Open Access Article
Open Access ArticleGold nanoparticle-coated apoferritin conductive nanowires†
Gloria Pelayo-Punzano a,
Rocío Juradoa,
Miguel López-Haro
a,
Rocío Juradoa,
Miguel López-Haro *b,
Rafael Cuestac,
José J. Calvinob,
José M. Domínguez-Veraa and
Natividad Gálvez
*b,
Rafael Cuestac,
José J. Calvinob,
José M. Domínguez-Veraa and
Natividad Gálvez *a
*a
aDepartment of Inorganic Chemistry, University of Granada, 18071 Granada, Spain. E-mail: ngalvez@ugr.es
bDepartment of Material Science and Metallurgy Engineering and Inorganic Chemistry, University of Cadiz, 11510, Cadiz, Spain
cDepartment of Organic and Inorganic Chemistry, EPS Linares, University of Jaen, 23700 Linares, Spain
First published on 27th June 2023
Abstract
Gold-metallic nanofibrils were prepared from three different iso-apoferritin (APO) proteins with different Light/Heavy (L/H) subunit ratios (from 0% up to 100% L-subunits). We show that APO protein fibrils have the ability to in situ nucleate and grow gold nanoparticles (AuNPs) simultaneously assembled on opposite strands of the fibrils, forming hybrid inorganic–organic metallic nanowires. The AuNPs are arranged following the pitch of the helical APO protein fiber. The mean size of the AuNPs was similar in the three different APO protein fibrils studied in this work. The AuNPs retained their optical properties in these hybrid systems. Conductivity measurements showed ohmic behavior like that of a continuous metallic structure.
Introduction
AuNPs are amongst the most promising nanomaterial candidates for a wide range of fields, such as medicine, biology, physics, chemistry and sensing, although it is in nanomedicine where they are reportedly responsible for an impressive resurgence.1–4 AuNP properties, including plasmonic properties, targeting, and bio-compatibility, have made them incredibly useful nanomaterials with a wide range of chemical and biological applications.5–10 In particular, AuNP assemblies exhibit unique optical functionalities, which depend largely on the local order of the nanoparticles. To be applied in novel optical devices, their optical properties must be fine-tuned which, in turn, calls for methods for precisely controlling their assembly.11–14Amyloid-like protein fibrils have been used successfully for templating metallic nanowires.11,15–17 Fabricating nanoscale amyloid-inorganic hybrid materials using amyloid protein fibers is interesting because these scaffolds present high aspect ratios and generally display multiple binding sites for small molecules, cations or nanoparticles along their surface for the design of nanocomposites through specific post-functionalization or in situ approaches.18–20 In addition to their regularity and ready production, these biological templates may have another key advantage, as they can grow directly on the required surfaces, thus guiding the assembly of the inorganic nanostructures straight onto the nanodevices and eliminating the positioning and micro-manipulation stage.
Benefitting from their fibrillar morphology, amyloid fibrils can be engineered or modified into conductive materials;21,22 these are currently in high demand both for constructing conductive circuits in nanoscale electronics and as the active components in electrochemical devices for sensing applications.23 With a rational design, high electron mobility can be achieved along the highly ordered protein fibrils, resulting in conductive wires that are also biocompatible and biodegradable. One strategy for improving electron mobility along amyloid fibrils is the incorporation of conductive nanoparticles such as AuNPs.24,25
The ferritin protein has an essential metabolic role involving iron storage and homeostasis, in practically all life forms.26,27 Genuine apoferritin (APO), the iron-free ferritin molecule, is a hollow nanocage protein composed of 24 polypeptide subunits (Mn ∼ 480 kDa). The APO organic shell is a product of the self-assembly of mainly two primary peptide subunits or chains: L “light” (∼20 kDa), and H “heavy” (∼21 kDa).28 Each subunit has been assigned a specific role: H-chains display ferroxidase activity able to catalyze the oxidation of Fe(II) to Fe(III), while L-chains provide nucleation sites for iron oxyhydroxide growth.29 The APO proteins used in this work are formed primarily of 90% L-subunits and 10% H-subunits (herein APO), as well as H-pure (APO-H), and L-pure proteins (APO-L), that is, with 100% of either the H or L subunits, respectively. The L- and H-subunits drive the final chirality and polymorphism of the APO fibrils formed.30 In a previous study, we described the formation of APO amyloid fibrils and their use as new templates for incorporating AuNPs with different morphologies.31 Herein, we demonstrate for the first time, the use of APO-derived fibrils as biotemplates to in situ grow helically arranged chains of AuNPs with conductive and optical activity in the visible range. The possible implications of these subunits in promoting AuNP formation or structuring is also investigated.
To characterize the structure of the 3D self-organization of the AuNPs in detail and quantify their distribution along the helix of the fibrils, Electron Tomography (ET) experiments were performed. In particular, the reconstruction of ET tilt series acquired using an imaging mode suitable for materials combining soft (beam sensitive) and hard matter, through Scanning-Transmission Electron Microscopy High Angle Annular Dark Field (STEM-HAADF),32 has allowed us to determine the 3D arrangement of the gold particles. This information is crucial for understanding the conductive and optical properties of these samples. Complementary, conductive atomic force microscopy (c-AFM) experiments were performed to determine the conductive properties of protein fibers containing AuNPs.
Materials and methods
Preparation of APO amyloid fibrils
Horse spleen apoferritin protein was purchased from MERCK LIFE SCIENCE SLU (Madrid, Spain). Purified protein solution (0.1 wt%) was adjusted to pH 2 (0.1 M HCl) before heat treatment (90 °C and 100 rpm, in hermetically sealed glass tubes) over an incubation time period of 9 hours. After heat treatment, the glass tube was cooled in an ice bath to quench the aggregation process.APO amyloid fibril-templated AuNPs
To prepare the AuNP hybrid system, we mixed previously synthesized APO amyloid fibrils (375 μL, 0.1 wt%) and chloroauric acid (HAuCl4·3H2O, 4 mM) from Sigma Aldrich using three different buffers: pH 2 miliQ water, 1 mM HEPES pH5 solution and 0.1 M citrate pH5 solution. The volume of chloroauric acid solution added increased from 0 to 60 μL. A 1 mM solution of NaBH4 was freshly prepared. We added 2 μL of the NaBH4 solution to the previous mixture after 15 minutes of incubation time.TEM and HAADF-STEM measurements
Samples were prepared by placing a drop onto a carbon-coated Cu grid. Electron micrographs were taken with a LIBRA 120 PLUS microscope operating at 80 and 120 KeV. High-resolution scanning transmission electron microscopy (HR-STEM), high-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM), and energy-dispersive X-ray spectroscopy (EDXS) maps were obtained with a FEI TITAN G2 microscope.Electron tomography (ET) measurements
The experiments were performed in a FEI Titan Themis 60–300 Double Aberration Corrected microscope operated at 80 kV. A convergence angle of 9 mrad was selected to improve the depth of focus. A camera length of 115 mm was used. Then, a series of STEM-HAADF images at different tilts were captured using the software FEI Explore3D v.4.1. The tracking, focusing and tilting were carried out automatically. The samples were tilted from −70° to +70° and images acquired every 5°. To denoise the tilt series, homemade scripts in MATLAB were developed using the WAVELAB850 toolbox (http://statweb.stanford.edu/%7Ewavelab/Wavelab_850/index_wavelab850.html) and the invansc package, downloaded from http://www.cs.tut.fi/%7Efoi/invansc. The denoising was performed using a combination of the Anscomb and Wavelet Transforms. The whole set of HAADF-STEM denoised images was aligned by combining a cross-correlation method, using FEI Inspect3D v 4.1, and the landmark-based alignment implemented in TomoJ. The tilt series images were also background-subtracted, normalized and binned to 512 × 512 pixels. Afterward, the images were reconstructed into a 3D volume using a Compressed Sensing algorithm based on Minimization of the Total Variation (TVM) of each single STEM-HAADF image. Specifically, a 3D implementation of the TVAL3 routine, using the AstraToolBox, was employed. FEI Avizo 7.0 software was used to visualize and further nanometrologically analyze the reconstructed volumes.UV-vis spectroscopy
UV-visible absorbance spectra of solutions were recorded at various times (30 min, 1 h, 2 h, 3 h, 5 h, 6 h) in a 24-well plate format using a Nanoquant Infinite M200 Pro multilabel plate reader.AFM measurements
All the measurements were performed with an atomic force microscope Park NX20 (Park Systems, South Korea) operating at 25 °C and at atmospheric pressure.Topographical images were obtained by scanning a 5 μm × 5 μm surface with probe ACTA at a resonant frequency of about 300 kHz and a force constant (K) of 40 N m−1.
Conductive AFM (c-AFM) measurements
CONTSPt probes were used. These probes are 225 μm long with a resonant frequency of about 25 kHz and a spring constant of 0.2 N m−1. Electrical contact was made by applying a drop of Ag paste to one corner of the crystal and to the metallic chuck. The current was measured directly using a preamplifier with a gain of 109 to 1011 V A−1 (I-AFM (internal variable gain current amplifier)). c-AFM imaging was performed with an applied bias of 3 V.Results and discussion
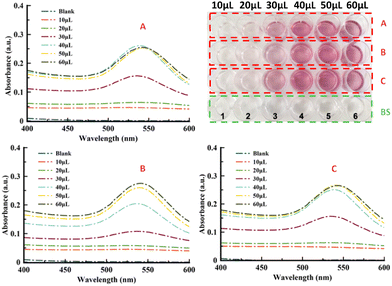
APO protein was brought to polymerization under low pH and elevated temperature conditions (Scheme 1a). This polymerization process resulted in helical APO fibrils with diameters of 6 to 9 nm and lengths of several micrometers. The fibrils had a complex substructure, consisting of 2 intertwining protofilaments.30 These APO fibrils served as templates for the in situ nucleation and growth of AuNPs along the 1D nanostructures, forming AuAPO hybrid nanowires.18 To determine the best chemical conditions for preparing AuAPO nanowires, we screened them, keeping the protein concentration constant at 2 μM, while varying the reducing reaction media and initial Au3+ concentration (Scheme 1b).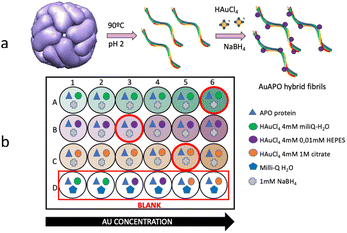 | ||
| Scheme 1 (a) Protein polymerization process illustrating amyloid-fibril formation. (b) Scheme of a 24-well plate in the experimental set-up. | ||
Fabrication of gold nanoparticle nanowires
Standard 24-well plates were used for UV-vis colorimetric determination of AuNP formation (Fig. 1). APO protein fibers were incubated with Au3+ ions for 15 min and subsequently reduced by NaBH4. The kinetic formation study of the AuAPO hybrid system at 30 min, 1, 2, 3, 5 and 6 h is presented in Fig. S1.† The absorbance spectrum of each well, with increasing concentrations of initial Au3+ ions, was collected (Fig. 1). The characteristic absorption wavelength at λ = 530 nm (ref. 33) for AuNPs was monitored in the kinetic study.In the optimization experiments, the volume of HAuCl4 salt (4 mM) added varied from 10 μl to 60 μl for 3 different reaction media: Milli-Q water pH 6 (medium A), 0.01 M HEPES pH 5 (medium B), and 1 M citrate pH 5 buffer (medium C). After adding NaBH4, the color of the APO fibril solution changed to light pink due to the Au3+ ions being reduced to Au0. As the concentration of added Au3+ increased, the color of the solution became gradually darker (Fig. 1, top-right). The UV-vis spectra for the Au-APO hybrid system showed no differences in absorption intensity after 5 h and 6 h of incubation time (Fig. S1†); also no differences were observed for the 50 and 60 μl (1 mM HAuCl4) samples (Fig. S2†), indicating that the reaction was complete. The blank solutions (BS) showed that Au3+ cannot be reduced without NaBH4.
X-ray photoelectron spectroscopy was used to determine the surface composition and chemical environment of the elements present in these hybrid inorganic–organic gold nanoparticle nanowires. An XPS complete scan (not shown) revealed the presence of carbon, nitrogen, oxygen (coming from APO protein) and gold core level. Fig. 2 shows the high-resolution XPS spectrum of the Au 4f core level of the sample prepared with an initial Au3+ added volume of 50 μl in citrate buffer (AuAPOC5). Two distinct peaks separated by 3.6 eV were observed, namely the Au 4f5/2 and Au 4f7/2 lines, which occur because of the spin–orbit splitting of the Au 4f level. The positions of these peaks were 87.5 eV and 83.9 eV, respectively, according to metallic gold (Au0 84.04 eV). XPS elemental composition (Table S1†) to determine the atomic ratio of Au metal vs. organic matter is included in ESI.† Similar spectra were obtained for AuAPOB3 and AuAPOA6.
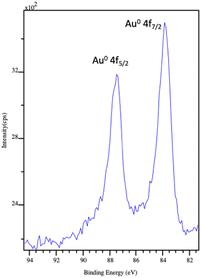 | ||
| Fig. 2 XPS spectrum of the Au 4f level of AuAPOC5 hybrid system. The two peaks at 87.5 eV and 83.9 eV correspond to the Au 4f5/2 and Au 4f7/2 core level, respectively. | ||
TEM, HAADF and ET characterization of AuAPO hybrid nanowires
A complete structural study was conducted using a TEM for those samples where the light pink color was revealed. TEM images of AuAPOA3, AuAPOB3, AuAPC3, AuAPOA5, AuAPOB5, AuAPOC5, AuAPOA6, AuAPOB6, and AuAPOC6 samples showed AuNP growth onto the APO fibers (Fig. S3†). For Au3+ added volumes below 30 μl, no AuNP formation took place, whereas AuNP aggregation was observed for volumes of more than 60 μl (Fig. S4†).Based on the optimized parameters, three samples for the 3 different media were selected for an in-depth HAADF-STEM and ET study: AuAPOB3, AuAPOC5, and AuAPOA6.
Fig. 3 (AuAPOB3 and AuAPOC5) and 4 (AuAPOA6) show TEM and STEM-HAADF micrographs of AuAPO hybrid systems and their corresponding elemental XEDS Au maps. Note that the AuNPs distributed on both sides of the fibrils (mean size 5 ± 2 nm). In this way, nearly continuous gold nanowires were successfully prepared. We were able to demonstrate the specific assembly of fairly homogeneous AuNPs onto APO based nanofibril scaffolds, suggesting a uniform growth process from the initial seeding to termination.
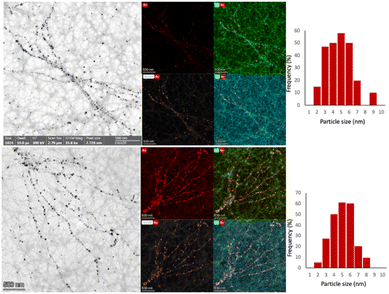 | ||
| Fig. 3 (left panels) BF-STEM images of (top) AuAPOB3 and (bottom) AuAPOC5 hybrid systems. The corresponding Au, O, and C EDX maps are also presented in the right-hand panels. | ||
To further investigate the morphology and to reveal the 3D organization of the AuAPO hybrid nanowires at the nanometric scale, HAADF-STEM electron tomography (ET) experiments were performed (Fig. 4). ET has proven to be a powerful tool for obtaining detailed 3D information for characterizing macro-molecular assemblies and different nanostructures at very high resolutions.34–39
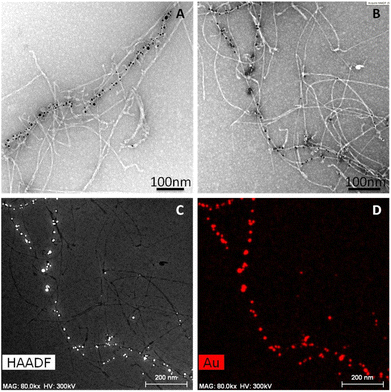 | ||
| Fig. 4 (A, B) TEM images of AuAPOA6 fibrils. (C) HAADF-STEM images and (D) the corresponding elemental mapping of gold. | ||
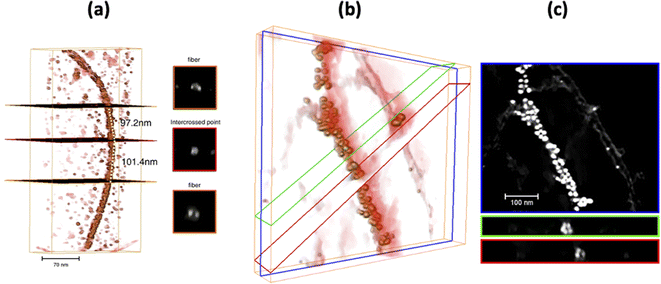
Careful analysis of the STEM images (Fig. 5) suggested a helical twist in the assembly of AuNPs on the APO fibril template. The amyloid template, like other biological templates, induces a helical AuNP arrangement following the pitch of the twisted fibrils.40–43 In the HAADF-STEM image (Fig. 5, left), the collected signal is directly related to the atomic number (Z). One question that arises when seeing these images relates to the intensity levels at which the different components of the system are imaged. Note how in the STEM-HAADF image three intensity levels are observed: (1) very intense, nanometer-sized, rounded regions, which can be assigned to the AuNPs, in which Z(Au) = 79; (2) a medium intensity, extended background, assignable to the uranyl-coated carbon film used as support on which to deposit the sample. The presence of a very thin uranyl coating, containing U (Z = 92), increases the signal intensity above that expected for a bare C-film (Z = 6); (3) a dark region with the shape expected for the APO fibrils. This suggests that the fibrils in this AuAPOA6 sample are not completely stained by uranyl.
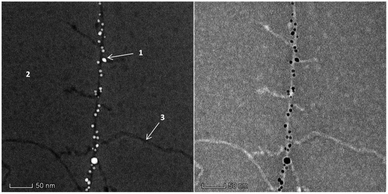 | ||
| Fig. 5 STEM-HAADF (left) and STEM-BF (right) images of the AuAPOA6 fibrils recorded in the FEI Titan3 60–300 Microscope at 80 kV. | ||
STEM tomography studies were performed on the AuAPOA6 sample to obtain 3D information about this type of material. In the STEM tomography experiments 3 different signals were simultaneously collected, namely HAADF (collection angle between 120 and 200 mrad); DF4 (collection angle between 24 and 100 mrad) and DF2 (collection angle between 12 and 24 mrad). The contrast in the DF4 and DF2 detectors, which collect electrons in the intermediate scattering angle range, mixes both diffraction and Z contrast information, with Z contrast predominating in DF4 and diffraction contrast in the case of DF2. In the case of STEM-DF2 images, the contrasts are therefore the opposite of those in HAADF images. This contrast effect allowed us to efficiently and accurately separate the signal from the AuNPs from that due to the fibrils. The former is detected as very bright areas in the HAADF image whereas the later correspond to the bright areas in the DF2 channel. Each component is therefore precisely separated from the background in one of the images. The two results can then be combined to construct the signal from the AuAPO fibril system.
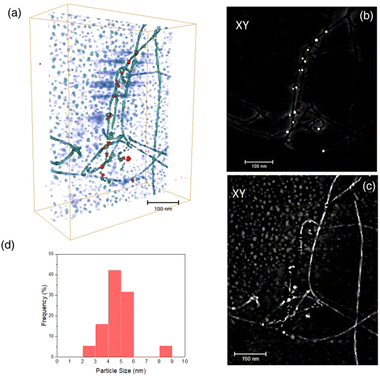
Fig. 6 presents the results of the ET study. In particular, Fig. 6a depicts the 3D rendered volume of the whole sample. This volume was obtained by combining the reconstruction of the HAADF and DF2 signals. It is therefore a composite volume. The 3D reconstruction of the HAADF signal allowed us to determine the 3D volume of the AuNPs. Fig. 6b shows an XY orthoslice of this volume, in which the AuNPs can be seen clearly. The section of these nanoparticles appears rounded, suggesting spherical-like NPs, as is evident in the 3D rendered image. It is also important to highlight the fact that the AuNPs are only observed attached to the APO fibrils, i.e., no isolated AuNPs were seen on the background.
A closer view of the 3D reconstruction clearly reveals that the AuNPs are bounded to the edge of the fibrils, suggesting their growth onto certain nucleation sites at specific locations along the APO protein, responsible for binding the reduced Au species. Fig. 6c shows an XY orthoslice of the same volume, reconstructed from the DF2 signal. In this case, note how the fibrils present greater contrast, with alternating bright and dark segments. This feature points to the helical nature of the fibrils. Fig. 6d presents the AuNP size distribution (diameter) as determined from the volumes obtained after segmentation of the reconstructed HAADF signal. According to this distribution, AuNPs ranging in size from roughly 2 to 9 nm grow onto the fibrils, with an average particle size close to 5 nm, as previously determined by TEM.
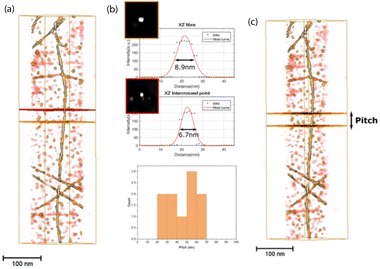
We also analyzed the 3D details of the fibrils in this sample. To do this, bare AuNP-free fibrils were chosen. Fig. 7a shows a perspective view of the rendered volume corresponding to the long fibril observed at the right side of Fig. 6a and c. There is clear alternation of greyish and yellowish zones along the fibril, which correspond to thicker and thinner areas of the fibril, respectively. At locations in the thick areas a rounded point, 6.7 nm in diameter, can be observed in the XZ orthoslice. These locations must therefore correspond to the intercrossed points existing in these APO helical nanostructures, already described in a previous work.30 The XZ orthoslices in the thin areas of the fibril show an elongated point with a larger diameter, about 9 nm, suggesting the presence of two very close strands. Although in this case it was not possible to clearly discriminate the two strands, the pitch along the fibril could still be estimated by locating the distance between the planes corresponding to two consecutive intercrossed points (Fig. 7c). From the distribution of the whole set of pitch measurements, at the bottom of Fig. 7b, an average pitch value of 50 nm was determined. This last result indicates right-handed fibrils, which is in line with the nature of the sample, comprising 90% helical right-handed structures,30 as previously described.
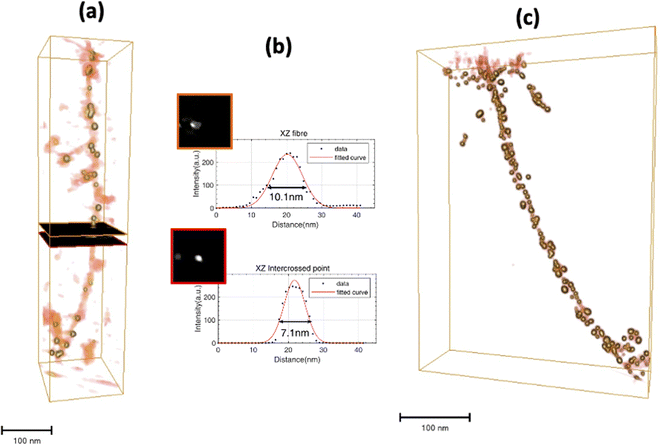
Using the same chemical reaction conditions as for AuAPOA6, AuAPO hybrid nanowires were also formed starting from apoferritin proteins comprising 100% L-subunits (APOL) and 100% H-subunits (APOH). AuAPOH and AuAPOL fibrils were additionally analyzed using ET (Fig. 8 and 9).
Fig. 8a illustrates a 3D rendering of the reconstructed volume of a single APOL fibril. Likewise, Fig. 8b and c show representative STEM results for this sample. The maximum and minimum apparent widths across the fibril are around 10 and 7 nm for AuAPOL (Fig. 8b). For the pitch, a value close to that observed in the Au-NP free fibrils, 50 nm, was measured. In this sample, it was sometimes possible to clearly discriminate the two strands (Fig. 8c). As in AuAPOA6, the AuNPs (mean size value 5 nm± 2 nm) grew on both sizes of the fibril, following the helical arrangement of the protein tem. ET tomography studies were also performed for the AuAPOH system (Fig. 9). No difference was detected in this case in terms of the maximum apparent width across the fibrils, which was also around 10 nm. However, a much larger value was observed for the pitch, which in this sample amounted, on average, to roughly 90 nm.
A closer inspection of the 3D reconstruction clearly reveals that the AuNPs bind to the edge of the fibrils in a helical arrangement and the AuNPs have a mean size of 5 nm± 2 nm. Here, we should highlight that the mean pitch values measured from the ET reconstructions are in excellent agreement with those obtained in previous work.30
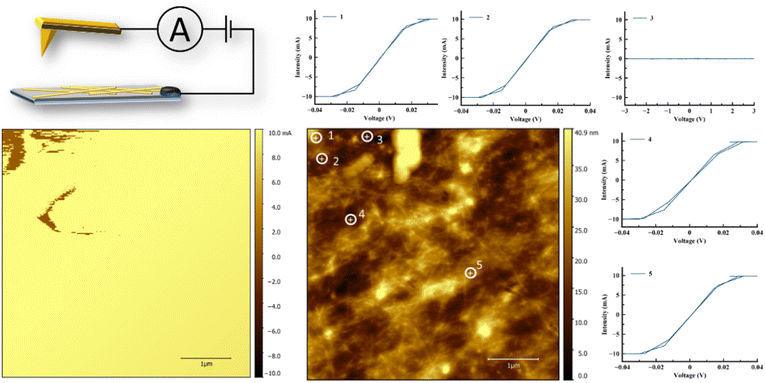
Conductivity measurements of AuAPOA3, AuAPOB5 and AuAPOC6 samples were performed through conductive atomic force microscopy (c-AFM) after evaporation on Si/SiO2 (200 nm) substrate surfaces. The electrical behavior of randomly deposited AuAPO metallic fibers was assessed by c-AFM and compared to other similar systems.44–50 The AFM images showed that the AuAPOA3 and AuAPOC6 nanofibers were very aggregated and it was not possible to close the circuit. The AuAPOB5 sample conductivity measurements are shown in Fig. 10. The current image and its corresponding topographical image are presented in Fig. 10a and b, respectively. Current–voltage measurements were taken while contacting various individual points on different locations (points are shown in Fig. 10a) to determine the nanowire conductivity. The results of the current–voltage (IV) measurements are shown in Fig. 10c. From the IV curve, it can be concluded that nanofibers exhibit ohmic behavior and the absence of hysteresis confirms a continuous metallic structure. This observation is consistent with 1-D electron transport. The measurements were made at a bias voltage in the −3 to 3 V range, with a maximum conductivity of 10 mA. The IV curve corresponding to point 3 did not show conductivity and served as proof of the non-conductive surface, according to the topographic image. The current maps of the sample indicate that while most of the top surface of the sample is not conductive, spots with high conductivity are spread over the surface. These observations confirm that conduction occurs through localized conduction pathways, revealing AuAPO to be a highly conductive hybrid system. Compared to similar systems, that is, AuNPs synthesis templated by DNA or other proteins our system present current intensity values an order of magnitude superior.44,45,47,48
The electrical properties of the material are rather stable. In fact, measurements of the I–V behavior in 3 consecutive cycles revealed that conductivity was fully retained.
Conclusions
In this study, we report how amyloid APO protein fibrils can serve as scaffolds onto which AuNPs can be grown, leading to AuAPO inorganic-organic hybrid systems. These systems facilitate the development of conductive nanoscaffolds that have a potential in a variety of both biological and non-biological applications.Through an easy, one-step chemical set-up, 1D Au-metallic nanofibrils were prepared starting from iso-apoferritin proteins, with three different L/H subunit ratios, ranging from 100%, through 90%, down to 0%. We showed that in all cases APO protein fibrils have the ability to nucleate and grow AuNPs, leading to AuAPO hybrid nanofibrils with improved conductivity and optical properties. In terms of AuNP formation, no significant differences were observed among the samples prepared using the three different reducing media studied in this work. An incubation time of 5 or 6 h and an Au3+ added volume of 30–60 μl from a HAuCl4 salt solution (4 mM) were the most suitable chemical conditions for forming a sol containing the AuAPO hybrid system.
Advanced electron microscopy techniques evidence a helical twist in the AuNP assembly on the APO fibril template. The amyloid template, like other biological templates, induces a helical arrangement of AuNPs, following the pitch of the twisted fibrils, in other words, 50 or 90 nm.
Spectroscopic studies of gold nanofibrils showed the characteristic LSPR bands of spherical AuNPs. There was no difference in the final mean size value obtained for AuNPs (5 nm± 2 nm) prepared in situ using APOs with different L or H subunit contents. Current–voltage measurements indicate that the nanofibers exhibit ohmic behavior and have a continuous metallic structure.
Author contributions
Conceptualization, N. G.; method, G. P. and M. L. H.; software, M. L. H. and J. J. C.; validation, J. J. C.; formal analysis, G. P. and M. L. H.; investigation, R. C.; resources, J. M. D. V. and N. G.; writing-original draft preparation, N. G.; all the authors participated in writing-reviewing and editing; all the authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Junta de Andalucía Project P18-FQM-1373 and MINECO Project PID2019-111461GB-100. The authors acknowledge the “Unidad de Excelencia de Quimica aplicada a Biomedicina y Medioambiente” at the University of Granada.References
- G. Chen, I. Roy, C. Yang and P. N. Prasad, Chem. Rev., 2016, 116, 2826–2885 CrossRef CAS PubMed.
- P. M. Tiwari, K. Vig, V. A. Dennis and S. R. Singh, Nanomaterials, 2011, 1, 31–63 CrossRef CAS PubMed.
- N. Elahi, M. Kamali and M. H. Baghersad, Talanta, 2018, 184, 537–556 CrossRef CAS PubMed.
- L. Dykman and N. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
- Y. Wu, M. R. K. Ali, K. Chen, N. Fang and M. A. El-Sayed, Nano Today, 2019, 24, 120–140 CrossRef CAS.
- S. Siddique and J. C. L. Chow, Appl. Sci., 2020, 10 Search PubMed.
- P. Sharma, S. Brown, G. Walter, S. Santra and B. Moudgil, Adv. Colloid Interface Sci., 2006, 123–126, 471–485 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- X. Hu, Y. Zhang, T. Ding, J. Liu and H. Zhao, Front. Bioeng. Biotechnol., 2020, 8, 1–17 CrossRef PubMed.
- N. Zhao, L. Yan, X. Zhao, X. Chen, A. Li, D. Zheng, X. Zhou, X. Dai and F. J. Xu, Chem. Rev., 2019, 119, 1666–1762 CrossRef CAS PubMed.
- O. Deschaume, B. De Roo, M. J. Van Bael, J. Locquet, C. Van Haesendonck and C. Bartic, Chem. Mater., 2014, 26(18), 5383–5393 CrossRef CAS.
- R. A. Alvarez-Puebla, A. Agarwal, P. Manna, B. P. Khanal, P. Aldeanueva-Potel, E. Carbo-Argibay, N. Pazos-Perez, L. Vigderman, E. R. Zubarev, N. A. Kotov and L. M. Liz-Marzan, Proc. Natl. Acad. Sci., 2011, 108, 8157–8161 CrossRef CAS PubMed.
- C.-L. Chen, P. Zhang and N. L. Rosi, J. Am. Chem. Soc., 2008, 130, 13555–13557 CrossRef CAS PubMed.
- H. Li, X. Gao, C. Zhang, Y. Ji, Z. Hu and X. Wu, Biosensors, 2022, 12, 957 CrossRef CAS PubMed.
- Z. S. Al-Garawi, J. R. Thorpe and L. C. Serpell, Angew. Chem., Int. Ed., 2015, 54, 13327–13331 CrossRef CAS PubMed.
- S. Bolisetty, J. Adamcik, J. Heier and R. Mezzenga, Adv. Funct. Mater., 2012, 22, 3424–3428 CrossRef CAS.
- S. Padalkar, J. D. Hulleman, S. M. Kim, J. C. Rochet, E. A. Stach and L. A. Stanciu, Nanotechnology, 2008, 19, 275602 CrossRef CAS PubMed.
- R. Jurado and N. Gálvez, Nanomaterials, 2021, 11, 1–12 CrossRef PubMed.
- R. Jurado, F. Castello, P. Bondia, S. Casado, C. Flors, R. Cuesta, J. M. Domínguez-Vera, A. Orte and N. Gálvez, Nanoscale, 2016, 8, 9648–9656 RSC.
- T. P. J. Knowles and R. Mezzenga, Adv. Mater., 2016, 28, 6546–6561 CrossRef CAS PubMed.
- T. Scheibel, R. Parthasarathy, G. Sawicki, X. Lin, H. Jaeger and S. L. Lindquist, PNAS, 2003, 100, 4527–4532 CrossRef CAS PubMed.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- C. A. E. Hauser, S. Maurer-Stroh and I. C. Martins, Chem. Soc. Rev., 2014, 43, 5326–5345 RSC.
- J. Zhou, A. Saha, J. Adamcik, H. Hu, Q. Kong, C. Li and R. Mezzenga, Adv. Mater., 2015, 27, 1945–1950 CrossRef CAS PubMed.
- C. Li, S. Bolisetty and R. Mezzenga, Adv. Mater., 2013, 25, 3694–3700 CrossRef CAS PubMed.
- P. Harrison and P. Arosio, Biochim. Biophys. Acta, 1996, 1275, 161–203 CrossRef PubMed.
- P. M. Harrison, Biochim. Biophys. Acta, 1996, 1275, 161–203 CrossRef PubMed.
- D. Finazzi and P. Arosio, Arch. Toxicol., 2014, 1787–1802 CrossRef CAS PubMed.
- D. M. Lawson, P. J. Artymiuk, S. J. Yewdall, J. M. A. Smith, J. C. Livingstone, A. Treffry, A. Luzzago, S. Levi, P. Arosio, G. Cesareni, C. D. Thomas, W. V. Shaw and P. M. Harrison, Nature, 1991, 349, 541–544 CrossRef CAS PubMed.
- R. Jurado, J. Adamcik, M. López-Haro, J. A. González-Vera, Á. Ruiz-Arias, A. Sánchez-Ferrer, R. Cuesta, J. M. Domínguez-Vera, J. J. Calvino, A. Orte, R. Mezzenga and N. Gálvez, J. Am. Chem. Soc., 2019, 141 Search PubMed.
- N. Gálvez, B. Fernandez, E. Valero, P. Sánchez, R. Cuesta and J. M. Domínguez-Vera, C. R. Chim., 2008, 11, 1207–1212 CrossRef.
- D. J. Stokes and E. Baken, Imaging Microsc., 2007, 9, 18–20 CrossRef.
- N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS PubMed.
- P. A. Midgley and R. E. Dunin-Borkowski, Nat. Mater., 2009, 8, 271–280 CrossRef CAS PubMed.
- M. Kollmer, K. Meinhardt, C. Haupt, F. Liberta, M. Wulff, J. Linder, L. Handl, L. Heinrich, C. Loos, M. Schmidt, T. Syrovets, T. Simmet, P. Westermark, G. T. Westermark, U. Horn, V. Schmidt, P. Walther and M. Fändrich, Proc. Natl. Acad. Sci., 2016, 113, 5604–5609 CrossRef CAS PubMed.
- S. Han, M. Kollmer, D. Markx, S. Claus, P. Walther and M. Fändrich, Sci. Rep., 2017, 7, 43577 CrossRef PubMed.
- E. Vázquez-Fernández, M. R. Vos, P. Afanasyev, L. Cebey, A. M. Sevillano, E. Vidal, I. Rosa, L. Renault, A. Ramos, P. J. Peters, J. J. Fernández, M. van Heel, H. S. Young, J. R. Requena and H. Wille, PLoS Pathog., 2016, 12, 1–21 Search PubMed.
- P. Rueda-Fonseca, E. Robin, E. Bellet-Amalric, M. Lopez-Haro, M. Den Hertog, Y. Genuist, R. André, A. Artioli, S. Tatarenko, D. Ferrand and J. Cibert, Nano Lett., 2016, 16, 1637–1642 CrossRef CAS PubMed.
- M. Lopez-Haro, L. Guétaz, T. Printemps, A. Morin, S. Escribano, P.-H. Jouneau, P. Bayle-Guillemaud, F. Chandezon and G. Gebel, Nat. Commun., 2014, 5, 5229 CrossRef CAS PubMed.
- C. L. Chen, P. Zhang and N. L. Rosi, J. Am. Chem. Soc., 2008, 130, 13555–13557 CrossRef CAS PubMed.
- J. Kumar, H. Eraña, E. López-Martínez, N. Claes, V. F. Martín, D. M. Solís, S. Bals, A. L. Cortajarena, J. Castilla and L. M. Liz-Marzán, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 3225–3230 CrossRef CAS PubMed.
- C. Song, M. G. Blaber, G. Zhao, P. Zhang, H. C. Fry, G. C. Schatz and N. L. Rosi, Nano Lett., 2013, 13, 3256–3261 CrossRef CAS PubMed.
- A. Kuzyk, R. Schreiber, Z. Fan, G. Pardatscher, E. M. Roller, A. Högele, F. C. Simmel, A. O. Govorov and T. Liedl, Nature, 2012, 483, 311–314 CrossRef CAS PubMed.
- T. Scheibel, R. Parthasarathy, G. Sawicki, X. M. Lin, H. Jaeger and S. L. Lindquist, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4527–4532 CrossRef CAS PubMed.
- M. Moreno-Moreno, P. Ares, C. Moreno, F. Zamora, C. Gómez-Navarro and J. Gómez-Herrero, Nano Lett., 2019, 19, 5459–5468 CrossRef CAS PubMed.
- A. Stern, G. Eidelshtein, R. Zhuravel, G. I. Livshits, D. Rotem, A. Kotlyar and D. Porath, Adv. Mater., 2018, 30, 1800433 CrossRef PubMed.
- R. S. Gill, R. F. Saraf and S. Kundu, ACS Appl. Mater. Interfaces, 2013, 5, 9949–9956 CrossRef CAS PubMed.
- S. Kundu, R. S. Gill and R. F. Saraf, J. Phys. Chem. C, 2011, 115, 15845–15852 CrossRef CAS.
- C. Li, S. Bolisetty and R. Mezzenga, Adv. Mater., 2013, 25, 3694–3700 CrossRef CAS PubMed.
- R. A. Taheri, Y. Akhtari, T. Tohidi Moghadam and B. Ranjbar, Sci. Rep., 2018, 8, 1–10 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: kinetic formation study of AuAPO hybrid system at different incubation times; Fig. S2: kinetic formation study of AuAPO hybrid system at different Au3+ concentrations. Fig. S3 and S4: TEM study of prepared AuAPO hybrid systems. See DOI: https://doi.org/10.1039/d3ra03186a |
| This journal is © The Royal Society of Chemistry 2023 |