Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dual activity of indolin-2-ones containing an arylidene motif: DNA and BSA interaction†
Syed Nasir Abbas Bukhari*a,
Tariq G. Alsahlib,
Hasan Ejazc,
Naveed Ahmedd,
Waqas Ahmade,
Mervat A. Elsheriff,
Nasser H. Alotaibig,
Kashaf Junaidh and
Nenad Janković *i
*i
aDepartment of Pharmaceutical Chemistry, College of Pharmacy, Jouf University, Sakaka, Al Jouf 72388, Saudi Arabia. E-mail: sbukhari@ju.edu.sa
bDepartment of Pharmacology, College of Pharmacy, Jouf University, Sakaka, Al Jouf 72388, Saudi Arabia. E-mail: TGAlsahli@ju.edu.sa
cDepartment of Clinical Laboratory Sciences, College of Applied Medical Sciences, Jouf University, Sakaka, Al Jouf 72388, Saudi Arabia. E-mail: hetariq@ju.edu.sa
dDepartment of Pharmaceutics, College of Pharmacy, Jouf University, Sakaka, Al Jouf 72388, Saudi Arabia. E-mail: nakahmad@ju.edu.sa
eDiscipline of Pharmaceutical Chemistry, School of Pharmaceutical Sciences, Universiti Sains Malaysia, Gelugor, 11800, Penang, Malaysia. E-mail: waqas@usm.my
fChemistry Department, College of Science, Jouf University, Sakaka, Al Jouf 72388, Saudi Arabia. E-mail: maelsherif@ju.edu.sa
gDepartment of Clinical Pharmacy, College of Pharmacy, Jouf University, Sakaka 72388, Saudi Arabia. E-mail: nhalotaibi@ju.edu.sa
hSchool of Biological and Behavioural Sciences, Queen Mary University of London, London, E1 4NS, UK. E-mail: kashaf.junaid@qmul.ac.uk
iUniversity of Kragujevac, Institute for Information of Technologies Kragujevac, Department of Science, Jovana Cvijića bb, 34000 Kragujevac, Serbia. E-mail: nenad.jankovic@kg.ac.rs
First published on 25th September 2023
Abstract
Applying a multistep approach, novel indolin-2-ones (IND) that possess an arylidene motif have been synthesized. Eight compounds were chosen for different biological tests (antimicrobial and cytotoxicity). IND containing 2-thienyl (4h) fragment have been found to exhibit good antimicrobial activity against B. cereus. Molecules that have 3-aminophenyl (4d) or 2-pyridyl (4g) groups have shown the best antifungal activities against all tested fungi. These compounds have also been noticed as promising pharmaceuticals against MCF-7 cancer cell lines. Experimental outcomes from the investigation of the interaction of 4d with DNA implied its moderate binding to DNA (KSV = 1.35 × 104 and 3.05 × 104 M−1 for EB and Hoechst binder, respectively). However, considerably stronger binding of 4d to BSA has been evidenced (Ka = 6.1 × 106 M−1). In summary, IND that contains m-aminobenzylidene fragment (4d) exhibits a good dual biological activity making it a promising candidate for further investigation in the drug discovery sector.
Introduction
Indole and its derivatives have been used as synthetic intermediates or even equivalents in the production of diverse pharmaceuticals.1 Its chemical properties and reactivity make it very important in the chemical industry, where it is used as a starting material for the synthesis of many organic compounds, including dyes and agrochemicals.2 From a synthetic point of view and for drug discovery, 1H-indole-2,3-dione (isatin) is particularly important. In addition to its industrial applications, isatin is also found in nature as a component of many natural products, such as alkaloids and plant pigments. Along these lines, isatin is found in the genus Isatis3 in Couroupita guianensis4 and Couroupita guianensis Aubl.5 Substituted isatin has also been isolated from plant bodies such as Melochia tomentosa6 and the bacteria Streptomyces albus.7 In humans, isatin was found to be a metabolic derivative of adrenaline.8 It has also been studied for its potential biological activities. Indolin-2-one (IND) derivatives prepared from isatin have been shown to have various biological activities involving antimicrobial, antitumor, and anti-inflammatory properties, which make them very important for drug discovery and development.1,2Some spiro-pyrazolo-3,3-oxindoles were synthesized by Abo-Salem and have shown promising anticancer activity against HCT-116 and MCF-7 cell lines, and the activities were like Doxorubicin.9 On the other hand, N-alkylated isatin connected with thiourea showed good potential against colon and multiple melanoma cancer cell lines. The investigated compound has shown ALDH inhibition potential and increased ROS activity.10 Interesting access was applied by Karthikeyan et al. Within this study, indol-2-one hybrids were interconnected to the chalcone pharmacologic motif. The achieved results were comparable to cis-platinum, with measured GI50 values in the range of 4.23–12.63 μM against MDA-MB468 and MCF-7 cell lines.11
Therefore, hydrazones of isatin have been shown to have promising anti-inflammatory activity at a dose level of 10 mg kg−1.12 Ibrahim et al. designed IND-based hydrazides with a diclofenac unit as part of the structure.13 In this study, diclofenac conjugates demonstrated a high percentage of inhibition of edema. Interesting antiviral activities against HIV1 and HIV2 were demonstrated by oxindole derivatives.14–16 3-Arylidene-2-oxindole was denoted as GSK-3β inhibitors performed in vitro and in vivo experiments. Even antidiabetic activity authors suggested that arylidene-2-oxindole could be useful for the simultaneous treatment of two diseases cancer and diabetes.17 Based on biological evaluations that were obtained for chromone-isatin derivatives similar opinions were given by Wang and Rahim et al.18,19 Generally, multistep synthetic approaches open access to antimicrobial agents based on isatin or oxindole. In that sense, Biginelli's hybrids connected with isatin moiety showed antifungal activity against A. niger and C. albicans (MIC = 6–200 μg ml−1).20 8-Methoxyl ciprofloxacin with isatin by linkage through the propylene chain was evaluated as an antibacterial agent. High activities comparable to vancomycin were achieved against Gram-positive bacteria with low MIC values (up to 0.25 μg ml−1). To date, the most useful IND-containing molecules that researchers have used as templates are the FDA-approved drugs Sunitinib and Nintedanib (Fig. 1).21–24 Sunitinib is the gold standard in the treatment of metastatic renal cell carcinoma, while Nintedanib is approved for use to treat idiopathic pulmonary fibrosis.25–28
During the last decade, significant progress has been made in the field of arylidene indolin-2-one (AIND) derivatives. The mentioned pharmacophores have been verified as antiplasmodial,29 tyrosine kinase inhibitors,30 radiotracers for Parkinson's disease detection,31 and antidepressants.32 Princiotto et al. investigated anticancer activity of selected 3-(hetero) AINDs against MCF-7 cell lines. Thiazole-containing 3-(hetero) AINDs have been the most promising activity (IC50 = 33 ± 2 μM).33 Very good selectivity followed with inhibitory potency against FGFR1 enzyme was declared for AINDs that contain morpholine or piperazine moiety.34 Furthermore, Senwar et al. pointed out the importance of the presence of an additional heterocyclic motif that positively affects the anticancer activity of AINDs. The presence of indolin-1-yl notably improves activity against prostate cancer cell lines (IC50 = 1.89 ± 0.6 μM) in comparison with morpholino, piperidin-1-yl, and pyrrolidine-1-yl fragments.35 Similar cytotoxicity study demonstrated that dimethylmorpholino and piperazin-1-yl tethered AINDs possessed good activity against breast cancer cell lines in the range 1.26–2.77 μM.36
The dual-activity principle in drug design usually refers to molecules that simultaneously have two different biological effects (e.g. anticancer and antimicrobial). The development of dual-active therapeutic entities has become a hot topic and emergency in the pharmaceutical industry. During the literature survey, we did not find that AIND has activity simultaneously against two different therapeutic targets. Considering, the main goal of this work was to synthesize a library of novel AIND derivatives and investigate their potential for dual biological activity. Our ongoing effort is to create novel potential bioactive compounds with benzylidene37,38 or indol-2-one units.22,39 The antimicrobial and antiproliferative effects of the IND-containing arylidene motif have been studied. In addition, for the most active compound under both biological tests, interactions with biomacromolecules (DNA and BSA) were investigated.
Results and discussion
Synthesis and characterization
Our initial experiments began with the synthesis of the precursor N-allyl isatin (1). Molecule 1 was prepared using an earlier described method with a small modification.40 In the next synthetic step, 1 was subjected to a reaction with aromatic ketones 2a–h under basic catalytic conditions (Et2NH). During this step, the significant issue was reaching the Knoevenagel adduct in a single step. None of the attempts gave the final product 4 even after 24 hours. The product of aldol addition 3 was isolated and structurally confirmed in only one case. Namely, in the reaction of acetophenone (2a) and 1, the product of aldol addition (3) precipitates from the solution. Applying other aromatic ketones 2b–h we failed to isolate the target adduct 3. Upon reaction between 1 and 2 solvent was evaporated, and then the crude mixture was stirred with glacial acetic acid and cc. HCl (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). After the work-up, 4a–h products were characterized using IR, NMR, elemental analysis, and MS. The yields of products 4a–h were in the range of 27–81%. The best yield was obtained for 4e (81%). Compounds 4c–4g are described for the first time (Scheme 1).
1, v/v). After the work-up, 4a–h products were characterized using IR, NMR, elemental analysis, and MS. The yields of products 4a–h were in the range of 27–81%. The best yield was obtained for 4e (81%). Compounds 4c–4g are described for the first time (Scheme 1).
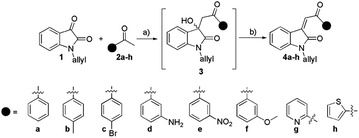 | ||
| Scheme 1 The synthesis of arylidene isatin-2-ones 4a–h: (a) Et2NH, abs. ethanol; (b) glacial acetic acid, cc. HCl. | ||
Generally, IR spectra typically exhibit strong absorption bands of unsaturated ketones in the range of 1700–1600 cm−1. In general, around 1710 cm−1 is the band for the stretching of the amide carbonyl (N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) group as part of a five-membered lactam. The middle bands located at higher frequencies 3100–3000 cm−1 are attributed to the –C
O) group as part of a five-membered lactam. The middle bands located at higher frequencies 3100–3000 cm−1 are attributed to the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H vibration. Especially, in the IR spectrum of 4d characteristic absorption for primary amines in the range 2800–2550 cm−1 were displayed.41 Free primary amine vibration frequencies were found at 2803 and 2572 cm−1. In proton NMR spectra of 4a–h typically sharp and intensive singlet was located at ≈7.8 ppm that originated from the resonance of exocyclic double bond proton
C–H vibration. Especially, in the IR spectrum of 4d characteristic absorption for primary amines in the range 2800–2550 cm−1 were displayed.41 Free primary amine vibration frequencies were found at 2803 and 2572 cm−1. In proton NMR spectra of 4a–h typically sharp and intensive singlet was located at ≈7.8 ppm that originated from the resonance of exocyclic double bond proton ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH. Carbon NMR of 4a–h showed characteristic resonance of keto carbonyl C
CH. Carbon NMR of 4a–h showed characteristic resonance of keto carbonyl C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in the range 183–193 ppm. Amide carbonyl and quaternary carbon of the exocyclic double bond (Cq
O in the range 183–193 ppm. Amide carbonyl and quaternary carbon of the exocyclic double bond (Cq![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) showed resonance at ≈167 ppm and ≈145 ppm, respectively. Additional characterization for all samples was done using ESI-MS techniques (Fig. S19–S26†). Characteristic molecular ions of 4a–h molecules were found. Also, in some specific cases, cluster ions were also found. For instance, molecule 4a had molecular ion [M + H]+ = 290 m/z, and cluster ion [M + M + H]+ = 579 m/z. Therefore, upon ionization in all samples fragment ions at 214 m/z were obtained. Most likely, mentioned arise because of the fragmentation of the O
CH) showed resonance at ≈167 ppm and ≈145 ppm, respectively. Additional characterization for all samples was done using ESI-MS techniques (Fig. S19–S26†). Characteristic molecular ions of 4a–h molecules were found. Also, in some specific cases, cluster ions were also found. For instance, molecule 4a had molecular ion [M + H]+ = 290 m/z, and cluster ion [M + M + H]+ = 579 m/z. Therefore, upon ionization in all samples fragment ions at 214 m/z were obtained. Most likely, mentioned arise because of the fragmentation of the O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–aromatic ketone bond.42
C–aromatic ketone bond.42
Biological activity
AIND derivatives 4a–h were exposed to different biological tests (antimicrobial and antifungal). For these experiments, we treated different Gram-positive (Bacillus subtilis, Bacillus cereus, Staphylococcus aureus, and Enterococcus faecalis) and Gram-negative bacteria (Escherichia coli, Proteus mirabilis, and Klebsiella pneumoniae). The antifungal potential was checked against six fungi Cladosporium cladosporioides, Penicillium italicum, Aspergillus niger, Candida albicans, Candida krusei and Candida parapsilosis. Among them, three species of Candida belong to the group of the most common fungi responsible for superficial and systemic infections.43,44 The two most active AINDs were subjected to cytotoxicity tests against cancer (A549, LS174, MCF-7, and PaCa-2) and normal cell lines (MRC-5).The results of antimicrobial and cytotoxicity testing are shown in Tables 1–3 and present the average of selected activities of indolin-2-ones 4a–h defined in triplicate.
| E. coli | P. mirabilis | K. pneumoniae | B. subtilis | S. aureus | E. faecalis | B. cereus | |
|---|---|---|---|---|---|---|---|
| 4a | 12.5 | 12.5 | 25 | 25 | 12.5 | 25 | 25 |
| 4b | 12.5 | 25 | 25 | 12.5 | 25 | 25 | 25 |
| 4c | 6.25 | 6.25 | 25 | 12.5 | 12.5 | 25 | 12.5 |
| 4d | 12.5 | 6.25 | 12.5 | 12.5 | 25 | 6.25 | 25 |
| 4e | 12.5 | 12.5 | 12.5 | 6.25 | 12.5 | 25 | 25 |
| 4f | 25 | 12.5 | 3.1 | 6.25 | 25 | 25 | 25 |
| 4g | 25 | 25 | 3.1 | 3.1 | 12.5 | 25 | 25 |
| 4h | 25 | 12.5 | 3.1 | 3.1 | 6.25 | 12.5 | 12.5 |
| Streptomycin | 0.062 | 0.062 | 0.031 | 0.016 | 0.031 | 0.031 | 0.016 |
| C. cladosporioides | C. albicans | C. krusei | C. parapsilosis | P. italicum | A. niger | |
|---|---|---|---|---|---|---|
| 4a | 6.25 | 6.25 | 12.5 | 25 | 12.5 | 6.25 |
| 4b | 12.5 | 6.25 | 12.5 | 25 | 6.25 | 6.25 |
| 4c | 12.5 | 6.25 | 12.5 | 6.25 | 12.5 | 12.5 |
| 4d | 0.36 | 0.36 | 6.25 | 6.25 | 0.36 | 0.36 |
| 4e | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| 4f | 12.5 | 12.5 | 3.1 | 6.25 | 12.5 | 12.5 |
| 4g | 6.25 | 3.1 | 3.1 | 3.1 | 6.25 | 3.1 |
| 4h | 12.5 | 25 | 25 | 25 | 6.25 | 3.1 |
| Fluconazole | 0.39 | 0.78 | 0.78 | 0.39 | 0.78 | 0.39 |
| A549 | LS174 | MCF-7 | PaCa-2 | MRC-5 | |
|---|---|---|---|---|---|
| 4d | 87.41 ± 1.59 | 121.38 ± 3.17 | 18.42 ± 0.45 | 140 ± 1.74 | 107.29 ± 1.51 |
| 4g | 143.75 ± 2.40 | 101.85 ± 1.06 | 25.19 ± 0.62 | 128 ± 2.35 | 104.12 ± 2.86 |
| cisPt | 11.59 ± 1.64 | 4.83 ± 0.35 | 18.05 ± 1.28 | 11.34 ± 1.36 | 9.35 ± 1.29 |
Results of antimicrobial and antiproliferative screening of indolin-2-one derivatives 4a–h are presented in Tables 1–3. According to the results of antimicrobial testing, all tested compounds were moderately active against both Gram-positive and Gram-negative bacteria (Table 1). For example, 4f, 4g, and 4h had noticeably greater efficacy against K. pneumoniae Gram-negative strains (MIC = 3.125 mg ml−1). Generally, experimental deliverables from the antifungal study implied that indolin-2-ones 4a–h possessed better antifungal than antibacterial potential. MIC values were in the range of 0.36–25 mg ml−1. 2-Pyridyl fragment (4g) has been crucial for very good anticandida activities. Indolin-2-one with pyridyl moiety showed the highest activity against C. albicans, C. krusei, and C. parapsilosis (Table 2). For all three Candida species have been achieved MIC values 3.1 mg ml−1. Compound 4d which contains 3-aminophenyl function has shown the highest activities against Cladosporium cladosporioides, C. albicans, P. italicum, and A. niger fungi strains. Considering results from antimicrobial screening, compounds 4d and 4g were used as the most active and were exposed to antiproliferative activity (Table 3).
The antiproliferative potential of selected molecules was investigated against both normal (MRC-5) and malignant cell lines (A549, LS174, MCF-7, and PaCa-2). Table 3 displays the experimental findings that are given as an IC50 value. While 4d and 4g have good activities against MCF-7, on other cancer cell lines they have a mild antiproliferative impact. 4d reached the lowest IC50 and consequently the highest antiproliferative action (18.42 ± 0.45 μM). Investigation on normal MRC-5 cell lines suggests no significant toxicity of both compounds 4d and 4g. A good selectivity index for 4d and 4g was achieved.
The antiproliferative evaluation of related AINDs revealed diverse actions that were strongly dependent on the studied compounds structures. The 6-methoxy group at the indolin-2-one ring that relates to the m-methoxybenzylidene motif showed very strong activity against MCF-7 cell lines (IC50 = 1.2 ± 0.4 μM), but without selectivity. Indolin-2-one with m-methoxy- or m-methylbenzylidene function has shown limited activity against Hepa1c1c7 cell lines (IC50 > 25 μM) due to the absence of an alkyl group at the nitrogen position.45 This is also in accordance with investigation of the Yang et al., where authors proposed that anticancer actions are strongly dependent by the presence of N-substituent.46 The addition of a N-aryl fragment to the structure of indolin-2-ones affects their anticancer activity. The activities against K562 cell lines were significantly improved in the presence of N-phenyl and -methoxy functions (IC50 = 31.68 μM). Despite this, the introduction of m-aminobenzylidene or 2-pyridyl and an allyl fragment in the indolin-2-one structure had a significant influence on biological activities (Tables 1–3). Compound 4d contains an m-aminobenzylidene motif and has demonstrated antibacterial and anticancer activity, particularly against MCF-7 cell lines (IC50 = 18.42 ± 0.45 μM). Furthermore, molecule 4g includes a pyridyl motif and demonstrated very good anticancer activity against the same cancer cell lines. In comparison to the positive control, cis-platinum, both compounds have higher and positive selectivity (Table 3). From structural point of view, m-aminobenzylidene (4d) or 2-pyridyl (4g) motif have the potential to make hydrogen bonds. It could be the reason for the significant difference between biological activities of 4d and 4g in comparison with other synthesized indolin-2-ones.
Interactions with biomacromolecules (DNA and BSA)
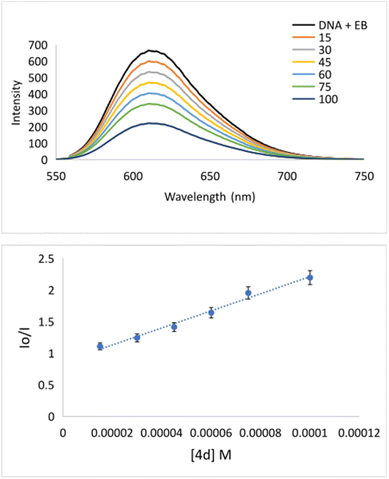
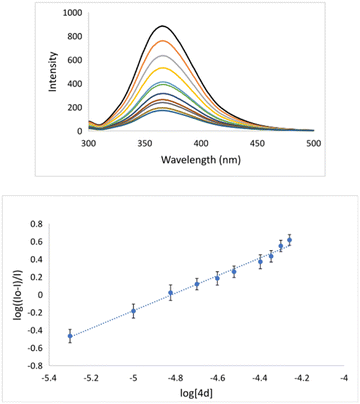
The interaction of potential pharmaceuticals with DNA can occur in several ways that generally depend on the type of structural motif. The most common interaction mode is intercalation. Under this process, molecules, or even ions are inserted between the base pairs of DNA. Many chemotherapeutics acts by intercalating into the DNA of cancer cells, disrupting the normal cell division process, and ultimately causing the death of cancer cell.47,48 Because of the importance of mode of action, the interaction of 4d with DNA was explored using fluorescence spectroscopy (Fig. 2). Apart from this, bovine serum albumin is a commonly used protein in biological research, and it is often used as a model carrier protein to study drug–protein interactions. The binding potential of pharmaceuticals to BSA can provide insights into how the molecule interacts with carrier proteins in the body. To bind a molecule to BSA, it is typically necessary to mix the potential drug with a solution of BSA and incubate the mixture for 12 h. During this time, the investigated compound will bind to the BSA forming drug–BSA complex species.The binding could be quantified using fluorescence spectroscopy.49 Considering the importance of understanding the binding of a drug to BSA, we have investigated the potential for the formation of a 4d–BSA complex employing the emission titration method (Fig. 4). All measurements were done in triplicate. During fluorescence titration of preformed DNA–EB species with quencher 4d emission spectra, they were recorded (Fig. 2). The fluorescence emissions of 4d were captured between 550 and 750 nm (Fig. 2). The intensity of the fluorescence emission line of DNA–EB significantly decreased during the addition of increasing concentrations of 4d to DNA–EB. This observed quenching implies that the molecules 4d and EB are competitive for DNA binding.
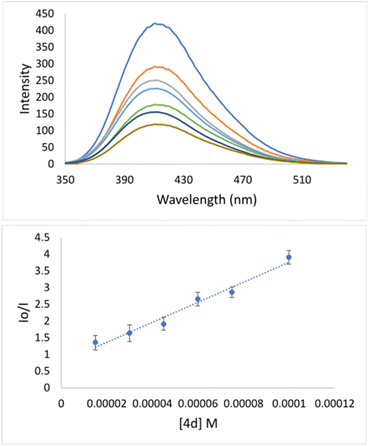
Specifically, the Stern–Volmer constant (KSV) indicates moderate binding to DNA. Even though 4d is quite a rigid and planar molecule, its surprisingly low KSV value indicates no intercalation action between base pairs on the major groove side. Therefore, low KSV combined with high kq indicates minor groove binding. The quenching constant (1.35 × 1012) has been higher than the maximum scattering collision quenching constant (2.0 × 1010 l mol−1) implying static quenching.50 Measured KSV of 4d implies its similar mechanism of binding to DNA as it was declared for well-known indole-based nonsteroidal anti-inflammatory drug Indomethacin.51 Going deeper inside of groove binding, minor groove binder Hoechst 33258 (Hoechst) has been applied.52 As can be seen from Fig. 3, after adding of 4d into DNA–Hoechst solution (blue line) emission intensity line decreased rapidly. Stern–Volmer constant is 3.05 × 104 M−1 (Table 4). The constant is higher in comparison with EB quencher (KSV = 1.35 × 104 M−1) indicating that Hoechst was replaced with 4d much easier then EB. The mentioned fact indicates minor groove binding of 4d.51
| KSV [M−1] | kq [M−1 s−1] | Ka [M−1] | n | ||
|---|---|---|---|---|---|
| EB | Hoechst | ||||
| 4d | 1.35 × 104 | 3.05 × 104 | 1.35 × 1012 | 6.1 × 106 | 1.1 |
The binding constant (Ka = 6.1 × 106 M−1) implies that 4d binds sufficiently strongly to BSA (Table 4 and Fig. 4). This experimental fact indicates that 4d can form the 4d–BSA cluster and should be successfully carried out by BSA to selected pharmacological targets.
Experimental
All necessary chemicals were supplied by Sigma Aldrich. The IR spectra were recorded by a PerkinElmer Spectrum One FT-IR spectrometer (USA) on a KBr pellet. Melting points (mp) were measured at Bibby Scientific Limited SMP10 (UK). Elemental analyses were performed using a Vario Elemental Analyser Leco CHNS-932 (UK). Fluorescence measurements were conducted at the RF-6000 PC spectrofluorometer (Shimadzu, Japan). NMR spectra were recorded on a Varian Gemini 200 MHz NMR spectrometer (USA) in DMSO-d6 as solvent. Signals are described as s = singlet, d = doublet, and m = multiplet. Proton and carbon NMR (Fig. S1–S18†), MS (Fig. S19–S26†), and IR spectra (Fig. S27–S34†) are given in ESI.† Bacteria, fungi, and cancer cell lines (A549, LS174, PaCa-2, and MCF-7) were from the American Type Culture Collection (ATCC). Antimicrobial activities were investigated by performing the broth dilution method.53 Concentrations of the investigated compounds were in the range of 25–0.0195 mg ml−1. The minimal inhibitory concentration (MIC) was established using 96-well microtiter plates.54 Cytotoxicity tests and MTT assays of selected compounds were done following the described protocol.22 All procedures were performed in triplicate. Cis-platinum, fluconazole, and streptomycin were used as references.Synthesis of indolin-2-one derivatives (4)
Allyl isatin (5 mmol) and an excess of various ketones (7 mmol) were dissolved in 30 ml of absolute ethanol. Diethylamine (0.5 ml) was used as a catalyst. Reactions were refluxed for 12–24 hours. Upon selected reaction time, reaction mixtures were evaporated. In the dark red reaction crude, 10 ml of freshly distilled glacial acetic acid and 1 ml of HCl (36%) were added and refluxed over 6 hours. The hot mixture was poured into the crushed ice/iPrOH mixture. The precipitated product was filtrated and washed with 50% iPrOH. 4a–h has been isolated in good yields. Short column was used for purification of samples 4a–f (eluent DCM). Experimental data for 4a–h are given below.Experimental data
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 5.39–5.16 (m, 2H,
CH), 5.39–5.16 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.31 (dt, J = 4.9, 1.7 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 183.2, 157.9, 150.5, 138.1, 131.4, 124.5, 123.3, 117.5, 111.2, 41.9 ppm.
CH2), 4.31 (dt, J = 4.9, 1.7 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 183.2, 157.9, 150.5, 138.1, 131.4, 124.5, 123.3, 117.5, 111.2, 41.9 ppm.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.77–7.68 (m, 1H, CHAr), 7.63–7.55 (m, 2H, CHAr), 7.44–7.35 (m, 1H, CHAr), 7.04–6.97 (m, 2H, CHAr), 5.97–5.78 (m, 1H,
CH), 7.77–7.68 (m, 1H, CHAr), 7.63–7.55 (m, 2H, CHAr), 7.44–7.35 (m, 1H, CHAr), 7.04–6.97 (m, 2H, CHAr), 5.97–5.78 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.24–5.14 (m, 2H,
CHallyl), 5.24–5.14 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.39–4.37 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 191.3, 166.5, 144.9, 136.9, 135.0, 134.2, 132.7, 131.8, 129.2, 128.7 (2C), 127.1 (2C), 126.4, 122.4, 119.4, 117.1, 109.8, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 290; elemental analysis (%): cacld for C19H15NO2: C 78.87; H 5.23; N 4.84; found: C 78.64; H 5.12; N 4.69.
CH2), 4.39–4.37 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 191.3, 166.5, 144.9, 136.9, 135.0, 134.2, 132.7, 131.8, 129.2, 128.7 (2C), 127.1 (2C), 126.4, 122.4, 119.4, 117.1, 109.8, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 290; elemental analysis (%): cacld for C19H15NO2: C 78.87; H 5.23; N 4.84; found: C 78.64; H 5.12; N 4.69.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.41–7.32 (m, 3H, CHAr), 7.03–6.96 (m, 2H, CHAr), 6.01–5.78 (s, 1H,
CH), 7.41–7.32 (m, 3H, CHAr), 7.03–6.96 (m, 2H, CHAr), 6.01–5.78 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.23–5.14 (m, 2H, CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2), 2.39 (s, 3H, CH3) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.8, 166.6, 144.9, 144.8, 134.7, 134.5, 132.6, 131.8, 129.8 (2C), 128.8 (2C), 127.4, 126.3, 122.3, 119.4, 117.1, 109.7, 41.9, 21.5 ppm; ESI-MS (m/z): [M + H]+ = 304; cacld for C20H17NO2: C 79.19; H 5.65; N 4.62; found: C 79.02; H 5.48; N 4.50.
CHallyl), 5.23–5.14 (m, 2H, CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2), 2.39 (s, 3H, CH3) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.8, 166.6, 144.9, 144.8, 134.7, 134.5, 132.6, 131.8, 129.8 (2C), 128.8 (2C), 127.4, 126.3, 122.3, 119.4, 117.1, 109.7, 41.9, 21.5 ppm; ESI-MS (m/z): [M + H]+ = 304; cacld for C20H17NO2: C 79.19; H 5.65; N 4.62; found: C 79.02; H 5.48; N 4.50.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH + CHAr), 7.45–7.35 (m, 1H, CHAr), 7.06–6.98 (m, 2H, CHAr), 6.02–5.78 (s, 1H,
CH + CHAr), 7.45–7.35 (m, 1H, CHAr), 7.06–6.98 (m, 2H, CHAr), 6.02–5.78 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.23–5.14 (m, 2H, CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.3, 166.5, 145.1, 136.0, 135.5, 132.9 (2C), 132.2 (2C), 131.8, 130.6, 128.4, 126.6, 126.4, 122.4, 119.4, 117.2, 109.8, 41.9 ppm; ESI-MS (m/z): [M]+ = 368; cacld for C19H14BrNO2: C 61.97; H 3.83; N 3.80; found: C 61.70; H 3.64; N 3.72.
CHallyl), 5.23–5.14 (m, 2H, CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.3, 166.5, 145.1, 136.0, 135.5, 132.9 (2C), 132.2 (2C), 131.8, 130.6, 128.4, 126.6, 126.4, 122.4, 119.4, 117.2, 109.8, 41.9 ppm; ESI-MS (m/z): [M]+ = 368; cacld for C19H14BrNO2: C 61.97; H 3.83; N 3.80; found: C 61.70; H 3.64; N 3.72.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.66–7.60 (m, 2H, CHAr), 7.45–7.37 (m, 1H, CHAr), 7.06–6.98 (m, 2H, CHAr), 5.96–5.78 (m, 1H,
CH), 7.66–7.60 (m, 2H, CHAr), 7.45–7.37 (m, 1H, CHAr), 7.06–6.98 (m, 2H, CHAr), 5.96–5.78 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.22–5.13 (m, 2H,
CHallyl), 5.22–5.13 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.38 (d, J = 5.0 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.4, 166.6, 145.1, 138.1, 136.0, 135.6, 133.0, 131.8, 130.6, 127.3, 126.6, 126.5, 126.2, 122.5, 121.4, 119.4, 117.2, 109.9, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 305; cacld for C19H16N2O2: C 74.98; H 5.30; N 9.20; found: C 74.82; H 5.15; N 9.28.
CH2), 4.38 (d, J = 5.0 Hz, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.4, 166.6, 145.1, 138.1, 136.0, 135.6, 133.0, 131.8, 130.6, 127.3, 126.6, 126.5, 126.2, 122.5, 121.4, 119.4, 117.2, 109.9, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 305; cacld for C19H16N2O2: C 74.98; H 5.30; N 9.20; found: C 74.82; H 5.15; N 9.28.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH + CHAr), 7.48–7.39 (m, 1H, CHAr), 7.08–6.99 (m, 2H, CHAr), 6.04–5.79 (m, 1H,
CH + CHAr), 7.48–7.39 (m, 1H, CHAr), 7.08–6.99 (m, 2H, CHAr), 6.04–5.79 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.24–5.15 (m, 2H,
CHallyl), 5.24–5.15 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.40–4.38 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 189.2, 166.5, 148.2, 145.4, 138.2, 136.5, 134.8, 133.4, 131.7, 131.0, 128.1, 127.0, 125.4, 122.9, 122.5, 119.3, 117.2, 109.8, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 335; cacld for C19H14N2O4: C 68.26; H 4.22; N 8.38; found: C 68.10; H 4.20; N 8.25.
CH2), 4.40–4.38 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 189.2, 166.5, 148.2, 145.4, 138.2, 136.5, 134.8, 133.4, 131.7, 131.0, 128.1, 127.0, 125.4, 122.9, 122.5, 119.3, 117.2, 109.8, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 335; cacld for C19H14N2O4: C 68.26; H 4.22; N 8.38; found: C 68.10; H 4.20; N 8.25.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.69–7.63 (m, 1H, CHAr), 7.54–7.26 (m, 4H, CHAr), 7.05–6.97 (m, 2H, CHAr), 6.02–5.76 (m, 1H,
CH), 7.69–7.63 (m, 1H, CHAr), 7.54–7.26 (m, 4H, CHAr), 7.05–6.97 (m, 2H, CHAr), 6.02–5.76 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.23–5.14 (m, 2H,
CHallyl), 5.23–5.14 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2), 3.84 (s, 3H, OCH3) ppm; 13C NMR (50 MHz, DMSO-d6) δ 191.0, 166.6, 159.7, 145.0, 138.3, 135.1, 132.8, 131.8, 130.4, 127.1, 126.4, 122.4, 121.5, 120.4, 119.4, 117.2, 112.6, 109.8, 55.6, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 320; cacld for C20H17NO3: C 75.22; H 5.37; N 4.39; found: C 75.12; H 5.26; N 4.21.
CH2), 4.37 (d, J = 5.0 Hz, 2H, N–CH2), 3.84 (s, 3H, OCH3) ppm; 13C NMR (50 MHz, DMSO-d6) δ 191.0, 166.6, 159.7, 145.0, 138.3, 135.1, 132.8, 131.8, 130.4, 127.1, 126.4, 122.4, 121.5, 120.4, 119.4, 117.2, 112.6, 109.8, 55.6, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 320; cacld for C20H17NO3: C 75.22; H 5.37; N 4.39; found: C 75.12; H 5.26; N 4.21.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 8.14–8.07 (m, 2H, CHAr), 7.74–7.72 (m, 1H, CHAr), 7.43–7.39 (m, 1H, CHAr), 7.06–6.96 (m, 2H, CHAr), 5.96–5.77 (m, 1H,
CH), 8.14–8.07 (m, 2H, CHAr), 7.74–7.72 (m, 1H, CHAr), 7.43–7.39 (m, 1H, CHAr), 7.06–6.96 (m, 2H, CHAr), 5.96–5.77 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.22–5.13 (m, 2H,
CHallyl), 5.22–5.13 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.38–4.35 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.0, 166.9, 153.4, 149.3, 145.5, 138.0, 136.5, 133.4, 131.8, 128.2, 127.6, 124.9, 122.6, 122.5, 119.7, 117.1, 109.7, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 291; cacld for C18H14N2O2: C 74.47; H 4.86; N 9.65; found: C 74.28; H 4.71; N 9.49.
CH2), 4.38–4.35 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 190.0, 166.9, 153.4, 149.3, 145.5, 138.0, 136.5, 133.4, 131.8, 128.2, 127.6, 124.9, 122.6, 122.5, 119.7, 117.1, 109.7, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 291; cacld for C18H14N2O2: C 74.47; H 4.86; N 9.65; found: C 74.28; H 4.71; N 9.49.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.43 (td, J = 7.8, 1.3 Hz, 1H, CHAr), 7.32 (dd, J = 4.9, 3.9 Hz, 1H, CHAr), 7.09–6.98 (m, 2H, CHAr), 5.97–5.78 (m, 1H,
CH), 7.43 (td, J = 7.8, 1.3 Hz, 1H, CHAr), 7.32 (dd, J = 4.9, 3.9 Hz, 1H, CHAr), 7.09–6.98 (m, 2H, CHAr), 5.97–5.78 (m, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHallyl), 5.23–5.13 (m, 2H,
CHallyl), 5.23–5.13 (m, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.40–4.36 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 182.6, 166.6, 145.2, 144.9, 137.0, 135.8, 134.7, 133.1, 131.8, 129.4, 127.4, 125.5, 122.4, 119.5, 117.1, 109.7, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 296; cacld for C17H14NO2S: C 68.90; H 4.76; N 4.73; found: C 68.82; H 4.57; N 4.63.
CH2), 4.40–4.36 (m, 2H, N–CH2) ppm; 13C NMR (50 MHz, DMSO-d6) δ 182.6, 166.6, 145.2, 144.9, 137.0, 135.8, 134.7, 133.1, 131.8, 129.4, 127.4, 125.5, 122.4, 119.5, 117.1, 109.7, 41.9 ppm; ESI-MS (m/z): [M + H]+ = 296; cacld for C17H14NO2S: C 68.90; H 4.76; N 4.73; found: C 68.82; H 4.57; N 4.63.MTT assay
The inhibitory action of selected compounds on the cell proliferation of the tested cancer cell lines was assessed using the MTT assay. For this testing, mitochondrial succinate dehydrogenase was used to convert yellow MTT to purple formazan in accordance with Mosmann55 and Ohno and Abe56 72 h after the addition of selected compounds. The cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum. In an incubator at 37 °C and 5% CO2, streptomycin, and penicillin (100 g ml−1 and 100 units per ml, respectively) were added. Cancer cells were grown in 96-well plates for two days in 5% CO2 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well). The cell lines were cultured for 24 hours after being exposed to various doses of the investigated chemicals. DMSO content never exceeded 0.5%, which was non-toxic to the cells. In an independent experiment, antineoplastic drug cis-platin was used as a positive control. The absorbance of treated samples was measured at 570 nm.
000 cells per well). The cell lines were cultured for 24 hours after being exposed to various doses of the investigated chemicals. DMSO content never exceeded 0.5%, which was non-toxic to the cells. In an independent experiment, antineoplastic drug cis-platin was used as a positive control. The absorbance of treated samples was measured at 570 nm.
Fluorescence titration of DNA and BSA
ctDNA, and BSA (bovine serum albumin) and ethidium bromide (EB) were purchased from Sigma, while buffer phosphate buffered saline (PBS) was secured by Fisher BioReagents. A stock solution of ctDNA and EB or Hoechst in double distilled water was prepared in a selected buffer (10 mM PBS) that provides pH = 7.4. Before titration started, it was necessary to check the concentration of the stock DNA solution. The solution gave a ratio of UV absorbances at 260 and 280 nm (A260/A280) of cca. 1.8–1.9, implying that the DNA was adequately free of proteins.DNA and EB or Hoechst concentrations were equal (100 μM). The concentration of 4d varied from 0 to 100 μM (15, 30, 45, 60, 75, and 100 μM). A series of solutions was prepared by mixing up equal volumes of DNA and EB or Hoechst, then adding an increasing volume of 4d, and finally, adding PBS to mark in a volumetric flask (10 ml). The incubation time was 24 hours. The fluorescence spectra were measured in the range 550–750 nm or 350–530 nm upon excitation at 520 or 340 nm for DNA–EB or DNA–Hoechst species, respectively. Results were analyzed using Stern–Volmer eqn (1):57
| I0/I = 1 + KSV[Q] | (1) |
A stock BSA solution (10 μM) was prepared in buffer (10 mM PBS). Serie of 4d–BSA solutions (5–50 μM) were prepared and stored in a dark place for 12 hours at room temperature. The molar ratio of 4d![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA was varied in order 0
BSA was varied in order 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 1
1; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 2.0
1; 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3.0
1, 3.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 4.0
1; 4.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 5.0
1; 5.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 6.0
1; 6.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 7.0
1; 7.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 8.0
1; 8.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 9.0
1; 9.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; and 10.0
1; and 10.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the volumetric flask of 10.0 ml. The concentration of BSA was 5.0 μM. The fluorescence spectra were measured between 300 and 500 nm upon excitation at 287 nm. Fluorescence emission titration data were evaluated utilizing eqn (2):58
1 in the volumetric flask of 10.0 ml. The concentration of BSA was 5.0 μM. The fluorescence spectra were measured between 300 and 500 nm upon excitation at 287 nm. Fluorescence emission titration data were evaluated utilizing eqn (2):58
log((I0 − I)/I) = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q]
| (2) |
Conclusions
Several indoline-2-one (4a–h) derivatives were synthesized and analysed using NMR, IR, ESI-MS, and elemental analysis. Isolated yields of 4a–h were moderate to good (up to 81%). The presence of a heteroaromatic core (thienyl or pyridyl) influenced the yield significantly. All indolin-2-ones were tested for antibacterial activity against Gram-positive and Gram-negative bacteria as well as fungi. Gram-negative K. pneumoniae strains were far more sensitive than other examined bacteria. The best antibacterial activity had molecules 4f–h (MIC = 3.1 mg ml−1). Despite this, the examined indolin-2-ones had better antifungal than antibacterial activity. C. albicans, P. italicum, and A. niger were particularly sensitive after treatment with molecules that have the m-aminophenyl (4d) or pyridin-2-yl (4g) functions. These indolin-2-ones were chosen for investigation of their antiproliferative effect on cancer (LS174, MCF-7, PaCa-2, and A549) and normal cell lines (MRC-5). Investigated molecules possessed moderate-to-good activity against MCF-7 cell lines. Compound 4d has the lowest IC50 value (18.42 ± 0.45 μM) and consequently the strongest antiproliferative potential with a selectivity index ∼6. 4d presents a promising compound with good potential for dual activity (antifungal and antiproliferative). Nevertheless, the potential of indolin-2-ones (4d and 4g) as anticandidal and anticancer agents is encouraging, and their further development may lead to the discovery of new dual-active drugs for the treatment of candidiasis combined with cancer. Overall evidence also suggests that indoline-2-ones containing arylidene and N-allyl fragments may be useful for the development of new dual-active pharmaceuticals or even dual-active polymers.Author contributions
Conceptualization, writing original draft preparation and supervision S. N. A. B., and N. J.; synthesis, characterization, and binding study S. N. A. B., and N. J.; methodology and visualization H. E., T. G. A., N. A., W. A., M. A. E., N. H. A., and K. J. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Deanship of Scientific Research at Jouf University under Grant Number (DSR2022-RG-0148).Notes and references
- R. Nath, S. Pathania, G. Grover and J. M. Akhtar, J. Mol. Struct., 2020, 1222, 128900 CrossRef CAS.
- V. Sonam and R. Kakkar, Med. Chem. Commun., 2019, 10, 351–368 RSC.
- M. A. Borad, M. N. Bhoi, N. P. Prajapati and H. D. Patel, Synth. Commun., 2014, 44, 1043–1057 CrossRef CAS.
- M. Yoshikawa, T. Murakami, A. Kishi, T. Sakurama, H. Matsuda, M. Nomura, H. Matsuda and M. Kubo, Chem. Pharm. Bull., 1998, 46, 886–888 CrossRef CAS PubMed.
- G. S. Singh, Z. Yibralign, D. Bahir and Z. Y. Desta, Chem. Rev., 2012, 112, 6104–6155 CrossRef CAS PubMed.
- G. J. Kapadia and Y. N. Shukla, Planta Med., 1993, 59, 568–569 CrossRef CAS PubMed.
- U. Gräfe and L. Radics, J. Antibiot., 1986, 39, 162–163 CrossRef.
- I. Chiyanzu, E. Hansell, J. Gut, P. J. Rosenthal, J. H. McKerrow and K. Chibale, Bioorg. Med. Chem. Lett., 2003, 13, 3527–3530 CrossRef CAS.
- H. M. Abo-Salem, A. Nassrallah, A. A. F. Soliman, M. S. Ebied, M. E. Elawady, S. A. Abdelhamid, E. R. El-Sawy, Y. A. Al-Sheikh and M. A. M. Aboul-Soud, Molecules, 2020, 25, 1124 CrossRef CAS PubMed.
- S. S. Dinavahi, R. Gowda, C. G. Bazewicz, M. B. Battu, J. M. Lin, R. J. Chitren, M. K. Pandey, S. Amin, G. P. Robertson and K. Gowda, Eur. J. Med. Chem., 2020, 187, 111962 CrossRef CAS PubMed.
- C. Karthikeyan, V. R. Solomon, H. Lee and P. Trivedi, Biomed. Prev. Nutr., 2013, 3, 325–330 CrossRef.
- S. Zeeshan, M. Naveed, A. Khan, A. Atiq, M. Arif, M. N. Ahmed, Y. S. Kim and S. Khan, Inflammation Res., 2019, 68, 613–632 CrossRef CAS PubMed.
- M. M. Ibrahim, T. Elsaman and M. Y. Al-nour, Int. J. Med. Chem., 2018, 2018, 1–11 Search PubMed.
- P. Selvam, M. Chandramohan, E. De Clercq, M. Witvrouw and C. Pannecouque, Eur. J. Pharm. Sci., 2001, 14, 313–316 CrossRef CAS.
- S. N. Pandeya, P. Yogeeswari, D. Sriram, E. De Clercq, C. Pannecouque and M. Witvrouw, Chemotherapy, 1999, 45, 192–196 CrossRef CAS PubMed.
- S. N. Pandeya, D. Sriram, G. Nath and E. De Clercq, Arzneimittelforschung, 2000, 50, 55–59 CAS.
- M. Efremov, M. D. Tsymlyakov, L. V. Anikina, O. Yu, V. Borisov, V. N. Perfilova, I. N. Tyurenkov and M. V. Proskurnina, Bioorg. Med. Chem., 2019, 27, 1804–1817 CrossRef.
- G. Wang, M. Chen, J. Qiu, Z. Xie and A. Cao, Bioorg. Med. Chem. Lett., 2018, 28, 113–116 CrossRef CAS PubMed.
- F. Rahim, F. Malik, H. Ullah, A. Wadood, F. Khan, M. T. Javid, M. Taha, W. Rehman, A. U. Rehman and K. M. Khan, Bioorg. Chem., 2015, 60, 42–48 CrossRef CAS PubMed.
- S. George, Acta Pharm., 2008, 58, 119–129 CAS.
- H. K. Mahmoud, T. A. Farghaly, H. G. Abdulwahab, N. T. Al-Qurashi and M. R. Shaaban, Eur. J. Med. Chem., 2020, 208, 112752 CrossRef CAS.
- M. A. Abdelgawad, A. M. Hayallah, S. N. A. Bukhari, A. Musa, M. Elmowafy, H. M. Abdel-Rahman and M. K. Abd El-Gaber, Pharmaceuticals, 2022, 15, 1416 CrossRef CAS.
- I. Skaraitė, E. Maccioni and V. Petrikaitė, Int. J. Mol. Sci., 2023, 24, 5422 CrossRef.
- M. Nalli, M. Puxeddu, G. La Regina, S. Gianni and R. Silvestri, Molecules, 2021, 26, 7463 CrossRef CAS.
- M. Moran, D. Nickens, K. Adcock, M. Bennetts, A. Desscan, N. Charnley and K. Fife, Target. Oncol., 2019, 14, 405–416 CrossRef.
- A. H. Case and P. Johnson, BMJ Open Respir. Res., 2017, 4, e000192 CrossRef PubMed.
- K. Mross, M. Stefanic, D. Gmehling, A. Frost, F. Baas, C. Unger, R. Strecker, J. Henning, B. Gaschler-Markefski, P. Stopfer, L. de Rossi and R. Kaiser, Clin. Cancer Res., 2009, 16, 311–319 CrossRef.
- T. Eisen, Y. Shparyk, N. MacLeod, R. Jones, G. Wallenstein, G. Temple, Y. Khder, C. Dallinger, M. Studeny, A.-B. Loembe and I. Bondarenko, Invest. New Drugs, 2013, 31, 1283–1293 CrossRef CAS PubMed.
- A. Luczywo, L. G. González, A. C. C. Aguiar, J. Oliveira de Souza, G. Oliva, L. F. Aguilar, J. J. Casal, R. V. C. Guido, S. E. Asís and M. Mellado, Nat. Prod. Res., 2022, 36, 3887–3893 CrossRef CAS PubMed.
- S. Soleimani-Amiri and M. Koohi, J. Phys. Org. Chem., 2019, 32, e3929 CrossRef.
- W. Chu, D. Zhou, V. Gaba, J. Liu, S. Li, X. Peng, J. Xu, D. Dhavale, D. P. Bagchi, A. d'Avignon, N. B. Shakerdge, B. J. Bacskai, Z. Tu, P. T. Kotzbauer and R. H. Mach, J. Med. Chem., 2015, 58, 6002–6017 CrossRef CAS PubMed.
- S. Kumar Suthar, S. Bansal, M. M. Alam, V. Jaiswal, A. Tiwari, A. Chaudhary, A. Treasa Alex and A. Joseph, Bioorg. Med. Chem. Lett., 2015, 25, 5281–5285 CrossRef.
- S. Princiotto, L. Musso, F. Manetti, V. Marcellini, G. Maga, E. Crespan, C. Perini, N. Zaffaroni, G. Luca Beretta and S. Dallavalle, J. Enzyme Inhib. Med. Chem., 2022, 37, 2382–2394 CrossRef CAS PubMed.
- G. Çoban and F. Aydın Köse, Saudi Pharm. J., 2019, 27, 952–967 CrossRef PubMed.
- K. R. Senwar, T. S. Reddy, D. Thummuri, P. Sharma, V. G. M. Naidu, G. Srinivasulu and N. Shankaraiah, Eur. J. Med. Chem., 2016, 118, 34–46 CrossRef CAS PubMed.
- K. R. Senwar, T. S. Reddy, D. Thummuri, P. Sharma, S. K. Bharghava, V. G. M. Naidu and N. Shankaraiah, Bioorg. Med. Chem. Lett., 2016, 26, 4061–4069 CrossRef CAS.
- N. Janković, J. Muškinja, Z. Ratković, Z. Bugarčić, B. Ranković, M. Kosanić and S. Stefanović, RSC Adv., 2016, 6, 39452–39459 RSC.
- S. N. A. Bukhari, M. A. Abdelgawad, N. Ahmed, M. W. Amjad, M. A. Hussain, M. A. Elsherif, H. Ejaz, N. H. Alotaibi, I. Filipović and N. Janković, Pharmaceuticals, 2023, 16, 281 CrossRef CAS PubMed.
- B. G. M. Youssif, M. H. Abdelrahman, A. H. Abdelazeem, M. A. Abdelgawad, H. M. Ibrahim, O. I. A. Salem, M. F. A. Mohamed, L. Treambleau and S. N. A. Bukhari, Eur. J. Med. Chem., 2018, 146, 260–273 CrossRef CAS PubMed.
- V. B. M. Brito, G. F. Santos, T. D. S. Silva, J. L. C. Souza, G. C. G. Militão, F. T. Martins, F. P. L. Silva, B. G. Oliveira, E. C. C. Araújo, M. L. A. A. Vasconcellos, C. G. Lima-Júnior and E. B. Alencar-Filho, Mol. Diversity, 2020, 24, 265–281 CrossRef CAS PubMed.
- K. Nakanishi, G. Toshio and O. Mamoru, Bull. Chem. Soc. Jpn., 1957, 30, 403–408 CrossRef CAS.
- N. Hu, Y.-P. Tu, Y. Liu, K. Jiang and Y. Pan, J. Org. Chem., 2008, 73, 3369–3376 CrossRef CAS.
- S. C. Turner and G. Butler, Cold Spring Harbor Perspect. Med., 2014, 4, a019778 CrossRef.
- P. G. Pappas, M. S. Lionakis, M. C. Arendrup, L. Ostrosky-Zeichner and B. J. Kullberg, Nat. Rev. Dis. Primers, 2018, 4, 18026 CrossRef.
- W. Zhang and M.-L. Go, Bioorg. Med. Chem., 2009, 17, 2077–2090 CrossRef CAS PubMed.
- Y. Yang, B. Song, K. Han, L. Zhang, D. Wu, X. Qu, H. Li, H. Sun, Y. Teng and P. Yu, J. Chem. Pharm. Res., 2014, 6, 140–145 CAS.
- https://atdbio.com/nucleic-acids-book/Nucleic-acid-drug-interactions#Introduction.
- A. Sánchez-González, T. G. Castro, M. Melle-Franco and A. Gil, Phys. Chem. Chem. Phys., 2021, 23, 26680–26695 RSC.
- A. Sulkovska, J. Mol. Struct., 2002, 614, 227–232 CrossRef.
- Y. Wu and G. Yang, Spectrosc. Lett., 2010, 43, 28–35 CrossRef CAS.
- M. A. Husain, H. M. Ishqi, T. Sarwar, S. Ur Rehman and M. Tabish, Med. Chem. Commun., 2017, 8, 1283–1296 RSC.
- L. H. Fornander, L. Wu, M. Billeter, P. Lincoln and B. Nordén, J. Phys. Chem. B, 2013, 117, 5820–5830 CrossRef CAS PubMed.
- H. Ejaz, M. Sultan, M. U. Qamar, K. Junaid, N. Rasool, A. Alanazi, M. W. Alruways, B. B. Z. Mazhari, Y. Alruwaili, S. N. A. Bukhari and S. Younasa, BMC Complementary Med. Ther., 2023, 23, 42 CrossRef CAS.
- S. D. Sarker, L. Nahar and Y. Kumarasamy, Methods, 2007, 42, 321 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS.
- M. Ohno and T. Abe, J. Immunol. Methods, 1991, 145, 199–203 CrossRef CAS PubMed.
- M. H. Gehlen, J. Photochem. Photobiol., C, 2020, 42, 100338 CrossRef CAS.
- L. L. He, X. Wang, B. Liu, J. Wang and Y. G. Sun, J. Solution Chem., 2010, 39, 654–664 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04997c |
| This journal is © The Royal Society of Chemistry 2023 |