Open Access Article
Open Access ArticleAlkyne-tethered oligodeoxynucleotides that allow simultaneous detection of multiple DNA/RNA targets using Raman spectroscopy†
Hikaru Watanabe,
Daigo Maehara,
Tatsuya Nishihara and
Kazuhito Tanabe *
*
Department of Chemistry and Biological Science, College of Science and Engineering, Aoyama Gakuin University, 5-10-1 Fuchinobe, Chuo-ku, Sagamihara, 252-5258, Japan. E-mail: tanabe.kazuhito@chem.aoyama.ac.jp
First published on 11th July 2023
Abstract
Detection of multiple DNA/RNA targets is essential for understanding cellular function. Herein, we propose a general method for the simultaneous detection of plural nucleic acids based on surface-enhanced Raman scattering (SERS) using gold nanoparticles bearing functional oligodeoxynucleotides (ODNs) on their surface. Modified ODNs bearing an acetylene tag hybridized with their complementary ODNs on the surface of the gold nanoparticles, inducing a strong SERS signal of the acetylene tag. The addition of the target nucleic acid to the system resulted in a spontaneous displacement of the strand on the particle and dissociation of the alkyne-tagged ODN from the particle, resulting in a dramatic decrease in signal intensity. By using an alkyne tag for each of the multiple target nucleic acids, each target could be detected simultaneously. In addition, we successfully detected cellular microRNA. Different targets showed changes with different wavenumbers in the Raman spectra, allowing for the detection of multiple nucleic acids.
Introduction
Cells contain a number of specific DNA and RNA components that are essential for their physiological functions.1–5 For an integrated understanding of the function of cells, it is ideal to be able to detect multiple RNAs and DNAs in parallel. Conventionally, fluorescently labeled artificial nucleic acids have been utilized as a method for detecting RNA and DNA.6–9 For example, fluorophore-labeled DNA microarrays have become a powerful analytical tool to detect target DNA or RNA, due to their efficient parallel analysis.10 In addition, a single-stranded oligonucleotide probe labeled with one or a few fluorescent dyes has been used to recognize the target RNA and to enable visualization of intracellular nucleic acids.11–13 However, these fluorescence detection methods are limited by the “color barrier” due to the broad fluorescence signals. In other words, they have the disadvantage of limiting the number of targets that can be separated simultaneously up to five or six.14,15 Thus, there are increasing demands for procedures to detect multiple nucleic acids simultaneously.To address this issue, surface-enhanced Raman scattering (SERS)-based detection of nucleic acids has been focused on, because the narrow Raman spectral linewidth enables the monitoring of multiple target molecules simultaneously.16–25 Recent studies showed that metal nanoparticles with reporter tags produced robust Raman signals when the target nucleic acids were hybridized to form second-order structures on the particles.26–32 More recently, we reported a method using Hoechst derivatives (AH) as a Raman probe.33 Binding and dissociation of AH on the particles led to a change in the Raman signal intensity, and thus allowed sensitive detection of the target nucleic acids. Although these conventional procedures are useful to detect specified nucleic acids, their application to the simultaneous detection of multiple targets is difficult because a single functional group is used as the signaling part.
These research contexts prompted us to construct a molecular system that can be used to identify multiple nucleic acid targets using Raman spectroscopy. Herein, we demonstrate a proof-of-concept study using ODNs bearing different acetylene tags as Raman signal reporters attached to gold nanoparticles. By the combination of modification of ODNs and their strand-displacement reaction, we successfully identified plural target oligonucleotides simultaneously. We also confirmed that the system could be applied to the detection of cellular microRNA.
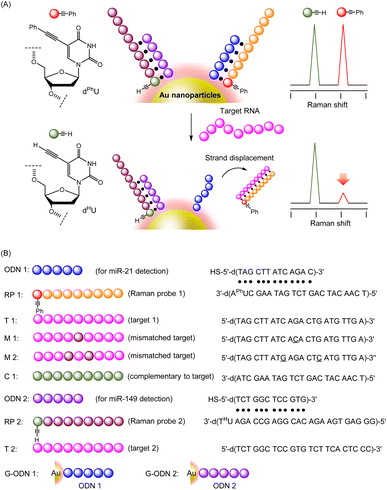
When Raman probe (RP: ODNs with alkyne-tag) binds to its corresponding DNA sequence tethered on gold nanoparticles, they show robust SERS signals. When target strand binds with RPs on nanoparticles in a sequence selective manner, the RPs dissociate from the particles, resulting in the decrease of signal intensity. The signal from RPs were changed in the presence of target strands, thereby enabling highly sensitive detection of each target RNA.
Results and discussion
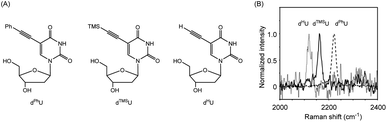
To construct a multiple detection system for DNA or RNA strands, we designed alkyne-tagged ODNs (Raman probes: RPs) that hybridize with complementary ODNs on gold nanoparticles. As shown in Fig. 1A, each RP has a tag portion that generates a unique signal, corresponding to the target strand. Since each RP binds to its corresponding DNA tethered on gold nanoparticles, RPs show robust SERS signals that are induced by the proximal gold nanoparticles. On the other hand, the signal intensity decreases dramatically when target strands that are complementary to each RP on the particles were added to the system. Strand-displacement reactions between the target strand and ODNs on the particles occur spontaneously, leading to the formation of a duplex of RPs and the target strand, which is released from the particles. Dissociation of RP/target duplex proceeds in a sequence-specific manner. Therefore, if RP strands with several different tag sites are prepared, signal changes corresponding to several specific target strands can be obtained. In the experiments reported here, model ODNs were first employed as targets instead of RNA strands because of their stability and ease of handling; actual microRNAs were subsequently detected with the present system using cell lysates.Initially, we prepared three alkyne-tagged uridine derivatives and characterized their Raman spectra (Fig. 2A). According to the reported procedure,34,35 we synthesized uridine derivatives with an acetylene substituent at the 5-position (dPhU, dTMSU and dHU). The Raman spectra of these compounds exhibited robust signal at 2221, 2161 and 2119 cm−1 for dPhU, dTMSU and dHU, respectively, arising from the acetylene group (Fig. 2B). Thus, these unsubstituted and substituted acetylene groups each gave signals at different wavenumbers. In light of these properties, we attempted to utilize uridine derivatives equipped with these functional groups as signal transmitters for the visualization of multiple target strands.
To validate the system, we prepared the 22-mer RP 1 and 25-mer RP 2, bearing dPhU and dHU in the strand, respectively. RP 1, bearing dPhU for detection of T 1 and RP 2, bearing dHU for detection of T 2, were designed. The dTMSU was not used to transmit RNA sequence information, but rather as an internal standard when measuring Raman spectra. The alkyne-tagged ODNs were synthesized using phosphoramidite methods; the sequences are shown in Fig. 1. We then characterized the strand-displacement reaction of ODNs. A partial duplex was created by mixing ODN 1, which is the DNA strand to be fixed on the gold nanoparticles, and RP 1, which is the Raman probe. Then, the target strand T 1 was added to the ODN 1/RP 1 duplex, and the progress of the strand-exchange reaction was investigated using an electrophoretic mobility shift assay. As clearly seen in Fig. 3, upon the addition of 1 equiv. of T 1, the band of ODN 1/RP 1 duplex disappeared, while the fully complementary duplex (RP 1/T 1) was formed. These results strongly indicate that the addition of the target strand induced strand displacement. We also measured melting temperature (Tm) of ODN 1/RP 1 (36.7 °C) and RP 1/T 1 duplexes (55.5 °C), and confirmed the stable duplex formation of RP 1/T 1 duplex. In a separate experiment, we confirmed that ODN 2/RP 2 duplex was also displaced to form the RP 2/T 2 duplex, after the addition of the target strand, T 2 (Fig. S1†). Thus, the expected reaction proceeded, indicating that the specific target strand could be detected using Raman spectroscopy.
In light of these reaction characteristics of RP 1 and RP 2, we applied the present strand-displacement reaction to the detection of specified targets using Raman spectra. For this purpose, we designed gold nanoparticles bearing ODNs on their surface (G-ODNs). Gold nanoparticles bearing ODN 1 (G-ODN 1) or ODN 2 (G-ODN 2) were prepared, according to reported protocols using thiolated ODNs.33 The amount of ODNs on the gold nanoparticles was estimated to be (5.8 ± 0.2) × 102 DNA/particle. We prepared a partial duplex on the gold nanoparticles (G-ODN 1/RP 1) by hybridization of G-ODN 1 and RP 1, and measured its Raman spectra using excitation at 532 nm in the presence of monomeric dTMSU as an internal standard. As shown in Fig. 4A, G-ODN 1/RP 1 showed a typical band at 2220 cm−1 arising from the acetylene group of dPhU in the strand. It is striking that the addition of the target T 1 strand led to the disappearance of the signal at 2220 cm−1, indicating that RP 1 was released from gold nanoparticles due to the strand displacement and consequent loss of the SERS effect. When the concentration of target T 1 was varied, the signal intensity changed proportionally (Fig. 4C and D). Similar behavior was observed in Raman spectra when T 2 as another target strand, was added to G-ODN 2/RP 2 (Fig. 4B). Thus, we could clearly detect two specified target strands using Raman spectroscopy. In a control experiment, to the G-ODN 1/RP 1, we added mismatched targets M 1 and M 2, which had one or two mismatched base pairs, respectively (Fig. S2†). The samples showed a strong signal in the presence of target strand with two mismatched base pairs (M 2), whereas a weak signal was observed in the presence of the target strand with a single mismatched base pair (M 1). The melting temperature of the M 2/RP 1 duplex (35.4 °C) was dramatically lower than that of the M 1/RP 1 duplex (43.5 °C) and T 1/RP 1 duplex (55.5 °C), indicating that the instability of duplex with two mismatched base pairs led weakened signal in Raman spectra. These results suggest that this system was able to distinguish between the target sequence and sequence with two mismatched base pairs. To discriminate multiple targets simultaneously, we mixed G-ODN 1/RP 1 and G-ODN 2/RP 2 and measured the Raman spectra in the presence or absence of their targets, T 1 and T 2. As shown in Fig. 4E, three signals attributed to G-ODN 1/RP 1, G-ODN 2/RP 2 and internal standard (dTMSU) were observed in the absence of targets. When target strand T 1 was added to the mixture, the signal of RP 1 disappeared, and two signals of G-ODN 2/RP 2 and the internal standard remained. Similarly, when target strand T 2 was added to the mixture in place of T 1, the signal of RP 2 was weakened. When both of T 1 and T 2 targets were added, both signals of G-ODN 1/RP 1 and G-ODN 2/RP 2 disappeared in response to the added targets. Thus, the present system could discriminate multiple targets, simultaneously.
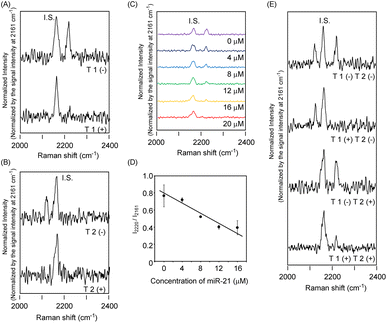 | ||
| Fig. 4 Change of Raman signal by strand displacement reaction of ODNs. The dTMSU (3 mM) was used as internal standard (IS). The Raman spectra were measured using 532 nm excitation. (A) Raman spectra of G-ODN 1/RP 1 (0.1 μM) in the presence or absence of T 1 (20 μM). (B) Raman spectra of G-ODN 2/RP 2 (0.1 μM) in the presence or absence of T 2 (20 μM). (C) Raman spectra of G-ODN 1/RP 1 (0.1 μM) after addition of T 1 (purple: 0 μM, black: 4 μM, blue: 8 μM, green: 12 μM, orange: 16 μM, red: 20 μM). (D) Plot of relative signal intensity obtained from G-ODN 1/RP 1 (I2220/I2161) after addition of T 1 strand. The data of signal intensity was obtained from Fig. 3C. (E) Raman spectra of the mixture consisted of G-ODN 1/RP 1 and G-ODN 2/RP 2 in the presence or absence of T 1 and T 2 targets. | ||
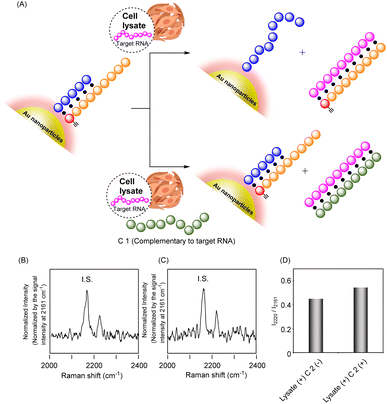
Further attempts were made to detect cellular RNA targets by the present system. To assess the function of the system, we measured the Raman spectra of the G-ODN/RP system in the presence of a cell lysate of a human lung adenocarcinoma cell line, A549, in which microRNA-21 (miR-21) was expressed (Fig. 5A). We employed G-ODN 1/RP 1, because, in addition to the T 1 strand, they were designed to hybridize with miR-21 as a cellular target.36 After the cell lysate was harvested, G-ODN 1/RP 1 was added to the lysate and Raman spectra were measured. As shown in Fig. 5C, G-ODN 1/RP 1 showed a strong signal at 2219 cm−1 in the presence of C 1 even after the addition of cell lysate. C 1 was complementary to miR-21, and formed a stable duplex structure with miR-21; thus, hybridization of miR-21 with RP 1 and dissociation of RP 1 from G-ODN 1 was prevented, leading to retention of the strong Raman signal (Fig. 5A). In contrast, in the absence of C 1, relatively weakened signal of G-ODN 1/RP 1 was observed after the addition of cell lysate. Although the amount of RNA present in the cells was small and the difference in signal intensity in Raman spectra was slight, these experimental results suggest the dissociation of RP 1 from the particles, leading to weakened signal. Thus, intracellular RNA could be identified using the present system.
Conclusions
A molecular system has been constructed to detect multiple target strands using Raman spectroscopy. Two types of alkyne-tethered ODNs were prepared and loaded on gold nanoparticles by hybridization with complementary strands on the particles. Because of the SERS effect, the alkyne unit on the ODNs showed a strong signal around 2100–2200 cm−1 in the Raman spectra. On the other hand, when target strand was added to the nanoparticles, a strand-displacement reaction occurred on the particles, leading to the dissociation of the alkyne-tethered ODNs from the particles and the consequent weakening of the signal of the acetylene unit. Thus, changes in the signal intensity attributed to alkyne-tethered ODNs allowed the specified targets to be detected. We successfully identified plural targets by the measurement of Raman spectra simultaneously. In addition, we detected a specified target strand in cell lysate. The advantage of the Raman spectroscopy-based identification method is that it can be used to detect multiple samples simultaneously. Taking advantage of this feature, this procedure is being developed further to detect several cellular RNAs.Experimental section
Preparation of gold nanoparticles bearing ODNs on their surface
To an aqueous solution of gold nanoparticles (40 nm diameter, 1 mL), ODNs with thiol modification (3 nmol) was added and the resulting mixture was incubated at r.t. for 24 h. To remove unreacted ODNs, the reaction mixture was centrifuged (16.1 × 103 rcf) for 10 min, then and supernatant was removed. The obtained particles were resuspended in water (100 μL) and the resulting solution was used for further experiments.To estimate the amount of ODNs modified on the surface of gold nanoparticles concentration, aqueous solution of gold nanoparticle (4 μL) was treated with AP, Pl and phosphodiesterase I at 37 °C for 24 h. After centrifugation, the supernatant was analyzed by HPLC to estimate the amount of ODNs ((5.8 ± 0.2) × 102 DNA/particle).
Melting temperature (Tm) of hybridized ODNs
2.5 μM of appropriate ODNs were dissolved in 10 mM sodium phosphate buffer (pH 7.0). Melting curves were obtained by monitoring the UV absorbance at 260 nm with elevating temperature from 20 °C to 90 °C at a rate of 1 °C min−1.Measurement of Raman spectra
Aqueous solution of ODNs and ODNs-modified gold nanoparticles were prepared. The resulting solution was subjected to the measurement of Raman spectra (Ex. 532 nm). The laser power was 50 mW, and exposure time was 20 s. The Raman spectra were measured in the presence of dTMSU as an internal standard (IS).Gel electrophoresis of ODNs
(General procedure for identification of strand displacement reaction, lane 6 in Fig. S1†). ODN 1 (5 μM) was hybridized with RP 1 (5 μM) to form duplex, and then T 1 (5 μM) was added to the sample. After incubation at ambient temperature for 1 h, the resulting sample was loaded into 10% polyacrylamide gel and analyzed by electrophoresis. The ODNs on the gels were dyed with Gelred. For the assay, excitation at 530 nm was used, and the emission above 550 nm was monitored.Cell culture
A549 cells were cultured in Dulbecco's Modified Eagle Medium (D-MEM) supplemented with 10% fetal bovine serum (FBS), and 1% penicillin–streptomycin. The cells were maintained at 37 °C in 5% CO2 and were kept in a logarithmic growth phase by routine passages every 3–4 days. Prior to the use of cells, the densities of cells were determined using a hemocytometer.Preparation of cell lysate
Human lung carcinoma cell line, A549, was purchased from JCRB cell bank (Japanese Collection of Research Bioresources Cell Bank). A549 cells were washed with PBS for two time, and then the cell were lysed by pH 7.8 lysate buffer (containing Tris HCl 100 mM, EDTA 2 mM and Triton X 0.05% v/v) and harvested. The cell lysate was subjected to measurement of Raman spectra in the presence of G-ODN 1/RP 1.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by Grant-in-Aid for Scientific Research (for K. T. Grant number 20H02863 and 23H02086). A graphic in Fig. 4 was created with https://BioRender.com. We thank Prof. Atsushi Maruyama and Dr Naohiko Shimada in Tokyo Institute of Technology for thermal denaturation studies.Notes and references
- K. H. Chen, A. N. Boettiger, J. R. Moffitt, S. Wang and X. Zhuang, Science, 2015, 348, aaa6090 CrossRef PubMed.
- C. H. L. Eng, M. Lauson, Q. Zhu, R. Dries, N. Koulena, Y. Takei, J. Yun, C. Cronin, C. Karp, G. C. Yuan and L. Cai, Nature, 2019, 86, 235–239 CrossRef.
- L. Yan, J. Zhou, Y. Zheng, A. S. Gamson, B. T. Roembke, S. Nakayama and H. O. Sintim, Mol. BioSyst., 2014, 10, 970–1003 RSC.
- Y. Cao, Z. Zheng and H. G. Monbouquette, Curr. Opin. Biotechnol., 2021, 71, 145–150 CrossRef CAS PubMed.
- M. Shen, Y. Zhou, J. Ye, A. A. A. Al-maskri, Y. Kang, S. Zeng and S. Cai, J. Pharm. Anal., 2020, 10, 97–101 CrossRef PubMed.
- J. Ge, L. L. Zhang, S. J. Liu, R. Q. Yu and X. A. Chu, Anal. Chem., 2014, 86, 1808–1815 CrossRef CAS.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS.
- J. Li, X. Li, Y. Li, H. Yang, L. Wang, Y. Qin, H. Liu, L. Fu and X. Y. Guan, PLoS One, 2013, 8, e53582 CrossRef CAS PubMed.
- J. Lu and A. Tsourkas, Nucleic Acids Res., 2009, 37, e100 CrossRef.
- B. H. Kim, A. Podder and H. J. Lee, Bull. Chem. Soc. Jpn., 2021, 94, 1010–1035 CrossRef.
- M. Ogata, G. Hayashi, A. Ichiu and A. Okamoto, Org. Biomol. Chem., 2020, 18, 8084–8088 RSC.
- D. Kosman, C. M. Mizutani, D. Lemons, W. G. Cox, W. McGinnis and E. Bier, Science, 2004, 305, 856 CrossRef PubMed.
- E. J. M. Speel, A. H. N. Hpman and P. Komminoth, J. Histochem. Cytochem., 1999, 47, 281–288 CrossRef CAS PubMed.
- H. Fujioka, J. Shou, R. Kojima, Y. Urano, Y. Ozeki and M. Kamiya, J. Am. Chem. Soc., 2020, 142, 20701–20707 CrossRef CAS PubMed.
- F. Hu, L. Shi and W. Min, Nat. Methods, 2019, 16, 830–842 CrossRef CAS.
- C. Zeng, F. Hu, R. Long and W. Min, Analyst, 2018, 143, 4844–4848 RSC.
- H. Yamakoshi, A. F. Palonpon, K. Dodo, J. Ando, S. Kawata, K. Fujita and M. Sodeoka, Chem. Commun., 2014, 50, 1341–1343 RSC.
- L. Shi, C. Zheng, Y. Shen, Z. Chen, E. S. Silveira, L. Zhang, M. Wei, C. Liu, C. de Sena-Tomas, K. Targoff and W. Min, Nat. Commun., 2018, 9, 2995 CrossRef.
- Z. Chen, D. W. Paley, L. Wei, A. L. Weisman, R. A. Friesner, C. Nuckolls and W. Min, J. Am. Chem. Soc., 2014, 136, 8027–8033 CrossRef CAS.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102–6105 CrossRef CAS.
- Y. Ozeki, F. Dake, S. Kajiyama, K. Fukui and K. Itoh, Opt. Express, 2009, 17, 3651–3658 CrossRef CAS PubMed.
- C. W. Freudiger, W. Min, B. G. Saar, S. Lu, G. R. Holtom, C. He, J. C. Tsai, J. X. Kang and X. S. Xie, Science, 2008, 322, 1857–1861 CrossRef CAS PubMed.
- Y. C. Cao, R. Jin and C. A. Mirkin, Science, 2002, 207, 1536–1540 CrossRef PubMed.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- A. Campion and P. Kambhampati, Chem. Soc. Rev., 1998, 27, 241–250 RSC.
- R. Ota, N. Takagi, Y. Imaizumi, T. Waku and A. Kobori, Nucleosides, Nucleotides Nucleic Acids, 2021, 40, 166–177 CAS.
- E. Eduardo Garcia-Rico, R. A. Alvarez-Puebla and L. Guerrini, Chem. Soc. Rev., 2018, 47, 4909–4923 RSC.
- R. Ota, Y. Fukushima, Y. Araki, K. Sasaki, T. Waku and A. Kobori, Chem. Lett., 2021, 50, 513–517 CrossRef CAS.
- Y. Qian, T. Fan, Y. Yao, X. Shi, X. Liao, F. Zhou and F. Gao, Sens. Actuators, B, 2018, 254, 483–489 CrossRef CAS.
- S. Ye, Y. Wu, X. Zhai and B. Tang, Anal. Chem., 2015, 87, 8242–8249 CrossRef CAS.
- H. Shi, N. Chen, Y. Su, H. Wang and Y. He, Anal. Chem., 2017, 89, 10279–10285 CrossRef CAS.
- Y. Wu, F. Xiao, Z. Wu and R. Yu, Anal. Chem., 2017, 89, 2852–2858 CrossRef CAS PubMed.
- H. Makanai, T. Nishihara and K. Tanabe, ACS Appl. Nano Mater., 2022, 5, 2935–2942 CrossRef CAS.
- R. Itaya, W. Idei, T. Nakamura, T. Nishihara, R. Kurihara, A. Okamoto and K. Tanabe, ACS Omega, 2021, 6, 31595–31604 CrossRef CAS.
- M. Slavíčková, M. Janoušková, A. Šimonová, H. Cahová, M. Kambová, H. Šanderová, L. Krásný and M. Hocek, Chem.–Eur. J., 2018, 24, 8311–8314 CrossRef.
- The sequences were designed according to the previous report. H.-N. Wang, B. M. Crawford, S. J. Norton and T. Vo-Dinh, J. Phys. Chem. B, 2019, 123, 10245–10251 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra03861k |
| This journal is © The Royal Society of Chemistry 2023 |