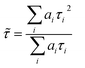Open Access Article
Open Access ArticleDesigning a green-emitting viscosity-sensitive 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY) probe for plasma membrane viscosity imaging†
Artūras Polita*a,
Milda Stancikaitėb,
Rokas Žvirblisc,
Karolina Maleckaitė b,
Jelena Dodonova-Vaitkūnienė
b,
Jelena Dodonova-Vaitkūnienė d,
Sigitas Tumkevičius
d,
Sigitas Tumkevičius d,
Arun Prabha Shivabalana and
Gintaras Valinčiusa
d,
Arun Prabha Shivabalana and
Gintaras Valinčiusa
aInstitute of Biochemistry, Life Sciences Center, Vilnius University, Saulėtekio Av. 7, Vilnius, LT-10257, Lithuania. E-mail: arturas.polita@gmc.vu.lt
bCenter of Physical Sciences and Technology, Saulėtekio Av. 3, Vilnius, LT-10257, Lithuania
cLife Sciences Center, Institute of Biotechnology, Vilnius University, Saulėtekio Av. 7, Vilnius, LT-10257, Lithuania
dInstitute of Chemistry, Faculty of Chemistry and Geosciences, Vilnius University, Naugarduko St. 24, Vilnius, LT-03225, Lithuania
First published on 26th June 2023
Abstract
Viscosity is a key characteristic of lipid membranes – it governs the passive diffusion of solutes and affects the lipid raft formation and membrane fluidity. Precise determination of viscosity values in biological systems is of great interest and viscosity-sensitive fluorescent probes offer a convenient solution for this task. In this work we present a novel membrane-targeting and water-soluble viscosity probe BODIPY-PM, which is based on one of the most frequently used probes BODIPY-C10. Despite its regular use, BODIPY-C10 suffers from poor integration into liquid-ordered lipid phases and lack of water solubility. Here, we investigate the photophysical characteristics of BODIPY-PM and demonstrate that solvent polarity only slightly affects the viscosity-sensing qualities of BODIPY-PM. In addition, with fluorescence lifetime imaging microscopy (FLIM), we imaged microviscosity in complex biological systems – large unilamellar vesicles (LUVs), tethered bilayer membranes (tBLMs) and live lung cancer cells. Our study showcases that BODIPY-PM preferentially stains the plasma membranes of live cells, equally well partitions into both liquid-ordered and liquid-disordered phases and reliably distinguishes lipid phase separation in tBLMs and LUVs.
Introduction
Viscosity is the essential physical characteristic of cell membranes – it controls diffusion coefficients of lipids and macromolecules and influences the passive transport of solutes across the plasma membrane.1 Furthermore, cell membranes are microscopically heterogeneous and are able to separate into liquid-ordered (Lo) and liquid-disordered (Ld) phases that possess distinct viscosities.2–5 Viscous Lo phases are of particular biological importance – ordered microdomains of lipids and proteins, so called lipid rafts, play a key role in immune signaling,6,7 host–pathogen interactions,8,9 cardiovascular diseases,10 and cancer.11–13 Thus, the ability to distinguish Lo and Ld phases in live cells would benefit studies of membrane dynamics and biochemical processes associated with lipid raft formation. Fortunately, lipids in the Lo phase are packed tightly, and the Lo phase is much more viscous compared to the Ld phase, thus the two phases can be distinguished on the basis of viscosity.14,15 Precise determination of viscosity values in Lo and Ld phases would allow comparison of lipid packaging efficiencies and molecular order changes in the plasma membrane.Accurate mechanical viscosity measurements in micron-scale objects are inherently difficult. Additionally, biological specimens are highly heterogeneous and viscosity values in protein-rich Lo domains will differ from Ld domains or the cytoplasm.16 Precise and quantitative viscosity measurements on the small scale can be made utilizing fluorescent class of compounds called molecular rotors.17–19 Molecular rotors, when paired with fluorescence lifetime imaging microscopy (FLIM), are able to produce spatial microviscosity maps.20–22 The term microviscosity here refers to the lipid packaging and molecular mobility of the probe's local environment.23
In the excited state, molecular rotors undergo intramolecular rotation, which results in a molecular rotor leaving the fluorescent state.24 As the rate of intramolecular rotation is directly influenced by microviscosity, molecular rotors display great fluorescence lifetime and fluorescence intensity sensitivity to local viscosity values.25 In high viscosity environments, intramolecular motion of molecular rotors is restricted and radiative decay dominates, leading to increased fluorescence lifetime values and high quantum yields.25 Conversely, in low viscosity environments, non-radiative decay dominates, causing the decrease in fluorescence lifetime and quantum yield. Since the fluorescence intensity is dependent on the local concentration of a fluorophore, it is particularly advantageous to evaluate viscosity values from the fluorescence lifetime, which is independent of the fluorophore concentration, and is largely unaffected by optical properties of the medium or the excitation setup. Fluorescence lifetimes have been used to quantify microviscosities of model lipid bilayer systems, as well as live eukaryotic and prokaryotic cells through FLIM.26–31
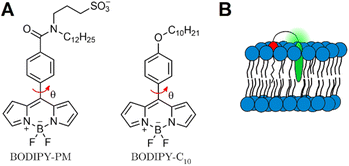
Currently, BODIPY-C10/12 (Fig. 1A) is one of the most extensively used molecular rotors for viscosity sensing.32,33 Previously, BODIPY-C10/12 and its derivatives were used to quantify viscosity in polymers,34 model lipid membranes,35 live cells,36,37 plasma membrane,38 mitochondria,39 and lysosomes.40 Although BODIPY-C10 has a monoexponential fluorescence decay,41 which greatly simplifies data analysis, this rotor lacks affinity for the higher ordered lipid phases,42 which decreases its ability to sense the Lo phase. Due to poor partitioning of BODIPY-C10 into Lo phases, small Lo domains (e.g. lipid rafts) are easily missed in biological samples, and especially in live cells.35
Moreover, BODIPY-C10 suffers from unclear localisation of the probe in higher ordered lipid bilayers42,43 – around 10–15% of BODIPY molecules are localised in such a way that viscosity changes in a bilayer do not alter their intramolecular rotation.35 Ultimately, this lowers the average lifetime values and reduces the probe's sensitivity to viscosity changes.
In this work, we present a novel BODIPY derivative probe, BODIPY-PM, which carries a long hydrophobic plasma membrane (PM) targeting group (C12H25) together with a polar sulfonate group SO3−. The combination of a charged sulfonate group with a hydrophobic tail and the non-polar BODIPY-core, mimics natural phospholipid structures and enables firm positioning of BODIPY-PM into the lipid bilayer (Fig. 1B). We explore the photophysics of BODIPY-PM and its ability to distinguish Lo and Ld membrane phases in large unilamellar vesicles (LUVs), tethered bilayer lipid membranes (tBLMs) and live cancer cells. LUVs and tBLMs represent one of the most frequently used model lipid bilayer systems and are often employed to study and mimic cell membranes.44 Specifically, by comparing fluorescence lifetimes of BODIPY-PM in solvents of different polarities and viscosities, we show that BODIPY-PM is affected mainly by the viscosity of the solvent. By performing time-resolved fluorescence measurements in LUVs with Lo and Ld lipid compositions, we demonstrate that BODIPY-PM successfully distinguishes lipid order and displays vastly different fluorescence lifetimes in Lo and Ld LUVs. Furthermore, with the help of FLIM, we investigate the potential of BODIPY-PM to distinguish separate Lo domains in tBLMs and LUVs. Finally, by carrying out FLIM measurements on live lung cancer cells, we show that BODIPY-PM preferentially localises in the plasma membrane and is efficient at recognising viscous Lo phases in the lipid bilayer. To our knowledge, among comparable viscosity probes, BODIPY-PM achieves the largest fluorescence lifetime contrast between the Lo and Ld phases. The narrow emission spectrum of BODIPY-PM allows for the multiple dye usage in biological samples and offers a convenient alternative to Kuimova's thiophene27,45 or Klymchenko's solvatochromic dyes.46
Results and discussion
Probe design
Our BODIPY-PM design was based on one of the most applicable and successful rotors up to date – BODIPY-C10. Our intent was to retain the same mechanism of viscosity sensitivity of BODIPY-C10, thus we decided to keep the rotation of the phenyl group versus the BODIPY unhindered and modified the para-position of the phenyl group. Our aim was to enable the firm positioning of the probe inside highly ordered lipid phases, increase the water solubility of the probe and decrease the possibility of dye aggregation in water. Consequently, we elongated the hydrophobic tail (C12H25) and introduced a sulfonate group – this combination of substituents mimics the structure of lipids, with polar sulfonate group emerging from the bilayer and hydrophobic tail serving as a membrane anchor. In addition to conferring increased water solubility, the sulfonate group also disrupts dye–dye interactions and confers resistance to probe aggregation. Finally, the C12H25 side chain grants the dye high affinity for ordered lipid bilayer phases and cell membranes. The synthesis of BODIPY-PM is described in the ESI.†Absorption, steady-state and time-resolved fluorescence
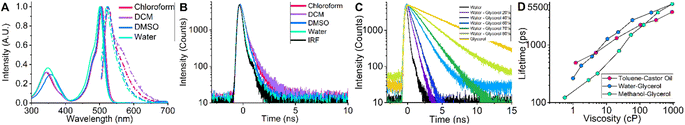
In order to investigate the spectroscopic properties of BODIPY-PM, we carried out absorption, steady-state, and time-resolved fluorescence experiments in different polarity solvents from non-polar [chloroform and dichloromethane (DCM)], to very polar [dimethyl sulfoxide (DMSO) and water] (Fig. 2). The absorption spectra of BODIPY-PM consist of a low intensity band at 300–400 nm and the main band in the 450–550 nm region with maximum positioned at 503 nm (Fig. 2A). The fluorescence spectra of BODIPY-PM feature a small bathochromic shift with the fluorescence peak at 523 nm and a Stokes shift of 760 cm−1 (Fig. 2A). Additionally, BODIPY-PM displays almost no fluorescence solvatochromism, resulting in a blue shift of only 3 nm when going from the least polar chloroform to the most polar water solvent (Fig. 2A). We have also observed that in very non-polar solvents, such as toluene and cyclohexane, BODIPY-PM does not completely dissolve and tends to aggregate, producing characteristic red bands in the fluorescence spectra and increased fluorescence lifetimes (Fig. S1†). In addition, we performed time-resolved fluorescence measurements in chloroform, DCM, DMSO, and water (Fig. 2B). In the high polarity solvents, the fluorescence decays of BODIPY-PM are monoexponential with fluorescence lifetimes close to 200 ps, while in the low-polarity solvents (cyclohexane, toluene, chloroform, and DCM) BODIPY-PM displays biexponential fluorescence decays with an insignificant long lifetime component (0.5–10%) (Fig. 2B and S1†). These data clearly show that the fluorescence lifetimes of BODIPY-PM are minimally influenced by the polarity of the solvent. To explore the viscosity sensitivity of BODIPY-PM, we measured the steady-state and time-resolved fluorescence responses in water–glycerol mixtures with varying water–glycerol compositions that cover a viscosity range from 1–930 cP, (Fig. 2C and S4†). The fluorescence decays were monoexponential in compositions from pure water to 40% water–60% glycerol. At higher glycerol concentrations, the decays become biexponential with low amplitude short-lifetime component. We note that BODIPY-PM displays very strong viscosity sensitivity, with fluorescence lifetimes ranging from 264 ps in pure water to 5127 ps in pure glycerol. Additionally, we quantified the fluorescence lifetime of BODIPY-PM in non-polar toluene–castor oil and methanol–glycerol calibration mixtures (Fig. 2D, S2 and S6†). In contrast to BODIPY-C10, which possessed fluorescence lifetimes ranging from 300 to 800 ps in the solvents of 0.5 cP viscosity,38 BODIPY-PM exhibits very similar fluorescence lifetimes in mixtures of different polarity and is mostly influenced by the viscosity of the mixture itself (Fig. 2D). The insensitivity of BODIPY-PM to solvent polarity is particularly important since biological systems, and especially cells, are highly heterogeneous and accurate viscosity measurements are only possible if the lifetimes are not affected by general solvent effects.47 Because of small fluorescence lifetime values in the non-viscous solvents, BODIPY-PM features greatly expanded dynamic viscosity range, which can be calculated from the lifetime ratio at high and low viscosities, compared to BODIPY-C10. In the methanol–glycerol mixtures, the dynamic range of BODIPY-PM is 30.0 compared to 11.2 of BODIPY-C10 in the same mixture.41To conclude, BODIPY-PM shows almost no sensitivity to solvent polarity and its fluorescence lifetime is mainly affected by the local microviscosity.
Determination of lipid order in LUVs and tBLMs
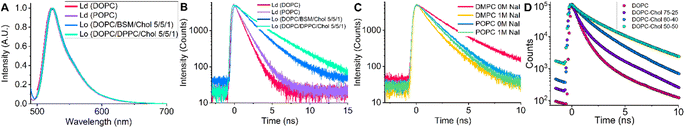
Next, we explored the viscosity sensitivity of BODIPY-PM in Lo and Ld lipid phases. We prepared LUVs with two distinct lipid packaging compositions – pure DOPC and pure POPC for Ld phases, and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSM
BSM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol and DOPC
Chol and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol (5
Chol (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs for Lo phases.48 The steady-state fluorescence spectra of BODIPY-PM in the Lo and Ld phases display no solvatochromism, with maxima located at 521 nm (Fig. 3A). We note that BODIPY-PM successfully integrates into both Lo and Ld LUVs without any observable aggregation. Most importantly, the time-resolved fluorescence decays in the Lo phases are remarkably longer compared to the Ld phases (Fig. 3B), with biexponential intensity-weighted fluorescence lifetimes being 877 and 1607 ps for DOPC and POPC LUVs, respectively. In contrast, the biexponential fluorescence lifetimes of BODIPY-PM in DOPC
1) LUVs for Lo phases.48 The steady-state fluorescence spectra of BODIPY-PM in the Lo and Ld phases display no solvatochromism, with maxima located at 521 nm (Fig. 3A). We note that BODIPY-PM successfully integrates into both Lo and Ld LUVs without any observable aggregation. Most importantly, the time-resolved fluorescence decays in the Lo phases are remarkably longer compared to the Ld phases (Fig. 3B), with biexponential intensity-weighted fluorescence lifetimes being 877 and 1607 ps for DOPC and POPC LUVs, respectively. In contrast, the biexponential fluorescence lifetimes of BODIPY-PM in DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol and DOPC
Chol and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSM
BSM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol LUVs were 3321 ps and 2167 ps, respectively. Although the biexponential nature of fluorescence decays likely reflects different positioning of the dye in the bilayer and complicates data analysis, the ability to determine lipid order and discern Lo and Ld phases is especially important for determination of biologically relevant Lo phases, and lipid rafts, via FLIM, particularly in heterogeneous lipid systems and live cells.
Chol LUVs were 3321 ps and 2167 ps, respectively. Although the biexponential nature of fluorescence decays likely reflects different positioning of the dye in the bilayer and complicates data analysis, the ability to determine lipid order and discern Lo and Ld phases is especially important for determination of biologically relevant Lo phases, and lipid rafts, via FLIM, particularly in heterogeneous lipid systems and live cells.
In addition, we tested BODIPY-PM in gel-phase DMPC and DPPC LUVs. Even though the lipid packaging density in the gel-phase is significantly higher than in Lo phase, the time-resolved fluorescence decays of BODIPY-PM were slightly shorter in DMPC and DPPC LUVs compared to Lo LUVs (Fig. S8†). It is likely that the BODIPY core is sterically too large and is unable to integrate deeply into densely-packed gel-phase bilayers. To test this hypothesis, we carried out time-resolved fluorescence measurements of gel-phase LUVs in the presence of the quencher NaI (Fig. 3C and S9†). The fluorescence lifetimes of DMPC LUVs sharply dropped from 2674 ps in NaI-free solution, to 1405 ps in solutions containing NaI (1 M). In contrast, the time-resolved fluorescence decays of Ld POPC LUVs remained almost unaffected by the presence of 1 M NaI (Fig. 3C), with fluorescence lifetimes decreasing from 1607 ps in NaI-free solution, to 1458 ps in the presence of NaI (1 M). This result indicates that the fluorescent core of BODIPY-PM is exposed to solvent in gel-phase LUVs and only the hydrophobic C12H25 side chain integrates into the bilayer. On the other hand, the minimal quenching of BODIPY-PM in Ld LUVs confirms that BODIPY-PM core is indeed deeply buried inside the bilayer.
Next, we explored how cholesterol in tBLMs (see Experimental section for tBLM composition) affects the lifetimes of BODIPY-PM, using FLIM. Cholesterol is one the most important lipids in eukaryotic cells and is particularly known for its ordering and condensing effects on lipid bilayers.49 For the preparation of tBLMs we chose pure DOPC (Ld), DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 72
Chol 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, DOPC
25, DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 60
Chol 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, and DOPC
40, and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 50
Chol 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 compositions. The time-resolved fluorescence decays of BODIPY-PM in tBLMs become significantly longer with increase in cholesterol concentration, resulting in the intensity weighted biexponential fluorescence lifetimes of 500 and 1900 ps for pure DOPC and DOPC
50 compositions. The time-resolved fluorescence decays of BODIPY-PM in tBLMs become significantly longer with increase in cholesterol concentration, resulting in the intensity weighted biexponential fluorescence lifetimes of 500 and 1900 ps for pure DOPC and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 50
Chol 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 tBLMs, respectively (Fig. 3D).
50 tBLMs, respectively (Fig. 3D).
Importantly, not only is BODIPY-PM capable of distinguishing between the Lo and Ld lipid compositions, it also gives valuable insights into the general lipid packaging efficiency and order of the lipids in the bilayer. The ability of BODIPY-PM to assess the degree of lipid order may be particularly useful in studying lipid rearrangement in membranes, the fusion of membranes, and in monitoring the biophysical changes in the bilayer upon binding of proteins. Moreover, the minimal quenching displayed by BODIPY-PM in liquid membranes is a very convenient property for live cell imaging, since the probe, buried deeply in the bilayer, is shielded from extracellular proteins, which may interfere with the viscosity measurements of the plasma membrane.
Imaging of Lo domains in LUVs, tBLMs and live cells
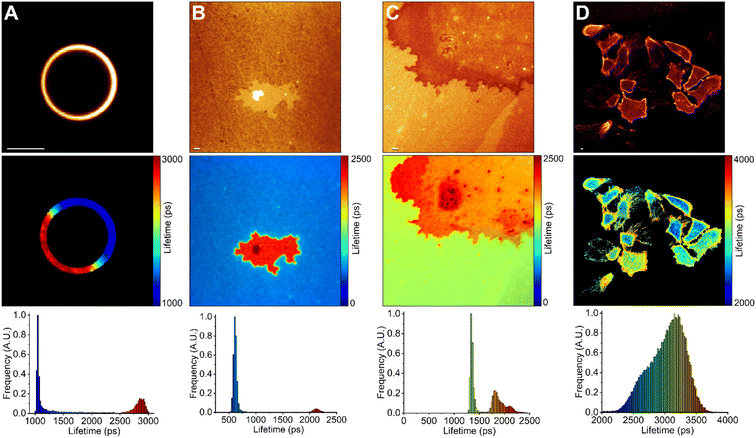
We used FLIM to test BODIPY-PM on LUVs and tBLMs in which Lo and Ld phase separation occurs (Fig. 4). For this purpose, we chose DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 1
Chol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 LUVs, DOPC
1 LUVs, DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 2
Chol 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and DOPC
1 and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSM
BSM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 1
Chol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 tBLMs, since large, micron-scale, Lo domains are easily formed in these compositions.50 The Lo domains in the tBLMs formed with characteristic geometrical features, analogous to the Lo domains observed with atomic force microscopy in supported lipid bilayers of similar compositions.51 We observed successful staining of both Lo and Ld phases with BODIPY-PM in both tBLMs and LUVs. The BODIPY-PM showed particularly high affinity for the Lo phase, with fluorescence intensities in the Lo phase being similar (or even 2-times higher in the DPPC case) compared to the Ld phase (Fig. 4 top panel). Despite the fact that the fluorescence decays in both LUVs and tBLMs were biexponential, the fluorescence lifetimes of BODIPY-PM in Lo and Ld phases are vastly different, allowing easy determination of lipid phases via FLIM (Fig. 4 middle panel). Curiously, the fluorescence lifetimes in DOPC
1 tBLMs, since large, micron-scale, Lo domains are easily formed in these compositions.50 The Lo domains in the tBLMs formed with characteristic geometrical features, analogous to the Lo domains observed with atomic force microscopy in supported lipid bilayers of similar compositions.51 We observed successful staining of both Lo and Ld phases with BODIPY-PM in both tBLMs and LUVs. The BODIPY-PM showed particularly high affinity for the Lo phase, with fluorescence intensities in the Lo phase being similar (or even 2-times higher in the DPPC case) compared to the Ld phase (Fig. 4 top panel). Despite the fact that the fluorescence decays in both LUVs and tBLMs were biexponential, the fluorescence lifetimes of BODIPY-PM in Lo and Ld phases are vastly different, allowing easy determination of lipid phases via FLIM (Fig. 4 middle panel). Curiously, the fluorescence lifetimes in DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 1
Chol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 LUVs were notably higher in both Lo and Ld phases compared to the fluorescence lifetimes in the corresponding phases of the DOPC
1 LUVs were notably higher in both Lo and Ld phases compared to the fluorescence lifetimes in the corresponding phases of the DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPC
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 2
Chol 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 tBLMs (Fig. 4A and B). We suppose that the increase in cholesterol concentration not only orders Ld phase, but also induces order in Lo domains, consequently making them more viscous. Similarly, tBLMs with high cholesterol content, such as DOPC
1 tBLMs (Fig. 4A and B). We suppose that the increase in cholesterol concentration not only orders Ld phase, but also induces order in Lo domains, consequently making them more viscous. Similarly, tBLMs with high cholesterol content, such as DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSM
BSM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 1
Chol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, possessed increased fluorescence lifetimes in the Ld phases (Fig. 4C).
1, possessed increased fluorescence lifetimes in the Ld phases (Fig. 4C).
Finally, we used FLIM to image BODIPY-PM in A549 live lung cancer cells (Fig. 4D). BODIPY-PM stained the cells in less than 5 minutes directly from the aqueous solution at 1 μM concentration. Notably, no signs of cell toxicity or BODIPY-PM induced morphology changes were observed after staining (Fig. S10†). We also note that BODIPY-PM preferentially partitions into the plasma membranes of the cells, although slow internalisation of the dye is also occurring. In addition, we observed slight clustering of highly ordered lipid domains in the plasma membranes of lung cancer cells. The ability to directly observe the heterogeneities of cell membranes, in combination with lipid order determination, could prove to be a powerful tool in studying and developing anticancer drugs that modulate the properties of the plasma membrane.52 The slow internalisation of BODIPY-PM allows for simultaneous plasma membrane and cytoplasm viscosity measurements. The large dynamic range of BODIPY-PM viscosity sensitivity is particularly convenient in detecting small viscosity changes in cell membranes.
In the case of human lung cancer cells, the intensity-weighted fluorescence lifetime of BODIPY-PM in the plasma membrane regions, where the clustering of Lo domains is not occurring, was about 3250 ps, which corresponds to the viscosity value of 300 cP in methanol–glycerol calibration mixtures. In comparison, similar plasma membrane viscosity values of 290 and 326 cP were obtained by Shimolina et al. in the colorectal cancer CT26 cells and cervical HeLa cells, respectively.53 In contrast, the fluorescence lifetime of BODIPY-PM in the more ordered plasma membrane regions of the lung cancer cells was about 3700 ps, corresponding to a viscosity value of about 400 cP. The intensity-weighted fluorescence lifetime of BODIPY-PM in the cytoplasm ranged from 3250 to 3700 ps, corresponding to the viscosity values of 130 and 180 cP, respectively. Similar cytoplasmic viscosity values were reported in ovarian cancer cells (SK-OV-3) by Kuimova et al., with an average intracellular viscosity of 140 cP.54
Conclusions
In conclusion, we have synthesised and explored the capabilities of a new BODIPY-based molecular rotor – BODIPY-PM (A, Fig. 1). We found that addition of a hydrophobic dodecyl group (–C12H25) allows the dye to position deeply in liquid-ordered and liquid-disordered phases of a lipid bilayer. Additionally, a polar sulfonate group (–SO3−) provides good water solubility and enables the easy, non-toxic, staining of the cells from aqueous solutions. We show that fluorescence decays of BODIPY-PM display minimal sensitivity to solvent polarity, but are sensitive to viscosity over a large range, from 0.5 to 1000 cP. Most importantly, FLIM measurements on tBLMs and LUVs, demonstrate that BODIPY-PM possesses vastly different fluorescence lifetimes in lipid-ordered and lipid-disordered environments, allowing this probe to successfully distinguish membrane phase heterogeneities. Additionally, the extent of lipid packaging in bilayers can be estimated based on the dynamic range of viscosity sensing of BODIPY-PM. We have also discovered that BODIPY-PM is localised deeply in non-gel phase bilayers and is not susceptible to external quenching effects. BODIPY-PM preferentially stains the plasma membranes of eukaryotic cells without causing any changes to the cell morphology.Overall, our results demonstrate that BODIPY-PM is capable of probing viscosities in physically distinct membranes, making the dye widely applicable in studying lipid packaging and membrane dynamics in live cells or artificial membranes.
Experimental section
Dyes, reagents, and solvents
The synthetic details of BODIPY-PM, with mass and NMR spectra, are presented in the ESI.† NMR spectra were recorded on a Bruker Ascend 400 spectrometer (400 MHz for 1H, 100 MHz for 13C, 128.4 MHz for 11B, 376.5 MHz for 19F). NMR spectra were referenced to residual solvent peaks. HRMS spectra were recorded on a quadrupole, time-of-flight mass spectrometer (microTOF-Q II, Bruker Daltonik GmbH, Germany). Column chromatography was performed using silica gel 60 (0.040–0.063 mm) (Merck). Thin layer chromatography (TLC) was performed using TLC-aluminium sheets with silica gel (Merck 60 F254). Visualisation was accomplished by UV light. Melting points were determined in open capillaries with a digital melting point IA9100 series apparatus (Thermo Fisher Scientific) and were not corrected. Reagents and solvents for the organic synthesis of the BODIPY compounds were purchased directly from commercial suppliers; solvents were purified by known procedures. Stock solutions of 2 mM BODIPY-PM were prepared in methanol or water and diluted for further experiments in solvents or their mixtures. All solvents used were spectroscopic grade obtained from Sigma-Aldrich. DOPC, DPPC, brain sphingomyelin, cholesterol were obtained from Avanti Polar Lipids (Alabaster, USA). Solvent mixtures were made by mixing reagents at different ratios.Formation of tBLMs and LUVs
Stock solutions of lipids were mixed in appropriate ratios and placed under gentle gaseous nitrogen stream for at least 2 hours to evaporate the chloroform. After complete evaporation, PBS buffer was added on top of lipid film and the mixture was slowly re-suspended until the characteristic milk colour appeared, indicating vesicle formation. The total concentration of lipids in the PBS solution was 1 mM. Afterwards, vesicle mixtures were poured on top of the 100 nm thick gold surface of mica and membranes were allow to form for 2 hours. 20-Tetradecyloxy-3,6,9,12,15,18,22 heptaoxahexatriacontane-1-thiol (WC14) was used as an anchoring compound in combination with β-mercaptoethanol in 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 molar ratios. For DOPC/DPPC/Chol and DOPC/BSM/Chol bilayer compositions, the lipids were re-suspended in hot PBS buffer (60 °C) and the membranes were formed in an oven, with slow decrease in temperature from 60 to 20 °C over the course of 2 h, after which, the membranes were gently washed in PBS buffer and an aqueous solution of BODIPY-PM (1 μM) was added. The membranes were stained (3 min), followed by removal of non-incorporated dye with a PBS wash.
7 molar ratios. For DOPC/DPPC/Chol and DOPC/BSM/Chol bilayer compositions, the lipids were re-suspended in hot PBS buffer (60 °C) and the membranes were formed in an oven, with slow decrease in temperature from 60 to 20 °C over the course of 2 h, after which, the membranes were gently washed in PBS buffer and an aqueous solution of BODIPY-PM (1 μM) was added. The membranes were stained (3 min), followed by removal of non-incorporated dye with a PBS wash.
LUVs were formed by mixing lipids in appropriate ratios, evaporating the chloroform under nitrogen stream for at least 2 hours. After evaporation, PBS buffer was added on top of the film. In the case of gel-phase or mixed-phase LUVs, PBS buffer was heated to 60 °C. After resuspension, mixtures were placed in an ultrasonic bath (1 h) and extruded through 100 nm or 1000 nm polycarbonate membranes with a syringe extruder (Avanti Polar Lipids Inc., USA). Extrusions were performed above the melting temperature of the lipids.
Absorption, steady-state, and time-resolved fluorescence
Absorption spectra were measured using a Jasco V-670 spectrophotometer. Fluorescence spectra were recorded with Edinburgh-F900 (Edinburgh Instruments) fluorimeter using Fianium white laser, together with bandpass filters (Thorlabs), emitting at 488 nm as an excitation source. Fluorescence decays were measured using time-correlated single-photon counting (TCSPC). Fluorescence decays had 5000 counts at the peak of the decay with 20 ns time window being used with 4096 channels in time domain. 10 mm quartz cuvettes were used for absorption and fluorescence measurements with BODIPY-PM concentration being up to 1 μM. Fluorescence decays of BODIPY-PM in solvent mixtures and pure solvents were taken at 20 °C.Imaging of live cells
Cell imaging experiments were done using the A549 human lung cancer cell line (ATCC). The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% foetal bovine serum (FBS), 100 IU mL−1 penicillin and 100 μg mL−1 streptomycin (Thermo Fisher). The cells were incubated at 37 °C with 5% CO2. Before imaging, cells were seeded into Ibidi μ-Dish (Ibidi) at seeding density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and allowed to grow for 24 hours. For cell imaging, cells were kept in the 1 μM solution of BODIPY-PM in PBS for 1 min at 37 °C and then washed with PBS. FLIM imaging was done using Leica SP8 with 63× objective (HC PL APO oil immersion, N.A. – 1.4, Leica).
000 cells per mL and allowed to grow for 24 hours. For cell imaging, cells were kept in the 1 μM solution of BODIPY-PM in PBS for 1 min at 37 °C and then washed with PBS. FLIM imaging was done using Leica SP8 with 63× objective (HC PL APO oil immersion, N.A. – 1.4, Leica).
Fluorescence lifetime imaging microscopy (FLIM)
FLIM was done with a Leica SP8 microscope using 63× objective (HC PL APO oil immersion, N.A. – 1.4, Leica). The fluorescence decay signal was measured over 505–550 nm range using 488 nm white light laser excitation line. The FLIM images were produced in 256 × 256 pixel resolution with 256 channels in time domain, pixels were binned in order to have at least 1000 counts at the peak of the decay curve for reliable biexponential fitting. The instrument response function (IRF) was measured by recording the laser reflection signal off a glass coverslip.Data analysis
FLIM images were analysed with FLIMFIT software (v4.6.1, Imperial College London).55 The biexponential fluorescence decay model with intensity-weighted mean lifetimes was applied for both FLIM and fluorimeter time-correlated single photon counting (TCSPC) measurements:where ai and τi are the amplitudes of the individual components. The goodness-of-fit parameter (χ2) was 1.3 or less for both FLIM images and fluorimeter TCSPC decays. Fluorimeter TCSPC decays of BODIPY-PM were fitted using Edinburgh-F900 software package F900.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors thank David J. Vanderah (Institute for Bioscience and Biotechnology Research at University of Maryland, Rockville MD, USA) for providing samples of anchor compound WC14 and critical reading of the manuscript.Notes and references
- M. S. Gh, Mol. Pharmaceutics, 2021, 18, 2122–2141 CrossRef.
- D. Lingwood and K. Simons, Science, 2010, 327, 46–50 CrossRef CAS.
- H.-J. Kaiser, D. Lingwood, I. Levental, J. L. Sampaio, L. Kalvodova, L. Rajendran and K. Simons, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 16645–16650 CrossRef CAS.
- A. T. Hammond, P. Sengupta, S. T. Hess, D. A. Holowka, B. A. Baird and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3165–3170 CrossRef.
- C. Eggeling, C. Ringemann, R. Medda, G. Schwarzmann, K. Sandhoff, S. Polyakova, V. N. Belov, B. Hein, C. v. Middendorff, A. Schönle and S. W. Hell, Nature, 2009, 457, 1159–1162 CrossRef CAS.
- K. A. Field, D. Holowka and B. Baird, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9201–9205 CrossRef CAS.
- P. Varshney, V. Yadav and N. Saini, Immunology, 2016, 149, 13–24 CrossRef CAS.
- K. Iwabuchi, Front. Biosci., 2015, 20, 325–334 CrossRef CAS.
- E. Teissier and E. Pecheur, Eur. Biophys. J., 2007, 36, 887–899 CrossRef CAS PubMed.
- F. J. O. Rios, M. Ferracini, M. Pecenin, M. M. Koga, Y. Wang, D. F. J. Ketelhuth and S. Jancar, PLoS One, 2013, 8, e76893 CrossRef CAS PubMed.
- J. B. Larsen, M. B. Jensen, V. K. Bhatia, S. L. Pedersen, T. Bjørnholm, L. Iversen, M. Uline, I. Szleifer, K. J. Jensen, N. S. Hatzakis and D. Stamou, Nat. Chem. Biol., 2015, 11, 192–194 CrossRef CAS PubMed.
- C. Gajate and F. Mollinedo, Blood, 2007, 109, 711–719 CrossRef CAS PubMed.
- Á. Cuesta-Marbán, J. Botet, O. Czyz, L. M. Cacharro, C. Gajate, V. Hornillos, J. Delgado, H. Zhang, F. Amat-Guerri, A. U. Acuña, C. R. McMaster, J. L. Revuelta, V. Zaremberg and F. Mollinedo, Nat. Chem. Biol., 2015, 11, 192–194 CrossRef.
- C. Dietrich, L. A. Bagatolli, Z. N. Volovyk, N. L. Thompson, M. Levi, K. Jacobson and E. Gratton, Biophys. J., 2001, 80, 1417–1428 CrossRef CAS.
- R. Žvirblis, K. Maleckaitė, J. Dodonova-Vaitkūnienė, D. Jurgutis, R. Žilėnaitė, V. Karabanovas, S. Tumkevičius and A. Vyšniauskas, J. Mater. Chem. B, 2023, 11, 3919–3928 RSC.
- A. S. Klymchenko, Acc. Chem. Res., 2017, 50, 366–375 CrossRef CAS.
- A. Vyšniauskas and M. K. Kuimova, Int. Rev. Phys. Chem., 2018, 37, 259–285 Search PubMed.
- M. A. Haidekker and E. A. Theodorakis, Org. Biomol. Chem., 2007, 5, 1669–1678 RSC.
- M. A. Haidekker and E. A. Theodorakis, J. Biol. Eng., 2010, 4, 11 CrossRef PubMed.
- L. E. Shimolina, A. A. Gulin, M. Paez-Perez, I. López-Duarte, I. N. Druzhkova, M. M. Lukina, M. V. Gubina, N. J. Brooks, E. V. Zagaynova, M. K. Kuimova and M. V. Shirmanova, J. Biomed. Opt., 2020, 25, 1–16 Search PubMed.
- L. Shimolina, M. Lukina, V. Shcheslavskiy, V. Elagin, V. Dudenkova, N. Ignatova, M. K. Kuimova and M. Shirmanova, J. Visualized Exp., 2021, 173, e62708 Search PubMed.
- A. Vyšniauskas, M. Qurashi and M. K. Kuimova, Chem. –Eur. J., 2016, 22, 13210–13217 CrossRef.
- M. R. Bittermann, M. Grzelka, S. Woutersen, A. M. Brouwer and D. Bonn, J. Phys. Chem. Lett., 2021, 12, 3182–3186 CrossRef CAS.
- M. K. Kuimova, Phys. Chem. Chem. Phys., 2012, 14, 12671–12686 RSC.
- S.-C. Lee, J. Heo, H. C. Woo, J.-A. Lee, Y. H. Seo, C.-L. Lee, S. Kim and O.-P. Kwon, Chem. –Eur. J., 2018, 24, 13706–13718 CrossRef CAS PubMed.
- A. Vyšniauskas, M. Qurashi and M. K. Kuimova, Chem. –Eur. J., 2016, 22, 13210–13217 CrossRef PubMed.
- M. R. Dent, I. López-Duarte, C. J. Dickson, P. Chairatana, H. L. Anderson, I. R. Gould, D. Wylie, A. Vyšniauskas, N. J. Brooks and M. K. Kuimova, Chem. Commun., 2016, 52, 13269–13272 RSC.
- M. Kubánková, I. López-Duarte, D. Kiryushko and M. K. Kuimova, Soft Matter, 2018, 14, 9466–9474 RSC.
- A. Vyšniauskas, I. López-Duarte, N. Duchemin, T.-T. Vu, Y. Wu, E. M. Budynina, Y. A. Volkova, E. P. Cabrera, D. E. Ramírez-Ornelas and M. K. Kuimova, Phys. Chem. Chem. Phys., 2017, 19, 25252–25259 RSC.
- Z. Yang, Y. He, J. H. Lee, W.-S. Chae, W. X. Ren, J. H. Lee, C. Kang and J. S. Kim, Chem. Commun., 2014, 50, 11672–11675 RSC.
- S.-C. Lee, J. Heo, J.-W. Ryu, C.-L. Lee, S. Kim, J.-S. Tae, B.-O. Rhee, S.-W. Kima and O.-P. Kwon, Chem. Commun., 2016, 52, 13695–13698 RSC.
- Y. Shi, P. A. Summers, M. K. Kuimova and H. S. Azevedo, Nano Lett., 2020, 20, 7375–7381 CrossRef CAS.
- R. Clark, M. A. Nawawi, A. Dobre, D. Pugh, Q. Liu, A. P. Ivanov, A. J. P. White, J. B. Edel, M. K. Kuimova, A. J. S. McIntosh and T. Welton, Chem. Sci., 2020, 11, 6121–6133 RSC.
- H. Doan, S. L. Raut, D. Yale, M. Balaz, S. V. Dzyuba and Z. Gryczynski, Chem. Commun., 2016, 52, 9510–9513 RSC.
- M. R. Dent, I. Lopez Duarte, C. J. Dickson, N. D. Geoghegan, J. M. Cooper, I. R. Gould, R. Krams, J. A. Bull, N. J. Brooks and M. K. Kuimova, Phys. Chem. Chem. Phys., 2015, 17, 18393–18402 RSC.
- E. Gatzogiannis, Z. Chen, L. Wei, R. Wombacher, Y.-T. Kao, G. Yefremov, V. W. Cornish and W. Min, Chem. Commun., 2012, 48, 8694–8696 RSC.
- M. K. Kuimova, S. W. Botchway, A. W. Parker, M. Balaz, H. A. Collins, H. L. Anderson, K. Suhling and P. R. Ogilby, Nat. Chem., 2009, 1, 69–73 CrossRef CAS PubMed.
- I. López-Duarte, T. T. Vu, M. A. Izquierdo, J. A. Bull and M. K. Kuimova, Chem. Commun., 2014, 50, 5282–5284 RSC.
- Z. Yang, Y. He, J.-H. Lee, N. Park, M. Suh, W.-S. Chae, J. Cao, X. Peng, H. Jung, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 9181–9185 CrossRef CAS PubMed.
- L. Wang, Y. Xiao, W. Tian and L. Deng, J. Am. Chem. Soc., 2013, 135, 2903–2906 CrossRef CAS PubMed.
- A. Polita, S. Toliautas, R. Žvirblis and A. Vyšniauskas, Phys. Chem. Chem. Phys., 2020, 22, 8296–8303 RSC.
- Y. Wu, M. Štefl, A. Olzyńska, M. Hof, G. Yahioglu, P. Yip, D. R. Casey, O. Ces, J. Humpolíčková and M. K. Kuimova, Phys. Chem. Chem. Phys., 2013, 15, 14986–14993 RSC.
- M. Olšinová, P. Jurkiewicz, M. Pozník, R. Šachl, T. Prausová, M. Hof, V. Kozmík, F. Teplý, J. Svobodab and M. Cebecauer, Phys. Chem. Chem. Phys., 2014, 16, 10688–10697 RSC.
- S. Rebaud, O. Maniti and A. P. Girard-Egrot, Biochimie, 2014, 107, 135–142 CrossRef CAS PubMed.
- M. Páez-Pérez, I. López-Duarte, A. Vyšniauskas, N. J. Brooks and M. K. Kuimova, Chem. Sci., 2021, 12, 2604–2613 RSC.
- Y. Niko and A. S. Klymchenko, J. Biochem., 2021, 170, 163–174 CrossRef CAS PubMed.
- E. Sezgin, T. Gutmann, T. Buhl, R. Dirkx, M. Grzybek, Ü. Coskun, M. Solimena, K. Simons, I. Levental and P. Schwille, PLoS One, 2015, 10, e0123930 CrossRef.
- G. W. Feigenson, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 47–52 CrossRef CAS PubMed.
- T. Róg, M. Pasenkiewicz-Gierula, I. Vattulainen and M. Karttunen, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 97–121 CrossRef.
- S. L. Veatch and S. L. Keller, Biophys. J., 2003, 85, 3074–3083 CrossRef CAS.
- S. D. Connell and D. A. Smith, Mol. Membr. Biol., 2006, 23, 17–28 CrossRef CAS PubMed.
- G. Preta, Front. Cell Dev. Biol., 2020, 8, 571237 CrossRef PubMed.
- L. E Shimolina, A. A. Gulin, M. Paez-Perez, I. López-Duarte, I. N. Druzhkova, M. M. Lukina, M. V. Gubina, N. J. Brooks, E. V. Zagaynova, M. K. Kuimova and M. V. Shirmanova, J. Biomed. Opt., 2020, 25, 126004 Search PubMed.
- M. K. Kuimova, G. Yahioglu, J. A. Levitt and K. Suhling, J. Am. Chem. Soc., 2008, 130, 6672–6673 CrossRef CAS PubMed.
- L. S. C. Warren, A. Margineanu, D. Alibhai, D. J. Kelly, C. Talbot, Y. Alexandrov, I. Munro, M. Katan, C. Dunsby and P. M. W. French, PLoS One, 2013, 8, e70687 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04126c |
| This journal is © The Royal Society of Chemistry 2023 |