 Open Access Article
Open Access ArticleEffect of “magic chlorine” in drug discovery: an in silico approach†
Sravani Joshi and
Ruby Srivastava *
*
Centre for Cellular and Molecular Biology-CSIR, Hyderabad, India. E-mail: amitruby1@gmail.com
First published on 30th November 2023
Abstract
The chlorine atom plays a vital role in drug design, yet the benefits of chlorine in 250 FDA-approved chlorine-containing drugs have not been studied properly. To see the “magic chloro” effect, computational studies have been carried out for 35 inhibitors, which are numbered as 12 complexes with (parent (–H), one chlorine, or two chlorine) substituents. The physicochemical properties are studied by conceptual density functional theory (CDFT). The pharmacokinetics, toxicity and metabolic properties of the studied inhibitors are estimated using chemoinformatics tools. SwissTargetPrediction is used to predict the multitarget activities of the studied inhibitors. Four FDA-approved drugs, diazepam, chloroquine, chloramphenicol, and bendamustine, are referenced to validate the studies. A higher HOMO–LUMO gap predicted high stability for the studied one and two chlorine-substituted analogues. Most of the studied inhibitors show “drug likeliness”, nontoxicity, and high gastrointestinal (GI) absorption. The addition of one or two chloro substituents has increased the physicochemical properties and stability of most of the inhibitors compared to the parent analogues, whereas the toxicity is not affected. No change in metabolic properties is observed on addition of one or two chlorine substituents. The multi-target activities of all the studied inhibitors are validated by the reference drugs and experimental results.
Introduction
Halogen atoms play a central role in many drug discovery applications as halogens increase the sensitivity and enhance physicochemical properties in small-molecule drugs. Halogen bond formation involving side-chain groups is essential for drug design. Halogen bonds with hit-to-lead candidate optimization have improved drug target binding affinity.1 The unique features of halogen bonds are their directionality, strength tunability, hydrophobicity, and donor atom dimensions which allow interaction for designing self-assembled systems.1–5 It is observed that a positively charged electrostatic region on the extension of C–X bonds (a σ hole) attracts a nucleophile due to the anisotropy of the charge distribution (halogen atom-X).6,7 Studies indicate that fluorine and chlorine atoms enhance the physicochemical properties of the complexes, whereas bromine and iodine are added to improve the selectivity of the complexes.2 Like chlorine and fluorine, sulfur is also used as a potential candidate for drugs,8 light harvesting,9 and non-linear optical activities.10–12 In medicinal chemistry, the potency of a drug against a biological target decides the daily dosage for patients.13 The potency of drugs can be improved by larger structural changes, but they unpredictably change parameters such as water solubility and cell membrane permeability. Pharmacokinetic parameters such as clearance, half-life, and distribution in vivo are also affected when the chemical structure is changed or modified. Presently 250 FDA-approved drugs containing chlorine are used to design novel small-molecule drugs.14,15 Drug interactions are easy to understand with the structure–activity relationship (SAR) and the pharmacophore of a compound. A chlorine substituent can act as a bioisostere not only by replacing another halide, but also as a monovalent substituent to replace OH or SH, or as a pseudohalide to replace CF3 or CN. A chlorine substituent can easily be added to an aromatic complex with hydrogen rather than replacing it with a –F, –CH3, or –CF3 group.16,17 Chlorine can act as either an electron-donating group or an electron-withdrawing group, or a quasielectron-neutral group depending on the scaffold.18,19 As it takes approximately 13 years to launch one new molecular entity (NME) with an average cost of $1778 million, consisting of the drug discovery stage (5.5 years) and the drug development stage (8 years), it is necessary to reduce this timespan and huge cost by exploiting advanced computational methods for drug design.20,21Computational tools provide a better option for drug design and development as these tools are cost effective and provide significant information in a limited timeframe. Computational methods are used to predict the geometries, structures, and physicochemical and biological properties of drugs. Density functional theory (DFT) techniques can extract useful information with high accuracy for biological systems in less time. A profound drug effect can also be anticipated with effective target-drug binding sites.22–26 Chemical reactivity (DFT-CR), often called conceptual DFT,27–30 is a well-established method31–37 that helps us to understand various physicochemical processes with several global and local reactivity descriptors. DFT-CR defined a quantitative scale for chemical hardness,38 which led to an increase in the understanding of hard–soft acid–base (HSAB) theory and an entirely new principle for chemical reactivity and molecular stability: the maximum hardness principle (MHP).39–48 Here it is necessary to mention that the principles of chemical reactivity may have exceptions. Therefore, instead of principles, “rules of thumb” can be applied for chemical reactivity. DFT-CR-based applications use a one-reagent approach. The computed response functions of a reactant molecule are used to predict its reactivity. Despite the usefulness of this approach, it sometimes fails and is sometimes insufficient for understanding the inherent reactivity of a molecule; an anticipation of how “well-matched two reagents are” is also needed.
To see the “magic chloro effect”, we selected 35 inhibitors with >1000-fold potency, which are divided into 12 groups. Groups 1–3 and 5–8 have two inhibitors (one parent (a) and the other one a chlorine-substituent analogue (b)); group 4 has parent –Cl (a) and (b), (c) with one more –Cl at a different position; groups 9–12 have three inhibitors (one parent (a), one chlorine substituent (b), and two chlorine substituent (c) analogues).49 Different groups are taken to study the effect of one and two chloro groups. Four FDA-approved drugs are taken as references to validate our results for the effect of one chloro (Ref 1 (diazepam) and Ref 2 (chloroquine)) and two chloro (Ref 3 (chloramphenicol) and Ref 4 (bendamustine)), respectively. Ref 1 (diazepam) is anxiolytic, Ref 2 (chloroquine) is anti-malarial, Ref 3 (chloramphenicol) is an antibiotic and Ref 4 (bendamustine) is an oncology drug.
Computational methods
Various conformational structures of the four Refs (1–4) and 35 inhibitors were optimized with the wB97XD50/6-31G(d,p) method in the G09 (ref. 51) software suite and lower minima structures with no negative vibrational frequency were considered for further study. The studies were performed with the integral equation formalism–polarized continuum model (IEF-PCM) SMD solvation model52 with water as a solvent, as the parent analogues of the studied inhibitors and reference drugs are taken orally.31–33 GaussView53 was used to draw the optimized structures, electrostatic potentials (ESP) and frontier molecular orbitals (FMOs) for all the studied complexes. FMOs play a vital role in various systems due to the location of chemically reactive bonds with their associated orbital energies.54–56 The physicochemical properties of the inhibitors were estimated with Molinspiration57 software. The Osiris software program58 was used to evaluate the pharmacokinetic properties and mutagenic toxicity for the viability of the inhibitors. The cardiotoxicity of the inhibitors was studied with the cardioToxCSM59 webserver, which uses the concepts of molecular fingerprints and graph-based signatures. There are various different cross-validation schemes which validate the cardioToxCSM results. The six types of cardiotoxicity of arrhythmia, cardiac failure, heart block, hERG toxicity, hypertension, and myocardial infarction are predicted with cardioToxCSM. Other pharmacokinetic properties, such as gastrointestinal (GI) absorption, permeation of the blood–brain barrier (BBB), and metabolic processes, are predicted with SwissADME BOILED-Egg60 tools. The toxicity, effectiveness, and clearance are studied for the majority of Phase I metabolic transformations with cytochrome P450 enzyme (CYP)-related parameters. Various potential macromolecular bioactive targets of all the studied inhibitors were studied with SwissTargetPrediction61 to determine the molecular properties related to druggability for a variety of drug targets (GPCR ligands, kinase inhibitors, ion channel modulators, enzymes, and nuclear receptors) through a homology modeling procedure. The targets were predicted from a combination of 2D and 3D similarity to a large number of 370![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 active complexes on more than 3000 proteins from different target species.
000 active complexes on more than 3000 proteins from different target species.
Results
The SMILES notation for the four reference drugs and 35 studied inhibitors is given in Table 1. The selected 35 inhibitors with >1000-fold potency were divided into 12 groups. Groups 1–3 and 5–8 have two inhibitors with –H (a) and –Cl (b) substituents; group 4 has –Cl attached (a), one more attached –Cl (b), and –Cl (c) at different positions; while groups 9–12 have –H (a), –Cl substituent (b), and two –Cl substituents (c). The optimized structures of the four reference drugs and 35 studied inhibitors are given in Fig. 1. The reactive sites of the four reference drugs and studied inhibitors were determined from the molecular electrostatic potential map (MEP), which is given in Fig. 2. For the studied inhibitors, the areas in red around the oxygen indicate the sites with the most substantial negative charge (more electron density); and the ones in blue correspond to sites that have a positive charge (less electron density). It can be seen in the studied inhibitors that there is an increase in the electron density at the sites where chlorine is substituted, which means that the addition of chlorine increases the ability to interact with electrophilic molecules and nucleophilic molecules (carbon atoms with low electron density). The ESP of reference drugs shows no correlation with regard to electron density towards the studied inhibitors, as only the ESP surfaces of Ref 2 and Ref 4 indicate an increase in electron density towards mono and dichloro substituents, respectively.| Complex | SMILES notation |
|---|---|
| Ref 1 | CN1C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CN O)CN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C2 C(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C C1C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)Cl)C3 C2)Cl)C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3 (diazepam) C3 (diazepam) |
| Ref 2 | Clc1cc2nccc(c2cc1)NC(C)CCCN(CC)CC (chloroquine) |
| Ref 3 | C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C(C(CO)NC( C1C(C(CO)NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C(Cl)Cl)O)[N+]( O)C(Cl)Cl)O)[N+](![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)[O–] (chloramphenicol) O)[O–] (chloramphenicol) |
| Ref 4 | CN1C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)N(CCCl)CCCl)N C2)N(CCCl)CCCl)N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1CCCC( C1CCCC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O (bendamustine) O)O (bendamustine) |
| 1a | COC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C1 O)C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC( C(NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)\C( O)\C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N\NC2 N\NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C#N)SC( C2)C#N)SC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)[N]3N C1)[N]3N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(O)C C(O)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3N C3N |
| 1b | COC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C1 O)C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC( C(NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)\C( O)\C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N\NC2 N\NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C C(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C#N)SC( C2)C#N)SC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)[N]3N C1)[N]3N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(O)C C(O)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3N C3N |
| 2a | CCOC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)c1c([O–])c2c(cc(OC)cc2)[nH+]c1c1ccccc1 O)c1c([O–])c2c(cc(OC)cc2)[nH+]c1c1ccccc1 |
| 2b | CCOC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)c1c([O–])c2c(cc(OC)cc2)[nH+]c1c1cc(ccc1)Cl O)c1c([O–])c2c(cc(OC)cc2)[nH+]c1c1cc(ccc1)Cl |
| 3-1a | Cc1c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c([nH]1)cccc2 O)O)c2c([nH]1)cccc2 |
| 3-1b | Cc1c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c([nH]1)cc(Cl)cc2 O)O)c2c([nH]1)cc(Cl)cc2 |
| 3-2a | Cn1c(c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cccc2)C O)O)c2c1cccc2)C |
| 3-2b | Cn1c(c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cc(Cl)cc2)C O)O)c2c1cc(Cl)cc2)C |
| 3-3a | Cn1ccc(n1)n1c(c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cccc2)C O)O)c2c1cccc2)C |
| 3-3b | Cn1ccc(n1)n1c(c(Sc2cccc(n2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cc(Cl)cc2)C O)O)c2c1cc(Cl)cc2)C |
| 3-4a | Cn1ccc(n1)n1c(c(Sc2cccc(c2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cccc2)C O)O)c2c1cccc2)C |
| 3-4b | Cn1ccc(n1)n1c(c(Sc2cccc(c2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O)c2c1cc(Cl)cc2)C O)O)c2c1cc(Cl)cc2)C |
| 4a | Oc1c(NNC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)c2c(O)ccc(c2)Cl)c2c([nH]1)cccc2 O)c2c(O)ccc(c2)Cl)c2c([nH]1)cccc2 |
| 4b | Oc1c(NNC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)c2c(O)ccc(c2)Cl)c2c([nH]1)cccc2 O)c2c(O)ccc(c2)Cl)c2c([nH]1)cccc2 |
| 4c | Oc1c(NNC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)c2c(O)ccc(c2)Cl)c2c([nH]1)cc(Cl)cc2 O)c2c(O)ccc(c2)Cl)c2c([nH]1)cc(Cl)cc2 |
| 5a | CCN(CCC1(O)CC(C1)NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C2 O)C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3C C3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC3 CC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C4CC4C5 C2)C4CC4C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5 C5 |
| 5b | CCN(CCC1(O)CC(C1)NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C2 O)C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3C C3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC3 CC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C4CC4C5 C2)C4CC4C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C C(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5 C5 |
| 6a | Oc1ccc(cc1)N1SCC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1O C1O |
| 6b | Oc1ccc(cc1)N1SC(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1O C1O |
| 7a | CN1CCN(CC1)C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C( C2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC3 O)NC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N[NH]C4 N[NH]C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NC( NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C34)C5 C34)C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5)O)O C5)O)O |
| 7b | CN1CCN(CC1)C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C( C2)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC3 O)NC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N[NH]C4 N[NH]C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NC( NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C34)C5 C34)C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C( C(Cl)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5Cl)O)O C5Cl)O)O |
| 8a | CCOc1c(cccc1)/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/c1nc2c(cccc2O)cc1 C/c1nc2c(cccc2O)cc1 |
| 8b | CCOc1c(cccc1)/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/c1nc2c(cc1)c(cc(c2O)Cl)Cl C/c1nc2c(cc1)c(cc(c2O)Cl)Cl |
| 9a | O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NCCCN1CCOCC1)C2 C(NCCCN1CCOCC1)C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 C2 |
| 9b | ClC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C( C1)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NCCCN2CCOCC2 O)NCCCN2CCOCC2 |
| 9c | ClC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C C(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C( C1)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NCCCN2CCOCC2 O)NCCCN2CCOCC2 |
| 10a | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C C(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C( C1C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC2 O)NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 C2 |
| 10b | Oc1ccc(Cl)cc1C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)Nc1ccccc1 O)Nc1ccccc1 |
| 10c | Oc1ccc(Cl)cc1C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)Nc1cc(c(Cl)cc1)Cl O)Nc1cc(c(Cl)cc1)Cl |
| 11a | C1CC(CCN1)c1c(Oc2ccccc2)nccc1 |
| 11b | Clc1c(Oc2c(cccn2)C2CCNCC2)cccc1 |
| 11c | Clc1cccc(c1Cl)Oc1c(cccn1)C1CCNCC1 |
| 12a | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(O)C( C(O)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C( C1)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC2 O)NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C([NH]C3 C([NH]C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)C( C3)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C4 O)C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4) C4) |
| 12b | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(O)C( C(O)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C( C1)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC2 O)NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C([NH]C3 C([NH]C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)Cl)C( C3)Cl)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C4 O)C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4)O C4)O |
| 12c | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(O)C( C(O)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C( C1)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC2 O)NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C([NH]C3 C([NH]C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)Cl)C( C3)Cl)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C4 O)C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(Cl)C C(Cl)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4)O C4)O |
 | ||
| Fig. 1 Structures for the reference drugs Ref (1–4) and 12 groups of studied inhibitors optimized with the DFT (wB97XD/6-31G(d,p)) method using G09 Software. | ||
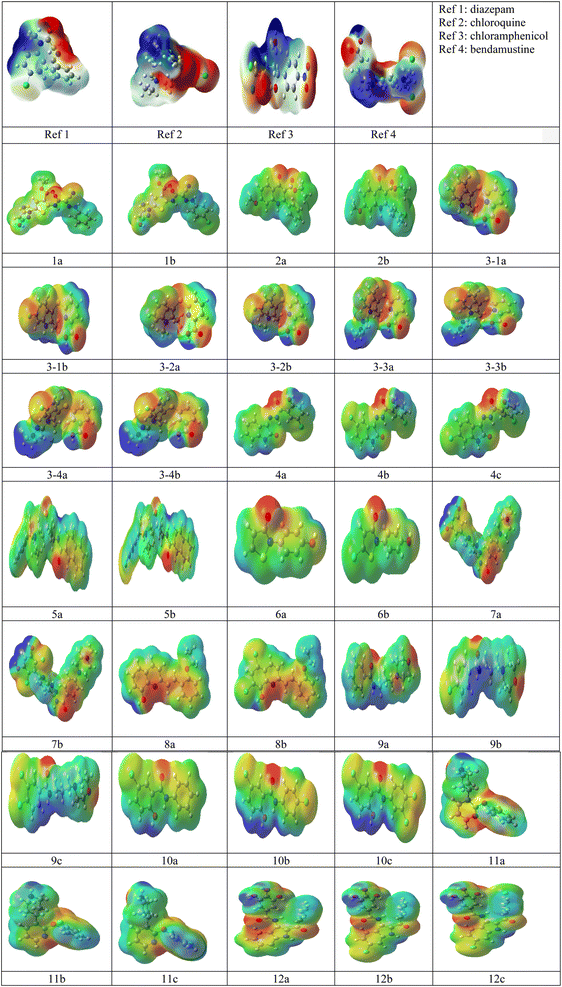 | ||
| Fig. 2 Electrostatic potential (ESP) diagrams for the reference drugs Ref (1–4) and 12 groups of studied inhibitors produced with G09 software using the DFT (wB97XD/6-31G(d,p)) method. | ||
The calculated descriptors (ionization energy (lP), electron affinity (EA), global hardness (η), electronegativity (χ), electrophilicity index (ω), chemical potential (μp), HOMO–LUMO (HL) gap, electron donating (ω−), electron accepting (ω+), net electrophilicity (Δω±), and softness (S)) for the four reference drugs and 12 groups of studied inhibitors are given in Table 2. The formulae used for calculating these parameters are given in the ESI.† IP represents the energy to remove an electron from a molecule which is computed from the energy difference between the cation and the neutral molecule. The lower IP energy to remove an electron from the molecule indicates easier ionization of the molecule. Generally, IP is affected by the size of the molecule. For higher molecular size, the IP is lower and for smaller molecular size the IP is higher. EA is the energy released when an electron is added to a neutral molecule. It is computed as the energy difference between the neutral form and the anion. In our studied inhibitors, higher IP values and lower EA values are observed for (one chloro) substituent compared to the –H parent analogues. The IP values are higher for 4a, 4b, and 4c, in which the parent analogue already has a –Cl substituent and the other two inhibitors have one more chlorine added at different positions, 4b and 4c. The change of chlorine at two different positions has not affected the IP values; rather EA values have increased on the addition of chlorine at different positions. The IP values of Ref 1 and Ref 2 are higher and are positively correlated with the studied complexes, but the EA values of the reference drugs are negatively correlated with the studied inhibitors.
| Complex | IP | EA | η | χ | ω | μp | HL gap | ω− | ω+ | Δω± | S |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref 1 | 8.36 | 0.14 | 4.11 | 4.25 | 2.20 | −4.25 | 8.22 | 6.31 | 2.20 | 8.51 | 0.240 |
| Ref 2 | 9.18 | 0.69 | 4.24 | 4.93 | 2.87 | −4.93 | 8.49 | 7.05 | 2.81 | 9.87 | 0.240 |
| Ref 3 | 7.65 | 0.43 | 3.61 | 4.04 | 2.26 | −4.04 | 7.22 | 5.84 | 2.23 | 8.08 | 0.280 |
| Ref 4 | 7.00 | 1.30 | 2.85 | 4.15 | 3.03 | −4.15 | 5.70 | 5.58 | 2.73 | 8.31 | 0.350 |
| 1a | 7.76 | 0.68 | 3.54 | 4.22 | 1.26 | −4.22 | 7.08 | 5.99 | 2.45 | 8.45 | 0.282 |
| 1b | 7.77 | 0.60 | 3.58 | 4.18 | 1.22 | −4.18 | 7.16 | 5.97 | 2.39 | 8.36 | 0.279 |
| 2a | 8.10 | 0.15 | 3.98 | 4.13 | 1.07 | −4.13 | 7.95 | 6.11 | 2.14 | 8.25 | 0.252 |
| 2b | 8.15 | 0.22 | 3.97 | 4.18 | 1.10 | −4.18 | 7.93 | 6.17 | 2.20 | 8.37 | 0.252 |
| 3-1a | 7.55 | 0.04 | 3.79 | 3.76 | 0.93 | −3.76 | 7.58 | 5.65 | 1.86 | 7.51 | 0.264 |
| 3-1b | 7.63 | 0.02 | 3.83 | 3.80 | 0.95 | −3.80 | 7.65 | 5.72 | 1.89 | 7.61 | 0.261 |
| 3-2a | 7.49 | 0.04 | 3.77 | 3.72 | 0.92 | −3.72 | 7.53 | 5.61 | 1.84 | 7.45 | 0.266 |
| 3-2b | 7.60 | 0.03 | 3.81 | 3.78 | 0.94 | −3.78 | 7.62 | 5.69 | 1.88 | 7.57 | 0.262 |
| 3-3a | 7.57 | 0.03 | 3.80 | 3.77 | 0.94 | −3.77 | 7.60 | 5.67 | 1.87 | 7.54 | 0.263 |
| 3-3b | 7.66 | 0.01 | 3.84 | 3.83 | 0.95 | −3.83 | 7.68 | 5.74 | 1.91 | 7.65 | 0.260 |
| 3-4a | 7.66 | 0.43 | 4.05 | 3.62 | 0.81 | −3.62 | 8.10 | 5.64 | 1.59 | 7.23 | 0.247 |
| 3-4b | 7.76 | 0.42 | 4.09 | 3.67 | 0.82 | −3.67 | 8.17 | 5.71 | 1.63 | 7.34 | 0.245 |
| 4a | 8.13 | 1.77 | 3.18 | 4.95 | 1.93 | −4.95 | 6.36 | 5.72 | 1.55 | 7.27 | 0.240 |
| 4b | 8.18 | 1.89 | 3.15 | 5.04 | 2.01 | −5.04 | 6.29 | 5.77 | 1.69 | 7.46 | 0.245 |
| 4c | 8.18 | 1.88 | 3.15 | 5.03 | 2.00 | −5.03 | 6.31 | 6.54 | 3.36 | 9.91 | 0.315 |
| 5a | 7.79 | 0.20 | 3.79 | 3.99 | 1.05 | −3.99 | 7.58 | 6.61 | 3.46 | 10.07 | 0.318 |
| 5b | 7.81 | 0.20 | 3.81 | 4.01 | 1.06 | −4.01 | 7.61 | 6.61 | 3.45 | 10.06 | 0.317 |
| 6a | 7.95 | 1.07 | 4.51 | 3.44 | 0.66 | −3.44 | 9.02 | 5.89 | 2.10 | 7.99 | 0.264 |
| 6b | 8.21 | 0.78 | 4.49 | 3.72 | 0.77 | −3.72 | 8.99 | 5.91 | 2.11 | 8.02 | 0.263 |
| 7a | 7.50 | 0.89 | 3.31 | 4.20 | 1.33 | −4.20 | 6.61 | 5.70 | 1.19 | 6.89 | 0.222 |
| 7b | 7.51 | 1.03 | 3.24 | 4.27 | 1.41 | −4.27 | 6.48 | 5.96 | 1.47 | 7.43 | 0.222 |
| 8a | 7.44 | 0.19 | 3.63 | 3.81 | 1.00 | −3.81 | 7.25 | 5.85 | 2.54 | 8.39 | 0.302 |
| 8b | 7.61 | 0.47 | 3.57 | 4.04 | 1.14 | −4.04 | 7.15 | 5.89 | 2.65 | 8.54 | 0.308 |
| 9a | 8.13 | 0.94 | 4.53 | 3.60 | 0.71 | −3.60 | 9.07 | 5.63 | 2.00 | 7.63 | 0.276 |
| 9b | 8.14 | 0.41 | 4.27 | 3.86 | 0.87 | −3.86 | 8.54 | 5.83 | 2.25 | 8.08 | 0.280 |
| 9c | 8.14 | 0.19 | 4.17 | 3.98 | 0.95 | −3.98 | 8.33 | 5.86 | 1.33 | 7.19 | 0.221 |
| 10a | 8.06 | 0.16 | 4.11 | 3.95 | 0.95 | −3.95 | 8.22 | 6.00 | 1.73 | 7.73 | 0.234 |
| 10b | 8.08 | 0.11 | 4.10 | 3.99 | 0.97 | −3.99 | 8.19 | 6.06 | 1.89 | 7.95 | 0.240 |
| 10c | 8.24 | 0.17 | 4.09 | 4.04 | 0.97 | −4.04 | 8.41 | 6.01 | 1.89 | 7.90 | 0.243 |
| 11a | 8.26 | 1.22 | 4.74 | 3.52 | 0.65 | −3.52 | 9.48 | 6.04 | 1.94 | 7.98 | 0.244 |
| 11b | 8.25 | 1.21 | 4.73 | 3.52 | 0.65 | −3.52 | 9.46 | 6.14 | 1.94 | 8.07 | 0.238 |
| 11c | 8.25 | 1.03 | 4.64 | 3.61 | 0.70 | −3.61 | 9.28 | 5.89 | 1.15 | 7.04 | 0.211 |
| 12a | 7.57 | 0.29 | 3.64 | 3.93 | 1.06 | −3.93 | 7.28 | 5.88 | 1.15 | 7.04 | 0.212 |
| 12b | 7.67 | 0.39 | 3.64 | 4.03 | 1.12 | −4.03 | 7.28 | 5.93 | 1.29 | 7.23 | 0.215 |
| 12c | 7.71 | 0.47 | 3.62 | 4.09 | 1.16 | −4.09 | 7.24 | 5.75 | 2.11 | 7.86 | 0.275 |
In complexes 9–12 where one chlorine (b) and two chlorines are added (c), no correlation pattern is observed for the IP and EA values. The IP values of 9, 10, and 11 are increased for one and two chlorine analogues compared to the parent analogue (9a, 10a, 12a). The EA values are decreased for one and two chloro substituents of 9 and 11, while the EA values are increased for one chlorine (12b) and two chlorine substituents (12c) compared to the parent analogue (12a). The EA values are lower for one chlorine inhibitor 10b, whereas inhibitor 10c has higher EA values. Similarly Ref 3 and Ref 4 have lower IP values and higher EA values compared to the two chlorine substituents. For inhibitors 1, 3, 10, and 11, IP and hardness display the same trend. Larger IP values and considerable η values show that the systems are hard and it is difficult to donate one electron. The energy for donation has the highest value corresponding to the acceptance of an electron charge.
The stability of a molecular system is also measured by the chemical hardness (η), as this global descriptor is the resistance of a molecule to intramolecular charge transfer. Chemical hardness allows a quantitative classification of the global electrophilic nature of a molecule within a relative scale. Chemical hardness is higher for most of the inhibitors with a chlorine substituent except for 2b, 4b, 6b, 7b, and 8b. Ref 1 and Ref 2 also have higher values for chemical hardness. In inhibitors 9–12, the hardness values are lower for the one (b) and two chlorine (c) substituents compared to the –H analogues (a). Chemical hardness is lower for Ref 3 and Ref 4, which correlates well with the hardness values for two chloro substituents. The electrophilicity index (ω) is a measure of energy lowering due to the maximum flow of electrons between the donor and acceptor, and measures the stabilization energy when a system attracts electronic charge.63 The electrophilicity index also measures the electron-accepting capacity by molecules. It predicts the responsiveness of the type of reaction a reagent undergoes.64 In the studied complexes, inhibitors with one chlorine and two chlorine substituent have a higher electrophilicity index compared to the –H analogues, except for 1. The reference drugs Ref 1 to Ref 4 have a larger electrophilicity index. The chemical potential is the negative of the electronegativity.65 It is an indicator for a chemical reaction to take place. This means that a reagent with high electronic chemical potential is a good electron donor, whereas a reagent with a small electronic chemical potential is a good electron acceptor. A molecule with a good electrophilicity index has low chemical potential and low chemical hardness.65 Inhibitors with one and two chlorine substituents have higher chemical potential compared to the –H analogue, which shows that these inhibitors are good electron donors. The higher chemical values of the studied inhibitors correlate well with the reference drugs.
The hardness and softness are measured by the HOMO–LUMO (HL) energy gap with the hard–soft acid–base (HSAB) principle as proposed by Pearson,62 which indicates a large energy gap for a hard molecule and a small energy gap for a soft molecule. The HSAB principle also states that soft systems could interact more easily with electrophiles or nucleophiles. As compared to the –H analogues, the –Cl inhibitors have higher values for the HL gap, meaning higher stability of the inhibitors with one chlorine attached (b). A similar trend is not observed for 9–12 with one chlorine and two chlorine substituents. The HL gaps for one chloro substituent are higher than the HL gap for two chloro substituents for reference drugs Ref 1–4. The (HL) energy gap is lower for the inhibitors with one chlorine and two chlorine substituents, except for 1 and 3, which have a higher energy gap for one chlorine substituent compared to their parent analogue. In 10, the trend is a little different, as the inhibitor with two chlorine substituents (10c) has a higher HL gap than 10a or 10b. The energy gap value indicates the hardness or softness of a molecule according to the hard–soft acid–base (HSAB) principle.65 An increase in the energy gap of these inhibitors predicts that they behave more as hard acids, so they should react more readily with hard bases. If we look at the FMO orbitals, for one chloro substituent, the HOMO is localized at the Cl-substituted region for some inhibitors and for the others the LUMO is localized at the Cl-substituted region. No particular trend is observed for the localization of HOMO and LUMO for inhibitors with two chloro substituents and the FMOs of the reference drugs. See ESI Fig. 1.†
Electron-withdrawing power indicates the ability of an atom to pull an electron away from another atom. An atom with higher electronegativity and usually a smaller radius will have greater electron-withdrawing power. All the reference drugs and studied inhibitors show lower electron-withdrawing power for the one chloro and two chloro substituents. The electron-donating power is higher for the reference drugs and studied inhibitors. The net electrophilicity, which is an electron-accepting power relative to its electron-donating power, has higher values. Another global descriptor, global chemical softness (S), is the reciprocal of hardness, and it measures the tendency of a system to react. Softness values are in the range of 0.240–0.318 for the reference drugs and the studied inhibitors.
The possible fate of the drug is predicted by the pharmacokinetics of the therapeutics. These properties play a vital role in the drug development process. The individual absorption, distribution, metabolism, and excretion (ADME) parameters can easily be obtained using various computational tools that can be used as an effective alternative to the experimental methods. The bioavailability, metabolic stability, toxicity, and transport drug-like properties of a viable candidate depend on the compound's size, molecular flexibility, hydrophobicity, and the distribution of electronic bonds. The estimated parameters of clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, molecular weight (MW), topological polar surface area (TPSA), hydrogen bond donors (HBDs), hydrogen bond acceptors (HBAs) and number of rotatable bonds (nrotb) are given in ESI Table 1.† The logarithm of the octanol–water partition coefficient of a compound, the log
P, molecular weight (MW), topological polar surface area (TPSA), hydrogen bond donors (HBDs), hydrogen bond acceptors (HBAs) and number of rotatable bonds (nrotb) are given in ESI Table 1.† The logarithm of the octanol–water partition coefficient of a compound, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is considered a reliable measure of the compound's hydrophilicity in drug design.66 Low hydrophilicity corresponds to a high log
P value is considered a reliable measure of the compound's hydrophilicity in drug design.66 Low hydrophilicity corresponds to a high log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value. Studies showed that a compound is likely to be well absorbed by an organism if its log
P value. Studies showed that a compound is likely to be well absorbed by an organism if its log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value is less than 5. All the inhibitors have log
P value is less than 5. All the inhibitors have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values < 5. The HBD (total number of nitrogen–hydrogen and oxygen–hydrogen bonds) is less than 5, HBA (total number of nitrogen and oxygen atoms) is less than 10 and nrotb is less than or equal to 7 for all the studied inhibitors. The value of nrotb determines the flexibility of the molecule and can predict its oral bioavailability. Previous studies in rats showed that 10 or fewer rotatable bonds in a molecule indicated good oral bioavailability for more than 1100 drugs.67,68 The molecular mass (150–500) g mol−1 shows that all the inhibitors are “drug like”. The total polar surface area (TPSA) predicts the transport of a drug through membranes, which includes its intestinal absorption and crossing of the BBB.69 TPSA < 90 Å2 is needed to penetrate the BBB, and thus acts on receptors in the central nervous system, which is reflected in complexes 2, 3, 5, 6, 8, 9, 10, and 11. All the other complexes with 90 Å2 < TPSA value > 140 Å2 or more show poor intestinal absorption, which is seen in 4, 6, and 12. FDA-approved drugs Ref 1 to Ref 4 follow the Lipinski rule, with TPSA < 60 Å2 for Ref 1, 3, and 4; and TPSA > 120 Å2 for Ref 2. Osiris Property Explorer also predicts the potential (mutagenic, tumorigenic, irritant, and reproductive) risks of the inhibitors. Other properties, such as hydrogen bonding, hydrophobicity, electronic distribution, flexibility, size of molecules, and the presence of different pharmacophores, affect the behaviour of drugs. The druglikeliness (DL), drug score (DS), solubility, and toxicity (MUT, TUM, IRRI, REP) of the reference drugs and studied inhibitors are given in ESI Table 2.† Toxicity is a very important factor, which sometimes dominates ADME behaviour. The drugs are failed at the clinical trial stage due to adverse toxic effects. Various computational tools offer toxicity prediction for drugs so that they help to accelerate the discovery of new targets and ultimately lead to compounds with effective biological activities. Most of the complexes showed nontoxic behaviour, except 9 and 10, which are highly toxic in terms of mutagenicity, tumorigenicity, irritant effect, and reproductive effects. Other inhibitors which showed toxicity are 1a (high-TUM), 1b (mild-REP), 6a (high-MUT), 7b (high-TUM, REP), 8b (high-MUT, TUM, REP), 11b (mild-IRR), and 11c (high-IRR, REP). Ref 1 and Ref 3 showed nontoxicity for (IRR) and Ref 3 is safe for use (TUM, REP).
P values < 5. The HBD (total number of nitrogen–hydrogen and oxygen–hydrogen bonds) is less than 5, HBA (total number of nitrogen and oxygen atoms) is less than 10 and nrotb is less than or equal to 7 for all the studied inhibitors. The value of nrotb determines the flexibility of the molecule and can predict its oral bioavailability. Previous studies in rats showed that 10 or fewer rotatable bonds in a molecule indicated good oral bioavailability for more than 1100 drugs.67,68 The molecular mass (150–500) g mol−1 shows that all the inhibitors are “drug like”. The total polar surface area (TPSA) predicts the transport of a drug through membranes, which includes its intestinal absorption and crossing of the BBB.69 TPSA < 90 Å2 is needed to penetrate the BBB, and thus acts on receptors in the central nervous system, which is reflected in complexes 2, 3, 5, 6, 8, 9, 10, and 11. All the other complexes with 90 Å2 < TPSA value > 140 Å2 or more show poor intestinal absorption, which is seen in 4, 6, and 12. FDA-approved drugs Ref 1 to Ref 4 follow the Lipinski rule, with TPSA < 60 Å2 for Ref 1, 3, and 4; and TPSA > 120 Å2 for Ref 2. Osiris Property Explorer also predicts the potential (mutagenic, tumorigenic, irritant, and reproductive) risks of the inhibitors. Other properties, such as hydrogen bonding, hydrophobicity, electronic distribution, flexibility, size of molecules, and the presence of different pharmacophores, affect the behaviour of drugs. The druglikeliness (DL), drug score (DS), solubility, and toxicity (MUT, TUM, IRRI, REP) of the reference drugs and studied inhibitors are given in ESI Table 2.† Toxicity is a very important factor, which sometimes dominates ADME behaviour. The drugs are failed at the clinical trial stage due to adverse toxic effects. Various computational tools offer toxicity prediction for drugs so that they help to accelerate the discovery of new targets and ultimately lead to compounds with effective biological activities. Most of the complexes showed nontoxic behaviour, except 9 and 10, which are highly toxic in terms of mutagenicity, tumorigenicity, irritant effect, and reproductive effects. Other inhibitors which showed toxicity are 1a (high-TUM), 1b (mild-REP), 6a (high-MUT), 7b (high-TUM, REP), 8b (high-MUT, TUM, REP), 11b (mild-IRR), and 11c (high-IRR, REP). Ref 1 and Ref 3 showed nontoxicity for (IRR) and Ref 3 is safe for use (TUM, REP).
The cardiotoxicity results showed the safe use of drugs except for a few inhibitors where toxic behaviour is observed. See ESI Table 3.† No correlation pattern is observed, which shows that the addition of one chlorine and two chlorines compared to the parent complex greatly affects cardiotoxicity; rather it shows severity for separate features, such as arrhythmia (6, 7, 9), cardiac failure (9b), heart block (6b, 7, 9a, 9c), herG toxicity (3-4b, 4, 5a, 9b, 11c), hypertension (2, 3b, 3-2a, 3-2b, 4c, 5b, 8c, 9b, 9c), and myocardial infarction (5b, 8a, 9, 11, 12a). These results show no conclusive results about toxicity on addition of one chloro and two chloro substituents. Ref 1 and Ref 2 are still safe to use, but Ref 3 and Ref 4 are highly toxic drugs.
One of the salient features of a good drug candidate is that it is absorbed in a required time and is well distributed throughout the system for effective metabolism and action. Drug metabolism is needed to define the pharmacological and toxicological profile of drugs, which is vital for drug discovery and development. Predictions for high human gastrointestinal (GI) absorption and no permeation of the BBB are seen in complexes 3, 4, 7, and 12; whereas high human gastrointestinal (GI) absorption and permeation of the BBB is seen in complexes 2, 5, 6, 8, 9, 10, 11a, and 11b. Complex 1 has low human gastrointestinal (GI) absorption and no permeation of the BBB, while complex 11c shows high human gastrointestinal (GI) absorption and no permeation of the BBB. The GI absorption is high for Ref (1–4) and permeation of the BBB is observed in Ref 1, Ref 2, and Ref 3. The role of permeability glycoprotein (P-gp) is to protect the central nervous system (CNS) from xenobiotics.66 In our studies 4, 5, 9a, 9b, and 11a are predicted as substrates of (P-gp) glycoprotein, but Ref (1–4) are not predicted as substrates of (P-gp) glycoprotein. It is essential to know the interaction of potential therapeutic drugs with cytochromes P450 (CYP) because this group of isoenzymes plays a key role in drug elimination through metabolic biotransformation. The estimated results indicated no correlation patterns for interaction with the CYP1A2, CYP2C19, CYP2C9, and CYP2D6 and CYP3A4 members of the isoenzyme family. See Table 3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (cm s−1) for the reference drugs Ref (1–4) and 12 groups of studied inhibitors using BOILED-Egg in the SwissADME webserver
Kp (cm s−1) for the reference drugs Ref (1–4) and 12 groups of studied inhibitors using BOILED-Egg in the SwissADME webserver
| Complex | GI abs | BBB | P-gp subs | CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (cm s−1) Kp (cm s−1) |
|---|---|---|---|---|---|---|---|---|---|
| Ref 1 | High | Yes | No | Yes | Yes | Yes | Yes | Yes | −5.91 |
| Ref 2 | High | Yes | No | Yes | No | No | Yes | Yes | −4.96 |
| Ref 3 | High | No | No | No | No | No | No | No | −7.46 |
| Ref 4 | High | Yes | No | No | Yes | No | Yes | No | −6.43 |
| 1a | Low | No | No | No | No | Yes | No | Yes | −5.92 |
| 1b | Low | No | No | No | No | No | No | Yes | −5.92 |
| 2a | High | Yes | No | Yes | Yes | Yes | No | Yes | −5.29 |
| 2b | High | Yes | No | Yes | Yes | Yes | Yes | Yes | −5.02 |
| 3-1a | High | No | No | Yes | No | No | Yes | No | −5.45 |
| 3-1b | High | No | No | Yes | Yes | Yes | No | No | −5.21 |
| 3-2a | High | No | No | Yes | No | Yes | No | No | −5.45 |
| 3-2b | High | No | No | Yes | Yes | Yes | No | No | −5.33 |
| 3-3a | High | No | No | Yes | No | Yes | No | No | −5.75 |
| 3-3b | High | No | No | Yes | No | Yes | No | No | −5.52 |
| 3-4a | High | No | No | Yes | Yes | Yes | No | No | −5.46 |
| 3-4b | High | No | No | Yes | Yes | Yes | No | No | −5.23 |
| 4a | High | No | Yes | No | No | No | Yes | No | −5.02 |
| 4b | High | No | Yes | No | No | No | Yes | No | −5.01 |
| 4c | High | No | Yes | No | Yes | No | Yes | No | −4.77 |
| 5a | High | Yes | Yes | No | No | Yes | Yes | Yes | −5.43 |
| 5b | High | Yes | Yes | No | No | Yes | Yes | Yes | −5.35 |
| 6a | High | Yes | No | Yes | No | No | No | No | −6.04 |
| 6b | High | Yes | No | Yes | No | No | No | No | −5.78 |
| 7a | High | No | Yes | Yes | No | No | No | No | −6.07 |
| 7b | High | No | No | No | Yes | Yes | Yes | Yes | −6.50 |
| 8a | High | Yes | No | Yes | Yes | Yes | Yes | Yes | −4.90 |
| 8c | High | Yes | No | No | Yes | Yes | Yes | No | −4.43 |
| 9a | High | Yes | Yes | No | No | No | No | No | −7.18 |
| 9b | High | Yes | Yes | No | No | No | No | No | −7.18 |
| 9c | High | Yes | No | Yes | Yes | No | Yes | No | −6.47 |
| 10a | High | Yes | No | Yes | Yes | Yes | No | No | −4.94 |
| 10b | High | Yes | No | Yes | Yes | Yes | No | No | −4.94 |
| 10c | High | Yes | No | Yes | Yes | Yes | No | No | −4.46 |
| 11a | High | Yes | Yes | Yes | No | No | Yes | Yes | −5.84 |
| 11b | High | Yes | No | No | No | No | No | No | −5.61 |
| 11c | High | No | No | No | No | No | No | No | −5.37 |
| 12a | High | No | No | Yes | No | Yes | Yes | No | −5.59 |
| 12b | High | No | No | Yes | No | Yes | No | No | −5.56 |
| 12c | Low | No | No | Yes | No | Yes | No | No | −5.33 |
The results predicted that the physiological actions of the studied reference drugs and studied inhibitors involve multiple target interactions with kinase (1, 3, 4, 6), enzyme (2, 5, 7, 9, 12), family AG protein coupled receptor (8), protease (10), and phosphodiesterase families (11). See Fig. 3. Ref 1 has ligand gated ion channel and family AG protein coupled receptor interactions and Ref 2 has major interactions with eraser and phosphodiesterase families. Ref 3 and Ref 4 interact with kinases and family AG protein coupled receptor families.
 | ||
| Fig. 3 Representation of biological targets for the reference drugs Ref (1–4) and 12 groups of studied inhibitors predicted with the SwissTargetPrediction webserver. | ||
Now we look into the experimental and synthesis results of the reference drugs and the studied inhibitors and try to validate our computational results. Ref 1 (diazepam) is anxiolytic, Ref 2 (chloroquine) is listed as an anti-malarial drug by the World Health Organization, Ref 3 (chloramphenicol) is antibiotic, and Ref 4 (bendamustine) is an oncology drug. The parent phenylhydrazone 1a showed micromolar inhibitory activity against cancer cell lines; however, a simple para-chloro substituent 1b created the “magic” effect of significantly improving potency against all three cancer cell lines MCF-7, NCI-H460, and SF-268.70 Inhibitors 2a and 2b showed antimalarial activity against chloroquine-sensitive Plasmodium falciparum strains (“D10” strains). These complexes showed a remarkable magic chloro effect, due to the lipophilicity and the specific van der Waals radius of the substituent. The change in lipophilicity of these inhibitors due to the chlorine substituent is seen in the theoretical results. See ESI Table 1.† The (3-1a) to (3-4b) series show consistent SAR trends with protein binding sites and ligand pharmacophore. The experimental results showed a “magic chloro effect” at the indole C6-position, improving the IC50 value toward ATX.71 The endocannabinoid system (ECS) is a retrograde lipid signaling pathway that regulates a variety of physiological functions in the body.72 Recent studies have indicated that the inhibition of endocannabinoid degrading enzymes exerts therapeutic effects in a few neurodegenerative diseases, such as Parkinson's, Alzheimer's, and Huntington's disease.73 A magic chloro effect was observed with an isatin-derived scaffold incorporating a hydrazone moiety in studies of MAGL inhibition. Compared to the unsubstituted isatin 4a, the 5-chloro analogue 4b was seen to be most active against MAGL, whereas 4c, with a Cl in position C6 of the isatin core, showed high potency compared to 4a. Compounds 4b and 4c are considered promising candidates for the treatment of neurological and mood disorders,74 as these complexes showed acceptable pharmacokinetic properties. The chlorine effect is observed in 5b, as it is considered the most potential competitor to the human dopamine-3 (D3) receptor. These targets are used for the treatment of various neurological diseases, depression, and disorders. Compound 5b has shown high potency with minimized side effects and enhanced therapeutics. 5a binds to a D2-like receptor, whereas 5b does not bind to a D2-like receptor because the addition of a hydrophobic substituent to the phenyl ring increases the binding affinity of 5b to the D3 receptor due to the attached chlorine substituent.75 A significant chloro effect was observed in compound 6b, as an inhibitor of Pseudomonas aeruginosa. Inhibitor 6b shows potency and exhibits antibacterial activities.76 The effect of two chlorines, which play an important role in protein–ligand interactions, can be observed for the fibroblast growth factor receptor 1 (FGFR1) kinase domain, in compound 7. Experimental results showed that the potency of 7b is due to the acidity and increased electrophilicity, which is seen in the theoretical results (electrophilicity).77 See ESI Table 1.† Compound 9 show inhibitory activities for a nicotinamide-based potent and selective monoamine oxidase A (MAO-A) target.78 The introduction of two chlorine atoms in 9c makes it more highly potent than 9b in which the chlorine atom is put at the R2-position, and π–π stacking interactions were proposed with one of the protein's tyrosine residues. Compound 9 (3,4-dichlorophenyl analogue) is used for antibacterial activities. The two chlorine substituents of 9c increase its lipophilicity and electron-withdrawing properties. The correlation between ring electron density and potency is further supported by the theoretical results.79 Compound 11 is a series of dual norepinephrine reuptake transporter (NET) inhibitors/5HT1A partial agonists. The potency of adding one chlorine or two chlorines is seen when chlorine atoms were introduced onto the phenyl ring of 11a at the C2- and C3-positions.80 Compound 12 showed in vitro cytotoxicity studies against the human cancer cell line T47D. It is observed that the parent compound indole 12a is inactive, but the activity is increased when two hydrogen atoms on different rings are substituted with chlorine atoms.81
Conclusions
Results for the 12 studied complexes predicted that the IP values are higher and the EA values are lower for one-chloro inhibitors compared to the –H analogues, which are positively correlated with the IP values of Ref 1 and Ref 2. The change of chlorine at two different positions has not affected the IP values. The chemical hardness for one chloro substituent is higher than the chemical hardness for two chloro substituents. All the reference drugs and studied inhibitors show high electrophilicity indexes for one chloro and two chloro substituents. A higher HL gap for the studied inhibitors showed stability and a “magic chloro effect” for the studied inhibitors. The pharmacokinetic parameters validate the “drug likeliness” for the studied inhibitors. Most of the complexes showed nontoxic behaviour towards four toxic effects (mutagenicity, tumorigenic, irritant, and reproductive) and cardiotoxicity. The reference drugs are also safe for use with few toxic effects. High gastrointestinal (GI) absorption is seen for the reference drugs and studied inhibitors. The chlorine bonds have increased the metabolic activities. The toxicity is not increased by adding one chloro or two chloro substituents. The multitarget activities of the inhibitors reflected the potency of the chlorine atom against various diseases, which is validated by the experimental results. In conclusion, we can say that the chlorine substituent plays a vital role not only in tuning pharmacokinetic properties but also in improving drug-target binding affinity. Studies should focus more on the ADME/T property optimization of –Cl substituents for drug design and discovery in future.Data availability
The optimized Cartesian coordinates of reference drugs and studied 35 inhibitors is given in the ESI.†Author contributions
SJ prepared all tables and figures; and the entire manuscript was written by RS.Conflicts of interest
There are no conflicts of interest to report.Acknowledgements
RS acknowledges the financial assistance by DST WOSA project (SR/WOS-A/CS-69/2018). RS is thankful to her Mentor Dr Shrish Tiwari, Bioinformatics, CSIR-Centre for Cellular and Molecular Biology, and Dr G. Narahari Sastry, Director, CSIR-NEIST for the technical support.References
- P. Metrangolo and G. Resnati, Halogen Bonding: A Paradigm in Supramolecular Chemistry, Chem.–Eur. J., 2001, 7, 2511–2519 CrossRef CAS PubMed.
- P. Metrangolo and G. Resnati, Metal-Bound Halogen Atoms in Crystal Engineering, Chem. Commun., 2013, 49, 1783–1785 RSC.
- G. R. Desiraju, P. S. Ho, L. Kloo, A. C. Legon, R. Marquardt, P. Metrangolo, P. Politzer, G. Resnati and K. Rissanen, Definition of the Halogen Bond (IUPAC Recommendations 2013), Pure Appl. Chem., 2013, 85, 1711–1713 CrossRef CAS.
- J. S. Murray, P. Lane and P. Politzer, Expansion of the σ-Hole Concept, J. Mol. Model., 2009, 15, 723–729 CrossRef CAS PubMed.
- P. Politzer, J. S. Murray and T. Clark, Halogen Bonding and Other σ-Hole Interactions: A Perspective, Phys. Chem. Chem. Phys., 2013, 15, 11178–11189 RSC.
- R. S. Mulliken, Structures of Complexes Formed by Halogen Molecules with Aromatic and with Oxygenated Solvents, J. Am. Chem. Soc., 1950, 72, 600–608 CrossRef CAS.
- O. Hassel, Structural aspects of interatomic charge-transfer bonding, Science, 1970, 170(3957), 497–502 CrossRef CAS PubMed.
- A. Saba, F. Sarwar, S. Muhammad, M. Ilyas, J. Iqbal, A. G. Al-Sehemi, K. Ayub, M. A. Gilani and M. Adnan, Insighting the inhibitory potential of novel modafinil drug derivatives against estrogen alpha (ERα) of breast cancer through a triple hybrid computational methodology, J. Mol. Liq., 2022, 366, 120234 CrossRef CAS.
- A. Bibi, S. Muhammad, S. UrRehman, S. Bibi, S. Bashir, K. Ayub, M. Adnan and M. Khalid, Chemically Modified Quinoidal Oligothiophenes for Enhanced Linear and Third-Order Nonlinear Optical Properties, ACS Omega, 2021, 6(38), 24602–24613 CrossRef CAS PubMed.
- A. Waheed, S. Muhammad, M. A. Gilani, M. Adnan and Z. Aloui, A Systematic and Comparative Analysis of Four Major Classes of DFT Functionals to Compute Linear and Nonlinear Optical Properties of Benchmark Molecules, J. Comput. Biophys. Chem., 2021, 20(05), 517–528 CrossRef CAS.
- S. Muhammad, F. Sarwar and S. S. Alarfaji, et al., A computational study for optical and nonlinear optical properties of distinctive V-shaped cyclopenta dithiophene derivatives, Opt. Quantum Electron., 2023, 55, 895 CrossRef CAS.
- M. Adnan and J. K. Lee, All Sequential Dip-Coating Processed Perovskite Layers from an Aqueous Lead Precursor for High Efficiency Perovskite Solar Cells, Sci. Rep., 2018, 8, 2168–2178 CrossRef PubMed.
- S. A. Waldman, Does potency predict clinical efficacy? Illustration through an antihistamine model, Ann. Allergy, Asthma, Immunol., 2002, 89(1), 7–11 CrossRef CAS PubMed ; quiz 11-2, 77..
- A. L. Hopkins, G. M. Keserü, P. D. Leeson, D. C. Rees and C. H. Reynolds, The role of ligand efficiency metrics in drug discovery, Nat. Rev. Drug Discovery, 2014, 13, 105–121 CrossRef CAS PubMed.
- W. Y. Fang, L. Ravindar, K. P. Rakesh, H. M. Manukumar, C. S. Shantharam, N. S. Alharbi and H. L. Qin, Synthetic approaches and pharmaceutical applications of chloro-containing molecules for drug discovery: a critical review, Eur. J. Med. Chem., 2019, 173, 117–153 CrossRef CAS PubMed.
- D. Chiodi and Y. Ishihara, “Magic Chloro”: Profound Effects of the Chlorine Atom in Drug Discovery, J. Med. Chem., 2023, 66(8), 5305–5331, DOI:10.1021/acs.jmedchem.2c02015.
- D. G. Brown, M. M. Gagnon and J. Bostrom, Understanding our love affair with p-chlorophenyl: present day implications from historical biases of reagent selection, J. Med. Chem., 2015, 58, 2390–2405 CrossRef CAS PubMed.
- J. G. Topliss, Utilization of operational schemes for analog synthesis in drug design, J. Med. Chem., 1972, 15, 1006–1011 CrossRef CAS PubMed.
- J. G. Topliss, A manual method for applying the Hansch approach to drug design, J. Med. Chem., 1977, 20, 463–469 CrossRef CAS PubMed.
- B. R. Smith, C. M. Eastman and J. T. Njardarson, Beyond C, H, O, and N! Analysis of the elemental composition of US FDAapproved drug architectures, J. Med. Chem., 2014, 57, 9764–9773 CrossRef CAS PubMed.
- W. Y. Fang, L. Ravindar, K. P. Rakesh, H. M. Manukumar, C. S. Shantharam, N. S. Alharbi and H. L. Qin, Synthetic approaches and pharmaceutical applications of chloro-containing molecules for drug discovery: a critical review, Eur. J. Med. Chem., 2019, 173, 117–153 CrossRef CAS PubMed.
- E. D. Jemmis, K. T. Giju and J. Leszczynski, Ionic to covalent bonding: a density functional theory study of linear and bent X2Y3 monomers (X=B, Al, Ga, In; Y=O, S, Se), Electron. J. Theor. Chem., 1997, 2, 130–138 CrossRef CAS.
- O. Mó, M. Yáñez, M. Eckert-Maksic, Z. B. Maksić, I. Alkorta and J. Elguero, Periodic trends in bond dissociation energies. A theoretical study, J. Phys. Chem. A, 2005, 109, 4359–4365 CrossRef PubMed.
- E. C. Allen and K. J. Beers, Ab initio study of the binding strength of POSS-cation complexes, Polymer, 2005, 46, 569–573 CrossRef CAS.
- C. Tuma, A. D. Boese and N. C. Handy, Predicting the binding energies of H-bonded complexes: A comparative DFT study, Phys. Chem. Chem. Phys., 1999, 1, 3939–3947 RSC.
- J. R. Reimers, Z.-L. Cai, A. Bilic and N. S. Hush, The appropriateness of density-functional theory for the calculation of molecular electronics properties, Ann. N. Y. Acad. Sci., 2003, 1006, 235–251 CrossRef CAS.
- R. G. Parr, and W. Yang, Density-Functional Theory of Atoms and Molecules, Oxford UP, New York, 1989 Search PubMed.
- H. Chermette, Chemical reactivity indexes in density functional theory, J. Comput. Chem., 1999, 20, 129–154 CrossRef CAS.
- P. Geerlings, F. De Proft and W. Langenaeker, Conceptual density functional theory, Chem. Rev., 2003, 103, 1793–1873 CrossRef CAS PubMed.
- P. W. Ayers, An elementary derivation of the hard/soft-acid/base principle, J. Chem. Phys., 2005, 122, 141102 CrossRef.
- R. Srivastava, Theoretical Studies on the Molecular Properties, Toxicity, and Biological Efficacy of 21 New Chemical Entities, ACS Omega, 2021, 6(38), 24891–24901 CrossRef CAS PubMed.
- R. Srivastava, Chemical Reactivity and Optical and Pharmacokinetics Studies of 14 Multikinase Inhibitors and Their Docking Interactions toward ACK1 for Precision Oncology, Front. Chem., 2022, 10, 843642 CrossRef CAS PubMed.
- R. Srivastava, Chemical reactivity theory (CRT) study of small drug-like biologically active molecules, J. Biomol. Struct. Dyn., 2021, 39(3), 943–952 CrossRef CAS PubMed.
- N. Flores-Holguín, J. Frau and D. Glossman-Mitnik, in Density Functional Theory, ed. S. R. De Lazaro, L. H. Da Silveira Lacerda and R. A. Pontes Ribeiro, IntechOpen, London, UK, 2021, vol. 3, pp. 57–67, ISBN 978-1-78985-167-0, eISBN 978-1-78985-168-7 Search PubMed.
- N. Flores-Holguín, J. Frau and D. Glossman-Mitnik, A Fast and Simple Evaluation of the Chemical Reactivity Properties of the Pristinamycin Family of Antimicrobial Peptides, Chem. Phys. Lett., 2020, 739, 137021, DOI:10.1016/j.cplett.2019.137021.
- N. Flores-Holguín, J. Frau and D. Glossman-Mitnik, Conceptual DFT Based Computational Peptidology of Marine Natural Compounds. Discodermins A–H, Molecules, 2020, 25, 4158, DOI:10.1016/j.cplett.2019.137021.
- N. Flores-Holguín, J. Frau and D. Glossman-Mitnik, Virtual Screening of Marine Natural Compounds by Means of Chemoinformatics and CDFT Based Computational Peptidology, Mar. Drugs, 2020, 18, 478, DOI:10.3390/md18090478.
- R. G. Pearson, Recent advances in the concept of hard and soft acids and bases, J. Chem. Educ., 1987, 64, 561–567 CrossRef CAS.
- Z. Zhou and R. G. Parr, New measures of aromaticity: absolute hardness and relative hardness, J. Am. Chem. Soc., 1989, 111, 7371–7379 CrossRef CAS.
- R. G. Parr and P. K. Chattaraj, Principle of maximum hardness, J. Am. Chem. Soc., 1991, 113, 1854–1855 CrossRef CAS.
- R. G. Pearson, The Principle of Maximum Hardness, Acc. Chem. Res., 1993, 26, 250–255 CrossRef CAS.
- R. G. Pearson, Principle of maximum physical hardness, J. Phys. Chem., 1994, 98, 1989–1992 CrossRef CAS.
- P. K. Chattaraj, The maximum hardness principle: an overview, Proc. Indian Natl. Sci. Acad., Part A, 1996, 62, 513–531 CAS.
- R. G. Pearson, Maximum chemical and physical hardness, J. Chem. Educ., 1999, 76, 267–275 CrossRef CAS.
- P. W. Ayers and M. Levy, Perspective on “Density functional approach to the frontier-electron theory of chemical reactivity”, Theor. Chem. Acc., 2000, 103, 353–360 Search PubMed.
- M. Torrent-Sucarrat, J. M. Luis, M. Duran and M. Sola, On the validity of the maximum hardness and minimum polarizability principles for nontotally symmetric vibrations, J. Am. Chem. Soc., 2001, 123, 7951–7952 CrossRef CAS PubMed.
- M. Torrent-Sucarrat, J. M. Luis, M. Duran and M. Sola, Are the maximum hardness and minimum polarizability principles always obeyed in nontotally symmetric vibrations?, J. Chem. Phys., 2002, 117, 10561–10570 CrossRef CAS.
- R. A. Miranda-Quintana, P. K. Chattaraj and P. W. Ayers, Finite temperature grand canonical ensemble study of the minimum electrophilicity principle, J. Chem. Phys., 2017, 147(12), 124103 CrossRef PubMed.
- D. Chiodi and Y. Ishihara, “Magic Chloro”: Profound Effects of the Chlorine Atom in Drug Discovery, J. Med. Chem., 2023, 66(8), 5305–5331, DOI:10.1021/acs.jmedchem.2c02015 , PMID: 37014977..
- J. D. Chai and M. Head-Gordon, Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery Jr, T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, W. J. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, K. D. Malick, D. A. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Hallacombe, C. P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez and J. A. Pople, J. A. C.02, Gaussian, Inc., Wallingford, CT, 2009 Search PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions, J. Phys. Chem. B, 2009, 113, 6378–6396, DOI:10.1021/jp810292n.
- R. Dennington, T. A. Keith and J. M. Millam, GaussView, version 6.1, Semichem Inc., Shawnee Mission, KS, 2016 Search PubMed.
- S. Riaz, R. Hussain, M. Adnan, M. U. Khan, S. Muhammad, J. Yaqoob, M. U. Alvi, M. Khalid, Z. Irshad and K. Ayub, Ab Initio Study of Two-Dimensional Cross-Shaped Non-Fullerene Acceptors for Efficient Organic Solar Cells, ACS Omega, 2022, 7(12), 10638–10648 CrossRef CAS PubMed.
- R. Hussain, M. Adnan, K. Atiq, M. U. Khan, Z. H. Farooqi, J. Iqbal and R. Begum, Designing of silolothiophene-linked triphenylamine-based hole transporting materials for perovskites and donors for organic solar cells-A DFT study, Sol. Energy, 2023, 253, 187–198 CrossRef CAS.
- R. Hussain, M. Adnan, S. Nawab, M. U. Khan, M. Khalid, Z. Irshad, Z. Ayub and L. Lim, Role of novel carbon-oxygen-bridged Z-shaped non-fullerene acceptors for high efficiency organic solar cells, Synth. Met., 2022, 290, 117159 CrossRef CAS.
- Molinspiration Chemoinformatics software, https://www.molinspiration.com Search PubMed.
- Osiris property explorer, https://www.organicchemistry.org/prog/peo/ Search PubMed.
- S. Iftkhar, A. G. C. de Sá, J. P. L. Velloso, R. Aljarf, D. B. Pires and D. E. V. Ascher, cardioToxCSM: A Web Server for Predicting Cardiotoxicity of Small Molecules, J. Chem. Inf. Model., 2022, 62(20), 4827–4836 CrossRef CAS PubMed.
- A. Daina and V. Zoete, A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules, ChemMedChem, 2016, 11(11), 1117–1121, DOI:10.1002/cmdc.201600182.
- A. Daina, O. Michielin and V. Zoete, SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules, Nucleic Acids Res., 2019, 47, W357–W364, DOI:10.1093/nar/gkz382.
- R. G. Parr and R. G. Pearson, Absolute Hardness: Companion Parameter to Absolute Electronegativity, J. Am. Chem. Soc., 1983, 105, 7512–7516 CrossRef CAS.
- R. G. Parr and W. Yang, Density Functional Approach to the Frontier-Electron Theory of Chemical Reactivity, J. Am. Chem. Soc., 1984, 106, 4049–4050 CrossRef CAS.
- R. G. Pearson, The principle of maximum hardness, Acc. Chem. Res., 1993, 26, 250–255 CrossRef CAS.
- P. W. Ayers, J. S. M. Anderson and L. J. Bartolotti, et al., Perturbative perspectives on the chemical reaction prediction problem, Int. J. Quantum Chem., 2005, 101(5), 520–534 CrossRef CAS.
- P. W. Ayers and R. G. Parr, Variational Principles for Describing Chemical Reactions: The Fukui Function and Chemical Hardness Revisited, J. Am. Chem. Soc., 2000, 122(9), 2010–2018 CrossRef CAS.
- D. F. Veber, S. R. Johnson, H.-Y. Cheng, B. R. Smith, K. W. Ward and K. D. Kopple, Molecular properties that influence the oral bioavailability of drug candidates, J. Med. Chem., 2002, 45(12), 2615–2623 CrossRef CAS.
- P. Ertl, B. Rohde and P. Selzer, Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties, J. Med. Chem., 2000, 43, 3714–3717 CrossRef CAS.
- N. Flores-Holguín, J. Frau and D. Glossman-Mitnik, Computational peptidology approach to the study of the chemical reactivity and bioactivity properties of Aspergillipeptide D, a cyclopentapeptide of marine origin, Sci. Rep., 2022, 12(1), 506, DOI:10.1038/s41598-021-04513-z.
- R. M. Mohareb, A. E. Abdallah and M. A. Abdelaziz, New approaches for the synthesis of pyrazole, thiophene, thieno [2,3-b] pyridine, and thiazole derivatives together with their anti-tumor evaluations, Med. Chem. Res., 2014, 23, 564–579 CrossRef CAS.
- Y. Zhang, J. A. Clark, M. C. Connelly, F. Zhu, J. Min, W. A. Guiguemde, A. Pradhan, L. Iyer, A. Furimsky, J. Gow, T. Parman, F. El Mazouni, M. A. Phillips, D. E. Kyle, J. Mirsalis and R. K. Guy, Lead optimization of 3-carboxyl-4(1H)-quinolones to deliver orally bioavailable antimalarials, J. Med. Chem., 2012, 55, 4205–4219 CrossRef CAS PubMed.
- K. Ahn, M. K. McKinney and B. F. Cravatt, Enzymatic pathways that regulate endocannabinoid signaling in the nervous system, Chem. Rev., 2008, 108, 1687–1707 CrossRef CAS PubMed.
- A. Papa, S. Butini, S. Pasquini, C. Contri, S. Gemma, G. Campiani, K. Varani and F. Vincenzi, Polypharmacological approaches for CNS diseases: focus on endocannabinoid degradation inhibition, Cells, 2022, 11, 471 CrossRef CAS PubMed.
- S. Jaiswal and S. R. Ayyannan, Discovery of isatin-based carbohydrazones as potential dual inhibitors of fatty acid amide hydrolase and monoacylglycerol lipase, ChemMedChem, 2022, 17, e202100559 CrossRef CAS PubMed.
- J. Chen, B. Levant, C. Jiang, T. M. Keck, A. H. Newman and S. Wang, Tranylcypromine substituted cis-hydroxycyclobutylnaphthamides as potent and selective dopamine D3 receptor antagonists, J. Med. Chem., 2014, 57, 4962–4968 CrossRef CAS PubMed.
- D. F. Manvich, A. K. Petko, R. C. Branco, S. L. Foster, K. A. Porter-Stransky, K. A. Stout, A. H. Newman, G. W. Miller, C. A. Paladini and D. Weinshenker, Selective D2 and D3 receptor antagonists oppositely modulate cocaine responses in mice via distinct postsynaptic mechanisms in nucleus accumbens, Neuropsychopharmacology, 2019, 44, 1445–1455 CrossRef CAS PubMed.
- W. Cieslik, R. Musiol, J. E. Nycz, J. Jampilek, M. Vejsova, M. Wolff, B. Machura and J. Polanski, Contribution to investigation of antimicrobial activity of styrylquinolines, Bioorg. Med. Chem., 2012, 20, 6960–6968 CrossRef CAS PubMed.
- L. Shi, Y. Yang, Z. L. Li, Z. W. Zhu, C. H. Liu and H. L. Zhu, Design of novel nicotinamides as potent and selective monoamine oxidase A inhibitors, Bioorg. Med. Chem., 2010, 18, 1659–1664 CrossRef CAS PubMed.
- E. A. Meyer, R. K. Castellano and F. Diederich, Interactions with aromatic rings in chemical and biological recognition, Angew. Chem., Int. Ed., 2003, 42, 1210–1250 CrossRef CAS PubMed.
- A. B. Dounay, N. S. Barta, B. M. Campbell, C. Coleman, E. M. Collantes, L. Denny, S. Dutta, D. L. Gray, D. Hou, R. Iyer, S. N. Maiti, D. F. Ortwine, A. Probert, N. C. Stratman, R. Subedi, T. Whisman, W. Xu and K. Zoski, Design, synthesis, and pharmacological evaluation of phenoxy pyridyl derivatives as dual norepinephrine reuptake inhibitors and 5-HT1A partial agonists, Bioorg. Med. Chem. Lett., 2010, 20, 1114–1117 CrossRef CAS PubMed.
- P. Chen, Y. Zhuang, P. Diao, F. Yang, S. Wu, L. Lv, W. You and P. Zhao, Synthesis, biological evaluation, and molecular docking investigation of 3-amidoindoles as potent tubulin polymerization inhibitors, Eur. J. Med. Chem., 2019, 162, 525–533 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06638j |
| This journal is © The Royal Society of Chemistry 2023 |
