Open Access Article
Open Access ArticleNanoengineering of gold nanoribbon-embedded isogenic stem cell-derived cardiac organoids†
Alejandra Patino-Guerrero a,
Hamid Esmaeili
a,
Hamid Esmaeili a,
Raymond Q. Migrino
a,
Raymond Q. Migrino bc and
Mehdi Nikkhah
bc and
Mehdi Nikkhah *ad
*ad
aSchool of Biological and Health Systems Engineering, Arizona State University, Tempe, AZ 8528, USA. E-mail: mnikkhah@asu.edu
bPhoenix Veterans Affairs Health Care System, Phoenix, AZ 85012, USA
cUniversity of Arizona College of Medicine, Phoenix, AZ 85004, USA
dCenter for Personalized Diagnostics Biodesign Institute, Arizona State University, Tempe, AZ 85281, USA
First published on 6th June 2023
Abstract
Cardiac tissue engineering is an emerging field providing tools to treat and study cardiovascular diseases (CVDs). In the past years, the integration of stem cell technologies with micro- and nanoengineering techniques has enabled the creation of novel engineered cardiac tissues (ECTs) with potential applications in disease modeling, drug screening, and regenerative medicine. However, a major unaddressed limitation of stem cell-derived ECTs is their immature state, resembling a neonatal phenotype and genotype. The modulation of the cellular microenvironment within the ECTs has been proposed as an efficient mechanism to promote cellular maturation and improve features such as cellular coupling and synchronization. The integration of biological and nanoscale cues in the ECTs could serve as a tool for the modification and control of the engineered tissue microenvironment. Here we present a proof-of-concept study for the integration of biofunctionalized gold nanoribbons (AuNRs) with hiPSC-derived isogenic cardiac organoids to enhance tissue function and maturation. We first present extensive characterization of the synthesized AuNRs, their PEGylation and cytotoxicity evaluation. We then evaluated the functional contractility and transcriptomic profile of cardiac organoids fabricated with hiPSC-derived cardiomyocytes (mono-culture) as well as with hiPSC-derived cardiomyocytes and cardiac fibroblasts (co-culture). We demonstrated that PEGylated AuNRs are biocompatible and do not induce cell death in hiPSC-derived cardiac cells and organoids. We also found an improved transcriptomic profile of the co-cultured organoids indicating maturation of the hiPSC-derived cardiomyocytes in the presence of cardiac fibroblasts. Overall, we present for the first time the integration of AuNRs into cardiac organoids, showing promising results for improved tissue function.
1. Introduction
Cardiovascular diseases (CVDs) are the primary cause of death around the world.1 Moreover, the risk factors associated with CVDs are expected to increase in the near future.2 Studying the mechanisms underlying CVDs is limited by the reduced availability of primary human cardiac cells. Therefore preclinical cardiac research has mainly relied on animal models.3 However, with the increased utilization of stem cell technologies (i.e., human induced pluripotent stem cells (hiPSCs)), it is now easy to source human cardiac cells for advanced in vitro studies.4 Protocols for the directed differentiation of human cardiomyocytes (CMs), cardiac fibroblasts (CFs), and other cell types have been developed and refined to produce these cells within a short period of time in the order of a few weeks.5,6 While these advancements have provided access to an unlimited number of cardiac cells, one remaining main disadvantage is that these stem cell-derived cardiac cells are usually in an immature state and phenotype.7The refinement of micro and nanoengineering technologies has allowed for the manufacturing of highly reproducible engineered tissues. This, in combination with stem cell technologies, has resulted in powerful protocols for the fabrication of complex and biomimetic human cardiac tissues in vitro.3,8,9 Different cardiac tissues have been engineered for specific applications such as disease modeling,10,11 drug screening,12 or myocardial regeneration.13 Regardless of a specific application, the following general principles still apply: (a) it is desirable that the hiPSC-derived cardiac cells reach an adult-like phenotype and genotype in order to better represent the adult human myocardium;7,14 (b) the design and control of microenvironmental cues is one of the key factors promoting cellular maturation and enhancing physiologic performance of the engineered tissues;15,16 and (c) the manipulation of the microenvironment within the engineered tissues can be achieved through physical, chemical, and biological cues, or more often, by a combination of two or more of these methods.15–17
The introduction of advanced biomaterials with unique properties has been proposed as a practical approach for the fabrication of engineered cardiac tissues (ECTs) within a controlled microenvironment.8,18,19 Specifically, nanoengineered biomaterials are of particular interest due to their unique physical and chemical properties.20 Their size allows for intimate interaction with cellular structures and the intercellular microenvironment. Recently, high aspect ratio nanoparticles, also known as one-dimensional (1D) nanoparticles, have been of special interest for tissue engineering applications due to their morphological features. On one hand, they may present attractive nanoscale features, such as electrical conductivity and surface plasmon resonance.21 Additionally, their larger size, compared to nanoparticles with smaller aspect ratios, allow them to remain and be integrated within the intercellular space while continuing to modify the cellular microenvironment.22 For example, it has been demonstrated that high aspect ratio silicon nanowires can improve the maturation of stem cell-derived cardiac spheroids.23–25 Moreover, filamentous gold nanoparticles (i.e. gold nanowires) have been recently integrated with scaffold-based cardiac patches to improve cell survival and engraftment.26,27 Thus, 1D nanoparticles present attractive features for the construction of ECTs. Specifically, the investigation of the effects of 1D gold nanoparticles, such as gold nanoribbons (AuNRs), in scaffold-free cardiac tissues is of special interest for the field as it has not been reported in the past, to the best of our knowledge.
One of the most relevant functions of the cardiac tissue relates to its electrical activity and the ability of cardiac cells to conduct electrical signals.28 This signal propagation is possible due to the formation of a syncytium that transmits electrical and mechanical pulses.29 Often, ECTs do not closely mimic, if at all, the electrophysiological features of the native myocardium.30 In this regard, electrically conductive nanoparticles could be of the interest for cardiac tissue engineering as it is believed that they can enhance the maturation and functionality of hiPSC-derived ECTs by creating a more electrically conductive extracellular microenvironment.8 Since ECTs fabricated with hiPSC-derived cardiac cells present an immature phenotype with poor cell–cell coupling and inefficient electrical activity, the incorporation of electrically conductive nanoparticles could also represent a promising approach to modulate the microenvironment of ECT and improve the physiologic performance of cardiac tissues.9 For example, it has been reported that the integration of Si nanowires into cardiac spheroids improved their gene expression profile and cell-to-cell connection.24,25 The integration of graphene and graphene oxide has also been found to upregulate the expression of cardiac-relevant genes.31,32 A more detailed description of those studies can be found elsewhere.8,9
The incorporation of different cardiac cell types has also been widely proposed as another key factor affecting the ECTs microenvironment.33–35 It was been demonstrated that CFs play an important role in the secretion of growth factors and cytokines that regulate the activity of CMs.36,37 Moreover, CFs are the main producers of extracellular matrix (ECM) proteins in the cardiac tissue.38 To that end, isogenic cardiac organoids are of special interest for tissue engineering due to the lack of exogenic biomaterials, recapitulation of cardiac tissue composition, and feasible application in disease modeling and regeneration through minimally invasive route.9 The benefits of co-culture of CMs and CFs for the formation of cardiac organoids have been widely studied and reported, and in our recent work, the CMs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFs ratio was optimized for the creation of isogenic cardiac organoids.33 Thus, the integration of hiPSC-CFs in ECTs serves as another relevant tool for the regulation of the microenvironment and the fabrication of cardiac organoids.
CFs ratio was optimized for the creation of isogenic cardiac organoids.33 Thus, the integration of hiPSC-CFs in ECTs serves as another relevant tool for the regulation of the microenvironment and the fabrication of cardiac organoids.
In this manuscript we present a proof-of-concept study for the integration of gold nanoribbons (AuNRs) with scaffold-free isogenic hiPSC-derived cardiac organoids. We hypothesize that the inclusion of 1D gold nanoparticles, namely AuNRs, in combination with the co-culture of isogenic hiPSC-derived cardiac cells (CMs and CFs), can lead to the improved maturation of the cells and an overall enhanced physiologic performance of the cardiac organoids. First, we adapted a seed-mediated growth synthesis method for the fabrication and characterization of the AuNRs with desired length and geometry. We further functionalized the surface of the AuNRs with polyethylene glycol (PEG) to render them biocompatible and suitable for cardiac tissue engineering applications. We differentiated CMs and CFs from a hiPSC line to generate isogenic lines of cells for tissue formation. By seeding these cells (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 CM
3 CM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CF ratio) along with AuNRs (10 μg mL−1) in agarose microwells, we induced cellular aggregation and the formation of scaffold-free cardiac organoids. We analyzed the effect of the AuNRs in the formation of the cardiac organoids by examining the integrity and viability of the cardiac organoids and evaluating electrophysiological features as well as the resulting transcriptomic profile.
CF ratio) along with AuNRs (10 μg mL−1) in agarose microwells, we induced cellular aggregation and the formation of scaffold-free cardiac organoids. We analyzed the effect of the AuNRs in the formation of the cardiac organoids by examining the integrity and viability of the cardiac organoids and evaluating electrophysiological features as well as the resulting transcriptomic profile.
2. Experimental methods
2.1 Materials
All the materials, as wells as their working concentrations, brands and identifier numbers are provided in the supplementary “Table S1† of resources” of this chapter.2.2 Synthesis of gold nanoribbons (AuNRs)
The protocol for the synthesis of AuNRs was adapted from Xu et al.39 This method is based in the seed-mediated aggregation of gold atoms, guided by a bisurfactant system. First, the gold seed solution was prepared as follows. In a 15 mL conical tube, 7.5 mL of 0.1 M of cetrimonium bromide (CTAB, pH: 5.0) solution were mixed with 250 μL of HAuCl4 (0.01 M, pH: 1.0). Then, 600 μL of ice-cold NaBH4 (0.01 M, pH: 8.0) was added to the seed solution and vigorously vortexed for two minutes. This solution was then incubated at 35 °C for 1 hour, resulting in a reddish-maroon solution. After the incubation period, the solution was diluted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio.
10 ratio.
The growth solution was prepared in a 50 mL conical tube. First 230 mg of CTAB and 48.5 mg of sodium oleate (NaOL) were dissolved in 38.4 mL of deionized water (DIW). Then, 250 μL of HAuCl4 (0.01 M) was added to this solution which immediately turned the growth solution color to bright yellow (pH: 6.0). This solution was incubated at 35 °C for 15 minutes or until it turned colorless. Finally, 3.8 mL of an ascorbic acid solution (0.1 M, pH: 1.0) were added to the growth solution to reduce all ionic gold to metallic gold (pH: 4.0). 20 μL of the diluted seed solution were added to the final growth solution and vortexed for 30 s. The solution was incubated undisturbed at 35 °C for 8 h. The synthesized AuNRs solution had a characteristic pink to maroon color after the incubation period.
2.3 Purification and characterization of the AuNRs
In order to purify and obtain a highly concentrated AuNRs solution, the synthesized AuNRs solution was washed three times. For this, the samples were centrifuged at 3000×g for 17 minutes to precipitate the AuNRs. The centrifugation settings were optimized through a series of speed and time swipes, and selected according to the combination that led to the highest optical density (O.D.), measured at 450 nm (data not shown). The supernatant was removed and discarded, and the AuNRs were subsequently resuspended in DIW. Gold nanoparticles present optical properties, namely surface plasmon resonance (SPR) phenomenon, that allows for their characterization using UV-visible spectrophotometry (UV-vis). A sample of the concentrated AuNRs solution was measured through UV-vis to corroborate that the synthesis was successful.To evaluate the morphology of the AuNRs the samples were mounted in a copper mesh grid (Ted Pella). The samples were visualized through transmission electronic microscope (TEM) (Phillips JEOL 2010). To determine the average size of the ribbon-like nanoparticles a total of 100 particles above 0.6 μM length were measured using FIJI ImageJ. The average size was calculated as the arithmetic mean of these particles.
A standard concentration curve was generated to measure the concentration of the AuNRs. For this, serial dilutions of the AuNRs were prepared and the concentration of each of the dilutions was measured using inductively coupled plasma mass spectrometry (ICP-MS). Each of the measured concentrations was correlated to the O.D. at 450 nm of the corresponding AuNRs dilution. Finally, the linear regression of the obtained curve was calculated. Subsequently, the concentration of AuNRs was obtained by inserting the O.D. in the equation generated from the linear regression.
2.4 PEGylation of AuNRs
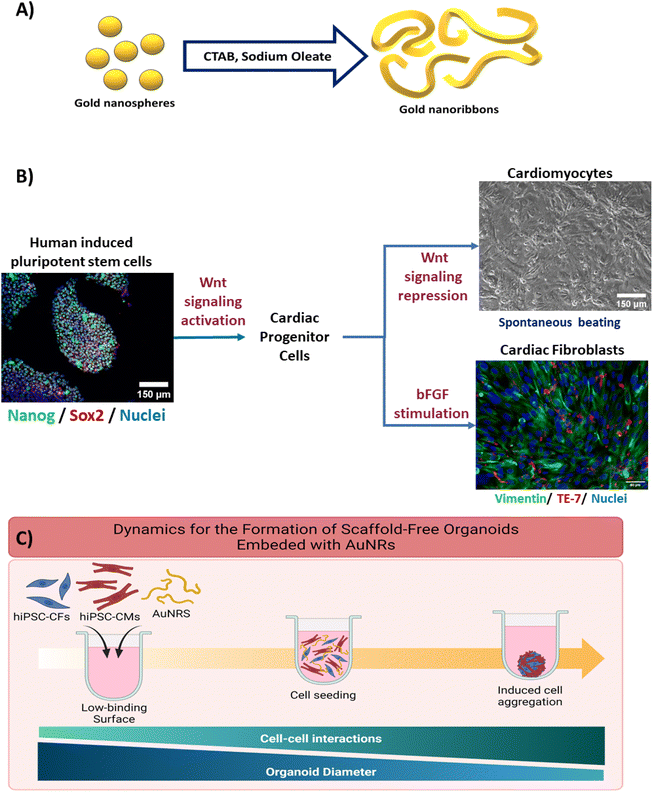
A surface molecule exchange was performed to substitute the residual CTAB and NaOL on the surface of the AuNRs with polyethylene glycol (PEG) (Fig. 1A). This PEGylation reaction was adapted from Zhang et al.40 For this purpose, the concentrated AuNRs solution was centrifuged at 3000×g for 17 minutes. The supernatant was removed and replaced with 400 μL of Tris buffer (pH: 3.0). Then, 30 μL of a solution of 2 mM PEG in Tris buffer was added to the AuNRs solution and vortexed vigorously for 2 minutes. The resultant solution was incubated undisturbed at room temperature for 24 hours. After this incubation period, the AuNRs solution was centrifuged again, and the supernatant was replaced with 400 μL of 20% ethanol (VWR). Then 30 μL of PEG (2 mM) in 20% ethanol was added to the solution and vortexed vigorously. The solution was incubated undisturbed at room temperature for 24 hours. Then, the samples were washed twice with sterilize 1× DPBS (Dulbecco's phosphate buffered solution). Finally, the AuNRs were resuspended in sterile 1× DPBS for concentration measurement analysis and stored at room temperature until their use.2.5 Human induced pluripotent stem cells maintenance
The hiPSCs (IMR90-4, WiCell) were maintained in mTeSR-1 media (StemCell Technologies) with daily changes. The cells were seeded on hESC-matrigel (corning)-coated plates to help to maintain the pluripotency state of the cells. The hiPSC colonies were allowed to reach ∼85% confluency before replating. When the cells reached confluency, they were incubated in a 0.5 mM EDTA (corning) in 1× DPBS solution for 6–7 minutes and mechanical dissociation by gently pipetting with 1 mL of mTeSR-1 media was performed to remove the cells from the plates. The cells were replated at a seeding density of 150–250 K cells per well, according to manual cell counting using hemocytometer. To ensure the maintenance of the hiPSCs, they were stained for the stemness factors Nanog and Sox2 (Fig. 1B).2.6 Directed differentiation of hiPSC-derived cardiomyocytes
The hiPSC-CMs differentiation protocol was adapted from the GiWi protocol,5 which is based on the regulation of the Wnt canonical pathway. Briefly, when the hiPSCs reached 80–95% confluency, the maintenance media was substituted with of RPMI 1640 supplemented with 2% B27 minus insulin (gibco), 1% pen/strep (gibco) (RPMI−) and 7–9 μM of CHIR99021 (BioVision) in order to induce the activation of the Wnt pathway in the cells (designated as day 0). After 24 hours, the media was substituted for RPMI− media only. Then, 72 hours after day 0, the media was substituted by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mix of conditioned media and fresh RPMI− supplemented with 5 μM IWP2 (sigma) in order to inhibit the Wnt signaling within the cells. Culture media was refreshed 48 hours after this. Finally, the hiPSC-CM were maintained in RPMI 1640 supplemented with 2% B27 plus insulin (gibco) and 1% pen/strep (gibco) (RPMI+) until day 13 after day 0, the media was refreshed every 48 hours. The cells typically presented spontaneous beating around day 10.
1 mix of conditioned media and fresh RPMI− supplemented with 5 μM IWP2 (sigma) in order to inhibit the Wnt signaling within the cells. Culture media was refreshed 48 hours after this. Finally, the hiPSC-CM were maintained in RPMI 1640 supplemented with 2% B27 plus insulin (gibco) and 1% pen/strep (gibco) (RPMI+) until day 13 after day 0, the media was refreshed every 48 hours. The cells typically presented spontaneous beating around day 10.
In order to increase the purity of the obtained hiPSC-CMs, the differentiated cells were submitted to a metabolic selection through glucose starvation period.41 As previously reported by our group, this metabolic selection consistently leads to highly purify populations of CMs (>90%hiPSC-CMs).42 On day 13 of differentiation, the cell culture media was exchanged to RPMI 1640 minus glucose supplemented with 2% B27 plus insulin (gibco), 1% pen/strep (gibco), and 4 mM sodium lactate (sigma). Media was refreshed every 72 hours up to day 19. On day 19, the media was substitute with RPMI+ to allow for the recovery of the cells. To eliminate the debris generated from the purification of the hiPSC-CMs, the cells were dissociated on day 21 using TrypLE Express (LifeTech) for 10–12 minutes, followed by mechanical dissociation. The hiPSC-CMs were replated on matrigel with a cell seeding density of 1–1.5 million cells per well (Fig. 1B). Cells were maintained with RPMI+ until their utilization for the experiments, and the media was refreshed every 48 hours.
2.7 Directed differentiation of hiPSC-derived cardiac fibroblasts
The differentiation of isogenic human cardiac fibroblast (CF) was adapted from Fan et al.43 It is based on the modulation of Wnt canonical pathway to produce cardiac progenitor cells, followed by the introduction of fibroblast growth factor (FGF). Briefly, the CFs differentiation was initiated when the hiPSCs reached around 80% confluency. On day 0, the hiPSCs basal media was changed to RPMI 1640 which was supplemented with 7 μM CHIR99021 (BioVision). After 24 hours (day 1), the media was replaced by RPMI−. Upon day 1, the hiPSC-CFs differentiation was carried out with a basal media, with changes every 48 hours. The basal media consisted of high-glucose (4.5 g L−1) DMEM with L-glutamine (4 mM) (gibco), supplemented with HLL supplement (human serum albumin (500 μg mL−1)), linoleic acid (0.6 μM), lecithin (0.6 μg mL−1), and β-fibroblast growth factor (β-FGF) (70 ng μL−1)); this stage was carried for 20 days. Then, the hiPSC-CFs were dissociated with 1× trypsin and replated in uncoated plates to remove cellular debris. After this, the cells were maintained with fibroblast growth media 3 (FGM-3), with media changes every 48 hours. The CFs were replated when reached ∼85% confluency. The hiPSC-CFs used for all of the experiments were between passage number P6 and P12 to avoid further differentiation into myofibroblasts.2.8 Fabrication of isogenic scaffold-free cardiac organoids embedded with AuNRs
Commercially available silicone molds (Microtissues, #35-24) were used to cast agarose microwells (800 μM diameter, 800 μM depth). Briefly, ∼350 μL of a 2% agarose in saline solution were deposited in the silicone molds. After the agarose was solidified, it was removed from the molds. The agarose microwells were conditioned with RPMI+ media prior their utilization for organoid fabrication.The hiPSC-CMs were incubated at 37 °C for 10–12 minutes with Tryple Express (LifeTech), followed by mechanical dissociation. The Tryple was neutralized with RPMI+. Additionally, the hiPSC-CFs were incubated with 1× trypsin (gibco) for 3–4 minutes at 37 °C followed by neutralization with FGM3. The cells were manually counted to determine the concentration and were centrifuged (300×g for 3 minutes) to remove the supernatant. The cells were resuspended in RPMI+ to a final concentration of 175 K cells per 75 μL (Fig. 1C). For the cardiac organoids integrated with the AuNRs, the cell suspension was mixed with PEG-AuNRs to a final concentration of 10 μg mL−1, prior their seeding in the agarose microwells.
The cells were seeded in the agarose microwells and were incubated undisturbed for 1 hour at 37 °C to allow the cells to settle in the microwells. The used cell seeding density led to cardiac organoids with ∼5000 cells per organoid. This density has been thoroughly optimized by our group,33 and is also based on other works found in the literature.34 Then, the culture wells were filled with RPMI+ and the media was changed every 48 hours until the end of the experiment.
2.9 Imaging and video signal acquisition and analysis
Phase-contrast images and videos were acquired with a Zeiss Axio Observer Z1 microscope. The Apotome2 complement was used for all fluorescent images. ZenPro and ImageJ (FIJI) software were used for image and video processing. A custom-made MATLAB code was used for extracting the spontaneous beating signals from the cardiac organoids. The beating rate of the cardiac organoids was calculated by detecting the peaks presented in the acquired signals. The inter-beat interval variability (IIV) was calculated as the standard deviation of the variation in time between the presented peaks.2.10 Viability assay
The viability assay was performed with the viability/cytotoxicity assay kit for animal live & dead cells (biotium), according to the instructions of the kit. Briefly, CFs were seeded on 6-well plates and were allowed to reach ∼80% confluency. Then, the cell culture media was replaced by media supplemented with AuNBs (PEGylated and non-PEGylated) at a concentration of 20 μg mL−1. The CFs were incubated for 96 hours without media changes. At the end of the 96 hours a solution of 2 μM calcein AM and 4 μM EthD-III in DPBS was prepared. The cells were washed twice with 1× DPBS followed by a 30 minutes incubation with the viability assay solution at 37 °C. Finally, the solution was replaced by 1× DPBS before imaging. For the quantification of the hiPSC-CFs viability, three fields of view per experimental condition (i.e. NoPEG-AuNRs and PEG-AuNRs) were evaluated, and two biological replicates of the experiment were performed.Additionally, on day 7 after seeding the cardiac cells in the agarose microwells, the viability assay solution was prepared, and the organoids were incubated for 30 minutes at 37 °C. The cardiac organoids were harvested from the agarose microwells and the solution was replaced by 1× DPBS before imaging.
2.11 TUNEL assay
In order to complete TUNEL assay within the cardiac organoids, cryosectioning of the tissues was performed prior the staining. The cryosectioning was performed on cardiac organoids seven days after seeding on the agarose microwells. First, the tissues were harvested and washed twice with 1× DPBS. Then a solution of 4% paraformaldehyde (PFA) was used to fix the organoids, which were incubated in the solution for 45 minutes at 37 °C. To prevent image artifacts from freezing the tissues, a cryoprotection stage was performed. This consisted of a 15% sucrose (VWR) incubation at room temperature for 1 hour, followed by a 30% sucrose incubation at 4 °C overnight. After this, the cardiac organoids were flash-frozen using OCT (Tissue-Tek). The tissues were sectioned at 10 μM thickness with the CryoStar NX70 cryostat (ThermoScientific) and preserved at −20 °C until their utilization.The clik-iT Plus TUNEL assay (invitrogen) was used for evaluating early apoptosis inside the cardiac organoids. The staining was performed according to the instructions of the kit without any modifications.
2.12 qRT-PCR
The qRT-PCR reaction was performed similar to our previous publication.33 Briefly, the organoids were collected from the agarose microwells seven days after seeding and washed with 1× DPBS twice to remove cellular debris. Then the tissues were digested and RNA was extracted utilizing the RNA microprep kit (Zymo). NanoDrop UV-vis spectrophotometer was utilized to evaluate the RNA quality and calculate RNA concentration. iScript reverse transcription supermix (BioRad) was used to performed the cDNA conversion with a normalized amount of RNA from each sample.Validated primers were used for the qRT-PCR reaction and 18 s was used as housekeeping gene for data normalization (Table S2†). The PCR master mix reaction consisted of 5 μL of iTaq Universal SYBR Green Supermix (BioRad), 1 μL of 8 μM forward and reverse primers in NF water, 3.9 μL of NF water, 0.1 μL of the cDNA sample. The reaction was carried on using the q-Tower real time thermocycler (Analitik Jena). dCT values were normalized to their corresponding day 0 values.
2.13 Statistical analysis
All the statistical analyses were performed using the GraphPad Prism software. T-test or one-way ANOVA test, with post hoc pairwise analysis using Tukey test, with alpha = 0.05 were utilized to determine statistical differences between the study groups. For all the assays presented in this manuscript, at least 3 biological replicates (N = 3) were performed.3. Results and discussion
3.1 Synthesis and characterization of gold nanoribbons (AuNRs)
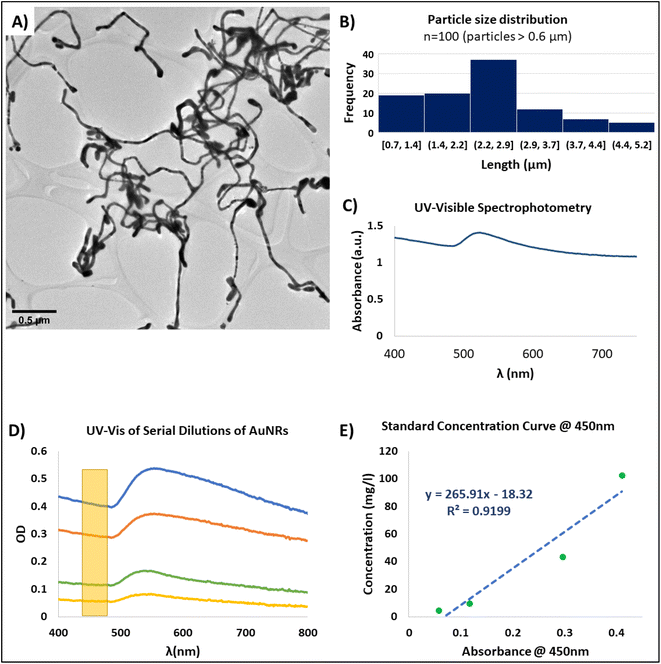
High aspect ratio one dimensional (1D) gold nanoparticles, such as AuNRs are often fabricated through complex processes, for example, electrospinning, lithography, and chemical reductions.21,44 However, some of these technologies are not widely accessible and require dedicated equipment. Thus, we have adapted a synthesis protocol based on the directed aggregation of gold, guided by a bi-surfactant template system39 to produce AuNRs with suitable features for integration into the scaffold-free organoids. The bisurfactant system is formed by NaOL and CTAB, and as reported by the authors,39 CTAB micelles are formed in the presence NaOL. These micelles have higher affinity for specific facets of the gold crystals, serving as template for the guided aggregation of gold in one direction, thus leading to the obtention nanoparticles with ribbon-like morphology. This protocol presents several advantages over other synthesis methods. First, all the reactions can be performed with benchtop equipment and reagents that are commercially available and easy to handle. Second, all the reactions are carried at either room temperature or temperatures that are easily achievable with conventional lab equipment. Finally, and most important, the length of the AuNRs can be tuned by controlling the reaction time and purification method.The length of the AuNRs is dictated by several factors throughout the synthesis process. For example, different template molecules can potentially limit the aggregation of gold atoms at determined lengths. Also, the availability of gold in the growth solution can act as a limiting factor. The growth of the AuNRs is a time-dependent reaction;39 therefore, by controlling the synthesis time, i.e., the incubation time of the growth solution after the gold seeds have been introduced, it is possible to tune the length of the AuNRs. Finally, the purification of the AuNRs is performed by precipitation through centrifugation, thus, it is possible to select particles of certain size based on the centrifugation settings. The original reported protocol for the synthesis of AuNRs yielded particles of about 40 μM in length. However, nanoparticles of this length can negatively impact the cellular aggregation and the formation of a cardiac syncytium as they can hinder cell–cell interactions and the formation of gap junctions in ECTs.24
The morphology of the synthesized nanoparticles herein was verified through TEM imaging. Ribbon-like particles where observed (Fig. 2A), and the morphology was consistent through different batches of AuNRs. In order to increase the yield of the reaction we performed a centrifugation swipe by varying the speed and time of the precipitation (not shown). We observed that the highest nanoparticle concentration, based on UV-vis spectrophotometry, was achieved when the nanoparticle solution was centrifuged at 3000×g for 17 minutes. Measurement of the length of ∼100 nanoparticles showed a range from 0.6 μM up to 6.0 μM and larger, with an average length of 2.4 ± 0.99 μM (Fig. 2B). While previous studies have demonstrated some advantages in the integration of nanoparticles for the fabrication of scaffold-free cardiac microtissues, there is a wide range of utilized nanoparticle materials, geometries, sizes, and concentrations.9 Despite these variations, there are two common factors for all of these studies. First, the nanomaterials are selected for their electroconductivity characteristics; and second, the size and geometry of the nanoparticles are tuned in order to prevent uptake by the cells. Additionally, it has been demonstrated that even small concentrations of gold nanoparticles have a positive effect in the electrophysiological behavior and formation of cardiac engineered tissues.45
1D gold nanoparticles typically present the surface plasmon resonance phenomenon (SPR). SPR is a complex phenomenon that occurs in some metals due to the availability of free electrons in the material lattice. This phenomenon is more easily observed in nanoparticles due to their high surface-to-volume ratio.46 Specifically, the resonance frequencies of gold nanoparticles are presented within the visible range of the electromagnetic spectrum.47 Therefore, when gold nanoparticles are found in suspension, optical methods can be used for detecting their SPR frequency. For example, UV-visible spectrophotometry can be used as an accessible method for evaluating SPR of AuNRs.
The SPR spectral signature of gold nanoparticles is dependent of their size, geometry, and concentration;46,48 and therefore, can be used for the characterization of the AuNRs. For instance, in suspensions of high aspect ratio nanoparticles usually only one peak is observed for the SPR phenomenon in the UV-visible spectrophotometric analysis. This peak corresponds to the cross-section of the AuNRs, that is within the nanometer scale. For our synthesized AuNRs, this peak was consistently presented at around 520 nm (Fig. 2C). While SPR have many applications,46 in our particular application, this phenomenon served as a tool for confirming the effective synthesis of AuNRs through UV-vis spectrophotometry. Moreover, the correlation of the absorbance of the AuNRs suspension with the nanoparticle concentration allowed us to create a standard concentration curve that permitted to calculate the concentration of any AuNRs solution based on its absorbance (Fig. 2D and E).
In order to obtain a concentration standard curve, we first performed serial dilutions of a colloidal suspension within the working range of AuNRs. These diluted suspensions where then analyzed by UV-vis spectrophotometry (Fig. 2D) and ICP-MS. While our AuNRs synthesis method proved to be a robust and consistent technique, based on the TEM images and UV-vis spectra, there were minor batch-to-batch variations in the readings of the SPR peak wavelength. Therefore, in order to standardize the concentration measurements based on the UV-vis spectrum, we selected the region localized at 450 nm as it has been reported to be a suitable wavelength for measuring the concentration of gold nanoparticles.49 Based on this approach, we were able to calculate the linear correlation between the reported absorbance at 450 nm and the nanoparticle concentration found by mass spectrometry (ICP-MS) (Fig. 2E). With this, we achieved a non-destructive and straightforward method for faithful measuring the concentration of the resultant AuNRs suspensions. Therefore, we were able to prepare working solutions with specific and desired concentrations for the integration of the AuNRs into the scaffold-free cardiac organoids.
3.2 PEGylation and cytotoxicity evaluation of AuNRs
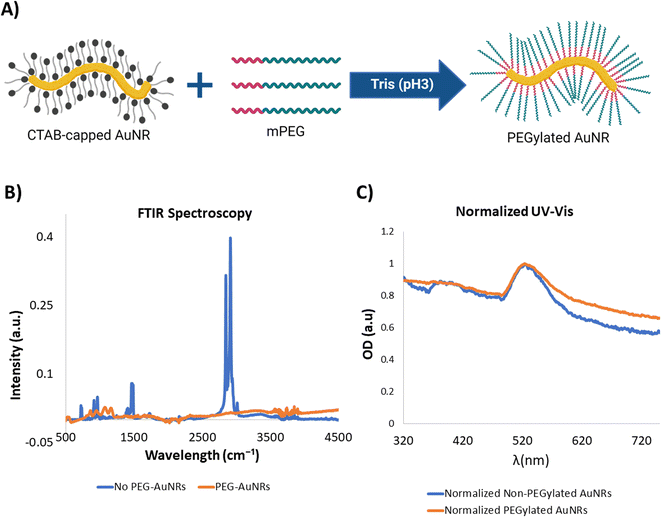
Cytotoxicity and bioaccumulation of gold nanoparticles has been studied previously due to their promising applications for various tissue engineering and therapeutics applications in vitro and in vivo.22 It has been found that gold nanoparticles are bioinert and biostable by themselves.50,51 However, the template molecules (CTAB and sodium oleate) used for the synthesis of the AuNRs may exhibit some cytotoxic effects.52,53 Thus, in order to render the AuNRs useable for our cardiac tissue engineering applications it was necessary to modify the surface of the nanoparticles and replace the original template molecules by biocompatible ones (Fig. 3A).Polyethylene glycol (PEG) has been widely accepted as a biocompatible biomaterial.54,55 Moreover, several protocols have been developed and adapted for its use as a replacing surface molecule specifically for gold nanoparticles.40 Besides being biocompatible, PEG has several other attractive features for its use in fabrication of nanoparticles. For example, it has been proven that the PEGylation of nanoparticles can affect their interaction with surrounding cells, by improving their biocompatibility54 and modifying their internalization rate.55 In this specific case, we were interested in performing the exchange of the surface molecules of the AuNRs.
Due to a similar surface chemistry of the AuNRs, developed in our work, to the nanoparticles used in another work by Zhang and Lin,40 we were able to adapt the methodology by modifying the concentrations of the reagents in order to ensure the availability of PEG molecules to perform a total surface exchange. In order to confirm the successful PEGylation of the AuNRs, we performed FTIR analysis. The FTIR spectra of CTAB and PEG were well characterized and presented distinctive peaks for both substances. One hand, the FTIR spectrum for CTAB was characterized by presence of two peaks around 2849.77 cm−1 and 2918.24 cm−1 corresponding to the stretching of C–H bonds. Additionally, several peaks were present around 1470 cm−1.56 The analyzed CTAB-capped AuNRs presented the same FTIR spectrum as those reported in the literature (Fig. 3B). Similarly, the PEGylated samples presented a FTIR pattern that correlated to reported studies,57 consisting of absorption bands between 840 cm−1 and 1466 cm−1 that correspond to C–C stretching and CH2 rocking57 (Fig. 3A). Thus, this analysis provided a clear fingerprint to determine the successful exchange of the molecules on the surface of the synthesized AuNRs.
Spectrophotometric analysis of gold nanoparticles has been deem extremely sensitive to changes in their morphology, surface chemistry, and physical aggregation.58 Notably, UV-vis spectrophotometry of AuNRs before and after PEGylation demonstrated that the SPR peak and overall UV-vis spectrum of the PEGylated nanoparticles remained almost unchanged, suggesting that significant changes to the morphology of the nanoparticles or particle aggregation did not occur during the process of surface modification through PEG exchange (Fig. 3C).
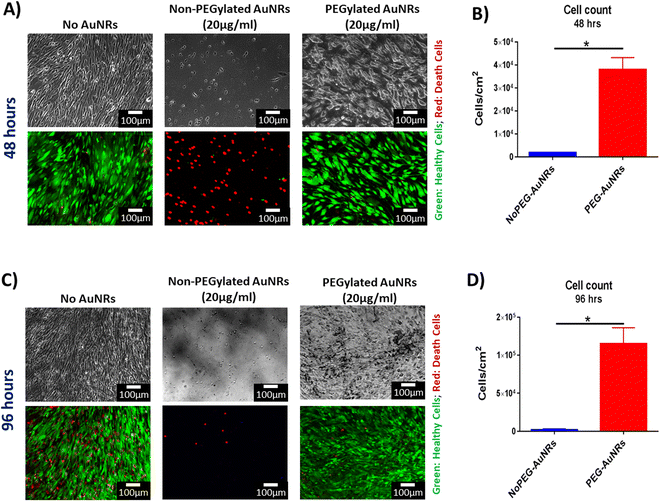
To investigate the effect of PEGylated and non-PEGylated AuNRs on the viability of cells, we performed an acute cytotoxicity assay on two dimensional (2D) monolayers of hiPSC-CFs. We carried the assay with a concentration that was double the concentration of the working solution for the fabrication of the cardiac organoids to ensure the feasibility of using PEG-AuNRs in the formation of cardiac organoids. To the best of our knowledge, this is the first study to present the integration of AuNRs with scaffold-free cardiac tissues. Thus, the selected working concentration of the PEG-AuNRs (10 μg mL−1) was based on similar studies that utilized nanoparticles embedded in cardiac organoids.31
We evaluated the viability of the cells through co-staining with calcein AM and ethidium homodimer III (EthD-III). Calcein AM is a cell-permeable molecule, that is retained in the cytoplasm of the cells and is cleaved by the action of esterases. When is cleaved, this causes the calcein dye to be released and in consequence the live healthy cells appear fluorescent green. On the other hand, EthD-III is membrane permeable and is fluorescent when binds to DNA molecules. Thus, EthD-III will only stain the nuclei of cells with defective cellular membranes, appearing as fluorescent red.
The acute cytotoxicity test for PEGylated and non-PEGylated AuNRs at 48 and 96 hours of exposure showed differences in the viability of the cells. Similar cellular density was observed for PEG-AuNRs and the control cellular populations for both timepoints (Fig. 4A and B). Moreover, the quantification of viable cells reflected statistical differences between non-PEG and PEG-AuNRs (Fig. 4C and D). It has been reported that the cytotoxicity of CTAB-capped gold nanoparticles is mainly due to traces of free CTAB found in the suspension of the nanoparticles.52 The desorption of CTAB molecules from the surface of the nanoparticles has also been proposed as a probable theory behind the toxic effect of gold nanoparticles,58 causing microtubule damage of the cells.59 However, IF images demonstrated that there is a significant difference between the PEGylated and non-PEGylated AuNRs, and that the cellular densities are similar between the groups with PEGylated AuNRs and without AuNRs (Fig. 4B).
Overall, primarily, we demonstrated the feasibility for the surface modification of AuNRs synthesized by seed-mediated growth method, which to the best of our knowledge, has not been reported before. Additionally, we showed that the PEGylation of AuNRs significantly reduced their acute cytotoxicity in human cardiac cells, rendering them safe for integration into cardiac engineered tissues after the surface molecule exchange is completed and the AuNRs have been biofunctionalized.
3.3 Viability evaluation of 3D scaffold-free cardiac organoids integrated with AuNRs
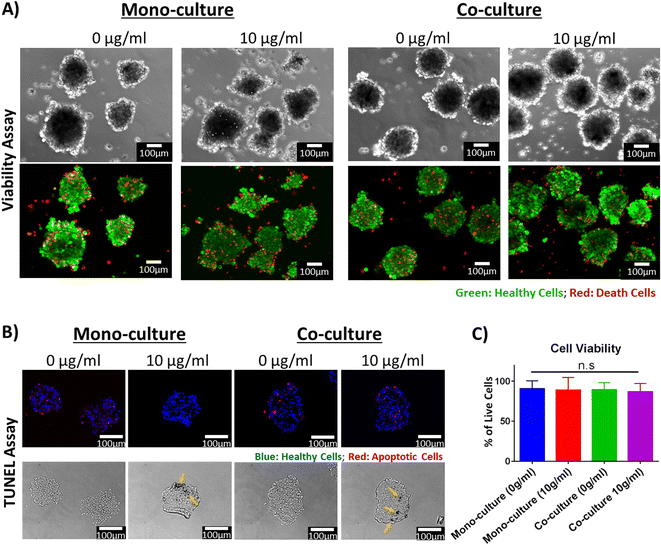
While the low cytotoxicity of the PEG-AuNRs was demonstrated on 2D monolayers of hiPSC-CFs (Fig. 4), it was still crucial to validate that the fabricated 3D cardiac organoids remained viable upon integration and exposure to PEG-AuNRs. Additionally, harvesting the cardiac organoids from the agarose microwells is a key factor for downstream analyses and applications. Therefore, it was important to evaluate that the integrity of the tissues was not influenced by their removal from the agarose microwells.To investigate the cellular viability and 3D tissue integrity, a viability assay was performed on the cardiac organoids. After staining, the microtissues were removed from the microwells by gently pipetting with culture media and washed with 1× DPBS. After rinsing, the vast majority of the cardiac cells appeared viable, denoted by the green staining after viability assay, with no apparent differences between the experimental conditions (Fig. 5A). Additionally, it is well known that one of the mechanisms that induce apoptosis in adherent cells is the anoikis phenomenon, driven by the lack of anchorage sites to the cells.60 Therefore, it is possible that some cell death is also generated during the formation of the cardiac organoids in the microwells. These dead cells are deposited at the bottom of the microwells and due to their depth (800 μM), it is not possible for these debris to be washed during routinely media changes, being present until the cardiac organoids are harvested. This cellular debris was washed when the organoids were removed from the microwells and does not seem to negatively affect in any of the parameters analyzed in this study.
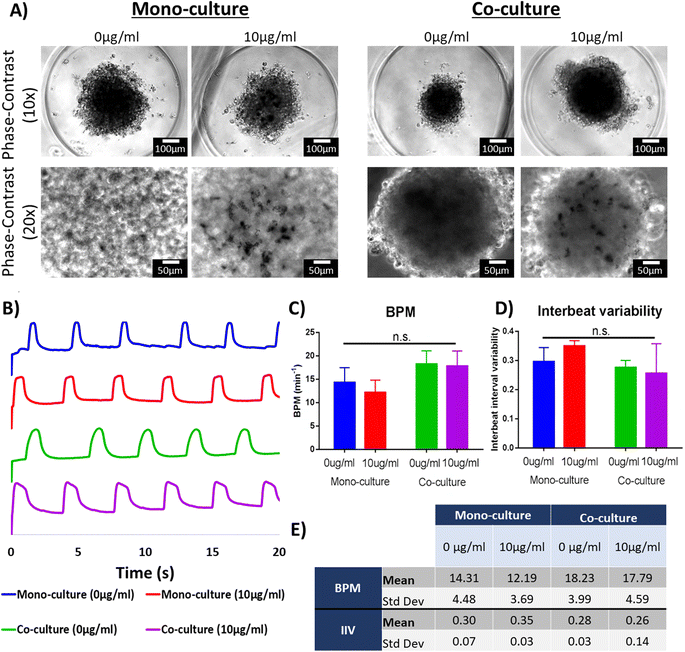
After harvesting, the morphology of the organoids was preserved and the cardiac microtissues presented spontaneous beating after harvesting (Videos S1–S4†), corroborating that the contractile activity of the tissues was also preserved. Many features of these organoids are dependent on their morphology. For example, it has been reported that oxygen and nutrient gradients generated within the organoids depends on their diameter.25,61 The oxygen and nutrient gradients within the microtissues have been leveraged to model pathologic conditions, such as hypoxia.61 Studies have proposed utilizing cardiac organoids for the manufacture of cardiac engineered tissues at bigger scales.62,63 These techniques rely on maintaining uniformity of the cardiac organoids. Therefore, creating and preserving cardiac organoids with specific diameters is important for their utilization in disease modeling and drug screening applications. We have demonstrated in this study that our proposed method leads to the formation of cardiac organoids with uniform size and morphology (Fig. S1 and S2†). Moreover, we have confirmed, for the first time, that it is feasible to integrate AuNRs in the fabrication of cardiac organoids. These organoids remained viable and maintained their morphology and electrophysiological activity after being removed from the agarose microwells.
Diffusion of oxygen and nutrients has been a critical concern in the tissue engineering field to facilitate the transport of these molecules. It has been reported that cellular density, cellular origin and organoid size can affect the oxygen consumption rate and can lead to the creation of hypoxic or necrotic cores.25,61 To that end, we further evaluated dearly apoptosis of the cells within the cryosectioned slices of the cardiac organoids using TUNEL (Fig. 5B). This is to demonstrate that the diameter and cellular density of the cardiac organoids were appropriate and that the AuNRs did not cause any cytotoxic effect in the hiPSC-CMs due to the 3D microenvironment. Based on our obtained images, we did not find significant cell death due to early apoptosis in any of the analyzed tissues (Fig. 5C). This indicates that the nutrient diffusion within the organoids was appropriate and hypoxic cores were not generated. These secondary analyses were indeed consistent with our primary viability assay, confirming that the synthesized and PEGylated AuNRs (PEG-AuNRs) did not induce any toxicity effects on cardiac organoids.
Opaque spots (Fig. 5B, yellow arrows) were found in the phase-contrast images of the organoids integrated with the AuNRs but were not seen in organoids without gold nanoparticles. These opaque areas are presumably regions were the AuNRs were concentrated. This suggests that the gold nanoparticles were integrated through the entire body of the organoids. However, it remains to be elucidated in our future work whether the AuNRs underwent aggregation due their integration in the cellular microenvironment.
3.4 Functional contractility analysis of isogenic cardiac organoids embedded with AuNRs
The integration of nanoparticles in cardiac organoids has been of keen interest in the cardiac tissue engineering field.9,24,25 One of the proposed hypotheses across these studies is that enriching the intercellular microenvironment with electrically conductive nanomaterials and nanoscale topographical cues can enhance the performance of the engineered microtissues.8,45,64,65 Additionally, it is well accepted that co-culture of cardiac cells in engineered tissues can improve the maturation of hiPSC-derived cardiac cells.33,66 The creation of isogenic engineered tissues for the purpose of drug screening and disease modeling has been extensively discussed.67 Specifically, scaffold-free engineered tissues present a particular complexity due to the lack of extracellular matrix (ECM) or ECM-like materials that can provide support to the cells. Therefore, cellular aggregation and tissue compaction would be relevant in the fabrication of cardiac organoids.33 First, the cellular ratio for co-culture groups was thoroughly optimized, as it was shown that it leads to improved tissue compaction.33 Additionally, it was necessary to corroborate the presence of the PEG-AuNRs in the engineered tissues after their fabrication. The phase-contrast images of the cardiac organoids taken seven days after seeding in the agarose microwells showed rounded and dense tissues that presented spontaneous beating (Fig. 6A, top row). For the organoids fabricated with PEG-AuNRs, opaque regions were observed in magnified images (Fig. 6A, bottom row) as also shown in Fig. 5B, that the gold nanoparticles were effectively integrated within the cardiac tissues.Contractile activity of the cardiac engineered tissues is one of the main features that needs to be evaluated, as most of their tissue engineering applications rely on an effective electromechanical function.68 After signal extraction from time-lapse images, it was found that all the experimental groups showed a similar spontaneous beating behavior (Fig. 6B). The organoids presented comparable beating rates (beat per minute; BPM) (Fig. 6C) ranging from 12.19 to 18.23 BPM (Fig. 6D). The inter-beat interval variability (IIV) has been described as an indirect measure of tissue synchronicity.42,69 Tissue synchronicity is a relevant biomarker of functionality of hiPSC-derived tissues,42 as it requires the formation of gap junctions and the capability of cardiac cells to be paced by endogenous or exogenous electrical stimulation.70 Thus, decreased IIV is a desirable feature of engineered cardiac tissues. Our analysis did not show a significant difference in the IIV of the four experimental groups (Fig. 6D and E). Overall, this suggests that the PEG-AuNRs had negligible effect in the contractility of the cardiac organoids.
Previous studies focused on the integration of nanoparticles in scaffold-free cardiac tissues demonstrated a wide range of concentrations. Moreover, the methods for measuring the concentrations of those nanoparticles and integrating them with cardiac organoids were not consistent across different studies. Therefore, moving forward with future studies, we aim to optimize the PEG-AuNRs concentration in order to maximize their interaction with cardiac cells.
3.5 Analysis of transcriptomic profile of cardiac organoids
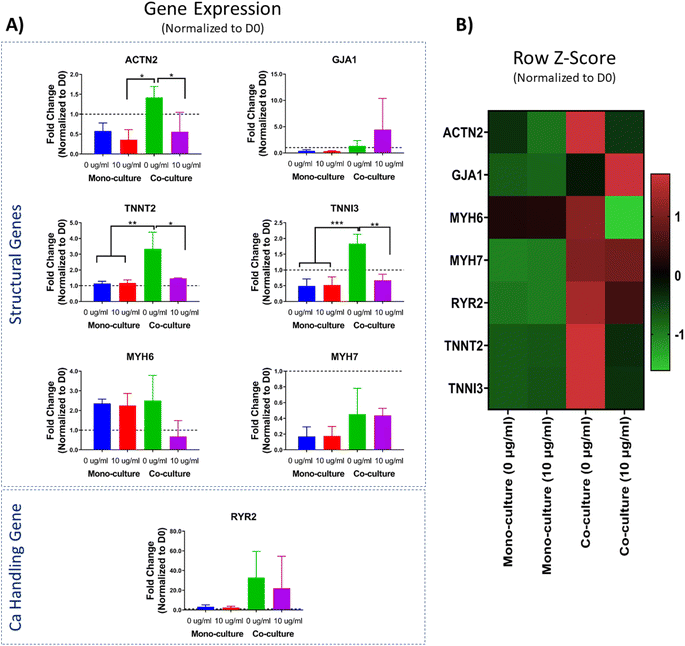
The evaluation of the transcriptomic profile has become one of the gold standard molecular-level methods for evaluating the overall performance of engineered tissues. The assessment of gene expression levels is of particular interest for cells and cardiac tissues derived from human stem cells to evaluate their maturity. Genes relevant for the cardiac function, such as those involved in the formation of the contractile machinery of the CMs (i.e., ACTN2, TNN2, TNNI3, MYH6, and MYH7) and calcium handling (i.e., RYR2) are of utmost interest as surrogate biomarkers for maturation and performance of engineered tissues.14 Herein, the transcriptomic profile of the scaffold-free cardiac organoids was evaluated through a panel of seven cardiac-relevant genes with the purpose of assessing the effect of the co-culture versus mono-culture conditions. In addition, we were interested in comparing the gene expression profile of the cardiac organoids in presence or absence of PEG-AuNRs.It has been reported that one of the main factors contributing to the maturation of hiPSC-CMs is the presence of non-cardiomyocyte cells (i.e., hiPSC-CFs).33,34 When CFs are integrated into ECTs, these cells secrete growth factors and cytokines that lead to cellular crosstalk with CMs, and ultimately lead to improved maturation of hiPSC-CMs in engineered tissues.71 Moreover, it has been demonstrated that hiPSC-CFs can potentially modulate electromechanical properties of 3D ECTs.72 Similar to those reports, in our study, we found that sarcomere-related genes, namely ACTN2, TNNT2, and TNNI3, were upregulated in the co-culture (0 μg mL−1) cardiac organoids (Fig. 7A). For four of the other evaluated genes (GJA1, RYR2, MYH6, and MYH7) we did not find any significant difference between any of the experimental conditions. However, there was a trend for higher expression of the analyzed gene panel for both co-culture conditions (0 μg mL−1 and 10 μg mL−1) (Fig. 7B). Thus, our results are consistent with previous findings in the literature, indicating the beneficial effect of the integration of hiPSC-CFs in ECTs.
Alternatively, the integration of the PEG-AuNRs overall appeared to be negligible for the effects of modifying the transcriptomic profile of the cardiac organoids. Specifically, we did not find any significant difference in the expression of any of the analyzed genes for the mono-culture conditions (0 μg mL−1 vs. 10 μg mL−1) (Fig. 7A). However, for the co-culture conditions (0 μg mL−1 vs. 10 μg mL−1) we found significant differences for the expression of ACTN2, TNNT2, and TNNI3, with higher expression in the group without PEG-AuNRs (co-culture, 0 g mL−1). These observations show the dominant effect of co-culture groups in maturation of the organoids. We suspect that the negligible effect of the PEG-AuNRs in the gene expression of the cardiac organoids is related to the possible particle aggregation observed previously (Fig. 6A). Overall, we have demonstrated the feasibility of the integration of PEG-AuNRs within cardiac organoids; however, variables such as nanoparticle concentration, aggregation, and other external factors (i.e., electrical stimulation) need to be further studied in future works.
4. Conclusion
The integration of nanomaterials has been widely proposed for various applications in tissue engineering. It is speculated that the electroconductive properties of some of the utilized nanoparticles, along with the biophysical cues that they provide when introduced in engineered tissues can enhance their structure and function. To the best of our knowledge, this is the first study to evaluate the integration of 1D gold nanoparticles with scaffold-free isogenic cardiac organoids.We first presented a modified method for the generation of 1D gold nanoparticles, namely gold nanoribbons (AuNRs). These nanoribbons have been extensively characterized and have been biofunctionalized by performing a surface molecule exchange through a PEGylation reaction. The feasibility for utilizing these nanoribbons was primarily demonstrated through cytotoxicity assays in 2D monolayers of hiPSC-CFs and 3D isogenic cardiac organoids derived from mono-culture and co-culture of and hiPSC-CMs and hiPSC-CFs. We verified the presence of the AuNRs within the engineered organoids by phase-contrast imaging of the surface and cryosectioned slices of the cardiac organoids. We then performed contractility evaluation, as well as analysis of the transcriptomic profile of the isogenic cardiac organoids embedded with the AuNRs. Although we did not find any significant difference in the spontaneous beating behavior of the engineered tissues, we found significant upregulation of some cardiac-relevant genes, specifically ACTN2, TNNT2, and TNNI3 for the co-culture (0 μg mL−1) cardiac organoids. These observations demonstrated the critical influence of hiPSC-CFs on maturation and function of isogenic cardiac organoids regardless of the presence of AuNRs.
We speculate that the lack of an observable effect from the PEG-AuNRs can be due to two main factors: (a) the selected concentration for this proof-of-concept needs to be optimized, and (b) there is possible aggregation of the AuNRs when they interact with the cardiac cells in the 3D microenvironment. Thus, our future studies will be focused on evaluating the overall distribution of the AuNRs within the cardiac organoids and elucidating the aggregation mechanisms of the AuNRs, in order to promote a more homogeneous distribution of the nanoparticles within organoids. Additionally, optimization of the concentration of the AuNRs may result in the enhanced cell–particle interactions and the overall function of hiPSC-derived isogenic cardiac organoids. Overall, the findings of this study have potential for applications in cardiac tissue engineering for the creation of physiologically relevant scaffold-free tissues for disease modeling and regenerative medicine applications.
Author contributions
APG contributed to the conceptualization, formal analysis, investigation, methodology, visualization, and writing – original draft. HE contributed to the formal analysis, investigation, visualization, and writing – review & editing. RQM contributed to the conceptualization, funding acquisition, and writing – review & editing. MN contributed to the conceptualization, funding acquisition, methodology, project administration, supervision, validation, and writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors would like to acknowledge the Arizona Biomedical Research Commission (ABRC) New Investigator Award #AWD32676, and the National Science Foundation (NSF) CAREER Award #1653193 and NANOSCALE Award #2016501, for providing funding sources for this project. The authors would like to thank Dr Barbara Smith for providing access to qTower 2.0., and Dr Jaimeson Veldhuizen for providing support in cell differentiation and cell sourcing.References
- K. Mc Namara, H. Alzubaidi and J. K. Jackson, Integr. Pharm. Res. Pract., 2019, 8, 1–11 CrossRef PubMed.
- G. A. Roth, G. A. Mensah, C. O. Johnson, G. Addolorato, E. Ammirati, L. M. Baddour, N. C. Barengo, A. Z. Beaton, E. J. Benjamin, C. P. Benziger, A. Bonny, M. Brauer, M. Brodmann, T. J. Cahill, J. Carapetis, A. L. Catapano, S. S. Chugh, L. T. Cooper, J. Coresh, M. Criqui, N. DeCleene, K. A. Eagle, S. Emmons-Bell, V. L. Feigin, J. Fernandez-Sola, G. Fowkes, E. Gakidou, S. M. Grundy, F. J. He, G. Howard, F. Hu, L. Inker, G. Karthikeyan, N. Kassebaum, W. Koroshetz, C. Lavie, D. Lloyd-Jones, H. S. Lu, A. Mirijello, A. M. Temesgen, A. Mokdad, A. E. Moran, P. Muntner, J. Narula, B. Neal, M. Ntsekhe, G. Moraes de Oliveira, C. Otto, M. Owolabi, M. Pratt, S. Rajagopalan, M. Reitsma, A. L. P. Ribeiro, N. Rigotti, A. Rodgers, C. Sable, S. Shakil, K. Sliwa-Hahnle, B. Stark, J. Sundstrom, P. Timpel, I. M. Tleyjeh, M. Valgimigli, T. Vos, P. K. Whelton, M. Yacoub, L. Zuhlke, C. Murray, V. Fuster and GBD-NHLBI-JACC Global Burden of Cardiovascular Diseases Writing Group, J. Am. College Cardiol., 2020, 76, 2982–3021 CrossRef PubMed.
- J. Veldhuizen, R. Q. Migrino and M. Nikkhah, J. Biol. Eng., 2019, 13, 29 CrossRef PubMed.
- M. E. Hartman, D.-F. Dai and M. A. Laflamme, Adv. Drug Delivery Rev., 2016, 96, 3–17 CrossRef CAS PubMed.
- X. Lian, J. Zhang, S. M. Azarin, K. Zhu, L. B. Hazeltine, X. Bao, C. Hsiao, T. J. Kamp and S. P. Palecek, Nat. Protoc., 2013, 8, 162–175 CrossRef CAS PubMed.
- J. Zhang, R. Tao, K. F. Campbell, J. L. Carvalho, E. C. Ruiz, G. C. Kim, E. G. Schmuck, A. N. Raval, A. M. da Rocha, T. J. Herron, J. Jalife, J. A. Thomson and T. J. Kamp, Nat. Commun., 2019, 10, 2238 CrossRef PubMed.
- F. B. Bedada, M. Wheelwright and J. M. Metzger, Biochim. Biophys. Acta, 2016, 1863, 1829–1838 CrossRef CAS PubMed.
- H. Esmaeili, A. Patino-Guerrero, M. Hasany, M. O. Ansari, A. Memic, A. Dolatshahi-Pirouz and M. Nikkhah, Acta Biomater., 2022, 139, 118–140 CrossRef CAS PubMed.
- A. Patino-Guerrero, J. Veldhuizen, W. Zhu, R. Q. Migrino and M. Nikkhah, J. Mater. Chem. B, 2020, 8, 7571–7590 RSC.
- J. Veldhuizen, H. F. Mann, N. Karamanova, W. D. Van Horn, R. Q. Migrino, D. Brafman and M. Nikkhah, Sci. Adv., 2022, 8, eabq6720 CrossRef CAS PubMed.
- J. Veldhuizen, R. Chavan, B. Moghadas, J. G. Park, V. D. Kodibagkar, R. Q. Migrino and M. Nikkhah, Biomaterials, 2022, 281, 121336 CrossRef CAS PubMed.
- J. Li, Y. Hua, S. Miyagawa, J. Zhang, L. Li, L. Liu and Y. Sawa, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- D. Yu, X. Wang and L. Ye, J. Cardiovasc. Dev. Dis., 2021, 8(11), 153 CrossRef CAS PubMed.
- R. E. Ahmed, T. Anzai, N. Chanthra and H. Uosaki, Front. Cell Dev. Biol., 2020, 8, 178 CrossRef PubMed.
- R. Augustine, P. Dan, A. Hasan, I. M. Khalaf, P. Prasad, K. Ghosal, C. Gentile, L. McClements and P. Maureira, Biomed. Pharmacother., 2021, 138, 111425 CrossRef CAS PubMed.
- M. Wanjare and N. F. Huang, Regener. Med., 2017, 12, 187–201 CrossRef CAS PubMed.
- R. R. Besser, M. Ishahak, V. Mayo, D. Carbonero, I. Claure and A. Agarwal, Theranostics, 2018, 8, 124–140 CrossRef CAS PubMed.
- J. Cutts, M. Nikkhah and D. A. Brafman, Biomarker Insights, 2015, 10, S20313 CrossRef PubMed.
- A. Dolatshahi-Pirouz, M. Nikkhah, K. Kolind, M. R. Dokmeci and A. Khademhosseini, J. Funct. Biomater., 2011, 2, 88–106 CrossRef CAS PubMed.
- M. Kharaziha, A. Memic, M. Akbari, D. A. Brafman and M. Nikkhah, Adv. Healthc. Mater., 2016, 5, 1533–1553 CrossRef CAS PubMed.
- X. Hong, C. Tan, J. Chen, Z. Xu and H. Zhang, Nano Res., 2015, 8, 40–55 CrossRef CAS.
- C. Lopez-Chaves, J. Soto-Alvaredo, M. Montes-Bayon, J. Bettmer, J. Llopis and C. Sanchez-Gonzalez, Nanomedicine, 2018, 14, 1–12 CrossRef CAS PubMed.
- Y. Tan, D. Richards, R. Xu, S. Stewart-Clark, S. K. Mani, T. K. Borg, D. R. Menick, B. Tian and Y. Mei, Nano Lett., 2015, 15, 2765–2772 CrossRef CAS PubMed.
- D. J. Richards, Y. Tan, R. Coyle, Y. Li, R. Xu, N. Yeung, A. Parker, D. R. Menick, B. Tian and Y. Mei, Nano Lett., 2016, 16, 4670–4678 CrossRef CAS PubMed.
- Y. Tan, D. Richards, R. C. Coyle, J. Yao, R. Xu, W. Gou, H. Wang, D. R. Menick, B. Tian and Y. Mei, Acta Biomater., 2017, 51, 495–504 CrossRef CAS PubMed.
- L. Tian, M. Wei, L. Ji, M. Zheng, G. Liu and L. Wang, J. Exp. Nanosci., 2021, 16, 212–228 CrossRef.
- X.-P. Li, K.-Y. Qu, F. Zhang, H.-N. Jiang, N. Zhang, C. Nihad, C.-M. Liu, K.-H. Wu, X.-W. Wang and N.-P. Huang, J. Mater. Chem. B, 2020, 8, 7213–7224 RSC.
- J. Sundnes, G. T. Lines, X. Cai, B. F. Nielsen, K.-A. Mardal and A. Tveito, Computing the Electrical Activity in the Heart, Springer Science & Business Media, 2007 Search PubMed.
- M. M. Slotvitsky, V. A. Tsvelaya, A. D. Podgurskaya and K. I. Agladze, Sci. Rep., 2020, 10, 7774 CrossRef CAS PubMed.
- C. C. Veerman, G. Kosmidis, C. L. Mummery, S. Casini, A. O. Verkerk and M. Bellin, Stem Cells Dev., 2015, 24, 1035–1052 CrossRef CAS PubMed.
- J. Park, Y. S. Kim, S. Ryu, W. S. Kang, S. Park, J. Han, H. C. Jeong, B. H. Hong, Y. Ahn and B.-S. Kim, Adv. Funct. Mater., 2015, 25, 2590–2600 CrossRef CAS.
- S. Ahadian, Y. Zhou, S. Yamada, M. Estili, X. Liang, K. Nakajima, H. Shiku and T. Matsue, Nanoscale, 2016, 8, 7075–7084 RSC.
- A. Patino-Guerrero, R. D. Ponce Wong, V. D. Kodibagkar, W. Zhu, R. Q. Migrino, O. Graudejus and M. Nikkhah, ACS Biomater. Sci. Eng., 2023, 9, 944–958 CrossRef CAS PubMed.
- G. Campostrini, V. Meraviglia, E. Giacomelli, R. W. J. van Helden, L. Yiangou, R. P. Davis, M. Bellin, V. V. Orlova and C. L. Mummery, Nat. Protoc., 2021, 16, 2213–2256 CrossRef CAS PubMed.
- H. Saini, A. Navaei, A. Van Putten and M. Nikkhah, Adv. Healthc. Mater., 2015, 4, 1961–1971 CrossRef CAS PubMed.
- W. Chen, W. Bian, Y. Zhou and J. Zhang, Front. Bioeng. Biotechnol., 2021, 9, 599928 CrossRef PubMed.
- C. A. Souders, S. L. Bowers and T. A. Baudino, Circ. Res., 2009, 105, 1164–1176 CrossRef CAS PubMed.
- M. J. Daseke, M. A. A. Tenkorang, U. Chalise, S. R. Konfrst and M. L. Lindsey, Matrix Biol., 2020, 91–92, 109–116 CrossRef CAS PubMed.
- Y. Xu, X. Wang, L. Chen, Y. Zhao, L. He, P. Yang, H. Wu, F. Bao and Q. Zhang, J. Mater. Chem. C, 2015, 3, 1447–1451 RSC.
- Z. Zhang and M. Lin, RSC Adv., 2014, 4, 17760–17767 RSC.
- A. Sharma, G. Li, K. Rajarajan, R. Hamaguchi, P. W. Burridge and S. M. Wu, J. Vis. Exp., 2015, e52628, DOI:10.3791/52628.
- J. Veldhuizen, J. Cutts, D. A. Brafman, R. Q. Migrino and M. Nikkhah, Biomaterials, 2020, 256, 120195 CrossRef CAS PubMed.
- C. Fan, Y. Tang, M. Zhao, X. Lou, D. Pretorius, P. Menasche, W. Zhu and J. Zhang, J. Mol. Cell. Cardiol., 2020, 141, 1–10 CrossRef CAS PubMed.
- C. Zhao, X. Xu, A. R. Ferhan, N. Chiang, J. A. Jackman, Q. Yang, W. Liu, A. M. Andrews, N. J. Cho and P. S. Weiss, Nano Lett., 2020, 20, 1747–1754 CrossRef CAS PubMed.
- A. Navaei, H. Saini, W. Christenson, R. T. Sullivan, R. Ros and M. Nikkhah, Acta Biomater., 2016, 41, 133–146 CrossRef CAS PubMed.
- V. Amendola, R. Pilot, M. Frasconi, O. M. Maragò and M. A. Iatì, J. Phys. Condens. Matter, 2017, 29, 203002 CrossRef PubMed.
- V. V. Apyari, S. G. Dmitrienko and Y. A. Zolotov, Sens. Actuators, B, 2013, 188, 1109–1115 CrossRef CAS.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- A. G. Shard, L. Wright and C. Minelli, Biointerphases, 2018, 13, 061002 CrossRef CAS PubMed.
- C. Rambanapasi, J. R. Zeevaart, H. Buntting, C. Bester, D. Kotze, R. Hayeshi and A. Grobler, Molecules, 2016, 21 Search PubMed.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Small, 2005, 1, 325–327 CrossRef CAS PubMed.
- A. M. Alkilany, P. K. Nagaria, C. R. Hexel, T. J. Shaw, C. J. Murphy and M. D. Wyatt, Small, 2009, 5, 701–708 CrossRef CAS PubMed.
- L. Wang, X. Jiang, Y. Ji, R. Bai, Y. Zhao, X. Wu and C. Chen, Nanoscale, 2013, 5, 8384–8391 RSC.
- M. J. Santos-Martinez, K. Rahme, J. J. Corbalan, C. Faulkner, J. D. Holmes, L. Tajber, C. Medina and M. W. Radomski, J. Biomed. Nanotechnol., 2014, 10, 1004–1015 CrossRef CAS PubMed.
- L. Shi, J. Zhang, M. Zhao, S. Tang, X. Cheng, W. Zhang, W. Li, X. Liu, H. Peng and Q. Wang, Nanoscale, 2021, 13, 10748–10764 RSC.
- R. R. Shettigar, N. M. Misra and K. Patel, J. Pet. Explor. Prod. Technol., 2018, 8, 597–606 CrossRef CAS.
- N. S. Vrandečić, M. Erceg, M. Jakić and I. Klarić, Thermochim. Acta, 2010, 498, 71–80 CrossRef.
- C. J. Murphy, A. M. Gole, J. W. Stone, P. N. Sisco, A. M. Alkilany, E. C. Goldsmith and S. C. Baxter, Acc. Chem. Res., 2008, 41, 1721–1730 CrossRef CAS PubMed.
- Q. Li, C. Huang, L. Liu, R. Hu and J. Qu, Nanomaterials, 2018, 8, 1063 CrossRef PubMed.
- S. M. Frisch and R. A. Screaton, Curr. Opin. Cell Biol., 2001, 13, 555–562 CrossRef CAS PubMed.
- D. J. Richards, Y. Li, C. M. Kerr, J. Yao, G. C. Beeson, R. C. Coyle, X. Chen, J. Jia, B. Damon, R. Wilson, E. Starr Hazard, G. Hardiman, D. R. Menick, C. C. Beeson, H. Yao, T. Ye and Y. Mei, Nat. Biomed. Eng., 2020, 4, 446–462 CrossRef CAS PubMed.
- Y. Kawai, S. Tohyama, K. Arai, T. Tamura, Y. Soma, K. Fukuda, H. Shimizu, K. Nakayama and E. Kobayashi, Front. Cardiovasc. Med., 2022, 8, 806215 CrossRef PubMed.
- K. Arai, D. Murata, A. R. Verissimo, Y. Mukae, M. Itoh, A. Nakamura, S. Morita and K. Nakayama, PLoS ONE, 2018, 13, e0209162 CrossRef CAS PubMed.
- A. Navaei, K. Rahmani Eliato, R. Ros, R. Q. Migrino, B. C. Willis and M. Nikkhah, Biomater. Sci., 2019, 7, 585–595 RSC.
- A. Navaei, N. Moore, R. T. Sullivan, D. Truong, R. Q. Migrino and M. Nikkhah, RSC Adv., 2017, 7, 3302–3312 RSC.
- P. Beauchamp, C. B. Jackson, L. C. Ozhathil, I. Agarkova, C. L. Galindo, D. B. Sawyer, T. M. Suter and C. Zuppinger, Front. Mol. Biosci., 2020, 7, 14 CrossRef CAS PubMed.
- S. Cho, C. Lee, M. A. Skylar-Scott, S. C. Heilshorn and J. C. Wu, J. Mol. Cell. Cardiol., 2021, 157, 56–65 CrossRef CAS PubMed.
- S. Cho, D. E. Discher, K. W. Leong, G. Vunjak-Novakovic and J. C. Wu, Nat. Methods, 2022, 19, 1064–1071 CrossRef CAS PubMed.
- H. Iseoka, S. Miyagawa, S. Fukushima, A. Saito, S. Masuda, S. Yajima, E. Ito, N. Sougawa, M. Takeda, A. Harada, J.-K. Lee and Y. Sawa, Tissue Eng. Part A, 2017, 24, 287–300 CrossRef PubMed.
- Y. Wu and L. Guo, IEEE Trans. Biomed. Eng., 2018, 65, 264–272 Search PubMed.
- A. Colliva, L. Braga, M. Giacca and S. Zacchigna, J. Physiol., 2020, 598, 2923–2939 CrossRef CAS PubMed.
- C. E. Rupert, T. Y. Kim, B.-R. Choi and K. L. K. Coulombe, Stem Cells Int., 2020, 2020, 9363809 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01811c |
| This journal is © The Royal Society of Chemistry 2023 |