Open Access Article
Open Access ArticleStable biogenic silver nanoparticles from Syzygium nervosum bud extract for enhanced catalytic, antibacterial and antifungal properties†
Thi Lan Pham‡
a,
Van Dat Doan‡ b,
Quang Le Danga,
Tuan Anh Nguyena,
Thi Lan Huong Nguyenc,
Thi Dieu Thuy Tranb,
Thi Phuong Lan Nguyend,
Thi Kieu Anh Voa,
Trung Huy Nguyena and
Dai Lam Tran
b,
Quang Le Danga,
Tuan Anh Nguyena,
Thi Lan Huong Nguyenc,
Thi Dieu Thuy Tranb,
Thi Phuong Lan Nguyend,
Thi Kieu Anh Voa,
Trung Huy Nguyena and
Dai Lam Tran *a
*a
aInstitute for Tropical Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay, Hanoi, Vietnam. E-mail: trandailam@gmail.com
bFaculty of Chemical Engineering, Industrial University of Ho Chi Minh City, No. 12 Nguyen Van Bao, Ward 4, Go Vap District, Ho Chi Minh City 70000, Vietnam
cInstitute of Biotechnology and Food Technology, Industrial University of Ho Chi Minh City, No. 12 Nguyen Van Bao, Ward 4, Go Vap District, Ho Chi Minh City 70000, Vietnam
dUniversity of Economics and Technology for Industries (UNETI), 456, Minh Khai, Vinh Tuy, Hai Ba Trung District, Ha Noi, Vietnam
First published on 12th July 2023
Abstract
In the present study, the biosynthesis of stable silver nanoparticles (BioAgNPs) was accomplished successfully for the first time by using an aqueous extract derived from the buds of Syzygium nervosum (SN) as both a reducing and a stabilizing agent. Transmission electron microscopy (TEM) and high-resolution transmission electron microscopy (HR-TEM) investigations revealed that the biosynthesized BioAgNPs were predominantly spherical with an average size of 10–30 nm. It was found that the outstanding stability of the BioAgNPs colloidal solution was assigned to the additive effect of the surrounding protective organic layer and the highly negatively charged surface of the nanoparticles. Consequently, good antibacterial activity was demonstrated by the colloidal BioAgNPs solution against four distinct bacterial strains, including Gram-positive S. aureus and B. subtilis as well as Gram-negative E. coli and S. typhi. Interestingly, the biosynthesized BioAgNPs displayed greater antibacterial activity even when tested at low doses against Gram-negative S. typhi. In addition, the biogenic AgNPs demonstrated a significant level of catalytic activity in the process of converting 2-NP, 3-NP, and 4-NP into aminophenols within 15 min, with reaction rate constants of 9.0 × 10−4, 10 × 10−4, and 9.0 × 10−4 s−1, respectively. BioAgNPs formulations were assessed against anthracnose disease in tea plants and were found to be as effective as the positive control at a dose of 20-fold dilution, but less effective at a dose of 30-fold dilution. Both doses of BioAgNPs formulations significantly suppressed Colletotrichum camelliae (anthracnose disease) without affecting the growth of the tea plants.
Introduction
Research on nanoparticles is a topic of active scientific investigation in the modern era, with an emphasis placed on the application of nanoparticles in a variety of fields including electronics, catalysis, biosensors, wastewater treatment, biomedicine, pharmaceuticals, and cosmetics.1 Nanoparticles are finding an increasing number of applications in industry, which has resulted in an increase in the volume of their production. The synthesis of nanoparticles is primarily based on physical and chemical processes, which either require a significant amount of energy to be expended or involve the utilization of potentially hazardous chemical compounds.2–4According to,5 the goal of nano-biotechnology is to develop technologies that are both inexpensive and environmentally friendly, with the end goal being the manufacture of various nanoparticles using biological components, live organisms, and the products of those organisms. It has been stated that bacteria, fungi, molds, and plants may be employed for the successful synthesis of nanoparticles in an environmentally acceptable manner.6
However, the utilization of certain natural resources in the synthesis of nanoparticles, such as microorganisms, carries with it the risk of potential adverse effects on human health as well as environmental damage.7–10 The microorganism-based synthesis of metal nanoparticles also has a number of limitations, the primary ones being the difficult stages involved, such as microbial sample, isolation, culturing, and storage.11
When compared to microorganisms, the potential for nanoparticle synthesis held by certain plant extracts and phytochemicals obtained from medicinal herbs and spices, is significantly higher. These plant-originated products may possess beneficial biological characteristics and transfer their properties to nanoparticles during synthesis.12,13 This is in addition to the fact that these plant-originated products provide environmentally friendly reducing and stabilizing agents for the formation of nanoparticles. Secondary metabolites such as phenolic compounds, tannins, and flavonoids are abundant in plant extracts. These secondary metabolites are well-known for their useful pharmacological activities and their accentuated redox features. Their redox capacity makes it possible for them to be used as agents for the reduction of metal ions during the plant-mediated synthesis of metal nanoparticles, and their structure is responsible for the stabilization of the nanoparticles that are produced as a result of this process.14,15
Silver nanoparticles (AgNPs) have a long history of application, primarily because they possess antimicrobial properties.16 AgNPs are used in cosmetic preparations, as a catalyst in the chemical industry, for biomedical purposes, and preparation of material with antimicrobial properties.17–19 The application of silver nanoparticles for catalytic or antibacterial effects is becoming more common, and their synthesis through the use of plant extracts could be a significant benefit when taking into account the ecological impact of the synthesis process as well as the possibility of the nanoparticles themselves being enriched with biologically significant compounds.20,21 Besides, the escalating utilization of AgNPs in consumer products raises concerns due to their detrimental impact on human health.22 The widespread use of AgNPs leads to increased human exposure through various routes, particularly oral consumption. Upon absorption, AgNPs can be dispersed throughout multiple bodily systems, including the dermis, respiratory, spleen, digestive, urinary, immune, nervous, and reproductive systems. They tended to accumulate primarily in the spleen, kidney, liver, and lungs, with minimal deposition in teeth and bones. Due to their minute size, AgNPs can easily penetrate the body and traverse biological barriers like the blood-testis barrier and blood–brain barrier, potentially inducing cytotoxic effects.23 Therefore, silver nanoparticles derived through a green synthesis approach, such as from plant extracts, hold significant promise in enhancing their safety during usage.
Antibiotic resistance among bacteria is turning into a global concern as a direct result of the extensive application of antibiotics. Among these are the following: (I) Escherichia coli, sometimes known as E. coli, is a Gram-negative bacterium that is frequently discovered in the digestive tracts of humans and other warm-blooded animals. Most strains of E. coli are harmless. However, certain strains, such as those that produce the Shiga toxin, E. coli (STEC), which has the potential to cause serious illness, is responsible for conditions such as cholecystitis, bacteremia, cholangitis, urinary tract infection, and diarrhea. Consumption of infected foods, such as ground beef products that are raw or undercooked, raw milk, and contaminated raw vegetables and sprouts, is the primary means by which it is passed from humans to other humans.24. (II) Gram-negative, facultative anaerobe Salmonella enterica serovar Typhimurium (referred to as S. typhi) mimics human typhoid fever in mice by causing a systemic infection. Humans infected with S. typhi typically get a mild case of gastroenteritis that resolves on its own. Salmonella infections continue to be a problem, particularly in underdeveloped regions, due to the wide variety of species and serovars within the genus. Headache, stomach pain, and persistent high fever are classic symptoms of typhoid. Rarely, neurological abnormalities have been observed in typhoid fever patients. Salmonella brain infections have a significant death rate, especially in babies. (III) Bacillus cereus (B. cereus) is a Gram-positive bacterium that is found in soil, vegetation, and food. It is anaerobic with a facultative anaerobic metabolism and produces toxins. It is a common factor in gastrointestinal ailments, including sickness, throwing up, and diarrhea. On the other hand, it has been linked to significant infections in immunocompromised hosts, including septicemia and endophthalmitis, both of which can result in irreversible eyesight loss if left untreated. This exercise provides an overview of the diagnosis and management of Bacillus cereus, as well as a discussion of the role of the interdisciplinary team in the care of patients affected by this ailment. (IV) Bacillus subtilis (B. subtilis) has been employed in a risk-free manner in the traditional fermented cuisines of many east Asian nations for generations. This practice has continued for decades. Despite the inclusion of the species on published lists of organisms generally regarded acceptable for use in nutritional supplements and food, it is important to note that not all strains of a single species should be considered safe for consumption. Bacillus subtilis is included on a list of biological agents that have a qualified presumption of safety (QPS) that is published by the European Food Safety Authority. Some Bacillus subtilis strains have been shown to produce the hemolytic enterotoxin Hbl.
It is also common knowledge that biogenic AgNPs are very efficient catalysts that may completely degrade toxic effluents as well as facilitate the hydrogenation of derivatives based on nitroaromatic chemicals.25–28 Dangerous pollutants of nitrophenol derivatives such as 4-nitrophenol (4-NP), 3-nitrophenol (3-NP), and 2-nitrophenol (2-NP), were found in industrial wastewater. This wastewater is discharged primarily because of petrochemical refining, the production of pesticides and fertilizer, and manufacturing activities related to dyes. Recently, the catalytic reduction of the nitroaromatic compounds utilizing BioAgNPs as catalysts has been the subject of extensive study.
Syzygium nervosum belonging to Myrtaceae plant family with the common names “Voi” in Vietnamese, “Shui weng” in Chinese is widely dispersed in Southeast Asian tropical regions. In Vietnam and China, S. nervosum leaves or flower buds brewed in hot water are often taken as a beverage. It is also well known for treating influenza and some stomach disorders. In addition, the leaves and flower buds of S. nervosum were applied topically to treat inflammatory disorders, such as bruises, acne, and skin ulcers. Phytochemical research on the flower buds of S. nervosum led to the isolation and identification over 80 chemicals, which can be divided into three main groups: terpenoids, flavonoids, and phloroglucinols,29 that could be excellent source for synthesizing AgNPs. The predominant chemical composition of the aqueous extract derived from SN buds primarily comprises of organic compounds characterized by the presence of carboxylic and polyhydroxy groups. These specific functional groups are recognized for their significant role in the biosynthesis and stabilization of silver nanoparticles.30
The stability of AgNPs solutions, which refers to their ability to maintain a homogeneous dispersion of nanoparticles over time, plays a crucial role in their catalytic and antibacterial properties. It is essential because any aggregation or precipitation of nanoparticles can significantly affect their catalytic and antibacterial efficiency. To enhance the stability of AgNPs solutions, various strategies can be employed, such as surface functionalization and stabilizing agents.31,32
In this study, for the first time, stable AgNPs were successfully synthesized by a green approach using an aqueous extract of SN buds and without using any additional chemicals. The antimicrobial activity of the biosynthesized AgNPs was investigated against four bacterial strains, including two Gram-positive bacteria (B. subtilis and S. aureus) and two Gram-negative bacteria (S. typhi and E. coli). Additionally, the catalytic degradation potential of the AgNPs towards 2-NP, 3-NP, and 4-NP in an aqueous medium was assessed. Finally, a field trial was conducted to evaluate the antifungal properties of the AgNPs against Colletotrichum camelliae, the causative agent of anthracnose disease in tea plants. Thus, the significant contribution of this study involves the successful biosynthesis and the stability of bioAgNPs, which highlighted as the crucial factors affecting their catalytic, antibacterial and antifungal properties. Furthermore, the application in the agricultural segment demonstrates the practical solution to combat anthracnose and other diseases in tea plants with economic and environmental benefits.
Materials and methods
Materials and chemicals
All of the compounds were of an analytical quality and did not require any additional purification before usage. Acros (Belgium) was the vendor for the acquisition of silver nitrate (AgNO3). Merck (India) was the company that provided the following chemicals: sodium tetrahydridoborate (NaBH4), 2-nitrophenol (C6H5NO3), 3-nitrophenol (C6H5NO3), and 4-nitrophenol (C6H5NO3). Syzygium nervosum buds were gathered in Vietnam's Gialai Province, which is known for its rugged terrain, suitable for growing conditions of SN buds. The Department of Biotechnology, Institute of Food and Biotechnology, Industrial University of Ho Chi Minh City, Vietnam, contributed four bacterial strains, including two Gram-positive bacteria (B. subtilis and S. aureus) and two Gram-negative bacteria (S. typhi and E. coli).Preparation of SN bud extract
Before being used, the fresh buds were given a thorough washing to eliminate all impurities, dried at a temperature of 50 °C for three days, and then ground into a fine powder. The powder of SN buds (ten grams) was steeped for 60 min at a temperature of 90 °C with reflux in distilled water (200 mL). After waiting for the obtained extract to reach room temperature, it was cooled further and filtered using Whatman filter paper No. 1 in order to eliminate any remaining plant debris. In preparation for subsequent tests, the extract was kept in the refrigerator at a temperature of 4 °C.Preparation of BioAgNPs
To produce biogenic AgNPs, an equal volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 between AgNO3 solution and an aqueous SN bud extract were used in the synthesis process. In an essence, 1 mL of the extract was combined with 1 mL of a solution containing Ag+ ions while the mixture was vigorously stirred in the dark. This was done to prevent any undesired photochemical reaction from occurring with the newly formed AgNPs.33,34 After the reaction was complete, a visible sign indicating that the synthesis process had been successful appeared in the form of a change in the color of the colloidal solution. To identify the best conditions, the factors that influence the synthesis process, such as the concentration of silver ions (0.5–2 mmol L−1), reaction duration (15–100 min), and reaction temperature (60–100 °C), were also studied. In order to carry out the optimization, UV-vis measurements were carried out on a Cary 60 UV-vis spectrophotometer. The characteristic maximum absorption peak of AgNPs was located somewhere about 425 nm. Before being used for the purpose of researching their physicochemical properties and catalytic activity, the acquired AgNPs were subjected to the ideal conditions of centrifugation and drying until they reached a consistent mass.
1 between AgNO3 solution and an aqueous SN bud extract were used in the synthesis process. In an essence, 1 mL of the extract was combined with 1 mL of a solution containing Ag+ ions while the mixture was vigorously stirred in the dark. This was done to prevent any undesired photochemical reaction from occurring with the newly formed AgNPs.33,34 After the reaction was complete, a visible sign indicating that the synthesis process had been successful appeared in the form of a change in the color of the colloidal solution. To identify the best conditions, the factors that influence the synthesis process, such as the concentration of silver ions (0.5–2 mmol L−1), reaction duration (15–100 min), and reaction temperature (60–100 °C), were also studied. In order to carry out the optimization, UV-vis measurements were carried out on a Cary 60 UV-vis spectrophotometer. The characteristic maximum absorption peak of AgNPs was located somewhere about 425 nm. Before being used for the purpose of researching their physicochemical properties and catalytic activity, the acquired AgNPs were subjected to the ideal conditions of centrifugation and drying until they reached a consistent mass.
Analysis of BioAgNPs
The Fourier transform infrared spectroscopy (FTIR) technique was utilized on a Bruker Tensor 27 (Germany) in the region of 4000–500 cm−1 in order to identify covalent bonds of potential functional groups that were present in the dried SN extract as well as the powdered BioAgNPs. Powder X-ray diffraction (XRD) analysis was performed on a Shimadzu 6100 X-ray diffractometer (Japan) operating at the voltage of 40 kV, the current of 30 mA with CuK radiation at the wavelength of 1.5406 nm, scanning speed of 0.05° s−1 and step size of 0.02° in the range 2 from 10° to 80° to determine the crystalline structure and composition of BioAgNPs. Transmission electron microscopy (TEM) and high-resolution transmission electron microscopy (HR-TEM) using a JEOL JEM-2100 (Japan) at accelerated voltages of 120 and 200 kV, respectively, were used to assess the morphology of the BioAgNPs in colloidal solution. The morphology of biosynthesized BioAgNPs in the aggregation state after centrifugation was further investigated using field-emission scanning electron microscopy (FE-SEM) on a Hitachi S-4800 HI-9057-0006 (Japan) with an accelerating voltage of 10 kV. In order to determine the chemical elemental composition of the powder nanoparticles, energy dispersive X-ray spectroscopy (EDX) was carried out using a Horiba EMAX Energy EX-400 analyzer (Japan). Dynamic light scattering (DLS) technology was utilized on a Horiba SZ-100 (Japan) to measure the dynamic diameter of AgNPs in colloidal solution. The amount of silver and organic matrix that acted as a capping agent for AgNPs was figured out by using thermogravimetric analysis (TG and DTA) on a Setaram Labsys Evo S60/58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988 thermoanalyzer from France in the temperature range of 30–800 °C and at a heating rate of 10 °C min−1 in atmosphere air. Eventually, UV-vis spectroscopy validated both the BioAgNPs characterization and the 4-NP reduction. The band gap value was obtained from UV-vis diffuse reflectance spectra (DRS) using a Shimadzu UV-vis 2550 spectrophotometer (Japan), covering a wavelength range of 200–800 nm with a resolution of 1 nm.
988 thermoanalyzer from France in the temperature range of 30–800 °C and at a heating rate of 10 °C min−1 in atmosphere air. Eventually, UV-vis spectroscopy validated both the BioAgNPs characterization and the 4-NP reduction. The band gap value was obtained from UV-vis diffuse reflectance spectra (DRS) using a Shimadzu UV-vis 2550 spectrophotometer (Japan), covering a wavelength range of 200–800 nm with a resolution of 1 nm.
Catalytic activity of BioAgNPs
In order to examine the catalytic performance of BioAgNPs, contaminated organic compounds (2-NP, 3-NP, and 4-NP) were subjected to a reduction process in a quartz cuvette 10 mm at room temperature. This reaction was carried out with a NaBH4 solution as the reducing agent. The contaminants (2.5 mL, 0.1 mmol L−1) were combined with excess NaBH4 (0.5 mL, 0.1 mol L−1) and BioAgNPs (3 mg). After the chemical reaction was finished, the BioAgNPs could be extracted using centrifugation and then carefully cleaned with ethanol before being reused. UV-vis spectroscopy at 380, 410, and 390 nm for 2-NP, 3-NP, and 4-NP, respectively, was used to assess catalytic performance and kinetics.35,36 Given the circumstances, the quantity of NaBH4 that was utilized was significantly higher than the concentration of the pollutants; hence, the concentration of NaBH4 could be assumed to be constant while the reaction was taking place. In this regard, the reaction could be a pseudo-first-order one, the kinetics of which are described by the linear equation ln(At) = −kt + ln(Ao), in which At and Ao are the concentrations of pollutants at the time t and initial concentration, k is the rate constant, and t is the reaction time, respectively. The slope of the straight line is used in conjunction with linear regression of the exponential function At over the reaction time t to derive the value of the rate constant k.Antibacterial activity of BioAgNPs
The agar disk diffusion method was utilized to investigate the BioAgNPs antibacterial effectiveness while in the form of an optimized and stable colloidal solution. Positive control was carried out using an antibiotic known as ampicillin at a concentration of 0.1 μg mL−1, whereas negative control was carried out using sterile distilled water. For BioAgNPs, the concentrations of AgNPs that were utilized were 1.5, 0.75, 0.375, and 0.1875 mM respectively. The antibacterial activity of the biosynthesized BioAgNPs was evaluated in this study using four different bacterial strains. They included two Gram-positive bacteria (B. subtilis and S. aureus) and two Gram-negative bacteria (S. typhi and E. coli). In an essence, aliquots of BioAgNPs suspension with a volume of 50 μL each were spread out over Mueller Hinton agar in Petri plates with a volume of 100 μL and a concentration of 106 CFU mL−1. The plates were stored at 37 °C for 24 h, after which the antibacterial activity was evaluated by measuring the diameter of the inhibitory zone produced by the tested bacteria.37,38Field trial and assessment of BioAgNPs
The concentration of the BioAgNPs formula used in this study was 1.5 mM. The trial was conducted in a 1000 m2 tea field (cultivar of PH11, on 5–6 year-old plants) in Tuyen Quang province, Vietnam, from July to August 2022. The weather on the day of the BioAgNPs application was sunny with occasional rainfall, and the temperature ranged between 28 and 35 °C. Green planthoppers, another pest in the experimental area, were controlled and had no effect on the disease control test.The test BioAgNPs and positive control were applied twice, with the second application sprayed 7 days after the first. When brown spot disease of anthracnose appeared and affected 3 to 4% of the leaves, the tea field was evenly sprayed with BioAgNPs at doses of 20-fold and 30-fold dilutions.
The assessment was conducted by selecting 10 random points on two diagonal lines in each survey plot. These points were located 0.5 m from the edge of the experimental plot, and one tree was selected at each point. From each tree, four top-level fixed branches were chosen in four directions, and all leaves on the branches were investigated. The disease severity index (I, %) was calculated using the formula below
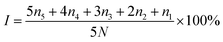 | (1) |
The analysis of variance was performed using the program IRRISATAT 4.0, and the Duncan multiple comparisons were examined for significant differences (P < 0.05).
Results and discussions
As a result of the production of BioAgNPs brought about by the addition of the AgNO3 solution to the SN bud extract, the color of the resulting mixture went from light yellow to dark yellow. It was proposed that the formation process of BioAgNPs was a reaction that was driven by phytochemicals. The phytochemicals that are contained in SN bud extracts, such as alkaloids, flavonoids, amides, lignans, and steroids, have the potential to act as reducing agents for the conversion of Ag+ ions into silver metal Ag0, which is then followed by the agglomeration process of silver atoms into nanosized particles. The phytochemical residues were able to rapidly stabilize the newly obtained AgNPs, resulting in the formation of spherical AgNPs. In order to exert size control over AgNPs, it is necessary to conduct research on three primary synthesis conditions: the concentration of silver ions, the reaction temperature, and the reaction duration. Visual examination was used as a strategy for optimization, and it was determined that the change in color of the BioAgNPs solution caused by the localized surface plasmon resonance (SPR) as well as the existence of the characteristic UV-vis peak at 425 nm were both important factors.39,40 Experimental results demonstrated that the optimal synthesis conditions for obtaining BioAgNPs particles are as follows: a silver ion concentration of 1.5 mmol L−1, a reaction duration of 90 minutes, and a reaction temperature of 100 °C (Fig. S1†).XRD patterns were used to determine the characteristics of the crystalline structures of the biosynthesized BioAgNPs, and the results are displayed in Fig. 1a. The XRD pattern of AgNPs reveals distinctive peaks at the 2θ of 38.12, 44.27, 64.42, and 77.47°. These peaks are indexed for the (111), (200), (220), and (311) planes of the face-centered cubic structure of Ag, corresponding with the ICDD PDF card number 00-004-0783. Specifically, the largest diffraction peak of AgNPs was observed at a 2θ of 38.12°, which suggested that the crystals have a preferential development direction in the plane (111). Therefore, the (111) plane corresponding to the diffraction peak of BioAgNPs at 2θ of 38.12° was used to estimate the d-spacing, lattice parameter, and average crystal size. The d-spacing for a specific set of Miller indices (hkl) can be calculated using Bragg's Law, d = λ/(2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin(θ)), where λ = 1.5406 nm is the wavelength of the X-ray used in the analysis. The lattice parameter (a) for an FCC structure can be determined using the formula a = d/√(h2 + k2 + l2), here, d is the calculated d-spacing value, and h, k, and l are the Miller indices. The Debye–Scherrer equation, D = 0.9λ/(β
sin(θ)), where λ = 1.5406 nm is the wavelength of the X-ray used in the analysis. The lattice parameter (a) for an FCC structure can be determined using the formula a = d/√(h2 + k2 + l2), here, d is the calculated d-spacing value, and h, k, and l are the Miller indices. The Debye–Scherrer equation, D = 0.9λ/(β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos(θ)), where D is the average crystal size, β is the full width at half maximum (FWHM) of the peak 38.12°, was used to compute the average crystal size of BioAgNPs. The d-spacing, lattice parameter, and average crystal size of BioAgNPs were found to be 2.35 Å, 4.07 Å, and 15.1 nm.41 The small crystal size of BioAgNPs offers significant advantages due to the increased surface area-to-volume ratio. It can enhance the nanoparticles's reactivity and efficiency, making them highly suitable for a wide array of applications. Specifically, the larger surface area allows for more active sites, facilitating efficient catalytic reactions and promoting effective antimicrobial activity.
cos(θ)), where D is the average crystal size, β is the full width at half maximum (FWHM) of the peak 38.12°, was used to compute the average crystal size of BioAgNPs. The d-spacing, lattice parameter, and average crystal size of BioAgNPs were found to be 2.35 Å, 4.07 Å, and 15.1 nm.41 The small crystal size of BioAgNPs offers significant advantages due to the increased surface area-to-volume ratio. It can enhance the nanoparticles's reactivity and efficiency, making them highly suitable for a wide array of applications. Specifically, the larger surface area allows for more active sites, facilitating efficient catalytic reactions and promoting effective antimicrobial activity.
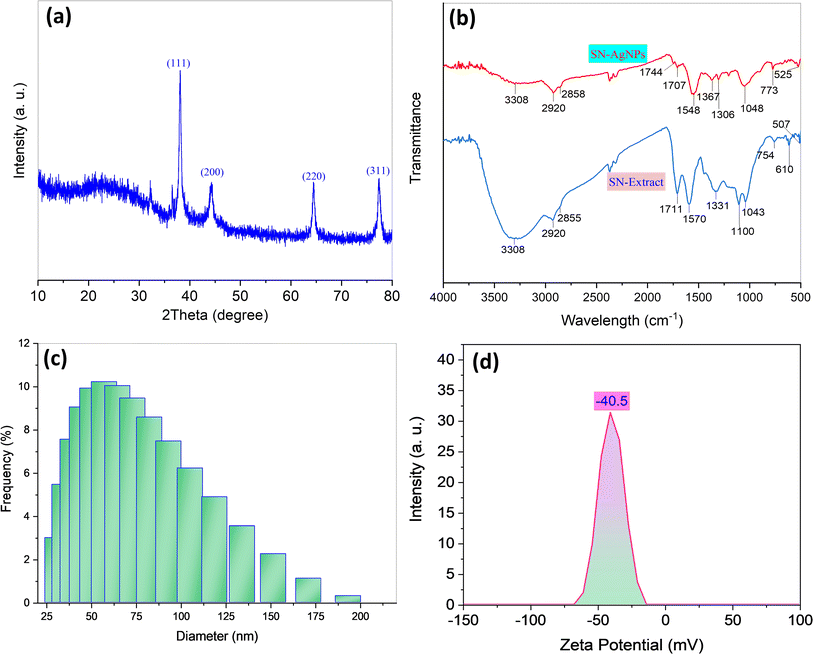 | ||
| Fig. 1 XRD pattern of BioAgNPs (a), FTIR spectra of SN bud extract and SN bud extract capped AgNPs (b), dynamic light scattering diameter distributions (c) and zeta potential (d) of BioAgNPs. | ||
Fig. 1b displays the FTIR spectra of dried extract from SN bud as well as the FTIR spectra of SN bud extract capped AgNPs. The FTIR spectra of both samples showed that the absorption bands were comparable to one another. According to published works, the broad bands that are centered at 3276 cm−1 can be attributed to the stretching vibrations of O–H groups42 present in the phloroglucinols that are found in the extract. C–H stretching vibrations of –CH3 and –CH2- bonds are responsible for the peaks that appear at 2933 and 2883 cm−1, respectively.43,44 The peaks at 1716 and 1627 cm−1 correspond, respectively, to an acyclic stretch in ketone and a stretch in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond of an aromatic molecule. The peak at 1412 cm−1 is associated with the stretching of aromatic bonds at the C
C bond of an aromatic molecule. The peak at 1412 cm−1 is associated with the stretching of aromatic bonds at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond level. The cyclic nature of ether and the carboxylic acid functional group are respectively responsible for the peaks that are located at 1242 and 1028 cm−1.45 These peaks are associated with aromatic C–O and aliphatic C–O band stretching, respectively.46,47 The FTIR analysis of dried plant extract and AgNPs confirmed the presence of an organic molecular layer surrounding the BioAgNPs, indicating the plant extract's function as a source of reducing agents, most likely organic compounds with polyhydroxy groups, and stabilizing agents, namely organic compounds with carboxyl groups.
C bond level. The cyclic nature of ether and the carboxylic acid functional group are respectively responsible for the peaks that are located at 1242 and 1028 cm−1.45 These peaks are associated with aromatic C–O and aliphatic C–O band stretching, respectively.46,47 The FTIR analysis of dried plant extract and AgNPs confirmed the presence of an organic molecular layer surrounding the BioAgNPs, indicating the plant extract's function as a source of reducing agents, most likely organic compounds with polyhydroxy groups, and stabilizing agents, namely organic compounds with carboxyl groups.
DLS measurements were used to examine the particle size distribution of BioAgNPs, and the results obtained from these measurements are displayed in Fig. 1c. Fig. 2c reveals that the particle size distribution diagrams of BioAgNPs suggested dynamic diameters of 46.7 nm. It was determined that the value of the colloidal AgNPs' zeta potential was the best indicator of their long-term stability (Fig. 1d). In fact, the significant zeta potential value of BioAgNPs was measured to be −40.5 mV, and it is possible that this was caused by the presence of polar functional groups of organic compounds such as fatty acids in the SN bud extract. It has been discovered that the generated AgNPs in the aqueous extract are stabilized by two primary mechanisms. The electrostatic contact between charged AgNPs is the basis for the first mechanism. In order for a colloidal AgNPs suspension to be physically stable, the zeta potential value must be greater than 30 mV.48 This is because there is a powerful force of repulsion between two charged particles that have the same sign. The steric effect of large organic molecules that are present in the aqueous extract is the second mechanism that comes into play here. These organic molecules serve as a barrier, preventing nanoparticles from adhering to one another and forming larger aggregates. After a period of three weeks of observation, it was found that the colloidal BioAgNPs solution did not undergo any discernible color shifts or coagulation. The zeta potential value is almost unchanged from the initial (Fig. 2a). From Fig. 2b and c, it can be seen that the BioAgNPs were stable for at least six months. Hence, it is possible to draw the outstanding stability of the BioAgNPs are established by the additive effect of both aforementioned mechanisms.
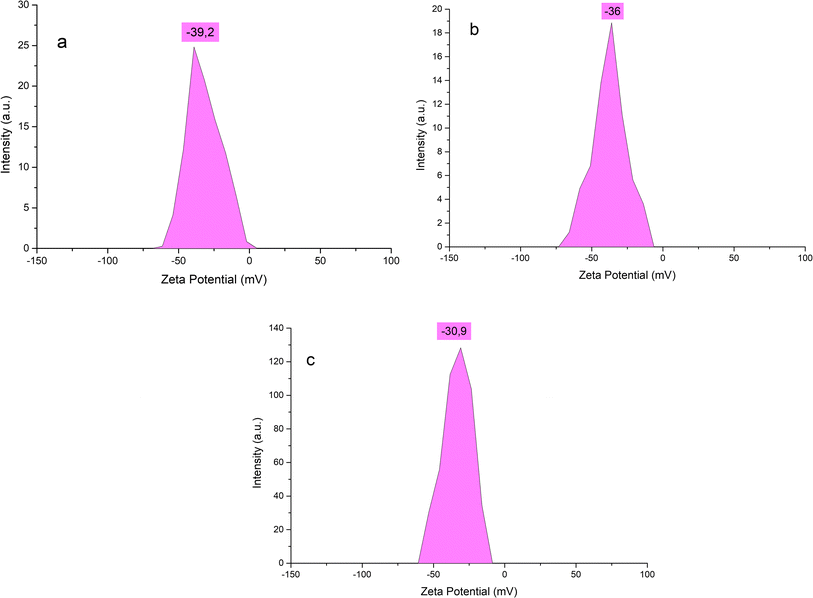 | ||
| Fig. 2 Zeta potential of BioAgNPs after three weeks (a), three months (b) and six months (c) of storage. | ||
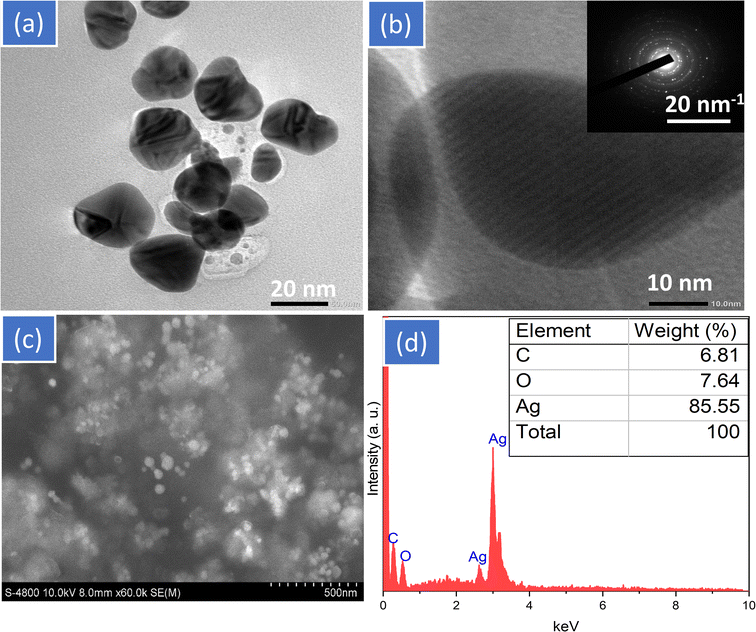
TEM and HR-TEM measurements were used to analyze the particle size and shape of AgNPs and the results of these analyses are displayed in Fig. 3. Fig. 3a reveals that the BioAgNPs crystals were created in the shape of homogeneous spheres, and the typical size range was found to be between 10 and 30 nm. It is possible to visibly observe the crystalline nature of BioAgNPs by the use of HR-TEM images (Fig. 3b). The d-spacing of the lattice fringe in BioAgNPs was measured to be 0.22 nm, corresponding to the (111) plane. It was consistent with the XRD data, confirming the priority development of (111) lattice planes in the cubic crystal structure of BioAgNPs. These results corroborate the crystalline nature of the synthesized BioAgNPs and provide valuable insights into the nanoparticles' crystal growth, their size and lattice parameters.
Once the biosynthesized AgNPs had been through the coagulation process, SEM images and EDX analyses were used to respectively express the surface morphology and element composition of the biosynthesized AgNPs (Fig. 3c and d). The produced AgNPs have an irregular shape but are somewhat homogeneous in size. The SEM image further revealed that there was no sign of true aggregation of the biosynthesized AgNPs because the surrounding matrix of SN bud extract worked as an excellent capping agent. This was owing to the fact that the matrix prevented the formation of aggregates. Fig. 3d displays the results of an EDX analysis performed on the chemical element composition of SN bud extract capped AgNPs. The characteristic peaks of the biosynthesized AgNPs can be found around 2.65, 2.85, and 2.95 keV, as shown by the EDX spectrum. The biosynthesized BioAgNPs are primarily made up of silver (85.55%). The existence of phytoconstituents of SN bud extract capped on AgNPs was confirmed by the observation of carbon (6.81%) and oxygen (7.64%) at energies of 0.256 and 0.54 keV, respectively.
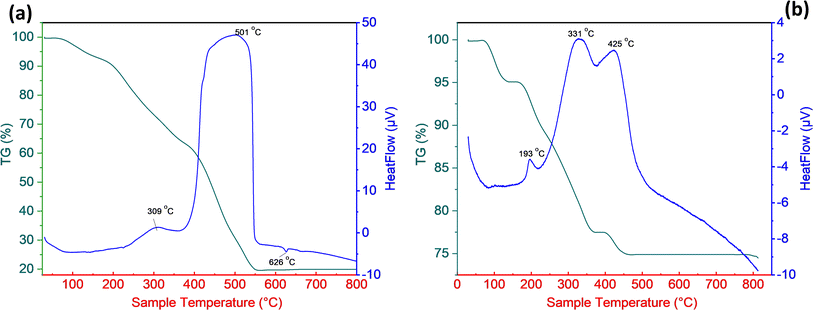
The overall quantity of phytochemical constituents served as a capping agent for the AgNPs, and the thermal behaviors of these constituents were investigated using TGA in the environment air. Fig. 4a and b show, respectively, the TG-DTA curves of dried extract from SN bud and SN bud extract capped AgNPs. These figures are given below. As can be shown in Fig. 4, the weight loss that was detected in the first temperature area spanned from room temperature to 235 °C was around 17.1% for the dried extract and 10.8% for the BioAgNPs. The initial weight loss occurred as a result of the evaporation of volatile chemicals that were contained in the SN bud extract. The second most significant weight loss, which was measured at 62.6% for the dried extract and 14.1% for the AgNPs, occurred in the temperature range between 220 and 560 °C. In this context, the thermal degradation of silver nanoparticles (AgNPs) can generally be linked to the pyrolysis of organic molecules that are capped on the surface of AgNPs. The TGA curve of AgNPs remained practically unaltered at temperatures over 560 °C for SB bud extract and 460 °C for BioAgNPs. This showed that the biosynthesized BioAgNPs consisted of roughly 75% silver and 25% phytochemical ingredients. Both the TGA and the EDX techniques produce results that have a greater proportion of silver when compared to the amount of organic matter present in the sample. This is one of the ways in which these two techniques are comparable. Hence, the thermal behavior of BioAgNPs suggests that the organic layer derived from SN bud extract is mostly responsible for the nanoparticles' stability.
Catalytic performance for reduction of nitrophenols
Many different applications make use of resistant substituted phenols, particularly nitrophenols, which are employed extensively in the chemical industry. Nonetheless, due to their high toxicity and difficulty in degrading, they can pose a significant risk to aquatic life even at low concentrations. This is because they are difficult to break down. The catalytic performance of BioAgNPs was tested in this study by degrading 2-NP, 3-NP, and 4-NP with NaBH4 as the reductant. It is common knowledge that the reduction of those organic substances by NaBH4 without a catalyst is a reaction that is favorable thermodynamically but unfavorable kinetically due to a kinetic barrier between the BH4− and nitrophenolate ions. This barrier prevents the reaction from occurring as quickly as it would otherwise. By employing AgNPs in conjunction with an electron transfer mechanism, this obstacle can be easily circumvented. Fig. 6 displays the outcomes of the reduction of 2-NP, 3-NP, and 4-NP brought about by NaBH4 in the presence of biosynthesized BioAgNPs.After adding NaBH4 to the yellowish solution containing 2-NP, 3-NP, and 4-NP, the color of the solution changed to a dark yellow, and the maximum absorbance peaks emerged at 410, 390, and 400 nm. This occurred despite the fact that the breakdown reactions had not yet begun at the time. As soon as the BioAgNPs were introduced into the solution, the color started to fade away gradually. The UV-vis spectra and first-order kinetics for the breakdown of 2-NP, 3-NP, and 4-NP by NaBH4 in the presence of BioAgNPs are presented in Fig. 5. The presence of a nitrophenol breakdown process was established by a steady decrease in UV-vis maximum absorbance at the wavelength of 410, 390, and 400 nm. According to the findings, the decomposition of 2-NP, 3-NP, and 4-NP in the presence of BioAgNPs was nearly finished after 15 min, as evidenced by characteristic absorbances that were close to zero (Fig. 5a, c and e). The linear relationship between ln(At) and reaction time (Fig. 5b, d and f) allows for the analysis of the calculated relatively similar first-order reaction rate constants k. Table 1 presents the k values for the degradation of 2-NP, 3-NP, and 4-NP using BioAgNPs, along with a comparison to other catalysts reported in the literature. Remarkably, the calculated first-order reaction rate constants k for the mentioned-above nitrophenols were determined to be 9.0 × 10−4, 10 × 10−4, and 9.0 × 10−4 s−1, respectively, using BioAgNPs as the catalyst. These values demonstrated significantly higher degradation rates compared to numerous catalysts documented in existing literature. The observed higher first-order reaction rate constant for the reduction of 3-NP compared to 2-NP and 4-NP can be explained by the presence of conjugation and steric effects.49 The conjugation effect plays a role in stabilizing the 2-NP and 4-NP isomers, allowing for the delocalization of the negative charge in the phenoxide ion onto the nitro group. This delocalization results in their lower reactivity compared to 3-NP. Additionally, the steric hindrance caused by the substituents in the ortho- and para-positions of 2-NP and 4-NP influences their accessibility to the reducing agent, NaBH4, on the catalyst's surface.
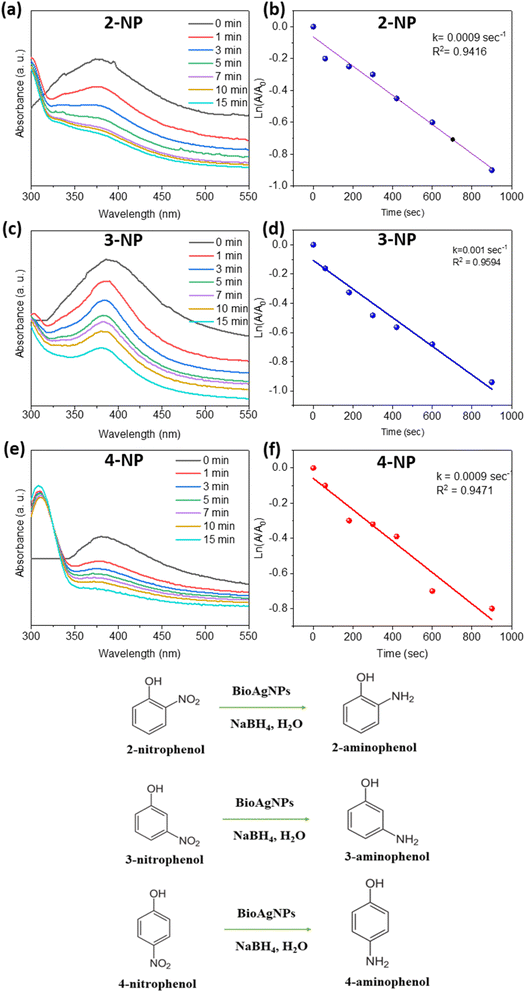 | ||
| Fig. 5 UV-vis spectra (a, c, e), first-order kinetics (b, d, f) of 2-NP, 3-NP, and 4-NP by NaBH4 in the presence of BioAgNPs and diagrams of chemical reactions. | ||
| Substrate | Catalyst | Particle size (nm) | Reduction conditions | k (s−1) | Ref. |
|---|---|---|---|---|---|
| 2-NP | NiFe2O4 | 20 | 20 mL of 0.036 M 2-NP; 1.36 g NaBH4; 2% mol Ni | 4.16 × 10−4 | 50 |
| Ag@cellulose filter paper | 25 | 3 mL of 0.05 mM 2-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 10 mg | 1.15 × 10−4 | 51 | |
| BioAgNPs | 10–30 | 2.5 mL of 0.1 mM 2-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 3 mg | 9.0 × 10−4 | This study | |
| 3-NP | Siraitia grosvenorii fruit extract capped AuNPs | 7.5 | 2.5 mL of 1 mM 3-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 3 mg | 4.0 × 10−4 | 34 |
| BioAgNPs | 10–30 | 2.5 mL of 0.1 mM 3-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 3 mg | 10 × 10−4 | This study | |
| 4-NP | AgNPs@cellulosic fiber | 50 | 2.5 mL of 0.01 mM 4-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 15 mg | 1.44 × 10−5 | 52 |
| Silver-poly(N-isopropylacrylamide-co-methacrylic acid) hybrid microgels | Not reported | 2.5 mL of 2.5 mM 4-NP; 25 mL of 250 mM NaBH4; V (AgNPs) = 0.2 mL | 5.9 × 10−4 | 53 | |
| Ginkgo biloba leaves extract based AgNPs | 20–40 | 25 mL of 0.01 mM 4-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 10 mg | 7.53 × 10−4 | 54 | |
| BioAgNPs | 10–30 | 2.5 mL of 0.1 mM 2-NP; 0.5 mL of 0.1 M NaBH4; mcatalyst = 3 mg | 9.0 × 10−4 | This study |
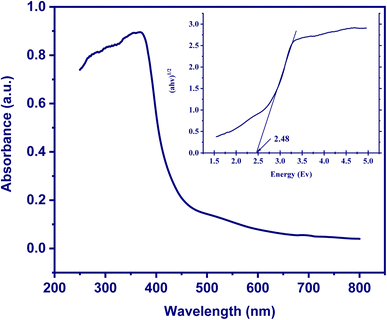
The band gap is a crucial parameter in the study of semiconductor materials, representing the energy required to move an electron from the valence band to the conduction band. It has significant implications for the electronic and chemical properties of materials. The band gap of BioAgNPs was determined utilizing the Tauc plot method based on the DRS spectra.55 By plotting (αhν)1/2 on the y-axis against the light frequency hν on the x-axis, the linear region associated with energies proximal to the band gap was identified. The band gap (Eg) value was estimated by drawing a straight line through this region and determining the intersection point with the x-axis. In this study, the band gap of BioAgNPs was estimated to be 2.48 eV, as shown in Fig. 6. Thus, the BioAgNPs exhibited semiconductor behavior with the ability to absorb and emit light within the ultraviolet range and a portion of the visible light spectrum, confirmed their potential application as catalysts with electron transfer mechanisms, including the reaction between nitrophenol and NaBH4.
Antibacterial assay
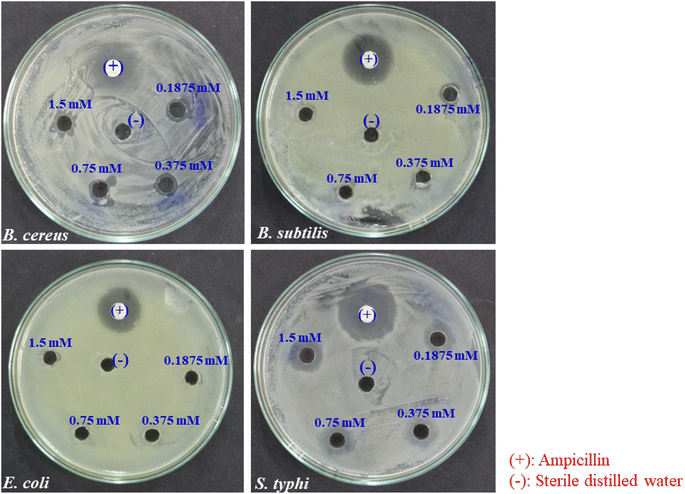
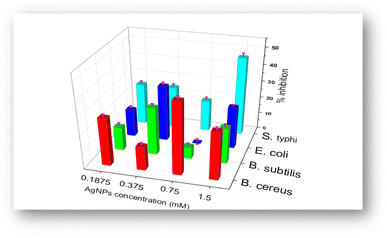
The antibacterial activity of biosynthesized BioAgNPs in the form of colloidal solutions was tested against four bacterial strains: two Gram-positive (S. aureus and B. subtilis) and two Gram-negative (E. coli and S. typhi) with varying concentrations of BioAgNPs. Fig. 7 and 8 exhibit, respectively, the antibacterial effects of BioAgNPs as well as the growth inhibition percentages of the bacteria at various doses of BioAgNPs. It was discovered that BioAgNPs demonstrated the antibacterial activity against all four of the strains of bacteria. Even at low concentrations, the colloidal sample of BioAgNPs demonstrated higher antibacterial activity against S. typhi. The observed difference in antibacterial activity among the strains can be attributed to several factors, such as variations in bacterial cell wall structures, membrane permeability, and susceptibility to the mechanisms of action of AgNPs. Additionally, differences in the specific metabolic pathways or defense mechanisms of each bacterial strain may contribute to variations in their response to the antibacterial effects of AgNPs.56–58 The BioAgNPs obtained from the extract of SN buds also exhibited a comparatively potent antibacterial effect when compared to silver nanoparticles synthesized using various other plant sources (Table 2).| Bacterial strains | Extracts | AgNPs size (nm) | AgNPs concentration | Zone of inhibition (mm) | Ref. |
|---|---|---|---|---|---|
| S. typhi | Polygonum glabrum Willd. leaves | 10–35 | 20 μL 1 mM | 6 | 59 |
| Nypa fruticans (NF) fruit husk | 10–15 | 1.24 μg/1 mL | 6 | 43 | |
| B. stracheyi roots | 25–73 | 30 μg/well | 11 | 60 | |
| Ctenolepis garcini L. leaves | 20–50 | 50 μg/1 mL | 12.4 | 61 | |
| Syzygium nervosum buds | 10–30 | 50 μL 1.5 mM | 13 | This study | |
| B. cereus | Nypa fruticans (NF) fruit husk | 10–15 | 1.24 μg/1 mL | 7 | 48 |
| R. hastatus roots | 25–73 | 30 μg/well | 7 | 60 | |
| Syzygium nervosum buds | 10–30 | 50 μL 1.5 mM | 12 | This study | |
| Leaves extract of U. dioica Linn | 20–30 | 0.05 mg/100 mL | 14 | 62 | |
| E. coli | B. stracheyi roots | 25–73 | 30 μg/well | 0 | 60 |
| Basella alba leaves | 40 | 10 mg mL−1 | 9 | 63 | |
| Moringa Oleifera leaves | 38.7 | 100 μL 1 mg mL−1 | 9 | 64 | |
| Ganoderma lucidum mushroom | 50 | 25 mg mL−1 | 9.8 | 65 | |
| Coccinia indica leaves | 8–48 | 50 μL 20 mg mL−1 | 10.3 | 66 | |
| Syzygium nervosum buds | 10–30 | 50 μL 1.5 mM | 10 | This study | |
| B. subtilis | Ocimum sanctum (L.) leaves | 15.0 ± 12.34 | 100 μL 1.96 mg mL−1 | 2 | 67 |
| Ocimum basilicum leaves | 17 ± 8.94 | 100 μL 1.96 mg mL−1 | 2 | 67 | |
| Ocimum gratissimum leaves | 12–60 | 20 μL 0.5 mM | 8 | 68 | |
| U. dioica Linn leaves | 20–30 | 0.05 mg/100 mL | 10 | 62 | |
| Syzygium nervosum buds | 10–30 | 50 μL 1.5 mM | 10 | This study |
Antifungal property and field trial assessment on tea plants
Colletotrichum camelliae is one of the most significant and serious phytopathogenic molds that regularly infects tea fields, causing anthracnose disease in tea plants (Fig. 9). In Tuyen Quang northeastern province of Vietnam, this disease is responsible for up to 7–8% of infections in tea plantation. Currently, synthetic fungicides such as chlorothalonil and Daconil are used to control the anthracnose of tea. However, due to the high demand for organic tea production and Vietnam's agricultural policies, reducing the use of synthetic fungicides is necessary. This has prompted scientists to investigate new biological and green fungicides to partly replace the current synthetic fungicides used in tea cultivation. | ||
| Fig. 9 Cultivating tea is very popular in many areas of northeastern Vietnam province Tuyen Quang (A); the normal tea leaf (B) and the tea leaf with Colletotrichum camelliae symtom (C). | ||
The control group was treated with water only, while the positive control group was treated with Daconil 75WP at a dose of 2.0 kg ha−1. The disease severity was assessed at three different time points: 7 days after the first treatment (7 DA1T), 7 days after the second treatment (7 DA2T), and 14 days after the second treatment (14 DA2T). Non-significant differences (P < 0.05) are indicated by identical lowercase letters in each column.
During the assessment periods, the test BioAgNPs formulation was sprayed at doses of 20-fold dilution and 30-fold dilution, and the positive control of Daconil 75WP was also used. All disease severity indices for the BioAgNPs formulations were lower than those for the control group.
In all assessments of the field study, the disease severity indices of the BioAgNPs at a dose of 20-fold dilution were equivalent to those of the control formula of Daconil 75WP (the difference was not significant). However, at a dilution ratio of 30-fold dilution, there was a significantly higher disease severity index than that of Daconil 75 WP (Table 3). The BioAgNPs formulations significantly suppressed brown spots caused by anthracnose disease in tea plants at both doses of 20-fold dilution and 30-fold dilution, without affecting the growth and development of the tea plants.
| No | Formulation | Dose (fold-dilution) | Disease severity index (%) | |||
|---|---|---|---|---|---|---|
| Before treatment | 7 DA1T | 7 DA2T | 14 DA2T | |||
| a Control was treated with water only. The positive control was treated with Daconil 75WP at the dose of 2.0 kg ha−1 7 DA1T: assessment at 7 days after the 1st treatment; 7 DA2T: assessment at 7 days after the 2nd treatment; 14 DA2T: assessment at 14 days after the 2nd treatment. Duncan multiple comparisons were examined for significant differences between mean values and a non-significant difference (P < 0.05) is indicated by identical lowercase letters (a, b, c) in each column. | ||||||
| 1 | BioAgNPs | 1/20 | 1.04a | 1.31b | 1.41c | 1.37c |
| 2 | BioAgNPs | 1/30 | 0.90a | 2.13a | 2.23b | 2.44b |
| 3 | Daconil 75WP | 2.0 (kg ha−1) | 1.00a | 1.22b | 1.12c | 1.00c |
| 4 | Control | — | 1.03a | 2.51a | 3.05a | 4.95a |
Conclusion
In this work, the synthesis of stable BioAgNPs was accomplished by employing an aqueous extract from the buds of Syzygium nervosum as both a reducing and a stabilizing agent, giving an additive effect to the nanoparticles' stability by forming an organic protective layer and highly negatively charged nanoparticles surface. The biosynthesized BioAgNPs that were produced were formed in the shape of spheres and ranged in size from around 10 to 30 nm on average. The colloidal BioAgNPs showed good antibacterial activity against four different bacterial strains, including Gram-positive S. aureus and B. subtilis, as well as Gram-negative E. coli and S. typhi. Interestingly, the synthesized BioAgNPs demonstrated higher antibacterial activity towards Gram-negative S. typhi even when evaluated at low concentrations. In addition, the biogenic AgNPs exhibited high catalytic activity in the process of degrading 2-NP, 3-NP, and 4-NP into aminophenols. Furthermore, the field study demonstrated that the BioAgNPs formulations significantly suppressed brown spots caused by anthracnose disease in tea plants at doses of 20-fold dilution and 30-fold dilution, without affecting the growth and development of the tea plants. Hence, BioAgNPs produced by the aqueous extract of Syzygium nervosum bud can be regarded as prospective nanomaterials for large-scale applications in biological and catalytic processes.Author contributions
Thi Lan Pham: investigation, writing-original draft. Van Dat Doan: methodology, writing-original draft. Quang Le Dang: resources, investigation. Tuan Anh Nguyen: investigation. Thi Lan Huong Nguyen: methodology, investigation. Thi Dieu Thuy Tran: investigation. Thi Phuong Lan Nguyen: data curation. Thi Kieu Anh Vo: investigation, visualization. Trung Huy Nguyen: data curation. Dai Lam Tran: conceptualization, writing – review & editing.Conflicts of interest
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.Acknowledgements
This work was funded by The Vietnam Academy of Science and Technology (VAST) under the grant number NCXS 01.01/22-24.References
- J. Yadav and P. Chauhan, Mater. Today: Proc., 2022, 62(P10), 6177–6181 CrossRef CAS.
- N. S. Alharbi, N. S. Alsubhi and A. I. Felimban, J. Radiat. Res. Appl. Sci., 2022, 15(3), 109–124 CAS.
- D. Bamal, A. Singh, G. Chaudhary, M. Kumar, M. Singh, N. Rani, P. Mundlia and A. Sehrawat, Nanomaterials, 2021, 11(8), 2086 CrossRef CAS PubMed.
- M. Gomathi, A. Prakasam, P. V. Rajkumar, S. Rajeshkumar, R. Chandrasekaran and P. M. Anbarasan, S. Afr. J. Chem. Eng., 2020, 32, 1–4 Search PubMed.
- S. Palithya, S. A. Gaddam, V. S. Kotakadi, J. Penchalaneni, N. Golla, S. B. N. Krishna and C. V. Naidu, Part. Sci. Technol., 2022, 40(1), 84–96 CrossRef CAS.
- S. M. Rakib-Uz-Zaman, E. Hoque Apu, M. N. Muntasir, S. A. Mowna, M. G. Khanom, S. S. Jahan, N. Akter, M. A. R. Khan, N. S. Shuborna, S. M. Shams and K. Khan, Challenges, 2022, 13(1), 18 CrossRef.
- M. I. Al-Zaban, M. A. Mahmoud and M. A. AlHarbi, Saudi J. Biol. Sci., 2021, 28(3), 2007–2013 CrossRef CAS PubMed.
- H. B. Herbin, M. Aravind, M. Amalanathan, M. S. M. Mary, M. M. Lenin, C. Parvathiraja, M. R. Siddiqui, S. M. Wabaidur and M. A. Islam, J. Inorg. Organomet. Polym. Mater., 2022, 32(3), 1103–1115 CrossRef CAS.
- M. Sharifi-Rad, P. Pohl and F. Epifano, Nanomaterials, 2021, 11(4), 1045 CrossRef CAS PubMed.
- E. Sreelekha, B. George, A. Shyam, N. Sajina and B. Mathew, Bionanoscience, 2021, 11(2), 489–496 CrossRef.
- H. Bahrulolum, S. Nooraei, N. Javanshir, H. Tarrahimofrad, V. S. Mirbagheri, A. J. Easton and G. Ahmadian, J. Nanobiotechnol., 2021, 19(1), 1–26 CrossRef PubMed.
- N. Garg, S. Bera, L. Rastogi, A. Ballal and M. V. Balaramakrishna, Spectrochim. Acta, Part A, 2020, 232, 118126 CrossRef CAS PubMed.
- Y. H. Gonfa, A. A. Gelagle, B. Hailegnaw, S. A. Kabeto, G. A. Workeneh, F. B. Tessema, M. G. Tadesse, S. M. Wabaidur, K. A. Dahlous, S. Abou Fayssal, P. Kumar, B. Adelodun, A. Bachheti and R. K. Bachheti, Sustainability, 2023, 15(1), 1–20 CrossRef.
- W. W. Melkamu and L. T. Bitew, Heliyon, 2021, 7(11), e08459 CrossRef CAS.
- S. Periasamy, U. Jegadeesan, K. Sundaramoorthi, T. Rajeswari, V. N. B. Tokala, S. Bhattacharya, S. Muthusamy, M. Sankoh and M. K. Nellore, J. Nanomater., 2022, 2022, 8123854 Search PubMed.
- R. Thiurunavukkarau, S. Shanmugam, K. Subramanian, P. Pandi, G. Muralitharan, M. Arokiarajan, K. Kasinathan, A. Sivaraj, R. Kalyanasundaram, S. Y. AlOmar and V. Shanmugam, Sci. Rep., 2022, 12(1), 1–11 CrossRef PubMed.
- M. Moond, S. Singh, S. Sangwan, P. Devi, A. Beniwal, J. Rani, A. Kumari and S. Rani, Molecules, 2023, 28(3), 1–20 CrossRef PubMed.
- N. Xu, S. Jin and L. Wang, Rev. Anal. Chem., 2020, 40(1), 1–11 CrossRef.
- V. H. Tran, D. L. Tran, T. C. Ba, D. H. Vu, N. T. Nguyen, G. D. Pham and X. P. Nguyen, Colloids Surf., A, 2010, 360(1–3), 32–40 CrossRef.
- S. K. Deivanathan and J. T. J. Prakash, Chem. Data Collect., 2022, 38, 100831 CrossRef CAS.
- Z. P. Qiao, M. Y. Wang, J. F. Liu and Q. Z. Wang, Inorg. Chem. Commun., 2022, 138, 109301 CrossRef CAS.
- M. Noga, J. Milan, A. Frydrych and K. Jurowski, Int. J. Mol. Sci., 2023, 24(6), 5133 CrossRef CAS PubMed.
- L. Xu, Y. Y. Wang, J. Huang, C. Y. Chen, Z. X. Wang and H. Xie, Theranostics, 2020, 10(20), 8996–9031 CrossRef CAS.
- I. Mohiuddin, S. Bhogal, A. Grover, A. K. Malik and J. S. Aulakh, Environ. Nanotechnol., Monit. Manage., 2021, 16, 100527 CAS.
- A. R. Deshmukh and B. S. Kim, J. Environ. Chem. Eng., 2021, 9(6), 106707 CrossRef CAS.
- S. Kapoor, H. Sood, S. Saxena and O. P. Chaurasia, Bioprocess Biosyst. Eng., 2022, 45(2), 365–380 CrossRef CAS PubMed.
- N. Z. Srećković, Z. P. Nedić, D. Liberti, D. M. Monti, N. R. Mihailović, J. S. Katanić Stanković, S. Dimitrijević and V. B. Mihailović, RSC Adv., 2021, 11(56), 35585–35599 RSC.
- S. Yadav, A. Asthana, R. Chakraborty, B. Jain, A. K. Singh, S. A. C. Carabineiro and Md. A. B. H. Susan, Nanomater, 2020, 10(1), 170 CrossRef CAS PubMed.
- G. N. Pham, T. T. T. Nguyen and N. H. Nguyen, J. Evidence-Based Complementary Altern. Med., 2020, 2020, 8263670 Search PubMed.
- T. Rasheed, M. Bilal, C. Li, F. Nabeel, M. Khalid and H. M. N. Iqbal, J. Photochem. Photobiol., B, 2018, 181, 44–52 CrossRef CAS.
- V. T. Hoang, M. Mai, T. T. Le, N. P. Vu, K. T. Nguyen, D. T. Nguyen, Q. H. Tran, A. T. Le, X. D. Ngo and V. H. Tran, Adv. Polym. Technol., 2020, 2020, 9437108 CrossRef.
- A. P. Z. Stevenson, D. B. Bea, S. Civit, S. A. Contera, A. I. Cerveto and S. Trigueros, Nanoscale Res. Lett., 2012, 7(1), 151 CrossRef PubMed.
- V. D. Doan, V. T. Le, T. D. Nguyen, T. L. H. Nguyen and H. T. Nguyen, Mater. Res. Express, 2019, 6(11), 1150g1 CrossRef.
- V. T. Le, T. G. Duong, V. T. Le, T. L. Phan, T. H. L. Nguyen, T. P. Chau and V. D. Doan, RSC Adv., 2021, 11(25), 15438–15448 RSC.
- T. M. T. Nguyen, T. T. T. Huynh, C. H. Dang, D. T. Mai, T. T. N. Nguyen, D. T. Nguyen, V. S. Dang, T. D. Nguyen and T. D. Nguyen, Res. Chem. Intermed., 2020, 46(3), 1975–1990 CrossRef CAS.
- T. T. Vo, C. H. Dang, V. D. Doan, V. S. Dang and T. D. Nguyen, J. Inorg. Organomet. Polym. Mater., 2020, 30(2), 388–399 CrossRef CAS.
- J. Khan, I. Naseem, S. Bibi, S. Ahmad, F. Altaf, M. Hafeez, M. M. Almoneef and K. Ahmad, Materials, 2023, 16(1), 1–13 Search PubMed.
- P. Velmurugan, M. Cho, S. S. Lim, S. K. Seo, H. Myung, K. S. Bang, S. Sivakumar, K. Cho and B. T. Oh, Mater. Lett., 2015, 138, 272–275 CrossRef CAS.
- N. Thangamani and N. Bhuvaneshwari, Chem. Phys. Lett., 2019, 732, 136587 CrossRef CAS.
- G. Abilash, P. Ramakrishna, M. Ramakrishna, K. Lohith, J. Chelli and M. R. Apparao, Langmuir, 2011, 8(1), 4007–4021 Search PubMed.
- A. Monshi, M. R. Foroughi and M. R. Monshi, World J. Nano Sci. Eng., 2012, 2(03), 154–160 CrossRef.
- V. Kumar, S. Singh, B. Srivastava, R. Bhadouria and R. Singh, J. Environ. Chem. Eng., 2019, 7(3), 103094 CrossRef CAS.
- V. D. Doan, A. T. Thieu, T. D. Nguyen, V. C. Nguyen, X. T. Cao, T. L. H. Nguyen and V. T. Le, J. Nanomater., 2020, 2020, 4548790 Search PubMed.
- Y. H. Yang, X. Z. Li and S. Zhang, RSC Adv., 2018, 8(52), 29980–29987 RSC.
- Y. M. Yukhin, A. I. Titkov, G. K. Kulmukhamedov and N. Z. Lyakhov, Theor. Found. Chem. Eng., 2015, 49(4), 490–496 CrossRef CAS.
- M. Irani, L. R. Rad, H. Pourahmad and I. Haririan, Microporous Mesoporous Mater., 2015, 206, 1–7 CrossRef CAS.
- M. Sokolsky-Papkov and A. Kabanov, Polymers, 2019, 11(10), 1553 CrossRef CAS PubMed.
- V. D. Doan, B. A. Huynh, T. D. Nguyen, X. T. Cao, V. C. Nguyen, T. L. H. Nguyen, H. T. Nguyen and V. T. Le, J. Nanomater., 2020, 2020, 8492016 Search PubMed.
- M. Gholinejad, Z. Naghshbandi and J. M. Sansano, Appl. Organomet. Chem., 2020, 34(4), e5522 CrossRef CAS.
- A. Goyal, S. Bansal and S. Singhal, Int. J. Hydrogen Energy, 2024, 39(10), 4895–4908 CrossRef.
- S. M. Albukhari, M. Ismail, K. Akhtar and E. Y. Danish, Colloids Surf., A, 2019, 577, 548–561 CrossRef CAS.
- F. Torkamani and S. Azizian, J. Mol. Liq., 2016, 214, 270–275 CrossRef CAS.
- S. R. Khan, Z. H. Farooqi, M. Ajmal, M. Siddiq and A. Khan, J. Dispersion Sci. Technol., 2013, 34(10), 1324–1333 CrossRef CAS.
- F. Wang, W. Zhang, X. Tan, Z. Wang, Y. Li and W. Li, Colloids Surf., A, 2019, 563, 31–36 CrossRef CAS.
- V. A. Tran, T. P. Nguyen, V. T. Le, I. T. Kim, S. W. Lee and C. T. Nguyen, J. Sci.: Adv. Mater. Devices, 2020, 6(1), 108–117 Search PubMed.
- P. Singh, K. R. B. Singh, R. Verma, P. Prasad, R. Verma, S. N. Das, J. Singh and R. P. Singh, New J. Chem., 2022, 46, 8805–8816 RSC.
- P. Singh, K. R. B. Singh, J. Singh, P. Prasad and R. P. Singh, RSC Adv., 2021, 11, 25752–25763 RSC.
- P. Singh, K. R. B. Singh, J. Singh, S. N. Das and R. P. Singh, RSC Adv., 2021, 11, 18050–18060 RSC.
- V. K. Rokhade and T. C. Taranath, J. Pharm. Biol. Sci., 2018, 13(3), 68–75 Search PubMed.
- S. Rashid, M. Azeem, S. A. Khan, M. M. Shah and R. Ahmad, Colloids Surf., B, 2019, 179, 317–325 CrossRef CAS PubMed.
- M. Narayanan, S. Divya, D. Natarajan, S. Senthil-Nathan, S. Kandasamy, A. Chinnathambi, T. A. Alahmadi and A. Pugazhendhi, Process Biochem., 2021, 107, 91–99 CrossRef CAS.
- K. Jyoti, M. Baunthiyal and A. Singh, J. Radiat. Res. Appl. Sci., 2016, 9(3), 217–227 CAS.
- S. Vinodhini, B. S. M. Vithiya and T. A. A. Prasad, J. King Saud Univ., Sci., 2022, 34(4), 101939 CrossRef.
- L. H. Abdel-Rahman, B. S. Al-Farhan, D. Abou El-ezz, M. A. Abd-El Sayed, M. M. Zikry and A. M. Abu-Dief, J. Inorg. Organomet. Polym. Mater., 2022, 32(4), 1422–1435 CrossRef CAS.
- T. H. A. Nguyen, V. C. Nguyen, T. N. H. Phan, V. T. Le, Y. Vasseghian, M. Trubitsyn, A. T. Nguyen, T. P. Chau and V. D. Doan, Chemosphere, 2022, 287(P3), 132271 CrossRef CAS PubMed.
- S. V. Chinni, S. C. B. Gopinath, P. Anbu, N. K. Fuloria, S. Fuloria, P. Mariappan, K. Krusnamurthy, L. V. Reddy, G. Ramachawolran, S. Sreeramanan and S. Samuggam, Crystals, 2021, 11(2), 97 CrossRef CAS.
- V. Malapermal, I. Botha, S. B. N. Krishna and J. N. Mbatha, Saudi J. Biol. Sci., 2017, 24(6), 1294–1305 CrossRef CAS PubMed.
- K. Sharma, S. Guleria and V. K. Razdan, J. Plant Biochem. Biotechnol., 2020, 29(2), 213–224 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra02754f |
| ‡ The authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |