D. Liu, M. R. McErlean, Z. Cui, A. L. Arlinghaus and J. S. Thorson, J. Nat. Prod., 2016, 79, 2731–2739 CrossRef CAS PubMed.
- N. V. Machushynets, S. S. Elsayed, C. Du, M. A. Siegler, M. de la Cruz, O. Genilloud, T. Hankemeier and G. P. van Wezel, Sci. Rep., 2022, 12, 2813 CrossRef CAS PubMed.
- W. Z. Zhao, G. F. Wang, L. Guo, J. M. Wang, C. C. Jing, B. Liu, F. Zhao, S. M. Zhang and Z. P. Xie, Org. Biomol. Chem., 2023, 21, 1737–1743 RSC.
- Q. Wang, Y. Zhang, M. Wang, Y. Tian, X. X. Hu, H. W. He, C. L. Xiao, X. F. You, Y. G. Wang and M. L. Gan, Sci. Rep., 2017, 7, 3591 CrossRef PubMed.
- Y. Q. Chen, J. J. Liu, B. Yuan, C. L. Cao, S. Qin, X. Y. Cao, G. K. Bian, Z. Wang and J. H. Jiang, Mol. Carcinog., 2013, 52, 983–996 CrossRef CAS PubMed.
- W. H. Jiao, W. Yuan, Z. Y. Li, J. Li, L. Li, J. B. Sun, Y. H. Gui, J. Wang, B. P. Ye and H. W. Lin, Tetrahedron, 2018, 74, 5914–5919 CrossRef CAS.
- M. Dong, P. Cao, Y. T. Ma, J. Y. Luo, Y. J. Yan, R. T. Li and S. X. Huang, Nat. Prod. Res., 2019, 33, 219–225 CrossRef CAS PubMed.
- D. D. Brown, M. J. M. Hitchcock and E. Katz, Arch. Biochem. Biophys., 1980, 202, 18–22 CrossRef CAS PubMed.
- D. Brown, M. J. M. Hitchcock and E. Katz, Can. J. Microbiol., 1986, 32, 465–472 CrossRef CAS PubMed.
- J. W. Foster and E. Katz, J. Bacteriol., 1981, 148, 670–677 CrossRef CAS PubMed.
- A. Haese and U. Keller, J. Bacteriol., 1988, 170, 1360–1368 CrossRef CAS PubMed.
- T. Troost, M. J. M. Hitchcock and E. Katz, Biochim. Biophys., 1980, 612, 97–106 CrossRef CAS PubMed.
- R. S. Li, A. E. Tao, R. M. Yang, M. Fan, X. C. Zhang, Z. F. Du, F. N. Shang, C. L. Xia and B. Z. Duan, Biomed. Pharmacother., 2020, 131, 110687 CrossRef CAS PubMed.
- X. F. Zhang, X. W. Ye, W. Y. Chai, X. Y. Lian and Z. Z. Zhang, Mar. Drugs, 2016, 14, 181 CrossRef PubMed.
- L. Lartigue, K. Oumzil, Y. Guari, J. Larionova, C. Guérin, J. L. Montero, V. Barragan-Montero, C. Sangregorio, A. Caneschi, A. C. Innocenti, T. Kalaivani, P. Arosio and A. Lascialfari, Org. Lett., 2009, 11, 2993–2995 CrossRef PubMed.
- R. Li, M. Wang, Z. Ren, Y. Ji, M. Yin, H. Zhou and S. K. Tang, Front. Microbiol., 2021, 12, 743116 CrossRef PubMed.
- K. Blin, S. Shaw, A. M. Kloosterman, Z. Charlop-Powers, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, W12–W35 CrossRef PubMed.
- M. Jiang, Y. Zhang, Y. X. Zhang, Z. J. Ma and J. H. Wang, J. Nat. Prod., 2022, 85, 1771–1778 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: 1D, 2D NMR and HRESIMS spectra of actinrhaters A and B, along with other details. See DOI: https://doi.org/10.1039/d3ra07277k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |