Open Access Article
Open Access ArticleCurrent trends in luminescence-based assessment of apoptosis
Cristian Moldovanab,
Anca Onaciua,
Valentin Tomaa,
Raluca A. Munteanua,
Diana Guleia,
Alin I. Moldovana,
Gabriela F. Stiufiucc,
Richard I. Federa,
Diana Cenariua,
Cristina A. Iugaad and
Rares I. Stiufiuc *abe
*abe
aMedfuture-Research Center for Advanced Medicine, “Iuliu Hatieganu” University of Medicine and Pharmacy, Marinescu 23/Louis Pasteur Street No. 4-6, 400337 Cluj-Napoca, Romania. E-mail: rares.stiufiuc@umfcluj.ro; Tel: +40-0726-34-02-78
bDepartment of Pharmaceutical Physics & Biophysics, Faculty of Pharmacy, “Iuliu Hatieganu” University of Medicine and Pharmacy, Louis Pasteur Street No. 4-6, 400349 Cluj-Napoca, Romania
cFaculty of Physics, “Babes Bolyai” University, Mihail Kogalniceanu Street No. 1, 400084 Cluj-Napoca, Romania
dPharmaceutical Analysis, Faculty of Pharmacy, “Iuliu Hatieganu” University of Medicine and Pharmacy, Louis Pasteur Street 6, Cluj-Napoca, 400349, Romania
eTRANSCEND Research Center, Regional Institute of Oncology, 700483 Iasi, Romania
First published on 30th October 2023
Abstract
Apoptosis, the most extensively studied type of cell death, is known to play a crucial role in numerous processes such as elimination of unwanted cells or cellular debris, growth, control of the immune system, and prevention of malignancies. Defective regulation of apoptosis can trigger various diseases and disorders including cancer, neurological conditions, autoimmune diseases and developmental disorders. Knowing the nuances of the cell death type induced by a compound can help decipher which therapy is more effective for specific diseases. The detection of apoptotic cells using classic methods has brought significant contribution over the years, but innovative methods are quickly emerging and allow more in-depth understanding of the mechanisms, aside from a simple quantification. Due to increased sensitivity, time efficiency, pathway specificity and negligible cytotoxicity, these innovative approaches have great potential for both in vitro and in vivo studies. This review aims to shed light on the importance of developing and using novel nanoscale methods as an alternative to the classic apoptosis detection techniques.
Introduction
From the very beginning, cell death identification was and continues to be an important approach in the field of drug discovery and molecular biology.1 Many of the classical techniques rely on fluorescent probes in order to point out regulated cell death (RCD) mechanisms by targeting specific cellular markers.2 At the same time, the development of new dyes or probes for cell structures and organelles examination has allowed the indirect association of cell morphology and cell death. These modifications are mainly explored through microscopy imaging and usually only offer a qualitative analysis of cellular behaviour.On the other hand, for the generation of statistically significant data regarding the incidence of RCD and/or abundancy of RCD specific markers, other approaches that allow quantitative analysis are used. In this regard, RCD is monitored using techniques such as: nuclear magnetic resonance,3 flow cytometry,4 imaging approaches,5,6 photothermy7 or ultrasound frequency evaluation.8 Additionally, there are other analytical procedures for performing molecular biology assays that are suitable for cell death evaluation9 like toxicology tests,10 viability assays11 and PCR assays.12,13 These involve the analysis of specific genes, RNA or proteins, as valuable indicators of cell death. In this case, many of the techniques are engaging spectrophotometry. Moreover, mathematical models are offering new perspectives in cell death analysis especially regarding the most likely cell death pathway to be engaged in specific conditions.14,15 Qualitative and quantitative examinations of cell death synergize to offer an accurate and valid result.
According to the recommendations of the Nomenclature Committee on Cell Death, in 2018, a novel classification for the RCD processes based on mechanistic and other important aspects was established.16 Cell death categories are described depending on the signal molecules involved in and are named as follows: intrinsic apoptosis, extrinsic apoptosis, mitochondrial permeability transition driven necrosis, necroptosis, ferroptosis, pyroptosis, parthanatos, entotic cell death, netotic cell death, lysosome-dependent cell death, autophagy-dependent cell death, non-lethal processes. This review aims to highlight the current and most innovative methods for the detection of different apoptosis pathways in vitro and in vivo, while briefly summarizing the most commonly used techniques, their drawbacks and the need of implementing new and more advanced approaches.
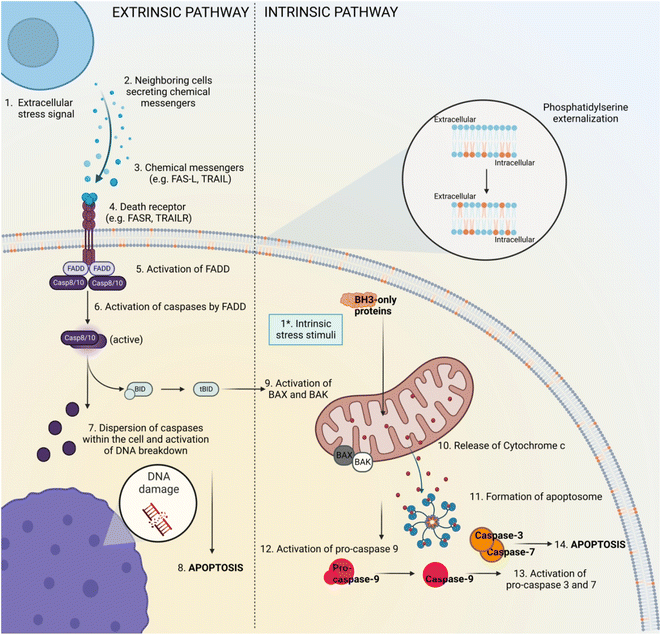
Apoptosis is a physiological process described as an active mechanism involving cell shrinkage, membrane blebbing, vacuolated cytoplasm, chromatin fragmentation and the production of apoptotic bodies.17 The main apoptosis signalling pathways are well described and can be seen in Fig. 1. This phenomenon happens relatively fast and its phases are difficult to capture. Beyond that, apoptotic bodies, if not cleared by phagocytes, can proceed to secondary necrosis18 and may influence neighbouring cell populations by inducing inflammation and/or cellular stress due to the release of intracellular contents. Even though secondary necrosis is not a RCD mechanism itself, it shares similar morphological features to apoptotic19 and necrotic cells and as a consequence, it can be challenging to decipher the exact cell death pathway. Moreover, other RCD mechanisms also resemble apoptosis when it comes to morphology and/or expression pattern of certain molecules involved in cell death establishment. This suggests the need of researchers to adapt their studies to the quickly developing field of cell death research by using novel and innovative techniques of RCD detection. Some of these methods, the targeted biomarkers correlated to the stages of apoptosis are presented in Table 1.
| Apoptosis stage | Event | Biomarker | Methods used for the detection | Sources |
|---|---|---|---|---|
| Early | Loss of membrane asymmetry | Phosphatidylserine (PS) externalization | Light microscopy | 20 |
| Carbon dots-annexin V probe & fluorescence spectroscopy | 21 | |||
| AI & PI staining | 22 | |||
| Flow cytometry | 23 and 24 | |||
| Electrochemical probes | 25 | |||
| Label free SERS sensing | 26 | |||
| Fluorometric methods | 27 | |||
| Iron oxide nanoparticles conjugated with Pisum sativum agglutinin & MR imaging & confocal microscopy | 24 | |||
| Imaging flow cytometry & convolutional autoencoder | 28 | |||
| Cell shrinkage and membrane damage | Decrease in cell size, presence of membrane blebs | Imaging techniques | 29 and 30 | |
| Flow cytometry | 4 and 29 | |||
| Dielectrophoretic assay | 31 | |||
| Chromatin changes | DNA condensation | Spectrofluorometric assay | 32 | |
| High resolution confocal imaging – single molecule localization microscopy | 33 | |||
| Monoclonal antibodies against single-stranded DNA | 34 and 35 | |||
| ELISA | 36–38 | |||
| TUNEL assay | 4,39 | |||
| DNA-specific fluorochromes & fluorescence microscopy | 40–42 | |||
| Haematoxylin & eosin staining | 43 | |||
| Mitochondrial changes | Depolarization of the mitochondrial membrane potential | Membrane permeable lipophilic and cationic fluorescent dyes & fluorescence microscopy | 44 and 45 | |
| Mitochondrial activity of streptolysin O permeabilised cells & luciferase luminescence assay | 46 | |||
| Release of cytochrome c and other pro-apoptotic factors from the mitochondria to the cytosol | Flow cytometry – anti-cytochrome c monoclonal antibodies & fluorochromes | 47 | ||
| Competitive immunoassay of ELISA, electrochemiluminescence ELISA | 48 and 49 | |||
| Western blot | 50 | |||
| Fluorometric assay based on aptamer/gold nanocluster probe | 51 | |||
| Confocal laser scanning microscopy using carbon dots | 52 | |||
| Activation of Bax proteins | Western blot, single molecule localization microscopy, confocal imaging, fluorescence cross correlation spectroscopy, protein-cross linking, and mass spectrometry | 53 | ||
| Immunocytochemistry | 54 | |||
| Activation of caspases and apoptosome formation | Caspase-8 activation by extracellular signals, like fas ligand or TNF | Colorimetric assay, Western blot | 55 | |
| ELISA | 56 and 57 | |||
| Flow cytometry, fluorometric activity assay | 58 | |||
| Immunofluorescence | 59 | |||
| Membrane permeant fluorescent-labelled inhibitors of caspases & fluorescence microscopy | 60 | |||
| Caspase-9 activation by cytosolic cytochrome c and the formation of the apoptosome together with Apaf-1 | Colorimetric assay (e.g. using unlabelled DEVD containing peptide substrate and unmodified gold nanoparticles) & Western blot | 55 and 61 | ||
| Mathematical simulation & computational methods | 62 and 63 | |||
| Split luciferase complementation assay (e.g. Lumiptosome, lentivirus transfected split luciferase fragment) | 64–66 | |||
| Calorimetry, NMR spectroscopy, site mutagenesis | 63 | |||
| Immunofluorescence | 59 | |||
| Mid | DNA damage | DNA fragmentation by caspase-activated DNases (CAD) into 180–200 bp long fragments (X) | SDS-PAGE | 67 |
| TUNEL & single stranded DNA assay | 68 | |||
| COMET assay | 69 | |||
| Laser scanning cytometry, Halo assay, combination assay | 22 | |||
| In situ ligation assay | 70 and 71 | |||
| In situ end labelling | 72 and 73 | |||
| Change in cell shape | Cell becomes spherical and detaches from neighbouring cells | Light microscopy, fluorescence microscopy TEM & SEM | 30 and 74 | |
| Apoptotic bodies formation | Activation of caspase-6 and caspase-7 | Electrochemiluminescence ELISA assay using a neo-epitope antibody against cleaved Lamin A | 75 | |
| Coumarin based fluorescent substrates, irreversible inhibitors, selective aggregation induced emission luminogens combined with mass cytometry, immunofluorescence, confocal microscopy, and western blot | 76 | |||
| Immuno blot, flow cytometry, luminescence & immunofluorescence | 59 and 76–79 | |||
| SDS-PAGE, Coomassie staining, fluorography analysis, caspase activity assay by means of fluorimetry | 79 | |||
| Membrane bound fragments called apoptotic bodies are formed in a controlled way, preventing the release of certain molecules in the extracellular matrix which could interfere with surrounding cells | Microscopy methods (phase contrast time lapse microscopy, confocal microscopy, TEM, SEM) | 74 and 80–82 | ||
| Dynamic light scattering, liquid chromatography tandem mass spectrometry | 74 and 80 | |||
| Fluorescence activated cell sorting, differential centrifugation & flow cytometry staining | 81 and 83 | |||
| Morphometric area analysis & morphometric cell count | 84 | |||
| Fluorescence labelling with Trp-BODIPY cyclic peptide | 85 | |||
| In situ DNA ligation | 86 | |||
| Late | Phagocytosis | Apoptotic bodies are engulfed and degraded by other cells | Microscopy methods (light microscopy, phase contrast, fluorescence, confocal, TEM, SEM) | 87–90 |
| Fluorescence activated cell sorting, differential centrifugation & flow cytometry staining | 83, 88 and 89 | |||
| Fluorescence labelling with Trp-BODIPY cyclic peptide | 85 | |||
| Degradation of cellular components | Organelles and other cellular structures are processed into component parts | Immunolabelling methods | 91–93 | |
| Microscopy methods (confocal microscopy, TEM &SEM) | 91, 92 and 94 |
Classic methods for apoptosis detection and their main drawbacks
As previously mentioned, many techniques for the detection of apoptosis were developed, many of which showed impressive results in vitro. One of the most common approaches is the use of fluorescent dyes that specifically bind to target molecules associated to apoptosis pathways. Microscopy or spectroscopy techniques are used to visualize morphological changes and/or quantify labelled biomarkers. However, through intensive research, new distinct mechanisms of cell death have lately been identified and many of the classic dyes (PI, DAPI, annexin V) lack an acceptable specificity or selectivity for their pathways. As a result, they may enter/label dead or dying cells, but the researcher cannot accurately discriminate one cell death pathway from another. For instance, DAPI and PI cannot distinguish between early apoptotic and necrotic cells leading to potential false-positive results when used to detect apoptosis. Moreover, annexin V often uses expensive recombinant proteins which can affect cell activity or introduce artifacts. The lack of real-time monitoring given by the fact the most classic methods use endpoint measurements, makes it difficult to study cell dynamics over time, which is necessary for several research issues. These are just a few drawbacks, which can be overcome with the aid of other molecular techniques. Even though this strategy can be time-consuming, it continues to be a good method for validating the results or for providing more in-depth answers.For the in vitro apoptosis detection, different methods are commonly used: microscopy, biochemical techniques, spectroscopy, electrochemistry and microfluidics.95 Among the first procedures, time-lapse electron microscopy was applied for morphological hallmarks monitorization of the cells.96
On the other hand, in vivo detection of apoptosis has additional limitations which makes it more challenging when it comes to designing fluorescent probes. Most of these are related to the luminescent properties of the probe, interaction of the light with different tissues or biocompatibility. Notably, scattering of both the excitation or the emitted light by the biological tissues can drastically impact the resolution or intensity of the signal. Moreover, the tissue penetration depth is highly dependent to the wavelength of the light used. One reason for this is that tissue molecules interact distinctly with different wavelengths when it comes to the magnitude of scattering or absorption events. Hence, longer wavelength light has lower energy, which besides the fact that it is less likely to be scattered or absorbed, also has the advantage of a reduced photobleaching effect on the fluorescent probe used. Other difficulties that need to be overcome in order to design effective fluorescent probes for apoptosis detection in vivo include the low signal to noise ratio given by the autofluorescence of the complex biological environments; cytotoxicity or other events triggered by the interaction of the exogenous probe with the cells; biological barriers (such as blood–brain-barrier) which sometimes need to be worked through by the fluorescent probe to analyse specific tissues. Another aspect that needs to be considered is that the utilization of in vivo research for the detection of apoptosis may give rise to ethical considerations in certain instances, as well as provide technical obstacles in terms of sample acquisition and observation of events within living organisms.
From a biochemical point of view, some specific proteins and genes that are activated during apoptosis are recognized as key effectors97 and can help investigate the cell death pathway with increased accuracy. Besides the simple quantification of these effectors, which remains a strong technique, the localization of particular molecules, such as the externalization of PS or cytochrome c release from the mitochondria, is also an appropriate method. Caspases are commonly studied key effectors of apoptosis. Their activity can be quantified by using fluorogenic caspase substrates via flow cytometry.98 On the other hand, cytosolic cytochrome c is often assessed via fluorescence microscopy52,99 or flow cytometry47,100 for apoptosis detection and, other biomarkers, such as oligonucleosomal multimers of 180–200 bp, or the ratio of proapoptotic to antiapoptotic proteins (ex. Bcl-2/Bax) can be quantified with the help of gel-based assays.101–103
Fluorescent dyes
Detection approaches are dependent on the fluorescence property of the probes used for apoptosis specific markers labelling engaging microscopy, flow cytometry or even genomic methods for more detailed investigations. In contrast, many widely used fluorescent dyes may exhibit limited specificity, since they have the potential to attach to other cellular structures or processes, hence resulting in potential instances of false positive results. The most used dye is annexin V conjugated to fluorescein isothiocyanate (FITC) which can detect middle to late stage apoptosis by binding to the exposed phosphatidylserine molecules on cell membrane.104 However, integrating these dyes with complementary techniques can mitigate some of the limitations. In many protocols, annexin V-FITC is used in combination with propidium iodide (PI) which discriminates between live and necrotic cells by displaying red fluorescence.105 In general, these two dyes are imaged using fluorescence microscopy, but for validation and numerical quantification, flow cytometry is usually required.106 This method is an analytical technology that ensures a rapid examination of cell populations offering flexibility for different particularities of cell cycle events research.107The combination of microscopy imaging and flow cytometry in one device makes the detection of apoptosis and necroptosis possible, as described by Pietkiewicz et al. 2015. They imaged the morphology of the nucleus at a single cell level using annexin V-FITC and propidium iodide staining protocol.108
TUNEL assay
TUNEL assay is another representative apoptosis assay that can be performed in vitro on cell cultures, but also in vivo on animal models109 and ex vivo on different tissues.110,111 The technique name comes from terminal deoxynucleotidyl transferase (Tdt)-mediated dUTP nick-end of DNA fragments labelling during chromatin condensation.112 The analysis have involved flow cytometry113 or microscopy.41 Being one of the firsts methods used for apoptosis detection, TUNEL assay had a big disadvantage, its accuracy in differentiating between necrotic cells and late-apoptotic cells.114 In the next years, an attempt to optimize these difficulties was made and the improvements consist in exploring other fluorophores115 or antibodies116 for labelling the DNA fragments or using different software platforms for the analysis.84Considering these aspects, the significance of developing novel techniques for apoptosis detection lies in their potential to address the challenges associated with conventional methods. By doing so, researchers can obtain more precise, targeted, and comprehensive insights into apoptotic mechanisms within both research and clinical settings. This phenomenon contributes to an enhanced comprehension of cellular apoptosis mechanisms, pathological pathways, and the potential of establishing improved therapeutic approaches.
Advanced methods for the detection of apoptosis
Caspases
Caspases are cysteine proteases that act as key components in certain types of programmed cell death, such as apoptosis, pyroptosis, necroptosis, necrosis, autophagy and mitotic catastrophe.117 Normally they are expressed as inactive zymogens but once stimulated, they become fully active subsequently autolytic cleavage.118 Apoptosis involves the activation of caspases which degrade cell components. They are known to cleave a broad spectrum of structural proteins like actin, tubulin, vimentin, ROCK1, filamin-A and fodrin which might contribute to the specific apoptotic cell morphology.119 This is followed by the formation of apoptotic bodies which are removed by surrounding cells via phagocytosis. Their active form and cleavage residue presence can be correlated to specific cell death types. Direct quantification methods for these cell death biomarkers that rely on antibody labelling are currently used, including ELISA, flow cytometry and Western blot. However, sample damage, long assay duration and the need of sophisticated instruments and expensive reagents, are some drawbacks that make them unsuitable for live cell assays, point-of-care applications and/or in vivo studies. Evaluation of apoptosis by monitoring the proteolytic activity of such caspases is a more reliable and less invasive approach that allows real-time monitoring of apoptosis both in vitro and in vivo. With hundreds of substrates identified so far,119 proteomic studies were able to highlight the dominant motifs cleaved by specific members of the caspase family of proteins.120 Since caspase 3 and 7 are the main proteases involved in cell death, many sensors for apoptosis rely on short peptides containing restriction sites for these two members. The optimal and most used sequence for caspase 3/7 activity sensor designing is a 4 amino acids peptide: DEVD (Asp-Glu-Val-Asp). Therefore, a wide variety of methods for apoptosis detection rely on linking a fluorophore to a quencher through a DEVD motif. The resulted construct will either switch from a fluorescent off state to an on state or significantly change its optic properties consequent to the specific proteolytic cleavage.A wide variety of methods to quantify caspase activity for the indirect evaluation of apoptosis have been reported.
Caspase cleavage of bioengineered substrate molecules as a fluorescence on/off switch. Since these techniques only show a fluorescent signal as a response to activated caspases, it is a reliable approach to distinguish apoptosis from other types of cell death. Therefore, they can be used to highlight the efficacy of treatments that involve the use of apoptosis-inducing compounds.
Genetically modified cells to express switch-on fluorescence-base indicators that are specifically cleaved by caspase-3 like proteases have been used to detect cell death in real-time.121 These indicators were designed by genetic engineering using Venus (a yellow fluorescent protein) as a template. The final genetically encoded construct is a cyclized protein, in a non-fluorescence state, that contains a caspase-3 cleavage site (DEVD). Fluorescence activity is only acquired after the caspase specific cleavage of DEVD. This method allows the evaluation of apoptosis in real-time in various conditions and is suitable both for 2D and 3D cell culture models.
One interesting and novel approach relies on aggregation induced emission (AIE) luminogens. AIE is a phenomenon that has gained significant interest in the domain of cellular imaging and theranostics. In contrast to other fluorescent molecules or nanoobjects, AIE luminogens (AIEgens) present a strong emission when they form aggregates due to a restriction in intramolecular movements.122 Since their discovery by Ben Zhong Tang's group,123 they have become a key component in many applications, including the evaluation of cellular processes. Moreover, AIE based probes suitable for both apoptosis evaluation (through caspase-3/-7 activity quantification) and apoptosis inducing compounds screening have been described. One such probe consists of an AIE hydrophobic luminogen (TPE) in a fluorescence off-state due to the conjugation with a caspase-specific peptide (DEVD). When apoptosis is triggered following caspase-3/-7 activation, the two subunits are freed through caspase cleavage and the TPE residues aggregate followed by a turn-on in their fluorescence.124
Fluorescence resonance energy transfer (FRET) between two proteins (CFP and YFP) linked together by DEVD has been used for apoptosis detection in living zebra fish at single cell resolution. The gene for this fusion protein was inserted after a skin specific keratin 4 promoter leading to a green fluorescence signal (specific to YFP) under the excitation of the donor molecules (CFP) in skin cells. The cleavage of DEVD by caspase 3/7 would reduce the energy transfer, shifting the overall emission from green to blue (specific to CFP). Single apoptotic cells could be tracked in real time in a live vertebrate animal model.125
Caspase cleavage of luciferase prosubstrates and bioluminescence evaluation
Besides fluorescence, bioluminescence is another non-invasive optical imaging technology that has shown huge potential for the detection of biological processes in vivo. This method relies on the enzymatic conversion of a non-luminescent substrate into a light producing molecule. As a result, bioengineered cells that express luciferase can metabolise exogenous luciferin leading to a luminescent signal, which can further be quantified. Lately, numerous bioluminescent probes for apoptosis detection in animal models have been developed.In one example, firefly luciferase positive cells were subcutaneously administered in SCID mice and tumours were allowed to grow. The authors used a modified substrate, Z-DEVD-amino luciferin, which could only be metabolised by luciferase after the initiation of RCD, the consequent activation of caspases and cleavage of the prosubstrate.126 Therefore, they were able to provide a biocompatible, preclinical method for the detection of apoptotic cells.
Cytochrome c
Cytochrome c (Cyt c) is a hemeprotein that is connected to the inner mitochondrial membrane and functions as an electron transporter required for ATP production.127The transfer of Cyt c from the intermembrane space of the mitochondria into the cytoplasm is an event unique to early apoptosis because, under normal circumstances, the outer mitochondrial membrane is only permeable to small metabolites up to ∼5 kDa.128
This process, which is a crucial step in the intrinsic apoptosis pathway, was frequently referred to as the irreversible point of apoptosis,99 even though more recent data suggest that cells might recover from late stages of programmed cell death through a mechanism called anastasis.129 However, quantifying cytochrome c release from the mitochondria remains a powerful tool for the evaluation of apoptosis.
In dying cells, a Cyt c concentration between 1 and 10 μM (ref. 99) can be identified because a little portion of Cyt c released from the mitochondria can stimulate various amplification loops, which lead to the release of all of Cyt c from the mitochondria to the cytosol.128,130 This will eventually lead to the formation of the apoptosome (Apaf-1/caspase-9/cytochrome c complex), which is in charge of cellular structure breakdown.
As a result, the quantification of Cyt c release is frequently employed to highlight mitochondria-dependent apoptosis.100 However, the most popular techniques are either laborious or call for complicated processes including immunolabeling, subcellular separation, and western blotting.47
As a result, nanoparticle-based assays for Cyt c quantification have gained popularity lately. Many of these rely on carbon dots (CDs), some carbon-based nanoparticles, due to their unique optical properties, increased stability, biocompatibility, selectivity and sensitivity. Certain quenching mechanisms of Cyt c towards CDs have made them ideal candidates for the quantification of this molecule. Based on these properties, recent studies report the successful usage of CDs for apoptosis detection in vitro.52,99 Moreover, Cyt c concentration was determined in human serum samples131 and in living zebrafish.99
Phosphatidylserine externalization sensors
Phosphatidylserine externalization acts as an “eat me” signal of apoptotic cells to neighbouring phagocytes and is considered a hallmark of apoptosis.132 However, recent studies suggest that the PS externalization process is inducible, reversible and not associated to other apoptosis specific events, such as mitochondrial Cyt c release, activation of caspases or DNA fragmentation.132,133 Nevertheless, PS export to the outer plasma membrane leaflet lays the foundation for Fluorescein isothiocyanate-conjugated annexin V binding assay for apoptosis detection.23 Notably, this process is regulated in a calcium dependent manner133 and a complete inhibition of PS externalization was reported in some hematopoietic cell lines in the absence of extracellular calcium.133 Moreover its externalization is also inhibited by calcium channel blockers,132 making the annexin V assay through flow cytometry or fluorescence microscopy rely on the addition of external calcium ions. Similar to this technique, more recent methods that focus on labelling PS have been developed. The detection of single apoptotic cells using luminol electrochemiluminescence has been reported.134 Other studies use nanomaterials such as carbon quantum dots to design annexin V conjugated apoptosis detection probes,21 or silver nanoparticles followed by SERS analysis to highlight externalized PS.26 Radiolabelled annexin V was also used to detect apoptotic cells in vivo.135,136 Although, there are lots of commercially available FITC-annexin V kits to highlight and distinguish apoptosis from other RCD types, and a wide diversity of methods have been developed and used to assess cell death based on the same principle, their efficacy and specificity have lately been questioned due to recent studies which report PS exposure in non-apoptotic cells, such as cells that undergo necroptosis or pyroptosis.137–140Computational tools
Label-free detection of apoptosis in live cells could help overcome the inconvenience of a potential interference between the fluorescent probe and the physiological functions of the cell. Although it can be difficult to achieve, deep learning- and activity recognition-based systems have allowed the detection and quantification of apoptotic events with high accuracy. The training model relies on large datasets of both apoptotic and nonapoptotic cells, multiple cell types and fluorescent labels and several imaging techniques. This computational tool can detect multiple apoptotic events in microscopy time-lapses and quantify cytotoxicity in vitro and in vivo based on live-cell imaging, such as intravital microscopy data.141Other recent studies describe the use of deep learning to discriminate apoptosis from necroptosis using label-free digital holographic microscopy,142 or to classify apoptosis and ferroptosis consequent to anti-transferrin receptor 1 antibody, DAPI and phalloidin-FITC staining.143 The aforementioned methods are able to predict or identify these cellular processes with increased accuracy.
Clinical evaluation of apoptosis
The last years have witnessed an increasing amount of newly approved therapeutic schemes, especially in the oncological domain. These treatments aim to disrupt the carcinogenesis process by interfering with the components from the cell death pathway and induce the apoptosis.144,145 While the main aspect behind these therapies is represented by efficient tumour clearance, a second goal is to achieve the installation of programmed cell death so that cancer cells can be eliminated by neighbouring phagocytic cells in a “quiet and quickly” manner.146 Therefore, it is critical to implement in clinical practice imagistic techniques that can specifically evaluate the efficiency of apoptosis-inducing therapies for disease monitoring, especially cancer. The different approaches for apoptosis detection, their advantages, drawbacks and some applications are presented in Table 2.| Method | Source | Advantages | Drawbacks | Applications |
|---|---|---|---|---|
| Bioluminescence | Bioluminescence proteins (e.g., firefly luciferase/Renilla luciferase) | High sensitivity | In some cases, the reporter gene must be introduced in the genome through genetic manipulation | In vitro, ex vivo and in vivo detection of apoptosis165 |
| Does not require external excitation light source | Lower resolution compared to other techniques | In vitro, ex vivo and in vivo drug screening of proapoptotic compounds166 | ||
| Bioluminescence resonance energy transfer (BRET) – based biosensor | Suitable for real-time detection | Transient signal | In vitro fluorescence-based detection and in vivo bioluminescence-based detection of apoptosis167 | |
| Suitable for in vivo studies | BRET-induced NIR emission-based detection of apoptotic cells168 | |||
| Fluorescence | Caspase activity reporters (fluorescent molecule (±quencher) bound to a peptidic substrate) | High sensitivity | The reporter gene must be introduced in the genome through genetic manipulation | Apoptosis monitoring and drug screening detection based on AIE124 |
| High specificity | Caspase kinetics alteration | Real-time monitoring of apoptosis in cultured cells169,170 | ||
| Suitable for real-time detection | Limited applicability for complex tissues and in vivo models | Caspase activity evaluation with potential applications in drug design and screening171 | ||
| Suitable for in vivo studies | Potential cytotoxicity | |||
| Challenging data interpretation | ||||
| Annexin V – conjugated probes | High sensitivity | In some cases, the reporter gene must be introduced in the genome through genetic manipulation | In vivo detection of cell death in murine cells and embryonic tissues172 | |
| Suitable for real-time detection | May require exogenous calcium ions133 | Vis/NIR-detection of cytotoxic compounds-induced apoptosis in vitro and in vivo173 | ||
| Suitable for ex vivo and in vivo studies | Specificity limitations (due to PS exposure in non-apoptotic cells)137–140 | In vitro and in vivo real-time imaging of apoptosis174 | ||
| Nanomaterials (e.g., carbon dots, hybrid nanoparticles) | High sensitivity | Potential cytotoxicity | Vis/NIR-detection of cytotoxic compounds-induced apoptosis in vitro and in vivo173 | |
| High specificity | Challenging data interpretation | In vitro monitoring of apoptosis inducing compounds52 | ||
| Suitable for real-time detection | Limited use in vivo | Real-time detection of apoptosis175 | ||
| Fluorescent antibodies | High sensitivity | Endpoint measurement | Flow cytometry evaluation of apoptosis | |
| High specificity | Challenging data interpretation | Immunofluorescence assays92 | ||
| Suitable for ex vivo studies | High cost | |||
| Other fluorescence dyes (e.g., DAPI/Hoechst/PI, FITC-annexin V, Mitotracker dyes) | Cost-effective | Limited use in vivo | In vitro detection of apoptosis106 | |
| Some of them have high specificity for target biomarkers | Challenging data interpretation | |||
| Versatility | Often – endpoint measurement | |||
| May interfere with other cellular processes, leading to false positive results | ||||
| Photobleaching | ||||
| Photoacoustic | Contrast agents (functionalized NPs, indocyanine green conjugates, apoptosis-specific nanobodies) | High sensitivity and spatial resolution | Limited specificity | In vivo real-time detection of tumour apoptosis176,177 |
| Suitable for real-time detection | May interfere with other cellular processes | In vivo evaluation of stem cell location and apoptosis178 | ||
| Suitable for in vivo studies | High costs of production | |||
| Photostability limitations | ||||
| Challenging acquisition setup and data interpretation | ||||
| MRI | Contrast agents, paramagnetic/ultrasmall superparamagnetic iron oxide NPs | High sensitivity | Potential cytotoxicity | Detection of drug-induced apoptosis in vitro, ex vivo and in vivo179 |
| Suitable for real-time detection | Limited specificity | In vivo real-time detection of tumour apoptosis177 | ||
| Suitable for ex vivo and in vivo studies | Long acquisition times | |||
| High-cost equipment | ||||
| Challenging data interpretation | ||||
| PET/SPECT | Specific radiotracers | Suitable for non-invasive, whole-body in vivo imaging | Limited specificity and resolution | In vivo evaluation of caspase activity153,180 |
| High sensitivity | High costs | In vivo monitoring of externalized PS during chemotherapy induced apoptosis181,182 | ||
| Quantitative | Radiation exposure | |||
| Versatility | Challenging data interpretation | |||
| Ultrasound | Contrast agents | Non-invasive | Limited specificity | Detection of drug-induced apoptosis in vitro, in situ and in vivo150 |
| Suitable for real-time detection | Limited contrast | Drug screening, in vitro detection of different forms of cell death183 | ||
| Low cost | Limited depth penetration | Pre-clinical in vitro monitorization of radiotherapy184 and photodynamic therapy effects149 | ||
| Versatility | Limited to structural information | |||
| High spatial resolution | Challenging acquisition setup and data interpretation |
Currently, in clinical setting, these methods primarily rely on magnetic resonance imaging (MRI) that can track the changes in the diffusive properties of a tissue or tumour mass (DW-MRI), where a tissue affected by cell death will have an increased water diffusion and diffusion coefficient respectively.147,148 In addition to MRI techniques, high-frequency ultrasound (10 MHz or greater) has shown promise in the detection of apoptotic cells, both in vitro and in vivo. This imaging modality utilizes the unique specular reflections produced by apoptotic cells, predominantly arising from the condensation and fragmentation of the cell nucleus during the later stages of apoptosis. Ultrasound transducers operating at frequencies of 10–60 MHz generate wavelengths of 25–150 μm, which closely match the size of individual cells and nuclei (10–20 μm), enabling sensitivity to change in cell size and nuclear morphology associated with apoptosis. Notably, the backscatter from apoptotic nuclei is significantly higher, up to six-fold, compared to non-apoptotic cellular nuclei. This distinctive signal can be observed in cultures treated with a variety of drugs and radiation. Consequently, the combination of high-frequency ultrasound with other imaging modalities, such as MRI, offers a comprehensive approach for evaluating apoptosis and provides valuable insights into the cellular and molecular changes occurring in pathological conditions.149,150
Magnetic resonance spectroscopy (MRS) is also used to measure lipid concentration and correlation of fat-water ratio for cell death quantification.151,152 However, these techniques are not capable to timely and directly quantify the induction of apoptosis and fail to offer specific indicators. Other intensive tested methods involve the radiolabelling of Annexin-V or aposense compounds and tracking of their uptake and biodistribution via positron emission tomography (PET) or single photon-emission tomography (SPECT). Although more specific, these methods are not yet efficient in specifically detecting and differentiating true cell death from growth arrest.146 Experimental imaging techniques are now relying on the specific labelling of active caspases or substrates that are cleaved by these enzymes153,154 and further quantification via PET imaging. Even so, there are issues related to the selectivity and specificity of the ligands or the uptake of the substrates.
Fluorescent imaging is one of the most commonly used technique for preclinical research involving early diagnosis, drug development, real-time therapeutic monitoring and prediction of prognosis. The reliability of this technique is supported by the sensibility of fluorescence, the stability of the fluorescent labels, advanced labelling techniques for specific molecular targets and ultimately, the reliable spatial resolution in comparison with the cellular dimensions.155 Most of the techniques currently used in the clinic involve ex vivo analysis for quantification of specific molecules or events based on fluorescence assay. Such an example is the evaluation of apoptosis through terminal deoxynucleotidyl transferase (TdT) dUTP nick and labelling (TUNEL). This assay is measuring the fluorescent labelled dUTPs that are integrated via TdT into the damaged DNA of a biopsy sample, where the intensity of the fluorescence is corelated with the degree of apoptosis.156,157 Similar principles are applied in the case of immunofluorescence techniques where specific tissue or circulatory molecules can be detected based on antibody – antigen affinity and use of fluorochromes to detect the location of the antibody. This technique can highlight the presence of specific molecules associated with the induction of apoptosis, or even multiple ones through simultaneous labelling with different fluorophores (e.g. FITC or rhodamine).158 These assays are valuable to quantify the molecular changes related to cell death in tissue samples before and after treatment; however, there is a significant disadvantage consisting in the necessity of multiple biopsies and inability to monitor patients in real-time. While serial tumours sampling (or other tissue) is not necessarily a proper strategy in clinical trials, an advantageous technique could be represented by specific measurement of circulating molecules associated with cell apoptosis, considering the minimally invasive character of blood sampling. One group of such molecules are cytokeratins that represent up to 5% of the intracellular proteins.159 Experimental evidence suggests that quantification of CK18 and/or CK19 in liquid biopsies could predict the cell death ratio in tumours, but are not able to differentiate apoptosis from necrosis: the release of soluble CKs takes place in both mechanisms.160,161 However, combination techniques like the one between M30 apoptosense ELISA directed toward a caspase-cleaved neo-epitope on CK18 and M65 ELISA for detection of cleaved and intact CK18 from liquid samples could differentiate between different mechanisms of cell death in patients.161 Currently used techniques for molecular diagnostics are based on ELISA assays that quantify the colorimetric reaction between an antibody directed toward a molecule of interest and linked to an enzyme, and a substrate that is processed by the specific enzyme.158 However, considering the low amounts of circulatory apoptotic biomarkers in the liquid samples of the patients, a more advantageous approach could consist in fluorescence-linked immunosorbent assay (FLISA) or fluorescent ELISA (FELISA), techniques that are a more sensitive variation from the standard colorimetric ELISA and also more suitable for high throughput screening due tominimal amounts of required antibodies and less manipulation. However, at the current time, there are no extensive clinical studies based on this approach.
Nevertheless, an ex vivo approach for dynamically measuring cell apoptosis in patients during treatment is not necessarily feasible compared to the imagistic approaches. Fluorescent evaluation in the clinical setting is hampered by the unavailability of fluorescent dyes approved for patients use.155 For the moment only two fluorophores, indocyanine green (ICG) and fluorescein, have been reported in the clinic for fluoresce based imaging of patients. ICG has been used to reconstruct in 3D an image of human breast cancer via fluorescence diffuse optical tomography (FDOT).162 Fluorescein has been experimentally used in ophthalmology for diagnosis of cystoid macular edema (CME) via fundus fluorescein angiography (FFA). Even so, the standard optical coherence tomography (OCT) has been assessed as more superior in demonstrating axial distribution of fluid.163 Although these studies are promising, ICG and fluorescein are non-specific exogenous fluorophores and thus are not suitable for targeted molecular imaging. However, these types of approaches are standing at the base of clinical trials based on targeted fluorophores for real life detection of specific biomarkers of apoptosis. Moreover, combination of fluorescence molecular tomography (FMT) with X-ray computed tomography (CT) or MRI will enhance the value of the imaging due to the possibility of specific mapping of molecular structures according to anatomical structures.155
Emerging data suggest that the dysregulation of apoptosis, through upregulation or downregulation, plays a significant role in the pathogenesis of various human diseases. However, the molecular mechanisms responsible for this dysregulation during disease initiation remain undefined. Consequently, future research should prioritize identifying the specific cellular and molecular entities that modulate apoptosis, with objectives centred on its inhibition or amplification based on therapeutic needs. It is crucial to emphasize functional assays to assess cellular viability, which provides a more precise evaluation of potential therapeutic agent efficacy compared to simply tracking cell retention or eradication.
A primary aim in biomedical research is harnessing insights into abnormal apoptosis to devise treatments enhancing patient outcomes in related diseases. Notably, in oncology, the observation that the majority of chemotherapeutic agents induce apoptosis in cancer cells has spurred investigations into treatments that target this dysregulated apoptosis. Furthermore, the integration of apoptosis imaging with imaging of other biological pathways, such as metabolism and angiogenesis, has potential to optimize clinical decision-making in diseases and interventions centred on apoptosis. Additionally, direct therapeutic interventions aiming at modulating apoptosis may exhibit reduced systemic toxicity compared to traditional chemotherapy. Such interventions could function as adjunctive treatments, augmenting the pro-apoptotic response of tumour cells during chemotherapeutic exposure. In autoimmune disease contexts, inducing apoptosis in auto-reactive lymphocyte subsets by introducing specific autoantigens without co-stimulatory survival signals offers promise. The efficacy of this strategy has been demonstrated in murine models of experimental autoimmune encephalomyelitis.164
Conclusions
Apoptosis detection involves specific equipment and reagents that sometimes require high costs, laborious work, and prolonged time. Moreover, the lack of sensitivity and/or specificity are common in conventional apoptosis detection assays, making them inadequate to establish cell death conclusively. Consequently, a combination of complementary yet distinct techniques is highly recommended, such as fluorogenic assays with molecular quantification of apoptosis specific proteins. However, conventional methods have evolved to be used in more general assays, such as routine and preliminary experiments. Given these circumstances, it is essential to use comprehensive apoptosis detection techniques which add considerable value to both in vitro and in vivo research to ensure thorough understanding of the RCD events and factors influencing them.The optical properties of various molecules or nanomaterials have been exploited to design improved probes with higher detection efficacy, increased signal to noise ratio and simplicity of approach, for example ON/OFF sensors and other probes which rely on event triggered changes in their optical properties. Furthermore, this allows to distinguish between RCD types that share similar characteristics and can investigate rapidly occurring cellular stages of such events. A novel trend is the use of computational tools such as deep learning in predicting apoptosis within a cell population, tissue or in vivo.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was granted by project “Development of the institutional capacity of UMFIH to support research, development and innovation activities in precision medicine (PrecisMED)” CNFIS-FDI-2023-0193. All figures were created using https://www.biorender.com/.References
- M. S. D'Arcy, Cell death: a review of the major forms of apoptosis, necrosis and autophagy, Cell Biol. Int., 2019, 43, 582–592 CrossRef PubMed.
- L. Galluzzi, S. A. Aaronson, J. Abrams, E. S. Alnemri, D. W. Andrews, E. H. Baehrecke, N. G. Bazan, M. V Blagosklonny, K. Blomgren, C. Borner, D. E. Bredesen, C. Brenner, M. Castedo, J. A. Cidlowski, A. Ciechanover, G. M. Cohen, V. De Laurenzi, R. De Maria, M. Deshmukh, B. D. Dynlacht, W. S. El-Deiry, R. A. Flavell, S. Fulda, C. Garrido, P. Golstein, M.-L. Gougeon, D. R. Green, H. Gronemeyer, G. Hajnóczky, J. M. Hardwick, M. O. Hengartner, H. Ichijo, M. Jäättelä, O. Kepp, A. Kimchi, D. J. Klionsky, R. A. Knight, S. Kornbluth, S. Kumar, B. Levine, S. A. Lipton, E. Lugli, F. Madeo, W. Malorni, J.-C. Marine, S. J. Martin, J. P. Medema, P. Mehlen, G. Melino, U. M. Moll, E. Morselli, S. Nagata, D. W. Nicholson, P. Nicotera, G. Nuñez, M. Oren, J. Penninger, S. Pervaiz, M. E. Peter, M. Piacentini, J. H. M. Prehn, H. Puthalakath, G. A. Rabinovich, R. Rizzuto, C. M. P. Rodrigues, D. C. Rubinsztein, T. Rudel, L. Scorrano, H.-U. Simon, H. Steller, J. Tschopp, Y. Tsujimoto, P. Vandenabeele, I. Vitale, K. H. Vousden, R. J. Youle, J. Yuan, B. Zhivotovsky and G. Kroemer, Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes, Cell Death Differ., 2009, 16, 1093–1107 CrossRef CAS PubMed.
- F. G. Blankenberg, P. D. Katsikis, R. W. Storrs, C. Beaulieu, D. Spielman, J. Y. Chen, L. Naumovski and J. F. Tait, Quantitative analysis of apoptotic cell death using proton nuclear magnetic resonance spectroscopy, Blood, 1997, 89, 3778–3786 CrossRef CAS PubMed.
- I. Vermes, C. Haanen and C. Reutelingsperger, Flow cytometry of apoptotic cell death, J. Immunol. Methods, 2000, 243, 167–190 CrossRef CAS PubMed.
- A. Lekshmi, S. N. Varadarajan, S. S. Lupitha, D. Indira, K. A. Mathew, A. Chandrasekharan Nair, M. Nair, T. Prasad, H. Sekar, A. K. Gopalakrishnan, A. Murali and T. R. Santhoshkumar, A quantitative real-time approach for discriminating apoptosis and necrosis, Cell Death Discovery, 2017, 3, 16101 CrossRef CAS PubMed.
- T. Vicar, M. Raudenska, J. Gumulec and J. Balvan, The Quantitative-Phase Dynamics of Apoptosis and Lytic Cell Death, Sci. Rep., 2020, 10, 1566 CrossRef CAS PubMed.
- S. Vasudevan, G. C. K. Chen, M. Andika, S. Agarwal, P. Chen and M. Olivo, Dynamic quantitative photothermal monitoring of cell death of individual human red blood cells upon glucose depletion, J. Biomed. Opt., 2010, 15, 057001 CrossRef PubMed.
- A. Sadeghi-Naini, S. Zhou, M. J. Gangeh, Z. Jahedmotlagh, O. Falou, S. Ranieri, M. Azrif, A. Giles and G. J. Czarnota, Quantitative evaluation of cell death response in vitro and in vivo using conventional-frequency ultrasound, Oncoscience, 2015, 2, 716–726 CrossRef PubMed.
- B. Méry, J.-B. Guy, A. Vallard, S. Espenel, D. Ardail, C. Rodriguez-Lafrasse, C. Rancoule and N. Magné, Vitro Cell Death Determination for Drug Discovery: A Landscape Review of Real Issues, J. Cell Death, 2017, 10, 117967071769125 CrossRef PubMed.
- B. S. Cummings and R. G. Schnellmann, Measurement of Cell Death in Mammalian Cells, Curr. Protoc. Pharmacol., 2004 DOI:10.1002/0471141755.ph1208s25.
- Ö. S. Aslantürk, in Genotoxicity – A Predictable Risk to Our Actual World, InTech, 2018 Search PubMed.
- B. Yu, B. Zhang, W. Zhou, Q. Wang, P. Yuan, Y. Yang and J. Kong, Assessing Apoptosis Gene Expression Profiling with a PCR Array in the Hippocampus of Ts65Dn Mice, BioMed Res. Int., 2015, 2015, 1–8 Search PubMed.
- D. J. Hooker, P. R. Gorry, A. M. Ellett, S. L. Wesselingh and C. L. Cherry, Measuring and monitoring apoptosis and drug toxicity in HIV patients by ligation-mediated polymerase chain reaction, J. Cell. Mol. Med., 2009, 13, 948–958 CrossRef CAS PubMed.
- B. Liu, Z. N. Oltvai, H. Bayır, G. A. Silverman, S. C. Pak, D. H. Perlmutter and I. Bahar, Quantitative assessment of cell fate decision between autophagy and apoptosis, Sci. Rep., 2017, 7, 17605 CrossRef PubMed.
- J. H. Buchbinder, D. Pischel, K. Sundmacher, R. J. Flassig and I. N. Lavrik, Quantitative single cell analysis uncovers the life/death decision in CD95 network, PLoS Comput. Biol., 2018, 14, e1006368 CrossRef PubMed.
- L. Galluzzi, I. Vitale, S. A. Aaronson, J. M. Abrams, D. Adam, P. Agostinis, E. S. Alnemri, L. Altucci, I. Amelio, D. W. Andrews, M. Annicchiarico-Petruzzelli, A. V. Antonov, E. Arama, E. H. Baehrecke, N. A. Barlev, N. G. Bazan, F. Bernassola, M. J. M. Bertrand, K. Bianchi, M. V. Blagosklonny, K. Blomgren, C. Borner, P. Boya, C. Brenner, M. Campanella, E. Candi, D. Carmona-Gutierrez, F. Cecconi, F. K.-M. Chan, N. S. Chandel, E. H. Cheng, J. E. Chipuk, J. A. Cidlowski, A. Ciechanover, G. M. Cohen, M. Conrad, J. R. Cubillos-Ruiz, P. E. Czabotar, V. D'Angiolella, T. M. Dawson, V. L. Dawson, V. De Laurenzi, R. De Maria, K.-M. Debatin, R. J. DeBerardinis, M. Deshmukh, N. Di Daniele, F. Di Virgilio, V. M. Dixit, S. J. Dixon, C. S. Duckett, B. D. Dynlacht, W. S. El-Deiry, J. W. Elrod, G. M. Fimia, S. Fulda, A. J. García-Sáez, A. D. Garg, C. Garrido, E. Gavathiotis, P. Golstein, E. Gottlieb, D. R. Green, L. A. Greene, H. Gronemeyer, A. Gross, G. Hajnoczky, J. M. Hardwick, I. S. Harris, M. O. Hengartner, C. Hetz, H. Ichijo, M. Jäättelä, B. Joseph, P. J. Jost, P. P. Juin, W. J. Kaiser, M. Karin, T. Kaufmann, O. Kepp, A. Kimchi, R. N. Kitsis, D. J. Klionsky, R. A. Knight, S. Kumar, S. W. Lee, J. J. Lemasters, B. Levine, A. Linkermann, S. A. Lipton, R. A. Lockshin, C. López-Otín, S. W. Lowe, T. Luedde, E. Lugli, M. MacFarlane, F. Madeo, M. Malewicz, W. Malorni, G. Manic, J.-C. Marine, S. J. Martin, J.-C. Martinou, J. P. Medema, P. Mehlen, P. Meier, S. Melino, E. A. Miao, J. D. Molkentin, U. M. Moll, C. Muñoz-Pinedo, S. Nagata, G. Nuñez, A. Oberst, M. Oren, M. Overholtzer, M. Pagano, T. Panaretakis, M. Pasparakis, J. M. Penninger, D. M. Pereira, S. Pervaiz, M. E. Peter, M. Piacentini, P. Pinton, J. H. M. Prehn, H. Puthalakath, G. A. Rabinovich, M. Rehm, R. Rizzuto, C. M. P. Rodrigues, D. C. Rubinsztein, T. Rudel, K. M. Ryan, E. Sayan, L. Scorrano, F. Shao, Y. Shi, J. Silke, H.-U. Simon, A. Sistigu, B. R. Stockwell, A. Strasser, G. Szabadkai, S. W. G. Tait, D. Tang, N. Tavernarakis, A. Thorburn, Y. Tsujimoto, B. Turk, T. Vanden Berghe, P. Vandenabeele, M. G. Vander Heiden, A. Villunger, H. W. Virgin, K. H. Vousden, D. Vucic, E. F. Wagner, H. Walczak, D. Wallach, Y. Wang, J. A. Wells, W. Wood, J. Yuan, Z. Zakeri, B. Zhivotovsky, L. Zitvogel, G. Melino and G. Kroemer, Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018, Cell Death Differ., 2018, 25, 486–541 CrossRef PubMed.
- N. J. McCarthy and G. I. Evan, Curr. Top. Dev. Biol., 1997, 36, 259–278 CrossRef PubMed.
- T. Vanden Berghe, N. Vanlangenakker, E. Parthoens, W. Deckers, M. Devos, N. Festjens, C. J. Guerin, U. T. Brunk, W. Declercq and P. Vandenabeele, Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features, Cell Death Differ., 2010, 17, 922–930 CrossRef CAS PubMed.
- M. Sachet, Y. Y. Liang and R. Oehler, The immune response to secondary necrotic cells, Apoptosis, 2017, 22, 1189–1204 CrossRef CAS PubMed.
- B. Fadeel, B. Gleiss, K. Högstrand, J. Chandra, T. Wiedmer, P. J. Sims, J.-I. Henter, S. Orrenius and A. Samali, Phosphatidylserine Exposure during Apoptosis Is a Cell-Type-Specific Event and Does Not Correlate with Plasma Membrane Phospholipid Scramblase Expression, Biochem. Biophys. Res. Commun., 1999, 266, 504–511 CrossRef CAS PubMed.
- M. Mahani, P. Karimi-Mazidi, F. Khakbaz and M. Torkzadeh-Mahani, Carbon quantum dots—Annexin V probe: photoinduced electron transfer mechanism, phosphatidylserine detection, and apoptotic cell imaging, Microchim. Acta, 2022, 189, 69 CrossRef CAS PubMed.
- Z. Bacso and J. F. Eliason, Measurement of DNA damage associated with apoptosis by laser scanning cytometry, Cytometry, 2001, 45, 180–186 CrossRef CAS PubMed.
- I. Vermes, C. Haanen, H. Steffens-Nakken and C. Reutellingsperger, A novel assay for apoptosis Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V, J. Immunol. Methods, 1995, 184, 39–51 CrossRef CAS.
- G. Lin, Q. Mu, R. Revia, Z. Stephen, M. Jeon and M. Zhang, A highly selective iron oxide-based imaging nanoparticle for long-term monitoring of drug-induced tumor cell apoptosis, Biomater. Sci., 2021, 9, 471–481 RSC.
- T. Liu, W. Zhu, X. Yang, L. Chen, R. Yang, Z. Hua and G. Li, Detection of Apoptosis Based on the Interaction between Annexin V and Phosphatidylserine, Anal. Chem., 2009, 81, 2410–2413 CrossRef CAS PubMed.
- H. Zhou, Q. Wang, D. Yuan, J. Wang, Y. Huang, H. Wu, J. Jian, D. Yang, N. Huang, C. Haisch, Z. Jiang and S. Chen, Early apoptosis real-time detection by label-free SERS based on externalized phosphatidylserine, Analyst, 2016, 141, 4293–4298 RSC.
- S. Morita, S. Shirakawa, Y. Kobayashi, K. Nakamura, R. Teraoka, S. Kitagawa and T. Terada, Enzymatic measurement of phosphatidylserine in cultured cells, J. Lipid Res., 2012, 53, 325–330 CrossRef CAS PubMed.
- J. Kranich, N. Chlis, L. Rausch, A. Latha, M. Schifferer, T. Kurz, A. Foltyn-Arfa Kia, M. Simons, F. J. Theis and T. Brocker, In vivo identification of apoptotic and extracellular vesicle-bound live cells using image-based deep learning, J. Extracell. Vesicles, 2020, 9, 1792683 CrossRef CAS PubMed.
- L. C. Crowley, B. J. Marfell, A. P. Scott and N. J. Waterhouse, Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry, Cold Spring Harb. Protoc., 2016, 2016, prot087288 CrossRef PubMed.
- C. Stadelmann and H. Lassmann, Detection of apoptosis in tissue sections, Cell Tissue Res., 2000, 301, 19–31 CrossRef CAS PubMed.
- J. Cheng, Z. Lv, X. Zhang, X. Mitchelson and J. Cheng, Systematic dielectrophoretic analysis of the Ara C induced NB4 cell apoptosis combined with gene expression profiling, Int. J. Nanomedicine, 2013, 2333 CrossRef PubMed.
- P. Majtnerova, J. Capek, F. Petira, J. Handl and T. Rousar, Quantitative spectrofluorometric assay detecting nuclear condensation and fragmentation in intact cells, Sci. Rep., 2021, 11, 11921 CrossRef PubMed.
- R. Rose, N. Peschke, E. Nigi, M. Gelléri, S. Ritz, C. Cremer, H. J. Luhmann and A. Sinning, Chromatin compaction precedes apoptosis in developing neurons, Commun. Biol., 2022, 5, 797 CrossRef CAS PubMed.
- O. S. Frankfurt and A. Krishan, Identification of Apoptotic Cells by Formamide-induced DNA Denaturation in Condensed Chromatin, J. Histochem. Cytochem., 2001, 49, 369–378 CrossRef CAS PubMed.
- O. S. Frankfurt, J. A. Robb, E. V. Sugarbaker and L. Villa, Monoclonal Antibody to Single-Stranded DNA Is a Specific and Sensitive Cellular Marker of Apoptosis, Exp. Cell Res., 1996, 226, 387–397 CrossRef CAS PubMed.
- M. Enari, H. Sakahira, H. Yokoyama, K. Okawa, A. Iwamatsu and S. Nagata, A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD, Nature, 1998, 391, 43–50 CrossRef CAS PubMed.
- C. Gabler, Extranuclear detection of histones and nucleosomes in activated human lymphoblasts as an early event in apoptosis, Ann. Rheum. Dis., 2004, 63, 1135–1144 CrossRef CAS PubMed.
- P. Salgame, An ELISA for detection of apoptosis, Nucleic Acids Res., 1997, 25, 680–681 CrossRef CAS PubMed.
- Z. Darzynkiewicz, D. Galkowski and H. Zhao, Analysis of apoptosis by cytometry using TUNEL assay, Methods, 2008, 44, 250–254 CrossRef CAS PubMed.
- W. G. Telford, L. E. King and P. J. Fraker, Comparative evaluation of several DNA binding dyes in the detection of apoptosis-associated chromatin degradation by flow cytometry, Cytometry, 1992, 13, 137–143 CrossRef CAS PubMed.
- Y. Gavrieli, Y. Sherman and S. A. Ben-Sasson, Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation, J. Cell Biol., 1992, 119, 493–501 CrossRef CAS PubMed.
- L. C. Crowley, B. J. Marfell and N. J. Waterhouse, Analyzing Cell Death by Nuclear Staining with Hoechst 33342, Cold Spring Harb. Protoc., 2016, 2016, prot087205 CrossRef PubMed.
- C. Charriaut-Marlangue, I. Margaill, A. Represa, T. Popovici, M. Plotkine and Y. Ben-Ari, Apoptosis and Necrosis after Reversible Focal Ischemia: An in Situ DNA Fragmentation Analysis, J. Cereb. Blood Flow Metab., 1996, 16, 186–194 CrossRef CAS PubMed.
- S. Sakamuru, M. S. Attene-Ramos and M. Xia, Mitochondrial Membrane Potential Assay, Methods Mol. Biol., 2016, 1743, 17–22 CrossRef PubMed.
- P. Nguyen, P. Doan, A. Murugesan, T. Ramesh, T. Rimpilainen, N. R. Candeias, O. Yli-Harja and M. Kandhavelu, GPR17 signaling activation by CHBC agonist induced cell death via modulation of MAPK pathway in glioblastoma, Life Sci., 2022, 291, 120307 CrossRef CAS PubMed.
- M. Fujikawa and M. Yoshida, A sensitive, simple assay of mitochondrial ATP synthesis of cultured mammalian cells suitable for high-throughput analysis, Biochem. Biophys. Res. Commun., 2010, 401, 538–543 CrossRef CAS PubMed.
- N. J. Waterhouse and J. A. Trapani, A new quantitative assay for cytochrome c release in apoptotic cells, Cell Death Differ., 2003, 10, 853–855 CrossRef CAS PubMed.
- P. Manickam, A. Kaushik, C. Karunakaran and S. Bhansali, Recent advances in cytochrome c biosensing technologies, Biosens. Bioelectron., 2017, 87, 654–668 CrossRef CAS PubMed.
- J. Radhakrishnan, R. Origenes, G. Littlejohn, S. Nikolich, E. Choi, S. Smite, L. Lamoureux, A. Baetiong, M. Shah and R. J. Gazmuri, Plasma Cytochrome c Detection Using a Highly Sensitive Electrochemiluminescence Enzyme-Linked Immunosorbent Assay, Biomarker Insights, 2017, 12, 117727191774697 CrossRef PubMed.
- X. Hu, Z. Li, R. Lin, J. Shan, Q. Yu, R. Wang, L. Liao, W. Yan, Z. Wang, L. Shang, Y. Huang, Q. Zhang and K. Xiong, Guidelines for Regulated Cell Death Assays: A Systematic Summary, A Categorical Comparison, A Prospective, Front. Cell Dev. Biol., 2021, 9 DOI:10.3389/fcell.2021.634690.
- H. Salmani-Zarchi, Y.-S. Borghei and M. Nikkhah, A turn-off fluorimetric -aptasensor for early detection of apoptosis inside the cells, Spectrochim. Acta, Part A, 2023, 300, 122933 CrossRef CAS PubMed.
- C. S. Moldovan, A. Onaciu, V. Toma, R. Marginean, A. Moldovan, A. B. Tigu, G. F. Stiufiuc, C. M. Lucaciu and R. I. Stiufiuc, Quantifying Cytosolic Cytochrome c Concentration Using Carbon Quantum Dots as a Powerful Method for Apoptosis Detection, Pharmaceutics, 2021, 13, 1556 CrossRef CAS PubMed.
- A. Jenner, A. Peña-Blanco, R. Salvador-Gallego, B. Ugarte-Uribe, C. Zollo, T. Ganief, J. Bierlmeier, M. Mund, J. E. Lee, J. Ries, D. Schwarzer, B. Macek and A. J. Garcia-Saez, DRP1 interacts directly with BAX to induce its activation and apoptosis, EMBO J., 2022 DOI:10.15252/embj.2021108587.
- T. Panaretakis, K. Pokrovskaja, M. C. Shoshan and D. Grandér, Activation of Bak, Bax, and BH3-only Proteins in the Apoptotic Response to Doxorubicin, J. Biol. Chem., 2002, 277, 44317–44326 CrossRef CAS PubMed.
- Y. Wu, D. Zhao, J. Zhuang, F. Zhang and C. Xu, Caspase-8 and Caspase-9 Functioned Differently at Different Stages of the Cyclic Stretch-Induced Apoptosis in Human Periodontal Ligament Cells, PLoS One, 2016, 11, e0168268 CrossRef PubMed.
- T. Wieder, F. Essmann, A. Prokop, K. Schmelz, K. Schulze-Osthoff, R. Beyaert, B. Dörken and P. T. Daniel, Activation of caspase-8 in drug-induced apoptosis of B-lymphoid cells is independent of CD95/Fas receptor-ligand interaction and occurs downstream of caspase-3, Blood, 2001, 97, 1378–1387 CrossRef CAS PubMed.
- M. T. Cencioni, S. Santini, G. Ruocco, G. Borsellino, M. De Bardi, M. G. Grasso, S. Ruggieri, C. Gasperini, D. Centonze, D. Barilá, L. Battistini and E. Volpe, FAS-ligand regulates differential activation-induced cell death of human T-helper 1 and 17 cells in healthy donors and multiple sclerosis patients, Cell Death Dis., 2015, 6, e1741 CrossRef CAS PubMed.
- P. Yang, J. J. Peairs, R. Tano, N. Zhang, J. Tyrell and G. J. Jaffe, Caspase-8–Mediated Apoptosis in Human RPE Cells, Investig. Opthalmology Vis. Sci., 2007, 48, 3341 CrossRef PubMed.
- E. Svandova, B. Vesela, A. S. Tucker and E. Matalova, Activation of Pro-apoptotic Caspases in Non-apoptotic Cells During Odontogenesis and Related Osteogenesis, Front. Physiol., 2018 DOI:10.3389/fphys.2018.00174.
- B. W. Lee, M. R. Olin, G. L. Johnson and R. J. Griffin, in Apoptosis and Cancer, Humana Press, Totowa, NJ, 2008, pp. 109–135 Search PubMed.
- Y. Pan, M. Guo, Z. Nie, Y. Huang, Y. Peng, A. Liu, M. Qing and S. Yao, Colorimetric detection of apoptosis based on caspase-3 activity assay using unmodified gold nanoparticles, Chem. Commun., 2012, 48, 997–999 RSC.
- M. L. Würstle and M. Rehm, A Systems Biology Analysis of Apoptosome Formation and Apoptosis Execution Supports Allosteric Procaspase-9 Activation, J. Biol. Chem., 2014, 289, 26277–26289 CrossRef PubMed.
- C. A. Elena-Real, A. Díaz-Quintana, K. González-Arzola, A. Velázquez-Campoy, M. Orzáez, A. López-Rivas, S. Gil-Caballero, M. Á. De la Rosa and I. Díaz-Moreno, Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition, Cell Death Dis., 2018, 9, 365 CrossRef PubMed.
- F. Sahebazzamani, S. Hosseinkhani, L. A. Eriksson and H. O. Fearnhead, Apoptosome Formation through Disruption of the K192-D616 Salt Bridge in the Apaf-1 Closed Form, ACS Omega, 2021, 6, 22551–22558 CrossRef CAS PubMed.
- E. S. Hosseini, M. Nikkhah, A. A. Hamidieh, H. O. Fearnhead, J.-P. Concordet and S. Hosseinkhani, The lumiptosome, an engineered luminescent form of the apoptosome can report cell death by using the same Apaf-1 dependent pathway, J. Cell Sci., 2020 DOI:10.1242/jcs.242636.
- K.-W. Hsu, C.-Y. Huang, K.-W. Tam, C.-Y. Lin, L.-C. Huang, C.-L. Lin, W.-S. Hsieh, W.-M. Chi, Y.-J. Chang, P.-L. Wei, S.-T. Chen and C.-H. Lee, The Application of Non-Invasive Apoptosis Detection Sensor (NIADS) on Histone Deacetylation Inhibitor (HDACi)-Induced Breast Cancer Cell Death, Int. J. Mol. Sci., 2018, 19, 452 CrossRef PubMed.
- B. B. Wolf, M. Schuler, F. Echeverri and D. R. Green, Caspase-3 Is the Primary Activator of Apoptotic DNA Fragmentation via DNA Fragmentation Factor-45/Inhibitor of Caspase-activated DNase Inactivation, J. Biol. Chem., 1999, 274, 30651–30656 CrossRef CAS PubMed.
- G. Barroso, M. Morshedi and S. Oehninger, Analysis of DNA fragmentation, plasma membrane translocation of phosphatidylserine and oxidative stress in human spermatozoa, Hum. Reprod., 2000, 15, 1338–1344 CrossRef CAS PubMed.
- K. Plaetzer, T. Kiesslich, C. Oberdanner and B. Krammer, Apoptosis Following Photodynamic Tumor Therapy: Induction, Mechanisms and Detection, Curr. Pharm. Des., 2005, 11, 1151–1165 CrossRef CAS PubMed.
- V. V. Didenko, DNA Damage Detection In Situ, Ex Vivo, and In Vivo, Methods Protoc., 2011, 65–75 CAS.
- A. Saraste, Morphologic and biochemical hallmarks of apoptosis, Cardiovasc. Res., 2000, 45, 528–537 CrossRef CAS PubMed.
- R. Gold, M. Schmied, G. Rothe, H. Zischler, H. Breitschopf, H. Wekerle and H. Lassmann, Detection of DNA fragmentation in apoptosis: application of in situ nick translation to cell culture systems and tissue sections, J. Histochem. Cytochem., 1993, 41, 1023–1030 CrossRef CAS PubMed.
- R. Hayashi, Y. Ito, K. Matsumoto, Y. Fujino and Y. Otsuki, Quantitative Differentiation of Both Free 3′-OH and 5′-OH DNA Ends Between Heat-induced Apoptosis and Necrosis, J. Histochem. Cytochem., 1998, 46, 1051–1059 CrossRef CAS PubMed.
- A. Khodavirdipour, M. Piri, S. Jabbari, S. Keshavarzi, R. Safaralizadeh and M. Y. Alikhani, Apoptosis Detection Methods in Diagnosis of Cancer and Their Potential Role in Treatment: Advantages and Disadvantages: a Review, J. Gastrointest. Cancer, 2021, 52, 422–430 CrossRef PubMed.
- D. E. Ehrnhoefer, N. H. Skotte, J. Savill, Y. T. N. Nguyen, S. Ladha, L.-P. Cao, E. Dullaghan and M. R. Hayden, A Quantitative Method for the Specific Assessment of Caspase-6 Activity in Cell Culture, PLoS One, 2011, 6, e27680 CrossRef CAS PubMed.
- K. M. Groborz, M. Kalinka, J. Grzymska, S. Kołt, S. J. Snipas and M. Poręba, Selective chemical reagents to investigate the role of caspase 6 in apoptosis in acute leukemia T cells, Chem. Sci., 2023, 14, 2289–2302 RSC.
- M. R. Mahib, S. Hosojima, H. Kushiyama, T. Kinoshita, T. Shiroishi, T. Suda and K. Tsuchiya, Caspase-7 mediates caspase-1-induced apoptosis independently of Bid, Microbiol. Immunol., 2020, 64, 143–152 CrossRef CAS PubMed.
- M. Zheng, R. Karki, P. Vogel and T.-D. Kanneganti, Caspase-6 Is a Key Regulator of Innate Immunity, Inflammasome Activation, and Host Defense, Cell, 2020, 181, 674–687.e13 CrossRef CAS PubMed.
- J. G. Walsh, S. P. Cullen, C. Sheridan, A. U. Lüthi, C. Gerner and S. J. Martin, Executioner caspase-3 and caspase-7 are functionally distinct proteases, Proc. Natl. Acad. Sci., 2008, 105, 12815–12819 CrossRef CAS PubMed.
- G. Serrano-Heras, I. Díaz-Maroto, B. Castro-Robles, B. Carrión, A. B. Perona-Moratalla, J. Gracia, S. Arteaga, F. Hernández-Fernández, J. García-García, O. Ayo-Martín and T. Segura, Isolation and Quantification of Blood Apoptotic Bodies, a Non-invasive Tool to Evaluate Apoptosis in Patients with Ischemic Stroke and Neurodegenerative Diseases, Biol. Proced. Online, 2020, 22, 17 CrossRef CAS PubMed.
- L. Jiang, S. Paone, S. Caruso, G. K. Atkin-Smith, T. K. Phan, M. D. Hulett and I. K. H. Poon, Determining the contents and cell origins of apoptotic bodies by flow cytometry, Sci. Rep., 2017, 7, 14444 CrossRef PubMed.
- J. D. Lane, V. J. Allan and P. G. Woodman, Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells, J. Cell Sci., 2005, 118, 4059–4071 CrossRef CAS PubMed.
- T. K. Phan, I. K. Poon and G. K. Atkin-Smith, Detection and Isolation of Apoptotic Bodies to High Purity, J. Visualized Exp., 2018 DOI:10.3791/58317.
- M. M. Garrity, L. J. Burgart, D. L. Riehle, E. M. Hill, T. J. Sebo and T. Witzig, Identifying and Quantifying Apoptosis: Navigating Technical Pitfalls, Mod. Pathol., 2003, 16, 389–394 CrossRef PubMed.
- R. Subiros-Funosas, L. Mendive-Tapia, J. Sot, J. D. Pound, N. Barth, Y. Varela, F. M. Goñi, M. Paterson, C. D. Gregory, F. Albericio, I. Dransfield, R. Lavilla and M. Vendrell, A Trp-BODIPY cyclic peptide for fluorescence labelling of apoptotic bodies, Chem. Commun., 2017, 53, 945–948 RSC.
- P. Hauser, S. Wang and V. V. Didenko, Apoptotic Bodies: Selective Detection in Extracellular Vesicles, Methods Mol. Biol., 2017, 1554, 193–200 CrossRef CAS PubMed.
- M. Archana, M. Bastian, T. Yogesh and K. Kumaraswamy, Various methods available for detection of apoptotic cells- A review, Indian J. Cancer, 2013, 50, 274 CrossRef CAS PubMed.
- E. Boero, I. Brinkman, T. Juliet, E. van Yperen, J. A. G. van Strijp, S. H. M. Rooijakkers and K. P. M. van Kessel, Use of Flow Cytometry to Evaluate Phagocytosis of Staphylococcus aureus by Human Neutrophils, Front. Immunol., 2021 DOI:10.3389/fimmu.2021.635825.
- J. J. Gaforio, M. J. Serrano, I. Algarra, E. Ortega and G. Alvarez de Cienfuegos, Phagocytosis of apoptotic cells assessed by flow cytometry using 7-Aminoactinomycin D, Cytometry, 2002, 49, 8–11 CrossRef CAS PubMed.
- Y. Horii, S. Matsuda, K. Watari, A. Nagasaka, H. Kurose and M. Nakaya, An Assay to Determine Phagocytosis of Apoptotic Cells by Cardiac Macrophages and Cardiac Myofibroblasts, Bio-Protoc., 2017 DOI:10.21769/BioProtoc.2553.
- K. Neikirk, Z. Vue, P. Katti, B. I. Rodriguez, S. Omer, J. Shao, T. Christensen, E. Garza Lopez, A. Marshall, C. B. Palavicino-Maggio, J. Ponce, A. F. Alghanem, L. Vang, T. Barongan, H. K. Beasley, T. Rodman, D. Stephens, M. Mungai, M. Correia, V. Exil, S. Damo, S. A. Murray, A. Crabtree, B. Glancy, R. O. Pereira, E. D. Abel and A. O. Hinton, Systematic Transmission Electron Microscopy-Based Identification and 3D Reconstruction of Cellular Degradation Machinery, Adv. Biol., 2023, 2200221 CrossRef CAS PubMed.
- M. Guo, B. Lu, J. Gan, S. Wang, X. Jiang and H. Li, Apoptosis detection: a purpose-dependent approach selection, Cell Cycle, 2021, 20, 1033–1040 CrossRef CAS PubMed.
- M. Bottone, G. Santin, F. Aredia, G. Bernocchi, C. Pellicciari and A. Scovassi, Morphological Features of Organelles during Apoptosis: An Overview, Cells, 2013, 2, 294–305 CrossRef PubMed.
- C. dos S. Vergilio and E. J. T. De Melo, Autophagy, apoptosis and organelle features during cell exposure to cadmiumč, Biocell, 2013, 37, 45–54 Search PubMed.
- M. M. Martinez, R. D. Reif and D. Pappas, Detection of apoptosis: A review of conventional and novel techniques, Anal. Methods, 2010, 2, 996 RSC.
- N. J. McCarthy, M. K. B. Whyte, C. S. Gilbert and G. I. Evan, Inhibition of Ced-3/ICE-related Proteases Does Not Prevent Cell Death Induced by Oncogenes, DNA Damage, or the Bcl-2 Homologue Bak, J. Cell Biol., 1997, 136, 215–227 CrossRef CAS PubMed.
- J. F. R. Kerr, C. M. Winterford and B. V. Harmon, Apoptosis. Its significance in cancer and cancer Therapy, Cancer, 1994, 73, 2013–2026 CrossRef CAS PubMed.
- H. Hug, M. Los, W. Hirt and K.-M. Debatin, Rhodamine 110-Linked Amino Acids and Peptides as Substrates To Measure Caspase Activity upon Apoptosis Induction in Intact Cells, Biochemistry, 1999, 38, 13906–13911 CrossRef CAS PubMed.
- H. H. Zhang, B. Zhang, C. Di, M. C. Ali, J. Chen, Z. Li, J. Si, H. H. Zhang and H. Qiu, Label-free fluorescence imaging of cytochrome c in living systems and anti-cancer drug screening with nitrogen doped carbon quantum dots, Nanoscale, 2018, 10, 5342–5349 RSC.
- C. B. L. Campos, B. A. Paim, R. G. Cosso, R. F. Castilho, H. Rottenberg and A. E. Vercesi, Method for monitoring of mitochondrial cytochrome c release during cell death: Immunodetection of cytochrome c by flow cytometry after selective permeabilization of the plasma membrane, Cytometry, Part A, 2006, 69, 515–523 CrossRef PubMed.
- P. D. Allen and A. C. Newland, Electrophoretic DNA analysis for the detection of apoptosis, Mol. Biotechnol., 1998, 9, 247–251 CrossRef CAS PubMed.
- Y. RahbarSaadat, N. Saeidi, S. Zununi Vahed, A. Barzegari and J. Barar, An update to DNA ladder assay for apoptosis detection, BioImpacts, 2017, 5, 25–28 CrossRef PubMed.
- M. H. Naseri, M. Mahdavi, J. Davoodi, S. H. Tackallou, M. Goudarzvand and S. H. Neishabouri, Up regulation of Bax and down regulation of Bcl2 during 3-NC mediated apoptosis in human cancer cells, Cancer Cell Int., 2015, 15, 55 CrossRef PubMed.
- G. Koopman, C. P. Reutelingsperger, G. A. Kuijten, R. M. Keehnen, S. T. Pals and M. H. van Oers, Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis, Blood, 1994, 84, 1415–1420 CrossRef CAS PubMed.
- J. Fried, A. G. Perez and B. D. Clarkson, Flow cytofluorometric analysis of cell cycle distributions using propidium iodide. Properties of the method and mathematical analysis of the data, J. Cell Biol., 1976, 71, 172–181 CrossRef CAS PubMed.
- A. M. Rieger, K. L. Nelson, J. D. Konowalchuk and D. R. Barreda, Modified Annexin V/Propidium Iodide Apoptosis Assay For Accurate Assessment of Cell Death, J. Vis. Exp., 2011 DOI:10.3791/2597.
- D. Wlodkowic, J. Skommer and Z. Darzynkiewicz, Flow cytometry-based apoptosis detection, Methods Mol. Biol., 2009, 559, 19–32 CrossRef CAS PubMed.
- S. Pietkiewicz, J. H. Schmidt and I. N. Lavrik, Quantification of apoptosis and necroptosis at the single cell level by a combination of Imaging Flow Cytometry with classical Annexin V/propidium iodide staining, J. Immunol. Methods, 2015, 423, 99–103 CrossRef CAS PubMed.
- T. Akiyama, T. Miyazaki, P. Bouillet, K. Nakamura, A. Strasser and S. Tanaka, In vitro and in vivo assays for osteoclast apoptosis, Biol. Proced. Online, 2005, 7, 48–59 CrossRef CAS PubMed.
- S. Fayzullina and L. J. Martin, Detection and Analysis of DNA Damage in Mouse Skeletal Muscle In Situ Using the TUNEL Method, J. Visualized Exp., 2014 DOI:10.3791/52211.
- J. T. Li, K. Bian, A. L. Zhang, D. H. Kim, W. W. Ashley, R. Nath, I. McCutcheon, B. Fang and F. Murad, Targeting different types of human meningioma and glioma cells using a novel adenoviral vector expressing GFP-TRAIL fusion protein from hTERT promoter, Cancer Cell Int., 2011, 11, 35 CrossRef CAS PubMed.
- K. Kyrylkova, S. Kyryachenko, M. Leid and C. Kioussi, Detection of apoptosis by TUNEL assay, Methods Mol. Biol., 2012, 41–47 CrossRef CAS PubMed.
- W. Gorczyca, S. Bruno, R. Darzynkiewicz, J. Gong and Z. Darzynkiewicz, DNA Strand Breaks Occurring During Apoptosis - Their Early Insitu Detection by the Terminal Deoxynucleotidyl Transferase and Nick Translation Assays and Prevention by Serine Protease Inhibitors, Int. J. Oncol., 1992 DOI:10.3892/ijo.1.6.639.
- B. G. Kraupp, B. Ruttkay-Nedecky, H. Koudelka, K. Bukowska, W. Bursch and R. Schulte-Hermann, In situ detection of fragmented DNA (tunel assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: A cautionary note, Hepatology, 1995, 21, 1465–1468 CrossRef PubMed.
- H. K. Choi, D. Yessayan, H. J. Choi, E. Schellenberger, A. Bogdanov, L. Josephson, R. Weissleder and V. Ntziachristos, Quantitative Analysis of Chemotherapeutic Effects in Tumors Using In Vivo Staining and Correlative Histology, Anal. Cell. Pathol., 2005, 27, 183–190 CrossRef CAS PubMed.
- S. K. Ray, K. E. Schaecher, D. C. Shields, E. L. Hogan and N. L. Banik, Combined TUNEL and double immunofluorescent labeling for detection of apoptotic mononuclear phagocytes in autoimmune demyelinating disease, Brain Res. Protoc., 2000, 5, 305–311 CrossRef CAS PubMed.
- S. Shalini, L. Dorstyn, S. Dawar and S. Kumar, Old, new and emerging functions of caspases, Cell Death Differ., 2015, 22, 526–539 CrossRef CAS PubMed.
- L. C. Crowley and N. J. Waterhouse, Detecting Cleaved Caspase-3 in Apoptotic Cells by Flow Cytometry, Cold Spring Harb. Protoc., 2016 DOI:10.1101/pdb.prot087312.
- A. U. Lüthi and S. J. Martin, The CASBAH: a searchable database of caspase substrates, Cell Death Differ., 2007, 14, 641–650 CrossRef PubMed.
- O. Julien and J. A. Wells, Caspases and their substrates, Cell Death Differ., 2017, 24, 1380–1389 CrossRef CAS PubMed.
- J. Zhang, X. Wang, W. Cui, W. Wang, H. Zhang, L. Liu, Z. Zhang, Z. Li, G. Ying, N. Zhang and B. Li, Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage, Nat. Commun., 2013, 4, 2157 CrossRef PubMed.
- Z. Zhao, B. He and B. Z. Tang, Aggregation-induced emission of siloles, Chem. Sci., 2015, 6, 5347–5365 RSC.
- J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, B. Z. Tang, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole, Chem. Commun., 2001, 1740–1741 RSC.
- H. Shi, R. T. K. Kwok, J. Liu, B. Xing, B. Z. Tang and B. Liu, Real-Time Monitoring of Cell Apoptosis and Drug Screening Using Fluorescent Light-Up Probe with Aggregation-Induced Emission Characteristics, J. Am. Chem. Soc., 2012, 134, 17972–17981 CrossRef CAS PubMed.
- H. Jia, Y. Song, B. Huang, W. Ge and K. Q. Luo, Engineered Sensor Zebrafish for Fast Detection and Real-Time Tracking of Apoptosis at Single-Cell Resolution in Live Animals, ACS Sens., 2020, 5, 823–830 CrossRef CAS PubMed.
- J. Hickson, S. Ackler, D. Klaubert, J. Bouska, P. Ellis, K. Foster, A. Oleksijew, L. Rodriguez, S. Schlessinger, B. Wang and D. Frost, Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin, Cell Death Differ., 2010, 17, 1003–1010 CrossRef CAS PubMed.
- N. S. Chandel, Mitochondria as signaling organelles, BMC Biol., 2014, 12, 34 CrossRef PubMed.
- G. Kroemer, L. Galluzzi and C. Brenner, Mitochondrial Membrane Permeabilization in Cell Death, Physiol. Rev., 2007, 87, 99–163 CrossRef CAS PubMed.
- H. L. Tang, H. M. Tang, K. H. Mak, S. Hu, S. S. Wang, K. M. Wong, C. S. T. Wong, H. Y. Wu, H. T. Law, K. Liu, C. C. Talbot, W. K. Lau, D. J. Montell and M. C. Fung, Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response, Mol. Biol. Cell, 2012, 23, 2240–2252 CrossRef CAS PubMed.
- C. Garrido, L. Galluzzi, M. Brunet, P. E. Puig, C. Didelot and G. Kroemer, Mechanisms of cytochrome c release from mitochondria, Cell Death Differ., 2006, 13, 1423–1433 CrossRef CAS PubMed.
- J. Zhu, H. Chu, J. Shen, C. Wang and Y. Wei, Nitrogen and fluorine co-doped green fluorescence carbon dots as a label-free probe for determination of cytochrome c in serum and temperature sensing, J. Colloid Interface Sci., 2021, 586, 683–691 CrossRef CAS PubMed.
- K. Balasubramanian, B. Mirnikjoo and A. J. Schroit, Regulated Externalization of Phosphatidylserine at the Cell Surface, J. Biol. Chem., 2007, 282, 18357–18364 CrossRef CAS PubMed.
- D. L. Bratton, V. A. Fadok, D. A. Richter, J. M. Kailey, L. A. Guthrie and P. M. Henson, Appearance of Phosphatidylserine on Apoptotic Cells Requires Calcium-mediated Nonspecific Flip-Flop and Is Enhanced by Loss of the Aminophospholipid Translocase, J. Biol. Chem., 1997, 272, 26159–26165 CrossRef CAS PubMed.
- R. Wang and D. Fang, Detection of phosphatidylserine in the plasma membrane of single apoptotic cells using electrochemiluminescence, RSC Adv., 2017, 7, 12969–12972 RSC.
- J. F. Tait, C. Smith, Z. Levashova, B. Patel, F. G. Blankenberg and J.-L. Vanderheyden, Improved detection of cell death in vivo with annexin V radiolabeled by site-specific methods, J. Nucl. Med., 2006, 47, 1546–1553 CAS.
- K. Ogawa and M. Aoki, Radiolabeled Apoptosis Imaging Agents for Early Detection of Response to Therapy, Sci. World J., 2014, 2014, 1–11 CrossRef PubMed.
- I. Shlomovitz, M. Speir and M. Gerlic, Flipping the dogma – phosphatidylserine in non-apoptotic cell death, Cell Commun. Signaling, 2019, 17, 139 CrossRef PubMed.
- S. Riedl, B. Rinner, M. Asslaber, H. Schaider, S. Walzer, A. Novak, K. Lohner and D. Zweytick, In search of a novel target — Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy, Biochim. Biophys. Acta, Biomembr., 2011, 1808, 2638–2645 CrossRef CAS PubMed.
- S. Nagata, J. Suzuki, K. Segawa and T. Fujii, Exposure of phosphatidylserine on the cell surface, Cell Death Differ., 2016, 23, 952–961 CrossRef CAS PubMed.
- M. A. Savitskaya, I. I. Zakharov and G. E. Onishchenko, Apoptotic Features in Non-Apoptotic Processes, Biochem, 2022, 87, 191–206 CAS.
- S. F. G. Alain Pulfer, D. U. Pizzagalli, P. A. Gagliardi, L. Hinderling, P. Lopez, R. Zayats, P. Carrillo-Barberà, P. Antonello, M. Palomino-Segura, A. Giusti, M. Thelen, L. M. Gambardella, T. T. Murooka and A. Vie, Transformer-based spatial-temporal detection of apoptotic cell death in live-cell imaging, bioRxiv, 2023, preprint, DOI:10.1101/2022.11.23.517318.
- J. Verduijn, L. Van der Meeren, D. V. Krysko and A. G. Skirtach, Deep learning with digital holographic microscopy discriminates apoptosis and necroptosis, Cell Death Discovery, 2021, 7, 229 CrossRef CAS PubMed.
- J. Jin, K. Schorpp, D. Samaga, K. Unger, K. Hadian and B. R. Stockwell, Machine Learning Classifies Ferroptosis and Apoptosis Cell Death Modalities with TfR1 Immunostaining, ACS Chem. Biol., 2022, 17, 654–660 CrossRef CAS PubMed.
- L. Nonnenmacher, S. Hasslacher, J. Zimmermann, G. Karpel-Massler, K. La Ferla-Brühl, S. E. Barry, T. Burster, M. D. Siegelin, O. Brühl, M.-E. Halatsch, K.-M. Debatin and M.-A. Westhoff, Cell Death Induction in Cancer Therapy - Past, Present, and Future, Crit. Rev. Oncog., 2016, 21, 253–267 CrossRef PubMed.
- J. C. Reed, Apoptosis-targeted therapies for cancer, Cancer Cell, 2003, 3, 17–22 CrossRef CAS PubMed.
- W. P. Bozza, J. D. Twomey, S.-R. Kim and B. Zhang, Detection of Apoptosis: From Bench Side to Clinical Practice, Apoptosis Methods in Toxicology, Methods in Pharmacology and Toxicology, ed. Muganda PM, Springer Science+Business Media, New York, 2016, Chapter 2, pp. 13–29, DOI:10.1007/978-1-4939-3588-8_2.
- F. Zhang, L. Zhu, G. Liu, N. Hida, G. Lu, H. S. Eden, G. Niu and X. Chen, Multimodality Imaging of Tumor Response to Doxil, Theranostics, 2011, 1, 302–309 CrossRef CAS PubMed.
- H. Wang, S. Galbán, R. Wu, B. M. Bowman, A. Witte, K. Vetter, C. J. Galbán, B. D. Ross, K. R. Cho and A. Rehemtulla, Molecular Imaging Reveals a Role for AKT in Resistance to Cisplatin for Ovarian Endometrioid Adenocarcinoma, Clin. Cancer Res., 2013, 19, 158–169 CrossRef CAS PubMed.
- B. Banihashemi, R. Vlad, B. Debeljevic, A. Giles, M. C. Kolios and G. J. Czarnota, Ultrasound Imaging of Apoptosis in Tumor Response: Novel Preclinical Monitoring of Photodynamic Therapy Effects, Cancer Res., 2008, 68, 8590–8596 CrossRef CAS PubMed.
- G. J. Czarnota, M. C. Kolios, J. Abraham, M. Portnoy, F. P. Ottensmeyer, J. W. Hunt and M. D. Sherar, Ultrasound imaging of apoptosis: high-resolution non-invasive monitoring of programmed cell death in vitro, in situ and in vivo, Br. J. Cancer, 1999, 81, 520–527 CrossRef CAS PubMed.
- M. Kumar, N. R. Jagannathan, V. Seenu, S. N. Dwivedi, P. K. Julka and G. K. Rath, Monitoring the therapeutic response of locally advanced breast cancer patients: Sequential in vivo proton MR spectroscopy study, J. Magn. Reson. Imaging, 2006, 24, 325–332 CrossRef PubMed.
- N. R. Jagannathan, M. Singh, V. Govindaraju, P. Raghunathan, O. Coshic, P. K. Julka and G. K. Rath, Volume localized in vivo proton MR spectroscopy of breast carcinoma: variation of water-fat ratio in patients receiving chemotherapy, NMR Biomed., 1998, 11, 414–422 CrossRef CAS PubMed.
- M. R. Hight, Y.-Y. Cheung, M. L. Nickels, E. S. Dawson, P. Zhao, S. Saleh, J. R. Buck, D. Tang, M. K. Washington, R. J. Coffey and H. C. Manning, A Peptide-Based Positron Emission Tomography Probe for In Vivo Detection of Caspase Activity in Apoptotic Cells, Clin. Cancer Res., 2014, 20, 2126–2135 CrossRef CAS PubMed.
- C.-F. Xia, G. Chen, U. Gangadharmath, L. F. Gomez, Q. Liang, F. Mu, V. P. Mocharla, H. Su, A. K. Szardenings, J. C. Walsh, T. Zhao and H. C. Kolb, In Vitro and In Vivo Evaluation of the Caspase-3 Substrate-Based Radiotracer [18F]-CP18 for PET Imaging of Apoptosis in Tumors, Mol. Imaging Biol., 2013, 15, 748–757 CrossRef PubMed.
- F. Stuker, J. Ripoll and M. Rudin, Fluorescence molecular tomography: principles and potential for pharmaceutical research, Pharmaceutics, 2011, 3, 229–274 CrossRef CAS PubMed.
- M. Cazzaniga, A. DeCensi, G. Pruneri, M. Puntoni, L. Bottiglieri, C. Varricchio, A. Guerrieri-Gonzaga, O. D. Gentilini, G. Pagani, P. Dell'Orto, M. Lazzeroni, D. Serrano, G. Viale and B. Bonanni, The effect of metformin on apoptosis in a breast cancer presurgical trial, Br. J. Cancer, 2013, 109, 2792–2797 CrossRef CAS PubMed.
- T. Etheridge, J. Liou, T. M. Downs, E. J. Abel, K. A. Richards and D. F. Jarrard, The impact of celecoxib on outcomes in advanced prostate cancer patients undergoing androgen deprivation therapy, Am. J. Clin. Exp. Urol., 2018, 6, 123–132 Search PubMed.
- C. Betterle and R. Zanchetta, The immunofluorescence techniques in the diagnosis of endocrine autoimmune diseases, Autoimmun. Highlights, 2012, 3, 67–78 CrossRef CAS PubMed.
- K. Bivén, H. Erdal, M. Hägg, T. Ueno, R. Zhou, M. Lynch, B. Rowley, J. Wood, C. Zhang, M. Toi, M. C. Shoshan and S. Linder, A novel assay for discovery and characterization of pro-apoptotic drugs and for monitoring apoptosis in patient sera, Apoptosis, 2003, 8, 263–268 CrossRef PubMed.
- S. Holdenrieder, P. Stieber, J. Von Pawel, H. Raith, D. Nagel, K. Feldmann and D. Seidel, Early and Specific Prediction of the Therapeutic Efficacy in Non-Small Cell Lung Cancer Patients by Nucleosomal DNA and Cytokeratin-19 Fragments, Ann. N. Y. Acad. Sci., 2006, 1075, 244–257 CrossRef CAS PubMed.
- G. Kramer, H. Erdal, H. J. M. M. Mertens, M. Nap, J. Mauermann, G. Steiner, M. Marberger, K. Bivén, M. C. Shoshan and S. Linder, Differentiation between cell death modes using measurements of different soluble forms of extracellular cytokeratin 18, Cancer Res., 2004, 64, 1751–1756 CrossRef CAS PubMed.
- A. Corlu, R. Choe, T. Durduran, M. A. Rosen, M. Schweiger, S. R. Arridge, M. D. Schnall and A. G. Yodh, Three-dimensional in vivo fluorescence diffuse optical tomography of breast cancer in humans, Opt. Express, 2007, 15, 6696 CrossRef PubMed.
- R. J. Antcliff, M. R. Stanford, D. S. Chauhan, E. M. Graham, D. J. Spalton, J. S. Shilling, T. J. Ffytche and J. Marshall, Comparison between optical coherence tomography and fundus fluorescein angiography for the detection of cystoid macular edema in patients with uveitis, Ophthalmology, 2000, 107, 593–599 CrossRef CAS PubMed.
- J. M. Critchfield, M. K. Racke, J. C. Zúñiga-Pflücker, B. Cannella, C. S. Raine, J. Goverman and M. J. Lenardo, T Cell Deletion in High Antigen Dose Therapy of Autoimmune Encephalomyelitis, Science, 1994, 263, 1139–1143 CrossRef CAS PubMed.
- T. Head, P. Dau, S. Duffort, P. Daftarian, P. M. Joshi, R. Vazquez-Padron, S. K. Deo and S. Daunert, An enhanced bioluminescence-based Annexin V probe for apoptosis detection in vitro and in vivo, Cell Death Dis., 2017, 8, e2826 CrossRef CAS PubMed.
- Y. Wang, B. Zhang, W. Liu, Y. Dai, Y. Shi, Q. Zeng and F. Wang, Noninvasive bioluminescence imaging of the dynamics of sanguinarine induced apoptosis via activation of reactive oxygen species, Oncotarget, 2016, 7, 22355–22367 CrossRef PubMed.
- Y. Zhang, J. Hu, M. Yu, Z. Wang, H. Qing, H. Fu, L. Yuan, F. Li and S. Zhao, A novel BRET based genetic coded biosensor for apoptosis detection at deep tissue level in live animal, Apoptosis, 2021, 26, 628–638 CrossRef CAS PubMed.
- S. Tsuboi and T. Jin, Bioluminescence Resonance Energy Transfer (BRET)-coupled Annexin V-functionalized Quantum Dots for Near-Infrared Optical Detection of Apoptotic Cells, ChemBioChem, 2017, 18, 2231–2235 CrossRef CAS PubMed.
- S. B. Nicholls, J. Chu, G. Abbruzzese, K. D. Tremblay and J. A. Hardy, Mechanism of a Genetically Encoded Dark-to-Bright Reporter for Caspase Activity, J. Biol. Chem., 2011, 286, 24977–24986 CrossRef CAS PubMed.
- X. Hu, H. Li, X. Huang, Z. Zhu, H. Zhu, Y. Gao, Z. Zhu and H. Chen, Cell membrane-coated gold nanoparticles for apoptosis imaging in living cells based on fluorescent determination, Microchim. Acta, 2020, 187, 175 CrossRef CAS PubMed.
- P. Wu, S. B. Nicholls and J. A. Hardy, A Tunable, Modular Approach to Fluorescent Protease-Activated Reporters, Biophys. J., 2013, 104, 1605–1614 CrossRef CAS PubMed.
- K. Martínez-Lagunas, Y. Yamaguchi, C. Becker, C. Geisen, M. C. DeRuiter, M. Miura, B. K. Fleischmann and M. Hesse, In vivo detection of programmed cell death during mouse heart development, Cell Death Differ., 2020, 27, 1398–1414 CrossRef PubMed.
- S. Tsuboi and T. Jin, In Vitro and In Vivo Fluorescence Imaging of Antibody–Drug Conjugate-Induced Tumor Apoptosis Using Annexin V–EGFP Conjugated Quantum Dots, ACS Omega, 2022, 7, 2105–2113 CrossRef CAS PubMed.
- H. Kim, H. Y. Kim, E. Y. Lee, B. K. Choi, H. Jang and Y. Choi, A Quenched Annexin V-Fluorophore for the Real-Time Fluorescence Imaging of Apoptotic Processes In Vitro and In Vivo, Adv. Sci., 2020 DOI:10.1002/advs.202002988.
- S. Le Gac, I. Vermes and A. van den Berg, Quantum Dots Based Probes Conjugated to Annexin V for Photostable Apoptosis Detection and Imaging, Nano Lett., 2006, 6, 1863–1869 CrossRef CAS PubMed.
- Y. Wang, X. Hu, J. Weng, J. Li, Q. Fan, Y. Zhang and D. Ye, A Photoacoustic Probe for the Imaging of Tumor Apoptosis by Caspase-Mediated Macrocyclization and Self-Assembly, Angew. Chem., Int. Ed., 2019, 58, 4886–4890 CrossRef CAS PubMed.
- H. Bai, Y. Wang, Y. Hu and D. Ye, A caspase-3-activatable bimodal probe for photoacoustic and magnetic resonance imaging of tumor apoptosis in vivo, Biosens. Bioelectron., 2022, 216, 114648 CrossRef CAS PubMed.
- A. Jhunjhunwala, J. Kim, K. P. Kubelick, C. R. Ethier and S. Y. Emelianov, Vivo Photoacoustic Monitoring of Stem Cell Location and Apoptosis with Caspase-3-Responsive Nanosensors, ACS Nano, 2023, 17, 17931–17945 CrossRef CAS PubMed.
- A. Nishie, O. Togao, C. Tamura, M. Yamato, K. Ichikawa, S. Nohara, Y. Ito, N. Kato, S. Yoshise and H. Honda, In Vitro and In Vivo Detection of Drug-induced Apoptosis Using Annexin V-conjugated Ultrasmall Superparamagnetic Iron Oxide (USPIO): A Pilot Study, Magn. Reson. Med. Sci., 2019, 18, 142–149 CrossRef CAS PubMed.
- Q.-D. Nguyen, G. Smith, M. Glaser, M. Perumal, E. Årstad and E. O. Aboagye, Positron emission tomography imaging of drug-induced tumor apoptosis with a caspase-3/7 specific [ 18 F]-labeled isatin sulfonamide, Proc. Natl. Acad. Sci., 2009, 106, 16375–16380 CrossRef CAS PubMed.
- F. Bulat, F. Hesse, D.-E. Hu, S. Ros, C. Willminton-Holmes, B. Xie, B. Attili, D. Soloviev, F. Aigbirhio, F. J. Leeper, K. M. Brindle and A. A. Neves, 18F-C2Am: a targeted imaging agent for detecting tumor cell death in vivo using positron emission tomography, EJNMMI Res., 2020, 10, 151 CrossRef CAS PubMed.
- Y. Kuge, S. Zhao, T. Takei and N. Tamaki, Molecular Imaging of Apoptosis with Radio-Labeled Annexin A5 Focused on the Evaluation of Tumor Response to Chemotherapy, Anticancer. Agents Med. Chem., 2009, 9, 1003–1011 CrossRef CAS PubMed.
- M. M. Pasternak, A. Sadeghi-Naini, S. M. Ranieri, A. Giles, M. L. Oelze, M. C. Kolios and G. J. Czarnota, High-frequency ultrasound detection of cell death: Spectral differentiation of different forms of cell death in vitro, Oncoscience, 2016, 3, 275–287 CrossRef PubMed.
- R. M. Vlad, N. M. Alajez, A. Giles, M. C. Kolios and G. J. Czarnota, Quantitative Ultrasound Characterization of Cancer Radiotherapy Effects In Vitro, Int. J. Radiat. Oncol., 2008, 72, 1236–1243 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |