Open Access Article
Open Access ArticleDiscovery of new pyridine heterocyclic hybrids; design, synthesis, dynamic simulations, and in vitro and in vivo breast cancer biological assays†
Menna M. Abdelshaheeda,
Hussein I. El Subbaghb,
Mohamed A. Tantawycdg,
Reem T. Attiae,
Khairia M. Yousseff and
Iten M. Fawzy *f
*f
aDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Horus University in Egypt, New Damietta, Egypt
bDepartment of Medicinal Chemistry, Faculty of Pharmacy, Mansoura University, P.O. Box 35516, Mansoura, Egypt
cHormones Department, Medical Research and Clinical Studies Institute, National Research Centre, Dokki, Giza, Egypt
dStem Cells Lab, Center of Excellence for Advanced Sciences, National Research Centre, Dokki, Cairo, Egypt
eDepartment of Pharmacology and Toxicology and Biochemistry, Faculty of Pharmacy, Future University in Egypt, 11835, Cairo, Egypt
fDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Future University in Egypt, 11835, Cairo, Egypt. E-mail: iten.mamdouh@fue.edu.eg; Tel: +201006064161
gDepartment of Orthopaedics and Rehabilitation, CORTS, Penn State University, College of Medicine, 500 University Drive, Hershey, PA 17033-0850, USA
First published on 24th May 2023
Abstract
Pyridine is a nitrogen bearing heterocyclic scaffold that shows a wide range of biological activities. The pyridine nucleus has become an interesting target for medicinal chemistry researchers worldwide. Several pyridine derivatives exhibited good anticancer effects against diverse cell lines. Therefore, to explore new anticancer pyridine entities, novel pyridine derivatives were designed and synthesized and evaluated for their anticancer abilities in vitro and in vivo. All of the target compounds were evaluated against three different human cancer cell lines (Huh-7, A549 and MCF-7) via MTT assay. Most of the compounds exhibited significant cytotoxic activities. Compounds 3a, 3b, 5a and 5b showed superior antiproliferative activities to Taxol. Where, compound 3b showed IC50 values of 6.54, 15.54 and 6.13 μM compared to Taxol (6.68, 38.05, 12.32 μM) against Huh-7, A549 and MCF-7, respectively. Also, tubulin polymerization assay was carried out. The most potent compounds 3a, 3b, 5a and 5b could significantly inhibit tubulin polymerization with IC50 values of 15.6, 4.03, 6.06 and 12.61 μM, respectively. Compound 3b exhibited the highest tubulin polymerization inhibitory effect with an IC50 value of 4.03 μM compared to combretastatin (A-4) (1.64 μM). Molecular modeling studies of the designed compounds confirmed that most of the compounds made the essential binding interactions compared to the reference compound which assisted in the prediction of the structure requirements for the detected anticancer activity. Finally, in vivo studies showed that compound 3b could significantly inhibit breast cancer.
1. Introduction
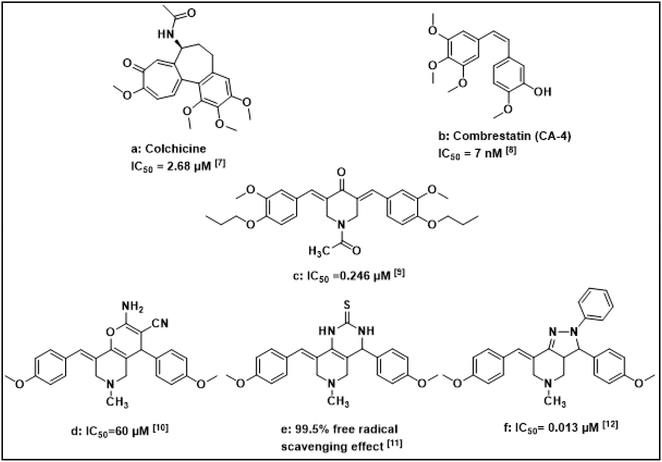
Cancer, uncontrolled cell growth, is a fatal disease. It is also one of the top leading causes of death worldwide. Cancer deaths are predicted to increase from 7.6 million in 2008 to 13 million in 2030.1,2 Despite improved diagnostic techniques, cancer still affects millions of patients worldwide.3 Although chemotherapy is the most common treatment of cancer, many chemotherapeutic agents cause severe side effects due to their cytotoxic and mutagenic effects on healthy cells. Notably, pyridine has been used as a key pharmacophore in drug design with an improved safety profile and less toxicity to non-tumorigenic cells. Several pyridine derivatives were reported to inhibit enzymes, receptors, and other targets to control and cure cancer. Therefore, researchers are currently focusing on the development of pyridine-based novel derivatives for cancer treatment.4–6Literature survey showed that different fused rings bearing pyridine moiety exhibited anticancer activity such as pyrano [3,2-c] pyridine, pyrido[4,3-d]-pyrimidine and pyrazolo[4,3-c]pyridine derivatives13 (Fig. 1). These reported fused rings were proved to be potent and selective antitumor agents.
 | ||
| Fig. 1 Reported antitumor agents with fused rings-pyridine bearing nucleus.7–12 | ||
Dynamic microtubules play an essential role in variable cellular processes such as intracellular trafficking, cell migration, cell skeleton development and cell division.14–16
Tubulin polymerization inhibitors are increasingly being investigated as anti-cancer drugs. Whereas microtubule targeting agents (MTAs) can interfere with microtubule dynamics and arrest cancer cells in the G2/M phase, therefore cancer cells undergo apoptosis.17–19
Compounds that bind to the colchicine site (Fig. 2) have been widely studied to find new agents that can deal with the limitations of existing tubulin-targeted drugs.20 Disruption of tubulin polymerization is an old anticancer target, however, this target is still one of the hit targets for discovery of anticancer agents. Several new anticancer drugs have been discovered via the microtubule/tubulin interaction-based drug design. Taxol has been a popular representative for tubulin-interactive targeting drugs which was used for many years as anticancer. Meanwhile, there have been emerging scaffolds of tubulin-interactive natural products as combretastatin A-4, epothilones A and B, halichondrin B, thiazole alkaloid, phenylahistin, diketopiperazine and dolastatin 10 peptide.21
Combretastatin A-4 (CA-4) (Fig. 1) is a strong antitumor agent which was isolated from Combretum caffrum family. CA-4 acts by inhibiting polymerisation of tubulin at colchicine binding site. Trimethoxyphenyl (TMP) moiety in CA-4 was found to be essential for activity as the methoxy group is responsible for the critical hydrogen bond with Cys 12.22–24 Therefore, methoxy groups were preserved in most anticancer agents targeting colchicine binding site.
In this study new pyridine heterocyclic hybrids were developed using pyran, pyrimidine and pyrazole rings. The synthesized hybrids were evaluated for their antiproliferative activity against three types of cancer cell lines: hepatocyte derived carcinoma, lung, and breast cancer cell lines. Meanwhile, the compounds were tested for their depolymerization ability towards tubulin. This was deeply illustrated via molecular docking and dynamic simulation studies. The most active compound was selected for in vivo activity investigation against breast cancer.
2. Experimental
The 1H NMR spectra were recorded on JEOL 500 MHz spectrometer at the faculty of science, Mansoura University whereas 13C NMR spectra were recorded on JEOL 125 MHz spectrometer in which TMS was used as an internal standard and chemical shifts were expressed as ppm and coupling constants (J) were given in Hz. Chemical shifts are given on the delta (d) scale in parts per million (ppm). Mass spectrometer (Hewlett Packard 5988) was utilised to record on spectrometer at Regional Centre for Mycology and Biotechnology, Al-Azhar University. Elemental analyses (%C, H, N, S) were also performed using (Thermo Scientific c Flash 2000 elemental analyser) at the Regional Center for Mycology and Biotechnology, Al-Azhar University and the results were in accordance with the proposed structures. Melting points were determined using capillary tubes with a Stuart SMP30 apparatus and are uncorrected. Progression of the reactions was monitored using TLC Merck Kieselgel 60 F254 aluminum packed plates.2.1. General procedure for synthesis of (2a–d) intermediates
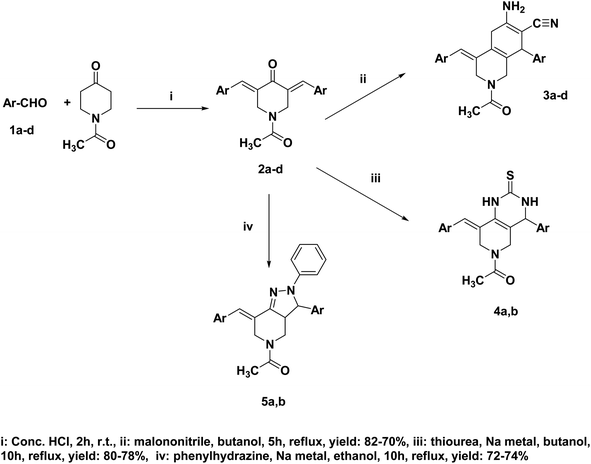
The intermediate compounds 2a–d were prepared with a procedure similar to a condensation reaction obtained from literature where (0.02 mol) of the appropriate aldehyde was added and condensed with (0.01 mol/1.41 ml) of N-acetyl-4-piperidone and then heated in a water bath at 25–30 °C until a clear solution was gained, then 4 ml of concentrated HCl was added dropwise while stirring for 5 min. The reaction mixture was eventually stirred at room temperature for 2 h. The mixture was left standing for 2 days, then it was treated with cold AcOH/water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and filtered. The solid obtained was then dried well and recrystallized from methanol/ethanol. The obtained intermediates were reported25 & their characterization coincided with those in literature.
1) and filtered. The solid obtained was then dried well and recrystallized from methanol/ethanol. The obtained intermediates were reported25 & their characterization coincided with those in literature.
2.2. General procedure for synthesis of (3a–d) target compounds
A mixture of diarylidene compounds 2a–d (0.01 mol) and malononitrile (0.7 g, 0.01 mol) in butanol (50 ml) was heated under reflux for 5 h. Evaporation of the solvent yielded the target compounds which were then washed, filtered off, dried, and recrystallized from ethanol.26![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1680 (alkenic C
N), 1680 (alkenic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z%: 475.50 [M+] (45.73%). 1H-NMR (400 MHz, DMSO-d6) δ 9.70 (s, 2H, OH, D2O exchangeable), 7.61 (s, 1H, CH
O of amide). Ms: m/z%: 475.50 [M+] (45.73%). 1H-NMR (400 MHz, DMSO-d6) δ 9.70 (s, 2H, OH, D2O exchangeable), 7.61 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.58 (dd, J = 7.5, 8.9 Hz, 2H, ArH), 7.11–7.09 (dd, J = 8 Hz, 2H, ArH), 7.00 (s, 1H, ArH), 6.87 (s, 2H, NH2 D2O exchangeable), 6.86 (s, 1H, ArH), 6.85 (s, 1H, C4–H), 4.77 (s, 2H, CH2–N), 4.75 (s, 2H, CH2–N), 3.80 (s, 3H, CH3–O), 3.78 (s, 3H, CH3–O), 1.87 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.76(C
C), 7.58 (dd, J = 7.5, 8.9 Hz, 2H, ArH), 7.11–7.09 (dd, J = 8 Hz, 2H, ArH), 7.00 (s, 1H, ArH), 6.87 (s, 2H, NH2 D2O exchangeable), 6.86 (s, 1H, ArH), 6.85 (s, 1H, C4–H), 4.77 (s, 2H, CH2–N), 4.75 (s, 2H, CH2–N), 3.80 (s, 3H, CH3–O), 3.78 (s, 3H, CH3–O), 1.87 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.76(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 168.75 (C–NH2), 148.60 (ArC), 148.58 (C
O), 168.75 (C–NH2), 148.60 (ArC), 148.58 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 136.71, 136.50, 129.97, 129.79, 126.02, 125.78, 124.47 (ArC), 124.42 (C
C), 136.71, 136.50, 129.97, 129.79, 126.02, 125.78, 124.47 (ArC), 124.42 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.84 (C
C), 115.84 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.80 (C
C), 115.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.09 (CN), 115.01 (C–CN), 55.72 (O–CH3) 47.11 (C4), 42.43 (CH2NCH2), 20.93 (CH3). Anal. calcd. for C26H25N3O6 (475.50): C, 65.68; H, 5.30; N, 8.84, found: C, 65.82; H, 5.41; N, 9.07.
C), 115.09 (CN), 115.01 (C–CN), 55.72 (O–CH3) 47.11 (C4), 42.43 (CH2NCH2), 20.93 (CH3). Anal. calcd. for C26H25N3O6 (475.50): C, 65.68; H, 5.30; N, 8.84, found: C, 65.82; H, 5.41; N, 9.07.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N),1680 (alkenic C
N),1680 (alkenic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z%: 504.21 [M+] (30.3%). 1H-NMR (400 MHz, DMSO-d6) δ 9.61 (s, 2H, OH, D2O exchangeable), 7.56 (s, 1H, CH
O of amide). Ms: m/z%: 504.21 [M+] (30.3%). 1H-NMR (400 MHz, DMSO-d6) δ 9.61 (s, 2H, OH, D2O exchangeable), 7.56 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.07 (dd, J = 7.9, 8.5 Hz, 2H, ArH), 6.98 (s, 2H, NH2, D2O exchangeable), 6.99 (s, 1H, ArH), 6.95 (s, 1H, ArH), 6.87 (dd, J = 8 Hz, 2H, ArH), 4.75 (s, 2H, CH2–N), 4.74 (s, 2H, CH2–N), 4.07 (s, 1H, C4–H), 4.01–4.06 (q, J = 8 Hz, 4H, O–CH2–CH3), 1.87 (s, 3H, CH3), 1.31 (t, J = 7.5 Hz, 6H, CH3–CH2–O). 13C-NMR (300 MHz, DMSO-d6): δ 185.69 (C
C), 7.07 (dd, J = 7.9, 8.5 Hz, 2H, ArH), 6.98 (s, 2H, NH2, D2O exchangeable), 6.99 (s, 1H, ArH), 6.95 (s, 1H, ArH), 6.87 (dd, J = 8 Hz, 2H, ArH), 4.75 (s, 2H, CH2–N), 4.74 (s, 2H, CH2–N), 4.07 (s, 1H, C4–H), 4.01–4.06 (q, J = 8 Hz, 4H, O–CH2–CH3), 1.87 (s, 3H, CH3), 1.31 (t, J = 7.5 Hz, 6H, CH3–CH2–O). 13C-NMR (300 MHz, DMSO-d6): δ 185.69 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 168.65 (C–NH2), 148.82 (ArC), 146.76 (C
O), 168.65 (C–NH2), 148.82 (ArC), 146.76 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 146.73, 136.66, 136.46, 129.87, 129.71, 125.96 (ArC), 125.72 (C
C), 146.73, 136.66, 136.46, 129.87, 129.71, 125.96 (ArC), 125.72 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 124.48 (C
C), 124.48 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 116.21 (C
C), 116.21 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 116.12(CN), 115.87 (C–CN), 63.92 (CH2–CH3), 63.86 (CH2–CH3) 47.05 (C4), 42.37 (CH2–N–CH2), 20.90 (CH3) 14.71 (CH3–CH2). Anal. calcd. for C28H29N3O6 (503.56): C, 66.79; H, 5.81; N, 8.34, found: C, 66.82; H, 5.77; N, 8.29.
C), 116.12(CN), 115.87 (C–CN), 63.92 (CH2–CH3), 63.86 (CH2–CH3) 47.05 (C4), 42.37 (CH2–N–CH2), 20.90 (CH3) 14.71 (CH3–CH2). Anal. calcd. for C28H29N3O6 (503.56): C, 66.79; H, 5.81; N, 8.34, found: C, 66.82; H, 5.77; N, 8.29.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1680 (alkenic C
N), 1680 (alkenic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1650 (C
C–) and 1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide), 850 (C–Cl). Ms: m/z%: 453.08 [M+] (63.9%). 1H-NMR (400 MHz, DMSO-d6) δ 7.63 (s, 1H, CH
O of amide), 850 (C–Cl). Ms: m/z%: 453.08 [M+] (63.9%). 1H-NMR (400 MHz, DMSO-d6) δ 7.63 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.58 (dd, J = 8, 7.7 Hz, 2H, ArH), 7.57 (dd, J = 7.5, 8.2 Hz, 2H, ArH), 7.55 (dd, J = 8, 7.4 Hz, 2H, ArH), 7.54 (dd, J = 8, 7.6 Hz, 2H, ArH), 7.53 (s, 2H, NH2 D2O exchangeable), 7.52 (s, 1H, C4–H) 4.75 (s, 2H, CH2–N), 4.74 (s, 2H, CH2–N), 1.84 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.3(C
C), 7.58 (dd, J = 8, 7.7 Hz, 2H, ArH), 7.57 (dd, J = 7.5, 8.2 Hz, 2H, ArH), 7.55 (dd, J = 8, 7.4 Hz, 2H, ArH), 7.54 (dd, J = 8, 7.6 Hz, 2H, ArH), 7.53 (s, 2H, NH2 D2O exchangeable), 7.52 (s, 1H, C4–H) 4.75 (s, 2H, CH2–N), 4.74 (s, 2H, CH2–N), 1.84 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.3(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.4 (C–NH2), 148.58 (ArC), 142.9 (C
O), 161.4 (C–NH2), 148.58 (ArC), 142.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C),142.4, 136.8, 134.4, 130.8, 129.1, 129.0, 128.7 (ArC), 124.42 (C
C),142.4, 136.8, 134.4, 130.8, 129.1, 129.0, 128.7 (ArC), 124.42 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.84 (C
C), 115.84 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.80 (C
C), 115.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 115.09 (CN), 115.01 (C–CN), 55.72 (CH2–CH3), 47.11 (C4), 42.43 (CH2–N–CH2), 20.93 (CH3). Anal. calcd. for C24H19Cl2N3O2 (452.34): C, 63.73; H, 4.23; Cl, 15.67; N, 9.29, found: C, 63.53; H, 4.25; Cl, 15.70; N, 9.20.
C), 115.09 (CN), 115.01 (C–CN), 55.72 (CH2–CH3), 47.11 (C4), 42.43 (CH2–N–CH2), 20.93 (CH3). Anal. calcd. for C24H19Cl2N3O2 (452.34): C, 63.73; H, 4.23; Cl, 15.67; N, 9.29, found: C, 63.53; H, 4.25; Cl, 15.70; N, 9.20.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1680 (alkenic C
N), 1680 (alkenic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z%: 436.20 [M+] (30.3%). 1H-NMR (400 MHz, DMSO-d6) δ 7.68 (dd, J = 7.4, 8 Hz, 2H, ArH), 7.41 (dd, J = 7.5, 8.4 Hz, 2H, ArH), 7.37 (t, J = 8 Hz, 4H, ArH), 7.34 (t, J = 7.4 Hz, 2H, ArH), 7.30 (t, J = 7.6 Hz, CH
O of amide). Ms: m/z%: 436.20 [M+] (30.3%). 1H-NMR (400 MHz, DMSO-d6) δ 7.68 (dd, J = 7.4, 8 Hz, 2H, ArH), 7.41 (dd, J = 7.5, 8.4 Hz, 2H, ArH), 7.37 (t, J = 8 Hz, 4H, ArH), 7.34 (t, J = 7.4 Hz, 2H, ArH), 7.30 (t, J = 7.6 Hz, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.28 (d, J = 8 Hz, 1H, CH
CH), 7.28 (d, J = 8 Hz, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.24 (d, J = 8 Hz, 1H, CH
CH), 7.24 (d, J = 8 Hz, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.22 (s, 2H, NH2, D2O exchangeable), 7.21 (d, J = 8 Hz, 1H, CH
CH), 7.22 (s, 2H, NH2, D2O exchangeable), 7.21 (d, J = 8 Hz, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.18 (t, J = 7.5 Hz, 1H, CH
CH), 7.18 (t, J = 7.5 Hz, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.15 (d, J = 7.5 Hz, 1H, C4–H), 4.72 (s, 2H, CH2–N), 4.71 (s, 2H, CH2–N), 2.10 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.23 (C
CH), 7.15 (d, J = 7.5 Hz, 1H, C4–H), 4.72 (s, 2H, CH2–N), 4.71 (s, 2H, CH2–N), 2.10 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 185.23 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 168.79 (C–NH2), 142.60, 142.49 (ArC), 136.27 (C
O), 168.79 (C–NH2), 142.60, 142.49 (ArC), 136.27 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 136.17 (ArC) 135.70 (C
C), 136.17 (ArC) 135.70 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 135.29, 131.78, 131.44 (ArC), 129.34, 128.88, 128.53 (C
C), 135.29, 131.78, 131.44 (ArC), 129.34, 128.88, 128.53 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 127.70, 127.31 (ArC), 123.04 (CN), 60.37 (C–CN), 45.62 (1C, C4) 41.58 (1C, CH2–N), 34.70 (CH2–N), 21.21 (1C, CH3). Anal. calcd. for C28H25N3O2 (435.53): C, 77.22; H, 5.79; N, 9.65 found: C, 67.05; H, 5.97; N, 8.50.
C), 127.70, 127.31 (ArC), 123.04 (CN), 60.37 (C–CN), 45.62 (1C, C4) 41.58 (1C, CH2–N), 34.70 (CH2–N), 21.21 (1C, CH3). Anal. calcd. for C28H25N3O2 (435.53): C, 77.22; H, 5.79; N, 9.65 found: C, 67.05; H, 5.97; N, 8.50.2.3. General procedure for synthesis of (4a, b) target compounds
To a mixture of the diarylidene compound 2a/b (0.01 mol) and thiourea (0.8 g, 0.01 mol), sodium butoxide was added (Na metal (0.5 g) in butanol (50 ml)) and heated under reflux for 10 h.27 The solvent used was evaporated in vacuo, water (20 ml) was then added, and the mixture was neutralized to pH 6. The separated solid was then filtered, washed, dried, and recrystallized from ethanol.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z: 468.15 [M+] (26.0%). 1H-NMR (400 MHz, DMSO-d6) δ 9.70 (s, 2H, OH, D2O exchangeable), 7.61 (s, 1H, NH–CH, D2O exchangeable), 7.58 (s, 1H, NH, D2O exchangeable), 7.10 (s, 1H, CH
O of amide). Ms: m/z: 468.15 [M+] (26.0%). 1H-NMR (400 MHz, DMSO-d6) δ 9.70 (s, 2H, OH, D2O exchangeable), 7.61 (s, 1H, NH–CH, D2O exchangeable), 7.58 (s, 1H, NH, D2O exchangeable), 7.10 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.08 (s, 1H, ArH), 7.01 (s, 1H, ArH), 6.99–6.97 (dd, J = 8, 7.1 Hz, 2H, ArH), 6.87–6.85 (dd, J = 8, 7.2 Hz, 2H, ArH), 4.77 (d, 1H, C4–H), 4.77 (s, 2H, CH2–N), 4.75 (s, 2H, CH2–N), 3.80 (s, 3H, CH3–O), 3.78 (s, 3H, CH3–O), 1.87 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6) δ 184.61 (C
C), 7.08 (s, 1H, ArH), 7.01 (s, 1H, ArH), 6.99–6.97 (dd, J = 8, 7.1 Hz, 2H, ArH), 6.87–6.85 (dd, J = 8, 7.2 Hz, 2H, ArH), 4.77 (d, 1H, C4–H), 4.77 (s, 2H, CH2–N), 4.75 (s, 2H, CH2–N), 3.80 (s, 3H, CH3–O), 3.78 (s, 3H, CH3–O), 1.87 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6) δ 184.61 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 169.04 (C
S), 169.04 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.14, 149.75, 149.58 (ArC), 137.60 (C–NH), 126.99, 126.87, 126.18 (ArC), 121.66 (C
O), 157.14, 149.75, 149.58 (ArC), 137.60 (C–NH), 126.99, 126.87, 126.18 (ArC), 121.66 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 121.01, 117.70, 117.52, 114.82, 114.69 (ArC), 55.76 (CH3–O), 55.71 (CH–NH), 47.72 (CH2–N), 43.09 (1C, CH2–N), 21.42 (1C, CH3). Anal. calcd for C24H25N3O5S (467.54): C, 61.66; H, 5.39; N, 8.99; S, 6.86 found: C, 63.24; H, 6.08; N, 8.70; S, 6.53.
C), 121.01, 117.70, 117.52, 114.82, 114.69 (ArC), 55.76 (CH3–O), 55.71 (CH–NH), 47.72 (CH2–N), 43.09 (1C, CH2–N), 21.42 (1C, CH3). Anal. calcd for C24H25N3O5S (467.54): C, 61.66; H, 5.39; N, 8.99; S, 6.86 found: C, 63.24; H, 6.08; N, 8.70; S, 6.53.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z: 495.71 [M+] (34.0%). 1H-NMR (400 MHz, DMSO-d6): δ 7.50 (s, 1H, OH, D2O exchangeable), 7.49 (s, 1H, OH, D2O exchangeable), 6.89 (s, 1H, NH–CH, D2O exchangeable), 6.86 (s, 1H, NH, D2O exchangeable), 6.85 (s, 1H, CH
O of amide). Ms: m/z: 495.71 [M+] (34.0%). 1H-NMR (400 MHz, DMSO-d6): δ 7.50 (s, 1H, OH, D2O exchangeable), 7.49 (s, 1H, OH, D2O exchangeable), 6.89 (s, 1H, NH–CH, D2O exchangeable), 6.86 (s, 1H, NH, D2O exchangeable), 6.85 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 6.57 (s, 1H, ArH), 6.55 (s, 1H, ArH), 6.53 (dd, J = 8, 7.2 Hz, 2H, ArH), 6.51 (dd, J = 8.2, 8.7 Hz, 2H, ArH) 4.73 (s, 1H, C4–H), 4.70 (s, 2H, CH2–N), 4.67 (s, 2H, CH2–N), 3.99–3.97 (dd, J = 7.0, 7.2 Hz, 4H, CH2–CH3),1.88 (s, 3H, CH3), 1.28 (t, J = 6.9 Hz, 6H, CH3–CH2). 13C-NMR (300 MHz, DMSO-d6): δ 184.61 (1C, C
C), 6.57 (s, 1H, ArH), 6.55 (s, 1H, ArH), 6.53 (dd, J = 8, 7.2 Hz, 2H, ArH), 6.51 (dd, J = 8.2, 8.7 Hz, 2H, ArH) 4.73 (s, 1H, C4–H), 4.70 (s, 2H, CH2–N), 4.67 (s, 2H, CH2–N), 3.99–3.97 (dd, J = 7.0, 7.2 Hz, 4H, CH2–CH3),1.88 (s, 3H, CH3), 1.28 (t, J = 6.9 Hz, 6H, CH3–CH2). 13C-NMR (300 MHz, DMSO-d6): δ 184.61 (1C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 169.04 (1C, C
S), 169.04 (1C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.14,149.75, 149.58 (ArC), 137.60 (C–NH), 126.99 126.87 126.18 (ArC), 121.66 (1C, C
O), 157.14,149.75, 149.58 (ArC), 137.60 (C–NH), 126.99 126.87 126.18 (ArC), 121.66 (1C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 121.01, 117.70, 117.52, 114.82, 114.69 (ArC), 55.76 (CH3–O), 55.71 (CH–NH), 47.72 (CH2–N), 43.09 (CH2–N), 21.42 (1C, CH3). Anal. calcd. for C26H29N3O5S (495.59): C, 63.01; H, 5.90; N, 8.48; S, 6.47 found: C, 73.25; H, 6.08; N, 9.16; S, 7.61.
C), 121.01, 117.70, 117.52, 114.82, 114.69 (ArC), 55.76 (CH3–O), 55.71 (CH–NH), 47.72 (CH2–N), 43.09 (CH2–N), 21.42 (1C, CH3). Anal. calcd. for C26H29N3O5S (495.59): C, 63.01; H, 5.90; N, 8.48; S, 6.47 found: C, 73.25; H, 6.08; N, 9.16; S, 7.61.2.4. General procedure for synthesis of 5a, b target compounds
In a flask containing phenyl hydrazine (4.3 g, 0.04 mol) and diarylidene compound 2a/b, (0.01 mol), sodium butoxide prepared (Na metal (0.5 g) in ethanol (50 ml)) was added and subjected to reflux for 10 h.28 The solvent was then removed under reduced pressure and the remaining residue was chromatographed on silica gel (CHCl3) to furnish compounds 5a, b.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1715 (C
C–) and 1715 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z%: 501.22 [M+] (2.0%). 1H-NMR (400 MHz, DMSO-d6): δ 9.69 (s, 2H, OH, D2O exchangeable), 7.11 (s, 1H, CH
O of amide). Ms: m/z%: 501.22 [M+] (2.0%). 1H-NMR (400 MHz, DMSO-d6): δ 9.69 (s, 2H, OH, D2O exchangeable), 7.11 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.09 (dd, J = 7.2, 8 Hz, 2H, ArH), 7.01 (t, J = 7.8 Hz, 1H, ArH), 6.99–6.99 (t, J = 8 Hz, 2H, ArH), 6.97 (dd, J = 8, 7.9 Hz, 4H, ArH), 6.88 (s, 2H, ArH), 6.85 (d, 1H, C3–H), 4.77 (s, 2H, CH2–N), 4.76 (d, 2H, CH2–N), 3.79 (s, 6H, CH3–O), 3.77 (q, J = 7.4 Hz, 1H, C3a–H), 1.88 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 186.19 (C
C), 7.09 (dd, J = 7.2, 8 Hz, 2H, ArH), 7.01 (t, J = 7.8 Hz, 1H, ArH), 6.99–6.99 (t, J = 8 Hz, 2H, ArH), 6.97 (dd, J = 8, 7.9 Hz, 4H, ArH), 6.88 (s, 2H, ArH), 6.85 (d, 1H, C3–H), 4.77 (s, 2H, CH2–N), 4.76 (d, 2H, CH2–N), 3.79 (s, 6H, CH3–O), 3.77 (q, J = 7.4 Hz, 1H, C3a–H), 1.88 (s, 3H, CH3). 13C-NMR (300 MHz, DMSO-d6): δ 186.19 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.15 (C
O), 169.15 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 137.11, 136.91 (ArC), 130.41 (C
N), 137.11, 136.91 (ArC), 130.41 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 130.23 (C
C), 130.23 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 126.45, 126.21, 124.90, 124.84, 116.27, 115.55 (ArC), 115.47 (C
C), 126.45, 126.21, 124.90, 124.84, 116.27, 115.55 (ArC), 115.47 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 56.17 (1C, C3), 47.53 (1C, C3a), 42.87 (CH2–N–CH2), 21.36 (CH3). Anal. calcd. for C29H29N3O5 (499.57): C, 69.72; H, 5.85; N, 8.41, found: C, 69.72; H, 5.85; N, 8.41.
C), 56.17 (1C, C3), 47.53 (1C, C3a), 42.87 (CH2–N–CH2), 21.36 (CH3). Anal. calcd. for C29H29N3O5 (499.57): C, 69.72; H, 5.85; N, 8.41, found: C, 69.72; H, 5.85; N, 8.41.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–) and 1650 (C
C–) and 1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide). Ms: m/z%: 528.25 [M+] (33.5%). 1H-NMR (400 MHz, DMSO-d6): δ 9.61 (s, 2H, OH, D2O exchangeable), 7.56 (s, 1H, CH
O of amide). Ms: m/z%: 528.25 [M+] (33.5%). 1H-NMR (400 MHz, DMSO-d6): δ 9.61 (s, 2H, OH, D2O exchangeable), 7.56 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.07 (dd, J = 8, 7.9 Hz, 2H, ArH), 7.08 (dd, J = 7.5, 7.9 Hz, 2H, ArH), 7.01 (t, J = 8.7 Hz, 2H, ArH), 6.99 (t, J = 8.2 Hz, 1H, ArH), 6.98 (dd, J = 8.1, 7.6 Hz, 2H, ArH), 6.96 (s, 2H, ArH), 6.86 (d, J = 8.1 Hz, 1H, C3–H), 4.75 (s, 2H, CH2–N), 4.74 (d, J = 8.5 Hz, 2H, CH2–N), 4.06 (s, 4H, CH2–CH3) 4.05 (q, J = 8 Hz, 1H, C3a–H), 1.87 (s, 3H, CH3), 1.31 (t, J = 7.5 Hz, 6H, CH3–CH2).
C), 7.07 (dd, J = 8, 7.9 Hz, 2H, ArH), 7.08 (dd, J = 7.5, 7.9 Hz, 2H, ArH), 7.01 (t, J = 8.7 Hz, 2H, ArH), 6.99 (t, J = 8.2 Hz, 1H, ArH), 6.98 (dd, J = 8.1, 7.6 Hz, 2H, ArH), 6.96 (s, 2H, ArH), 6.86 (d, J = 8.1 Hz, 1H, C3–H), 4.75 (s, 2H, CH2–N), 4.74 (d, J = 8.5 Hz, 2H, CH2–N), 4.06 (s, 4H, CH2–CH3) 4.05 (q, J = 8 Hz, 1H, C3a–H), 1.87 (s, 3H, CH3), 1.31 (t, J = 7.5 Hz, 6H, CH3–CH2).13C-NMR (300 MHz, DMSO-d6): δ 186.11 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.13 (C
O), 169.13 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 136.94, 130.30, 130.15 (ArC), 126.21(C
N), 136.94, 130.30, 130.15 (ArC), 126.21(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 124.93 (C
C), 124.93 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 116.66, 116.56 (ArC), 116.35 (C
C), 116.66, 116.56 (ArC), 116.35 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 64.41 (1C, C3), 64.35 (CH2–CH3), 47.51 (1C, C3a), 42.86 (CH2–N–CH2), 21.31 (CH3), 15.15 (CH3–CH2). Anal. calcd. for C31H33N3O5 (527.62):C, 70.57; H, 6.30; N, 7.96, found: C, 70.50; H, 6.33; N, 7.99.
C), 64.41 (1C, C3), 64.35 (CH2–CH3), 47.51 (1C, C3a), 42.86 (CH2–N–CH2), 21.31 (CH3), 15.15 (CH3–CH2). Anal. calcd. for C31H33N3O5 (527.62):C, 70.57; H, 6.30; N, 7.96, found: C, 70.50; H, 6.33; N, 7.99.
2.5. Biological evaluation
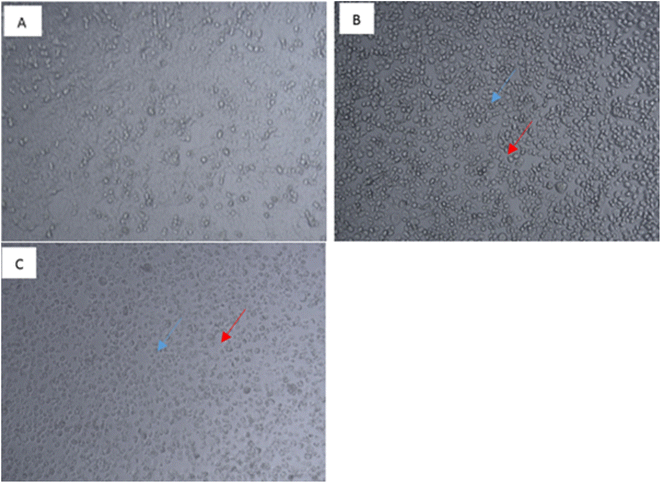
The assay volume was 100 μl and the spectrophotometer pathlength was 0.5 cm. The tubulin concentration was 4 mg ml−1. The spectrophotometer was set in kinetic absorbance mode at 340 nm wavelength. The spectrophotometer was set at 37 °C as polymerization reaction was started by the increase in temperature from 4 °C to 37 °C upon transfer of the reaction to a pre-warmed plate.31
2.5.3.1. Animals and experimental design. Eighteen Wistar rats (180–200 g) were obtained from the National Research Centre (Cairo, Egypt). Then rats were housed under normal laboratory conditions (22 ± 2 °C; 12 h/12 h light/dark cycle with free access to food and water). Mammary tumors were induced as previously described. Once the tumors volume reached 1 cm3, the rats were divided randomly into 2 different groups as follows:
• Group 1: untreated control group (C) n = 6.
• Group 2: breast induced cancer group (BC) n = 12.
To induce breast cancer, 7,12-dimethylbenz[a]anthracene (DMBA) supplied by Sigma-Aldrich company (St. Louis, MO, USA) was used. Rats in (Group 2) were injected intraperitoneally each with a single dose of 20 mg kg−1 body weight of DMBA dissolved in corn oil and given in a volume of 0.5 ml.32,33
Rats were palpated weekly to check for tumor appearance; the first detection of tumor was after three months (90 days). After 120 days, (Group 2) was divided into 2 subgroups, one was treated with Taxol and the other was treated with the synthetic drug (3b) at the same dose.
At the end of treatment period, rats from all groups were killed using a lethal dose of thiopental (IP 200 mg kg−1).
2.5.3.2. Histopathology. Samples from autopsy were taken from breast tissues of rats in different groups and fixed in 10% formal saline for twenty-four hours. Washing was done using tap water then serial dilutions of ethyl alcohol were used for dehydration. Specimens were cleared in xylene and embedded in paraffin at 56° in hot air oven for twenty-four hours. Paraffin bees wax tissue blocks were prepared for sectioning at 4 μm thickness by sledge microtome. The obtained tissue sections were collected on glass slides, deparaffinized, stained by hematoxylin and eosin (H&E) stain for examination through light electric microscope.34
2.5.3.3. Tumor volume and body weight. Tumor volume and body weight was recorded 3 times throughout the treatment, once after the end of the induction (day 0), once at 3 weeks of treatment (day 21) and at the end of treatment after 6 weeks of treatment (day 42). Animal handling followed the Guide for the Care and Use of Laboratory Animals.35 The experiments were approved by the ethics committee in the Future University in Egypt (REC-FPSPI-14/110).
2.6. Computational studies
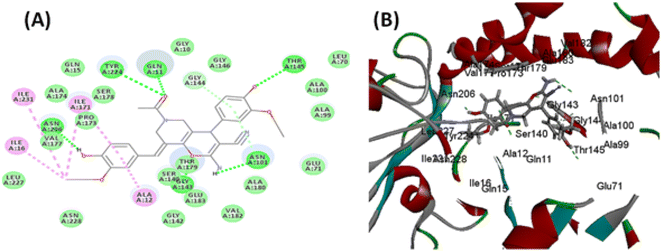
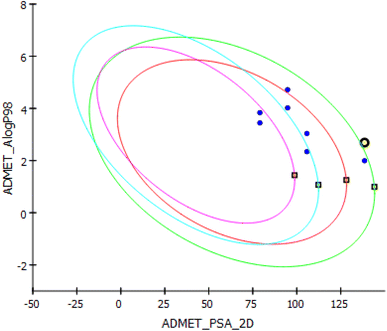
In search for the mechanism of inhibition behavior presented by the newly synthesized compounds for tubulin in the in vitro assays, molecular docking studies were performed on tubulin-podophyllotoxin: stathmin-like domain complex using Discovery Studio 4.1. Using the same software, dynamic simulation studies were also performed to check the stability of this interaction inhibition. Moreover, to validate the pharmacokinetic properties from the points of absorption, distribution, metabolism, and excretion ADME studies were performed side by side to in silico toxicity studies to inform the safety of the new structural compounds.Similarly, TOPKAT toxicity protocol was applied to previously prepared ligands after selection of criteria to be measured; Ames applicability, prediction, score, probability, skin irritancy, rat oral LD50, Rat Max. Tolerated Dose Feed Carcinogenic Potency TD50 for both mouse and rat.
3. Results and discussion
3.1. Rational & chemistry
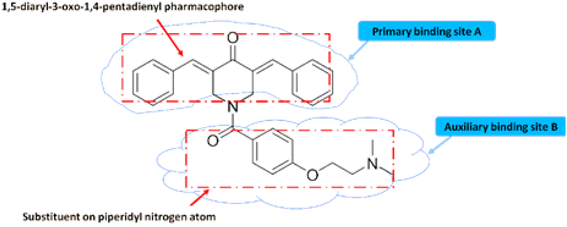
Literature studies exposed the structural activity relationship carried out on curcumin piperidone analogues which revealed that the 1,5-diaryl-3-oxo-1,4-pentadienyl pharmacophore is very essential in activity of the compound as this group reacts with cellular components at a specific binding site (primary binding site)38 (Fig. 3). Alignment of the 1,5-diaryl-3-oxo-1,4-pentadienyl group at the primary site may be affected by the nature of the group attached to the heterocyclic nitrogen atom.39–41 This group increased the cytotoxic potencies due to either extra binding with cellular constituents at the auxiliary binding site or by easing the interaction of the cytotoxin at the primary site.42 On the contrary, the group attached on the nitrogen atom may result in potency reduction because of the repulsion between this bulky group and the auxiliary site therefore, preventing interaction between the 1,5-diaryl-3-oxo-1,4-pentadienyl moiety and the primary binding site.43 Hence, bulky groups were avoided in the novel compounds and acetyl group was added on nitrogen instead.44It was also discovered that the charged molecules may be incapable of penetrating cell membranes and making a cytotoxic effect. Therefore, N-acylation was deemed a route to pursue making nitrogen atom in a non-basic form. An oxygen atom of the acyl group linked to the heterocyclic nitrogen atom may enable hydrogen bonding with the auxiliary site.45–48
The synthesis of the target compounds followed the pathway outlined in Scheme 1. The intermediate compounds 2a–d were prepared in one step. Briefly, aldol condensation of the appropriate aldehydes was carried out with N-acetyl-4-piperidone. Subsequently, the target compounds 3a–d, 4a, 4b, 5a and 5b were produced via cyclization of intermediate compounds using either malononitrile for series 3, thiourea for series 4 or phenylhydrazine for series 5. The chemical structures of newly synthesized compounds were confirmed and characterized via performing various elemental and spectral data.
3.2. Biological evaluation
| Compounds | Human hepatoma (Huh-7) IC50 in μM | Lung cell cancer adenocarcinoma (A549) IC50 in μM | Michigan cancer foundation-7 breast cancer (MCF-7) IC50 in μM |
|---|---|---|---|
| a ND = not defined. | |||
| 3a | 3.88 | 31.59 | 8.87 |
| 3b | 6.54 | 15.54 | 6.13 |
| 3c | 17.70 | 61.25 | 6.38 |
| 3d | 20.72 | ND | ND |
| 4a | 2.48 | 46.90 | 8.01 |
| 4b | 7.27 | 42.34 | 15.97 |
| 5a | 5.47 | 20.26 | 5.27 |
| 5b | 2.23 | 15.15 | 9.04 |
| Taxol | 6.68 | 38.05 | 12.32 |
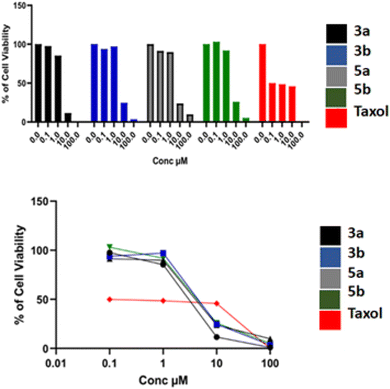
According to the above analysis, some structure–activity relationship (SAR) could be concluded: the introduction of fused-heterocyclic moiety was found effective and almost all the tested compounds showed moderate to potent antiproliferative activity. Also, it could be observed that methoxy and ethoxy groups attached to phenyl ring were an essential pharmacophore for retaining bioactivities, which is consistent with previously reported compounds.9,13
To investigate whether the antiproliferative activities of compounds; 3a, 3b, 5a and 5b is linked to the mechanism of tubulin depolymerization, these agents were evaluated for their inhibition of tubulin polymerization in comparison to CA-4 as a reference depolymerizing agent.
The assay yielded results displayed in Table 2. All compounds showed good ability to inhibit tubulin polymerisation with IC50 values ranging from 4.03 to 15.60 μg ml−1 and inhibition % ranging from 69% to 81%. Compound 3b had the most inhibitory effect with IC50 of 4.03 μg ml−1 and 81% inhibition which is comparable to the reference drug CA-4 that displayed inhibition of 86% and IC50 = 1.64 μg ml−1.
| Compound | Inhibition (%) | IC50 in μg ml−1 |
|---|---|---|
| 3a | 79 | 15.6 |
| 3b | 81 | 4.03 |
| 5a | 76 | 6.06 |
| 5b | 70 | 12.61 |
| CA-4 | 86 | 1.64 |
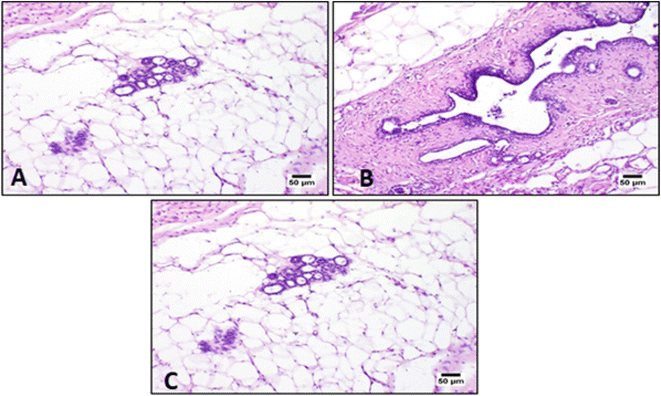
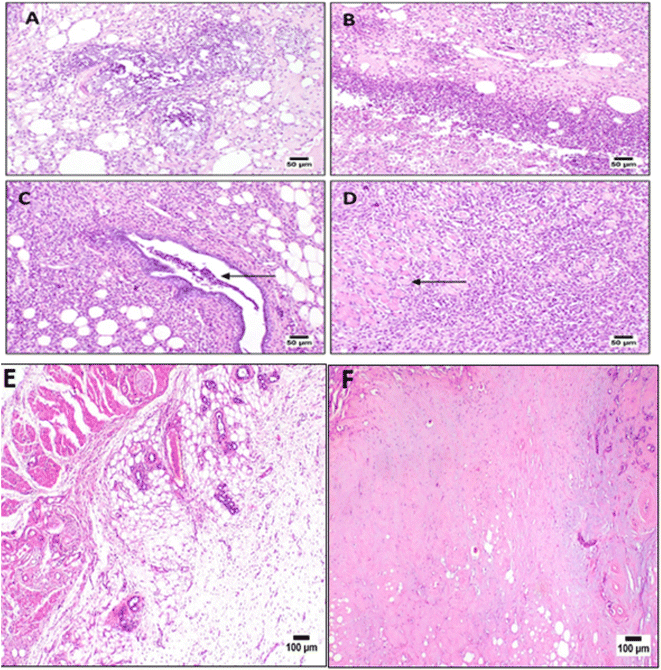
Microscopic examination of the control group (C) using hematoxylin and eosin (H&E) stain showed normal histology structure of mammary gland lobules which consisted of tubule-alveolar glands that were formed of branched system of ducts and secretory alveoli arranged in multiple lobules. Examination of covering skin showed normal epidermis and dermis with intact subcutaneous connective tissue (Fig. 6). The breast cancer group (BC) showed necrosis of the epithelial covering mammary gland. The mammary epithelia showed dysplastic changes that revealed numerous proliferating neoplastic cells that invaded surrounding tissues. The tumor cells showed hyperchromatic nuclei with few prominent nucleoli which were arranged in multilayer structure that narrowed the ductal lumen.
Histopathology results showed that the intensity of inflammatory cells infiltrates decreased after treatment with both Taxol and 3b (synthetic drug) as shown in (Fig. 7).
Mammary adenocarcinoma was observed in some examined sections and was characterized by solid aggregations of neoplastic cells associated with numerous mononuclear inflammatory cells infiltration34 (Fig. 7).
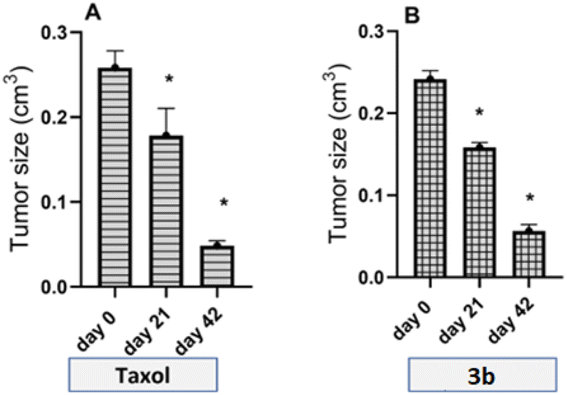
Tumor size and body weight of Wistar rats were measured and recorded 3 times throughout the treatment, once after the end of the induction (day 0), once at 3 weeks of treatment (day 21) and once at the end of treatment/after 6 weeks of treatment (day 42) to determine whether drug 3b could target breast cancer cell growth. Both Taxol and 3b did not have a significant effect on rats' body weights.
On the other hand, for the antitumor study, tumor size of the 3b-treated rats was significantly decreased by 79% at the end of the treatment compared to the tumor size before initiation of the treatment at P < 0.05 as shown in (Fig. 8) which was comparable to that of Taxol which showed 81% inhibition.
3.3. In silico studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150–12
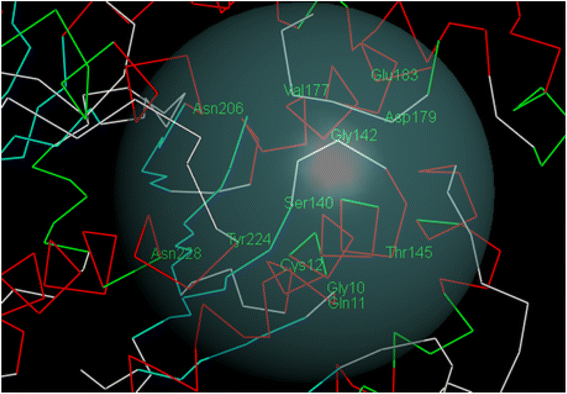
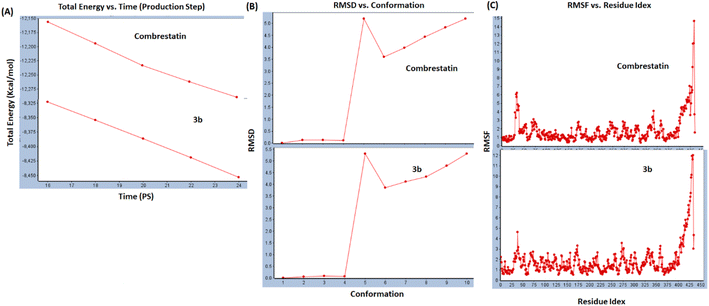
150–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 275) kcal mol−1 over time range (16–24) ps and 9 new hydrogen bonds were formed between CA-4 and tubulin. While over similar timeline 3b produced comparable stable total energy (8325–8450) kcal mol−1 and 8 new hydrogen bonds were formed between 3b and tubulin. The total energy decreases overtime which indicated interaction stability. On the other hand, RMSD showed that the deviations of both CA-4 and 3b were similar within the accepted. Steady fluctuations for both CA-4 and 3b were similar and within accepted ranges, where most fluctuations were below 3.5.
275) kcal mol−1 over time range (16–24) ps and 9 new hydrogen bonds were formed between CA-4 and tubulin. While over similar timeline 3b produced comparable stable total energy (8325–8450) kcal mol−1 and 8 new hydrogen bonds were formed between 3b and tubulin. The total energy decreases overtime which indicated interaction stability. On the other hand, RMSD showed that the deviations of both CA-4 and 3b were similar within the accepted. Steady fluctuations for both CA-4 and 3b were similar and within accepted ranges, where most fluctuations were below 3.5.
| Compound | ADMET solubility level | Absorption level | ADMET BBB level | Hepatotoxicity | PPB |
|---|---|---|---|---|---|
| 3a | 3 | 1 | 4 | FALSE | FALSE |
| 3b | 2 | 2 | 4 | FALSE | TRUE |
| 3c | 2 | 0 | 2 | TRUE | TRUE |
| 3d | 2 | 0 | 2 | FALSE | TRUE |
| 4a | 3 | 0 | 3 | FALSE | FALSE |
| 4b | 2 | 0 | 4 | FALSE | TRUE |
| 5a | 2 | 0 | 4 | FALSE | TRUE |
| 5b | 2 | 1 | 4 | FALSE | TRUE |
| Compound | TOPKAT Ames applicability | TOPKAT Ames prediction | TOPKAT Ames score | TOPKAT Ames probability | TOPKAT skin irritancy | TOPKAT rat oral LD50 g per kg body weight | TOPKAT Rat max. tolerated dose feed g per kg body weight | TOPKAT carcinogenic potency TD50 mouse g per kg body weight per day | TOPKAT carcinogenic potency TD50 Rat g per kg body weight per day |
|---|---|---|---|---|---|---|---|---|---|
| 3a | All properties and OPS components are within expected ranges | Non-mutagen | −18.63 | 0.071 | Non-irritant | 0.19 | 0.07 | 53.09 | 3.29 |
| 3b | −19.73 | 0.052 | 0.40 | 0.11 | 36.50 | 22.30 | |||
| 3c | −15.80 | 0.141 | 0.05 | 0.03 | 6.52 | 1.26 | |||
| 3d | −14.25 | 0.196 | 0.19 | 0.02 | 25.70 | 45.55 | |||
| 4a | −15.50 | 0.151 | 0.25 | 0.09 | 75.43 | 3.20 | |||
| 4b | −16.68 | 0.115 | 0.52 | 0.15 | 51.90 | 21.70 | |||
| 5a | −12.45 | 0.270 | 0.41 | 0.11 | 44.64 | 5.55 | |||
| 5b | −13.59 | 0.222 | 0.86 | 0.18 | 30.60 | 37.53 |
4. Conclusion
A series of novel pyridine heterocyclic hybrids were designed, synthesized, and evaluated for their in vitro anticancer activity against human hepatoma (Huh-7), lung cell cancer adenocarcinoma (A549), and breast cancer (MCF-7). Most of the novel pyridine hybrids exhibited potent cytotoxicity. Moreover, tubulin inhibitory effect of the most potent compounds was assessed, and they showed good tubulin inhibition activity. Compound 3b was found to be the most potent compound possessing cytotoxic activity on cancer cell lines with IC50 = 6.54 μM against (Huh-7) liver cancer cell line and IC50 = 6.13 μM against (MCF-7) breast cancer cell line. Also, compound 3b showed great ability to inhibit tubulin polymerization with superior value of IC50 = 4.03 μM (81% inhibition) compared to CA-4 the reference drug. These results also coincide with the docking interaction energy score (−48.22 kcal mol−1) which was obtained from the in silico study. Compound 3b was also the least toxic compound against the non-cancerous cells with IC50 of 8.11 μM. Also, compound 3b was able to suppress tumour size in experimental rats by 79% according to (in vivo studies). These findings have proven that pyrano-pyridine ring could be an essential feature for further new anticancer scaffolds.Ethical statement
The experiments were approved by the ethics committee in the Future University in Egypt (REC-FPSPI-14/110).Author contributions
Menna M. Abdelshaheed: contributed to the synthesis of the compounds, collecting literature studies and data, interpretation of the results and writing the scientific article. Hussein I. El Subbagh: contributed to the idea of work, scheme suggestion, writing and revision of the scientific article. Mohamed A. Tantawy: contributed to the in vitro biological assays and writing the results. Reem T. Attia: contributed to the in vivo biological assays and writing the results. Khairia M. Youssef: contributed to the idea of work, scheme suggestion, writing and revision of the scientific article. Iten M. Fawzy: contributed to the idea of work, scheme suggestion, molecular modeling, dynamic studies, toxicity and ADME studies, writing and revision of the scientific article.Conflicts of interest
We declare that the authors have no competing interests.References
- H. Q. Daniel, P. Mathurin, H. Cortez-Pinto and R. Loomba, Global epidemiology of alcohol-associated cirrhosis and HCC: trends, projections and risk factors, Nat. Rev. Gastroenterol. Hepatol., 2023, 20(1), 37–49, DOI:10.1038/s41575-022-00688-6.
- K. Peter, A. Ahmed and D. Kim, Causes and risk profiles of mortality among individuals with nonalcoholic fatty liver disease, Clin. Mol. Hepatol., 2023, 29, S43, DOI:10.3350/cmh.2022.0351.
- M. Berkman Amy, N. Mittal and M. E. Roth, Adolescent and young adult cancers: unmet needs and closing the gaps, Curr. Opin. Pediatr., 2023, 35(1), 84–90, DOI:10.1097/MOP.0000000000001200.
- R. Sahu, R. Mishra, R. Kumar, C. Majee, A. Mazumder and A. Kumar, Pyridine moiety: an insight into recent advances in the treatment of cancer, Mini-Rev. Med. Chem., 2022, 22(2), 248–272, DOI:10.2174/1389557521666210614162031.
- Y. Ling, Z. Y. Hao, D. Liang, C. L. Zhang, Y. F. Liu and Y. Wang, The expanding role of pyridine and dihydropyridine scaffolds in drug design, Drug Des., Dev. Ther., 2021, 15, 4289–4338, DOI:10.2147/DDDT.S329547.
- G. Bérubé, An overview of molecular hybrids in drug discovery, Expert Opin. Drug Discovery, 2016, 11(3), 281–305, DOI:10.1517/17460441.2016.1135125.
- Y. Lu, J. Chen, M. Xiao, W. Li and D. D. Miller, An overview of tubulin inhibitors that interact with the colchicine binding site, Pharm. Res., 2020, 29, 2943–2971, DOI:10.1007/s11095-012-0828-z.
- G. Ş. Karatoprak, E. Küpeli Akkol, Y. Genç, H. Bardakcı, Ç. Yücel and E. Sobarzo-Sánchez, Combretastatins: an overview of structure, probable mechanisms of action and potential applications, Mol, 2020, 25(11), 2560, DOI:10.3390/molecules25112560.
- I. M. Fawzy, K. M. Youssef, N. S. Ismail, J. Gullbo and K. A. Abouzid, Newly designed and synthesized curcumin analogs with in vitro cytotoxicity and tubulin polymerization activity, Chem. Biol. Drug Des., 2015, 86(1), 80–90, DOI:10.1111/cbdd.12464.
- M. Mahdavi, S. Asghari, M. Rahnamay, G. Dehghan, M. A. H. Feizi and S. Balalaie, Cytotoxicity, oxidative stress, and apoptosis in K562 leukemia cells induced by an active compound from pyrano-pyridine derivatives, Hum. Exp. Toxicol., 2018, 37(10), 1105–1116, DOI:10.1177/0960327118756719.
- K. M. Youssef, M. A. Al-Omar, H. I. El-Subbagh, L. A. Abou-Zeid, A. G. M. Abdel-Gader, N. G. Haress and A. S. Al-Tuwaijri, Synthesis, antiplatelet aggregation activity, and molecular modeling study of novel substituted-piperazine analogues, Med. Chem. Res., 2011, 20, 898–911, DOI:10.1007/s00044-010-9411-5.
- I. W. Cheney, S. Yan, T. Appleby, H. Walker, T. Vo, N. Yao, R. Hamatake, Z. Hong and J. Z. Wu, Identification and structure–activity relationships of substituted pyridones as inhibitors of Pim-1 kinase, Bioorg. Med. Chem. Lett., 2007, 17(6), 1679–1683, DOI:10.1016/j.bmcl.2006.12.086.
- H. I. El-Subbagh, S. M. Abu-Zaid, M. A. Mahran, F. A. Badria and A. M. Al-Obaid, Synthesis and biological evaluation of certain α, β-unsaturated ketones and their corresponding fused pyridines as antiviral and cytotoxic agents, J. Med. Chem., 2000, 43(15), 2915–2921, DOI:10.1021/jm000038m.
- A. Farce, C. Loge, S. Gallet, N. Lebegue, P. Carato, P. Chavatte, P. Berthelot and D. Lesieur, Docking study of ligands into the colchicine binding site of tubulin, J. Enzyme Inhib. Med. Chem., 2004, 19(6), 541–547, DOI:10.1080/14756360412331280545.
- N. Mizuno, S. Toba, M. Edamatsu, J. Watai-Nishii, N. Hirokawa, Y. Y. Toyoshima and M. Kikkawa, Dynein and kinesin share an overlapping microtubule-binding site, EMBO J., 2004, 23(13), 2459–2467, DOI:10.1038/sj.emboj.7600240.
- D. E. Pryor, A. O'Brate, G. Bilcer, J. F. Díaz, Y. Wang, M. Kabaki, M. K. Jung, J. M. Andreu, A. K. Ghosh and P. Giannakakou, The microtubule stabilizing agent laulimalide does not bind in the taxoid site, kills cells resistant to paclitaxel and epothilones, and may not require its epoxide moiety for activity, Biochemistry, 2002, 41(29), 9109–9115, DOI:10.1021/bi020211b.
- Y. T. Wang, T. Q. Shi, H. L. Zhu and C. H. Liu, Synthesis, biological evaluation and molecular docking of benzimidazole grafted benzsulfamide-containing pyrazole ring derivatives as novel tubulin polymerization inhibitors, Bioorg. Med. Chem., 2019, 27(3), 502–515, DOI:10.1016/j.bmc.2018.12.031.
- R. Gaspari, A. E. Prota, K. Bargsten, A. Cavalli and M. O. Steinmetz, Structural basis of cis-and trans-combretastatin binding to tubulin, Chem, 2017, 2(1), 102–113, DOI:10.1016/j.chempr.2016.12.005.
- A. S. Negi, Y. Gautam, S. Alam, D. Chanda, S. Luqman, J. Sarkar, F. Khan and R. Konwar, Natural antitubulin agents: Importance of 3, 4, 5-trimethoxyphenyl fragment, Bioorg. Med. Chem., 2015, 23(3), 373–389, DOI:10.1016/j.bmc.2014.12.027.
- W. Li, H. Sun, S. Xu, Z. Zhu and J. Xu, Tubulin inhibitors targeting the colchicine binding site: a perspective of privileged structures, Future Med. Chem., 2017, 9(15), 1765–1794, DOI:10.4155/fmc-2017-0100.
- K. Prasat, Anticancer drugs and potential anticancer leads inspired by natural products, Stud. Nat. Prod. Chem., 2015, 44, 251–307, DOI:10.1016/B978-0-444-63460-3.00005-5.
- B. Onel, M. Carver, G. Wu, D. Timonina, S. Kalarn, M. Larriva and D. Yang, A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription, J. Am. Chem. Soc., 2016, 138(8), 2563–2570, DOI:10.1021/jacs.5b08596.
- C. D. Churchill, M. Klobukowski and J. A. Tuszynski, The unique binding mode of laulimalide to two tubulin protofilaments, Chem. Biol. Drug Des., 2015, 86(2), 190–199, DOI:10.1111/cbdd.12475.
- M. A. Jordan and L. Wilson, Microtubules as a target for anticancer drugs, Nat. Rev. Cancer, 2004, 4(4), 253–265, DOI:10.1038/nrc1317.
- I. Mamdouh, K. Youssef, N. Ismail, J. Gullbo and K. Abouzid, Design and Synthesis and Biological evaluation of Novel Curcumin Analogs with anticipated anticancer activity, Future J. Pharm. Sci., 2015, 1, 22, DOI:10.1016/j.fjps.2015.06.001.
- U. Das, J. Alcorn, A. Shrivastav, R. K. Sharma, E. De Clercq, J. Balzarini and J. R. Dimmock, Design, synthesis and cytotoxic properties of novel 1-[4-(2-alkylaminoethoxy) phenylcarbonyl]-3, 5-bis (arylidene)-4-piperidones and related compounds, Eur. J. Med. Chem., 2007, 42(1), 71–80, DOI:10.1016/j.ejmech.2006.08.002.
- H. A. Soliman, M. N. M. Yousif, M. M. Said, N. A. Hassan, M. M. Ali, H. M. Awad and F. M. Abdel-Megeid, Synthesis of novel 1, 6-naphthyridines, pyrano [3, 2-c] pyridines and pyrido [4, 3-d] pyrimidines derived from 2, 2, 6, 6-tetramethylpiperidin-4-one for in vitro anticancer and antioxidant evaluation, Der Pharma Chem., 2014, 6(3), 394–410 Search PubMed.
- M. S. Nafie, A. M. Amer, A. K. Mohamed and E. S. Tantawy, Discovery of novel pyrazolo[3,4-b]pyridine scaffold-based derivatives as potential PIM-1 kinase inhibitors in breast cancer MCF-7 cells, Bioorg. Med. Chem., 2020, 28(24), 115828, DOI:10.1016/j.bmc.2020.115828.
- J. van Meerloo, G. J. Kaspers and J. Cloos, Cell sensitivity assays: the MTT assay, Methods Mol. Biol., 2011, 731, 237–245, DOI:10.1007/978-1-61779-080-5_20.
- H. Ali-Boucetta, K. T. Al-Jamal and K. Kostarelos, Cytotoxic assessment of carbon nanotube interaction with cell cultures, Methods Mol. Biol., 2011, 726, 299–312, DOI:10.1007/978-1-61779-052-2_19.
- T. Zhu, S. H. Wang, D. Li, S. Y. Wang, X. Liu, J. Song, Y. T. Wang and S. Y. Zhang, Progress of tubulin polymerization activity detection methods, Bioorg. Med. Chem. Lett., 2021, 37, 127698, DOI:10.1016/j.bmcl.2020.127698.
- N. El-Azem, M. Pulido-Moran, C. L. Ramirez-Tortosa, J. L. Quiles, F. E. Cara, P. Sanchez-Rovira, S. Granados-Principal and M. Ramirez-Tortosa, Modulation by hydroxytyrosol of oxidative stress and antitumor activities of paclitaxel in breast cancer, Eur. J. Nutr., 2019, 58(3), 1203–1211, DOI:10.1007/s00394-018-1638-9.
- K. Batcioglu, A. B. Uyumlu, B. Satilmis, B. Yildirim, N. Yucel, H. Demirtas, R. Onkal, R. M. Guzel and M. B. Djamgoz, Oxidative Stress in the in vivo DMBA Rat Model of Breast Cancer: Suppression by a Voltage-gated Sodium Channel Inhibitor (RS 100642), Basic Clin. Pharmacol. Toxicol., 2012, 111(2), 137–141, DOI:10.1111/j.1742-7843.2012.00880.x.
- J. D. Bancroft, A. Stevens, and D. R. Turner, Theory and Practice of Histological Techniques, Churchill Livingstone, London, Toronto, 4th edn, 1996 Search PubMed.
- National Research Council (US), Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, Washington (DC), National Academies Press (US), 8th edn, 2011 Search PubMed.
- S. Saxena, M. Abdullah, D. Sriram and L. Guruprasad, Discovery of novel inhibitors of Mycobacterium tuberculosis MurG: Homology modelling, structure based pharmacophore, molecular docking, and molecular dynamics simulations, J. Biomol. Struct. Dyn., 2018, 36(12), 3184–3198, DOI:10.1080/07391102.2017.1384398.
- S. Sahu, S. K. Ghosh, P. Gahtori, U. P. Singh, D. R. Bhattacharyya and H. R. Bhat, In silico ADMET study, docking, synthesis and antimalarial evaluation of thiazole-1, 3, 5-triazine derivatives as Pf-DHFR inhibitor, Pharmacol. Rep., 2019, 71(5), 762–767, DOI:10.1016/j.pharep.2019.04.006.
- U. Das, R. K. Sharma and J. R. Dimmock, 1, 5-diaryl-3-oxo-1, 4-pentadienes: a case for antineoplastics with multiple targets, Curr. Med. Chem., 2009, 16(16), 2001–2020, DOI:10.2174/092986709788682218.
- N. Edraki, U. Das, B. Hemateenejad, J. R. Dimmock and R. Miri, Comparative QSAR Analysis of 3,5-bis (Arylidene)-4-Piperidone Derivatives: The Development of Predictive Cytotoxicity Models, Iran. J. Pharm. Res., 2016, 15(2), 425–437 CAS.
- V. M. Muzalevskiy, O. V. Serdyuk, and V. G. Nenajdenko, Chemistry of fluorinated indoles, Fluorine in Heterocyclic Chemistry, 2014, vol. 1, pp. 117–156 Search PubMed.
- M. Helal, Investigation of Some Molecular Mechanisms of Cytotoxic 1, 5-diaryl-3-oxo-1, 4-pentadienes, Doctoral dissertation, University of Saskatchewan, Canada, 2012 Search PubMed.
- K. M. Lakhani, Synthesis and Bioactivities of Novel N1-Acylhydrazides, Doctoral dissertation, University of Saskatchewan, Canada, 2018 Search PubMed.
- Y. Lu, J. Chen, M. Xiao, W. Li and D. D. Miller, An overview of tubulin inhibitors that interact with the colchicine binding site, Pharm. Res., 2012, 29(11), 2943–2971, DOI:10.1007/s11095-012-0828-z.
- J. Sun, S. Zhang, C. Yu, G. Hou, X. Zhang, K. Li and F. Zhao, Design, Synthesis and Bioevaluation of Novel N-Substituted-3, 5-Bis (Arylidene)-4-piperidone Derivatives as Cytotoxic and Antitumor Agents with Fluorescent Properties, Chem. Biol. Drug Des., 2014, 83(4), 392–400, DOI:10.1111/cbdd.12254.
- M. Hossain, C. E. Enci, J. R. Dimmock and U. Das, Discovery and investigation of 1-[4-(2-aminoethoxy) phenylcarbonyl]-3, 5-bis-(benzylidene)-4-piperidones as candidate antineoplastic agents: Our last 15 years study, Curr. Med. Chem., 2021, 28(13), 2453–2464, DOI:10.2174/0929867327666200730215752.
- E. Robles-Escajeda, U. Das, N. M. Ortega, K. Parra, G. Francia, J. R. Dimmock, A. Varela-Ramirez and J. R. Aguilera, A novel curcumin-like dienone induces apoptosis in triple-negative breast cancer cells, Cell. Oncol., 2016, 39, 265–277, DOI:10.1007/s13402-016-0272-x.
- M. Hossain, U. Das, N. Umemura, H. Sakagami, J. Balzarini, E. De Clercq, M. Kawase and J. R. Dimmock, Tumour-specific cytotoxicity and structure–activity relationships of novel 1-[3-(2-methoxyethylthio) propionyl]-3, 5-bis (benzylidene)-4-piperidones, Bioorg. Med. Chem., 2016, 24(10), 2206–2214, DOI:10.1016/j.bmc.2016.03.056.
- M. Nakhjiri, M. Safavi, E. Alipour, S. Emami, A. F. Atash, M. Jafari-Zavareh, S. K. Ardestani, M. Khoshneviszadeh, A. Foroumadi and A. Shafiee, Asymmetrical 2,6-bis(benzylidene)cyclohexanones: Synthesis, cytotoxic activity and QSAR study, Eur. J. Med. Chem., 2012, 50, 113–123, DOI:10.1016/j.ejmech.2012.01.045.
- C. Dyrager, M. Wickström, M. Fridén-Saxin, A. Friberg, K. Dahlén, E. A. Wallén, J. Gullbo, M. Grøtli and K. Luthman, Inhibitors and promoters of tubulin polymerization: Synthesis and biological evaluation of chalcones and related dienones as potential anticancer agents, Bioorg. Med. Chem., 2011, 19(8), 2659–2665, DOI:10.1016/j.bmc.2011.03.005.
- N. El-Azem, M. Pulido-Moran, C. L. Ramirez-Tortosa, J. L. Quiles, F. E. Cara, P. Sanchez-Rovira, S. Granados-Principal and M. Ramirez-Tortosa, Modulation by hydroxytyrosol of oxidative stress and antitumor activities of paclitaxel in breast cancer, Eur. J. Nutr., 2019, 58, 1203–1211, DOI:10.1007/s00394-018-1638-9.
- K. Batcioglu, A. B. Uyumlu, B. Satilmis, B. Yildirim, N. Yucel, H. Demirtas, R. Onkal, R. M. Guzel and M. B. Djamgoz, Oxidative stress in the in vivo DMBA rat model of breast cancer: suppression by a voltage-gated sodium channel inhibitor (RS100642), Basic Clin. Pharmacol. Toxicol., 2012, 111(2), 137–141, DOI:10.1111/j.1742-7843.2012.00880.x.
- R. B. Ravelli, B. Gigant, P. A. Curmi, I. Jourdain, S. Lachkar, A. Sobel and M. Knossow, Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain, Nature, 2004, 428(6979), 198–202, DOI:10.1038/nature02393.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra02875e |
| This journal is © The Royal Society of Chemistry 2023 |