Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Insulin–cobalt core–shell nanoparticles for receptor-targeted bioimaging and diabetic wound healing†
Deepinder Shardaa and
Diptiman Choudhury *ab
*ab
aSchool of Chemistry and Biochemistry, Thapar Institute of Engineering and Technology, Patiala, 147004, Punjab, India. E-mail: diptiman@thapar.edu; Tel: +91-8196949843
bThapar Institute of Engineering and Technology-Virginia Tech (USA) Center of Excellence in Emerging Materials, Thapar Institute of Engineering and Technology, Patiala, Punjab-147004, India
First published on 6th July 2023
Abstract
Diabetic wounds represent a major issue in medical care and need advanced therapeutic and tissue imaging systems for better management. The utilization of nano-formulations involving proteins like insulin and metal ions plays significant roles in controlling wound outcomes by decreasing inflammation or reducing microbial load. This work reports the easy one-pot synthesis of extremely stable, biocompatible, and highly fluorescent insulin–cobalt core–shell nanoparticles (ICoNPs) with enhanced quantum yield for their highly specific receptor-targeted bioimaging and normal and diabetic wound healing in vitro (HEKa cell line). The particles were characterized using physicochemical properties, biocompatibility, and wound healing applications. FTIR bands at 670.35 cm−1, 849.79, and 973.73 indicating the Co–O bending, CoO–OH bond, and Co–OH bending, respectively, confirm the protein–metal interactions, which is further supported by the Raman spectra. In silico studies indicate the presence of cobalt binding sites on the insulin chain B at 8 GLY, 9 SER, and 10 HIS positions. The particles exhibit a magnificent loading efficiency of 89.48 ± 0.049% and excellent release properties (86.54 ± 2.15% within 24 h). Further, based on fluorescent properties, the recovery process can be monitored under an appropriate setup, and the binding of ICoNPs to insulin receptors was confirmed by bioimaging. This work helps synthesize effective therapeutics with numerous wound-healing promoting and monitoring applications.
1. Introduction
Wound healing is a complex process involving a sequential overlapping cascade of events that comes into action in response to some external chemical or physical stimuli and eventually causes healing by restoring lost tissue.1 The healing process is categorized into four phases, hemostasis, inflammation, proliferation, and remodeling.2 The hemostasis is initiated by clot formation due to platelets' activation, which prevents microbial infestation and promotes matrix organization. In proliferation, the accumulation of cells, connective tissue, growth factors, and angiogenesis factors across the wound occurs. The remodeling involves the resynthesis of the extracellular matrix to maintain the balance between the death of existing cells and the formation of new cells.3,4 However, the progress monitoring of recovery of wounds always remains a significant challenge. In some instances, normal healing gets disrupted. It gets arrested in one of the phases due to the loss of balance in the physiological mechanism of healing due to infection, chronic irritation, trauma, the persistence of microbes or other foreign bodies, and ischemia, making the wound a chronic one5–7 which can be prevented by keeping the wound moist, removing the dead tissues, covering the injury to avoid bacterial infection, and removing the excess tissue fluid.8Apart from these issues, the major problem for delayed healing is diabetes mellitus or long-term hyperglycemia, which alters the secretion of cytokines, making wound healing cumbersome.9 In diabetic conditions, there is the prolonged proinflammatory phase due to the persistent release of proteases, proinflammatory cytokines, and reactive oxygen species and the delayed anti-inflammatory phase because of the low secretion of anti-inflammatory cytokines.10 Further, the transport of nutrients to the wound site is prevented by atherosclerosis caused by diabetes.11 It also results in the dysfunction of endothelial cells due to vasodilation induced by pressure12 and disrupts the processes essential for re-epithelialization; the proliferation of keratinocytes and fibroblasts, synthesis of proteins, and cell migration.13–15 Further, it impairs the body's immune response, making the wounds prone to infection and leading to damaged structural components of the extracellular matrix.7 It also causes free radical damage due to the reduced activity of certain antioxidant enzymes such as glutathione peroxidase and superoxide dismutase.16 Various advanced bandages are developed for treating diabetic wound healing and overcoming the issues faced in effective healing.17,18
Recent advancements in nanotechnology, such as specific physicochemical properties and small size, allow the intracellular delivery of different drugs and biomolecules, protect them from degradation, increase the penetration ability of drugs into the wound, allow the topical application, and enhance the half-life of this agents.19,20 It results in a decline in repetitive drug application, thus lowering the cost and making them cost-effective.21,22 The metallic nanoparticles possess a high surface-to-volume ratio, high stability, biocompatibility, safety, and economical rates.22,23 Several gold, silver, copper, zinc, and cobalt metallic nanoparticles were prepared to study wound healing activities in vitro and in vivo models.24 Protein-based nanoparticles are gaining attention due to their high biodegradability, easily metabolizable nature, and the possibility to alter or modify their surface for the drugs to attach efficiently. The proteins can be of plant or animal origin, such as bovine serum albumin,25 insulin,26 transferrin,27 lactoferrin, and so on, for nano-formulation synthesis.28,29 The selection of nanoparticles in treating wounds is based on three parameters; antimicrobial action, role as a delivery agent, and role in the repair process.30
The use of insulin protein is increasing due to the enormous presence of insulin receptors on the membranes of all mammalian cells, and there is a significant variation in the number of receptors from just 40 in erythrocytes to 200–300 × 103 for adipocytes and hepatocytes; however, the number is much higher in cancerous cells compared to normal cells.26,31 It plays a crucial role as a growth factor that facilitates chemotaxis and pinocytosis or phagocytosis by macrophages, promotes the secretion of cytokines critical for inflammation, and helps in re-epithelialization, essential for wound healing. It works by transitioning proinflammatory cytokines into anti-inflammatory cytokines and promotes wound repair and regeneration.32,33 The nano-formulations involving insulin as a protein template exhibit promising applications in bioimaging, super-resolution microscopy, normal wound healing, and diabetic wound healing.32–35 The metal ions used earlier to form different formulations with insulin include silver, copper, zinc, nickel, etc.31 Apart from metal–insulin nanoparticles, other nano-formulations involving insulin as a wound-healing agent have been synthesized. Insulin-containing chitosan nanoparticles were synthesized to stimulate inflammatory cell proliferation, angiogenesis, and wound healing in diabetic rats.36 Lee et al. reported accelerated diabetic wound repair by promoting epithelialization using core–shell insulin-loaded scaffolds.37 The synthesis of human keratin conjugated insulin was reported by Li et al. for their role in full-thickness wound repair in rats by promoting cellular migration.38 Clinical studies were also performed to confirm the role of insulin further. The insulin injection was used to study the effect on systemic blood glucose levels and healing of diabetic foot ulcers in patients and found that insulin promotes the formation of vessels and granulation tissue.39 The impact of topical insulin was studied in 110 patients having diabetic foot ulcers using a double-blind placebo-controlled trial and found that patients treated with insulin gauze dressings had a wound diameter of 2.46 ± 0.57 cm after two weeks, while patients treated with saline gauze had a diameter of 3.90 ± 0.76 cm indicating the significant effect of insulin.40 Similarly, a mucoadhesive liposomal gel of insulin was explored on patients with chronic wounds in different body parts, demonstrating a more significant impact on test groups than control groups.41
The cobalt ions also help in wound healing and act as an alternative to hypoxia-mimicking agents. It inhibits the activity of PHDs (prolyl 4-hydroxylases), artificially stabilizes the HIF-1α (hypoxia-inducible factor-1α) under normal conditions, and activates the HIF pathway,42 allowing the formation of the HIF complex, which gets translocated to the nucleus and causes upregulation in the expression of genes such as VEGF, which are responsible for adaptation to hypoxia.43 As a result, it promotes angiogenesis during the tissue regeneration process.44 It was also found that incorporating cobalt ions in titanium-based bone implants enhances their antibacterial activity, indicating the role of cobalt as an antimicrobial agent.45 Further, the release of cobalt ions from the bioactive glass increases the VEGF protein secretion and osteogenic expression of genes by human bone marrow stromal cells.46,47 Moreover, if the cobalt ions are delivered as cobalt chloride, they enhance the secretion of VEGF by endothelial cells,48 fibroblasts,49 and human mesenchymal stem cells.50 The synthesis of silicate glass fibers for delivering the cobalt ions was done for their chronic wound healing activity by activating the HIF pathway and enhancing the expression of angiogenic genes.51 Similarly, a bioactive glass was made using the sol–gel method and doped with two different metal ions, including silver and cobalt, for their antibacterial activity in the case of wound healing applications.52
In this work, we are reporting the synthesis of novel protein-coated metal core–shell nanoparticles for their in vitro normal and diabetic wound healing and bioimaging application for the first time. Here, by using one-pot synthesis, we have prepared insulin–cobalt core–shell nanoparticles to check the synergistic effect of insulin and cobalt for treating wounds when applied in the form of particles in the nano range. The effective and efficient healing of wounds is essential for maintaining a person's life quality. Still, in some instances, the absence of proper internal wound monitoring makes the process even more complex, increasing the cost of treatment and putting risk to a patient's life. To overcome such challenges, the utilization of developed nanoparticles with dual healing and bioimaging capabilities offers a promising solution for enhanced wound management.
2. Materials and methods
2.1 Chemicals and cell line
The metal salt, cobalt chloride (CoCl2·6H2O), tryptophane, formaldehyde, HCl, and NaOH were of analytical grade and were purchased from Sigma Aldrich, India. Recombinant human insulin was purchased from Elli Lilly, India. For cell culture, DMEM cell culture media, Fetal Bovine Serum (FBS), 100× penicillin–streptomycin, and phosphate buffer solution (PBS) having pH 7.4 were purchased from HiMedia, India. Human Primary Epithelial Keratinocytes (HEKa cells) ATCC-PCS-200-011 were procured from Himedia, India, cultured, maintained, and treated in DMEM containing 5% FBS at 37 °C and 5% CO2.2.2 Synthesis of insulin-protected cobalt nanoparticles
At first, the insulin-protected core–shell cobalt nanoparticles were prepared by following a previously reported one-pot method in which the pH of the insulin solution having a final concentration of 1.82 μM, was adjusted to 10.5 using a NaOH solution (0.1 N) and kept in the dark. Afterward, a salt solution (CoCl2·6H2O) of the same molarity was prepared, followed by mixing two solutions in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by volume, which was further followed by adjustment of the pH of the final solution to a physiological pH of 7.4, using HCl (0.1 N). The resulting solution was kept in the incubator at slow stirring (240 rpm) for 24 h at 37 °C.53–55 The final solution was dialyzed using a 10 kDa cut-off dialysis membrane, stored at 4 °C, and used further for characterization. Thus, produced insulin–cobalt nanoparticles were termed as ICoNPs.
1 by volume, which was further followed by adjustment of the pH of the final solution to a physiological pH of 7.4, using HCl (0.1 N). The resulting solution was kept in the incubator at slow stirring (240 rpm) for 24 h at 37 °C.53–55 The final solution was dialyzed using a 10 kDa cut-off dialysis membrane, stored at 4 °C, and used further for characterization. Thus, produced insulin–cobalt nanoparticles were termed as ICoNPs.
2.3 Study of particle size, morphology, and elemental analysis
The DLS (dynamic light scattering) was done to determine the hydrodynamic size of ICoNPs formed using a Malvern DLS-Zeta size analyzer. Thereafter, to find out the morphology of insulin-linked cobalt nanoparticles, Scanning Electron Microscopy (SEM JEOL, JSM-6300) and High-Resolution Transmission Electron Microscopy (HRTEM) (Talos F200S G2, Thermo Scientific) were used. For this, the samples were centrifuged at 240 rpm for 15 minutes, then thoroughly washed the pellet to remove unbound metal salt or impurities associated with the sample. Furthermore, the sample pellet was investigated to determine the percentage of the elements present by the Energy dispersive X-ray Spectrometer (EDS) (Bruker QUANTAX 200).2.4 In silico studies
To determine the potential binding sites for different transition metal ions with the desired protein, MIB (Metal Ion Binding), an online docking tool, can be used as a user-friendly and integrated approach for finding the residues in metal binding sites. For operating this online server, protein chains were required, which were extracted from the protein data bank (PDB). The first chain consists of the template protein (T) of length m containing metal-ion, and the second chain consists of query protein (S) of length n. These chains were then aligned properly so the metal ion binding protein template could be converted into the query protein structure after alignment. Several different parameters were followed to obtain the desired protein structures. The first and the most crucial parameter is the prediction of binding residues with the twelve transition metal ions, including Ni2+, Cu2+, Mg2+, Ca2+, Hg2+, Cd2+, Co2+, Zn2+, Fe2+, Mn2+, Cu+, and Fe3+. The second factor is that the polypeptide chain length in the protein structures should have 50 residues; otherwise, it will be excluded.56 The third parameter is the presence of at least two metal ion binding residues having the minimum possible distance between the PDB coordinates and the metal center.57 The fourth crucial parameter is the allotment of a binding score to the residue, which should be greater than a specified threshold value assigned and calculated using the root mean square deviation of C-alpha carbons of local structural alignment and BLOSUM62 substitution. If so, only then will there be binding between the residue and the particular transition metal ion. Further, the binding score is denoted as CI and depends upon the target protein's sequence along with the protein's structural conservation.58,59 Hence, the binding site of metal ions was determined using the bioinformatics tool.In this manner, the binding site of a particular metal with that of a protein chain can be determined using the MIB tool. The PDB (Protein Data Bank) database was used to extract human insulin protein (PDB ID: 4EWW), a polypeptide with two distinct chains denoted as A and B, respectively. In the MIB tool, the Co2+ metal ion was independently docked with chains A and B of human insulin, followed by a comparison of the metal ion-binding score with that of the target protein.60
2.5 Study of cobalt–insulin interaction using spectroscopic techniques
To study the metal–protein interactions in the ICoNPs, FTIR was performed using an Agilent Cary 600 series Spectrophotometer. The potassium bromide (KBr) method was used to prepare the samples, and scanning was done from 400 cm−1 to 4000 cm−1.Furthermore, the same samples were analyzed using Surface Enhanced Raman Scattering (SERS) Spectra to study the structural changes in insulin protein.63 All the samples were prepared on a silicon wafer ten minutes before the measurement. The samples were scanned from 500 cm−1 to 1800 cm−1. The LabRam Hr Evolution Horiba, equipped with a detector and microscope, was used to record the Raman spectra of insulin and ICoNPs at 785 nm.
In order to monitor the changes in the secondary structure of the protein molecule, CD (Circular Dichroism) is the most reliable technique. In addition to the above studies, CD studies were conducted to confirm the interaction between insulin and cobalt chloride salt solution (freshly prepared and after 1 month). We have performed the Circular Dichroism (CD) spectroscopy using the instrument Mos500 CD biologic. The CD studies were performed at 25 °C with 1 ml of sample and scanned in the 200–260 nm wavelength range for pure insulin and insulin cobalt nanoparticles using phosphate buffer (pH 7.4) as a solvent.
2.6 Drug-loading and release kinetics
To monitor the release kinetics, 1 ml of the ICoNPs was centrifuged for 15 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The Bradford reagent determined insulin concentrations from the supernatant and the pellet. Further, the insulin release rate was measured at specific time intervals for 40 hours. The absorption values (at 595 nm) were later plotted to note the trend of drug release using BSA standard curves.61,62 In addition, absorbance values were also measured for the same samples at 272 nm (excitation of tryptophan) to support the data further.
000 rpm. The Bradford reagent determined insulin concentrations from the supernatant and the pellet. Further, the insulin release rate was measured at specific time intervals for 40 hours. The absorption values (at 595 nm) were later plotted to note the trend of drug release using BSA standard curves.61,62 In addition, absorbance values were also measured for the same samples at 272 nm (excitation of tryptophan) to support the data further.
2.7 Absorption and fluorescence spectroscopic study
For obtaining the UV-visible absorbance, the UV-2600 spectrophotometer of Shimadzu was operated between 200–800 nm, and a 4000 μl quartz cuvette with a 1 cm path was used, and the absorbance for insulin, salt solution (CoCl2), and insulin linked cobalt core–shell nanoparticles (ICoNPs) were measured.For measuring the fluorescence data of ICoNPs, Agilent technologies, a Cary Eclipse fluorescence spectrophotometer was used, which helps in determining the fluorescence intensity of protein-linked metal nanoparticles (ICoNPs), insulin, and CoCl2 solution, all having the same concentrations, at an excitation wavelength of 272 nm, coupled with an emission scan from 280 nm to 800 nm with excitation and emission slit of 10 mm.
In order to calculate and compare the percentage increase in intrinsic fluorescence intensity of both insulin and ICoNPs, the given eqn (a) was used.
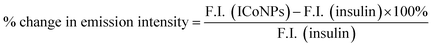 | (a) |
The standard tyrosine fluorescence quantum yield was used to calculate the quantum yield of insulin as well as insulin-protected cobalt nanoparticles (ICoNPs), and the given eqn (b) was used for further calculations.
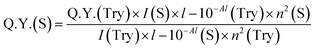 | (b) |
2.8 Stoichiometry ratio of Co2+ ions![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) insulin protein
insulin protein
To understand the interactions between metal–ligand complexes and complex proteins, the stoichiometry of cobalt ions to insulin protein is calculated using the following eqn (c).
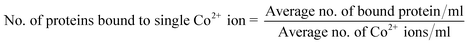 | (c) |
2.9 In vitro studies
| % inhibition = [1 − (At/Ac) × 100]% | (d) |
2.10 Determination of combination index (CI) for cobalt–insulin
The quantitative measure that provides the effect of a single drug in combination with another drug is Combination Index (CI). The level of synergism or antagonism is quantified by estimating the drug combination index in investigating synergistic or antagonistic drug combinations. The combination index of less than 1 (CI < 1) indicates that when different drugs are administered together, they work together to enhance each other's activity, called the synergistic effect. CI values equal to 1 (CI = 1) show that one drug does not interfere with the action of the other hence additive effect, and CI values greater than 1 (CI > 1) indicates the inhibition of drug action by the other drug, thus called the antagonistic effect. For the calculation of the combination index, the cell viability of HEKa cells was determined at varying concentrations of cobalt and insulin, and then calculations were done using eqn (e).| CI = (D)1/(Dx)1 + (D)2/(Dx)2 | (e) |
| Dx = Dm[fa/fu]1/m | (f) |
2.11 Statistical analysis
The data here is presented as the mean ± SD of at least three independent experiments. Using the one-way ANOVA, the statistical data analysis was done using MS Excel, and the corresponding p-value was calculated to check if the data was statistically significant.3. Results and discussions
3.1 Structure, composition, and stoichiometry of metal insulin nanoparticles
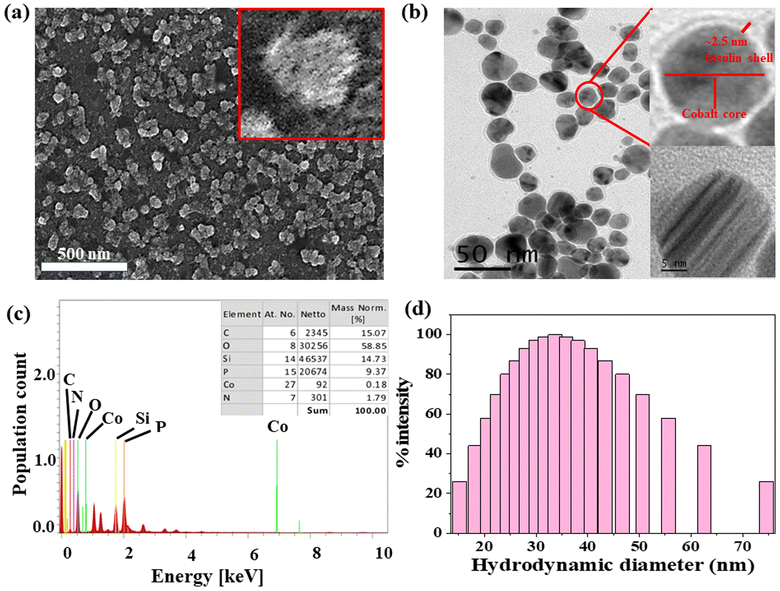
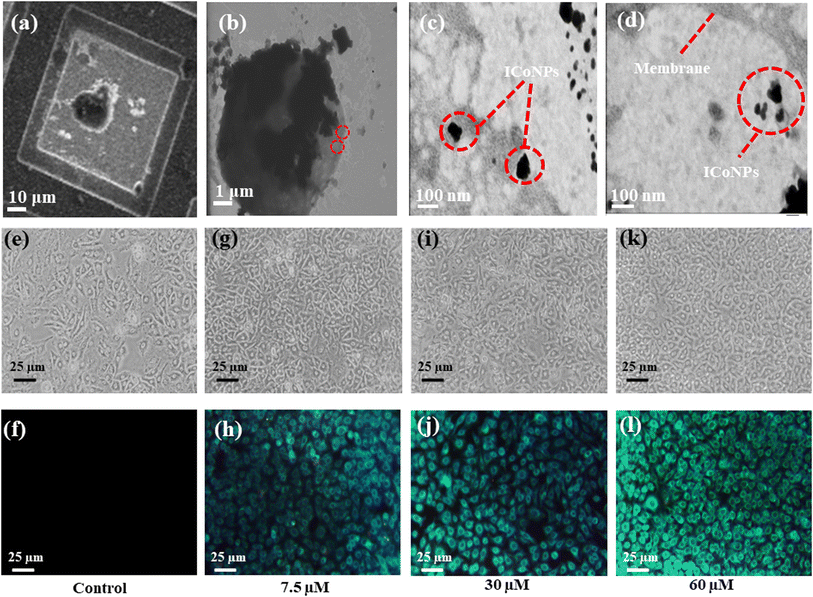
The formation of spherical nanoparticles of ICoNPs having size 13 ± 2 nm was confirmed by scanning electron microscopic images (at a scale of 500 nm) as shown in Fig. 1a (inset shows the magnified image of core–shell particle which shows the rough surface of nanoparticle due to an outer insulin shell) and transmission electron microscopic images (at a scale of 50 nm) as shown in Fig. 1b and inset of Fig. 1b (at a scale of 5 nm to show the lattice fringes and the insulin coating of ∼2.5 nm around the cobalt core). The nanoparticles are widely distributed over the protein matrix. The presence of different elements like Co, C, O, N, P, and Si is shown in the energy-dispersive X-ray spectroscopy. The even distribution and percentage of Co in the particles come out to be 0.18% as analyzed by EDS studies as indicated in Fig. 1c. The hydrodynamic size of ICoNPs was found to be 35 ± 5 nm as shown in Fig. 1d. Thereafter, the stoichiometric ratio between the insulin protein and Co2+ ions was calculated. It was found that ∼3792 insulin proteins encapsulate one nanoparticle. The detailed calculation is provided in ESI.†3.2 In silico studies to monitor the interactions between insulin and cobalt ions using metal-ion binding residue templates
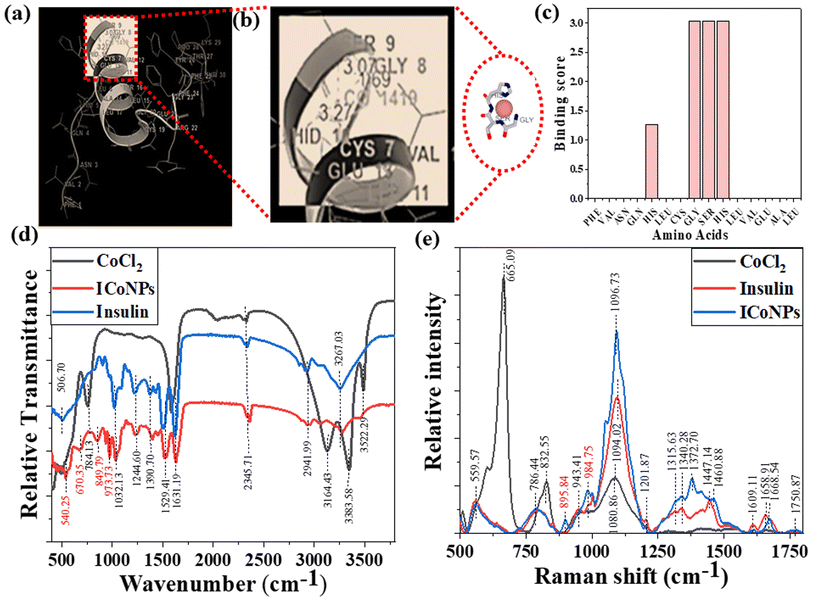
An online docking server determined the interaction sites between protein and metal ions, MIB (Metal Ion Binding). With this tool's help, we found the binding sites on chain B of insulin with which the transition metal Co2+ can bind. The interaction of the protein with metals is based mainly on the structure and sequence of the amino acids in a protein. Firstly, the sequence of human insulin was extracted from the Protein Data Base (PDB ID: 4EWW) and then inserted in the MIB (Metal Ion Binding) tool to dock the Co2+ ions with both the chains (chain A & B) of insulin. As a result, a binding score was assigned to each residue in the insulin protein. If the binding score exceeds the threshold, that particular residue is considered the binding site for that specific metal ion. Therefore, the binding sites for metal ions Co2+ were determined. The data indicates that chain A does not show any template indicating the absence of binding sites of Co2+ ions with chain A, whereas, in chain B, a binding site was found for Co2+ ions. Then we worked with another software called Maestro. Using this, we calculated the distance between the metal ion and its binding site, that is, amino acids, namely 8 GLY, 9 SER, and 10 HIS, present on chain B of insulin. The distance was found to be 1.69, 3.07, and 3.27, respectively. The binding score was also determined using MIB with different amino acids in chain B of insulin with Co2+ ions, thus finding the best possible sites for attachment. The binding of cobalt ions with different amino acids is determined and visualized through different orientations of the metal-binding amino acids in proteins. The possible binding sites of cobalt ions with different amino acids in insulin are shown in Fig. 2a and b, the binding score of amino acids with insulin protein is shown in Fig. 2c, and the binding score values are given in Table S1.†3.3 FTIR, Raman spectra, and CD spectroscopy for the interactions between protein and metal ions after the formation of nanoparticles
To figure out the interactions involved in metal–protein binding, FTIR was used for CoCl2, insulin, and ICoNPs. The peaks appeared due to intermolecular interactions of insulin and CoCl2 at different wavenumbers. Firstly, the peak appeared at 506.70 cm−1, which indicates the S–S stretching in insulin and ICoNPs.32 Then a peak at 540.25 cm−1 in ICoNPs indicates Co–O vibrations.67 In addition to the second peak, a peak at 670.35 cm−1 shows the Co–O bending, confirming the interaction between insulin and CoCl2.68 A peak at 784.13 cm−1 in CoCl2 shows librational vibrations.69 Two other peaks at 849.79 and 973.73 indicate the CoO–OH band and Co–OH bending, respectively, in ICoNPs.67,70 C–N stretching was observed in insulin and ICoNPs at 1032.13 cm−1.31 Amide I, amide II, and amide III are found at 1631.19 cm−1, 1529.41 cm−1, and 1244.60 cm−1, respectively, in insulin and ICoNPs.31,32,34 C–H bond was observed in insulin and ICoNPs at 1390.70 cm−1.31 Deformational scissor water vibrations were observed at 1573.72 cm−1 in CoCl2 only.69 Nitrile stretch was there at 2345.71 cm−1 in all three solutions.34 Intermolecular O–H stretching peaks at 2941.99 cm−1 in insulin and ICoNPs, but the peak was observed at 3164.13 cm−1 in CoCl2.31 A single peak in insulin and nanoparticles indicated amine N–H stretching at 3267.03 cm−1, which was absent in the CoCl2.71 For symmetrical and asymmetrical valence vibrations of water, peaks were observed at 3383.58 cm−1 and 3522.29 cm−1, both in CoCl2 but absent in insulin and ICoNPs,69 which is shown in Fig. 2d.Similarly, Raman spectral analysis was used to study the significant conformational changes in insulin after binding with cobalt ions. The S–S stretch and O–C–N bend were observed at 553.29 cm−1 and 786.44 cm−1, respectively, in both insulin and ICoNPs.32,72 The CoO–OH and Co–OH bond was observed at 895.84 cm−1 and 984.75 cm−1, respectively, only in ICoNPs but was absent in insulin.67,70 A weak C–N stretch was observed at 1094.02 cm−1 in insulin and 1096.73 cm−1.32 The C–H bend was found in insulin at 1343.21 cm−1 and in ICoNPs at 1372.70 cm−1.32 The peak that is responsible for amide-III random coils shifts slightly from 1315.63 cm−1 in insulin to 1332.38 cm−1 in ICoNPs.73 However, no such change was observed for amide-III (α-helix), as the peak appears at 1201.87 cm−1 in insulin and ICoNPs.74 A slight shift was observed for amide-II from 1447.14 cm−1 in insulin to 1460.88 cm−1 in ICoNPs.75 The peak for amide-I was observed at 1658.91 cm−1 and 1668.54 cm−1, respectively, in insulin and ICoNPs.76 The peak for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch remains unchanged and is found at 1750.73 cm−1 in insulin and ICoNPs,32 and this is shown in Fig. 2e. A comparative data of FTIR and Raman spectra are shown in Table 1.
O stretch remains unchanged and is found at 1750.73 cm−1 in insulin and ICoNPs,32 and this is shown in Fig. 2e. A comparative data of FTIR and Raman spectra are shown in Table 1.
| FTIR | Raman | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Functional groups | CoCl2 | Insulin | ICoNPs | References | Functional group | CoCl2 | Insulin | ICoNPs | References |
| S–S stretch | — | 506.70 | 506.70 | 32 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch O stretch |
— | 1750.73 | 1750.73 | 32 |
| Co–O vibrations | — | — | 540.25 | 67 | Amide I | — | 1658.91 | 1668.54 | 76 |
| Co–O stretching | — | — | 670.35 | 68 | Amide II | — | 1447.14 | 1460.88 | 75 |
| Librational vibrations | 784.13 | — | — | 69 | Amide III (α – helix) | — | 1201.87 | 1201.87 | 74 |
| CoO–OH band | — | — | 849.79 | 67 | Amide III (random coils) | — | 1315.63 | 1332.38 | 73 |
| Co–OH bending | — | — | 973.73 | 70 | C–H bend | — | 1343.21 | 1372.70 | 32 |
| C–N stretching | — | 1032.13 | 1032.13 | 31 | C–N (weak stretch) | — | 1094.02 | 1096.73 | 32 |
| Amide III | — | 1244.60 | 1244.60 | 31 | Co–OH | 1080.66 | — | 984.75 | 70 |
| C–H bond | — | 1390.70 | 1390.70 | 31 | CoO–OH | 832.55 | — | 895.84 | 67 |
Amide II C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch O stretch |
— | 1529.41 | 1529.41 | 34 | O–C–N (bend) | — | 786.44 | 786.44 | 32 |
| Deformational scissor vibration of water | 1573.72 | — | — | 69 | S–S stretch | 665.09 | 553.29 | 553.29 | 68 |
| Amide I | — | 1631.19 | 1631.19 | 32 | |||||
| Nitrile stretch | 2345.71 | 2345.71 | 2345.71 | 34 | |||||
| O–H intermolecular stretching | 3164.13 | 2941.99 | 2941.99 | 31 | |||||
| Amine N–H stretch | — | 3267.03 | 3267.03 | 71 | |||||
| Symmetrical and asymmetrical valence vibrations of water | 3383.58 | — | — | 69 | |||||
| 3522.29 | |||||||||
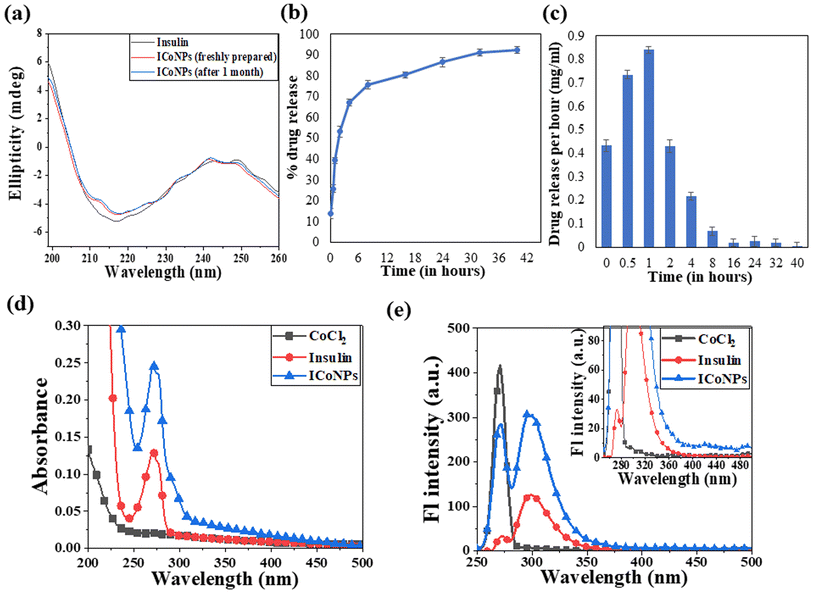
Circular dichroism was performed using insulin protein and ICoNPs (freshly prepared and after 1 month) to monitor the interactions between protein and salt solution and find the stability of the secondary structure of protein after binding with cobalt chloride. The three significant far UV signals were recorded in both the solutions, including a positive peak at ∼200 nm and two negative peaks at ∼208 nm (α-helix) and 218 nm (β sheet), which were indicative of the secondary protein structure. The % variation in the peak at 200 nm is 5.23% and 6.21%, at 208 nm is 3.56% and 4.24% and at 218 nm is 4.82% and 5.71% in freshly prepared ICoNPs and ICoNPs after 1 month; respectively and confirmed the protein stability with no significant structural changes were observed, as shown in Fig. 3a.
3.4 Drug loading and release kinetic studies
The drug loading capacity and release kinetics of ICoNPs were studied, and the encapsulation efficiency of insulin over the cobalt nanoparticles was found to be 89.48 ± 0.049%. The release kinetic studies were performed at physiological conditions that are pH of 7.4 and a physiological temperature of 37 °C. Burst drug release was observed for the initial 8 h, followed by a sustained drug release afterward. Most of the drug was released by the 40 h. The particles were suspended in a dialysis membrane with a concentration of 3.47 mg ml−1, and the OD values were observed at 595 nm after particular intervals of time. It was found that 92.21 ± 0.03% of the drug was released from ICoNPs by the end of 40 hours, making it an efficient drug delivery system in wound healing applications and is shown in Fig. 3b and c. For further confirmation of the release kinetics of the particles, absorbance value was taken at 272 nm (tryptophan absorbance) for all the samples after specific time intervals which shows the gradual increase in OD values with increasing time, indicating the sustained release of insulin from particles and is shown in Fig. S1a.†3.5 Spectroscopic changes after synthesis of core–shell nanoparticles using absorbance and fluorescence spectra
After synthesizing ICoNPs, its spectroscopic characterization was done to observe the absorption and fluorescence intensity of the formed nanoparticles. We obtained a sharp peak for insulin without any added metal salts at 271.61 with an absorbance value of 0.129, whereas, after incubating the insulin with metal salts for 48 h, an absorbance value of 0.245 was obtained for nanoparticles having a peak at 271.50 nm. This confirms the synthesis of ICoNPs, as shown in Fig. 3d. The absorption peak is also compared with tyrosine, as it was selected as a standard and demonstrated in Fig. S1b.†The ICoNPs were then excited at an excitation wavelength of 270 nm to determine the fluorescence intensity. An emission peak ranging from 280–440 nm was obtained with maxima at ∼303 nm with an intensity of 304.89 a.u when monitored from 200–800 nm, as shown in Fig. 3e. The % change in intrinsic fluorescence intensity was found to be 58.88%. Further, to determine the quantum yield of ICoNPs, tyrosine was used as a standard as its quantum yield value is known. The quantum yield of insulin was found to be 0.1798, and that of ICoNPs turned out to be 0.6825, indicating the fluorescent behavior of formulations synthesized and is shown in Fig. S1c.†
3.6 In vitro cell studies
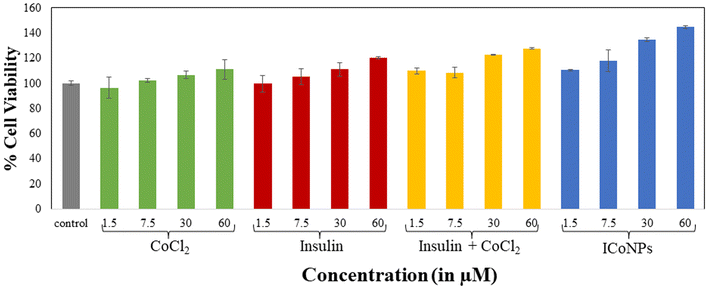
| % Of change in mitochondrial reductase activity | ||||
|---|---|---|---|---|
| Dose | CoCl2 | Insulin | Insulin + CoCl2 | ICoNPs |
| 1.5 μM | 96.54 ± 6.43 | 99.65 ± 8.14 | 109.97 ± 2.30 | 110.44 ± 0.70 |
| 7.5 μM | 102.12 ± 6.36 | 105.02 ± 1.48 | 108.32 ± 4.17 | 117.83 ± 8.76 |
| 30 μM | 106.75 ± 5.58 | 110.99 ± 2.89 | 122.78 ± 0.28 | 134.72 ± 1.76 |
| 60 μM | 110.99 ± 0.77 | 120.26 ± 7.70 | 127.57 ± 0.56 | 144.69 ± 0.98 |
| Combination index (CI) | |||
|---|---|---|---|
| The concentration of Co salt and insulin | (Dx)1 (Co salt) = Dm[fa/fu]1/m | (Dx)2 (insulin) = Dm [fa/fu]1/m | CI = (D)1/(Dx)1 + (D)2/(Dx)2 |
| 1.5 μM | 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 118.86 118.86 |
171![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 484.6 484.6 |
0.0000265 |
| 7.5 μM | 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 757.25 757.25 |
128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844.6 844.6 |
0.00018188 |
| 30 μM | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 284.49 284.49 |
96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861.06 861.06 |
0.00098882 |
| 60 μM | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936.43 936.43 |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800.79 800.79 |
0.00329352 |
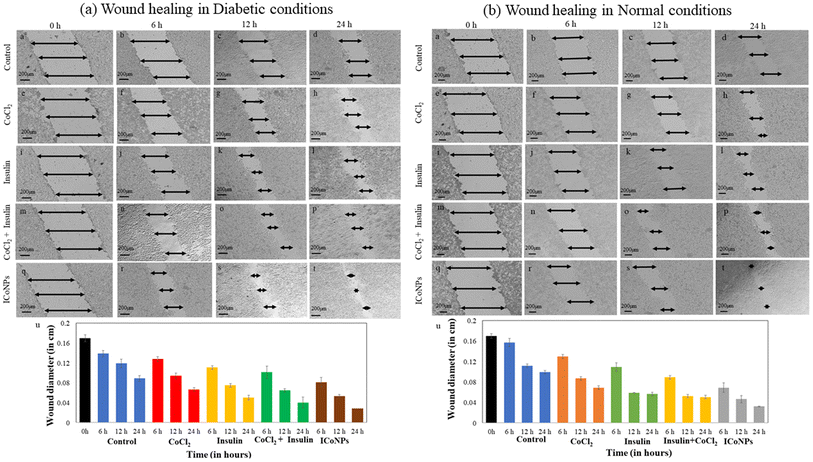
Similarly, in the normal wound, the variation in scratch diameter for control cells is shown in Fig. 6(b)a–d. The % change in wound diameter is monitored after the same time interval in normal conditions and found to be 14.84 ± 0.60%, 31.75 ± 0.35%, and 34.7 ± 0.70% in the cells treated with CoCl2 (Fig. 6(b)e–h). In the cells treated with insulin, insulin + CoCl2, the % variation in migration with time is more as compared to CoCl2 and is equal to 30.33 ± 0.80%, 41.27 ± 0.70%, and 48.98 ± 0.35% in insulin (Fig. 6(b)i–l), 40.65 ± 0.35%, 50.8 ± 0.35% and 59.19 ± 0.35% in insulin + CoCl2 (Fig. 6(b)m–p). The core–shell ICoNPs show the maximum migration, and the % wound diameter changed is 56.13 ± 0.92%, 63.50 ± 0.70%, and 71.43 ± 0.35%, respectively, indicating the significance of prepared nanoparticles in the healing application of normal wounds (Fig. 6(b)q–t). The graph in Fig. 6(b)u shows the variation in normal wound diameter as a comparative study of all the formulations after particular time intervals. A comparative data table of change in wound migration in normal and diabetic wounds is shown in Table S2.† The statistical significance of data was determined by calculating p values for the scratch assay in diabetic and normal wound conditions using one-way ANOVA. The comparative data are presented in Table S3.†
3.7 Combination index of cobalt and insulin
To calculate CI, Dm was calculated using m and y from Fig. S2a† for cobalt and Fig. S2b† for insulin. It was found from the CI's calculated values that the cobalt chloride salt and insulin show a synergistic effect. The values calculated for varying combinations of insulin and cobalt chloride concentrations come out to be less than one indicating the synergism between the two and, thus, work together to enhance each other's activity. This is in accordance with the results obtained for cell viability and cell migration assay indicating the significant role of insulin cobalt nanoparticles in faster wound healing applications. The data is shown in Table 2.4. Conclusion
The wounds were sometimes reopened to maintain the proper healing environment and eliminate any complications in the wound recovery process, paving the way for improper healing, enhanced healthcare costs, and decreased life quality. Thus, suitable agents are always required, which assist in selectively binding to the wound site and guide in monitoring the progress of wound recovery. We formulated novel core–shell ICoNPs containing insulin and cobalt and evaluated their role in wound healing activity. Insulin promotes cellular growth by modulating inflammation, reduces blood glucose concentration, and exhibits anti-inflammatory activity. Cobalt is known to reduce the microbial contamination load. Thus, the formulations were checked for a synergistic effect in wound treatment. Different techniques were used to confirm their formation, including TEM micrography, elemental analysis, FTIR, Raman spectroscopy, and metal ion binding site prediction and docking server (MIB) to ensure Co–metal interaction with the insulin protein. From FTIR and Raman spectra, we got peaks showing the intermolecular interaction between Co and insulin and exhibiting moderate structural changes in amide regions of the protein. The metal–ion binding site prediction and docking server were used to find possible sites for the metal ion to bind with insulin amino acid chains A and B. No site was observed in chain A, while in chain B, there was a binding site for Co2+ ions, and the amino acids involved in groove formation were 8 GLY, 9 SER, and 10 HIS. The loading efficiency and release kinetic studies were done to determine the effective loading of insulin drugs over the nanoparticles. The synthesized core–shell nanoparticles were tested for their effect on cell viability to be used further for diabetic and normal wound healing and bioimaging applications. The calculated values for the MTT assay and combination index values indicate the synergism between the cobalt chloride salt and insulin protein, making them suitable for further utilization in biological activities. The formed core–shell nanoparticles, ICoNPs, show excellent target-specificity. Due to the insulin consumption in their synthesis, they exhibit the property of binding to insulin receptors, making these particles unique from other particles in various respects, mainly in target-specificity properties. Further, they have enhanced fluorescent properties when compared with insulin alone which can be easily detected using fluorescence microscopy and fluorescence bioimaging. They also possess an enormous tendency to heal both diabetic and normal wounds. Thus, it has a wide range of applications in healing, bioimaging, and material and biomaterial sciences. Therefore, it can be further explored for in vivo applications in diverse fields.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
The authors are thankful to Thapar Institute of Engineering and Technology for providing them with all the facilities. Authors are grateful to TIET, the library, for providing access to journals. DS is willing to thank TIET-VT CEEMS and ICMR-Govt of India (Project No. 17X(3)/Adhoc/63/2022-ITR) for the fellowship. No. specific funding has been received for this work.References
- I. Pastar, O. Stojadinovic, N. C. Yin, H. Ramirez, A. G. Nusbaum, A. Sawaya, S. B. Patel, L. Khalid, R. R. Isseroff and M. Tomic-Canic, Epithelialization in Wound Healing: A Comprehensive Review, Adv. Wound Care, 2014, 3, 445–464 CrossRef PubMed.
- G. A. Duque and A. Descoteaux, Macrophage cytokines: involvement in immunity and infectious diseases, Front. Immunol., 2014, 5, 491, DOI:10.3389/fimmu.2014.00491.
- J. E. Janis and B. Harrison, Wound Healing: Part I. Basic Science, Plast. Reconstr. Surg., 2016, 138, 9S–17S CrossRef CAS PubMed.
- P. H. Wang, B. S. Huang, H. C. Horng, C. C. Yeh and Y. J. Chen, Wound Healing, J. Chin. Med. Assoc., 2018, 81, 94–101 CrossRef PubMed.
- L. Teot and N. Ohura, Challenges and Management in Wound Care, Plast. Reconst. Surg., 2021, 147, 9S–15S CrossRef CAS.
- K. L. Heras, M. Igartua, E. Santos-Vizcaino and R. M. Hernandez, Chronic wounds: Current status, available strategies and emerging therapeutic solutions, J. Controlled Release, 2020, 328, 532–550 CrossRef PubMed.
- T. N. Demidova-Rice, M. R. Hamblin and I. M. Herman, Acute and impaired wound healing: pathophysiology and current methods for drug delivery, part 1: normal and chronic wounds: biology, causes and approaches to care, Adv. Skin Wound Care, 2012, 25, 304–314 CrossRef.
- A. Sood, M. S. Granick and N. L. Tomaselli, Wound dressings and Comparative Effectiveness Data, Adv. Wound Care, 2014, 3, 511–529 CrossRef.
- J. L. Burgess, W. A. Wyant, B. Abdo Abujamra, R. S. Kirsner and I. Jozic, Diabetic wound-healing Science, Medicina, 2021, 57, 1072 CrossRef PubMed.
- R. G. Frykberg and J. Banks, Challenges in the Treatment of Chronic Wounds, Adv. Wound Care, 2015, 4, 560–582 CrossRef.
- J. A. Beckman, M. A. Creager and P. Libby, Diabetes and atherosclerosis: epidemiology, pathophysiology and management, JAMA, J. Am. Med. Assoc., 2002, 287, 2570–2581 CrossRef CAS PubMed.
- A. Koïtka, P. Abraham, B. Bouhanick, D. Sigaudo-Roussel, C. Demiot and J. L. Saumet, Impaired pressure-induced vasodilation at the foot in young adultswith type 1 diabetes, Diabetes, 2004, 53, 721–725 CrossRef.
- A. Lima, T. Illing, S. Schliemann and P. Elsner, Cutaneous Manifestations of Diabetes Mellitus: A review, Am. J. Clin. Dermatol., 2017, 18, 541–553 CrossRef PubMed.
- T. A. M. Andrade, D. S. Masson-Meyers, G. F. Caetano, V. A. Terra, P. P. Ovidio, A. A. Jordão-Júnior and M. A. C. Frade, Skin changes in streptozotocin-induced diabetic rats, Biochem. Biophys. Res. Commun., 2017, 490, 1154–1161 CrossRef CAS PubMed.
- J. H. Kim, N. Y. Yoon, D. H. Kim, M. Jung, M. Jun, H. Y. Park, C. H. Chung, K. Lee, S. Kim, C. S. Park, K. H. Liu and E. H. Choi, Impaired permeability and antimicrobial barriers in type 2 diabetes skin are linked to increased serum levels of advanced glycation end-product, Exp. Dermatol., 2018, 27, 815–823 CrossRef CAS PubMed.
- J. Dworzański, M. Strycharz-Dudziak, E. Kliszczewska, M. Kiełczykowska, A. Dworzańska, B. Drop and M. Polz-Dacewicz, Glutathione peroxidase (GPx) and Superoxide Dismutase (SOD) activity in patients with diabetes mellitus type 2 infected with Epstein- Barr virus, PLoS One, 2020, 15, e0230374, DOI:10.1371/JOURNAL.PONE.0230374.
- S. Matoori, A. Veves and D. J. Mooney, Advanced bandages for diabetic wound healing, Sci. Transl. Med., 2021, 13, eabe4839 CrossRef CAS.
- D. Sharda, K. Attri and D. Choudhury, Future research directions of antimicrobial wound dressings, Antimicrob. Dressings, 2023, pp. 229–246 Search PubMed.
- N. Naderi, D. Karponis, A. Mosahebi and A. M. Seifalian, Nanoparticles in wound healing; from hope to promise, from promise to routine, Front. Biosci., 2018, 23, 1038–1059 Search PubMed.
- E. T. Goh, G. Kirby, R. Jayakumar, X.-J. Liang and A. Tan, Accelerated wound healing using nanoparticles, Nanoscience in dermatology, Elsevier, 2016, pp. 287–306 Search PubMed.
- N. K. Rajendran, S. S. D. Kumar, N. N. Houreld and H. Abrahamse, A review on nanoparticle based treatment for wound healing, J. Drug Delivery Sci. Technol., 2018, 44, 421–430 CrossRef CAS.
- B. Blanco-Fernandez, O. Castaño, M. Á. Mateos-Timoneda, E. Engel and S. Pérez-Amodio, Nanotechnology Approaches in Chronic Wound Healing, Adv. Wound Care, 2021, 10, 234–256 CrossRef PubMed.
- C. Mendes, A. Thirupathi, M. E. A. B. Corrêa, Y. Gu and P. C. L. Silveira, The Use of Metallic Nanoparticles in Wound Healing: New Perspectives, Int. J. Mol. Sci., 2023, 23(23), 15376 CrossRef.
- S. K. Nethi, S. Das, C. R. Patra and S. Mukherjee, Recent advances in inorganic nanomaterials for wound-healing applications, Biomater. Sci., 2019, 7, 2652–2674 RSC.
- J. S. Mohanty, P. L. Xavier, K. Chaudhari, M. S. Bootharaju, N. Goswami, S. K. Pal and T. Pradeep, Luminescent, bimetallic, AuAg alloy quantum clusters in protein templates, Nanoscale, 2012, 4, 4255–4262 RSC.
- P. Kaur and D. Choudhury, Functionality of receptor targeted zinc-insulin quantum clusters in skin tissue augmentation and bioimaging, J. Drug Targeting, 2021, 29, 541–550 CrossRef CAS.
- P. L. Xavier, K. Chaudhari, P. K. Verma, S. K. Pal and T. Pradeep, Luminescent quantum clusters of gold in transferrin family protein, lactoferrin exhibiting FRET, Nanoscale, 2010, 2, 2769–2776 RSC.
- M. Tarhini, H. Greige-Gerges and A. Elaissari, Protein-based nanoparticles: From preparation to encapsulation of active molecules, Int. J. Pharm., 2017, 522, 172–197 CrossRef CAS PubMed.
- K. Chaudhari, P. L. Xavier and T. Pradeep, Understanding the evolution of luminescent gold quantum clusters in protein templates, ACS Nano, 2011, 5, 8816–8827 CrossRef CAS.
- I. Negut, V. Grumezescu and A. M. Grumezescu, Treatment Strategies for Infected Wounds, Molecules, 2018, 23(9), 2392 CrossRef.
- D. Sharda, K. Attri, P. Kaur and D. Choudhury, Protection of lead-induced cytotoxicity using nickel-insulin quantum clusters, RSC Adv., 2021, 11, 24656–24668 RSC.
- P. Kaur, A. K. Sharma, D. Nag, A. Das, S. Datta, A. Ganguli, V. Goel, S. Rajput, G. Chakrabarti, B. Basu and D. Choudhury, Novel nano-insulin formulation modulates cytokine secretion and remodeling to accelerate diabetic wound healing, Nanomedicine, 2019, 15, 47–57 CrossRef CAS.
- A. Oryan and E. Alemzadeh, Effects of insulin on wound healing: A review of animal and human evidences, Life Sci., 2017, 174, 59–67 CrossRef CAS.
- P. Kaur, S. Sharma, S. D. Choudhury, D. Singh, S. Sharma, K. Gadhave, N. Garg and D. Choudhury, Insulin-copper quantum clusters preparation and receptor targeted bioimaging, Colloids Surf., B, 2020, 188, 110785, DOI:10.1016/j.colsurfb.2020.110785.
- P.-F. Chen, C.-L. Liu, W.-K. Lin, K.-C. Chen, P.-T. Chou and S.-W. Chu, Fluorescence depletion properties of insulin-gold nanoclusters, Biomed. Opt. Express, 2015, 6, 3066–3073 CrossRef CAS PubMed.
- M. C. Ribeiro, V. L. R. Correa, F. K. L. da Silva, A. A. Casas, A. de L. das Chagas, L. P. de Oliveira, M. P. Miguel, D. G. A. Diniz, A. C. Amaral and L. B. de Menezes, Wound healing treatment using insulin within polymeric nanoparticles in the diabetes animal model, Eur. J. Pharm. Sci., 2020, 150, 105330, DOI:10.1016/J.EJPS.2020.105330.
- C.-H. Lee, K.-C. Hung, M.-J. Hsieh, S.-H. Chang, J.-H. Juang, I.-C. Hsieh, M.-S. Wen and S.-J. Liu, Core-shell insulin-loaded nanofibrous scaffolds for repairing diabetic wounds, Nanomedicine, 2020, 24, 102123 CrossRef CAS PubMed.
- W. Li, F. Gao, J. Kan, J. Deng, B. Wang and S. Hao, Synthesis and fabrication of a keratin-conjugated insulin hydrogel for the enhancement of wound healing, Colloids Surf., B, 2019, 175, 436–444 CrossRef CAS PubMed.
- Z. Zhang and L. Lv, Effect of local insulin injection on wound vascularization in patients with diabetic foot ulcer, Exp. Ther. Med., 2016, 11, 397–402 CrossRef CAS.
- M. K. Bhittani, M. Rehman, H. N. Altaf and O. S. Altaf, Effectiveness of topical insulin dressings in management of diabetic foot ulcers, World J. Surg., 2020, 44, 2028–2033 CrossRef PubMed.
- M. H. S. Dawoud, G. E. Yassin, D. M. Ghorab and N. M. Morsi, Insulin mucoadhesive liposomal gel for wound healing: a formulation with sustained release and extended stability using quality by design approach, AAPS PharmSciTech, 2019, 20, 1–15 CrossRef CAS.
- L. O. Simonsen, H. Harbak and P. Bennekou, Cobalt metabolism and toxicology - a brief update, Sci. Total Environ., 2012, 432, 210–215 CrossRef CAS.
- M. A. C. Déry, M. D. Michaud and D. E. Richard, Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators, Int. J. Biochem. Cell Biol., 2005, 37, 535–540 CrossRef.
- J. Li, F. Lv, J. Li, Y. Li, J. Gao, J. Luo, F. Xue, Q. Ke and H. Xu, Cobalt-based metal-organic framework as a dual cooperative controllable release system for accelerating diabetic wound healing, Nano Res., 2020, 13, 2268–2279 CrossRef CAS.
- J. Zhou and L. Zhao, Multifunctional Sr, Co and F co-doped microporous coating on titanium of antibacterial angiogenic and osteogenic activities, Sci. Rep., 2016, 6, 1–14 CrossRef PubMed.
- C. Wu, Y. Zhou, W. Fan, P. Han, J. Chang, J. Yuen, M. Zhang and Y. Xiao, Hypoxia- mimicking mesoporous bioactive glass scaffolds with controllable cobalt ion release for bone tissue engineering, Biomaterials, 2012, 33, 2076–2085 CrossRef CAS.
- Q. Shi, X. Luo, Z. Huang, A. C. Midgley, B. Wang, R. Liu, D. Zhi, T. Wei, X. Zhou, M. Qiao, J. Zhang, D. Kong and K. Wang, Cobalt-mediated multi-functional dressings promote bacteria - infected wound healing, Acta Biomater., 2019, 86, 465–479 CrossRef CAS.
- A. Hoppe, A. Brandl, O. Bleiziffer, A. Arkudas, R. E. Horch, B. Jokic, D. Janackovic and A. R. Boccaccini, In vitro cell response to Co-containing 1, 393 bioactive glass, Mater. Sci. Eng., C, 2015, 57, 157–163 CrossRef CAS PubMed.
- A. K. Solanki, F. V. Lali, H. Autefage, S. Agarwal, A. Nommeots-Nomm, A. D. Metcalfe, M. M. Stevens and J. R. Jones, Bioactive glasses and electrospun composites that release cobalt to stimulate the HIF pathway for wound healing applications, Biomater. Res., 2021, 25, 1–16 CrossRef CAS PubMed.
- M. M. Azevedo, O. Tsigkou, R. Nair, J. R. Jones, G. Jell and M. M. Stevens, Hypoxia inducible factor-stabilizing bioactive glasses for directing mesenchymal stem cell behavior, Tissue Eng., Part A, 2015, 21, 382–389 CrossRef CAS PubMed.
- A. K. Solanki, H. Autefage, A. R. Rodriguez, S. Agarwal, J. Penide, M. Mahat, T. Whittaker, A. Nommeots-Nomm, E. Littmann and D. J. Payne, Cobalt conatining glass fibres and their synergistic effect on the HIF-1 pathway for wound healing applications, Front. Bioeng. Biotechnol., 2023, 11, 1125060 CrossRef PubMed.
- D. Moura, M. T. Souza, L. Liverani, G. Rella, G. M. Luz, J. F. Mano and A. R. Boccaccini, Development of bioactive glass-polymer composite for wound healing applications, Mater. Sci. Eng., C, 2017, 76, 224–232 CrossRef CAS PubMed.
- J. Xie, Y. Zheng and J. Y. Ying, Protein-directed synthesis of highly fluorescent gold nanoclusters, J. Am. Chem. Soc., 2009, 131, 888–889 CrossRef CAS PubMed.
- J. Lin, Z. Zhou, Z. Li, C. Zhang, X. Wang, K. Wang, G. Gao, P. Huang and D. Cui, Biomimetic one-pot synthesis of gold nanoclusters/nanoparticles for targeted tumor cellular dual-modality imaging, Nanoscale Res. Lett., 2013, 8, 1–7 CrossRef PubMed.
- C.-L. Liu, H.-T. Wu, Y.-H. Hsiao, C.-W. Lai, C.-W. Shih, Y.-K. Peng, K.-C. Tang, H.-W. Chang, Y.-C. Chien, J.-K. Hsiao, J.-T. Cheng and P.-T. Chou, Insulin-directed synthesis of fluorescent gold nanoclusters: preservation of insulin bioactivity and versatility in cell imaging, Angew. Chem., Int. Ed., 2011, 50, 7056–7060 CrossRef CAS PubMed.
- A. G. Murzin, S. E. Brenner, T. Hubbard and C. Chothia, SCOP: a structural classification of proteins database for the investigation of sequences and structures, J. Mol. Biol., 1995, 247, 536–540 CAS.
- G. Monnier, E. Frahm, B. Luo and K. Missal, Developing FTIR microspectroscopy for analysis of plant residues on stone tools, J. Archaeol. Sci., 2017, 78, 158–178 CrossRef CAS.
- C.-H. Lu, Y.-F. Lin, J.-J. Lin and C.-S. Yu, Prediction of metal ion-binding sites in proteins using the fragment transformation method, PLoS One, 2012, 7, 39252, DOI:10.1371/journal.pone.0039252.
- J. C. Gower and G. J. S. Ross, Minimum spanning trees and single linkage cluster analysis, Appl. State, 1969, 18, 54–64 CrossRef.
- C.-H. Lu, Y.-S. Lin, Y.-C. Chen, C.-S. Yu, S.-Y. Chang and J.-K. Hwang, The fragment transformation method to detect the protein structural motifs, Proteins: Struct., Funct., Bioinf., 2006, 63, 636–643 CrossRef CAS PubMed.
- S. Razavi, R. Seyedebrahimi and M. Jahromi, Biodelivery of nerve growth factor and gold nanoparticles encapsulated chitosan nanoparticles for Schwann like cells differentiation of human adipose-derived stem cells, Biochem. Biophys. Res. Commun., 2019, 513, 681–687 CrossRef CAS PubMed.
- M. J. Oviedo, K. Quester, G. A. Hirata and R. Vazquez-Duhalt, Determination of conjugated protein on nanoparticles by an adaptation of the Coomassie blue dye method, Methods X, 2019, 6, 2134–2140 Search PubMed.
- D. Choudhury, P. L. Xavier, K. Chaudhari, R. John, A. K. Dasgupta, T. Pradeep and G. Chakrabarti, Unprecedented inhibition of tubulin polymerization directed by gold nanoparticles inducing cell cycle arrest and apoptosis, Nanoscale, 2013, 5, 4476–4489 RSC.
- Y. Shi, C. Tian, X. Yu, Y. Fang, X. Zhao, X. Zhang and D. Xia, Protective effects of Smilax glabra Roxb. Against Lead-induced Renal Oxidative Stress, inflammation and apotosis in Weaning rats and HEK-293 cells, Front. Pharmacol., 2020, 11, 556248, DOI:10.3389/fphar.2020.556248.
- O. M. Ali, A. A. Bekhit, S. N. Khattab, M. W. Helmy, Y. S. Abdel-Ghany, M. Teleb and A. O. Elzoghby, Synthesis of lactoferrin mesoporous silica nanoparticle for pemetrexed/ellagic acid synergistic breast cancer therapy, Colloids Surf., B, 2020, 188, 110824 CrossRef CAS PubMed.
- D. Choudhury, A. Ganguli, D. G. Dastidar, B. R. Acharya, A. Das and G. Chakrabarti, Apigenin shows synergistic anticancer activity with curcumin by binding at different sites of tubulin, Biochimie, 2013, 95, 1297–1309 CrossRef CAS PubMed.
- C. W. Tang, C. Bin Wang and S. H. Chien, Characterization of cobalt oxides studied by FT-IR, Raman, TPR and TG-MS, Thermochim. Acta, 2008, 473, 68–73 CrossRef CAS.
- M. H. Habibi and Z. Rezvani, Nanostructure copper oxocobaltate fabricated by co-precipitation route using copper and cobalt nitrate as precursors: characterization by combined diffuse reflectance and FT infrared spectra, Spectrochim. Acta, Part A, 2014, 130, 309–312 CrossRef CAS PubMed.
- O. V. Netskina, A. A. Pochtar, O. V. Komova and V. I. Simagina, Solid-state NaBH4 Composites as Hydrogen Generation Material: Effect of Thermal Treatment of a Catalyst Precursor on the Hydrogen Generation Rate, Catal, 2020, 10, 201, DOI:10.3390/catal10020201.
- F. Zhang, C. Yuan, X. Lu, L. Zhang, Q. Che and X. Zhang, Facile growth of mesoporous Co3O4 nanowire arrays on Ni foam for high performance electrochemical capacitors, J. Power Sources, 2012, 203, 250–256 CrossRef CAS.
- C. Joseph, A. Daniels, S. Singh and M. Singh, Histidine-Tagged Folate-Targeted Gold nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro, Pharm, 2021, 14, 53 Search PubMed.
- P. Maloň, L. Bedna'rova', M. Straka, L. Krejč I ', L. Luka', L. Kumprecht, T. Kraus, M. KubanOva' and V. Baumruk, Disulfide chromophore and its optical activity, Chirality, 2010, 22, E47–E55, DOI:10.1002/chir.20851.
- S. Luber, Solvent effects in calculated vibrational Raman optical activity spectra of α-helices, J. Phys. Chem. A, 2013, 117, 2760–2770 CrossRef CAS PubMed.
- M. Beg, A. Maji, A. K. Mandal, S. Das, M. N. Aktara, P. K. Jha and M. Hossain, Green synthesis of silver nanoparticles using Pongamia pinnata seed: Characterization, antibacterial property, and spectroscopic investigation of interaction with human serum albumin, J. Mol. Recognit., 2017, 30, e2565 CrossRef PubMed.
- C. M. V. Stålhandske, C. I. Stålhandske, I. Persson, M. Sandström and F. Jalilehvand, Crystal and solution structures of N, N-dimethylthioformamide-solvated copper (I), silver (I), and gold (I) ions studied by X-ray diffraction, X-ray absorption, and vibrational spectroscopy, Inorg. Chem., 2001, 40, 6684–6693 CrossRef PubMed.
- S. J. Roeters, C. N. Van Dijk, A. Torres-Knoop, E. H. G. Backus, R. K. Campen, M. Bonn and S. Woutersen, Determining In Situ Protein Conformation and Orientation from the Amide-I Sum-Frequency Generation Spectrum: Theory and Experiment, J. Am. Chem. Soc., 2013, 117, 6311–6322 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01473h |
| This journal is © The Royal Society of Chemistry 2023 |