Open Access Article
Open Access ArticleSynthesis and evaluation of potent (iso)ellipticine-based inhibitors of MYLK4 accessed via expeditious synthesis from isoquinolin-5-ol†
Szu Lee‡
a,
Min-Wu Chao‡bcd,
Yi-Wen Wue,
Chia-Min Hsue,
Tony Eight Linef,
Kai-Cheng Hsuefghi,
Shiow-Lin Panefghi and
Hsueh-Yun Lee *aij
*aij
aSchool of Pharmacy, College of Pharmacy, Taipei Medical University, Taiwan. E-mail: hyl@tmu.edu.tw; Tel: +886-2-7361661
bSchool of Medicine, College of Medicine, National Sun Yat-sen University, Kaohsiung, Taiwan
cInstitute of Biopharmaceutical Sciences, College of Medicine, National Sun Yat-sen University, Kaohsiung, Taiwan
dThe Doctoral Program of Clinical and Experimental Medicine, College of Medicine, National Sun Yat-sen University, Kaohsiung, Taiwan
eGraduate Institute of Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei, Taiwan
fPhD Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei, Taiwan
gTMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan
hTMU Research Center for Drug Discovery, Taipei Medical University, Taipei, Taiwan
iPhD Program in Drug Discovery and Development Industry, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
jMaster Program in Clinical Genomics and Proteomics, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
First published on 30th October 2023
Abstract
The K2S2O8-mediated generation of p-iminoquinone contributed to the regioselective substitution of isoquinolin-5,8-dione. This hydroxyl group-guided substitution was also applied to selected heterocycles and addressed the regioselectivity issue of quinones. This study has provided an expeditious pathway from isoquinolin-5-ol (5) to ellipticine (1) and isoellipticine (2), which benefits the comprehensive comparison of their activity. Compounds 1 and 2 displayed marked MYLK4 inhibitory activity with IC50 values of 7.1 and 6.1 nM, respectively. In the cellular activity of AML cells (MV-4-11 and MOLM-13), compound 1 showed better AML activity than compound 2.
Introduction
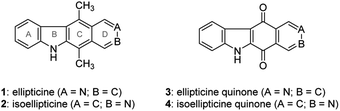
The pyrido[4,3-b]carbazole alkaloid ellipticine (1) was isolated by Goodwin et al.1 in 1959 from Ochrosia elliptica Labill. Ellipticine and its relevant analogues were reported to exhibit anticancer activity, possibly by DNA intercalation and inhibition of topoisomerase II.2–4 Because of its biological activity, it was of interest for researchers to develop synthetic approaches to ellipticine. The approaches are simply summarized into four methodologies according to late-stage ring formation of the B-, C-, D- and B/C-rings. The B-ring can be constructed from an aryl triazene5,6 with a Pd-catalyzed cross-dehydrogenative coupling reaction.7 Much efforts has been devoted to the formation of the C-ring of ellipticine through a Diels–Alder reaction between furo[3,4-b]indole and 3,4-pyridyne8,9 or between pyrano[3,4-b]indol-3-one and pyridyltriazene.10 The lateral D-ring can be built from 3-formyl-1,4-dimethylcarbazole by a Pomeranz–Fritsch isoquinoline synthesis.11–13 Differding et al. reported the intramolecular Diels–Alder cycloaddition of vinylketenimine, which assembled the B and C rings of ellipticine simultaneously.14 In addition, Pedersen showed that the B/C rings of ellipticine could also be constructed by an imidoyl radical cascade reaction (Fig. 1).15In addition to the methodologies cited above, ellipticine quinone (3) is considered to be a precursor of ellipticine, to which it could be converted by reaction of the carbonyl groups with methyl lithium followed by treatment with sodium borohydride.16 Ellipticine quinone (3) was obtained by a tandem directed metalation reaction between indole-3-carboxaldehyde and N,N-diethylisonicotinamide,17 Friedel–Crafts hydroxyalkylation of the resulting ethyl indole-3-carboxylate followed by oxidation and ortho-lithiation,18 or lead tetraacetate-mediated oxidative rearrangement of an acyl hydrazone followed by a Friedel–Crafts reaction.19 The common ground of these methodologies is the use of indole and pyridine derivatives as starting materials in the construction of the C-ring of ellipticine quinone (3). Naciuk et al. conducted substitution of isoquinolin-5,8-dione followed by cross dehydrogenative coupling to obtain an isomer of ellipticine quinone (3).20 This strategy of B-ring formation attracted our attention due to its unprecedented route to ellipticine quinone (3) and ellipticine (1). However, the regioselective substitution of isoquinolin-5,8-dione would appear to hamper the synthesis of ellipticine (1), because isoquinolin-5,8-dione favors C7 substitution.21 Considering that the synthesis of ellipticine (1) and isoellipticine (2) started from different starting materials and through different synthetic pathways. This study was aimed to develop a convergent pathway to ellipticine (1) and isoellipticine (2), beginning with the same starting material. This benefits the simultaneous comparison of their biological activity in the same study. For instance, this study tested the enzymatic activity of 1 and 2 against myosin light chain kinase family member 4 (MYLK4), which is correlated to the development of acute myeloid leukemia. In addition, this feasible synthetic methodology could be applied to understand the structure–activity relationship of (iso)ellipticine analogues in further study.
Results and discussion
Chemistry
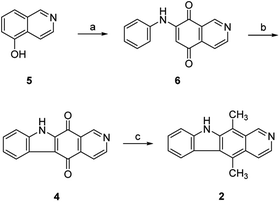
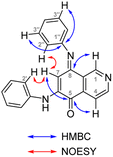
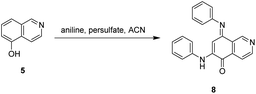
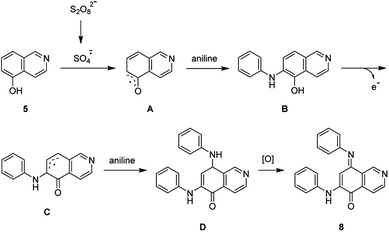
The synthesis of isoellipticine (2) is shown in Scheme 1. The reaction of isoquinolin-5-ol (5) with [bis(trifluoroacetoxy)iodo]benzene (PIFA) followed by the addition of aniline generated predominantly compound 6 together with trace amounts of its regioisomer (9). This regioselective substitution of isoquinolin-5,8-dione was also observed by Naciuk20 and Shin.22 Following reported methodology, isoellipticine quinone (4) was converted into isoellipticine (2).16,20 In comparison with Naciuk's work, this pathway is expeditious, generating isoellipticine (2) in four steps with a 19% overall yield.Due to the predominant selective C7 substitution of isoquinolin-5,8-dione, an alternative synthetic route is required to achieve C6 substitution. Halogen atoms and methoxy substituent are helpful in the regioselective substitution of (iso)quinolinediones. A trial reaction of 6-bromoisoquinolin-5,8-dione with aniline failed to form the anticipated product (9). In 2016, Zhao et al. reported a methodology fulfilling regioselective amination of phenol, which encouraged us to reorder the synthetic sequence.23 We tried to obtain 6-(phenylamino)isoquinolin-5-ol (7) using Zhao's methodology, and then to oxidize 7 to obtain 6-(phenylamino)isoquinoline-5,8-dione (9, Scheme 2). Interestingly, the reaction of isoquinolin-5-ol (5) with aniline in the presence of potassium persulfate (K2S2O8) under the irradiation of blue light gave unexpectedly a p-iminoquinone (8) containing two anilino moieties as judged by NMR and mass spectra. We hydrolyzed 8 with acetic acid and H2O24 under reflux, yielding the anticipated compound (9). This result revealed that one aniline group had indeed been introduced at C6 of the isoquinoline-5,8-dione, and p-iminoquinone (8) was the product of the K2S2O8-mediated reaction. Heteronuclear Multiple Bond Coherence (HMBC) and Nuclear Overhauser Effect Spectroscopy (NOESY) experiments were conducted to confirm the structure of the proposed p-iminoquinone (8), specifically the position of the imine moiety (Fig. 2 and ESI†). The proton at δH 9.81 ppm (H-1) in the NMR spectrum of 8 showed correlation with the 13C-NMR spectra reported by Pradhan et al.25 with a 13C-NMR signal at δC 154.10 ppm which is from the carbon atom (C-8) of the imine moiety. Correlations between the carbonyl carbon at δC 181.56 ppm (C-6) and protons at δH 7.96 ppm (H-4) signified the existence of a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group at the C-6 position. The quinone proton signal in 8 at δH 6.72 ppm displayed NOESY signals between H-2′ and H-2′′. This result revealed that this quinone proton is H-7 and is located between the two phenyl moieties. Together with the results of hydrolysis product, HMBC and NOESY, we concluded that the K2S2O8-mediated reaction generated the p-iminoquinone (8). The following Pd-catalyzed cyclization converted compound 9 to the ellipticine quinone (3), which was subjected to reaction with CH3Li and then NaBH4 to generate ellipticine (1).
O group at the C-6 position. The quinone proton signal in 8 at δH 6.72 ppm displayed NOESY signals between H-2′ and H-2′′. This result revealed that this quinone proton is H-7 and is located between the two phenyl moieties. Together with the results of hydrolysis product, HMBC and NOESY, we concluded that the K2S2O8-mediated reaction generated the p-iminoquinone (8). The following Pd-catalyzed cyclization converted compound 9 to the ellipticine quinone (3), which was subjected to reaction with CH3Li and then NaBH4 to generate ellipticine (1).
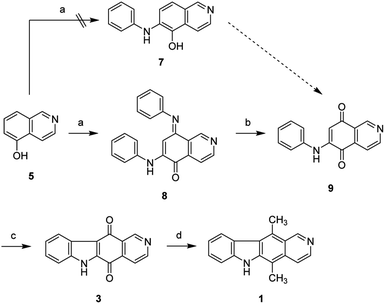 | ||
| Scheme 2 Synthetic approaches to ellipticine (1): (a) aniline, K2S2O8, CH3CN, 24W blue LED, rt; or conditions shown in Table 1; (b) glacial acetic acid, H2O, reflux; (c) Pd(OAc)2, Cu(OAc)2, PivOH, K2CO3, DMA, 130 °C in sealed tube; (d) (i) CH3Li, TMEDA, sealed tube, THF, 0 °C to 100 °C; (ii) NaBH4, EtOH, reflux. | ||
The scope and mechanism of the unexpected generation of p-iminoquinone (8) attracted our attention and we conducted the experiments shown in Table 1. Results from entries 1 and 3 in Table 1 indicated that this reaction proceeds independently of visible light. The increase of equivalence of aniline resulted in an increase in the reaction yield (entry 2). No conversion was observed in the presence of free radical scavengers such as TEMPO, BHT and benzoquinone (entries 4–6), which suggests a free radical mechanism for the formation of 8. In thermal reaction conditions the reaction time is dramatically shortened (entries 7 and 8), and 8 was obtained in a reaction time of 1 h, indicating that the reaction can be conducted through either a photo- or thermal-mediated pathway.
| Entry | Conditions | Additive | Time | Yield |
|---|---|---|---|---|
| a TEMPO: 2,2,6,6-tetramethyl-1-piperidinyloxy.b BQ: benzoquinone.c BHT: butylated hydroxytoluene. | ||||
| 1 | Aniline (2 equiv.), persulfate (3 equiv.), rt, blue LED | — | 64 h | 4.5% |
| 2 | Aniline (20 equiv.), persulfate (3 equiv.), rt, blue LED | — | 40 h | 12.5% |
| 3 | Aniline (2 equiv.), persulfate (3 equiv.), rt, in the dark | — | 64 h | 2.1% |
| 4 | Aniline (1 equiv.), persulfate (3 equiv.), rt, in the dark | TEMPOa | 24 h | 0 |
| 5 | Aniline (1 equiv.), persulfate (3 equiv.), rt, in the dark | BQb | 24 h | 0 |
| 6 | Aniline (1 equiv.), persulfate (3 equiv.), rt, in the dark | BHTc | 24 h | 0 |
| 7 | Aniline (10 equiv.), persulfate (3 equiv.), reflux | — | 1 h | 10.2% |
| 8 | Aniline (10 equiv.), persulfate (6 equiv.), reflux | — | 1 h | 19.2% |
Pradhan et al. reported the synthesis of p-iminoquinone using nitrosobenzene in the presence of hexafluoroisopropanol (HFIP).25 This study led us to examine the reaction of aniline with K2S2O8 in the absence of isoquinolin-5-ol (5). This reaction failed to generate nitrosobenzene, revealing the involvement of a pathway different from the route proposed by Pradhan et al. In light of the results in Table 1, a radical mechanism for the generation of p-iminoquinone (8) was proposed and is shown in Scheme 3. The potassium persulfate was photochemically or thermally cleaved to generate the corresponding sulfate radical which subsequently converted 5 into the radical (A). The addition of an aniline molecule followed by loss of a single electron resulted in another radical (C). A second aniline molecule was introduced and the subsequent oxidation generated the p-iminoquinone (8).
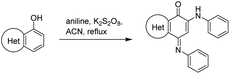
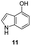
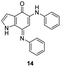
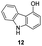
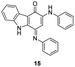
In an attempt to understand the scope of the K2S2O8-mediated synthesis of p-iminoquinones, we selected three heterocyclic compounds (10–12) each bearing one –OH group attached to the carbon adjacent to the ring fusion bond. Under the reaction conditions (Table 1, entry 8), compounds 10–12 were converted into the corresponding p-iminoquinones (13–15, Table 2). The resulting products produce the corresponding quinone derivatives through acetic acid-mediated hydrolysis. This methodology is helpful for regioselective synthesis of anilinoquinones because the position of the –OH determines the regioselectivity. A survey of the literature showed that reaction of aniline with persulfate yields polyaniline,26 which could explain the very low yield of the reaction.
Derivatives of (iso)ellipticine, such as 7-hydroxyisoellipticine and 9-methoxyellipticine, have been reported having AML activity.27,28 According to data from Gene Expression Profiling Interactive Analysis (GEPIA) database (Fig. S1†),29 the expression of myosin light chain kinase family member 4 (MYLK4) is significantly higher in acute myeloid leukemia tissues (T) when compared to normal tissues (N) and is considered as a potential therapeutic target for treatment of AML. In addition, some (iso)ellipticine derivatives were reported displaying kinase inhibition,30,31 which encouraged us to examine the effect of compounds 1–4 on MYLK4 and AML cells. Table 3 shows the enzymatic activity of compounds 1–4 against MYLK4. The result revealed that ellipticine (1) and isoellipticine (2) showed marked MYLK4 inhibitory activity as compared with their synthetic precursors (3 and 4), with IC50 values of 7.1 and 6.1 nM, respectively. Due to their distinct MYLK4 inhibitory activity, compounds 1 and 2 were tested for their cellular activity against MV-4-11 and MOLM-13 cells. Despite the appearance of comparable kinase activity of compounds 1 and 2, 1 showed better AML activity than 2. Ellipticine (1) inhibited the growth of MV-4-11 and MOLM-13 cells with IC50 values of 1.19 and 1.0 μM, respectively. These results suggested that (iso)ellipticine could pave ways to the development of MYLK4 inhibitors for the treatment of AML.
| Compound | Kinase activity | Cellular activity | |
|---|---|---|---|
| MYLK4 | MV-4-11 | MOLM-13 | |
| 1 | 7.1 | 1.19 ± 0.18 | 1.00 ± 0.33 |
| 2 | 6.1 | 4.18 ± 0.61 | 5.11 ± 0.30 |
| 3 | 477.6 | — | — |
| 4 | 285.6 | — | — |
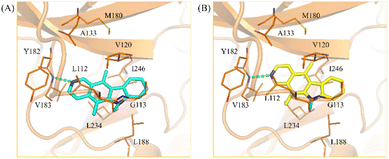
Kinases contain a binding site pocket sandwiched by two lobes connected by a hinge loop.32 Incidentally, many kinase inhibitors targeting the binding site generate hydrogen bonds to the hinge loop residues.33 As a member of the human kinome, MYLK4 would contain similar features. Molecular docking was used to determine if the MYLK4 inhibitors in this study can favorably bind to MYLK4. The binding poses of 1 and 2 showed the occupation of areas associated with the adenosine ring of ATP. Both 1 and 2 were observed to form hydrogen bonds to V183 (Fig. 3). V183 forms part of the hinge loop, suggesting favorable occupation of the binding site. An additional hydrogen bond to the carbonyl backbone of residue L112 was also observed. These hydrogen bonds were facilitated by the nitrogen atoms present in the ring structures. The core structure of 1 and 2 provided more ‘rigidity’ to the compound and formed hydrophobic interactions with amino acids with aliphatic side chains, such as residues L112, V120, V183, L188, L234, and I246. Compound 2 generated additional hydrophobic interactions. This may be due to the location of its nitrogen atom as it formed a hydrogen bond to the hinge residue V183, which caused compound 2 to occupy a slightly different orientation toward residues G113, A133, and M180. This hydrophobic region also explains the distinct lose of MYLK4 activity of 3 and 4, due to appearance of two carbonyl groups. In addition, it guides the structural optimization by introducing hydrophobic substituents on A-ring in the future. Together, the docking results show favorable occupation of the MYLK4 binding site, and their interactions may facilitate their inhibitory activity.
Conclusions
This study attempted to provide pathways to ellipticine (1) and isoellipticine (2), starting from a common starting material. The modified four-step synthetic route provided isoellipticine (2) in 19% overall yield from compound 5. To address the problematic substitution of isoquinolin-5,8-dione, a K2S2O8-mediated ortho-amination was conducted and unexpectedly generated a p-iminoquinone (8) which could be converted into the ellipticine quinone (3) and further transformed to ellipticine (1). As a result, this study reported four-step synthetic routes to ellipticine (1) and isoellipticine (2). The K2S2O8-mediated synthesis of p-iminoquinone was also studied for selected heterocycles (10–12), revealing that the –OH group was able to direct substituents to its ortho position, thus providing an approach to address the issue of regioselective substitution of quinones. Given that biological activity of compounds 1 and 2 was discussed separately in the past, the achievement of this study is to fulfil comprehensive comparison of 1 and 2 as well as their synthetic precursors. The result revealed that (iso)ellipticine (1 and 2) displayed distinct inhibitory against MYLK4, which is highly expressed in acute myeloid leukemia tissues, as compared with (iso)ellipticine quinone (3 and 4). Despite of the appearance of comparable MYLK4 activity, compound 1 showed more potent AML activity than that of 2. Taken together, this study not only provided a regioselective methodology toward (iso)ellipticine (1 and 2) but also found 1 as a potential hit to develop MYLK4 inhibitors for AML treatment. In addition, our facile synthetic methodology would help us fulfil the structure–activity relationship study of (iso)ellipticine derivatives.Experiment
Chemistry
Nuclear magnetic resonance spectra were obtained with an Agilent DD2 600 MHz NMR spectrometer operating at 600 MHz, with chemical shifts reported in parts per million (ppm, δ). High-resolution mass spectra (HRMS) were measured with a JEOL (JMS-700) electrospray ionization (ESI) mass spectrometer. The purity of the final compounds was determined using a Shimadzu LC-2030C LT and was found in all cases to be ≥95%. Flash column chromatography was carried out using silica gel (Merck Kieselgel 60, no. 9385, 230–400 mesh ASTM). All reactions were conducted under an atmosphere of dry N2.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 95
MeOH = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) on silica gel to give compound 1 as a yellow solid (8.1 mg, 48.4%). mp 281.7 °C (dec), 1H NMR (600 MHz, DMSO-d6) δ 11.37 (br, 1H), 9.70 (s, 1H), 8.43 (d, J = 6.0 Hz, 1H), 8.38 (d, J = 7.8 Hz, 1H), 7.92 (d, J = 6.0 Hz, 1H), 7.57 (d, J = 8.4 Hz, 1H), 7.53 (t, J = 7.5 Hz, 1H), 7.26 (t, J = 7.5 Hz, 1H), 3.26 (s, 3H), 2.79 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 149.6, 142.7, 140.6, 140.4, 132.5, 128.0, 127.1, 123.8, 123.4, 123.1, 121.9, 119.2, 115.9, 110.7, 108.0, 14.3, 11.9. HRMS (ESI) for C17H15N2 [M + H]+ calculated 247.1235, found 247.1236. HPLC purity: 99.7%.
5) on silica gel to give compound 1 as a yellow solid (8.1 mg, 48.4%). mp 281.7 °C (dec), 1H NMR (600 MHz, DMSO-d6) δ 11.37 (br, 1H), 9.70 (s, 1H), 8.43 (d, J = 6.0 Hz, 1H), 8.38 (d, J = 7.8 Hz, 1H), 7.92 (d, J = 6.0 Hz, 1H), 7.57 (d, J = 8.4 Hz, 1H), 7.53 (t, J = 7.5 Hz, 1H), 7.26 (t, J = 7.5 Hz, 1H), 3.26 (s, 3H), 2.79 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 149.6, 142.7, 140.6, 140.4, 132.5, 128.0, 127.1, 123.8, 123.4, 123.1, 121.9, 119.2, 115.9, 110.7, 108.0, 14.3, 11.9. HRMS (ESI) for C17H15N2 [M + H]+ calculated 247.1235, found 247.1236. HPLC purity: 99.7%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 95
MeOH = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) on silica gel and washed with MeOH to give compound 2 as an yellow solid (80 mg, 32.1%). mp 257.5 °C (dec). 1H NMR (600 MHz, DMSO-d6) δ 11.35 (br, 1H), 9.59 (s, 1H), 8.41–8.40 (m, 2H), 8.13 (dd, J = 6.0, 0.6 Hz, 1H), 7.59–7.54 (m, 2H), 7.25 (td, J = 7.2, 1.2 Hz, 1H), 3.14 (s, 3H), 2.96 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 148.7, 143.1, 138.4, 138.4, 128.2, 127.6, 125.7, 125.3, 124.8, 124.2, 122.6, 118.9, 116.8, 110.7, 110.6, 14.5, 11.8. HRMS (ESI) for C17H15N2 [M + H]+ calculated 247.1235, found 247.1237. HPLC purity: 97.9%.
5) on silica gel and washed with MeOH to give compound 2 as an yellow solid (80 mg, 32.1%). mp 257.5 °C (dec). 1H NMR (600 MHz, DMSO-d6) δ 11.35 (br, 1H), 9.59 (s, 1H), 8.41–8.40 (m, 2H), 8.13 (dd, J = 6.0, 0.6 Hz, 1H), 7.59–7.54 (m, 2H), 7.25 (td, J = 7.2, 1.2 Hz, 1H), 3.14 (s, 3H), 2.96 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 148.7, 143.1, 138.4, 138.4, 128.2, 127.6, 125.7, 125.3, 124.8, 124.2, 122.6, 118.9, 116.8, 110.7, 110.6, 14.5, 11.8. HRMS (ESI) for C17H15N2 [M + H]+ calculated 247.1235, found 247.1237. HPLC purity: 97.9%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 2
EtOAc = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica gel and washed with EtOAc to give compound 3 as an orange solid (15 mg, 12.1%). mp 336.8 °C (dec), 1H NMR (600 MHz, DMSO-d6) δ 13.21 (br, 1H), 9.25 (s, 1H), 9.07 (d, J = 4.8 Hz, 1H), 8.21 (d, J = 8.4 Hz, 1H), 7.93 (d, J = 4.8 Hz, 1H), 7.61 (d, J = 8.4 Hz, 1H), 7.49–7.47 (m, 1H), 7.39 (td, J = 7.5, 0.6 Hz, 1H). 13C NMR (150 MHz, DMSO-d6) δ 180.2, 176.8, 155.1, 147.4, 138.5, 138.4, 136.8, 127.5, 126.5, 124.3, 123.6, 122.4, 118.3, 117.4, 113.9. HRMS (ESI) for C15H9N2O2 [M + H]+ calculated 249.0664, found 249.0664. HPLC purity: 89.7%.
1) on silica gel and washed with EtOAc to give compound 3 as an orange solid (15 mg, 12.1%). mp 336.8 °C (dec), 1H NMR (600 MHz, DMSO-d6) δ 13.21 (br, 1H), 9.25 (s, 1H), 9.07 (d, J = 4.8 Hz, 1H), 8.21 (d, J = 8.4 Hz, 1H), 7.93 (d, J = 4.8 Hz, 1H), 7.61 (d, J = 8.4 Hz, 1H), 7.49–7.47 (m, 1H), 7.39 (td, J = 7.5, 0.6 Hz, 1H). 13C NMR (150 MHz, DMSO-d6) δ 180.2, 176.8, 155.1, 147.4, 138.5, 138.4, 136.8, 127.5, 126.5, 124.3, 123.6, 122.4, 118.3, 117.4, 113.9. HRMS (ESI) for C15H9N2O2 [M + H]+ calculated 249.0664, found 249.0664. HPLC purity: 89.7%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 1.5
EtOAc = 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica gel and washed with MeOH to give compound 4 as an orange solid (0.19 g, 27.9%). mp 311.3–313.3 °C. 1H NMR (600 MHz, DMSO-d6) δ 13.23 (br, 1H), 9.22 (s, 1H), 9.08 (d, J = 4.4 Hz, 1H), 8.19 (d, J = 7.8 Hz, 1H), 7.95 (d, J = 4.8 Hz, 1H), 7.61 (d, J = 8.4 Hz, 1H), 7.47 (t, J = 7.5 Hz, 1H), 7.39 (t, J = 7.5 Hz, 1H). 13C NMR (150 MHz, DMSO-d6) δ 179.2, 177.2, 155.9, 147.1, 139.7, 138.2, 137.0, 127.3, 125.9, 124.4, 123.7, 122.3, 118.7, 117.5, 114.0. HRMS (ESI) for C15H9N2O2 [M + H]+ calculated 249.0664, found 249.0667. HPLC purity: 99.4% (a 7.4
1) on silica gel and washed with MeOH to give compound 4 as an orange solid (0.19 g, 27.9%). mp 311.3–313.3 °C. 1H NMR (600 MHz, DMSO-d6) δ 13.23 (br, 1H), 9.22 (s, 1H), 9.08 (d, J = 4.4 Hz, 1H), 8.19 (d, J = 7.8 Hz, 1H), 7.95 (d, J = 4.8 Hz, 1H), 7.61 (d, J = 8.4 Hz, 1H), 7.47 (t, J = 7.5 Hz, 1H), 7.39 (t, J = 7.5 Hz, 1H). 13C NMR (150 MHz, DMSO-d6) δ 179.2, 177.2, 155.9, 147.1, 139.7, 138.2, 137.0, 127.3, 125.9, 124.4, 123.7, 122.3, 118.7, 117.5, 114.0. HRMS (ESI) for C15H9N2O2 [M + H]+ calculated 249.0664, found 249.0667. HPLC purity: 99.4% (a 7.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of regioisomers).
1 mixture of regioisomers).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 THF-H2O solution (24 mL). After stirring at 0 °C for 2 h, aniline (0.60 mL, 6.57 mmol) was added to the resulting mixture at 0 °C. After stirring at rt for 18 h, the mixture was diluted with DCM and stirred at rt for 0.5 h. The resulting mixture was neutralized with saturated aqueous NaHCO3 solution and filtered through Celite. The filtrate was extracted with DCM, dried by MgSO4, and concentrated in vacuo. The residue was purified by flash column chromatography (Hex
1 THF-H2O solution (24 mL). After stirring at 0 °C for 2 h, aniline (0.60 mL, 6.57 mmol) was added to the resulting mixture at 0 °C. After stirring at rt for 18 h, the mixture was diluted with DCM and stirred at rt for 0.5 h. The resulting mixture was neutralized with saturated aqueous NaHCO3 solution and filtered through Celite. The filtrate was extracted with DCM, dried by MgSO4, and concentrated in vacuo. The residue was purified by flash column chromatography (Hex![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 4
EtOAc = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica gel to give compound 6 as a red solid (0.45 g, 21.7%), mp 216.7–218.3 °C, 1H NMR (600 MHz, DMSO-d6) δ 9.47 (br, 1H), 9.20 (d, J = 0.6 Hz, 1H), 9.06 (d, J = 4.8 Hz, 1H), 7.81 (dd, J = 4.9, 0.8 Hz, 1H), 7.48–7.45 (m, 2H), 7.39 (dd, J = 8.4, 1.2 Hz, 2H), 7.27–7.24 (m, 1H), 6.15 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 181.1, 181.1, 156.0, 147.2, 146.6, 138.1, 137.6, 129.3, 125.7, 124.5, 124.0, 118.1, 102.1. HRMS (ESI) for C15H11N2O2 [M + H]+ calculated 251.0821, found 251.0822. HPLC purity: 97.5% (a 14.5
1) on silica gel to give compound 6 as a red solid (0.45 g, 21.7%), mp 216.7–218.3 °C, 1H NMR (600 MHz, DMSO-d6) δ 9.47 (br, 1H), 9.20 (d, J = 0.6 Hz, 1H), 9.06 (d, J = 4.8 Hz, 1H), 7.81 (dd, J = 4.9, 0.8 Hz, 1H), 7.48–7.45 (m, 2H), 7.39 (dd, J = 8.4, 1.2 Hz, 2H), 7.27–7.24 (m, 1H), 6.15 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 181.1, 181.1, 156.0, 147.2, 146.6, 138.1, 137.6, 129.3, 125.7, 124.5, 124.0, 118.1, 102.1. HRMS (ESI) for C15H11N2O2 [M + H]+ calculated 251.0821, found 251.0822. HPLC purity: 97.5% (a 14.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of regioisomers).
1 mixture of regioisomers).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 3
EtOAc = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica gel and washed with EtOH to give compound 8 as a red solid (0.26 g, 20%). mp 228.0–228.8 °C, 1H NMR (600 MHz, CDCl3) δ 9.81 (d, J = 0.6 Hz, 1H), 8.94 (d, J = 5.4 Hz, 1H), 7.96 (dd, J = 5.1, 0.9 Hz, 1H), 7.38–7.36 (m, 2H), 7.29–7.26 (m, 3H), 7.14–7.12 (m, 1H), 7.08–7.04 (m, 3H), 6.92 (dd, J = 8.1, 0.9 Hz, 2H), 6.72 (s, 1H). 13C NMR (150 MHz, CDCl3) δ 181.6, 154.1, 151.8, 150.9, 148.7, 140.4, 138.3, 135.2, 129.8, 129.1, 128.5, 124.9, 124.8, 121.4, 120.9, 118.0, 97.8. HRMS (ESI) for C21H16N3O [M + H]+ calculated 326.1293, found 326.1296. HPLC purity: 99.2%.
1) on silica gel and washed with EtOH to give compound 8 as a red solid (0.26 g, 20%). mp 228.0–228.8 °C, 1H NMR (600 MHz, CDCl3) δ 9.81 (d, J = 0.6 Hz, 1H), 8.94 (d, J = 5.4 Hz, 1H), 7.96 (dd, J = 5.1, 0.9 Hz, 1H), 7.38–7.36 (m, 2H), 7.29–7.26 (m, 3H), 7.14–7.12 (m, 1H), 7.08–7.04 (m, 3H), 6.92 (dd, J = 8.1, 0.9 Hz, 2H), 6.72 (s, 1H). 13C NMR (150 MHz, CDCl3) δ 181.6, 154.1, 151.8, 150.9, 148.7, 140.4, 138.3, 135.2, 129.8, 129.1, 128.5, 124.9, 124.8, 121.4, 120.9, 118.0, 97.8. HRMS (ESI) for C21H16N3O [M + H]+ calculated 326.1293, found 326.1296. HPLC purity: 99.2%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 2
EtOAc = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica gel and washed with EtOH to give compound 9 as a dark red solid (0.19 g, 61.7%). mp 239.6–240.3 °C, 1H NMR (600 MHz, DMSO-d6) δ 9.37 (br, 1H), 9.13 (s, 1H), 9.04 (d, J = 5.4 Hz, 1H), 7.89 (d, J = 4.8 Hz, 1H), 7.47–7.45 (m, 2H), 7.39 (d, J = 7.2 Hz, 2H), 7.24 (t, J = 7.5 Hz, 1H), 6.10 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 182.2, 181.4, 154.3, 146.9, 146.5, 137.8, 136.1, 129.3, 125.5, 124.8, 123.8, 118.0, 102.0. HRMS (ESI) for C15H11N2O2 [M + H]+ calculated 251.0821, found 251.0823. HPLC purity: 99.9%.
1) on silica gel and washed with EtOH to give compound 9 as a dark red solid (0.19 g, 61.7%). mp 239.6–240.3 °C, 1H NMR (600 MHz, DMSO-d6) δ 9.37 (br, 1H), 9.13 (s, 1H), 9.04 (d, J = 5.4 Hz, 1H), 7.89 (d, J = 4.8 Hz, 1H), 7.47–7.45 (m, 2H), 7.39 (d, J = 7.2 Hz, 2H), 7.24 (t, J = 7.5 Hz, 1H), 6.10 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 182.2, 181.4, 154.3, 146.9, 146.5, 137.8, 136.1, 129.3, 125.5, 124.8, 123.8, 118.0, 102.0. HRMS (ESI) for C15H11N2O2 [M + H]+ calculated 251.0821, found 251.0823. HPLC purity: 99.9%.Cytotoxicity analysis
The cell viability was analyzed by MTT assay. MV-4-11 and MOLM-13 cells were seeded into 24-well plates at a density of 2 × 105 cells per well and subsequently treated with the specified concentrations of compounds for a duration of 72 hours. Following treatment, MTT reagent (0.5 mg mL−1 in PBS) was added and allowed to incubate with the cells for 1 hour. The resulting crystal formazan dyes were then dissolved in 1 mL of sodium acetate buffer (10% SDS with 0.01 N HCl). The absorbance was determined spectrophotometrically at 550 nm using an ELISA reader (Synergy HTX ELISA reader, Biotek, CA, USA). The IC50 values of the compounds were calculated as the mean ± standard deviation (SD).Computational study
The MYLK4 protein structure (PDB ID: 2X4F) was downloaded from the Protein Data Bank repository.34 Structures of the protein and small molecules were prepared with default settings using LeadIT (LeadIT version 2.2.0; BioSolveIT GmbH, https://www.biosolveit.de/LeadIT). This includes the removal of water molecules and the addition of hydrogen atoms. The active binding site was designated with a radius of 10 Å with the co-crystal ligand as the centroid. Molecular docking was performed within the LeadIT software suite. The number of solutions for fragmentation and iteration was set to 300. All other settings were set to default. Protein–ligand interactions were generated using Pipeline Pilot35 and rendered using PyMOL.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science and Technology Council, Taiwan (grant no. MOST111-2320-B-038-046 and NSTC 112-2320-B-110-002-MY3).References
- S. Goodwin, A. F. Smith and E. C. Horning, J. Am. Chem. Soc., 1959, 81, 1903 CrossRef CAS.
- D. Thompson, C. Miller and F. O. McCarthy, Biochemistry, 2008, 47, 10333–10344 CrossRef CAS PubMed.
- M. Stiborová, J. Poljaková, E. Martínková, J. Ulrichová, V. Simánek, Z. Dvořák and E. Frei, Toxicology, 2012, 302, 233–241 CrossRef PubMed.
- E. C. O'Sullivan, C. M. Miller, F. M. Deane and F. O. McCarthy, Stud. Nat. Prod. Chem., 2013, 39, 189–232 Search PubMed.
- S. Miyake, A. Sasaki, T. Ohta and K. Shudo, Tetrahedron Lett., 1985, 26, 5815–5818 CrossRef CAS.
- C. Y. Liu and P. Knochel, J. Org. Chem., 2007, 72, 7106–7115 CrossRef CAS PubMed.
- S. Rasheed, D. N. Rao and P. Das, Asian J. Org. Chem., 2016, 5, 1499–1507 CrossRef CAS.
- G. W. Gribble, M. G. Saulnier, M. P. Sibi and J. A. Obaza-Nutaitis, J. Org. Chem., 1984, 49, 4518–4523 CrossRef CAS.
- D. A. Davis and G. W. Gribble, Tetrahedron Lett., 1990, 31, 1081–1084 CrossRef CAS.
- C. May and C. J. Moody, J. Chem. Soc., Perkin Trans. 1, 1988, 2, 247–250 RSC.
- P. A. Cranwell and J. E. Saxton, J. Chem. Soc., 1962, 3482–3487 RSC.
- L. K. Dalton, S. Demerac, B. C. Elmes, J. W. Loder, J. M. Swan and T. Teitei, Aust. J. Chem., 1967, 20, 2715–2727 CrossRef CAS.
- H. Y. Lee, G. S. Chen, C. S. Chen and J. W. Chern, J. Heterocycl. Chem., 2010, 47, 454–458 CrossRef CAS.
- E. Differding and L. Ghosez, Tetrahedron Lett., 1985, 26, 1647–1650 CrossRef CAS.
- J. M. Pedersen, W. R. Bowman, M. R. Elsegood, A. J. Fletcher and P. J. Lovell, J. Org. Chem., 2005, 70, 10615–10618 CrossRef CAS PubMed.
- M. G. Saulnier and G. W. Gribble, J. Org. Chem., 1983, 48, 2690–2695 CrossRef CAS.
- M. Watanabe and V. Snieckus, J. Am. Chem. Soc., 1980, 102, 1457–1460 CrossRef CAS.
- N. Ramkumar and R. Nagarajan, J. Org. Chem., 2014, 79, 736–741 CrossRef CAS PubMed.
- D. H. Dethe and G. M. Murhade, Eur. J. Org Chem., 2014, 6953–6962 CrossRef CAS.
- F. F. Naciuk, J. A. M. Castro, B. K. Serikava and P. C. M. L. Miranda, ChemistrySelect, 2018, 3, 436–439 CrossRef CAS.
- F. F. Naciuk, J. C. Milan, A. Andreão and P. C. M. L. Miranda, J. Org. Chem., 2013, 78, 5026–5030 CrossRef CAS PubMed.
- N. N. Shin, H. Jeon, Y. Jung, S. Baek, S. Lee, H. C. Yoo, G. H. Bae, K. Park, S. H. Yang, J. M. Han, I. Kim and Y. Kim, ACS Chem. Neurosci., 2019, 10, 3031–3044 CrossRef PubMed.
- Y. Zhao, B. Huang, C. Yang and W. Xia, Org. Lett., 2016, 18, 3326–3329 CrossRef CAS PubMed.
- L. M. Gornostaev, T. A. Rukovets, T. I. Lavrikova, Y. G. Khalyavina and G. A. Stashina, Russ. Chem. Bull., 2017, 66, 1007–1010 CrossRef CAS.
- S. Pradhan, S. Roy, S. Ghosh and I. Chatterjee, Adv. Synth. Catal., 2019, 361, 4294–4301 CrossRef CAS.
- I. Y. Sapurina and M. A. Shishov, Intech, 1989, 32, 137–144 Search PubMed.
- E. G. Russell, E. C. O'Sullivan, C. M. Miller, J. Stanicka, F. O. McCarthy and T. G. Cotter, Invest. New Drugs, 2014, 32, 1113–1122 CrossRef CAS PubMed.
- B. M. Ansari and E. N. Thompson, Postgrad. Med. J., 1975, 51, 103–105 CrossRef CAS PubMed.
- Z. Tang, C. Li, B. Kang, G. Gao, C. Li and Z. Zhang, Nucleic Acids Res., 2017, 45, W98–W102 CrossRef CAS PubMed.
- X. Jin, D. R. Gossett, S. Wang, D. Yang, Y. Cao, J. Chen, R. Guo, R. K. Reynolds and J. Lin, Br. J. Cancer, 2004, 91, 1808–1812 CrossRef CAS PubMed.
- J. Vendôme, S. Letard, F. Martin, F. Svinarchuk, P. Dubreuil, C. Auclair and M. Le Bret, J. Med. Chem., 2005, 48, 6194–6201 CrossRef PubMed.
- M. M. Attwood, D. Fabbro, A. V. Sokolov, S. Knapp and H. B. Schiöth, Nat. Rev. Drug Discovery, 2021, 20, 839–861 CrossRef CAS PubMed.
- Z. S. Derewenda, I. Hawro and U. Derewenda, IUBMB Life, 2020, 72, 1233–1242 CrossRef CAS PubMed.
- P. D. B. C. ww, Protein Data Bank: the single global archive for 3D macromolecular structure data, Nucleic Acids Res., 2019, 47, D520–D528, DOI:10.1093/nar/gky949.
- Dassault Systèmes BIOVIA Pipeline Pilot, Release 2017, Dassault Systemes, San Diego Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: HPLC purity data, 1H NMR spectra and 13C NMR spectra of compounds 1–4, 8–9, and 13–15. See DOI: https://doi.org/10.1039/d3ra06600b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |