DOI:
10.1039/D3RA05412H
(Paper)
RSC Adv., 2023,
13, 34012-34019
Kinetic study of NADPH activation using ubiquinone-rhodol fluorescent probe and an IrIII-complex promoter at the cell interior
Received
10th August 2023
, Accepted 14th November 2023
First published on 20th November 2023
Abstract
Nicotine adenine dinucleotide derivatives NADH and NADPH are intimately involved in energy and electron transport within cells. The fluorescent ubiquinone-rhodol (Q-Rh) probe is used for NADPH activation monitoring. Q-Rh reacts with NADPH yielding its quenched hydroquinone-rhodol (H2Q-Rh) form with concurrent NADPH activation (i.e. NADP+ formation). NADPH activation can be enhanced by the addition of an IrIII-complex (i.e. [(η5-C5Me5)Ir(phen)(H2O)]2+) as a promoter. The rate of the Q-Rh fluorescence quenching process is proportional to the NADPH activation rate, which can be used to monitor NADPH. Experiments were performed in phosphate-buffered saline (PBS) solution and on HeLa cell cultures to analyze the kinetics of Q-Rh reduction and the influence of the IrIII-complex promoter on the activation of NADPH (in PBS) and of other intracellular reducing agents (in HeLa cells). There is a substantial increase in Q-Rh reduction rate inside HeLa cells especially after the addition of IrIII-complex promoter. This increase is partly due to a leakage process (caused by IrIII-complex-induced downstream processes which result in cell membrane disintegration) but also involves the nonspecific activation of other intracellular reducing agents, including NADH, FADH2, FMNH2 or GSH. In the presence only of Q-Rh, the activation rate of intracellular reducing agents is 2 to 8 times faster in HeLa cells than in PBS solution. When both Q-Rh and IrIII-complex are present, the rate of the IrIII-complex catalyzed reduction reaction is 7 to 23 times more rapid in HeLa cells. Concentration- and time-dependent fluorescence attenuation of Q-Rh with third-order reaction kinetics (reasonably approximated as pseudo-first-order in Q-Rh) has been observed and modelled. This reaction and its kinetics present an example of “bioparallel chemistry”, where the activation of a molecule can trigger a unique chemical process. This approach stands in contrast to the conventional concept of “bioorthogonal chemistry”, which refers to chemical reactions that occur without disrupting native biological processes.
1 Introduction
The bioorthogonal chemistry approach is used to examine biomolecules in their native environment using chemical reactions that do not interfere with the biological processes.1–5 In contrast to biorthogonal chemistry, “bioparallel chemistry” is attributed to chemical reactions involving artificial molecules interacting with native biological processes, and has been introduced by Komatsu et al. in 2014.6 The intracellular activation of acetyl coenzyme A (acetyl-CoA) by tributylphosphine (PBu3), and its fluorescent detection, is considered the first successful example of the bioparallel chemistry concept.7 In a further study, an artificial reaction promoter (PBu3) was used to control ATP concentration and acetylation of mitochondrial proteins.8 These results effectively illustrate that novel artificial reaction promoters can be excellent candidates for intracellular imaging and are promising for the modulation of cellular functions. In 2013, Sadler and coworkers reported the reduction of quinone by reduced coenzyme NADH involving a cyclopentadienyl–IrIII catalyst complex in aqueous media.9,10 Subsequently, in 2014, Komatsu et al. reported the use of a fluorescent ubiquinone-rhodol (Q-Rh) conjugate containing a biocompatible rhodol fluorophore11 for intracellular activation and imaging of nicotinamide adenine dinucleotide (NAD) derivatives NADH and NADPH. Both NADH and NADPH act as electron transporters in living cells and play a crucial role in metabolism.12,13 Here, a kinetic study has been undertaken to understand the reaction mechanism of NADPH in the presence of Q-Rh fluorescent dye and an IrIII-complex (i.e. [(η5-C5Me5)Ir(phen)(H2O)]2+) promoter (Fig. 1a). Information regarding rates of chemical reactions (including in the biological system), reaction order, and rate-determining steps14,15 is essential for the further development of quinone reduction processes that mimic the action of reductases, such as NADH ubiquinone oxidoreductase,16 NADH cytochrome-b5,17 and NADPH cytochrome P-450 reductase.18
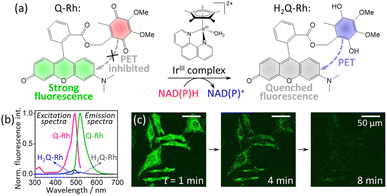 |
| | Fig. 1 (a) Reduction reaction of ubiquinone-rhodol (Q-Rh) to hydroquinone-rhodol (H2Q-Rh) by nicotinamide adenine dinucleotide derivatives (NAD(P)H) in the presence of [(η5-C5Me5)Ir(phen)(H2O)]2+ complex (IrIII-complex). (b) Normalized fluorescence excitation (at λem = 520 nm) and fluorescence emission spectra (at λex = 488 nm) of Q-Rh and its reduced (by Na2S2O4) form H2Q-Rh in phosphate-buffered saline (PBS) at pH = 7.4 and 25 °C. (c) Micrographs of fluorescence quenching of Q-Rh stained (0.01 mM) HeLa cells at 1, 4 and 8 min following the addition of IrIII-complex. | |
In this study, we present an analysis of the IrIII-complex catalyzed reduction of Q-Rh by NADPH yielding hydroquinone-rhodol (H2Q-Rh) and activated NADP+ (Fig. 1a). This reaction is important as an one of the leading examples of bioparallel chemical processes in living organisms.
2 Results and discussion
2.1 Kinetics in phosphate-buffered saline (PBS) solution
The Q-Rh fluorescent probe (synthesized according to Komatsu et al.6) has absorbance and fluorescence maxima at 492 and 518 nm, respectively (Fig. 1b), with a fluorescence quantum yield of 0.73 in phosphate-buffered saline (PBS) at pH = 7.4. The typical fluorescence lifetime of rhodol-type dyes is around 4 ns in aqueous phosphate buffer.19 The reduced form of Q-Rh (i.e. H2Q-Rh) obtained using sodium dithionite (Na2S2O4)20 has significantly attenuated UV-vis absorption and its fluorescence emission is strongly quenched involving photoinduced-electron transfer (PET) mechanisms, as shown in Fig. 1b. Micrographs illustrating the time dependence of fluorescence emission of Q-Rh at the interior of HeLa cells following addition of the IrIII-complex are shown in Fig. 1c. Also in this study, we elucidate the kinetics behind the time dependence of fluorescence emission intensity. However, first we will consider the normal kinetics of Q-Rh to H2Q-Rh conversion under simple conditions (in PBS, pH = 7.4) in the presence of NADPH and in the absence/presence of the IrIII-complex promotor (which activates NADPH).
The following equation describes the reduction reaction of Q-Rh by NADPH.
| |
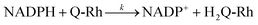 | (1) |
It should be noted that the overall reaction in
eqn (1) consists of the following two processes.
21| |
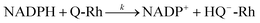 | (2) |
| |
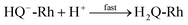 | (3) |
The reaction in
eqn (2) describes the direct hydride transfer leading to the formation of hydroquinone anion (HQ
−-Rh) and is followed by the reaction in
eqn (3), where hydroquinone H
2Q-Rh is formed due to the rapid protonation of HQ
−-Rh by H
+ from the medium.
21 This indicates that the reaction shown in
eqn (2) is the rate-limiting step. Therefore, the reaction rate constant,
k, for the overall reaction (
eqn (1)) is the same as that for the reaction in
eqn (2).
In the presence of the IrIII-complex reaction promoter, the following reversible association process between NADPH and IrIII-complex is assumed to occur.
| |
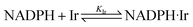 | (4) |
where Ir represents the promoter Ir
III-complex, NADPH·Ir is the complexed form of NADPH reactant with the Ir
III-complex, and
KIr = [NADPH·Ir]/([NADPH][Ir]) is the equilibrium association constant (square brackets denote concentrations of the species). Kinetics of the reaction in the presence of reactant-promoter NADPH·Ir complex is governed by the following reaction.
| |
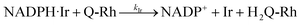 | (5) |
where
kIr is the reaction rate constant of this process (
i.e. the process in which the NADPH·Ir complex reduces Q-Rh). Thus, in the presence of Ir
III-complex promoter, all three processes described by the reactions in
eqn (1),
(4) and
(5) are simultaneous. The solution of these kinetics is done using the following approach. The reaction progress was monitored by fluorescence emission from the Q-Rh probe (at 518 nm). The reactions shown in
eqn (1) and
(5) lead to the following differential rate equation for the decrease of Q-Rh concentration.
| |
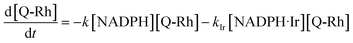 | (6) |
Substituting the [NADPH·Ir] term using the definition of
KIr (given in the context of the reaction in
eqn (4)) followed by rearrangement yields a differential rate equation for Q-Rh in the following form.
| |
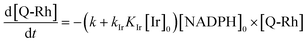 | (7) |
In this study, the concentration of Q-Rh fluorescent probe is always significantly lower than those of the NADPH reactant and Ir
III-complex promoter. Therefore, they can be assumed constant,
i.e. [NADPH] = [NADPH]
0 and [Ir] = [Ir]
0, where [NADPH]
0 and [Ir]
0 are initial concentrations of NADPH and Ir
III-complex promoter, respectively. This situation is denoted by ‘0’ subscripts in
eqn (7). The above assumptions reduce the initially third-order rate
eqn (7) (
i.e., first-order in [Ir], [NADPH] and [Q-Rh]) to pseudo-first-order in Q-Rh concentration with a pseudo-first-order rate constant
| | |
k′ = (k + kIrKIr[Ir]0)[NADPH]0.
| (8) |
The
eqn (7) can then be solved analytically in the form of
eqn (9).
14| | |
[Q-Rh] = [Q-Rh]0exp(−k′t)
| (9) |
where [Q-Rh]
0 is the initial concentration of Q-Rh probe. The time dependency of hydroquinone H
2Q-Rh concentration can be readily derived considering the mass balance equation [Q-Rh]
0 = [Q-Rh] + [H
2Q-Rh].
| | |
[H2Q-Rh] = [Q-Rh]0(1 − exp(−k′t))
| (10) |
Taking into account that the fluorescence emission of reacted quenched H
2Q-Rh (at 518 nm) is 1/30 (=
q) of the Q-Rh starting fluorescence intensity (
i.e. ca. 97% quenching efficiency) due to the operation of PET mechanism (see
Fig. 1b and Experimental section for more details), we can assume that the time dependence of the normalized fluorescence intensity
In(
t) at 518 nm is proportional to [Q-Rh] +
q[H
2Q-Rh]. Then the resulting
In(
t) can be expressed as
eqn (11), where normalization means that
In(
t = 0) = 1.
| | |
In(t) = q + (1 − q)exp(−k′t)
| (11) |
In order to extract the kinetic parameters, the time-dependent normalized fluorescence of Q-Rh (0.01 mM) with NADPH (1 mM) was measured at different IrIII-complex promoter concentrations in PBS solution, as shown in Fig. 2a. In the absence of IrIII-complex, the pseudo-first-order rate constant reduces to k′ = k[NADPH]0, and fitting of eqn (11) to experimental data (Fig. 2a) yields a value of the reaction rate constant k = 0.28 ± 0.06 M−1 s−1 describing the kinetics in eqn (1). After the addition of IrIII-complex (0.5 and 1 mM) into the solution (and using already-known k), the fitting procedure (Fig. 2a) further yields the value of the product kIrKIr = 764 ± 85 M−2 s−1 contained in the unreduced pseudo-first-order rate constant k′ in eqn (8). Table 1 summarises the kinetic parameters obtained. It can be seen that the presence of a small quantity of IrIII-complex promoter (for example, 1 mM) enhances the rate of the Q-Rh reduction reaction by a factor of 4; i.e. the promoting effect of IrIII-complex (1 mM) expressed as “enhancement ratio” is 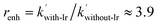 . Note that the enhancement ratio is independent of NADPH concentration and has the general formula shown in eqn (12).
. Note that the enhancement ratio is independent of NADPH concentration and has the general formula shown in eqn (12).
| |
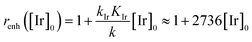 | (12) |
The enhancement ratio (plotted in
Fig. 2b) can be used to determine how many times (
i.e. renh-times) faster is the Q-Rh reduction reaction in the presence of Ir
III-complex promoter ([Ir]
0 in the units of M) compared to the uncatalyzed (unpromoted) reaction in
eqn (1). The individual constants in the product
kIrKIr cannot be extracted from the available kinetic data, although this does not preclude further analysis of the intracellular fluorescence behaviour of Q-Rh. It is clear that the non-zero
KIr constant (
i.e. NADPH·Ir complex formation in
eqn (4)) plays a substantial role in the overall Q-Rh reduction kinetics. This is also emphasized by the relatively large value of the catalytic “boost” factor
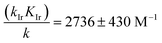
in
eqn (12), which highlights the activity of the Ir
III-complex.
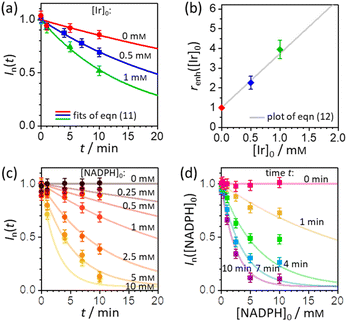 |
| | Fig. 2 (a) Plot of Q-Rh time-dependent normalized fluorescence intensity In(t) (at 520 nm) as a function of IrIII-complex promoter concentration: [Ir]0 = 0 mM (red data points), 0.5 mM (blue data points), and 1 mM (green data points). Experiments were performed in PBS solution (100 mM, pH = 7.4, 25 °C) at constant [NADPH]0 = 1 mM and [Q-Rh]0 = 0.01 mM. Solid lines are the best fits using eqn (11). (b) Plot of enhancement ratio renh as a function of IrIII-complex promoter concentration. The data points are obtained from the evaluation of renh independently from each experiment in (a). The grey solid line is a plot of eqn (12). (c) Plot of time-dependent Q-Rh normalized fluorescent intensity In(t) (at 520 nm) recorded after Q-Rh addition ([Q-Rh]0 = 0.01 mM) at t = 0 min into the PBS solutions (100 mM, pH = 7.4, 25 °C) containing constant IrIII-complex concentration ([Ir]0 = 0.5 mM) and varying NADPH concentration ([NADPH]0 = 0–10 mM). Solid lines are plots (not fits) of eqn (11) with k and kIrKIr values from Table 1 and t is an independent variable. (d) Plot of Q-Rh normalized fluorescent intensity In([NADPH]0) (at 520 nm) as a function of [NADPH]0 concentration recorded at various times (t = 0–10 min) after Q-Rh addition ([Q-Rh]0 = 0.01 mM) into the PBS solutions (100 mM, pH = 7.4, 25 °C) containing constant IrIII-complex concentration ([Ir]0 = 0.5 mM). Solid lines are plots (not fits) of eqn (11) with k and kIrKIr values from Table 1 and [NADPH]0 as an independent variable. | |
Table 1 Kinetic data (reaction rate constants) of Q-Rh reduction in PBS solution at 25 °C as obtained from time-dependent fluorescence measurements in Fig. 2a
| [Ir]0 / mM |
k′ / s−1 a |
Parameterb |
Value |
| Value k′ as obtained from fitting of eqn (11). The values of k, kMKM, and the “boost” factor kIrKIr/k are obtained from the definition of pseudo-first-order rate constant k′ in eqn (8) using known concentrations of [NADPH]0 and [Ir]0. |
| 0.0 |
(2.8 ± 0.4) × 10−4 |
k / M−1 s−1 |
0.28 ± 0.04 |
| 0.5 |
(6.3 ± 0.2) × 10−4 |
kIrKIr / M−2 s−1 |
764 ± 85 |
| 1.0 |
(11.0 ± 0.5) × 10−4 |
kIrKIr/k / M−1 |
2736 ± 430 |
The pseudo-first-order kinetics represented in eqn (7) was further tested by experiments where the initial concentration of NADPH was varied (at constant [Q-Rh]0 = 0.01 mM and [Ir]0 = 0.5 mM), with Q-Rh fluorescence emission monitored over time after the addition of Q-Rh to the solution at t = 0 min (Fig. 2c). The solid lines represent calculated (not fitted) behaviour as obtained using eqn (7) with k and kIrKIr values taken from Table 1, [Q-Rh]0 = 0.01 mM, [NADPH]0 as a parameter (0–10 mM) and t as the independent variable. Data from Fig. 2c can also be used to generate the Q-Rh fluorescence decay as a function of NADPH concentration at a given constant time of 0–10 min (Fig. 2d). The solid lines again represent calculated (not fitted) behaviour using eqn (7) (with k and kIrKIr from Table 1), [Q-Rh]0 = 0.01 mM, t as a parameter (0–10 min) and [NADPH]0 as the independent variable. It can be seen that the presented kinetic model describes well the experimental data.
Overall, from the above analyses, the kinetic model as introduced in eqn (1), (4) and (5), with the analytical solution represented by eqn (9) and (8) together with values in Table 1, yields a good description of the Q-Rh reduction process (i.e. formation of H2Q-Rh) and NADPH activation process (i.e. formation of NADP+) in the presence of IrIII-complex promoter in PBS solution (at pH = 7.4).
2.2 Kinetics at the interiors of HeLa cells
A series of experiments were performed using HeLa cells in order to analyze the intracellular kinetics of Q-Rh reduction with simultaneous NADPH activation. HeLa cells (with cellular passage number in the range 5–10) were incubated (30 min., 37 °C) with Q-Rh (0.01 mM) in Hanks' balanced salt solutions (HBSS) at pH = 7.4. Prior to fluorescence observation, HeLa cells placed in a glass bottom dish were rinsed twice with HBSS, and then the dish was filled with fresh HBSS. The average fluorescence intensity of each individual cell of the sample was obtained using a confocal laser scanning microscope on a cell culture maintained at 25 °C. Micrographs of Q-Rh fluorescence quenching in HeLa cells prior to and following the addition of IrIII-complex ([Ir]0 = 0.1 mM) are shown in Fig. 3a. Time dependence of the average fluorescence intensity as obtained from individual cells is plotted in Fig. 3b (for [Ir]0 = 0.1 mM) and Fig. 3c (for [Ir]0 = 0.5 mM). The sudden increase in fluorescence intensity following the addition of IrIII-complex is most likely due to injection shock (to some extent, an effect similar to addition of hypertonic solution and the corresponding change in the osmotic pressure). For further analyses, it is convenient to average and normalize the data before and after the addition of IrIII-complex, as shown in Fig. 3d and e (black lines). We have also observed that upon addition of IrIII-complex into the HeLa cell culture (in the absence of Q-Rh), the subsequent NADPH activation (or activation of other species) causes cell membrane disintegration, which is probably caused by the intracellular downstream signalling cascade (which regulates cell growth, proliferation, differentiation, and it can also trigger cell apoptosis22). We have used calcein acetoxymethyl (calcein-AM), a green fluorescent dye useful for cell viability monitoring, to stain the cytosol and then observed the dye leakage through HeLa cell membranes following the addition of IrIII-complex, as shown in Fig. 3d and e (orange lines) for addition of 0.1 mM and 0.5 mM of IrIII-complex, respectively. Due to its considerable effect, the leakage rate (kleak) has to be included in the overall Q-Rh reduction kinetics in HeLa cells. The leakage process was modelled as a release process in a confined space (i.e. the glass bottom dish), which is governed by the following differential equation.| |
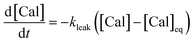 | (13) |
where [Cal] is the time-dependent concentration of calcein in the HeLa cells and [Cal]eq (≠0) is the equilibrium calcein concentration at infinite time introduced due to confined space. The solution of eqn (13) can be expressed in terms of normalized intensity (similarly as for eqn (11)) as| | |
In,Cal(t) = p + (1 − p)exp(−kleak(t − t0)),
| (14) |
where p = [Cal]eq/[Cal]0 is the fraction of remaining calcein fluorescence due to the confined space, and [Cal]0 is the initial calcein concentration at t = t0. Normalization means that In,Cal(t = t0) = 1, where t0 can be interpreted as a lag time in fluorescence decrease after the addition of IrIII-complex in the HeLa cell culture. Eqn (14) is then fitted into experimentally observed calcein fluorescence data, as shown in Fig. 3d and e (blue lines; values of fitted parameters are shown in the caption). The leakage rate (kleak) has a power law type dependence on added IrIII-complex concentration kleak = 0.216[Ir]01/2, as shown in Fig. 4a (and the inset).
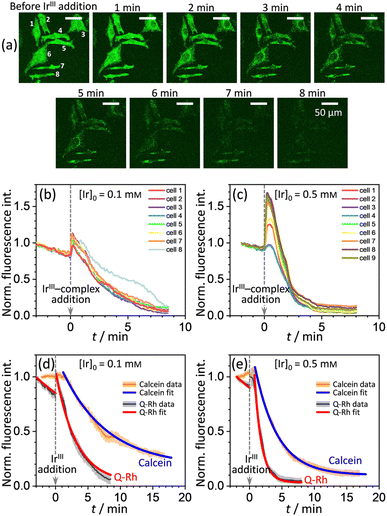 |
| | Fig. 3 (a) Micrographs of fluorescence quenching of Q-Rh stained (0.01 mM) HeLa cells after addition of IrIII-complex (0.1 mM) at 25 °C. Normalized fluorescence time profiles from individual cells are shown in (b) and (c), and averaged data are shown in (d) and (e). (b) and (c) Plots of normalized fluorescence time profiles as obtained from individual HeLa cells (λex = 488 nm, observed at 500–600 nm, 25 °C) stained with Q-Rh (0.01 mM) after the addition (at t = 0 min) of (b) 0.1 mM and (c) 0.5 mM of IrIII-complex. (d) and (e) Black lines show averaged and normalized Q-Rh fluorescence data from (b) and (c). Data are normalized prior to and following the addition of IrIII-complex (at t = 0 min). Orange lines show averaged and normalized fluorescence data of calcein stained (0.01 mM) HeLa cells after the addition (at t = 0 min) of (d) 0.1 mM and (e) 0.5 mM of IrIII-complex depicting the HeLa cell membrane disintegration (note that in this case, the Q-Rh is not present in HeLa cells). Grey and light orange backgrounds correspond to standard deviations. Blue and red lines are fitted according to eqn (14) and (16), respectively. Calcein data fitted parameters: (d) kleak = (2.18 ± 0.34) × 10−3 s−1, p = 0.14 ± 0.03, t0 = 1.5 min; (e) kleak = (4.82 ± 0.21) × 10−3 s−1, p = 0.10 ± 0.01, t0 = 1.2 min. | |
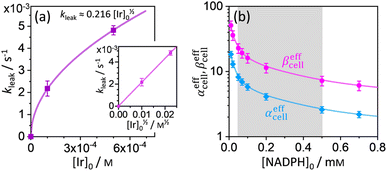 |
| | Fig. 4 (a) Plot of leakage rate (kleak) against the concentration of added IrIII-complex [Ir]0 in HeLa cell cultures at 25 °C. The inset shows linear dependence of kleak on [Ir]01/2 indicating a power law dependence. (b) Effective rate increase coefficients αeffcell and βeffcell for uncatalyzed and IrIII-complex catalyzed Q-Rh reduction in HeLa cells, respectively, as a function of intracellular NADPH concentration. The grey region denotes HeLa cell available NADPH concentration range. | |
Using the above analysis of the HeLa cell membrane leakage rate after the addition of IrIII-complex, we can construct the overall pseudo-first-order rate constant for Q-Rh reduction kinetics inside the HeLa cells in the following form:
| |
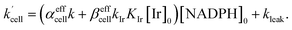 | (15) |
where
kleak can be well approximated as
kleak = 0.216[Ir]
01/2 (see
Fig. 4a). The
αeffcell and
βeffcell are dimensionless coefficients, which account for the increase of the Q-Rh reduction rate inside HeLa cells due to the presence of other intracellular reduction agents, such as flavin adenine dinucleotide (FADH
2),
23,24 flavin mononucleotide (FMNH
2)
16,23 or glutathione (GSH).
25–28
The αeffcell and βeffcell coefficients express effective rate increase of uncatalyzed Q-Rh reduction (shown in eqn (1)) and IrIII-complex catalyzed reduction (shown in eqn (5)), respectively, in HeLa cells as compared to the experiments in PBS solution (where of αeffcell = βeffcell = 1). It can also be noted that the αeffcell and βeffcell coefficients refer to the increase of Q-Rh reduction rate for t < 0 min and t > 0 min, respectively, as shown in Fig. 3d and e.
The rate constant 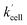 is then used in eqn (16) (which is derived from eqn (7) analogously as eqn (11)) and fitted into normalized and averaged experimental data, as shown in Fig. 3d and e (red lines).
is then used in eqn (16) (which is derived from eqn (7) analogously as eqn (11)) and fitted into normalized and averaged experimental data, as shown in Fig. 3d and e (red lines).
| |
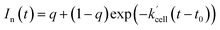 | (16) |
The
t0 is a lag time in Q-Rh fluorescence decrease after the addition of Ir
III-complex. In
eqn (16) αeffcell,
βeffcell and
t0 are fitted parameters. The
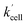
rate constant (
eqn (15)) contains [NADPH]
0 as a known parameter, however, the NADPH concentration in the HeLa cells can range from about 0.05 mM to 0.5 mM (from here on, referred to as HeLa cell available NADPH concentration range).
29–33 Therefore, the fitting of the experimental data in
Fig. 3d and e (red lines) has been performed for various NADPH concentrations, and
αeffcell and
βeffcell coefficients were obtained as a function of [NADPH]
0 (
Fig. 4b). The data in
Fig. 4b indicate that in the HeLa cell available NADPH concentration range (grey zone), the coefficients are within the following ranges: 2 <
αeffcell < 8 and 7 <
βeffcell < 23. From the fitting procedure, it can also be noted that in equilibrium, the Ir
III-complex fluorescence quenching efficiency on Q-Rh seems to be the same (within the experimental error) as in PBS solution,
i.e. 97%.
This large increase in reaction rates is due to the presence of the abovementioned reducing species in the cytosol, which apparently contribute to the Q-Rh reduction process before and, even more significantly, after the addition of IrIII-complex promoter. This can be quantified by the “enhancement ratio” renh,cell, from which the component of leakage process has been removed, as
| |
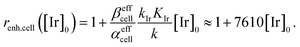 | (17) |
The ratio
βeffcell/
αeffcell ≈ 2.8 over the whole NADPH admissible concentration region (
Fig. 4b), which indicates an about 2.8 times higher Ir
III-complex activity for the Q-Rh reduction process in HeLa cells over that in PBS solution (as seen by comparison with
eqn (12)). Since the values of
αeffcell and
βeffcell are larger than 1 in the whole admissible region, it also suggests that other intracellular reducing agents besides NADPH (
e.g. NADH, FADH
2, FMNH
2 or GSH) are also activated by the presence of the Q-Rh fluorescence probe alone (2 <
αeffcell < 8) and more strongly by the further addition of Ir
III-complex (7 <
βeffcell < 23). It is worth mentioning that GSH is the main agent in the reduction process of intracellular quinones.
28 The GSH contribution to the increase in Q-Rh reduction rate is probably dominant due to its high intracellular concentrations from 0.5 to 10 mM compared to other reducing agents.
25–28 The high values of
αeffcell and
βeffcell coefficients (and accordingly increased
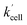
) most likely reflect this situation. There might also be a small contribution to the value of
αeffcell (and presumably also to the value of
βeffcell) originating from the lower intracellular pH in HeLa cells (pH from
ca. 5.7 to 7.1).
34–38 Lower pH (less than 5.7) increases the reaction rate
k in
eqn (1).
39,40 However, other research
21 has shown that there is almost no effect on the reaction rate
k in the pH range from 6 to 8. Therefore, there might be a limited contribution to the increase of the overall Q-Rh reduction rate constant in HeLa cells
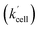
, which can be assigned to lower intracellular pH.
These results imply that the Q-Rh reduction process, which is accompanied by fluorescence quenching, can be used for the estimation of the relative rate of NADPH activation (i.e. the rate of NADP+ formation). The addition of IrIII-complex promoter can further increase this activation rate. In PBS solution, this process can be well controlled with an enhancement factor due to the IrIII-complex concentration expressed in eqn (12). In HeLa cells, the kinetics can also be analyzed, although the Q-Rh fluorescence quenching rate should be rather interpreted as the relative rate of activation involving several other intracellular reducing agents (not only NADPH). The presence of IrIII-complex also has a profound effect on the rate of the activation process, as expressed in eqn (17). Moreover, this activation caused by IrIII-complex inside the HeLa cells initiates a downstream signalling cascade, which results in cell membrane disintegration and cell death. This cascade is perhaps caused by the generation of reactive oxygen species, such as H2O2, which has been reported for similar organoiridium complex systems.10,41,42 The intrinsic cytotoxicity of the IrIII-complex is also evident from monitoring of calcein dye fluorescence after the addition of IrIII-complex (in the absence of Q-Rh), which initiates the cytosol leakage and eventual cell death. On the other hand, HeLa cells incubated only with Q-Rh (without the addition of IrIII-complex promoter) showed no signs of decreased cell viability.
3 Conclusions
We have shown that in the controlled environment of PBS solution, the fluorescent ubiquinone-rhodol (Q-Rh) probe reacts with NADPH leading to its quenched hydroquinone-rhodol (H2Q-Rh) form with simultaneous NADPH activation. This activation can be further increased by the addition of IrIII-complex (i.e. [(η5-C5Me5)Ir(phen)(H2O)]2+) promoter. The rate of Q-Rh fluorescence quenching process is proportional to NADPH activation rate. The kinetics of this process can be well-modelled by first-order kinetics for Q-Rh concentration with the pseudo-first-order rate constant involving the concentrations of IrIII-complex and NADPH.
Furthermore, we performed experiments on HeLa cells to analyze the intracellular kinetics of Q-Rh reduction and the influence of IrIII-complex promoter on the activation of intracellular reducing agents. We found that this process can also be modelled by modified first-order kinetics for Q-Rh. However, the IrIII-complex stimulates downstream intracellular processes, which result in HeLa cell membrane disintegration and leakage of the cytosol. Our kinetic model accounts for this process. Therefore, the actual fluorescence quenching of Q-Rh caused by reduction reactions can be quantified and their kinetic parameters extracted. There is a substantial increase in the Q-Rh reduction rate (accompanied by a corresponding increase of fluorescence quenching) inside the HeLa cells, especially after the addition of IrIII-complex promoter. This increase is partially due to the leakage process but also due to the nonspecific activation of other intracellular reducing agents other than NADPH, such as NADH, FADH2, FMNH2 or GSH (which might have the dominant contribution due to high intracellular GSH concentrations at mM levels). In the presence only of Q-Rh, the activation rate of the intracellular reducing agents is about 2 to 8 times greater in HeLa cells than in PBS solution. In the presence of both Q-Rh and IrIII-complex, the IrIII-complex catalyzed reduction reaction is about 7 to 23 times faster in HeLa cells.
The activation of NADPH or other intracellular species with simultaneous monitoring of this process can be used to exploit unique chemical reactions. This concept stands in contrast to the conventional, widely recognized concept of bioorthogonal chemistry. We have coined the term “bioparallel chemistry” to differentiate this approach. The analyses of IrIII-complex promoted NADPH activation, and its monitoring by Q-Rh fluorescence probe given in this study represents the first attempt to analyze the kinetics of a bioparallel reaction at the interiors of cells.
4 Experimental
4.1 General
Fluorescence spectra were measured on a JASCO FP-8500 spectrofluorophotometer using a quartz cuvette with a 1 cm path length. Phosphate-buffered saline (100 mM, pH 7.4) was used as a solvent. HeLa cells were obtained from RIKEN (Tsukuba, Japan), and cultured in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS), 50 U mL−1 of penicillin and 50 μg mL−1 streptomycin at 37 °C under a humidified atmosphere of 5% CO2.
4.2 Time-dependent fluorescence measurements in PBS (IrIII-complex concentration dependence)
Time-dependent fluorescence intensity of Q-Rh (0.01 mM) reduction in PBS solution (at pH 7.4) was measured in the absence of the promoter (blank measurement), and with 0.5 mM and 1 mM of the IrIII-complex promoter, and with 1 mM NADPH which was added at time t = 0 min. During the measurements (with excitation wavelength λex = 488 nm), the fluorescence was observed at its maximum at λem = 518 nm. Quenched fluorescence of Q-Rh reduced by sodium dithionite (Na2S2O4)20 to H2Q-Rh in PBS buffer was lowered to 1/30 of its original value (at 518 nm).6
4.3 Time-dependent fluorescence measurements in PBS (NADPH concentration dependence)
Time-dependence of fluorescence intensity during Q-Rh (0.01 mM) reduction was measured in PBS solution (at pH 7.4) with 0.5 mM of the IrIII-complex promoter following the addition (at t = 0 min) of various concentrations of NADPH (0, 0.25, 0.5, 1, 2.5, 5, and 10 mM). For these measurements, the excitation wavelength was λex = 488 nm, and the fluorescence maximum was observed at λem = 519 nm.
4.4 Time-dependent fluorescence imaging of Q-Rh in HeLa cells
For fluorescent imaging experiments, a confocal laser scanning microscope system (FluoView FV1000; Olympus, Tokyo, Japan) mounted on an inverted microscope (IX81; Olympus) with a 40× or 60× oil-immersed objective lens was used. The fluorescence imaging measurements were performed on HeLa cells cultured on glass-bottomed dishes (Iwaki, Tokyo, Japan). For Q-Rh dye loading, the cells were incubated for 30 min at 37 °C in Hanks' balanced salt solutions (HBSS) containing NaCl (137 mM), KCl (5.4 mM), CaCl2 (1.3 mM), MgCl2 (0.5 mM), MgSO4 (0.4 mM), Na2HPO4 (0.3 mM), KH2PO4, (0.4 mM), NaHCO3 (4.2 mM), D-glucose (5.6 mM), HEPES (5.0 mM) (pH was adjusted to 7.4 using NaOH) in the presence of 0.01 mM Q-Rh. HeLa cells were washed twice with HBSS solution to remove the remaining extracellular dye, and fluorescence imaging measurements were subsequently performed. Q-Rh was excited at λex = 488 nm, with the signal being observed at 500–600 nm. Fluorescence images were acquired and processed in the FluoView software package (Olympus). The fluorescence intensities were determined by calculating the average intensity within a defined region of interest that encompassed the cell body of each cell. Fluorescence imaging indicates that Q-Rh is uniformly distributed in the cytosol with partial accumulation in mitochondria.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was supported by World Premier International Research Center Initiative (WPI Initiative), MEXT, Japan, and KAKENHI Grant-in-Aid for Scientific Research No. 19K05229 (for J. L.). N. V. is also grateful for financial support from the Japan Society for the Promotion of Science (JSPS) for a JSPS postdoctoral fellowship (P21764) supported by JSPS KAKENHI Grant Number JP22KF0385.
Notes and references
- H. C. Hang, C. Yu, D. L. Kato and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14846–14851 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- J. Idiago-López, E. Moreno-Antolín, J. M. De La Fuente and R. M. Fratila, Nanoscale Adv., 2021, 3, 1261–1292 RSC.
- W. Mao, J. Tang, L. Dai, X. He, J. Li, L. Cai, P. Liao, R. Jiang, J. Zhou and H. Wu, Angew. Chem., Int. Ed., 2021, 60, 2393–2397 CrossRef CAS PubMed.
- H. Komatsu, Y. Shindo, K. Oka, J. P. Hill and K. Ariga, Angew. Chem., Int. Ed., 2014, 53, 3993–3995 CrossRef CAS PubMed.
- H. Komatsu, Y. Shindo, S. A. Kawashima, K. Yamatsugu, K. Oka and M. Kanai, Chem. Commun., 2013, 49, 2876–2878 RSC.
- Y. Shindo, H. Komatsu, K. Hotta, K. Ariga and K. Oka, Sci. Rep., 2016, 6, 29224 CrossRef CAS PubMed , 1–9..
- Z. Liu, R. J. Deeth, J. S. Butler, A. Habtemariam, M. E. Newton and P. J. Sadler, Angew. Chem., Int. Ed., 2013, 52, 4194–4197 CrossRef CAS PubMed.
- Z. Liu, I. Romero-Canelón, B. Qamar, J. M. Hearn, A. Habtemariam, N. P. E. Barry, A. M. Pizarro, G. J. Clarkson and P. J. Sadler, Angew. Chem., Int. Ed., 2014, 53, 3941–3946 CrossRef CAS PubMed.
- L. G. Lee, G. M. Berry and C. H. Chen, Cytometry, 1989, 10, 151–164 CrossRef CAS PubMed.
- X. Luo, R. Li and L. J. Yan, J. Diabetes Res., 2015, 2015, 512618 Search PubMed , 1–12..
- D. E. Metzler, Biochemistry The chemical reaction of Living Cells, Academic Press, 2nd edn, 2001 Search PubMed.
- K. A. Connors, Chemical Kinetics: The Study of Reaction Rates in Solution, VCH Publishers, Inc., New York, 1990 Search PubMed.
- J. E. House, Principles of chemical kinetics, Elsevier Inc., USA, 2nd edn, 2007 Search PubMed.
- J. Hirst, Biochem. J., 2010, 425, 327–339 CrossRef CAS PubMed.
- J. B. Schenkman and I. Jansson, Pharmacol. Ther., 2003, 97, 139–152 CrossRef CAS PubMed.
- R. Li, M. A. Bianchet, P. Talalay and L. M. Amzel, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 8846–8850 CrossRef CAS PubMed.
- J. E. Whitaker, R. P. Haugland, D. Ryan, P. C. Hewitt, R. P. Haugland and F. G. Prendergast, Anal. Biochem., 1992, 207, 267–279 CrossRef CAS PubMed.
- A. Pezzella, O. Crescenzi, A. Natangelo, L. Panzella, A. Napolitano, S. Navaratnam, R. Edge, E. J. Land, V. Barone and M. d’Ischia, J. Org. Chem., 2007, 72, 1595–1603 CrossRef CAS PubMed.
- B. W. Carlson and L. L. Miller, J. Am. Chem. Soc., 1985, 107, 479–485 CrossRef CAS.
- F. J. Bock and S. W. G. Tait, Nat. Rev. Mol. Cell Biol., 2020, 21, 85–100 CrossRef CAS PubMed.
- N. R. Bachur, S. L. Gordon, M. V. Gee and H. Kon, Proc. Natl. Acad. Sci. U. S. A., 1979, 76, 954–957 CrossRef CAS PubMed.
- A. L. Pey, C. F. Megarity and D. J. Timson, Biosci. Rep., 2019, 39, BSR20180459 CrossRef PubMed.
- G. Wu, Y.-Z. Fang, S. Yang, J. R. Lupton and N. D. Turner, J. Nutr., 2004, 134, 489–492 CrossRef CAS PubMed.
- M. P. Gamcsik, M. S. Kasibhatla, S. D. Teeter and O. M. Colvin, Biomarkers, 2012, 17, 671–691 Search PubMed.
- L. Kennedy, J. K. Sandhu, M.-E. Harper and M. Cuperlovic-Culf, Biomolecules, 2020, 10, 1429 Search PubMed.
- N. Watanabe, D. A. Dickinson, R.-M. Liu and H. Jay Forman, Methods Enzymol., 2004, 378, 319–340 CAS.
- R. Tao, Y. Zhao, H. Chu, A. Wang, J. Zhu, X. Chen, Y. Zou, M. Shi, R. Liu, N. Su, J. Du, H.-M. Zhou, L. Zhu, X. Qian, H. Liu, J. Loscalzo and Y. Yang, Nat. Methods, 2017, 14, 720–728 CrossRef CAS PubMed.
- S. A. J. Trammell and C. Brenner, Comput. Struct. Biotechnol. J., 2013, 4, e201301012 CrossRef PubMed.
- A. A. Heikal, Biomarkers Med., 2010, 4, 241–263 CrossRef CAS PubMed.
- Q. Yu and A. A. Heikal, J. Photochem. Photobiol., B, 2009, 95, 46–57 CrossRef CAS PubMed.
- N. Ma, M. A. Digman, L. Malacrida and E. Gratton, Biomed. Opt. Express, 2016, 7, 2441–2452 CrossRef CAS PubMed.
- M. Tafani, J. A. Cohn, N. O. Karpinich, R. J. Rothman, M. A. Russo and J. L. Farber, J. Biol. Chem., 2002, 277, 49569–49576 CrossRef CAS PubMed.
- I. Druzhkova, M. Lukina, V. Dudenkova, N. Ignatova, L. Snopova, A. Gavrina, L. Shimolina, V. Belousov, E. Zagaynova and M. Shirmanova, Cell Cycle, 2021, 20, 1540–1551 CrossRef CAS PubMed.
- H. Hou, Y. Zhao, C. Li, M. Wang, X. Xu and Y. Jin, Sci. Rep., 2017, 7, 1759 CrossRef PubMed.
- L. Lin, J. Zhao, L. Zhang, Y. Huang, F. Ye and S. Zhao, Chem. Commun., 2018, 54, 9071–9074 RSC.
- M. H. Lee, J. H. Han, J. H. Lee, N. Park, R. Kumar, C. Kang and J. S. Kim, Angew. Chem., Int. Ed., 2013, 52, 6206–6209 CrossRef CAS PubMed.
- Y. Song and G. R. Buettner, Free Radical Biol. Med., 2010, 49, 919–962 CrossRef CAS PubMed.
- N. K. Bridge and G. Porter, Proc. R. Soc. London, Ser. A, 1958, 244, 276–288 CAS.
- H. Nguyen and L. H. Do, Chem. Commun., 2020, 56, 13381–13384 RSC.
- T. Suenobu, S. Shibata and S. Fukuzumi, Inorg. Chem., 2016, 55, 7747–7754 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article af,
Jonathan P. Hill
af,
Jonathan P. Hill a and
Jan Labuta
a and
Jan Labuta *a
*a
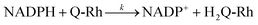
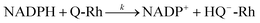
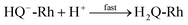

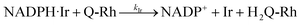
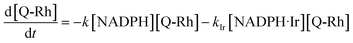
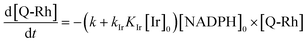
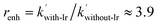 . Note that the enhancement ratio is independent of NADPH concentration and has the general formula shown in eqn (12).
. Note that the enhancement ratio is independent of NADPH concentration and has the general formula shown in eqn (12).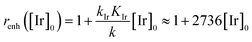
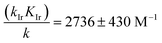 in eqn (12), which highlights the activity of the IrIII-complex.
in eqn (12), which highlights the activity of the IrIII-complex.

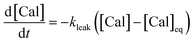

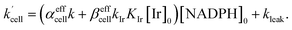
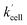 is then used in eqn (16) (which is derived from eqn (7) analogously as eqn (11)) and fitted into normalized and averaged experimental data, as shown in Fig. 3d and e (red lines).
is then used in eqn (16) (which is derived from eqn (7) analogously as eqn (11)) and fitted into normalized and averaged experimental data, as shown in Fig. 3d and e (red lines).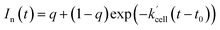
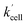 rate constant (eqn (15)) contains [NADPH]0 as a known parameter, however, the NADPH concentration in the HeLa cells can range from about 0.05 mM to 0.5 mM (from here on, referred to as HeLa cell available NADPH concentration range).29–33 Therefore, the fitting of the experimental data in Fig. 3d and e (red lines) has been performed for various NADPH concentrations, and αeffcell and βeffcell coefficients were obtained as a function of [NADPH]0 (Fig. 4b). The data in Fig. 4b indicate that in the HeLa cell available NADPH concentration range (grey zone), the coefficients are within the following ranges: 2 < αeffcell < 8 and 7 < βeffcell < 23. From the fitting procedure, it can also be noted that in equilibrium, the IrIII-complex fluorescence quenching efficiency on Q-Rh seems to be the same (within the experimental error) as in PBS solution, i.e. 97%.
rate constant (eqn (15)) contains [NADPH]0 as a known parameter, however, the NADPH concentration in the HeLa cells can range from about 0.05 mM to 0.5 mM (from here on, referred to as HeLa cell available NADPH concentration range).29–33 Therefore, the fitting of the experimental data in Fig. 3d and e (red lines) has been performed for various NADPH concentrations, and αeffcell and βeffcell coefficients were obtained as a function of [NADPH]0 (Fig. 4b). The data in Fig. 4b indicate that in the HeLa cell available NADPH concentration range (grey zone), the coefficients are within the following ranges: 2 < αeffcell < 8 and 7 < βeffcell < 23. From the fitting procedure, it can also be noted that in equilibrium, the IrIII-complex fluorescence quenching efficiency on Q-Rh seems to be the same (within the experimental error) as in PBS solution, i.e. 97%.
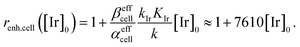
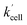 ) most likely reflect this situation. There might also be a small contribution to the value of αeffcell (and presumably also to the value of βeffcell) originating from the lower intracellular pH in HeLa cells (pH from ca. 5.7 to 7.1).34–38 Lower pH (less than 5.7) increases the reaction rate k in eqn (1).39,40 However, other research21 has shown that there is almost no effect on the reaction rate k in the pH range from 6 to 8. Therefore, there might be a limited contribution to the increase of the overall Q-Rh reduction rate constant in HeLa cells
) most likely reflect this situation. There might also be a small contribution to the value of αeffcell (and presumably also to the value of βeffcell) originating from the lower intracellular pH in HeLa cells (pH from ca. 5.7 to 7.1).34–38 Lower pH (less than 5.7) increases the reaction rate k in eqn (1).39,40 However, other research21 has shown that there is almost no effect on the reaction rate k in the pH range from 6 to 8. Therefore, there might be a limited contribution to the increase of the overall Q-Rh reduction rate constant in HeLa cells 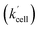 , which can be assigned to lower intracellular pH.
, which can be assigned to lower intracellular pH.


