Open Access Article
Open Access ArticleElucidating arsenic-bound proteins in the protein data bank: data mining and amino acid cross-validation through Raman spectroscopy†
Upendra Nayek,
Sudarshan Acharya and
Abdul Ajees Abdul Salam *
*
Department of Atomic and Molecular Physics, Manipal Academy of Higher Education, Manipal, Karnataka 576 104, India. E-mail: abdul.ajees@manipal.edu; Tel: +91 8147966458
First published on 12th December 2023
Abstract
The International Agency for Research on Cancer has unequivocally classified inorganic arsenic as a Group 1 carcinogen, definitively establishing its potential to induce cancer in humans. Paradoxically, despite its well-documented toxicity, arsenic finds utility as a chemotherapeutic agent. Notable examples include melarsoprol and arsenic trioxide, both employed in the treatment of acute promyelocytic leukemia. In both therapeutic and hazardous contexts, arsenic can accumulate within cellular environments, where it engages in intricate interactions with protein molecules. Gaining a comprehensive understanding of how arsenic compounds interact with proteins holds immense promise for the development of innovative inhibitors and pharmaceutical agents. These advancements could prove invaluable in addressing a spectrum of arsenic-related diseases. In pursuit of this knowledge, we undertook a systematic exploration of the Protein Data Bank, with a focus on 902 proteins intricately associated with 26 arsenic compounds. Our comprehensive investigation reveals insights into the interactions between these arsenical compounds and amino acids located within a 4.0 Å molecular distance from arsenic-binding sites. Our findings identify that cysteine, glutamic acid, aspartic acid, serine, and arginine frequently engage with arsenic. In complement to our computational analyses, we conducted rigorous Raman spectroscopy studies on the top five amino acids displaying robust interactions with arsenic. The results derived from experimental Raman spectroscopy were meticulously compared with our computational assessments, thereby enhancing the reliability and depth of our investigations. The current study presents a multidimensional exploration into the elaborate interplay between arsenic compounds and proteins. By elucidating the specific amino acids that preferentially interact with arsenic, this study not only contributes to the fundamental understanding of these molecular associations but also lays the foundation for future endeavors in drug design and therapeutic interventions targeting arsenic-related illnesses. Our work at the convergence of toxicology, medicine, and molecular biology carries profound implications for advancing our knowledge of arsenic's dual nature as both a poison and a potential cure.
1 Introduction
Metals and metalloids play significant roles in protein architecture and biological processes, contributing to the structure and function of proteins in various ways. Metals can coordinate with specific amino acid residues in proteins to form metal-binding sites, which help stabilize the protein's tertiary structure. For example, zinc ions often bind to cysteine (Cys) and histidine (His) residues, providing structural support to zinc finger motifs commonly found in transcription factors and DNA-binding proteins.1 Many proteins, known as metalloenzymes, contain metal ions as essential cofactors for catalytic activity. These metal ions often serve as Lewis acids, facilitating chemical reactions by acting as electron acceptors or donors. Examples include zinc in carbonic anhydrase, iron in hemoglobin, and magnesium in ATPases. Metals, particularly transition metals like iron and copper, can participate in electron transfer reactions within proteins. In redox proteins, they help shuttle electrons between molecules, enabling essential processes like respiration and photosynthesis. Hemoglobin, which contains iron, is a well-known example of a protein involved in electron transfer.2 Hemoglobin and myoglobin contain iron atoms at the center of a heme group. These proteins are responsible for binding and transporting oxygen in the bloodstream. The iron in heme binds to oxygen, allowing for efficient oxygen uptake in the lungs and its release to tissues.3 Metal ions can serve as regulatory elements in metalloregulatory proteins. These proteins control the intracellular concentration of specific metals and respond to changes in metal availability. Selenium is essential for certain enzymes, such as glutathione peroxidase, which helps protect cells from oxidative damage.4 Some proteins contain metal-binding sites that act as sensors for metal ions. These sensors can trigger signaling pathways in response to changes in metal ion concentrations. For example, metal-sensing transcription factor regulates the expression of genes involved in metal homeostasis.5Arsenic, a naturally occurring metalloid, is renowned for its human toxicity and carcinogenicity.6,7 A recent study by Paul et al. provides an overview of the historical, present, and future applications of arsenic-based medicines.7 The term “arsenic” finds its origins in the Greek word “arsenikon,” signifying potency. Throughout history, arsenicals have been employed in various fields including agriculture, cosmetics, electronics, metallurgy, medicine, and industry.6 Arsenic, classified as a human carcinogen, belongs to the category of toxic metalloids known for inducing substantial environmental pollution and has been linked to various health issues. Arsenic compounds such as melarsoprol and arsenic trioxide are chemotherapeutic agents in acute promyelocytic leukemia.8,9 The use of arsenic as an essential ingredient in pharmacology is widespread due to its mode of action in many disorders. The research focused on detoxifying and bioaccumulating trace elements present in the biological system, and it has been extensively reviewed based on metal-binding proteins.10–12 Arsenic is recognized as a hazardous element and has attracted significant research interest owing to its carcinogenic properties in humans.
Research has unveiled complex biological properties and toxicity mechanisms of arsenic. Various variations of the arsenic molecule's oxidative biological state (−3, 0, +3, and +5) and their interference with cellular activities have been slowly uncovered in arsenic sciences. Arsenite (As(III)) and arsenate (As(V)) are two essential forms of arsenic commonly chosen for research due to their distinct chemical properties, biological relevance, toxicity, environmental significance, and various applications in fields such as toxicology, environmental science, biochemistry, and health research. Arsenite, the reduced form of arsenic, is frequently employed in studies involving redox reactions. It serves as an electron donor in enzymatic processes and is one of the most toxic forms of arsenic, associated with adverse health effects. Understanding its mechanisms of toxicity is crucial for public health.13,14 Arsenite's toxicity makes it a focus in cancer therapy research, given its potential to harm rapidly dividing cells.15 On the other hand, arsenate, the oxidized form, shares structural similarities with phosphate (PO43−) and is used to investigate competitive inhibition and binding to phosphate-binding sites. While less toxic than arsenite, arsenate is of interest due to its potential health implications when present in drinking water and food.13,14 Both forms are biologically relevant and found in the environment, especially in groundwater. As per the WHO, over 50 countries and 140 million individuals face the risk of exposure to high levels of arsenic-contaminated water through drinking or groundwater, compromising their overall well-being. Approximately 39 million people in Bangladesh are affected by high levels of arsenic through contaminated water (more than 50 μg L−1), contributing to a 21.4% increase in the death rate.16 A recent study determined that chronic exposure to arsenic in drinking water significantly elevates mortality risk, irrespective of sex. This risk is particularly pronounced among the younger generation.17,18 Understanding how organisms transport, metabolize, and detoxify these arsenic forms is crucial for basic biological research and environmental science. Arsenite and arsenate are also valuable tools for structural and biochemical studies.10,13,19 Their interactions with proteins, enzymes, and nucleic acids provide insights into biological processes and facilitate the design of experiments investigating these interactions.
Inorganic arsenic forms, such as arsenite and arsenate, disrupt vital biochemical processes by binding to thiol (–SH) groups in proteins, leading to structural and functional changes and enzyme interference. Arsenic exposure triggers the generation of reactive oxygen species (ROS), causing cellular oxidative stress. ROS damages DNA, proteins, and lipids, contributing to health issues, including cancer. Arsenic compounds induce DNA damage, mutations, and genomic instability, all linked to cancer. Altered DNA methylation patterns from arsenic exposure silence tumor suppressor genes and activate oncogenes, further promoting carcinogenesis. Arsenic interferes with cellular signaling pathways controlling growth, apoptosis, and DNA repair, leading to uncontrolled cell proliferation and resistance to programmed cell death, both cancer hallmarks.20 Chronic arsenic exposure weakens the immune system, reducing the body's ability to detect and defend against cancer cells. Arsenic's carcinogenicity varies between tissues, with strong associations in skin, lung, and bladder cancers. Genetic factors play a role in individual susceptibility to arsenic's toxic effects, necessitating investigation to understand varying vulnerabilities. As discussed earlier, arsenic contamination in drinking water and food sources is a pressing public health concern, urging research to inform regulations, reduce exposure, and develop treatments for arsenic-induced diseases.21 Understanding arsenic's mechanisms of toxicity and carcinogenicity is critical for establishing safe exposure limits, implementing water treatment solutions, educating at-risk populations, and promoting targeted treatments.
Investigation into a few arsenical protein structures shows that the functional elementary attraction is offensive and unaltered by the evolving amino acids in active protein locations.22 A nontoxic pretreatment of heavy metal doses ensured enhanced Cys-rich metalloprotein expression. Metallothionein is a metal-binding protein synthesized in specific sites within tissues in response to induction by various metal ions.23 In 1999, D. B. Menzel et al. reported that human proteins had a high affinity for arsenical ions in the form of the As(III) oxidation state in lymphoblastoid cells.24,25 Knowing the detoxification process of different proteins, such as heme oxygenase 1, actin, and tubulin, may provide information about the risk associated with a specific dose of heavy metals. The arsenic-caused hepatocarcinogenesis also involves aberrant gene expression, and its entire phenomenon is predicated on the effects of hypomethylation, the enzyme involved in the DNA (deoxyribonucleic acid) repair mechanism.26 It is critically rich in thiol and is less sensitive to arsenical ions. According to Zhang et al., arsenic has been found to stabilize mRNA functions through a cell arrest mechanism, which helps protect the DNA damage of gene 45α.27 This stabilization also leads to a significant increase in DNA binding protein (DBP) activity. Furthermore, specific cofactors such as Zn, Fe and Cu play a vital role in facilitating the functionalization of proteins in various oxidation states. Cofactors within the protein's structural framework often serve as a critical factor in stabilizing the protein and significantly enhancing its functional capabilities.
A nanomolar quantity of arsenical ions can activate various cellular healing mechanisms by activating enzymes that are involved in cellular toxicity in the human system. Applying an arsenical substance to the cell cycle regulator, which releases the guanine nucleotide protein, results in a 32% difference in esophageal squamous carcinoma CdC25.28 The cell cycle regulator releases the guanine nucleotide protein as part of its normal function. However, when an arsenical substance is added to the cell cycle regulator, it disrupts the regulation process and removes the guanine nucleotide protein. This interference in cell cycle regulation can significantly affect cellular processes and functions. In the As(III) structural analysis with α-helical peptides, As(III) with low and high pH can form a trigonal pyramidal structure with three cysteines.29 Determining what happens when arsenic binds to Cys residues exposed to the surface in native protein helical sections is a fascinating topic to research in arsenic-bound proteins.30 Understanding how various proteins, peptides and amino acids interact with arsenic can help us better understand how arsenic affects human health.7,22 By continuously elucidating the structures of arsenic-binding proteins, it is possible to understand how arsenic affects cells.31 Various data from biomolecular studies show that arsenic binding to macromolecules causes heavy metal sequestration in bacterial proteins. However, the precise mechanism of arsenic transfer into human cells is currently unknown. There seems to be little research on structural pieces of evidence despite an increased number of studies on the toxicity of arsenic metal in humans. Based on this scientific background, structural studies of arsenite and arsenate are chosen for this research due to their unique properties and their contributions to understanding arsenic behavior and effects in various contexts. Understanding the interactions between these arsenic compounds and proteins will provide insight into arsenic detoxification, which will help design novel inhibitors and drug molecules to treat various arsenic-related diseases. PDB is an archive of experimentally determined 3-D structures of macromolecules such as proteins, DNA, and RNA in the presence or absence of multiple inhibitors. Currently, PDB holds 1068577 crystal structures. 902 arsenic-bound crystal structures were taken for this study to understand the environment of arsenic binding with proteins.
2 Methods
2.1 Structural data preparation
PDB has 902 structures with different arsenical compounds. The effects of amino acid interactions with arsenical molecules related to structural and functional protein consequences are significantly illustrated. The PDB is a fundamental resource for acquiring detailed 3-D structural protein information. The protein structures containing arsenic molecules as ligands have been extracted within the PDB. The 3-D structures of 26 types of arsenic ligands (Fig. 1 and 3) co-crystallized with 902 protein structures were downloaded and saved as *.pdb files; details are shown in ESI Tables 1 and 2.† When dealing with multiple chains (having the same amino acid sequence) of protein molecules, chain A is typically selected to investigate the interactions between the protein and its ligands. The protein–ligand interactions were calculated within a 4.0 Å distance using the CCP4 graphical suite, version 7.0.073.32 The bond distance between the arsenic ligands and the amino acids was tabulated using Microsoft Excel, and graphs were plotted using Origin (OriginPro version 2018; https://www.originlab.com/).33 The interactions between the carbon atoms and arsenic species were omitted due to the abundance of carbon atoms. The two-dimensional (2D) figures of ligands were prepared using MarvinSketch (version 19.10; https://chemaxon.com/products/marvin).34 https://pymol.org/2. R programming was employed for performing graphical and statistical studies of environmental data. The Ligplot figures were downloaded from PDBSum (https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/) and used without modification.2.2 Raman spectroscopy studies
The five amino acids, cysteine (Cys), arginine (Arg), aspartic acid (Asp), glutamic acid (Glu) and serine (Ser), which showed good arsenic interactions from the PDB database studies, were selected for Raman spectroscopic studies. Raman spectra were recorded for (i) sodium arsenite [As(III)], (ii) sodium arsenate [As(V)], (iii) amino acid powder, (iv) amino acid solution, (v) amino acid-As(III) mixed solution and (vi) amino acid-As(V) mixed solution. The commercial Raman setup i-Raman® Plus with an excitation wavelength of 785 nm was used for the experiment. Amino acids, sodium arsenite and sodium arsenate were purchased from Merck and used with any further purification. The amino acids and arsenic species were dissolved in water. In the case of a Cys–arsenic mixture, the solutions precipitated. In such a case, the precipitate was separated using a centrifuge, and Raman spectra were recorded separately for the supernatant and the sediment. The Raman spectra plotted the intensity (a.u.) of the scattered light (Y-axis) for each energy (frequency) of the laser (X-axis). The frequency is traditionally called a wavenumber unit, measured by the Raman shift (cm−1).3 Results and discussion
3.1 Protein structures contain As(III) and As(V) based ligands
Arsenic can exist in −3, 0, +3 and +5 oxidation states, in which arsenite-As(III) and arsenate-As(V) are the predominant states. Inorganic trivalent arsenic, such as arsenic trioxide, arsenic trichloride and sodium arsenite, and inorganic pentavalent compounds, such as arsenic pentoxide, arsenic acid, lead arsenate and calcium arsenate, are the most commonly available biologically relevant arsenic molecules. Among organic arsenic compounds, arsanilic acid, monomethyl arsenic acid, dimethyl arsenic acid and arsenobetaine are the most frequently occurring compounds.35 In PDB, all these varieties of compounds are found. ESI Table 1† contains the details of arsenic-based trivalent ligands in the PDB. The three-letter id, the chemical name, the chemical formula, the molecular weight of the ligand, and the respective PDB id of each ligand are summarized in ESI Table 1.† Similarly, the pentavalent arsenic ligands and their details are outlined in ESI Table 2.† In structural biology and molecular modeling, a common practice involves calculating molecular interactions between amino acids and drugs within a 4 Å radius.19,36 This radius approximates the distance at which metal or metalloid ions, like arsenic, directly interact with specific amino acid residues in proteins, primarily through coordination bonds. This choice of radius is vital in the context of arsenic-binding proteins as it captures the most relevant atomic interactions.10 These proteins, often metalloenzymes or metalloregulatory proteins, have specific binding sites designed to accommodate arsenic, with coordinating residues typically within a few angstroms of the arsenic atom. The 4 Å radius also accounts for solvation effects, including water molecules and solvent molecules associated with the arsenic-binding site. This is crucial because water molecules can influence the stability and geometry of metal binding. While larger radii could capture more distant interactions, a 4 Å radius strikes a balance between accuracy and computational efficiency, simplifying calculations. It defines the active or binding site in metalloproteins and metalloregulatory proteins, ensuring that only the most relevant atomic interactions are considered, focusing investigations on biologically significant interactions. Thus the 4 Å radius choice is significant for its alignment with coordination chemistry principles, biological relevance, and modeling accuracy. It captures essential interactions, accounting for both coordination bonds and solvent molecules, while maintaining computational efficiency in structural investigations.3.2 Interaction of trivalent arsenic ligands
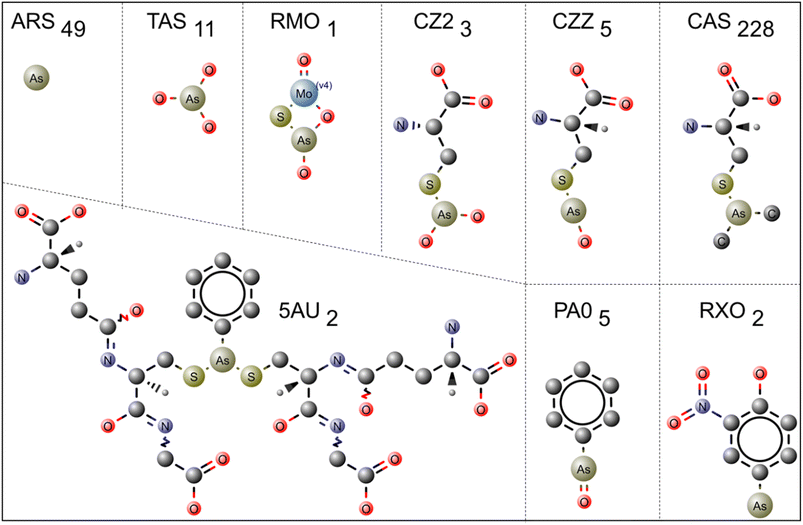
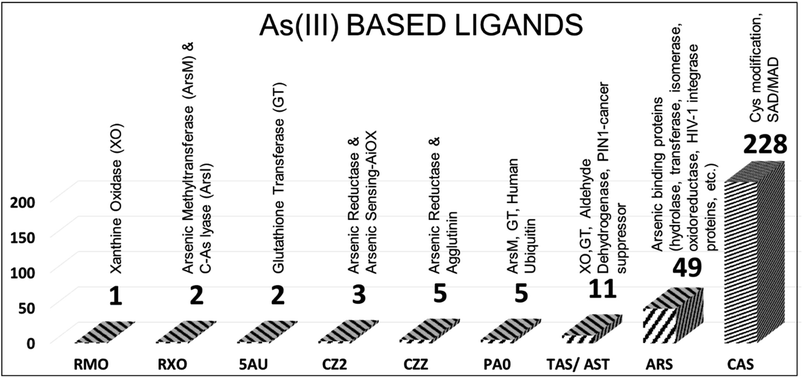
In the PDB database, 306 protein structures are cocrystallized with nine types of trivalent arsenic [As(III)] ligands (Fig. 1). Arsenothioito(2-)kappa-2-O,S(oxo)molybdenum (RMO – 1 entry, PDB id: 3SR6), 4-arsanyl-2-nitrophenol (RXO – 2 entries: PDB IDs: 4RSR, 5V0F), di-glutathione-phenylarsine (5AU – 2 entries: 5DDL, 5DAL), s-(dihydroxyarsino)-cysteine (CZ2 – 3 entries: PDB IDs: 1SJZ, 1SK0, 6EU7), phenylarsine oxide (PA0 – 5 entries: PDB IDs: 3E3Z, 4KW7, 5EG5, 5DAL, 6QHK), trihydroxy arsenite (TAS/AST – 11 entries), thiarsahydroxy-cysteine (CZZ – 5 entries, PDB IDs: 1J9B, 6YH0, 6TSQ, 6TSR, 6TSN), arsenic ion (ARS – 49 entries) and s-(dimethyl arsenic)cysteine (CAS – 228 entries) are the As(III)-based ligands present in the PDB (ESI Table 1†).The RMO ligand is co-crystallized with xanthine oxidase, which oxidizes hypoxanthine to xanthine in mammals. Arsenite is known for inhibiting xanthine oxidase and tightly binding to the reduced protein form. The soaking method introduces arsenic to protein crystals, and the final crystal structure of reduced (arsenic bound) and oxidized crystals did not show any significant difference. Arsenic is bonded between sulfur and oxygen atoms of the molybdenum complex structure.37
The RXO ligand is co-crystallized with arsenic methyltransferase (PDB id: 4RSR), which converts toxic inorganic arsenic to nontoxic organic arsenic.38,39 In this case, the monomethyl arsenite ligand RXO binds to two of the Cys174 and Cys224 residues of arsenic methyltransferase. The other crystal structure belongs to ArsI, a C–As lyase, which breaks the bond between carbon and arsenic and converts trivalent organoarsenicals to inorganic arsenate [As(III)].40
The 5AU ligand is co-crystallized with human glutathione transferase (PDB ids: 5DAL, 5DDL), and di-glutathione phenylarsine binds to the enzyme's active site. Interestingly, the PDB id 5DAL has another dimethyl arsenite derivative, PA0, which is also co-crystallized with this enzyme and is covalently attached to one of Cys residues (Cys101) near the enzyme's active site. CZ2 and CZZ are stereospecific compounds, and based on DrugBank, both are drug targets for arsenate reductase protein. CZ2 has three entries in PDB (PDB ids: 1SJZ, 1SK0, 6EU7). Two belong to the arsenate reductase R773-ArsC crystal structure, where CZ2 covalently binds to the sulfur atom of Cys12 and makes the protein more stable.41 The other structure (PDB id: 6EU7) belongs to the arsenic sensing protein (AioX), where CZ2 is bound to the sulfur atom of Cys106, an active site Cys residue.42
Interestingly, arsenite-bound protein brought significant conformational changes in one of the loop regions (residues 53–61) and the backbone shift in a nearby region (residues 83–88) compared with native protein. Thus, these arsenic derivatives are specific arsenic-binding molecules. CZZ is used to modify the Cys residues and has five entries. Out of five entries, four structures belong to a sugar-binding protein named Marasmius oreades agglutinin (PDB ids: 6TSR, 6YH0, 6TSQ, 6TSN),43 and one belongs to arsenate reductase (PDB id: 1J9B) protein. For example, in arsenate reductase (PDB id: 1J9B), Cys12 is modified to CZZ. Interestingly, TAS is also bound near CZZ in this structure.43,44 In the case of Marasmius Oreades agglutinin protein, the catalytic thiol group of Cys215, was chemically modified to selenocysteine.43
The PA0 ligand is a dimethyl phenyl arsenite derivative that co-crystallized with five proteins. Two of the enzymes (PDB ids: 5EG5, 4KW7) are arsenic methyltransferase (ArsM), and arsenic is attached to two of the catalytic Cys residues (Cys174, Cys224), which is similar to the RXO ligand discussed earlier. Another one is bovine coupling factor B (PDB id: 3E3Z), a metalloprotein. The ligand is coupled to the sulfur atom of Cys71 and a water molecule in the protein, and PA0 is an inhibitor of the coupling activity of factor B.45 The human ubiquitin-conjugating protein UBE2S is another enzyme co-crystallized with PA0 (PDB id: 6QHK). UBE2S is crystallized as a dimer (chains A and B), and PA0 connects the dimer with Cys118, which is situated in the dimer interface of both chains.46 The PDB id 5DAL is crystallized with the ligand 5AU.
AST (arsenite) and TAS (trihydroxy arsenite(III)) are stereospecific isomers. The AST has four entries (PDB ids: 1SIJ, 3L4P, 3NVV, 6CZ9) in PDB, and TAS (PDB ids: 1IHU, 1II0, 1II9, 1J9B, 1SJZ, 1SKO, 6DUN) has seven entries. The PDB IDs 1SIJ and 3L4P are aldehyde dehydrogenases, PDB id 3NVV is bovine xanthine oxidase and PDB id 6CZ9 is arsenate respiratory reductase. The TAS crystallized with R773-ArsA (PDB ids: 1IHU, 1II0, 1II9), R773-ArsC (PDB ids: 1J9B, 1SJZ, 1SKO) and P1N1 (PDB id: 6DUN) proteins. Here, R773-ArsA is an ATPase attached to the membrane protein ArsB and extrudes arsenite from the cell.47 R773-ArsC is an arsenic reductase that converts pentavalent arsenate to trivalent arsenite, and ATS is attached to one of the Cys12.41 A similar mechanism has been discussed for CZ2 previously. The PDB id 6DUN belongs to the PIN1 protein, an isomerase involved in leukemia, liver and breast cancer. In this work, ATS cooperatively works with another small molecule called ATRA (all-trans retinoic acid) to inhibit and degrade the PIN1 protein. The oncogenic function of PIN1 is suppressed by ATS by noncovalent binding to the active site of PIN1.48 In this case, Cys does not mediate ATS. In contrast, it forms hydrophobic interactions with glycine (Gly), leucine (Leu), methionine (Met), glutamine (Gln), and histidine (His) residues surrounded by the ATS.
According to PDB statistical data, ARS is co-crystallized with hydrolase, transferase, isomerase, oxidoreductase, membrane protein, transcription, signaling protein and HIV-1 integrase proteins. In total, ARS has 49 entries in the PDB. In general, ARS binds to three Cys residues. For example, in the arsenic binding protein ArsR (PDB IDs: 6J05 and 6J0E), which is an As(III)-responsive transcriptional repressor, ARS binds to three Cys residues (Cys95, Cys96, and Cys102). When arsenic binds to ArsR, it induces conformational changes and dissociates from the operator DNA. Once ArsR is dissociated from DNA, ars genes such as ArsA, ArsC, ArsB and ArsD are expressed to extrude arsenic from cells.49 Similarly, in arsenic methyltransferase (PDB id: 4FSD), the ARS binds to catalytic Cys residues (Cys174, Cys224).38 In the case of Leishmania siamensis triosephosphate isomerase (PDB id: 4GNJ), the ARS ion is bound to the active site Cys77. Regarding a three-stranded coiled coil of a synthetic peptide, arsenic ions bind to three Cys residues in a trigonal pyramidal geometry with an average As–S bond distance of 2.28 Å (PDB Id: 2JGO).50 Thus, the ARS predominantly binds to arsenic-binding proteins and plays a significant role in the arsenic detoxification mechanism.
The trivalent ligand CAS has the highest number of entries, totaling 228. CAS generally improves crystal quality by modifying the Cys amino acids of a particular protein. For example, in the methionyl-tRNA synthetase crystal structure, CAS is used to alter the Cys amino acids Cys318 and Cys470 (PDB id: 4EG4). According to DrugBank (https://go.drugbank.com/), CAS is also considered to be a drug target for toll-like receptor 2, capsid scaffolding protein, gag-poi poly protein, nitric oxide synthase-endothelial, coatomer subunit gamma-1, genome polyprotein, DNA repair protein XRCC4 and peptide methionine sulfoxide reductase MsrA. This ligand is co-crystallized in the PDB database with transferase, ligase inhibitor, oxidoreductase, RNA binding protein, protein transport transcription, and viral protein structures. Fig. 2 is the representative figure of As(III) ligands' interaction with protein molecules.
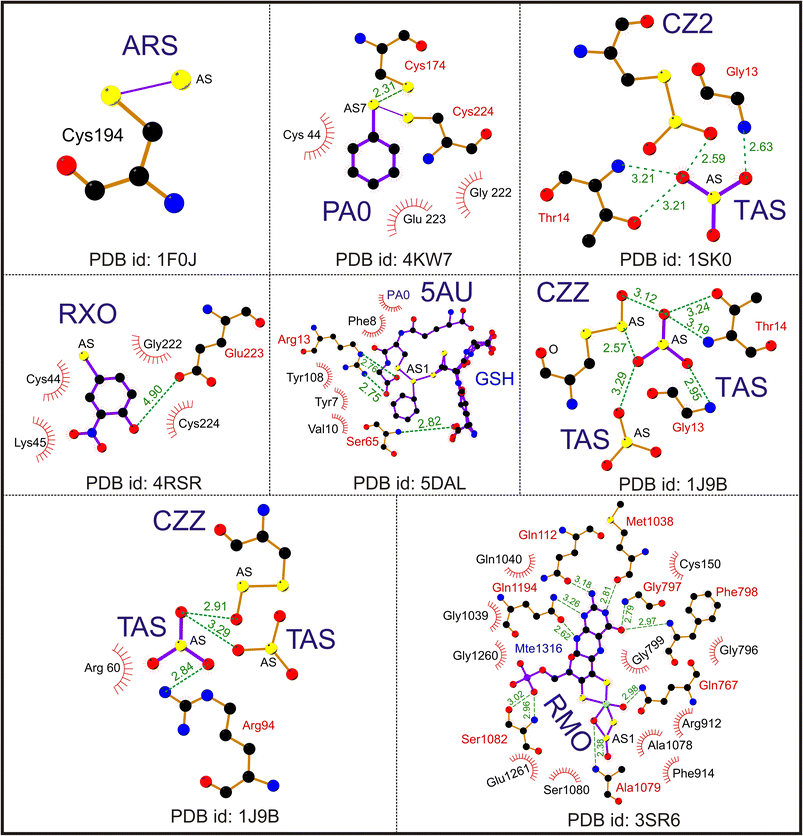 | ||
| Fig. 2 Interaction of trivalent arsenical [As(III)] ligands. A representative Ligplot showing the interaction of ligands ARS (PDB id: 1F0J), PA0 (PDB id: 4KW7), CZ2 (PDB id: 1SK0), RXO (PDB id: 4RSR), TAS (PDB id: 1J9B), CZZ (PDB id: 1J9B), 5AU (PDB id: 5DAL) and RMO (PDB id: 3SR6). The solid lines represent covalent bonds; the dotted lines represent hydrogen bonds. The hydrophobic residues are shown as spike circles. The hydrogen bond distances are marked. | ||
3.3 Interaction of pentavalent arsenic ligands
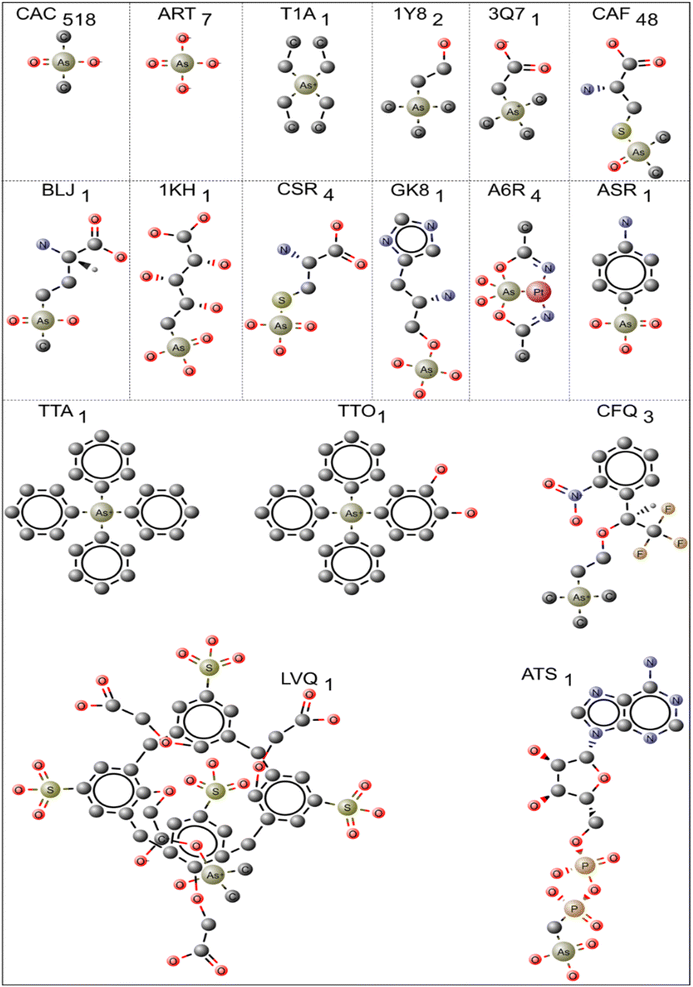
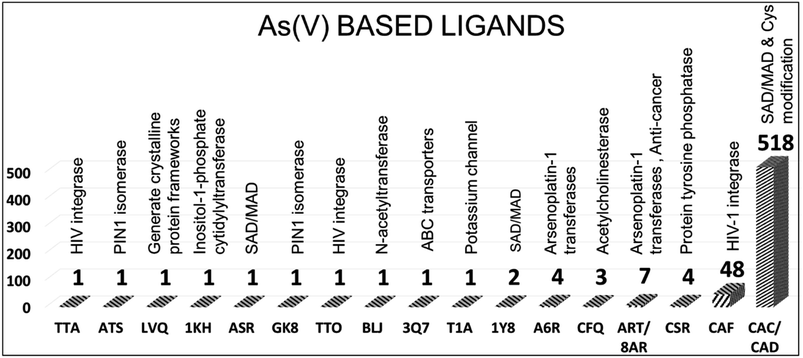
In PDB, 596 protein structures have 17 different pentavalent arsenical [As(V)] ligands. The 2D chemical structure of the ligands is shown in Fig. 3. The names of the ligands, three-letter codes and molecular formulas are listed in ESI Table 2.† Out of 17 arsenic compounds, ten small molecules, namely, tetraethylarsonium ion (T1A, PDB id: 2BOC), (trimethylarsonio)acetate (3Q7, PDB id: 5NXX), tetraphenyl-arsonium (TTA, PDB id: 1HYV), (3,4-dihydroxy-phenyl)-triphenyl-arsonium (TTO, PDB id: 1HYZ), 2S-2-amino-4-[hydroxy (methyl)arsoryl]butanoic acid or arsinothricin (BLJ, PDB id: 5 WPH), arsenic-ribose (1KH, PDB id: 4JD0), gamma-arsono-beta, gamma-methyleneadenosine-5′-diphosphate (ATS, PDB id: 1GLJ), octa-anionic calixarene (LVQ, PDB id: 6SUV), {[(2S)-2-amino-3-(1H-imidazol-5-yl)propyl]oxy}(trihydroxy)-lambda-5-arsanyl (GK8, PDB id: 4GK8) and 4-aminophenylarsonic acid (ASR, PDB id: 1N4F), have one entry.The remaining seven compounds are CAC (CAC has 457 entries, and CAD has 61 entries, both combined and presented under CAC, resulting in a total of 518 entries), CAF (48 entries), ART (7 entries, PDB ids: 1TA4, 3ENZ, 3 WE3, 6CZ8, 6XL2, 4F18, 4F19), CSR (4 entries, PDB ids: 1OKG, 1SK1, 1LJU, 1JZW), A6R (4 entries, PDB ids: 5NJ1, 7BD7, 5NJ7, 6ZS8), CFQ (3 entries, PDB ids: 2V96, 2V97, 2V98) and 1Y8 (2 entries, PDB ids: 4LLH, 5NXY) have more than one entry. The CAC and CAD represent cacodylic acid. The tetraphenyl-based arsenic small molecules (TTA, TTO) are co-crystallized with HIV integrase protein (PDB ids: 1HYV, 1HYZ). They are bound at the dimer interface of the catalytic domain of the protein.51 In these cases, the arsenic derivatives are also used to collect the single-wavelength anomalous diffraction (SAD) data and help model the structure of HIV integrase.
Similarly, the ASR (PDB id: 1N4F) molecule was also used to demonstrate the phasing power of arsenic in hen egg white lysozyme as a model protein.52 The T1A ligand is co-crystallized with prokaryotic potassium channel KcsA protein.53 The small molecule tetraethylammonium (TEA) blocks the ion conduction of KcsA, tetraethylarsonium is an analog of TEA, and it is chosen to co-crystallize with KcsA instead of TEA to obtain unambiguous electron density due to the anomalous scattering properties of arsenic. The arsenic molecule T1A helped identify the external entryway of the potassium channel. The recent crystal structure of ArsN, N-acetyltransferase (PDB id: 5 WPH), was crystallized with arsinothricin (BLJ), an analog of glutamate amino acid. BLJ is isolated from soil bacteria and identified as an antibiotic against Gram-positive and Gram-negative bacteria.14 BLJ occupies the binding site of ArsN and interacts with ArsN active site amino acids such as Ile31, Phe33, Arg75, Arg77, Ala124 and Val158. In the case of protein structure, T. maritima inositol-1-phosphate cytidylyltransferase, arsenosugar 1KH (PDB id: 4JD0), was accidentally found in the electron density, and the author did not include it in the crystallization buffer. The arsenoribose molecule 1KH occupies the protein's binding site and make intense interactions with several amino acids, which helps the authors identify the active site and deduce the nature of the catalytic mechanism.54
3.4. Arsenic-mediated cysteine residue modification in SAD/MAD phasing for protein crystallography
Arsenite (As(III)) can modify Cys residues in proteins through the formation of covalent bonds, primarily in the form of arsenic–thiol (As–S) complexes. This modification can have several biological implications, including alterations in protein structure and function. Arsenite reacts with cysteine residues in proteins through the nucleophilic attack of the thiol (–SH) group of Cys on the arsenic atom of As(III).55 This results in the formation of an As–S bond, which can modify the structure and function over 200 enzymes.56 The formation of As–S bonds can disrupt the native protein structure, potentially leading to misfolding or aggregation. This can affect the protein's stability and function. Some enzymes contain Cys at their active sites. Modification of these Cys residues by As(III) can lead to the inhibition of enzyme activity, disrupting critical cellular processes.20 Proteins involved in cellular signaling often contain Cys residues that play a role in redox signaling. Arsenic modification of these Cys residues can perturb redox signaling pathways. Arsenic modification of Cys residues can lead to the production of reactive oxygen species (ROS), contributing to oxidative stress and potential damage to cellular components. Arsenic can interfere with the activity of metalloenzymes that contain essential metal cofactors, such as zinc or iron, by replacing these cofactors with As(III) ions.1,20,56 Arsenic is well-known for its toxicity and carcinogenicity. Arsenic-induced modifications of Cys residues and disruption of cellular processes can contribute to these adverse effects, including the development of cancer.56Arsenic derivatives are also routinely used to mutate Cys and de novo phasing of protein structures using arsenic as the anomalous scatter for the SAD phasing method.52,57 Arsenic derivatives are more effective in collecting SAD data than other anomalous scatters (traditionally soaked heavy atoms such as selenium, iodine, or metals such as platinum and mercury) due to their covalent attachment with Cys residues of the proteins. Arsenic compounds also improve crystallization and increase the chance of good-quality protein crystals by eliminating unfavorable disulfide between the surface Cys residues. In addition, the arsenic derivatives did not change the secondary structure of the proteins and did not alter the protein structure. Selenium is commonly used to collect SAD data, and its K absorption edge is ∼0.98 Å. Arsenic is located beside selenium in the periodic table with a K absorption edge of ∼1.04 Å.
Arsenic molecules such as cacodylic acid (dimethylarsinate-CAC-C2H6AsO2/dimethylarsinic acid-CAD-C2H7AsO2) are mixed with protein samples during crystallization. CAD has a pKa value of 6.3 and is one of the most effective buffers. It modifies the Cys residues on the surface, and in most cases, it forms a more stable covalent bond between the arsenic and sulfur atom of the Cys. The pentavalent [As(V)] ligand CAC/CAD is attached to Cys and helps to generate the anomalous signal. The predominant anomalous signals of arsenic are sufficient to phase the protein structures alone. For example, in the crystal structure of pyrazinamidase/nicotinamidase from streptococcus (PDB id: 3S2S), the CAD is attached to Cys residue 170. In the human caspase-6 protein structure (PDB id: 3S70), CAD is reduced to a trivalent arsenic group and binds to Cys residues in four different positions of the protein (Cys68, Cys87, Cys264 and Cys277) in the presence of the reducing agent dithiothreitol (DTT). The arsenic derivatives modify the Cys residues and obtain the SAD phases in both cases. A similar case is also reported for HIV-1 integrase protein (PDB ids: 1B9F, 1B92, 1B9D), in which the CAC buffer covalently modifies two Cys residues 65, 130 and arsenic is used to collect the multiple anomalous scattering data to solve the protein structure.58 In a recent study, CAD was used to collect anomalous data to solve the structure of a biodegradable plastic-degrading cutinize-like enzyme (PDB id: 7CW1). However, in this case, arsenic does not form any covalent bonds.
The PDB holds sixty-one protein structures bound with cacodylic acid (CAD). Among these protein structures, 52 belong to nitric oxide synthase (EC: 1.14.13.39) and have specific gene functions (GO:0004517). The nine structures belong to the group of acetyltransferases, Bcl-2-related protein A1, esterase, and putative pyrazinamide/nicotinamides. The interaction between the arsenic atoms of CAD with the amino acids shows that Cys is the only amino acid near the arsenic atom for all the structures except two (PDB IDs: 3TO9 and 3TO7).
Interestingly, CAC buffer was used to crystallize all the nitric oxide synthase structures. Although the literature does not specify any role of CAC in nitric acid synthase function, it may help obtain crystals. As discussed earlier, arsenic is also used as an additive in CAC/CAD for the SAD/Multiple Anamalous Diffraction (MAD) experiment to collect anomalous scattering data. Arsenic has an advantage due to its wavelength (∼1.04 Å), which is close to that of Se (∼0.98 Å). In addition, CAC was also involved in the chemical reaction. Liu et al. have used CAC, and according to their reports, CAC is employed for similar chemical notifications involving an amino acid.57 Some structures of CAC are bound at the protein's active site, such as phosphate-binding protein (PDB ids: 3HTW, 4GY1, 1PQU, 2ACQ). Currently, the PDB holds 228 deposited protein structures bound with S-(dimethyl arsenic) cysteine (CAS). Among these structures, 32% belong to nitric-oxide synthase (Bos taurus), 12% to methionyl-tRNA synthetase (Trypanosoma brucei), 15% to elongin-B (Homo sapiens) and 10% to RNA polymerase (Enterovirus C). CAS, as a ligand, is used in various biochemical functions.
For instance, covalent binding between Cys and CAS helps improve the structure, and the dimethyl arsenic group has an average temperature of 31.3 Å2, similar to other amino acid atoms (30.0 Å2).55 This modification facilitates the orientation of the two methyl groups of CAS toward the protein core, allowing them to establish van der Waals interactions with side chain atoms. These modified Cys residues imply that the active site conformation, as observed in this study, remains consistent regardless of the crystal environment. Furthermore, these modified residues play a role in stabilizing the overall conformation of the protein (PDB id: 1BHL). The electron density of the modified Cys23 looks clear, it is covalently connected by dimethylarsenite, and the bond distance is approximately 2.4 Å between the arsenic and Cys23 sulfur.59 Cacodylate acid, an organoarsenic ligand, also helps stabilize the protein structure. It occupies the protein binding sites, replacing phosphate and citrate ions and enabling the covalent modification of surface Cys residues. A representative As(V) ligand interaction with protein molecules is shown in Fig. 4. The other primary pentavalent compound CAF has 48 entries and 28 crystal structures belonging to HIV-1 integrase; the remaining compounds belong to DNA/RNA binding, hydrolase, transferase and oxidoreductase proteins. CAF is the stereospecific compound of the trivalent arsenic molecule CAS. Thus, CAF has similar interactions to CAS, as discussed earlier.
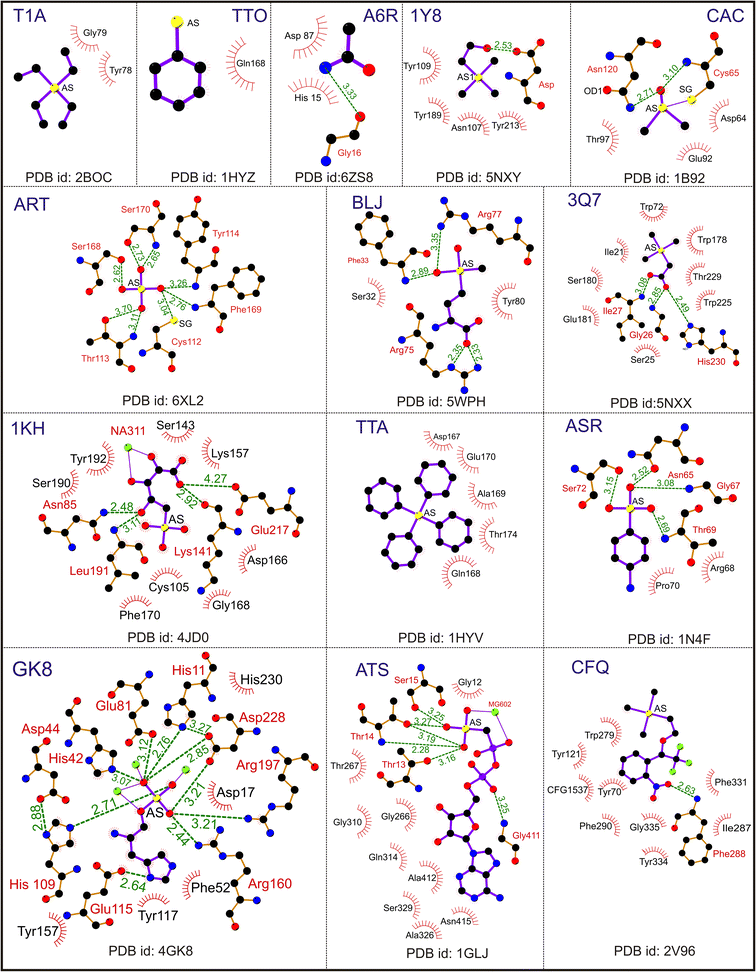 | ||
| Fig. 4 Interaction of pentavalent arsenical [As(V)] ligands. A representative Ligplot showing the interaction of ligands T1A (PDB id: 2BOC), TTO (PDB id: 1HYZ), A6R (PDB id: 6ZS8), 1Y8 (PDB id: 5NXY), CAC (PDB id: 1B92), ART (PDB id: 6XL2), BLJ (PDB id: 5 WPH), 3Q7 (PDB id: 5NXX), 1 KH (PDB id: 4JD0), TTA (PDB id: 1HYV), ASR (PDB id: 1N4F), GK8 (PDB id: 4GK8), ATS (PDB id: 1GLJ) and CFQ (PDB id: 2V96). The solid lines represent covalent bonds, and the dotted lines represent hydrogen bonds. The hydrophobic residues are shown as spike circles. The hydrogen bond distances are marked. | ||
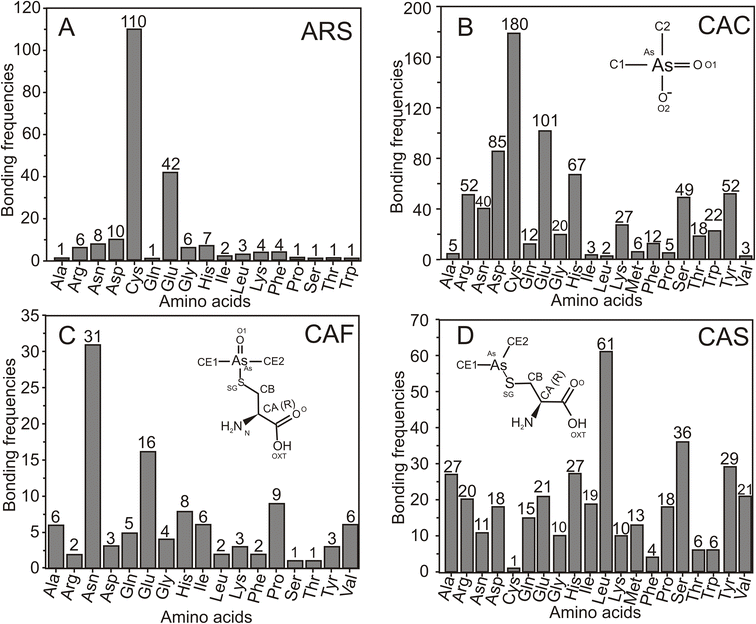
3.5 Interactions of arsenical ligands ARS, CAC, CAF and CAS
Among the 26 arsenic derivatives discussed in Section 2.1, four ligands, ARS, CAC/CAD, CAF and CAS, are frequently co-crystallized with various proteins. This analysis was conducted to determine the protein interactions of amino acids with these four ligands. The interaction profile of arsenical ligands ARS, CAC, CAF and CAS with amino acids within the proximity of 4.0 Å of arsenic items is calculated, and the results are summarized in Fig. 5.From the 49 protein structures co-crystallized with the ARS, the amino acids within the proximity of 4.0 Å of the arsenic atom are identified. The analysis revealed that the ARS predominantly interacts with Cys, followed by Glu, as shown in Fig. 5A. Compared with these two amino acids, all other amino acid interactions are more or less negligible. Cacodylic acid (CAC/CAD) is the most predominant arsenic derivative co-crystallized with various protein molecules, with 518 entries. The frequency of amino acid interactions with CAD shows that it prefers to bind with Cys, followed by Glu (Fig. 5B). Unlike the ARS, CAD is also closely associated with Asp, His, Arg, Tyr, Ser and Asn amino acids. Glu and Asp have close interactions regarding acidic amino acids, whereas the essential amino acids His and Arg interact better than Lys. The uncharged polar amino acids Tyr, Ser and Asn interact better than Thr and Gln. It is evident that nine hydrophobic amino acids, namely, Gly, Ala, Val, Leu, Ile, Pro, Phe, Met and Trp, mostly do not interact with CAD. From the literature, it is evident that when a reducing agent such as DTT is not involved in protein crystallization or purification, As(V)-based arsenic compounds, such as CAD, recruit nearby Asp and His amino acids and may catalyze the thiol exchange chemical interactions between As(V) and Cys.57,60 As discussed in Section 2.1, the CAD molecule is mainly used (i) to modify the surface/active site Cys residues of the proteins and (ii) to collect SAD/MAD data to solve the new protein structures. The amino acid interaction analysis supports the interaction of Cys. It also provides insight that the hydrophobic-rich active site is not a favorable binding site for CAD and does not help collect arsenic-based anomalous scattering data.
Similarly, the CAC molecule shows a predominant interaction with Cys and Glu. CAC also interacts with Asp, His, Arg, Tyr, Ser, and Asn, as depicted in Fig. 5B. On the other hand, the ligand CAF primarily interacts with Asn, Glu, His, and Pro, as shown in Fig. 5C. Furthermore, CAS predominantly interacts with Leu, followed by Ser, Tyr, alanine (Ala), and His (Fig. 5D). Interestingly, CAF and CAS have relatively fewer interactions with Cys than with other amino acids.
3.6 Comprehensive analyis of CAC/CAD with amino acids
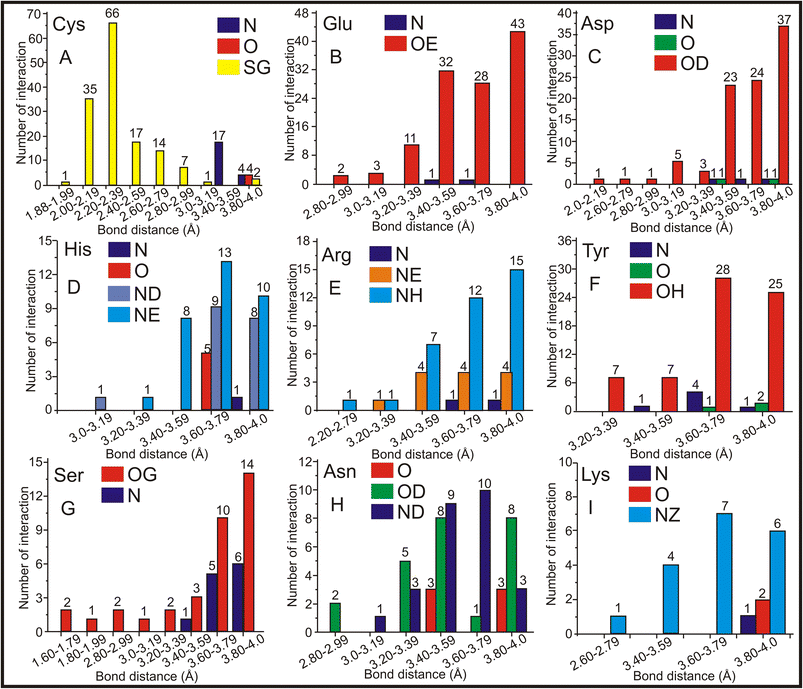
Hence, CAC/CAD has the highest entries (518 standalone entries), and a comprehensive analysis of the molecular interactions involving protein molecules associated with CAC was conducted. The distances between CAC and amino acids were calculated, and the findings are presented in Fig. 6. As discussed earlier, CAC/CAD predominantly interacts with Cys (Fig. 5B). The bond-length frequency between the Cys amino acid's main chain and the side chain with the CAC ligand was calculated. The interactions involving carbon atoms have been excluded from the figure to enhance clarity. The findings indicate that the sulfur atom in the side chain of the Cys amino acid frequently interacts with the arsenic atom of CAC. The sulfur atom of Cys interacts with the arsenic atom of CAC from 1.8–3.0 Å, and the maximum interaction is observed in the 2.0–2.4 Å range (Fig. 6A). In addition to the sulfur atom, Cys's primary chain nitrogen atom has shown moderate interaction at approximately 3.0–3.6 Å.The negatively charged amino acids Glu and Asp are the other two amino acids that primarily interact with the arsenic atom of CAC. The sidechain oxygen atoms (OE) of Glu are the only atoms predominantly interacting with an arsenic atom of CAC ranging from 2.8–4.0 Å and more frequently within 3.4–4.0 Å (Fig. 6B). On average, Glu–As interactions are found to be within 3.6 Å, which may be noncovalent interactions. Like Glu, Asp's side chain oxygen atom (OD) interacts closely with an arsenic atom of CAC with an average distance of 3.7 Å (Fig. 6C). Other than side chain oxygen atoms, there is not much arsenic interaction with other Glu and Asp atoms. The positively charged amino acid His has approximately 67 exchanges with CAC. The nitrogen atoms of the side chains of His residues frequently interact with an arsenic atom of CAC within an average bond distance of 3.7 Å (Fig. 6D). The NE atom is comparatively more regularly interacting with arsenic, followed by ND. Thus, the nitrogen atoms of His play a significant role in the arsenic interaction. Another positively charged amino acid, Arg, also interacts with side atoms of nitrogen atoms NH and NE. The best interaction range is between 3.4 and 4.0 Å (Fig. 6E).
The oxygen atom OH of Tyr showed good interaction with the arsenic atom in the range of 3.4–4.0 Å (Fig. 6F). The Ser side chain oxygen atom OG (Fig. 6G) showed a better interaction with arsenic; in this case, the main chain N atom also showed a moderate interaction with arsenic. The side chain oxygen atom (OD) and the nitrogen atom (ND) of Asn showed better interaction, and rarely, the central oxygen atom (O) also showed some interaction with arsenic (Fig. 6H). Finally, arsenic prefers to interact with Lys's side chain nitrogen atom (NZ) within 3.4–4.0 Å (Fig. 6I). Thus, the sulfur atom of Cys and side chain oxsygen atoms of Glu, Asp, Tyr, and Ser more frequently interact with arsenic. In the case of His, Arg, Asn, and Lys, the side chain nitrogen atoms closely interact with arsenic. Regarding bond distance, the Cys amino acid has a predominant interaction at 2.2–2.4 Å, and all other amino acids in the interaction range are between 3.4 – 4.0 Å. The frequency of interactions between the arsenic atom of CAC and these amino acids at different distances provides insights into their interaction nature and strength.
3.7 Arsenic interaction with amino acids
The study is further extended to understand the detailed interactions between the arsenic atom of all 26 arsenical ligands and all 20 amino acids. The results reveal that the amino acids Cys, Glu, Asp, Ser and Arg frequently interact with arsenic atoms (Fig. 7). These amino acids play a crucial role in forming complexes and interactions with arsenic, influencing the behavior and properties of arsenic-bound compounds.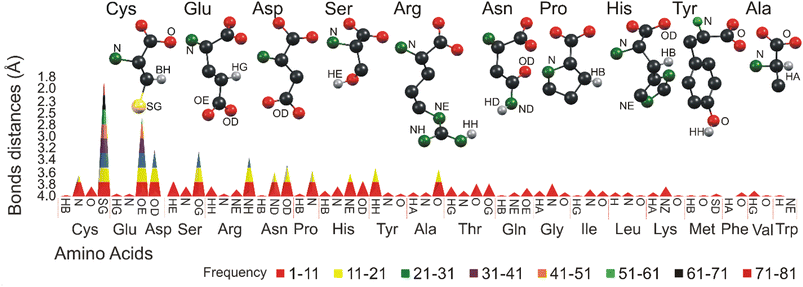 | ||
| Fig. 7 Graphical representation of the molecular contact of arsenic at 4.0 Å. Different colors represent different bonding frequencies between various amino acids and arsenic functional groups. | ||
In general, the study reflects a similar interaction of CAC. For example, the Cys sulfur atoms interact with arsenic more than any other atoms, and the arsenic-sulfur average bond length is 3.05 Å. Sulfur atoms (SG) present in Cys make 333 contacts, followed by nitrogen atoms with 60 contacts and oxygen atoms with 42 interactions. Glu makes 241 entries total with arsenic atoms. The side chain oxygen atom (OE) of Glu dominates the interactions with 209 entries from 2.86 to 4.0 Å. In general, the average bond length between oxygen (O) and arsenic (As) is reported to be approximately 3.05 Å. This value represents the average distance between the oxygen and arsenic atoms in a chemical bond involving these two elements. It is important to note that bond lengths can vary depending on the specific molecular environment and the chemical compounds involved. Asp makes 141 interactions in total, Asp's sidechain oxygen atom (OD) often interacts (104 entries) with arsenic, and the bond distances vary from 3.32 to 4.0 Å. The Ser amino acid makes 104 interactions in total.
The oxygen (OG) and a hydrogen atom (HA) of Ser dominate with a distance from 3.5 to 4.0 Å. The Arg side chain nitrogen atom (NH) significantly interacted with arsenic, with a bond distance ranging from 3.6 to 4.0 Å. Thus, the results show that side chain atoms such as sulfur, oxygen and nitrogen prefer interacting with the arsenic atoms more often than any other, similar to the CAD/CAC interaction with arsenic. Apart from Cys, Glu, Asp, Ser and Arg, the amino acids Asn, Pro, His, Tyr, and Ala are the other top five amino acids that interact with arsenic. Their interaction frequencies and the distance ranges are shown in ESI Table 3,† and the atoms involved in these interactions are shown in brackets.
The arsenic interaction between the top 10 amino acids with their atom details was calculated, and the statistical information is tabulated in ESI Table 3.† The minimum, maximum and mean interaction ranges with standard deviation provide a clear idea of the arsenic environment with different atoms of amino acids. The peak bond distance (Bond in ESI Table 3†) and the frequency help in designing new molecules and may help solve the arsenic-based structures unambiguously.
3.8 Raman analysis of amino acids with arsenic
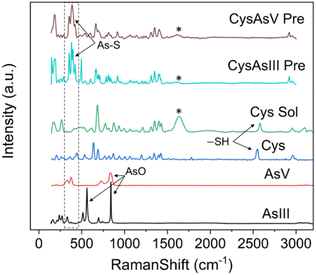
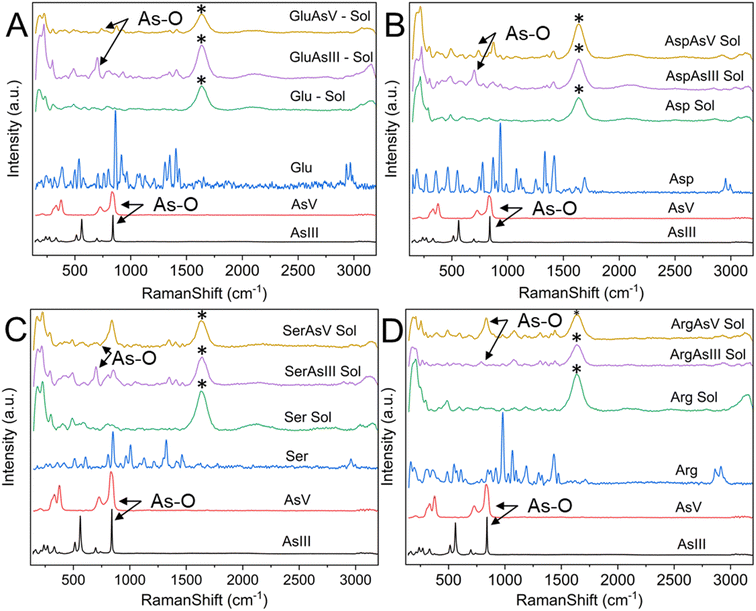
Raman spectroscopy is routinely used to examine the intermolecular bonding and molecular conformation properties of biomolecules such as amino acids, proteins and nucleic acids. Raman spectroscopy analysis of amino acids in the powder, solution and crystal forms are reported in the literature.61,62 Raman spectroscopy of specific amino acids, such as Cys interacting with arsenic, was also reported.63 However, binding between arsenic and other amino acids has yet to be established.22 We attempted to understand the interaction of amino acids with the biologically active arsenic species As(III) and As(V). The amino acids Cys, Glu, Asp, Ser and Arg, which have shown good interaction with arsenic based on the current PDB studies, were taken for this experimental study. Initially, Raman spectra of As(III) and As(V) in solid (powder) form were recorded. Then, each amino acid dissolved in water and mixed with As(III) and As(V) separately was recorded. When mixed with arsenic species, Cys immediately formed a white precipitate, which is well documented.22,64 In this study, precipitation was observed when Cys was mixed with arsenic species. In this case, the solution was centrifuged and dried, and Raman spectra of carefully removed dried precipitate and supernatant solution were recorded. Hence, the supernatant solution does not show any Raman spectra; only Raman spectra of precipitations were taken for further study.In Fig. 8, six types of Raman spectra, such as the Raman spectra of As(III), As(V) solid form (powder), Cys in solid form (powder), Cys mixed with water (Cys Sol), Cys and As(III) mixture precipitate (CysAsIII Pre) and Cys and As(V) mixture precipitate (CyasAsV Pre), are shown. The Raman peak profile was analyzed by As(III) and As(V) powders displayed in all the Raman assigned graphical plots. The As(III) and As(V) Raman profiles match the literature.65,66 The peak profile of As(III) has As–O characteristic peaks. The Raman spectra of Cys solid and solution fit well with the literature (ESI Table 4†).63 The S–H stretching vibration observed for both the solid (2550 cm−1) and solution (2557 cm−1) forms of Cys. In the case of CysAsIII Pre and CysAsV Pre, the S–H peak disappeared (Fig. 8). The signature peaks of the quartet (354 cm−1, 381 cm−1, 396 cm−1, and 424 cm−1) for CysAsIII Pre and triplet (332 cm−1, 375 cm−1, and 389 cm−1) for CysAsV Pre are observed, which is the formation of As–S covalent bonds (Fig. 8) and well supported by the literature.63 Thus, Raman spectra confirm that Cys interacts with the sulfur groups of As(III) and As(V) and forms complex materials. Fig. 9 shows the Raman spectra of Glu (Fig. 9A), Asp (Fig. 9B), Ser (Fig. 9C), and Arg (Fig. 9D) amino acids. Fig. 9A shows the spectra of As(III), As(V), and Glu alone and in the presence of As(III) and As(V). The Raman spectra of Glu, Asp, Ser and Arg match the literature.61,62 The peak assignments of Glu (ESI Table 5†), Asp (ESI Table 6†), Ser (ESI Table 7†), and Arg (ESI Table 8†) amino acids are summarized.
The peak at 863 cm−1 in the Raman spectrum of Glu corresponds to the rocking motion of C–C vibrations. Similarly, the C–C stretching vibrational peaks corresponding to Asp and Ser are observed at 935 cm−1 and 850 cm−1, respectively. The peak at 980 cm−1 in Arg corresponds to C–N/C–C stretching vibrations. CH2 stretching vibrations are generally observed in the 2928 cm−1 and 2970 cm−1 spectral range. In this study, CH2 stretching was observed at 2952 cm−1 and 2995 cm−1 for Asp, 2958 cm−1 and 2995 cm−1 for Ser, and 2863 cm−1 and 2914 cm−1 for Arg. NH3+ rocking motion vibrations are observed at 1128 cm−1 for Glu, 1250 cm−1 for Asp, 1127 cm−1 for Ser and 1189 cm−1 for Arg. These vibrational assignments are observed and reported in the literature.61 Compared with Glu, the GluAsIII Raman spectra showed a prominent peak at 702 cm−1, corresponding to the A1 mode of H3AsO3. Similarly, the obtained peak at 873 cm−1 in GluAsV corresponds to As-OH vibrational modes, and both peaks are well supported by the literature.67 These two peaks are also observed for Asp, Ser and Arg in the presence of As(III) and As(V), representing hydrate formation. Thus, As–O vibrations are predominantly observed for all four amino acids (Glu, Asp, Ser and Arg). These similar interactions are well documented in the literature.67
3.9 Arsenic–protein interactions and its potential applications
Arsenic has been utilized as a medicinal agent for numerous human ailments since 2000 BC. It has featured prominently in the traditional medical practices of various ancient civilizations such as the Greeks, Romans, Chinese, Persians, and Indians. In both inorganic and organic forms, arsenic-based drugs have emerged as essential components of modern chemotherapy, with compounds like arsenic trioxide (ATO), realgar, Salvarsan, arsphenamine, neoarsphenamine, and oxophenarsine hydrochloride demonstrating anticancer potential.6,68 Notably, ATO, marketed as Trisenox and approved by the U.S. Food and Drug Administration for treating acute promyelocytic leukemia (APL) and hematological disorders, has rekindled interest in the use of arsenic-based drugs. ATO, a trivalent arsenical, is known to bind thiol groups in arsenic-binding proteins, such as lipoamide dehydrogenase, and metabolites like glutathione. This action inhibits cellular energy production while increasing intracellular reactive oxygen species.69 Treatment with ATO has been found to cause DNA demethylation and affect the promoters of numerous genes. It also binds to oncoproteins and transcription factors, ultimately resulting in cell cycle arrest, apoptosis, and the transition from a mesenchymal to epithelial state.69 Moreover, ATO has been reported to target promyelocytic leukemia (PML) and protein-retinoic acid receptor alpha (RARα), leading to apoptosis.70 It is also a target of Wip1 phosphatase, which activates Chk2 and/or p38 MAPK apoptotic pathways. Inhibition of phosphatase activity amplifies Wip1, offering potential therapeutic benefits in breast, papillary thyroid, colorectal, prostate cancers, and various other tumor cells.71 Recent reports indicate that ATO is also targeted by peptidyl-prolyl cis–trans isomerase NIMA-interacting 1 (Pin1) protein, which regulates multiple cancer signaling networks and cancer stem cells.48Additionally, the inorganic arsenic compound realgar has been tested against multiple myeloma cell lines and patients with myeloma, as demonstrated.72 Beyond inorganic arsenicals, various organic arsenicals, such as dimethyl arsenic conjugates and darinaparsin, are currently undergoing testing for their anti-tumor properties against cancer cell lines like HL-60, SGC 7901, and MCF-7. These compounds are also progressing through phase 1 and/or phase 2 clinical trials.73 Furthermore, polyorganoarsenicals like arsenicin A, extracted from marine sources, have shown superior anti-tumor activities. In addition to their well-known applications in cancer chemotherapy, arsenic compounds have demonstrated antiviral properties against a range of viruses. These include the Hepatitis C Virus, the human immunodeficiency virus (HIV)-1, human adenovirus, human T-cell leukemia virus type 1, Epstein-Barr virus, and even SARS-CoV-2.7 Furthermore, natural arsenic-based products, such as methylarsenite and arsinothricin extracted from the soil bacterium Streptomyces griseus, have been found to possess antibacterial properties, effectively serving as antibiotics. Recently, arsinothricin has been newly recognized as an organoarsenical antibiotic with the ability to inhibit M. bovis BCG, a bacterium closely related to M. tuberculosis.14 Moreover, synthetic pentavalent aromatic arsenic molecules like atoxyl, carbarsone, roxarsone, and nitrarsone have found extensive use in treating infectious diseases in poultry, turkeys, and swine.39 However, it is essential to acknowledge that the primary drawback of using arsenical compounds as medicine lies in their inherent toxicity. To overcome these limitations and advance the field, there is a demanding need to develop a new generation of arsenic-based drugs while comprehensively understanding their physical, chemical, and biological properties. Numerous proposed arsenic-based drugs have been withdrawn from use due to insufficient scientific research. The current structural study, based on the PDB, represents a significant step towards elucidating the molecular interactions of various arsenic compounds. This research aims to mitigate toxicity concerns and provide valuable structural insights for the design and development of arsenic-based drugs with reduced toxicity and enhanced therapeutic efficacy.
Similarly, understanding the binding kinetics of arsenic within proteins is significant for several reasons, and it can have implications for the functionality of critical amino acids within protein motifs. Arsenic is a well-known toxic element, thus understanding its binding kinetics within proteins is essential for elucidating the mechanisms of arsenic detoxification in living organisms.56 Some organisms have evolved specialized proteins that can sequester and detoxify arsenic compounds.38 Arsenic can interfere with the activity of enzymes by binding to specific amino acid residues. Understanding the kinetics of this binding is critical for assessing the impact on enzyme function and, in turn, cellular processes.74 Arsenic binding within proteins can disrupt protein structure and affect function. This has implications for various cellular processes, including signaling, metabolism, and DNA repair.20 Studying arsenic binding kinetics can provide insights into the evolution of arsenic resistance mechanisms in microorganisms and plants.74 Some organisms have developed unique strategies to cope with arsenic exposure. Knowledge of arsenic binding kinetics can have applications in biotechnology, such as the development of biosensors for arsenic detection and bioremediation of arsenic-contaminated environments.75 Arsenic binding within proteins often involves interactions with specific amino acids, such as Cys, Glu, Asp, Ser, Arg, and His, which contain sulfur or oxygen or nitrogen atoms that can coordinate with arsenic.24 When arsenic binds to these amino acids, it can disrupt their normal functions, which may include catalysis, metal binding, or redox reactions. This disruption can have a cascading effect on the functionality of other amino acids within the protein, leading to altered enzymatic activity or structural changes.
Manipulating arsenic–protein interactions for enhanced binding characteristics or creating an environment conducive to binding arsenic with multiple proteins is an area of research with various potential applications. Researchers can use rational design to engineer proteins with enhanced arsenic-binding characteristics. This involves modifying the amino acid residues at or near the binding site to increase affinity and specificity for arsenic. Through techniques like phage display or yeast surface display, proteins with improved arsenic-binding properties can be evolved in the laboratory. This allows for the selection of proteins with higher arsenic affinity. Proteins with high affinity for arsenic can be used as the basis for biosensors. These biosensors can detect arsenic in environmental samples, food, or clinical specimens, offering a practical and rapid method for arsenic detection.75 Proteins or engineered microorganisms that can sequester or transform arsenic can be used for bioremediation of arsenic-contaminated environments. This approach can help reduce arsenic levels in water sources, soils, and industrial effluents.76 Arsenic-binding proteins or engineered proteins with arsenic-binding domains can also be used in drug delivery systems. These systems can carry arsenic-based drugs to specific cancer cells, enhancing the targeted therapy's efficacy while reducing off-target effects. Engineering plants to express arsenic-binding proteins or peptides can enhance their ability to accumulate and sequester arsenic from contaminated soils. This approach is relevant for phytoremediation.77 Understanding arsenic–protein interactions can inform the development of antidotes for acute arsenic poisoning by creating molecules that compete with arsenic for binding sites on critical. Engineering microbes to resist arsenic can be valuable in biotechnology and environmental remediation. This involves manipulating arsenic transporters, efflux pumps, and intracellular binding proteins.
4 Conclusion
Metals and metalloids play crucial roles in protein architecture and participate in various biological processes, often serving as protein cofactors that contribute significantly to their structure and function. The PDB offers a comprehensive repository of structural information, encompassing atomic coordinates and relevant experimental data. This study delves into the elaborate molecular interactions between arsenic and essential elements such as nitrogen (N), sulfur (S), and oxygen (O) extracted from the PDB. Arsenic's toxicity and carcinogenicity remain complex due to its diverse biological properties and mechanisms. This investigation sheds light on the molecular interactions involving two arsenic forms: arsenite [As(III)] and arsenic-arsenate [As(V)], co-crystallized with 902 proteins sourced from the PDB. Specifically, we calculated the proximity-based interactions between arsenic and arsenic-binding proteins within a 4 Å radius. The ligands were categorized based on their oxidation states as As(III) and As(V). Nine As(III)-based ligands and their interactions with various proteins have been identified and are summarized in Fig. 10.For instance, ARS emerges as a frequently employed As(III)-based ligand binding to arsenic-binding proteins, signifying the specificity of these proteins for arsenic binding (see Fig. 10). Simultaneously, the primary As(III) ligand CAS and its As(V) derivatives, CAC and CAD, find utility in SAD/MAD data collection. As(III) ligands primarily serve to modify Cys residues and form bonds with the thiols of arsenic-binding proteins, while As(V)-based ligands are typically employed for SAD/MAD data collection (see Fig. 11). Several As(V) ligands also interact with phosphate-utilizing enzymes, which holds biological significance (see Fig. 11). For example, the TTA ligand attaches to the dimer interface and is used for SAD data collection. Furthermore, this study identifies the five amino acids that frequently interact with arsenic: Cys, Glu, Asp, Ser, and Arg (CEDSR). The sulfur and oxygen atoms of these amino acids are frequent points of interaction with arsenic-based ligands.
Raman spectroscopy of amino acids in the presence of arsenate As(V) and arsenite As(III) validates covalent interactions with these five amino acids, including Arg, Asp, Glu, Ser, and Cys. This insight underscores the importance of arsenic's binding kinetics within a protein, as it influences the functionality of other critical amino acids within protein motifs. Therefore, a comprehensive structural analysis of atomic and molecular interactions involving arsenic has been conducted. Such a study holds the potential to inspire the development of new arsenic-based molecules, drugs, or innovative inhibitors for conditions like cancer. Manipulating the arsenic–protein interactions could be explored to enhance binding characteristics or create an environment conducive to binding arsenic with multiple proteins. As the identification of additional arsenic-binding proteins is anticipated in the future, this study can serve as a valuable resource for selecting suitable ligands based on the amino acids constituting the binding pockets of these proteins.
Thus, the current comprehensive structural analysis of arsenic's atomic and molecular interactions offers several potential implications, primarily inspiring the development of new arsenic-based molecules and drugs for conditions such as cancer. This analysis can provide insights into designing and optimizing arsenic-based compounds, facilitating the creation of molecules with improved pharmacological properties. Structural analysis can uncover specific protein targets and cellular pathways that interact with arsenic compounds, guiding the development of targeted therapies for conditions like cancer. Arsenic-based drugs can be designed to modulate the function of these targets for therapeutic benefit, enhancing their specificity and efficacy. Arsenic's high toxicity, which limits its therapeutic use, can be mitigated through a comprehensive structural analysis. The knowledge acquired from this study can guide the development of less toxic arsenic-based drugs while retaining their therapeutic benefits. Furthermore, structural insights can inform strategies for precise drug delivery, minimizing off-target effects, and improving bioavailability. Understanding the interactions between arsenic and cancer-related proteins can help develop strategies to mitigate resistance and enhance the long-term effectiveness of arsenic-based therapies. Additionally, structural analysis can identify synergistic interactions between arsenic-based compounds and other drugs or inhibitors, leading to the development of combination therapies targeting multiple pathways. This detailed structural information supports the development of personalized medicine approaches by characterizing individual patient profiles, including genetic factors, guiding the selection of effective arsenic-based therapies for specific cancer subtypes or patient populations.
Conflicts of interest
The authors assert that they have not received any remuneration from commercial organizations or industries for conducting this research.Acknowledgements
Mr Upendra Nayek extends his appreciation to the Indian Council of Medical Research (ISRM/11(69)/2019), while Dr Abdul Ajees Abdul Salam acknowledges the research funding provided by the Manipal Academy of Higher Education (MAHE) through intramural financing (MAHE/DREG/PhD/IMF/2019).References
- S. M. Quintal, Q. A. dePaula and N. P. Farrell, Metallomics, 2011, 3, 121–139 CrossRef CAS PubMed.
- H. Beinert, R. H. Holm and E. Münck, Science, 1997, 277, 653–659 CrossRef CAS PubMed.
- M. F. Perutz, Annu. Rev. Physiol., 2003, 52, 1–26 CrossRef PubMed.
- W. B. Minich, Biochemistry, 2022, 87, S168–S177 CAS.
- A. Changela, K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran and A. Mondragón, Science, 2003, 301, 1383–1387 CrossRef CAS PubMed.
- R. Bentley and T. G. Chasteen, Chem. Educat., 2002, 7, 51–60 CrossRef CAS.
- N. P. Paul, A. E. Galván, K. Yoshinaga-Sakurai, B. P. Rosen and M. Yoshinaga, BioMetals, 2023, 36, 283–301 CrossRef CAS PubMed.
- N. Baker, H. P. de Koning, P. Mäser and D. Horn, Trends Parasitol., 2013, 29, 110–118 CrossRef CAS PubMed.
- T. J. R. D. Reeves, A. J. M. Baker, G. Echevarria, P. D. Erskine and A. van der Ent, New Phytol., 2017, 218, 407–411 CrossRef PubMed.
- A. A. Ajees and B. P. Rosen, Geomicrobiol. J., 2015, 32, 570–576 CrossRef PubMed.
- C. H. Lu, Y. F. Lin, J. J. Lin and C. S. Yu, PLoS One, 2012, 7, e39252 CrossRef CAS PubMed.
- T. Dudev and C. Lim, Chem. Rev., 2014, 114, 538–556 CrossRef CAS PubMed.
- B. P. Rosen, A. A. Ajees and T. R. Mcdermott, BioEssays, 2011, 33, 350–357 CrossRef CAS PubMed.
- V. S. Nadar, J. Chen, D. S. Dheeman, A. E. Galván, K. Yoshinaga-Sakurai, P. Kandavelu, B. Sankaran, M. Kuramata, S. Ishikawa, B. P. Rosen and M. Yoshinaga, Commun. Biol., 2019, 2, 1–12 CrossRef PubMed.
- K. Jomova, Z. Jenisova, M. Feszterova, S. Baros, J. Liska, D. Hudecova, C. J. Rhodes and M. Valko, J. Appl. Toxicol., 2011, 31, 95–107 CrossRef CAS PubMed.
- M. Argos, T. Kalra, P. J. Rathouz, Y. Chen, B. Pierce, F. Parvez, T. Islam, A. Ahmed, M. Rakibuz-Zaman, R. Hasan, G. Sarwar, V. Slavkovich, A. van Geen, J. Graziano and H. Ahsan, Lancet, 2010, 376, 252–258 CrossRef CAS PubMed.
- M. Rahman, N. Sohel, F. M. Yunus, N. Alam, Q. Nahar, P. K. Streatfield and M. Yunus, Environ. Int., 2019, 123, 358–367 CrossRef CAS PubMed.
- M. F. Hughes, Arsenic toxicity and potential mechanisms of action, 2002, vol. 133 Search PubMed.
- J. Ye, A. A. Ajees, J. Yang and B. P. Rosen, Biochemistry, 2010, 49, 5206–5212 CrossRef CAS PubMed.
- L. M. Tam, N. E. Price and Y. Wang, Chem. Res. Toxicol., 2020, 33, 709–726 Search PubMed.
- P. Ravenscroft, H. Brammer and K. Richards, Arsenic Pollution: A Global Synthesis, 2009, vol. 1 Search PubMed.
- S. Shen, X.-F. Li, W. R. Cullen, M. Weinfeld and X. C. Le, Chem. Rev., 2013, 113, 7769–7792 CrossRef CAS PubMed.
- M. Vašák and D. W. Hasler, Curr. Opin. Chem. Biol., 2000, 4, 177–183 CrossRef PubMed.
- H. Bhattacharjee and B. P. Rosen, J. Bioenerg. Biomembr., 2001, 33, 459–468 CrossRef CAS PubMed.
- D. B. Menzel, H. K. Hamadeh, E. Lee, D. M. Meacher, V. Said, R. E. Rasmussen, H. Greene and R. N. Roth, Toxicol. Lett., 1999, 105, 89–101 CrossRef CAS PubMed.
- K. Piatek, T. Schwerdtle, A. Hartwig and W. Bal, Chem. Res. Toxicol., 2008, 21, 600–606 Search PubMed.
- Y. Zhang, D. Bhatia, H. Xia, V. Castranova, X. Shi and F. Chen, Nucleic Acids Res., 2006, 34, 485–495 CrossRef CAS PubMed.
- Y. C. Hu, K. Y. Lam, S. Law, J. Wong and G. Srivastava, Clin. Cancer Res., 2001, 7, 2213–2221 CAS.
- M. Matzapetakis, B. T. Farrer, T. C. Weng, L. Hemmingsen, J. E. Penner-Hahn and V. L. Pecoraro, J. Am. Chem. Soc., 2002, 124, 8042–8054 CrossRef CAS PubMed.
- D. J. Cline, C. Thorpe and J. P. Schneider, J. Am. Chem. Soc., 2003, 125, 2923–2929 CrossRef CAS PubMed.
- A. L. Femia, C. F. Temprana, J. Santos, M. L. Carbajal, M. S. Amor, M. Grasselli and S. V. Del Alonso, Protein J., 2012, 31, 656–666 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr. Sect. D Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- E. Seifert, J. Chem. Inf. Model., 2014, 54, 1552 CrossRef CAS PubMed.
- MarvinSketch 19.10, https://chemaxon.com/products/marvin Search PubMed.
- IARC Monographs series, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, International Agency for Research on Cancer, Lyon, France, 100 C., 2012, vol. 100C Search PubMed.
- M. Capdevila-Cortada, J. Castelló and J. J. Novoa, CrystEngComm, 2014, 16, 8232–8242 RSC.
- H. Cao, J. Hall and R. Hille, J. Am. Chem. Soc., 2011, 133, 12414–12417 CrossRef CAS PubMed.
- A. A. Ajees, K. Marapakala, C. Packianathan, B. Sankaran and B. P. Rosen, Biochemistry, 2012, 51, 5476–5485 CrossRef CAS PubMed.
- K. Marapakala, C. Packianathan, A. A. Ajees, D. S. Dheeman, B. Sankaran, P. Kandavelu and B. P. Rosen, Acta Crystallogr. Sect. D Biol. Crystallogr., 2015, 71, 505–515 CrossRef CAS PubMed.
- V. S. Nadar, P. Kandavelu, B. Sankaran, B. P. Rosen and M. Yoshinaga, J. Inorg. Biochem., 2022, 232, 111836–111857 CrossRef CAS PubMed.
- S. DeMel, J. Shi, P. Martin, B. P. Rosen and B. F. P. Edwards, Protein Sci., 2004, 13, 2330–2340 CrossRef CAS PubMed.
- C. Badilla, T. H. Osborne, A. Cole, C. Watson, S. Djordjevic and J. M. Santini, Sci. Rep., 2018, 8, 6282–6294 CrossRef PubMed.
- D. Manna, G. Cordara and U. Krengel, Curr. Res. Struct. Biol., 2020, 2, 56–67 CrossRef PubMed.
- P. Martin, S. DeMel, J. Shi, T. Gladysheva, D. L. Gatti, B. P. Rosen and B. F. P. Edwards, Structure, 2001, 9, 1071–1081 CrossRef CAS PubMed.
- J. K. Lee, G. I. Belogrudov and R. M. Stroud, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13379–13384 CrossRef CAS PubMed.
- A. K. L. Liess, A. Kucerova, K. Schweimer, L. Yu, T. I. Roumeliotis, M. Diebold, O. Dybkov, C. Sotriffer, H. Urlaub, J. S. Choudhary, J. Mansfeld and S. Lorenz, Structure, 2019, 27, 1195–1210 CrossRef CAS PubMed.
- T. Zhou, EMBO J., 2000, 19, 4838–4845 CrossRef CAS PubMed.
- S. Kozono, Y. M. Lin, H. S. Seo, B. Pinch, X. Lian, C. Qiu, M. K. Herbert, C. H. Chen, L. Tan, Z. J. Gao, W. Massefski, Z. M. Doctor, B. P. Jackson, Y. Chen, S. Dhe-Paganon, K. P. Lu and X. Z. Zhou, Nat. Commun., 2018, 9, 3069 CrossRef PubMed.
- C. Prabaharan, P. Kandavelu, C. Packianathan, B. P. Rosen and S. Thiyagarajan, J. Struct. Biol., 2019, 207, 209–217 CrossRef CAS PubMed.
- D. S. Touw, C. E. Nordman, J. A. Stuckey and V. L. Pecoraro, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11969–11974 CrossRef CAS PubMed.
- V. Molteni, J. Greenwald, D. Rhodes, Y. Hwang, W. Kwiatkowski, F. D. Bushman, J. S. Siegel and S. Choe, Acta Crystallogr. Sect. D Biol. Crystallogr., 2001, 57, 536–544 CrossRef CAS PubMed.
- P. Retailleau and T. Prangé, Acta Crystallogr. Sect. D Biol. Crystallogr., 2003, 59, 887–896 CrossRef PubMed.
- M. J. Lenaeus, M. Vamvouka, P. J. Focia and A. Gross, Nat. Struct. Mol. Biol., 2005, 12, 454–459 CrossRef CAS PubMed.
- O. V. Kurnasov, H.-J. D. Luk, M. F. Roberts and B. Stec, Acta Crystallogr. Sect. D Biol. Crystallogr., 2013, 69, 1808–1817 CrossRef CAS PubMed.
- S. Maignan, J. P. Guilloteau, Q. Zhou-Liu, C. Clément-Mella and V. Mikol, J. Mol. Biol., 1998, 282, 359–368 CrossRef CAS PubMed.
- P. B. Tchounwou, C. G. Yedjou, A. K. Patlolla and D. J. Sutton, in Molecular, Clinical and Environmental Toxicology, 2012, vol. 101, pp. 133–164 Search PubMed.
- X. Liu, H. Zhang, X. J. Wang, L. F. Li and X. D. Su, PLoS One, 2011, 6, e24227 CrossRef CAS PubMed.
- J. Greenwald, V. Le, S. L. Butler, F. D. Bushman and S. Choe, Biochemistry, 1999, 38, 8892–8898 CrossRef CAS PubMed.
- J. P. Bacik and B. Hazes, J. Mol. Biol., 2007, 365, 545–558 CrossRef PubMed.
- A. Dawson, F. Gibellini, N. Sienkiewicz, L. B. Tulloch, P. K. Fyfe, K. McLuskey, A. H. Fairlamb and W. N. Hunter, Mol. Microbiol., 2006, 61, 1457–1468 CrossRef CAS PubMed.
- P. T. C. Freire, F. M. Barboza, J. A. Lima, F. E. A. Melo and J. M. Filho, in Raman Spectroscopy and Applications, 2017, vol. 1, p. 171 Search PubMed.
- G. Zhu, X. Zhu, Q. Fan and X. Wan, Spectrochim. Acta, Part A, 2011, 78, 1187–1195 CrossRef PubMed.
- M. C. Teixeira, V. S. T. Ciminelli, M. S. S. Dantas, S. F. Diniz and H. A. Duarte, J. Colloid Interface Sci., 2007, 315, 128–134 CrossRef CAS PubMed.
- J. M. Johnson and C. Voegtlin, J. Biol. Chem., 1930, 89, 27–31 CrossRef CAS.
- J. Čejka, J. Sejkora, S. Bahfenne, S. J. Palmer, J. Plášil and R. L. Frost, J. Raman Spectrosc., 2011, 42, 214–218 CrossRef.
- M. Mulvihill, A. Tao, K. Benjauthrit, J. Arnold and P. Yang, Angew. Chem., Int. Ed., 2008, 47, 6456–6460 CrossRef CAS PubMed.
- C. Sudhakar, A. Anil Kumar, R. G. Bhuin, S. Sen Gupta, G. Natarajan and T. Pradeep, ACS Sustain. Chem. Eng., 2018, 6, 9990–10000 CrossRef CAS.
- N. C. Lloyd, H. W. Morgan, B. K. Nicholson and R. S. Ronimus, Angew. Chem., Int. Ed., 2005, 44, 941–944 CrossRef CAS PubMed.
- A. Emadi and S. D. Gore, Blood Rev., 2010, 24, 191–199 CrossRef CAS PubMed.
- X.-W. Zhang, X.-J. Yan, Z.-R. Zhou, F.-F. Yang, Z.-Y. Wu, H.-B. Sun, W.-X. Liang, A.-X. Song, V. Lallemand-Breitenbach, M. Jeanne, Q.-Y. Zhang, H.-Y. Yang, Q.-H. Huang, G.-B. Zhou, J.-H. Tong, Y. Zhang, J.-H. Wu, H.-Y. Hu, H. de Thé, S.-J. Chen and Z. Chen, Science, 2010, 328, 240–243 CrossRef CAS PubMed.
- A. Emelyanov and D. V. Bulavin, Oncogene, 2015, 34, 4429–4438 CrossRef CAS PubMed.
- D. Cholujova, Z. Bujnakova, E. Dutkova, T. Hideshima, R. W. Groen, C. S. Mitsiades, P. G. Richardson, D. M. Dorfman, P. Balaz, K. C. Anderson and J. Jakubikova, Br. J. Haematol., 2017, 179, 756–771 CrossRef CAS PubMed.
- X. Y. Fan, X. Y. Chen, Y. J. Liu, H. M. Zhong, F. L. Jiang and Y. Liu, Sci. Rep., 2016, 6, 1–12 CrossRef PubMed.
- G. Abbas, B. Murtaza, I. Bibi, M. Shahid, N. K. Niazi, M. I. Khan, M. Amjad, M. Hussain and Natasha, Int. J. Environ. Res. Public Health, 2018, 15, 59 CrossRef PubMed.
- A. S. Maghsoudi, S. Hassani, K. Mirnia and M. Abdollahi, Int. J. Nanomed., 2021, 16, 803–832 CrossRef PubMed.
- R. S. Oremland and J. F. Stolz, Science, 2003, 300, 939–944 CrossRef CAS PubMed.
- O. P. Dhankher, B. P. Rosen, E. C. McKinney and R. B. Meagher, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5413–5418 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra05987a |
| This journal is © The Royal Society of Chemistry 2023 |