Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Therapeutic potential of 1,3,4-oxadiazoles as potential lead compounds for the treatment of Alzheimer's disease†
Saira Naseem‡
a,
Ahmed Temirak‡ b,
Aqeel Imran
b,
Aqeel Imran cd,
Saquib Jalilc,
Shamool Fatimaa,
Parham Taslimi
cd,
Saquib Jalilc,
Shamool Fatimaa,
Parham Taslimi e,
Jamshed Iqbal
e,
Jamshed Iqbal *cg,
Mussarat Tasleem
*cg,
Mussarat Tasleem a,
Muhammad Nawaz Tahir
a,
Muhammad Nawaz Tahir f and
Zahid Shafiq
f and
Zahid Shafiq *a
*a
aInstitute of Chemical Sciences, Bahauddin Zakariya University, Multan, 60800, Pakistan. E-mail: zahidshafiq@bzu.edu.pk
bNational Research Centre, Chemistry of Natural and Microbial Products Department, Pharmaceutical and Drug Industries Research Institute, Dokki, Cairo P.O. Box 12622, Egypt
cCentre for Advanced Drug Research, COMSATS University Islamabad, Abbottabad Campus, Abbottabad-22060, Pakistan. E-mail: drjamshed@cuiatd.edu.pk
dDepartment of Pharmacy, COMSATS University Islamabad, Lahore Campus, Punjab 54000, Pakistan
eDepartment of Biotechnology, Faculty of Science, Bartin University, 74100 Bartin, Turkey
fDepartment of Physics, University of Sargodha, Sargodha, Pakistan
gDepartment of Chemistry, COMSATS University Islamabad, Abbottabad Campus, Abbottabad-22060Pakistan
First published on 9th June 2023
Abstract
Monoamine oxidase and cholinesterase enzymes are important targets for the treatment of several neurological diseases especially depression, Parkinson disease and Alzheimer's. Here, we report the synthesis and testing of new 1,3,4-oxadiazole derivatives as novel inhibitors of monoamine oxidase enzymes (MAO-A and MAO-B) and cholinesterase enzymes (acetyl and butyryl cholinesterase (AChE, BChE). Compounds 4c, 4d, 4e, 4g, 4j, 4k, 4m, 4n displayed promising inhibitory effects on MAO-A (IC50: 0.11–3.46 μM), MAO-B (IC50: 0.80–3.08 μM) and AChE (IC50: 0.83–2.67 μM). Interestingly, compounds 4d, 4e and 4g are multitargeting MAO-A/B and AChE inhibitors. Also, Compound 4m displayed promising MAO-A inhibition with IC50 of 0.11 μM and high selectivity (∼25-fold) over MAO-B and AChE enzymes. These newly synthesized analogues represent promising hits for the development of promising lead compounds for neurological disease treatment.
1. Introduction
Mental and neurological disorders are often highly disabling and are associated with increased premature mortality rates. Other than the health consequences, these disorders have also their negative impacts on society and the economy. There are many reported neurological disorders including; Parkinson's disease, Alzheimer's disease, stroke, epilepsy, migraine, meningitis, spinal cord injuries and many others.1Alzheimer's disease (AD) is a complicated neurological disorder.2 The number of AD patients worldwide in 2019 was around 57 million and it is expected that this number will triple by 2050. Many hypotheses have been reported explaining the onset and progression of AD pathology including cholinergic neuron damage, inflammation, oxidative stress and the abnormal deposition of amyloid β (Aβ) protein in the neurons, etc.3 The two major hallmarks of AD diagnosis are β-amyloid peptide (Aβ) and the generation of neurofibrillary tangles of the axon-enriched microtubule-associated protein tau.4
The brain can only function well when there is an equilibrium of the neurotransmitter systems for example: acetylcholine (ACh), dopamine, gamma-aminobutyric acid (GABA), serotonin, and others.5 Monoamine oxidase enzyme (MAO) is a flavin-containing membrane-bounded enzyme located particularly in brain and liver.6 It catalyzes the endogenous and exogenous oxidative deamination of monoamine neurotransmitters resulting in the formation of hydrogen peroxide which is a negotiator of oxidative stress.5 This affects the concentration of many xenobiotic and neurotransmitter amines leading to several neurological diseases.7
Two distinct forms of monoamine oxidase (MAO-A and MAO-B) are present in most tissues of mammals and displays different structure, regulation, and function. MAO-A mainly deaminates neurotransmitters such as serotonin, adrenaline and noradrenaline which are aromatic in nature. MAO-B which is the main isoform located in the brain preferentially oxidizes benzylamines, β-phenylethylamine (PEA) and polyamines.8 Tryptamine and dopamine which are basically dietary amines are generally effected by both enzyme isoforms.9 MAO plays an important role in the progression of several neurological diseases including AD. They increase the amyloid-beta (Aβ) deposition, impair the cognitive functions as a result of neuronal loss and causing the generation of neurofibrillary tangles. Therefore, monoamine oxidase inhibitors (MAOIs) are widely used in the treatment of several neurological and psychiatric conditions.10
Cholinesterase enzymes (ChEs) are involved by the degradation of the neurotransmitter acetyl choline (ACh) in the brain. The decrease in Ach levels in brain is involved with the cognitive dysfunction and memory loss in AD patients. Two ChEs are reported, acetylcholinesterase (AChE) and butyrylcholinesterase (BChE).11 AChE are more expressed than BChE in the cerebral cortex and in the hippocampus of the brain, whereas, during the progress of AD, a great increase in the activity of BChE was reported.12 Based on the important roles of cholinesterase enzymes in the pathophysiology of AD, their inhibitors (AChE and BuChE inhibitors) are FDA approved for the symptomatic treatment of AD.13
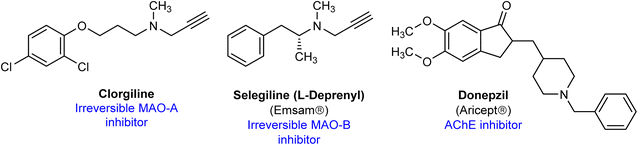
Several MAO inhibitors were approved by FDA for the treatments of several neurological disorders or psychiatric diseases.6 The irreversible MAO-A/B inhibitor tranylcypromine is used as antidepressant. Similarly, the highly selective and irreversible MAO-A inhibitor clorgyline demonstrated antidepressant effects in human, however, it is not used clinically because of dietary interactions (Fig. 1).14 L-Deprenyl, the irreversible MAO-B inhibitor is approved for the treatment of Parkinson disease with trade name Selegiline.15 Currently, several cholinesterase inhibitors, for example donepezil, neostigmine, galantamine and others, are used for the treatment of AD, myasthenia gravis and other disorders.13
Oxadiazole ring is a highly versatile building block in several medicines and in the scope of future drug development.16 Several activities were reported with 1,3,4-oxadiazole containing compounds including; anti-hypertensive (Tiodazosin), anti-inflammatory, anti-fungal, antibacterial, antiviral (Raltegravir), anticonvulsant, hypnotic (Fenadiazole), anticancer (Zibotentan), anti-AD, and many others.7,16–21 Also, several oxadiazole derivatives were reported in literature as MAO and ChE inhibitors (Fig. 2).9,22–25
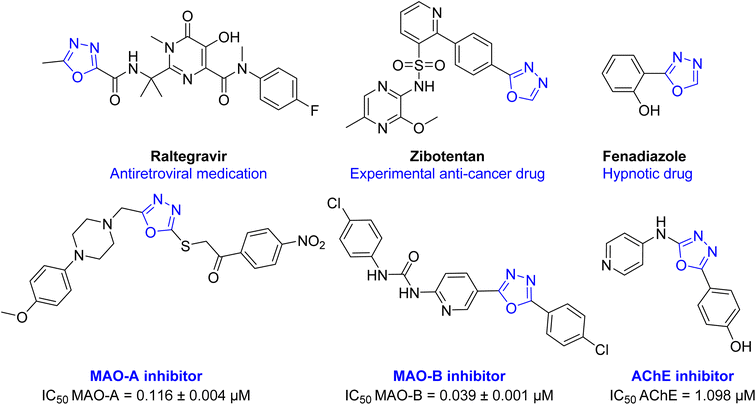 | ||
| Fig. 2 Chemical structure of some drugs and test compounds having 1,3,4-oxadiazole ring in their structure. | ||
In the light of the above discussion, oxadiazole is a versatile ring involved in several MAO and ChE inhibitors. We herein report the synthesis of a new library of 1,3,4-oxadiazol analogues using unique and effective synthetic strategies that resulted in structural diversity of unsymmetrical aryl/alkyl-substitutions. These new compounds were further evaluated against monoamine oxidase (MAO-A and MAO-B) and cholinesterase (AChE and BChE) enzymes. To validate the results, in silico docking studies have been conducted to assess the binding interaction of the synthesized compounds inside the active site of MAO- and ChE enzymes.
2. Results and discussion
2.1 Chemistry
The synthesis of the target compounds 4a–o is outlined in Scheme 1. We started with the synthesis of carbothioamides 3a–o through the reaction of various isothiocyanates 1a–h with acid hydrazides 2a–h.26 The cyclization of carbothioamides 3a–o using mercuric chloride as a catalyst and triethylamine as a base afforded the 1,3,4-oxadiazole derivatives 4a–o (Table 1).18 The procedure as previously reported involves desulfurization of thiosemicarbazides followed by cyclization reactions using mercuric chloride to obtain the desired oxadiazole derivatives.20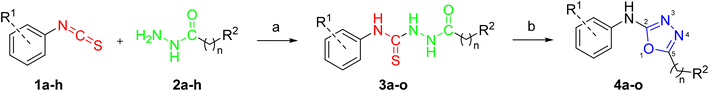 | ||
| Scheme 1 Synthesis of 1,3,4-oxadiazole derivatives 4a–o. Reagents and conditions: (a) benzene, 80 °C, 6 h; (b) HgCl2, TEA, DMF, rt, 8 h, 71–83%. | ||
| Compound | R1 | R2 | n | Yield (%) |
|---|---|---|---|---|
| 4a | 3-Cyano | 4-Trifluorophenyl | 0 | 83 |
| 4b | 3-Cyano | 2-Trifluromethylbenzyl | 1 | 78 |
| 4c | 2-Fluoro | 2-Trifluromethylbenzyl | 1 | 72 |
| 4d | H | 4-Pyridyl | 0 | 80 |
| 4e | 2,4-Dimethyl | 3-Fluorophenyl | 0 | 73 |
| 4f | 2-Fluoro | 3,5-Difluorobenzyl | 1 | 71 |
| 4g | 2,4-Difluoro | 3,5-Difluorobenzyl | 1 | 82 |
| 4h | 2-Fluoro | 2-Fluoro-5-methylphenyl | 0 | 75 |
| 4i | 2-Fluoro | 4-Trifluoromethylphenyl | 0 | 74 |
| 4j | 3-Cyano | 4-Pyridyl | 0 | 75 |
| 4k | 2,4-Dimethyl | 4-Pyridyl | 0 | 75 |
| 4l | 4-Bromo | 4-Trifluoromethylphenyl | 0 | 74 |
| 4m | 4-Isopropyl | 4-Trifluoromethylphenyl | 0 | 73 |
| 4n | 2-Methyl | Phenyl | 0 | 83 |
| 4o | H | 3-Fluoro-2-methyl | 0 | 81 |
The progress of the reaction was monitored by thin layer chromatography (TLC). Upon completion of the reaction, chloroform was added to the reaction mixture and the suspension was filtered and washed carefully to remove the side product mercuric sulfide. The obtained compounds were recrystallized from dichloromethane and their structure and purity were confirmed by different spectroscopic methods including; nuclear magnetic resonance (NMR), infrared (IR), mass spectroscopy and elemental analysis.
2.2 Single crystal X-ray analysis
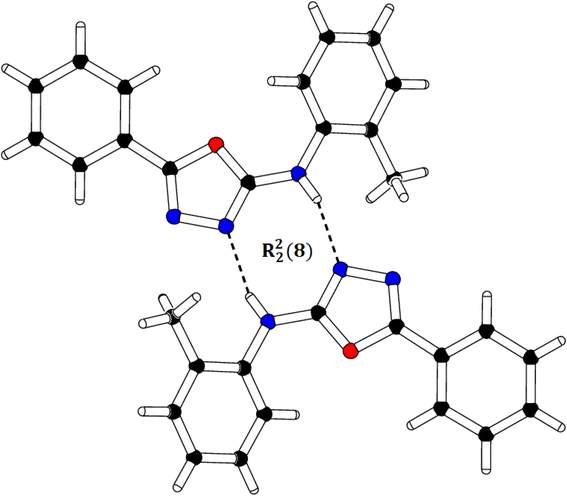
We successfully resolved the X-ray structure of compound 4n (N-(2-methylphenyl)-5-phenyl-1,3,4-oxadiazol-2-amine) using Bruker Kappa Apex-II CCD diffractometer having Mo-Kα X-rays source as depicted in Fig. 3. The o-toluidine ring A (C1–C7/N1), 1,3,4-oxadiazole ring B (C8/C9/N2/N3/O1) and attached phenyl ring C (C10–C15) are planar with root mean square deviation of 0.0060, 0.0037 and 0.0101 Å, respectively. The dihedral angles A/B, A/C and B/C are 38.1 (4)°, 39.7 (4)° and 15.3 (7)°, respectively (please check ESI file for further details displayed in Table S1†). Hence, the overall molecule is non-planar. The molecules are connected with the form of dimers through N–H⋯N bonding to form R22 (8) loop as shown in Fig. 4 and given in Table S2.† The acceptor N-atom is that of nearest to NH group i.e., N2. The crystal packing is further stabilized by C–H⋯π and π⋯π stacking interactions.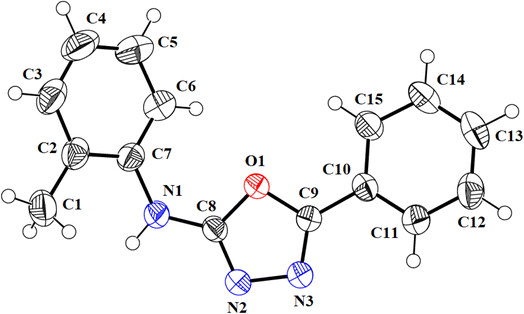 | ||
| Fig. 3 ORTEP diagram of compound 4n that is drawn at probability level of 50%. H-atoms are shown by small circles of arbitrary radii. | ||
2.3 Biological activities
| Compound | MAO-A | MAO-B | AChE | BChE |
|---|---|---|---|---|
| aIC50 ± SEMb (μM)/% age inhibition | ||||
| a IC50 half-maximal half inhibitory concentration in micromolar range.b SEM standard error mean.c %age inhibition.d Positive control. | ||||
| 4a | 33.23c | 37.06c | 42.32%c | 20.54%c |
| 4b | 2.86 ± 1.51b | 35.73c | 31.34%c | 23.65%c |
| 4c | 1.90 ± 0.25b | 0.80 ± 0.27c | 36.12%c | 14.76%c |
| 4d | 1.44 ± 0.22b | 1.04 ± 0.22c | 0.83 ± 0.007a | 23.67%c |
| 4e | 1.47 ± 0.45b | 1.91 ± 0.89 | 1.96 ± 0.176a | 10.54%c |
| 4f | 34.82c | 39.46c | 2.03 ± 0.434a | 22.61%c |
| 4g | 1.21 ± 0.04b | 2.02 ± 1.26 | 1.11 ± 0.080a | 15.23%c |
| 4h | 21.87c | 29.36c | 1.51 ± 0.462a | 12.43%c |
| 4i | 38.11c | 32.53c | 37.12%c | 21.45%c |
| 4j | 1.02 ± 0.09b | 18.57c | 1.56 ± 0.309a | 20.55%c |
| 4k | 3.46 ± 2.58b | 1.10 ± 1.16 | 39.65%c | 14.67%c |
| 4l | 36.81c | 24.86c | 44.56%c | 26.91%c |
| 4m | 0.11 ± 0.001b | 3.08 ± 0.06c | 2.67 ± 0.029a | 15.78%c |
| 4n | 21.07c | 18.47c | 1.23 ± 0.969a | 28.87%c |
| 4o | 29.98c | 22.65c | 2.32 ± 0.080a | 32.98%c |
| Clorgylined | 0.005 ± 0.03 | 61.35 ± 1.13 | — | — |
| Deprenyld | 67.25 ± 1.02 | 0.019 ± 0.01 | — | — |
| Donepezild | — | — | 0.032 ± 0.003a | 6.41 ± 0.34a |
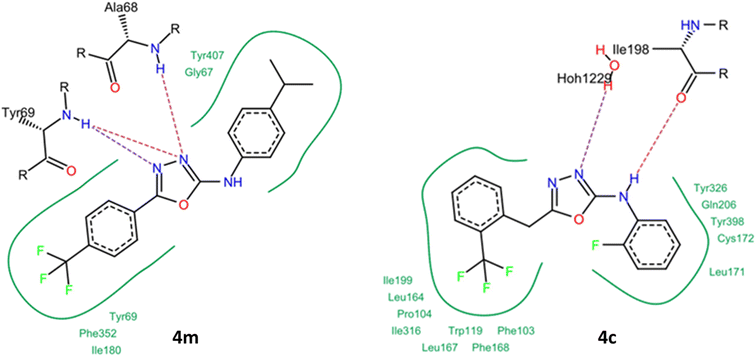
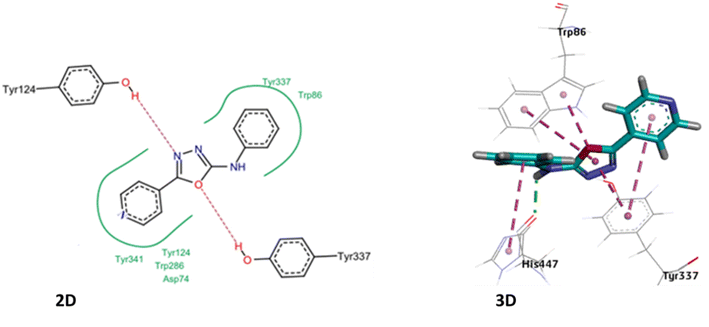
It was observed that several compounds exhibited strong inhibition against MAO-A/MAO-B and AChE, however, all the synthesized compounds did not show strong inhibition against BChE. Among the tested compounds 4c, 4d, 4e, 4g, 4j, 4k and 4m displayed potent inhibition against MAO-A with IC50 in lower micromolar range. Compound 4m exhibited potent inhibition against MAO-A with IC50 value of 0.11 μM whereas 4a, 4f, 4h, 4i, 4l and 4n showed weak inhibition against MAO-A. Inhibitory potential of the synthesized derivatives against MAO-B elaborated in Table 2 which displayed that compounds 4c, 4d, 4e, 4g, 4k and 4m exhibited strong inhibition against MAO-B with IC50 value in sub micromolar range, amongst these 4c exhibited potent inhibition of MAO-B with IC50 value of 0.80 μM.
The majority of tested compounds against AChE have shown potent inhibition with IC50 values in lower micromolar range. The compounds 4d, 4e, 4f, 4h, 4i, 4j, 4m and 4n and exhibited potent inhibition against AChE, amongst all identified inhibitors 4d was promising inhibitor with IC50 value of 0.83 μM. On the contrary, compounds 4a, 4b, 4c, 4i, 4k and 4l exhibited moderate to weak inhibition of AChE. However, all the tested compounds have shown weak inhibition against BChE (Table 2).
Substitution on the phenyl ring of the aniline moiety with bulky groups such as bromo as in compound 4l demolished the activity towards all MAO and ChE enzymes. On the other hand, replacement of the bromo with the isopropyl group as in compound 4m increased dramatically the activity and the selectivity towards the MAO-A enzyme with IC50 value of 0.11 μM. Compound 4d having the terminal unsubstituted phenyl and pyridyl rings displayed promising inhibitory effects against MAO-A, MAO-B and AchE enzymes. Upon substitution on the phenyl ring with m-cyano group as in compound 4j, the compound lost its inhibitory effects on MAO-B. On another example, having 2.4-dimethyl groups on the phenyl ring as in compound 4k, the compound restored its inhibitory effects on MAO-B, however, it lost its inhibitory effects on AchE. From these examples, we can conclude that small structural modifications can have great impact on the inhibitory effects of the developed compounds. Also, having multi-targeting compounds such as compounds 4d, 4e and 4g would be an interesting starting point for development of new chemical entities.
2.4 Molecular docking studies
2.5 In silico pharmacokinetic analysis and computational toxicological evaluation
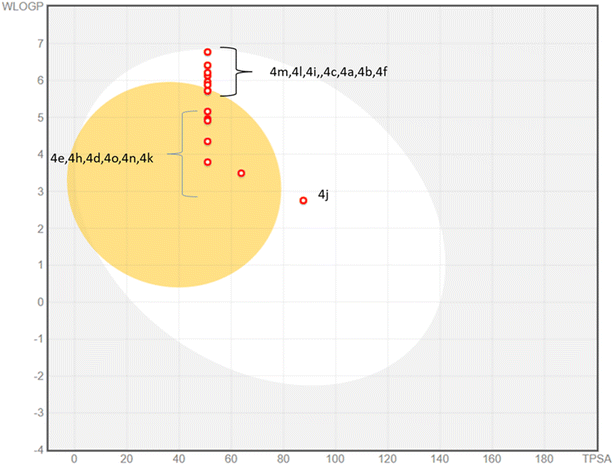
The physicochemical properties and drug likeness were determined through SwissADME web server. Newly synthesized compounds exhibited strict compliance towards rule of Lipinski. The boiled egg plot displayed prominent two portions; white part depicted the good gastrointestinal absorption properties of the tested compounds whereas yellow yolk of the boiled egg showed the permeation ability to blood–brain barrier. Among 4a–o compounds, majority of compounds have displayed excellent gastrointestinal absorption properties whereas some of them (4e, 4h, 4d, 4o, 4n, 4k) exhibited blood–brain barrier permeation ability (Fig. 7). None of the 4a–o compounds exhibited Lipinski violation. Due to their druggable nature these compounds were tested against monoamine oxidase A and B in search of new leads to treat Alzheimer diseases. The detailed parameters for in silico pharmacokinetic are shown in the Table 3.| No. | MW | H-bond acceptor | H-bond donors | TPSA | WLOGP | GI absorption | BBB permeant | Lipinski #violations | PAINS #alerts |
|---|---|---|---|---|---|---|---|---|---|
| 4a | 319.28 | 6 | 1 | 50.95 | 5.96 | High | No | 0 | 0 |
| 4b | 333.31 | 6 | 1 | 50.95 | 5.88 | High | No | 0 | 0 |
| 4c | 337.27 | 7 | 1 | 50.95 | 6.13 | High | No | 1 | 0 |
| 4d | 297.33 | 4 | 1 | 50.95 | 4.96 | High | Yes | 0 | 0 |
| 4e | 291.23 | 6 | 1 | 50.95 | 5.16 | High | Yes | 0 | 0 |
| 4f | 309.22 | 7 | 1 | 50.95 | 5.72 | High | Yes | 1 | 0 |
| 4g | 309.22 | 7 | 1 | 50.95 | 5.72 | High | Yes | 1 | 0 |
| 4h | 287.26 | 5 | 1 | 50.95 | 4.91 | High | Yes | 0 | 0 |
| 4i | 323.25 | 7 | 1 | 50.95 | 6.21 | High | No | 0 | 0 |
| 4j | 263.25 | 5 | 1 | 87.63 | 2.75 | High | No | 0 | 0 |
| 4k | 266.3 | 4 | 1 | 63.84 | 3.49 | High | Yes | 0 | 0 |
| 4l | 384.15 | 6 | 1 | 50.95 | 6.41 | High | No | 1 | 0 |
| 4m | 347.33 | 6 | 1 | 50.95 | 6.77 | High | No | 1 | 0 |
| 4n | 251.28 | 3 | 1 | 50.95 | 3.79 | High | Yes | 0 | 0 |
| 4o | 269.27 | 4 | 1 | 50.95 | 4.35 | High | Yes | 0 | 0 |
In the early phases of drug discovery and lead optimization, computational toxicity parameters cab be predicted through various software. In present study, a software named ProTox-II – Prediction of Toxicity of Chemicals was applied to determine toxicity of the compounds (https://tox-new.charite.de/protox_II/index.php?site=home). The prediction of various toxicity end points like cytotoxicity, immunogenicity, carcinogenicity, mutagenesis were computed for the compounds 4a–o (Table S3†). Among the tested compounds, majority of the compounds exhibited toxicity class 4 and could be considered as druggable compounds. The predicted median lethal dose (LD50) for the 4i and 4o was 1190 mg kg−1 whereas 4b, 4c have shown 800 mg kg−1. All other compounds exhibited LD50 above 500 mg kg−1 which corresponds with toxicity class 4. Among the toxicity classes, class I, II(fetal), whereas class III considered as toxic. However, class IV and class V may be considered as harmful but class VI belongs to non-toxic chemicals (https://www.osha.gov/hazcom). All the tested compounds exhibited no activity for the mutagenicity, carcinogenicity, immunogenicity, hence these compounds could further be explored for the treatment of Alzheimer's disease. Details for the predicted toxicity parameters can be seen in the ESI.†
3. Conclusion
A series of N-phenyl-5-aryl-1,3,4-oxadiazol-2-amine derivatives were synthesized bearing different substituents. Compounds 4a–o were further tested against monoamine oxidase enzymes (MAO-A and MAO-B), cholinesterase enzymes (acetylcholine esterase (AChE) and butyryl cholinesterase (BChE)). All compounds showed promising inhibition in the lower micro molar range against the four target enzymes. Several compounds showed promising inhibitory effects on MAO-A (IC50: 0.11–3.46 μM), MAO-B (IC50: 0.80–3.08 μM) and AChE (IC50: 0.83–2.67 μM). Remarkably, compounds 4d, 4e and 4g are multitargeting MAO-A/B and AChE inhibitors. Compound 4m displayed promising inhibition against MAO-A enzyme with IC50 value of 0.11 μM and high selectivity (∼25-fold) over MAO-B and AChE enzymes. Furthermore, the docking studies further open the window for binding site interactions of the inhibitors with the four studied enzymes. These newly developed multi-targeting compounds represent promising hits for the future treatments of neurological diseases.4. Experimental
4.1 Chemistry
The crystal data was collected on Bruker Kappa Apex-II CCD diffractometer having Mo-Kα X-rays source. SHEXT-2014 and SHELXL-2019/2 softwares were used for the structure solution and refinement, respectively. Anisotropic displacement parameters were assigned to the non-hydrogen atoms whereas isotropic displacement parameters were assigned to hydrogen atoms. The H-atoms are placed by using riding model. ORTEP-III, PLATON software were used for the graphical representations.4.2 General procedure for the synthesis of 1,3,4-oxadiazol derivatives (4a–o)
In a round-bottom flask, a suspension of carbothioamides (3a–o, 1.0 mmol) and mercuric chloride (1.25 mmol) were dispersed in dry DMF (5 mL), and a catalytic amount of triethylamine (0.1 mmol) was added dropwise. The reaction mixture was stirred at rt for 8 hours. Upon completion of the reaction, the mixture was filtered under reduced pressure and washed with 20 mL chloroform. The filtrate was extracted with distilled water (3× 10 mL), then the organic layers were collected, dried over MgSO4 and concentrated under vacuum. The obtained residues were further dissolved in dichloromethane and recrystallized to obtain nice powder of the products 4a–o.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2, 7.6 Hz, Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.15, 112.42, 119.18, 120.10, 126.08, 126.84, 126.91, 127.84, 131.15, 139.80, 157.65, 160.36, 207.02. Anal. calcd. for C16H9F3N4O (330.26): C, 60.19; H, 3.79; N, 13.66; found: C, 60.29; H, 3.55; N, 13.49.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2, 7.6 Hz, Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.15, 112.42, 119.18, 120.10, 126.08, 126.84, 126.91, 127.84, 131.15, 139.80, 157.65, 160.36, 207.02. Anal. calcd. for C16H9F3N4O (330.26): C, 60.19; H, 3.79; N, 13.66; found: C, 60.29; H, 3.55; N, 13.49.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 4.38 (s, 2H, CH2), 7.43–7.46 (m, 1H, Ar–H), 7.53–7.60 (m, 3H, Ar–H), 7.69–7.71 (m, 1H, Ar–H), 7.76–7.80 (m, 2H, Ar–H), 7.95–7.96 (m, 1H, Ar–H) 10.88 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.15, 112.31, 119.72, 122.05, 125.71, 126.10, 126.70, 128.01, 128.66, 131.00, 131.77, 132.77, 133.47, 139.98, 158.72, 159.97, 207.03. Anal. calcd. for C17H11F3N4O (344.29): C, 61.26; H, 4.23; N, 16.12; found: C, 61.13; H, 4.35; N, 16.29.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 4.38 (s, 2H, CH2), 7.43–7.46 (m, 1H, Ar–H), 7.53–7.60 (m, 3H, Ar–H), 7.69–7.71 (m, 1H, Ar–H), 7.76–7.80 (m, 2H, Ar–H), 7.95–7.96 (m, 1H, Ar–H) 10.88 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.15, 112.31, 119.72, 122.05, 125.71, 126.10, 126.70, 128.01, 128.66, 131.00, 131.77, 132.77, 133.47, 139.98, 158.72, 159.97, 207.03. Anal. calcd. for C17H11F3N4O (344.29): C, 61.26; H, 4.23; N, 16.12; found: C, 61.13; H, 4.35; N, 16.29.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (DMSO- d6) δ ppm; 4.35 (s, 2H, CH2), 7.06–7.09 (m, 1H, Ar–H), 7.18 (m, 2H, Ar–H), 7.56 (d, 2H, J = 7.6 Hz Ar–H), 7.71 (t, 1H, J = 8.0 Hz Ar–H), 7.78 (d, 1H, J = 8.0 Hz Ar–H), 8.01 (t, 1H, J = 8.0 Hz Ar–H), 10.22 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.17, 100.50, 115.69, 116.30, 121.25, 123.98, 124.82, 126.47, 128.60, 132.69, 140.78, 149.83, 158.64, 160.85, 207.07. Anal. calcd. for C16H11F4N3O (337.27): C, 56.98; H, 3.29; N, 12.46; found: C, 56.79; H, 3.55; N, 12.38.
N); 1H-NMR (DMSO- d6) δ ppm; 4.35 (s, 2H, CH2), 7.06–7.09 (m, 1H, Ar–H), 7.18 (m, 2H, Ar–H), 7.56 (d, 2H, J = 7.6 Hz Ar–H), 7.71 (t, 1H, J = 8.0 Hz Ar–H), 7.78 (d, 1H, J = 8.0 Hz Ar–H), 8.01 (t, 1H, J = 8.0 Hz Ar–H), 10.22 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.17, 100.50, 115.69, 116.30, 121.25, 123.98, 124.82, 126.47, 128.60, 132.69, 140.78, 149.83, 158.64, 160.85, 207.07. Anal. calcd. for C16H11F4N3O (337.27): C, 56.98; H, 3.29; N, 12.46; found: C, 56.79; H, 3.55; N, 12.38.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 1.44–1.53 (m, 1H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2, 7.6 Hz Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 117.75, 119.75, 122.71, 129.64, 131.38, 138.79, 151.26, 156.66, 161.03. Anal. calcd. for C13H10N4O (238.24): C, 65.54; H, 4.23; N, 23.52; found: C, 65.42; H, 4.09; N, 23.39.
N); 1H-NMR (500 MHz, DMSO-d6) δ 1.44–1.53 (m, 1H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2, 7.6 Hz Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 117.75, 119.75, 122.71, 129.64, 131.38, 138.79, 151.26, 156.66, 161.03. Anal. calcd. for C13H10N4O (238.24): C, 65.54; H, 4.23; N, 23.52; found: C, 65.42; H, 4.09; N, 23.39.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.26 (s, 6H, CH3), 2.30 (s, 3H, Ar–H), 7.03 (d, 2H, J = 8.0 Hz, Ar–H), 7.48 (t, 1H, J = 8.0 Hz, Ar–H), 7.52–7.55 (m, 1 Hz, Ar–H), 7.58–7.60 (m, 2H, Ar–H), 9.59 (s, 1H, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 14.68, 18.27, 20.83, 112.02, 112.27, 121.78, 122.07, 127.42, 129.88, 131.66, 133.63, 134.43, 157.47, 159.91, 162.33, 206.08. Anal. calcd. for C16H14FN3O (283.31): C, 67.83; H, 4.91; N, 14.83; found: C, 67.71; H, 5.05; N, 14.99.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.26 (s, 6H, CH3), 2.30 (s, 3H, Ar–H), 7.03 (d, 2H, J = 8.0 Hz, Ar–H), 7.48 (t, 1H, J = 8.0 Hz, Ar–H), 7.52–7.55 (m, 1 Hz, Ar–H), 7.58–7.60 (m, 2H, Ar–H), 9.59 (s, 1H, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 14.68, 18.27, 20.83, 112.02, 112.27, 121.78, 122.07, 127.42, 129.88, 131.66, 133.63, 134.43, 157.47, 159.91, 162.33, 206.08. Anal. calcd. for C16H14FN3O (283.31): C, 67.83; H, 4.91; N, 14.83; found: C, 67.71; H, 5.05; N, 14.99.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 4.25 (s, 2H, CH3), 7.50–7.13 (m, 3H, Ar–H), 7.17–7.28 (m, 3H, Ar–H), 8.03 (ddd, 1H, J = 1.2, 8.4 Hz, Ar–H), 10.22 (s, 1H, Ar–H), 8.04 (m, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.16, 103.26, 112.66, 112.91, 115.84, 116.02, 120.92, 123.91, 125.18, 139.42, 150.96, 154.01, 158.81, 160.79, 207.08. Anal. calcd. for C15H10F3N3O (305.25): C, 59.02; H, 3.30; N, 13.77; found: C, 58.89; H, 3.55; N, 13.59.
N); 1H-NMR (500 MHz, DMSO-d6) δ 4.25 (s, 2H, CH3), 7.50–7.13 (m, 3H, Ar–H), 7.17–7.28 (m, 3H, Ar–H), 8.03 (ddd, 1H, J = 1.2, 8.4 Hz, Ar–H), 10.22 (s, 1H, Ar–H), 8.04 (m, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 31.16, 103.26, 112.66, 112.91, 115.84, 116.02, 120.92, 123.91, 125.18, 139.42, 150.96, 154.01, 158.81, 160.79, 207.08. Anal. calcd. for C15H10F3N3O (305.25): C, 59.02; H, 3.30; N, 13.77; found: C, 58.89; H, 3.55; N, 13.59.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 4.29 (s, 2H, CH3), 7.08–7.21 (m, 4H, Ar–H), 7.30–7.36 (m, 1H, Ar–H), 7.98–8.04 (m, 1H, Ar–H), 10.20 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 31.16, 103.26, 103.52, 104.72, 111.61, 112.83, 112.90, 139.49, 158.83, 160.93, 161.69, 164.14, 207.01. Anal. calcd. for C15H9F4N3O (323.25): C, 55.73; H, 2.81; N, 13.00; found: C, 55.56; H, 2.55; N, 13.19.
N); 1H-NMR (500 MHz, DMSO-d6) δ 4.29 (s, 2H, CH3), 7.08–7.21 (m, 4H, Ar–H), 7.30–7.36 (m, 1H, Ar–H), 7.98–8.04 (m, 1H, Ar–H), 10.20 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 31.16, 103.26, 103.52, 104.72, 111.61, 112.83, 112.90, 139.49, 158.83, 160.93, 161.69, 164.14, 207.01. Anal. calcd. for C15H9F4N3O (323.25): C, 55.73; H, 2.81; N, 13.00; found: C, 55.56; H, 2.55; N, 13.19.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.37 (s, 3H, CH3), 7.90–7.17 (m, 1H, Ar–H), 7.23–7.27 (m, 1H, Ar–H), 7.29–7.36 (m, 2H, Ar–H), 7.41–7.45 (m, 1H, Ar–H), 8.06 (dd, 1H, J = 6.8 Hz, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 20.52, 11.98, 112.10, 115.98, 116.17, 117.34, 121.39, 124.37, 125.27, 126.89, 129.17, 134.94, 156.29, 158.80, 160.87. Anal. calcd. for C15H11F2N3O (287.26): C, 62.72; H, 3.86; N, 14.63; found: C, 62.58; H, 3.65; N, 14.79.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.37 (s, 3H, CH3), 7.90–7.17 (m, 1H, Ar–H), 7.23–7.27 (m, 1H, Ar–H), 7.29–7.36 (m, 2H, Ar–H), 7.41–7.45 (m, 1H, Ar–H), 8.06 (dd, 1H, J = 6.8 Hz, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 20.52, 11.98, 112.10, 115.98, 116.17, 117.34, 121.39, 124.37, 125.27, 126.89, 129.17, 134.94, 156.29, 158.80, 160.87. Anal. calcd. for C15H11F2N3O (287.26): C, 62.72; H, 3.86; N, 14.63; found: C, 62.58; H, 3.65; N, 14.79.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 7.10–7.15 (m, 1H, CH3), 7.24 (t, 1H, J = 8.0 Hz, Ar–H), 7.28–7.33 (m, 1H, Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H), 8.08–8.13 (m, 3H, Ar–H), 10.62 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 116.20, 121.43, 124.49, 125.28, 125.32, 126.64, 126.74, 126.89, 127.98, 128.90, 157.77, 161.13. Anal. calcd. for C15H9F4N3O (323.25): C, 55.73; H, 2.88; N, 13.00; found: C, 55.64; H, 2.81; N, 13.13.
N); 1H-NMR (500 MHz, DMSO-d6) δ 7.10–7.15 (m, 1H, CH3), 7.24 (t, 1H, J = 8.0 Hz, Ar–H), 7.28–7.33 (m, 1H, Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H), 8.08–8.13 (m, 3H, Ar–H), 10.62 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 116.20, 121.43, 124.49, 125.28, 125.32, 126.64, 126.74, 126.89, 127.98, 128.90, 157.77, 161.13. Anal. calcd. for C15H9F4N3O (323.25): C, 55.73; H, 2.88; N, 13.00; found: C, 55.64; H, 2.81; N, 13.13.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 7.50 (d, 1H, J = 7.6 Hz), 7.61 (t, 1H, J = 7.6 Hz, Ar–H), 7.83 (d, 2H, J = 6.0 Hz, Ar–H), 7.88 (d, 1H, J = 7.6, 8.4 Hz, Ar–H), 8.05 (t,1H, J = 1.6 Hz), 8.80 (d, 2H, J = 4.4 Hz), 11.33 (s, 1H); 13C-NMR (125 MHz, DMSO-d6) δ 117.42, 119.62, 122.10, 129.55, 131.35, 139.78, 154.35, 156.32, 160.86, 207.03. Anal. calcd. for C14H9N5O (263.25): C, 63.87; H, 3.45; N, 26.60; found: C, 63.77; H, 3.55; N, 26.89.
N); 1H-NMR (500 MHz, DMSO-d6) δ 7.50 (d, 1H, J = 7.6 Hz), 7.61 (t, 1H, J = 7.6 Hz, Ar–H), 7.83 (d, 2H, J = 6.0 Hz, Ar–H), 7.88 (d, 1H, J = 7.6, 8.4 Hz, Ar–H), 8.05 (t,1H, J = 1.6 Hz), 8.80 (d, 2H, J = 4.4 Hz), 11.33 (s, 1H); 13C-NMR (125 MHz, DMSO-d6) δ 117.42, 119.62, 122.10, 129.55, 131.35, 139.78, 154.35, 156.32, 160.86, 207.03. Anal. calcd. for C14H9N5O (263.25): C, 63.87; H, 3.45; N, 26.60; found: C, 63.77; H, 3.55; N, 26.89.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2 Hz 7.6 Hz Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H) 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 18.31, 20.77, 119.54, 121.82, 122.61, 127.48, 130.26, 131.11, 134.16, 151.01, 151.25, 161.97, 164.74. Anal. calcd. for C15H14N4O (266.30): C, 67.65; H, 5.30; N, 21.04; found: C, 67.44; H, 5.13; N, 21.01.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.09 (s, 3H, CH3), 7.51 (d, 1H, J = 8.0 Hz, Ar–H), 7.61 (t, 1H, J = 8.0 Hz, Ar–H), 7.88 (dd, 1H, J = 1.2 Hz 7.6 Hz Ar–H), 7.95 (d, 2H, J = 8.4 Hz, Ar–H) 8.04 (m, 1H, Ar–H) 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 11.27 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 18.31, 20.77, 119.54, 121.82, 122.61, 127.48, 130.26, 131.11, 134.16, 151.01, 151.25, 161.97, 164.74. Anal. calcd. for C15H14N4O (266.30): C, 67.65; H, 5.30; N, 21.04; found: C, 67.44; H, 5.13; N, 21.01.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 7.43 (dd, 2H, J = 4.8 Hz), 7.66 (dd, 2H, J = 2.0, 7.2 Hz, Ar–H), 7.96 (d, 2H, J = 8.4 Hz, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 10.98 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 119.25, 125.69, 126.21, 126.81, 126.86, 127.98, 129.50, 131.25, 137.90, 157.40, 160.57. Anal. calcd. for C15H9BrF3N3O (384.15): C, 46.90; H, 2.36; N, 10.96; found: C, 46.82; H, 2.45; N, 10.89.
N); 1H-NMR (500 MHz, DMSO-d6) δ 7.43 (dd, 2H, J = 4.8 Hz), 7.66 (dd, 2H, J = 2.0, 7.2 Hz, Ar–H), 7.96 (d, 2H, J = 8.4 Hz, Ar–H), 8.10 (d, 2H, J = 8.0 Hz, Ar–H), 10.98 (s, 1H, Ar-NH); 13C-NMR (125 MHz, DMSO-d6) δ 119.25, 125.69, 126.21, 126.81, 126.86, 127.98, 129.50, 131.25, 137.90, 157.40, 160.57. Anal. calcd. for C15H9BrF3N3O (384.15): C, 46.90; H, 2.36; N, 10.96; found: C, 46.82; H, 2.45; N, 10.89.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 1.19 (d, 6H, J = 6.4 Hz, CH3), 2.83–2.88 (m, 1H, Ar–H), 7.24 (d, 2H, J = 8.4 Hz, Ar–H), 7.53 (dd, 2H, J = 1.6, 8.0 Hz, Ar–H), 7.94 (d, 2H, J = 8.4 Hz, Ar–H) 8.08 (d, 2H, J = 8.4 Hz, Ar–H),10.69 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 24.47, 33.26, 117.81, 123.04, 125.74, 126.68, 126.85, 127.35, 128.03, 136.66, 142.72, 157.13, 160.95. Anal. calcd. for C18H16F3N3O (347.33): C, 62.24; H, 4.64; N, 12.10; found: C, 62.02; H, 4.42; N, 12.01.
N); 1H-NMR (500 MHz, DMSO-d6) δ 1.19 (d, 6H, J = 6.4 Hz, CH3), 2.83–2.88 (m, 1H, Ar–H), 7.24 (d, 2H, J = 8.4 Hz, Ar–H), 7.53 (dd, 2H, J = 1.6, 8.0 Hz, Ar–H), 7.94 (d, 2H, J = 8.4 Hz, Ar–H) 8.08 (d, 2H, J = 8.4 Hz, Ar–H),10.69 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 24.47, 33.26, 117.81, 123.04, 125.74, 126.68, 126.85, 127.35, 128.03, 136.66, 142.72, 157.13, 160.95. Anal. calcd. for C18H16F3N3O (347.33): C, 62.24; H, 4.64; N, 12.10; found: C, 62.02; H, 4.42; N, 12.01.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.31 (s, 3H, CH3), 7.05 (t, 1H, J = 5.6 Hz, Ar–H), 7.22–7.24 (m, 2H, Ar–H), 7.56–7.58 (m, 3H, Ar–H), 7.77 (d, 1H, J = 8.4 Hz, Ar–H), 7.87–7.89 (m, 2H, Ar–H), 9.65 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 18.62, 123.42, 123.84, 126.10, 126.58, 127.54, 128.71, 129.04, 129.25, 131.33, 142.65, 164.36, 169.32. Anal. calcd. for C15H13N3O (251.28): C, 71.70; H, 5.21; N, 16.72; found: C, 71.51; H, 5.09; N, 16.89.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.31 (s, 3H, CH3), 7.05 (t, 1H, J = 5.6 Hz, Ar–H), 7.22–7.24 (m, 2H, Ar–H), 7.56–7.58 (m, 3H, Ar–H), 7.77 (d, 1H, J = 8.4 Hz, Ar–H), 7.87–7.89 (m, 2H, Ar–H), 9.65 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 18.62, 123.42, 123.84, 126.10, 126.58, 127.54, 128.71, 129.04, 129.25, 131.33, 142.65, 164.36, 169.32. Anal. calcd. for C15H13N3O (251.28): C, 71.70; H, 5.21; N, 16.72; found: C, 71.51; H, 5.09; N, 16.89.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H-NMR (500 MHz, DMSO-d6) δ 2.41 (s, 3H, CH3), 7.07–7.23 (m, 1H, Ar–H), 7.23–7.29 (m, 2H, Ar–H), 7.46–7.48 (m, 3H, Ar–H), 7.87 (d, 1H, J = 4.8 Hz, Ar–H) 7.95 (d, 2H, J = 5.7 Hz, Ar–H), 10.42 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 12.85, 115.42, 117.18, 123.00, 123.08, 124.84, 129.91, 138.46, 138.92, 160.00, 164.55, 169.36. Anal. calcd. for C15H12FN3O (269.27): C, 66.91; H, 4.49; N, 15.60; found: C, 66.78; H, 4.55; N, 15.47.
N); 1H-NMR (500 MHz, DMSO-d6) δ 2.41 (s, 3H, CH3), 7.07–7.23 (m, 1H, Ar–H), 7.23–7.29 (m, 2H, Ar–H), 7.46–7.48 (m, 3H, Ar–H), 7.87 (d, 1H, J = 4.8 Hz, Ar–H) 7.95 (d, 2H, J = 5.7 Hz, Ar–H), 10.42 (s, 1H, Ar–H); 13C-NMR (125 MHz, DMSO-d6) δ 12.85, 115.42, 117.18, 123.00, 123.08, 124.84, 129.91, 138.46, 138.92, 160.00, 164.55, 169.36. Anal. calcd. for C15H12FN3O (269.27): C, 66.91; H, 4.49; N, 15.60; found: C, 66.78; H, 4.55; N, 15.47.4.3 Monoamine oxidase A and B inhibition assay
Newly synthesized compounds were tested on monoamine oxidase A and B (extracted). Freshly prepared enzyme extracts were used in fluorescence-based assays via estimating the change in fluorescence intensity in white colored 96 well plate with an excitation at 544 nm and an emission at 590 nm. To determine the enzyme activity of MAO and MAO-B. Clorgyline 6 mM and Deprenyl 10 mM were employed as standard inhibitors of MAO-A and MAO-B, respectively. Reaction mixture was composed of 100 μL of total assay volume having 30 μL buffer (pH 7.4), 10 μL test compounds, 10 μL of extracted enzyme (MAO-A: 13 μg mL−1, MAO-B: 15 μg mL−1), 30 μL of amplex red and 20 μL of substrate (p-tyramine). Initially reaction mixture having buffer + test compound + enzyme was incubated for 15 minutes for MAO-A And MAO-B. After Incubation the amplex red reagent was added followed by addition of substrate p-tyramine. The change in the fluorescence was determined by using fluorescence plate reader: FLUOstar® Omega (BMG Labtech GmbH, orten berg Germany). Those compounds exhibited greater than 50% inhibition of either the MAO-A or MAO-B activity, were further subjected for the estimation of IC50 values. All experiments were performed in triplicate. IC50 values were calculated by non-linear curve fitting program PRISM 5.0 (GraphPad, San Diego, California, USA) where log inhibitor vs. response curve was generated. The details of the enzyme inhibition assay for AChE and BChE are inserted in the ESI.†4.4 Docking studies protocol
Molecular docking studies were performed by using FlexX utility of BioSolveIT's LeadIT. The purpose was to analyze the binding behavior of identified inhibitors of MAO-A/MAO-B, AChE/BChE at the active site of enzyme. Crystal structures of MAO-A (2Z5X), MAO-B (2V5Z) and AChE (4BDT) were downloaded from protein data bank (http://www.rcsb.org).227–29 For the revalidation of docking protocol, initially the co-crystalized ligand was redocked at active site and its RMSD value was compared. The scoring and ranking of conformational poses were carried out via adopting enthalpy entropy hybrid approach of the FlexX utility. The highest scoring poses were further subjected to HYDE assessment in order to assess their binding affinities.30,31Conflicts of interest
The authors declare that they have no significant conflict of interest.Acknowledgements
Z. S. is thankful to the Higher Education Commission of Pakistan (HEC) via NRPU project no. 6975. The authors gratefully acknowledge the financial support for this research provided by the Higher Education Commission of Pakistan (HEC) via NRPU project no 20-15846/NRPU/R&D/HEC/2021, German-Pakistani Research Collaboration Programme and Equipment Grant funded by DAAD, Germany.References
- V. L. Feigin and T. Vos, Neuroepidemiology, 2019, 52, 1–2 Search PubMed.
- F. Rahim, M. T. Javed, H. Ullah, A. Wadood, M. Taha, M. Ashraf, M. A. Khan, F. Khan, S. Mirza and K. M. Khan, Bioorg. Chem., 2015, 62, 106–116 CrossRef CAS PubMed.
- X. Du, X. Wang and M. Geng, Transl. Neurodegener., 2018, 7, 2 CrossRef PubMed.
- M. P. Murphy and H. LeVine, 3rd, J. Alzheimer's Dis., 2010, 19, 311–323 Search PubMed.
- P. B. Watkins, H. J. Zimmerman, M. J. Knapp, S. I. Gracon and K. W. Lewis, J. Am. Med. Assoc., 1994, 271, 992–998 CrossRef CAS PubMed.
- A. W. K. Yeung, M. G. Georgieva, A. G. Atanasov and N. T. Tzvetkov, Front. Mol. Neurosci., 2019, 143 CrossRef CAS PubMed.
- R. R. Somani and P. Y. Shirodkar, ChemInform, 2011, 42 DOI:10.1002/chin.201110251.
- T. Behl, D. Kaur, A. Sehgal, S. Singh, N. Sharma, G. Zengin, F. L. Andronie-Cioara, M. M. Toma, S. Bungau and A. G. Bumbu, Molecules, 2021, 26, 3724 CrossRef CAS PubMed.
- S. Distinto, R. Meleddu, M. Yanez, R. Cirilli, G. Bianco, M. L. Sanna, A. Arridu, P. Cossu, F. Cottiglia, C. Faggi, F. Ortuso, S. Alcaro and E. Maccioni, Eur. J. Med. Chem., 2016, 108, 542–552 CrossRef CAS PubMed.
- T. Thomas, Neurobiol. Aging, 2000, 21, 343–348 CrossRef CAS PubMed.
- G. A. Reid, N. Chilukuri and S. Darvesh, Neuroscience, 2013, 234, 53–68 CrossRef CAS PubMed.
- A. Nordberg, C. Ballard, R. Bullock, T. Darreh-Shori and M. Somogyi, The primary care companion for CNS disorders, 2013, vol. 15, p. 26731 Search PubMed.
- M. Pohanka, Int. J. Mol. Sci., 2014, 15, 9809–9825 CrossRef PubMed.
- M. Bortolato, K. Chen and J. C. Shih, Adv. Drug Delivery Rev., 2008, 60, 1527–1533 CrossRef CAS PubMed.
- S. E. Lakhan, Mol. Neurodegener., 2007, 2, 13 CrossRef PubMed.
- N. Desai, J. Monapara, A. Jethawa, V. Khedkar and B. Shingate, Arch. Pharm., 2022, 355, e2200123 CrossRef PubMed.
- A. Vaidya, S. Jain, P. Jain, P. Jain, N. Tiwari, R. Jain, R. Jain, A. K. Jain and R. K. Agrawal, Mini-Rev. Med. Chem., 2016, 16, 825–845 CrossRef CAS PubMed.
- D. Szulczyk, P. Tomaszewski, M. Jozwiak, A. E. Koziol, T. Lis, D. Collu, F. Iuliano and M. Struga, Molecules, 2017, 22, 409 CrossRef PubMed.
- S. B. Kokkiligadda, S. Musunuri, B. Maiti, M. Rao and G. Sridhar, Russ. J. Gen. Chem., 2020, 90, 1331–1335 CrossRef CAS.
- J. I. Gavrilyuk, A. J. Lough and R. A. Batey, Tetrahedron Lett., 2008, 49, 4746–4749 CrossRef CAS.
- J. Janardhanan, M. Chang and S. Mobashery, Curr. Opin. Microbiol., 2016, 33, 13–17 CrossRef CAS PubMed.
- H. Uslu, B. N. SaĞlik, D. Osmanİye and K. Benklİ, J. Res. Pharm., 2022, 26(1), 020–027 CAS.
- F. Tok, Z. Uğraş, B. N. Sağlık, Y. Özkay, Z. A. Kaplancıklı and B. Koçyiğit-Kaymakçıoğlu, Bioorg. Chem., 2021, 112, 104917 CrossRef CAS PubMed.
- P. Mishra, P. Sharma, P. N. Tripathi, S. K. Gupta, P. Srivastava, A. Seth, A. Tripathi, S. Krishnamurthy and S. K. Shrivastava, Bioorg. Chem., 2019, 89, 103025 CrossRef CAS PubMed.
- A. Shetnev, S. Baykov, S. Kalinin, A. Belova, V. Sharoyko, A. Rozhkov, L. Zelenkov, M. Tarasenko, E. Sadykov, M. Korsakov and M. Krasavin, Int. J. Mol. Sci., 2019, 20, 1699 CrossRef CAS PubMed.
- A. Cansiz, M. Koparir and A. Demirdag, Molecules, 2004, 9, 204–212 CrossRef CAS PubMed.
- S.-Y. Son, J. Ma, Y. Kondou, M. Yoshimura, E. Yamashita and T. Tsukihara, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 5739–5744 CrossRef CAS PubMed.
- C. Binda, J. Wang, L. Pisani, C. Caccia, A. Carotti, P. Salvati, D. E. Edmondson and A. Mattevi, J. Med. Chem., 2007, 50, 5848–5852 CrossRef CAS PubMed.
- F. Nachon, E. Carletti, C. Ronco, M. Trovaslet, Y. Nicolet, L. Jean and P.-Y. Renard, Biochem. J., 2013, 453, 393–399 CrossRef CAS PubMed.
- I. Reulecke, G. Lange, J. Albrecht, R. Klein and M. Rarey, ChemMedChem, 2008, 3, 885–897 CrossRef CAS PubMed.
- N. Schneider, G. Lange, S. Hindle, R. Klein and M. A. Rarey, J. Comput. Aided Mol. Des., 2013, 27, 15–29 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2251452. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3ra01953e |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |