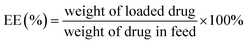Open Access Article
Open Access ArticlePreparation of reversible cross-linked amphiphilic polymeric micelles with pH-responsive behavior for smart drug delivery
Liu Tan ab,
Jinling Fana,
Yuqing Zhoua,
Di Xiongc,
Manzhen Duana,
Ding Hu*ab and
Zhimin Wu*ab
ab,
Jinling Fana,
Yuqing Zhoua,
Di Xiongc,
Manzhen Duana,
Ding Hu*ab and
Zhimin Wu*ab
aSchool of Chemical Engineering, Xiangtan University, Xiangtan 411105, China. E-mail: huding@hnu.edu.cn; xdwuzm@xtu.edu.cn
bNational & Local United Engineering Research Centre for Chemical Process Simulation and Integration, Xiangtan University, Xiangtan 411105, China
cSchool of Mechanical & Automotive Engineering, South China University of Technology, Guangzhou 510640, China
First published on 25th September 2023
Abstract
A new type of reversible cross-linked and pH-responsive polymeric micelle (PM), poly[polyethylene glycol methacrylate-co-2-(acetoacetoxy)ethyl methacrylate]-b-poly [2-(dimethylamino)ethyl methacrylate] [P(PEGMA-co-AEMA)-b-PDMAEMA], was synthesized for targeted delivery of curcumin. After reversible cross-linking of the micellar shell, the PMs with a typical core–shell structure exhibited excellent stability against extensive dilution and good reversibility of pH-responsiveness in solutions with different pH values. P(PEGMA9-co-AEMA6)-b-PDMAEMA10 has the lowest critical micelle concentration (CMC) value (0.0041 mg mL−1), the highest loading capacity (13.86%) and entrapment efficiency (97.03%). A slow sustained drug release at pH 7.4 with 12.36% in 108 h, while a fast release (42.36%) was observed at pH 5.0. Furthermore, a dissipative particle dynamics (DPD) simulation method was employed to investigate the self-assembly process and pH-responsive behavior of PMs. The optimal drug-carrier ratio (2%) and fraction of water (92%) were confirmed by analyzing the drug distribution and morphology of micelles during the self-assembly process of the block copolymer. The simulation results were consistent with experimental results, indicating DPD simulation shows potential to study the structure properties of reversible cross-linked micelles. The present findings provide a new method for the development of SDDS with good structural stability and controlled drug release properties.
Introduction
Smart drug delivery systems (SDDS) have attracted much attention in nanomedicine anticancer therapeutics.1–6 Polymeric micelles (PMs) formed by the self-assembly of amphiphilic block copolymers in selective solvents have been recognized as one of the most promising modalities in drug delivery systems for their unique properties, including highly loading capacity, long circulation time in blood, and good therapeutic efficacy.7–11Despite these advantages and the significant progress already made in achieving efficient drug delivery, PMs employed as SDDS are limited owing to poor water solubility and slow drug release at targeted tumor sites.12–14 The PMs are extensively diluted during circulation in the bloodstream because of dynamic shuttling of polymer chains between the micelles and bulk phase. In addition, statistics revealed that administered nanoparticle dose that is delivered to solid tumors is only 0.7% (median).15,16
To address these challenges, various stimuli-responsive PMs have been developed. Temperature,17–20 pH,21–27 light,28–30 electric,31,32 enzyme,33–35 etc. are the driving forces for controlling drug release. Among them, the pH-stimulus PMs have been investigated and used as a main trigger for inducing drug release. It is well known that micro-environments of tumor cells show a relatively lower pH than normal tissues because of their high preference of glycolysis only at aerobic conditions, which leads to lactic acid accumulation.36 Therefore, differences in pH value can be used to design PMs to realize the controllable release of drug. On the other hand, to reinforce the stability of micelles during circulation in the blood, conventional chemical cross-linking is an effective way by forming a barrier between shell and core to reduce the micelle dissociation and prevent drug leak.37–40 However, the covalent linkage of conventional chemical cross-linking is irreversible, which have a negative effect on the intracellular drug release and reduce the therapeutic effect.41
Many studies have proved that reversibly cross-linked micelles (RCLMs), which are cross-linked and decross-linked via external stimuli, is one of an ideal delivery system.42,43 RCLMs display good stability and restrained the drug release during blood circulation and in normal physiological environment (pH 7.4). Once it arrives acidic conditions at the tumor site (pH 5.0), de-crosslinking of the micelles occurred. The hydrophobic blocks become into hydrophilic ones, which was in favor of drug release. Driven by the goal of an ideal delivery system, we have previously synthesized a series of RCLMs, including poly(methyl methacrylate-co-methacrylic acid)-b-poly(aminoethyl methacrylate) and poly(ε-caprolactone)-b-poly(polyethylene glycol methacrylate-co-p-(2-methacryloxyethoxy)benzaldehyde).44–46 While the RCLMs showed an excellent stability in blood circulation, it is still highly desirable to develop a novel polymeric micelle with enhanced loading capacity and controlled drug release properties.
In this context, a novel reversible cross-linked and pH responsive polymeric micelle was prepared from a well-defined block copolymer which was synthesized via reversible addition–fragmentation chain transfer (RAFT) polymerization of polyethylene glycol methacrylate (PEGMA), 2-(acetoacetoxy)ethyl methacrylate (AEMA) and 2-(dimethylamino)ethyl methacrylate (DMAEMA). As illustrated in Scheme 1, PEGMA can introduce hydrophilic group into the PMs and keep the stability of the micelle structure. The tertiary amino groups in PDMAEMA blocks can protonize as a pH-responsive linkage. The β-keto ester containing in AEMA blocks provided the reversible cross-linked site. Hexamethylenediamine (EDA) was used as the cross-linker. Curcumin (CUR), a polyphenolic compound derived from Curcuma longa L., was selected to evaluate drug loading capacity (LC) and entrapment efficiency (EE). Dissipative particle dynamics (DPD) simulation was employed to study the self-assembly process and pH-responsive behavior of PMs. Reversible cross-linked amphiphilic PMs meet the required design principles for drug carriers and address the challenges in drug delivery systems to achieve highly tunable PMs with high stability and controlled release.
 | ||
| Scheme 1 Structural design and application of pH-responsive hydrophobic monomer for amphiphilic polyether micelles with high stability and pH controlled release. | ||
Experimental and methods
Materials
2-(dimethylamino)ethyl methacrylate (DMAEMA, 99%, Aldrich, Shanghai, China) and 2-(acetoacetoxy)ethyl methacrylate (AEMA, 99%, Aldrich) were passed through alkaline alumina before use to removal the inhibitor. Polyethylene glycol methacrylate (PEGMA, Mn = 500 Da, 99%, Aldrich) was passed through neutral alumina before use to removal the inhibitor. Azo diisobutyronitrile (AIBN, AR, Alfa Aesar, Tianjin, China) was refined with ethanol before use. Curcumin (CUR, 98%, Alfa Aesar), pyrene (99%, Aldrich), para-toluenesulfonic acid (PTsOH, 99%, Aldrich) and ethylenediamine (EDA, GC, Aldrich) were used as received without further treatment. 2-(Dodecyl trithiocarbonate)-2-methylpropionic acid (DDMAT) was prepared according to the literature.47 Other reagents were commercially available chemicals and used as received.Characterizations
1H NMR (400 MHz) spectra of block copolymers were recorded on a Bruker AVANCE III 400 (Zurich, Switzerland) spectrometer at room temperature (ca. 25 °C). Gel permeation chromatography (GPC) measurements were performed on an Agilent 1200 series GPC system (Agilent Technologies Inc., Santa Clara, CA, USA). THF was selected as the mobile phase with a flow rate of 1.0 mL min−1 at 30 °C. The number average, the weight average and polymer dispersity index (PDI, Mw/Mn) were confirmed by GPC. The morphology of PMs were studied by transmission electron microscopy (TEM, Hitachi H-7650, Tokyo, Japan) operating at 80 kV.Synthesis of the polymer PDMAEMA
Typically, DDMAT (183 mg, 0.5 mmol), AIBN (5.2 mg, 0.05 mmol) and DMAEMA (5.0 mL, 30 mmol) were dissolved in 10 mL anhydrous toluene at room temperature (ca. 25 °C). The flask was sealed after triple freeze–pump–thaw cycles to remove oxygen from the solution. Then, the reaction started at 70 °C for 24 h. After the reaction, the mixture was maintained in an ice bath for another 0.5 h. The crude polymer was dissolved in 25 mL THF followed by adding dropwise to tenfold excess of petroleum ether to precipitate the product. The hydrophobic block PDMAEMA was obtained as a white solid after dried under vacuum for 24 h.Synthesis of the copolymers P(PEGMA-co-AEMA)-b-PDMAEMA
Typically, PDMAEMA (1.17 g, 0.5 mmol), AIBN (2.04 mg, 0.05 mmol), PEGMA (1.91 mL, 5 mmol) and AEMA (0.42 mL, 10 mmol) were dissolved in 10 mL anhydrous toluene at room temperature (ca. 25 °C), and then the solution was degassed under reduced pressure by triple freeze–pump–thaw cycles. The polymerization was carried out at 70 °C for 24 h. The reaction was stopped by rapid cooling with ice bath. The crude polymer was dissolved in 25 mL THF followed by adding dropwise to tenfold excess of petroleum ether to precipitate the product. The amphiphilic block copolymer P(PEGMA-co-AEMA)-b-PDMAEMA was obtained as a faint yellow solid after dried under vacuum for 24 h.Critical micelle concentration (CMC) measurement
The CMC values of the amphiphilic block copolymer were confirmed by the fluorescence probe technique using pyrene as a probe. Pyrene dissolved in acetone was added into deionized water (pH 7.4) to make a concentration of 12 × 10−3M following by removed acetone 2 h through evaporation. The final concentration of pyrene was adjusted to 6 × 10−5 M. A series of polymer solution with different concentrations between 0.0001 mg mL−1 and 0.1 mg mL−1 were prepared. 0.1 mL diluted pyrene acetone solution was added to the solution by using a syringe pump. Then, the above solutions were equilibrated at room temperature overnight. The CMC value was obtained based on the fluorescence excitation spectra of the mixed solution. The emission wavelength was set at 373 nm, and the excitation spectrum was scanned from 300 to 350 nm.48 The ratio between the fluorescence intensities at the wavelengths of 336 and 334 nm (I336/I334) was plotted against the polymer concentration, and the CMC was determined from the inflection point.Preparation of blank and CUR-loaded micelles
The blank and drug-loaded micelles were self-assembled by dialysis method.46,49 Typically, 30 mg P(PEGMA-co-AEMA)-b-PDMAEMA was dissolved in 30 mL dimethyl sulfoxide (DMSO). The solution was transferred into a dialysis bag (MWCO = 3500 Da) and dialyzed against deionized water for 24 h. The deionized water for dialysis was changed every hour for the first 12 h and then replaced every 2 h for the next 12 h. The blank micelles were obtained by freeze-drying. CUR-loaded micelles were prepared by the same method as described in the blank micelle preparation. CUR-loaded micelles (1 mg) were dissolved in 10 mL DMSO under vigorous vortexing, and the absorbance at 433 nm was studied by using a UV-vis spectrophotometer (UV-2450, Shimadzu, Japan). The CUR loading content was calculated according to the standard curve of CUR/DMSO solution with different CUR concentrations. Drug loading capacity (LC) and entrapment efficiency (EE) are average values of three tests. LC and EE were obtained according to the following equations:Preparation of crosslinked micelles
Typically, 50 mg P(PEGMA-co-AEMA)-b-PDMAEMA and PTsOH (5 mol%) were dissolved in 30 mL dimethyl sulfoxide (DMSO). EDA (1.21 mL) was added in the solution and stirred at room temperature for 6 h. Then, the solution was transferred into dialysis bag (MWCO 3500 Da) and dialyzed against deionized water for 24 h to remove unreacted crosslinker. The crosslinked micelles were collected by freeze-drying to obtain dried product. CUR loaded cross-linked micelles were prepared by the same method as described in the blank cross-linked micelles.Stability study of micelles
The stability of mixed micelles was studied by Dynamic light scattering (DLS, ZS90, Malvern Instruments, UK). The micellar solution (1 mg mL−1, 2 mL) was extensively diluted by deionized water (pH 7.4) at room temperature. The solution was balanced for 24 h, and then the particle size and distribution were recorded by DLC measurement.In vitro drug release
The drug release profiles of micelles were investigated in different pH media (pH 5.5 and pH 7.4). The PBS buffer (pH 7.4) was used to simulate normal physiological environment of human body, and in acetate buffer (pH 5.0) simulated intracellular condition in tumor cells. Briefly, drug-loaded micelles (5 mg) immersed in 5 mL PBS solution (pH 7.4) or 5 mL acetate buffer (pH 5.0). Then, the solution was transferred to dialysis bags (MWCO 3500 Da) and placed in 45 mL of PBS or acetate buffer with bath thermostatic oscillator at 37 °C. At certain time intervals, 5 mL of dialysate was collected for measurement of CUR concentration and fresh PBS or acetate buffer of equal volume was supplement after each collection.The amount of CUR released in different buffers was quantified by using UV-visible spectrophotometer. The cumulative drug release (Er) was obtained according to the following equations:
Coarse-grained model and parameter calculation
The dissipative particle dynamics (DPD) simulation is a useful tool to study dynamic behavior of fluid by establishing a coarse granulation model.44,50–52 In this study, DPD simulation method was used to investigate the self-assembly process and pH-responsive behavior of RCLMs. Coarse-grained models of block copolymer P(PEGMA-co-AEMA)-b-PDMAEMA, CUR and waters are shown in Fig. 1. The molecular structure of the polymer was divided into four types of soft interacting beads (MAA, PEG, AAE and DMA). The drug CUR was divided into two types of soft interacting beads (JHS1 and JHS2), and three water molecules were denoted as one bead. Note that DMA beads will change into DMA+ when the system environment changes to weak acid (pH < pKb) because of the protonation of tertiary amino groups. | ||
| Fig. 1 Coarse-grained models of (a) the block copolymer P(PEGMA-co-AEMA)-b-PDMAEMA, (b) CUR and (c) waters. | ||
The interaction parameters between different beads as shown in following equation. The aij was the maximum repulsion between bead i and bead j, depending on the underlying atomistic interaction, and was linearly related to the Flory–Huggins parameters (χij).44,53,54
| aij = aii + 3.27χij |
where δi and δj are solubility parameters of the two beads, respectively. R is the ideal gas constant, T is the thermodynamic temperature, and Vbead is the total volume of the beads.
In this study, the box size in our simulation was set as 250 Å × 250 Å × 250 Å, the integration time step was set as 0.05 ns, and the total number of simulation step was 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps to ensure a table phase. DPD repulsion parameter aij between the different beads at 298.15 K was calculated and are given in Table 1. All the simulations were studied by using DPD program incorporated in the Materials Studio 8.0 software (Accelry Inc).
000 steps to ensure a table phase. DPD repulsion parameter aij between the different beads at 298.15 K was calculated and are given in Table 1. All the simulations were studied by using DPD program incorporated in the Materials Studio 8.0 software (Accelry Inc).
| aij | AAE | DMA | JHS1 | JHS2 | MAA | PEG | Water | DMA+ |
|---|---|---|---|---|---|---|---|---|
| AAE | 25.00 | |||||||
| DMA | 31.06 | 25.00 | ||||||
| JHS1 | 28.23 | 25.64 | 25.00 | |||||
| JHS2 | 28.01 | 25.21 | 25.40 | 25.00 | ||||
| MAA | 30.40 | 28.99 | 27.98 | 28.13 | 25.00 | |||
| PEG | 27.56 | 37.93 | 30.12 | 31.36 | 35.02 | 25.00 | ||
| Water | 103.86 | 135.08 | 114.69 | 118.85 | 69.13 | 58.26 | 25.00 | |
| DMA+ | 186.66 | 243.71 | 156.43 | 207.62 | 128.77 | 111.46 | 28.06 | 25.00 |
Results and discussion
Synthesis and characterization of copolymers
Amphiphilic block copolymers, P(PEGMA-co-AEMA)-b-PDMAEMA, were synthesized via two step RAFT method. PDMAEMA polymer was first synthesized. Fig. 2 shows the characteristic signals of 1H NMR spectrum of P(PEGMA-co-AEMA)-b-PDMAEMA, indicating the successful synthesis of the copolymer. The characteristic signals at 0.86 (g), 1.24 (a), and 1.82 (h) ppm were assigned to –CH3, –CH2-and –CCH2– on the copolymer backbone, respectively. The characteristic peak at 3.84 ppm (f), 2.56 ppm (d) and 4.07 ppm (e) were assigned to the protons of the –CH2–, –N–CH2– and –CH2–CH2–. The signal at 2.27 ppm (c) was assigned to the –N–CH3 in PDMAEMA unit and –C–CH3 in AEMA unit. The signal at 3.83 ppm (b) was assigned to O-CH3 in PEGMA unit.The degree of polymerization of DMAEMA(z), AEMA(y) and PEGMA(x) was calculated by comparing the integration ratio of the proton signal d, e, b to a, respectively. The number average molecular weight (Mn,NMR) of the copolymer P(PEGMA-co-AEMA-b-PDMAEMA) was calculated, and the results are listed in Table 2.
| Samplea | Mnb,NMR | Mnc,GPC | Mw/Mn |
|---|---|---|---|
| a The subscripts of PEGMA, AEMA and PDMAEMA were the DP of PEGMA(x), AEMA(y) and PDMAEMA (z) calculated from 1H NMR spectrum.b Measured by equation by 1H NMR.c Measured by GPC in THF. | |||
| P(PEGMA9-co-AEMA6)-b-PDMAEMA10 | 3687 | 5745 | 1.43 |
| P(PEGMA20-co-AEMA23)-b-PDMAEMA13 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 694 694 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 926 926 |
1.67 |
| P(PEGMA16-co-AEMA23)-b-PDMAEMA8 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185 185 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 591 591 |
1.52 |
As shown in Fig. 3, GPC results of the block copolymers showed monomodal distributions with narrow dispersity. In addition, Mn,GPC values were close to the Mn,NMR values, as shown in Table 2, indicating the controlled behavior of the RAFT polymerization of P(PEGMA-co-AEMA)-b-PDMAEMA.
Properties of polymeric micelles
The CMC value of self-assembled PMs is a crucial parameter to predict the stability of the micelles. To further investigate the self-assembled behaviors and to determine the CMC values with three different mass ratio of P(PEGMA-co-AEMA)-b-PDMAEMA block copolymers in aqueous solution, the micelle dispersion was investigated by fluorescence probe technique with pyrene as hydrophobic fluorescence probe. When PMs were formed, pyrene molecules preferably located inside or closed to the hydrophobic core of micelles, accompanied with large difference in photo-physical characteristics. A red shift of the major peak from 334 to 336 nm was observed in the fluorescence excitation spectra with increasing concentration.As shown in Fig. 4, the ratios of I336 to I334 were plotted against copolymer concentrations. The CMC values were 0.0041, 0.0067 and 0.0057 mg mL−1 for P(PEGMA9-co-AEMA6)-b-PDMAEMA10,P(PEGMA20-co-AEMA23)-b-PDMAEMA13, P(PEGMA16-co-AEMA23)-b-PDMAEMA8, respectively. The CMC values increased with increasing the hydrophilic block. Among the copolymer mixtures, the P(PEGMA9-co-AEMA6)-b-PDMAEMA10 had the lowest CMC value and show good stability among these three samples.
The hydrodynamic diameter distribution of the self-assembled PMs were obtained from DLS measurement, as shown in Table 3. DLS measurement revealed that the sizes of formed P(PEGMA-co-AEMA)-b-PDMAEMA micelles were in the range of 111.6–186 nm. The micelle size of P(PEGMA-co-AEMA)-b-PDMAEMA increased with increasing hydrophobic PDMAEMA block length, which mainly originated from the increase of hydrophobic property by the longer hydrophobic PDMAEMA chain in aqueous solution.
| Sample | Polymer (mg) | CUR (mg) | Dh (nm) | PDI | ξ (mV) | LC (%) | EE (%) |
|---|---|---|---|---|---|---|---|
| P(PEGMA9-co-AEMA6)-b-PDMAEMA10 | 30 | 0 | 126.4 | 0.69 | 21.7 | — | — |
| P(PEGMA20-co-AEMA23)-b-PDMAEMA13 | 30 | 0 | 111.6 | 0.46 | 24.2 | — | — |
| P(PEGMA16-co-AEMA23)-b-PDMAEMA8 | 30 | 0 | 186.1 | 0.43 | 10.8 | — | — |
| P(PEGMA9-co-AEMA6)-b-PDMAEMA10 | 30 | 5 | 143.4 | 0.25 | 14.8 | 13.86 | 97.03 |
| P(PEGMA20-co-AEMA23)-b-PDMAEMA13 | 30 | 5 | 149.3 | 0.42 | 9.0 | 13.61 | 95.29 |
| P(PEGMA16-co-AEMA23)-b-PDMAEMA8 | 30 | 5 | 255.3 | 0.29 | 2.5 | 13.50 | 94.53 |
As shown in Fig. 5, after drug loading, the CUR-loaded micelles showed a larger size than the empty micelles with Dh in the range of 143–255 nm. Hydrophobic CUR can promote hydrophobic interaction among the PEGMA chains and thus led to an increase in aggregation. The PDI is in the range of 0.25–0.42, indicating good physical performance of the assembled micelles. In addition, the P(PEGMA9-co-AEMA6)-b-PDMAEMA10 had the highest drug loading capacity (13.86%), entrapment efficiency (97.03%) and charged character (14.8 mV). The highly charged character of P(PEGMA9-co-AEMA6)-b-PDMAEMA10 can prevent the aggregation of micelles, extend blood circulation times, increase the interactions between micelles and cell membranes which can facilitate penetrating of cell membranes. Since P(PEGMA9-co-AEMA6)-b-PDMAEMA10 had the lowest CMC value and highest drug loading capacity, it was used to further prepare reversible cross-linked micelles.
 | ||
| Fig. 5 DLS particle size and distribution of (a–f) PMs with different copolymeric mass rations and PMs with different copolymeric mass rations after CUR loading. | ||
A cross-linked shell structure was built with the cross-linker hexamethylenediamine. As shown in Table 4, the hydrodynamic diameter (Dh) was 149.1 nm for the blank micelle after crosslinking, and the PDI is 0.11, indicating a good micelle size distribution. After drug loading, the particle size increased with Dh around 168.8 nm. It was because the hydrophobic CUR molecules promoted hydrophobic–hydrophobic interactions among the PEGMA blocks and thus led to an increase in aggregation. The sizes of CUR-loaded RCLMs confirmed by TEM measurement (Fig. 6). It is worthwhile to note that the average sizes shown in TEM images were almost in accordance with the DLS results. In addition, the CUR-loaded polymeric micelle after crosslinking can form a typical core–shell shape, indicating a good control of the micellar crosslinking process. The thickness of shell is about 30 nm. The PDMAEMA acted as a shell that provided micellar architecture stability. The LC and EE of CUR-loaded RCLMs were 12.7% and 90.0%. The PEGMA core provided a container for hydrophobic drug molecules, such as CUR. It was some lower than that before crosslinking, which was due to the leakage of CUR from micelles during crosslinking progress.
| Dh (nm) | PDI | ξ (mV) | LC (%) | EE (%) | |
|---|---|---|---|---|---|
| RCLMs | 149.1 | 0.11 | 14.0 | — | — |
| RCLMs (CUR) | 168.8 | 0.22 | 5.76 | 12.7 | 90.04 |
The DLS was employed to study the micellar stability against dilution. The original concentration of solution was both 1 mg mL−1. As shown in Fig. 7, the peak-shaped of non-crosslinked micelles (NCMs) changed from unimodal distribution to bimodal non-uniform distribution after 1000-fold dilution by water for 12 h. The particle size of NCMs increased to 1000 nm, indicating a large disassembly and aggregation of the micelles. On the contrary, the particle size distribution of cross-linked micelles is basically unchanged and show a good stability against dilution, proving that the micelle structure can be fixed by sufficient crosslinking. The results indicate that cross-linking is an efficient way to reinforce the stability of the micelles and prevent disintegration in the bloodstream.
 | ||
| Fig. 7 Size distribution of non-crosslinked P(PEGMA9-co-AEMA6)-b-PDMAEMA10 and reversibly cross-linked P(PEGMA9-co-AEMA6)-b-PDMAEMA10 before and after the 1000-fold water dilution. | ||
In vitro drug release profiles
In vitro drug release study was performed in PBS solution over 108 h under different simulated physiological conditions (pH 7.4, pH 5.0) to explore the effects of pH-responsive behavior on controlled drug delivery. As shown in Fig. 8, the CUR release was slow in the normal physiological condition (pH 7.4), and the cumulative drug release percentage of the NCMs reached 14.94% within 10 h, while that of the RCLMs was only 8.83%. The cumulative drug release percentages of the NCMs and RCLMs were 24.56% and 12.36% after 108 h. The core–shell structure formed by PEGMA, AEMA and PDMAEMA exhibited excellent stability against extensive dilution. In addition, the β-keto ester containing in AEMA blocks provided the reversible cross-linked site. Cross-linking further consolidates core–shell structure. The results indicated that the reversible cross-linked structure was an efficient barrier to prevent CUR leakage from micellar core. The release rates significantly accelerated as the pH decreased from 7.5 to 5.0. The cumulative drug release percentage of the NCMs and RCLMs were 29.13% and 26.88% within 10 h, and the final release amount were 44.63% and 42.36% at 108 h. The release of drug molecules from micelle was quite fast, accompanied by an obvious burst release, which was mainly for the breakage of the cross-linked structure with hydrolysis of imine. Moreover, tertiary amino groups in PDMAEMA blocks had been protonated, the distinctly decreased hydrophobicity of the micellar core contributed to degradation of micelles. | ||
| Fig. 8 In vitro drug release profiles of CUR-loaded RCLMs (P(PEGMA9-co-AEMA6)-b-PDMAEMA10) and NCMs (P(PEGMA9-co-AEMA6)-b-PDMAEMA10) at different simulated physiological condition. | ||
The results of the in vitro drug release prove that the cross-linked shell structures efficiently prevent CUR leakage from micellar core. Meanwhile, when arriving at the tumor site, the micelles responded to low pH and de-crosslinking of the micelles occurred. The hydrophobic blocks become into hydrophilic ones, which was in favor of drug release. Therefore, the pH-responsive PMs have attractive prospects for delivering drugs to specific sites.
DPD simulation
To obtain a more clear understanding on drug delivery system, optimize experimental conditions and design micelle structure, DPD simulation method was used to investigate the micellar morphology, distribution and self-assembly process in aqueous solution at different simulated steps.Self-assembly ability
The P(PEGMA9-co-AEMA6)-b-PDMAEMA10 was selected to explore the self-assembly process by DPD simulation, and the results are shown in Fig. 9. In the simulation, the volume fractions of polymer and water were 6% and 94%, respectively. As can be observed from the snapshots, many clusters were randomly distributed in water, and the polymer form a disordered system at the beginning of the self-assembly (10 steps). With the evolution of simulation (5000–30000 steps), the clusters began to form large aggregates via the hydrophobic interactions among the beads. Several large spherical aggregates were gradually formed after 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps. With the continuing increase of the simulation step, a stable and complete polymeric micelle was formed after 250
000 steps. With the continuing increase of the simulation step, a stable and complete polymeric micelle was formed after 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps. Owing to the hydrophobic characters of PDMAEMA and the hydrophilic characters of PEGMA, polymeric micelle shows a typical core–shell structure with the outer shell (green and blue beads) forming by p(PEGMA-co-AEMA) and the inner core (red beads) forming by PDMAEMA. The hydrophilic shell of the polymer micelles acts as a stabilizing and dispersing function. In general, the findings indicate that the P(PEGMA9-co-AEMA6)-b-PDMAEMA10 can self-assemble into polymeric micelle by coalescence of small clusters.
000 steps. Owing to the hydrophobic characters of PDMAEMA and the hydrophilic characters of PEGMA, polymeric micelle shows a typical core–shell structure with the outer shell (green and blue beads) forming by p(PEGMA-co-AEMA) and the inner core (red beads) forming by PDMAEMA. The hydrophilic shell of the polymer micelles acts as a stabilizing and dispersing function. In general, the findings indicate that the P(PEGMA9-co-AEMA6)-b-PDMAEMA10 can self-assemble into polymeric micelle by coalescence of small clusters.
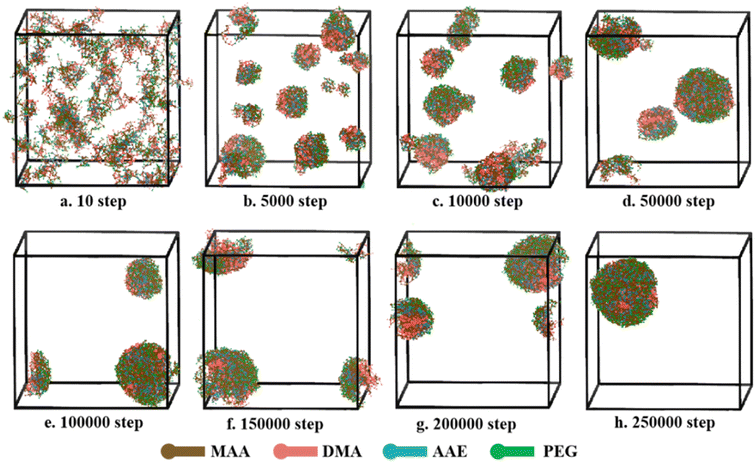 | ||
Fig. 9 Self-assembly process of P(PEGMA9-co-AEMA6)-b-PDMAEMA10 in aqueous solution at different simulated steps, (a–h) from 10 to 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, during DPD simulation. 000 steps, during DPD simulation. | ||
The self-assembly process of CUR-loaded micelle was also studies by DPD simulation. The volume fractions of polymer, drug and water were 6%, 2% and 92%, respectively. As shown in Fig. 10, the self-assembly process of CUR loaded polymeric micelle is similar to that of the blank sample. The simulation is started from homogeneous states. With the continuing increase of the simulation step (Fig. 10b), some micelle and CUR form small clusters due to the repulsion of water. At 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, most CUR molecules (yellow beads) were loaded into the micellar core, but there were still an amount of drug particles and drug-polymers clusters dispersed in aqueous medium. The hydrophobic core of the polymer micelles provides a loading space for hydrophobic drugs. Compact and stable CUR-loaded micelle were formed at 250
000 steps, most CUR molecules (yellow beads) were loaded into the micellar core, but there were still an amount of drug particles and drug-polymers clusters dispersed in aqueous medium. The hydrophobic core of the polymer micelles provides a loading space for hydrophobic drugs. Compact and stable CUR-loaded micelle were formed at 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps. PDMAEMA and CUR are both hydrophobic, which causes the CUR and PDMAEMA to form clusters, and clusters will be surrounded by hydrophilic groups. All CUR molecules are dispersed into micelle. Like the NCMs, the morphology of CUR-loaded NCMs show a good spherical structure. The results indicated that the CUR can co-self-assemble with copolymer molecules and have little influence on the self-assembly ability of polymeric micelle.
000 steps. PDMAEMA and CUR are both hydrophobic, which causes the CUR and PDMAEMA to form clusters, and clusters will be surrounded by hydrophilic groups. All CUR molecules are dispersed into micelle. Like the NCMs, the morphology of CUR-loaded NCMs show a good spherical structure. The results indicated that the CUR can co-self-assemble with copolymer molecules and have little influence on the self-assembly ability of polymeric micelle.
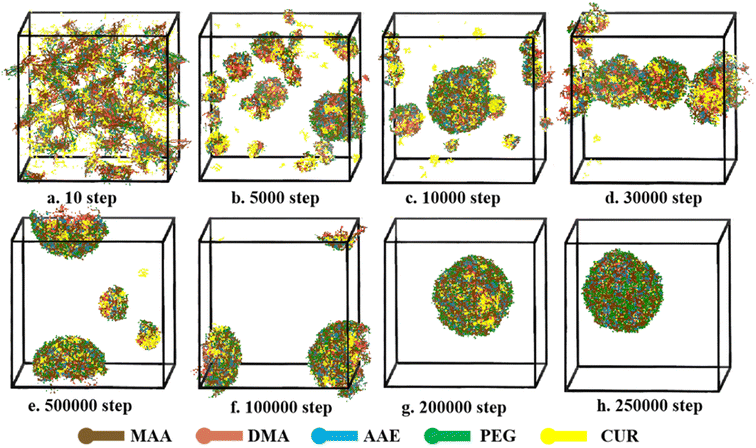 | ||
Fig. 10 Self-assembly process of CUR loaded P(PEGMA9-co-AEMA6)-b-PDMAEMA10 at different simulated steps, (a–h) from 10 to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, during DPD simulation. 000 steps, during DPD simulation. | ||
Effect of mole fraction of drug in feed
Water and drug are two important elements during the self-assembly process of PMs. Herein, the effect of water content on the self-assembly of PMs was studied by DPD simulation. The volume ration between polymer and drug were 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Fig. 11 shows the morphology snapshots and cross-section view of the mixtures, where the fraction of water varies from 52% to 92%. The morphology snapshots and cross-section view can help us to visualize the self-assembly process and location of the different beads. According to the morphology snapshots, the morphologies and bead distributions of the copolymers change significantly with the mole fraction of water. When the mole fraction of water was 52%, beads distributed disorderly at the interfaces, and the system formed an irregular polyhedron as shown in the Fig. 11a. This indicates that the system has a poor entrapment efficiency. When water content was increased to 60%, the clusters formed a sandwich-like structure from irregular polyhedron, and most drug self-assembled into the middle layer. When the mole fraction of water was increased to 67%, the sandwich-like structure began to rupture. A columnar structure was gradually formed as the mole fraction of water increased to 76%. When the mole fraction of water was increased from 76% to 84%, the system still maintains a columnar structure, but it is worth noting that the volume of system is significantly reduced. This result indicates that the same type of copolymers beads aggregate more intensively and thus the copolymer segments become more orderly across the interfaces. As the mole fraction of water further increases to 92%, the copolymers were completely separated from water and formed well sphere morphology. The results indicated that the water content will influence the balance between the water and block copolymer. As the water content increases, the copolymer structure will change from irregular polyhedron to sandwich-like structure, then to columnar structure and finally from a spherical structure.
1. Fig. 11 shows the morphology snapshots and cross-section view of the mixtures, where the fraction of water varies from 52% to 92%. The morphology snapshots and cross-section view can help us to visualize the self-assembly process and location of the different beads. According to the morphology snapshots, the morphologies and bead distributions of the copolymers change significantly with the mole fraction of water. When the mole fraction of water was 52%, beads distributed disorderly at the interfaces, and the system formed an irregular polyhedron as shown in the Fig. 11a. This indicates that the system has a poor entrapment efficiency. When water content was increased to 60%, the clusters formed a sandwich-like structure from irregular polyhedron, and most drug self-assembled into the middle layer. When the mole fraction of water was increased to 67%, the sandwich-like structure began to rupture. A columnar structure was gradually formed as the mole fraction of water increased to 76%. When the mole fraction of water was increased from 76% to 84%, the system still maintains a columnar structure, but it is worth noting that the volume of system is significantly reduced. This result indicates that the same type of copolymers beads aggregate more intensively and thus the copolymer segments become more orderly across the interfaces. As the mole fraction of water further increases to 92%, the copolymers were completely separated from water and formed well sphere morphology. The results indicated that the water content will influence the balance between the water and block copolymer. As the water content increases, the copolymer structure will change from irregular polyhedron to sandwich-like structure, then to columnar structure and finally from a spherical structure.
The self-assembly of CUR-loaded PMs with different drug contents was studied by the DPD method. As shown in Fig. 12, the system maintained a spherical structure as the drug contents increased from 1% to 12%, but it should note that the volume of system was increased. When the mole fraction of CUR in the range of 1-2%, the hydrophilic shell is mainly consisted by PEG (green beads), AAE (blue beads) and MAA (orange beads), few CUR (yellow beads) was found in the shell. The drug molecules could well load into the core of micelles by hydrophobic effect. When drug content was further increased, more and more CUR molecules were exposed in the shell, indicating the CUR molecules cannot well encapsulate in the core during the process of self-assembly. The results indicated that 2% CUR in the feed was the optimal formulation during preparation of CUR-loaded PMs.
Crosslinking mode of polymeric micelles
In this work, the polymeric micelle was cross-linked by EDA at the interface region.The β-keto ester containing in AEMA blocks provided the reversible cross-linked site. Before cross-linking, polymeric micelle shows a core–shell structure. It can be seen from the cross-section that most CUR molecules (yellow beads) distribute inside in the core, but a small amount of CUR exposed on the surface of the shell (Fig. 13a). The CUR-loaded PMs may not stable and CUR molecule is easily released from the micelle before reaching targeted sites. After cross-linking, polymeric micelle shows a typical core–shell structure, indicating the cross-linking process did not affect the self-assembly process of block copolymer. In addition, from the density profiles of beads (Fig. 13b), it can be clearly observed that DMA beads tend to move towards the outer surface after cross-linking, which increases the attractive interaction of the shell and interface layer. Drug molecules are well entrapped in the micellar core. The results indicate that reversible cross-linked structure can improve the stability of micelle and was an efficient barrier to prevent the release of drug molecules from the micellar core. | ||
| Fig. 13 Section views of P(PEGMA9-co-AEMA6)-b-PDMAEMA10 micelles and density profile of beads inside micelle (a) before cross-linking and (b) after cross-linking. | ||
pH responsiveness of polymeric micelles
The aggregate morphologies of CUR-loaded PMs at pH 5.0 were further studied by DPD simulation to better understanding the drug release process. The volume fractions of polymer, drug and water were 6%, 2% and 92%, respectively. As shown in Fig. 14, at the initial steps of simulation, a single spherical aggregate is formed, where hydrophilicity beads are located around the shell. As the simulation proceeds, pH sensitive groups are protonated under the acid condition, DMA beads will change into DMA+. This structure transformation from hydrophobicity to hydrophilicity broke the hydrophilicity–hydrophobicity balance. The shape of spherical aggregates began to deform, the hydrophobic inner core begins to distribute towards the hydrophilic outer shell after 5000 steps. At 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, a small part of clusters was found. As the polymers continues to swell, the typical core–shell structure was destroyed from 30
000 steps, a small part of clusters was found. As the polymers continues to swell, the typical core–shell structure was destroyed from 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 100
000 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, and the drug was released from the PMs. At 250
000 steps, and the drug was released from the PMs. At 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, lot clusters with different particle sizes were randomly distributed in water, and the amount of drug release has gradually increased. These findings indicated that drug release process was triggered by acidic condition, suggesting that the PMs have a good pH-sensitivity.
000 steps, lot clusters with different particle sizes were randomly distributed in water, and the amount of drug release has gradually increased. These findings indicated that drug release process was triggered by acidic condition, suggesting that the PMs have a good pH-sensitivity.
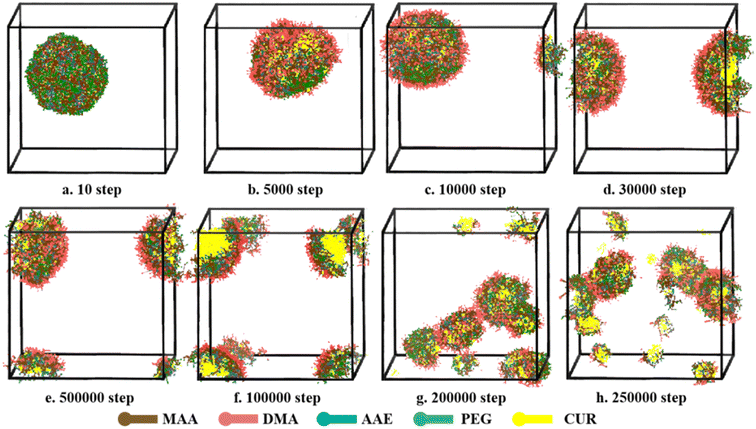 | ||
Fig. 14 Typical simulated snapshots of CUR-loaded micelles, (a–h) from 10 to 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps, at simulated physiological condition (pH 5) during DPD simulation. 000 steps, at simulated physiological condition (pH 5) during DPD simulation. | ||
Conclusions
In summary, serial pH-responsive amphiphilic PMs were successfully synthesized via RAFT polymerization and then self-assembled into micelles. P(PEGMA9-co-AEMA6)-b-PDMAEMA10 has the lowest CMC (0.0041 mg mL−1), the highest LC (13.86%) and EE (97.03%). After reversible crosslinking of the micellar shell, the RCLMs exhibited excellent stability against extensive dilution and good reversibility of pH responsiveness in solutions with different pH values. DPD simulation results show that the self-assembly of micelles follow a process from disordered states, small clusters, aggregates, and stable spherical structures. CUR have little influence on the self-assembly ability of polymeric micelle. In addition, the optimal preparation method of CUR-loaded PMs in aqueous solutions was confirmed by analysis drug distribution and morphology of PMs. 2% CUR in the feed was the optimal formulation during preparation of CUR-loaded PMs. The simulation results were consistent with experimental results, indicating that the DPD simulation could be a useful method to study the structure property of PMs. The pH-responsive amphiphilic PMs with imine cross-linked structures have great potential as drug carriers for smart drug delivery.Author contributions
Liu TAN: writing-original draft, visualization, conceptualization, investigation. Jinglin Fan, Yuqing Zhou, Manzhen Duan: data curation, validation, software. Di Xiong: validation, data curation. Ding Hu, Zhimin Wu: resources, supervision, funding acquisition, writing-review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 22208272), the Hunan Provincial Natural Science Foundation of China (No. 2022JJ40445; No. 2020JJ5132; No. 2020JJ4091), Hunan Provincial Education Department Project (20C0649) and the PhD Research Startup Foundation of Xiangtan University (No. 21QDZ58), Training Program of Innovation and Entrepreneurship for Undergraduates of Xiangtan University.References
- Z. Fang, L. Y. Wan, L. Y. Chu, Y. Q. Zhang and J. F. Wu, Expert Opin. Drug Delivery, 2015, 12, 1943–1953 CrossRef CAS.
- S. Hossen, M. K. Hossain, M. K. Basher, M. N. H. Mia, M. T. Rahman and M. J. Uddin, J. Adv. Res., 2019, 15, 1–18 CrossRef CAS.
- R. V. Kalaydina, K. Bajwa, B. Qorri, A. Decarlo and M. R. Szewczuk, Int. J. Nanomed., 2018, 13, 4727–4745 CrossRef CAS.
- P. Rathee, Pharm. Nanotechnol., 2017, 5, 242 CAS.
- B. R. Tian, Y. M. Liu and J. Y. Liu, Carbohydr. Polym., 2021, 251, 116871 CrossRef CAS.
- G. Geyik and N. Isiklan, Colloids Surf., A, 2022, 634, 127960 CrossRef CAS.
- Y. J. Chen, H. Chen, M. R. Feng and Y. X. Dong, Eur. Polym. J., 2016, 85, 489–498 CrossRef CAS.
- D. A. Filatov and E. N. Govorun, Soft Matter, 2021, 17, 90–101 RSC.
- Y. Sakamoto and T. Nishimura, Polym. Chem., 2022, 13, 6343–6360 RSC.
- P. Das Karmakar and S. Pal, Int. J. Biol. Macromol., 2021, 183, 718–726 CrossRef.
- Y. Bobde, T. Patel, M. Paul, S. Biswas and B. Ghosh, Colloids Surf., B, 2021, 204, 111833 CrossRef CAS PubMed.
- J. Bai, J. Wang, Y. C. Feng, Y. F. Yao and X. B. Zhao, Colloids Surf., A, 2022, 639, 128353 CrossRef CAS.
- Y. M. Li, A. H. Yu, L. B. Li and G. X. Zhai, J. Drug Targeting, 2018, 26, 753–765 CrossRef CAS PubMed.
- S. Patel, V. Patel, M. Yadav, D. Panjwani, P. Ahlawat, A. Dharamsi and A. Patel, J. Polym. Res., 2022, 30, 14 CrossRef.
- Q. T. H. Shubhra, K. Guo, Y. X. Liu, M. Razzak, M. S. Manir and A. Alam, Acta Biomater., 2021, 131, 493–507 CrossRef CAS.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Nat. Rev. Mater., 2016, 1, 16014 CrossRef CAS.
- E. L. Cyphert, H. A. von Recum, M. Yamato and M. Nakayama, J. Biomed. Mater. Res., Part A, 2018, 106, 1552–1560 CrossRef CAS PubMed.
- S. Ghasemi, L. Ahmadi and F. Farjadian, J. Mater. Sci., 2022, 57, 17433–17447 CrossRef CAS.
- Z. L. Kou, D. T. Dou, H. Q. Mo, J. Y. Ji, L. H. Lan, X. D. Lan, J. Y. Zhang and P. Lan, Int. J. Biol. Macromol., 2020, 165, 995–1001 CrossRef CAS PubMed.
- T. S. Wang, C. Wu, Y. G. Hu, Y. Zhang and J. K. Ma, RSC Adv., 2023, 13, 16488–16511 RSC.
- Y. Li, M. T. Leng, M. T. Cai, L. Huang, Y. W. Chen and X. L. Luo, Colloids Surf., B, 2017, 154, 397–407 CrossRef CAS PubMed.
- R. S. Kalhapure and J. Renukuntla, Chem.-Biol. Interact., 2018, 295, 20–37 CrossRef CAS PubMed.
- J. Dong, Y. N. Wang, J. Zhang, X. W. Zhan, S. Q. Zhu, H. Yang and G. J. Wang, Soft Matter, 2013, 9, 370–373 RSC.
- C.-W. Hsu, M.-H. Hsieh, M.-C. Xiao, Y.-H. Chou, T.-H. Wang and W.-H. Chiang, Int. J. Biol. Macromol., 2020, 163, 1106–1116 CrossRef CAS PubMed.
- S. Porrang, N. Rahemi, S. Davaran, M. Mahdavi and B. Hassanzadeh, Colloids Surf., A, 2021, 623, 126719 CrossRef CAS.
- A. Nk, B. Su, A. Sjl, J. Yu, A. Ra, M. Kang, C. Ikp and A. Ik, React. Polym., 2021, 166, 104966 CrossRef.
- C. Yu, L. Wang, Z. Z. Xu, W. Q. Teng, Z. M. Wu and D. Xiong, J. Polym. Res., 2020, 27, 111–120 CrossRef CAS.
- Y. F. Li, Y. L. Niu, D. Hu, Y. W. Song, J. W. He, X. Y. Liu, X. N. Xia, Y. B. Lu and W. J. Xu, Macromol. Chem. Phys., 2015, 216, 77–84 CrossRef CAS.
- Y. Zhang, X. B. Zhang, W. W. Chen, Y. L. He, Y. Liu and H. X. Ju, J. Controlled Release, 2021, 336, 469–479 CrossRef CAS PubMed.
- J. E. Yap, L. Zhang, J. T. Lovegrove, J. E. Beves and M. H. Stenzel, Macromol. Rapid Commun., 2020, 41, 2000236 CrossRef CAS PubMed.
- Y. Zhao, A. C. Tavares and M. A. Gauthier, J. Mater. Chem. B, 2016, 4, 3019–3030 RSC.
- D. Kalafatovic, M. Nobis, N. Javid, P. Frederix, K. I. Anderson, B. R. Saunders and R. V. Ulijn, Biomater. Sci., 2015, 3, 246–249 RSC.
- G. Slor, S. Tevet and R. J. Amir, ACS Polym. Au, 2022, 2, 380–386 CrossRef CAS.
- A. Barve, A. Jain, H. Liu, Z. Zhao and K. Cheng, Acta Biomater., 2020, 113, 501–511 CrossRef CAS PubMed.
- Y. J. Wang, W. J. Song, L. J. Bao, J. W. Wei, Y. Y. Qian and Y. M. Bi, RSC Adv., 2023, 13, 22079–22087 RSC.
- G. H. Gao, Y. Li and D. S. Lee, J. Controlled Release, 2013, 169, 180–184 CrossRef CAS PubMed.
- J. Hu, J. L. He, D. L. Cao, M. Z. Zhang and P. H. Ni, Polym. Chem., 2015, 6, 3205–3216 RSC.
- I. A. Isoglu, Y. Ozsoy and S. D. Isoglu, Curr. Top. Med. Chem., 2017, 17, 1469–1489 CrossRef CAS PubMed.
- Y. Kim, M. H. Pourgholami, D. L. Morris and M. H. Stenzel, Biomacromolecules, 2012, 13, 814–825 CrossRef CAS.
- S. Yadav, P. Kumar, S. H. Jo, S. H. Park, W. K. Lee, S. I. Yoo and K. T. Lim, React. Funct. Polym., 2022, 175, 105271 CrossRef CAS.
- X. Zhao and P. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 166–174 CrossRef CAS.
- Y. Shi, C. V. Nostrum and W. E. Hennink, ACS Biomater. Sci. Eng., 2015, 1, 393–404 CrossRef CAS.
- Y. C. Liu, F. Chen, K. Zhang, Q. Wang, Y. W. Chen and X. L. Luo, J. Mater. Chem. B, 2019, 7, 3884–3893 RSC.
- Z. M. Wu, M. Z. Duan, D. Xiong and C. Y. Zhang, Pharmaceutics, 2019, 11, 11120620 Search PubMed.
- W. Y. Hu, Z. M. Wu, Q. Q. Yang, Y. J. Liu, J. Li and C. Y. Zhang, Colloids Surf., B, 2019, 183, 110443 CrossRef CAS.
- D. Xiong, L. Y. Wen, S. Y. Peng, J. C. Xu and L. J. Zhang, Pharmaceutics, 2020, 12, 12030258 CrossRef.
- J. Skey and R. K. O'Reilly, Chem. Commun., 2008, 35, 4183–4185 RSC.
- S. M. Davis, D. Reichel, Y. Bae and K. R. Pennypacker, Pharm. Res., 2018, 35, 6–15 CrossRef.
- W. Lin, S. Nie, D. Xiong, X. Guo, J. Wang and L. Zhang, Nanoscale Res. Lett., 2014, 9, 243 CrossRef PubMed.
- Y. Wang, Q. Y. Li, X. B. Liu, C. Y. Zhang, Z. M. Wu and X. D. Guo, ACS Appl. Mater. Interfaces, 2015, 7, 25592–25600 CrossRef CAS.
- J. Koelman and P. J. Hoogerbrugge, Europhys. Lett., 1993, 21, 363–368 CrossRef CAS.
- C. Yang, L. Yin, C. Yuan, W. Liu, J. Guo, P. S. Shuttleworth, H. Yue and W. Lin, Colloids Surf., B, 2021, 204, 111797 CrossRef CAS PubMed.
- A. Maiti and S. Mcgrother, J. Chem. Phys., 2004, 120, 1594–1601 CrossRef CAS PubMed.
- G. Rd and W. Pb, J. Chem. Phys., 1997, 107, 4423–4435 CrossRef.
| This journal is © The Royal Society of Chemistry 2023 |