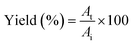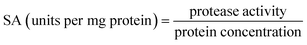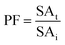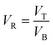Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Partitioning and recovery of proteases from lizardfish (Saurida micropectoralis) stomach using combined phase partitioning systems and their storage stability
Sakonwat Kuepethkaewa,
Sappasith Klomklao *b,
Yi Zhang
*b,
Yi Zhang c and
Benjamin K. Simpsond
c and
Benjamin K. Simpsond
aBiotechnology Program, Faculty of Agro and Bio Industry, Thaksin University, Phatthalung Campus, Pa-Phayom, Phatthalung 93210, Thailand
bDepartment of Food Science and Technology, Faculty of Agro and Bio Industry, Thaksin University, Phatthalung Campus, Pa-Phayom, Phatthalung 93210, Thailand. E-mail: sappasith@tsu.ac.th; Fax: +66 7460 9618; Tel: +66 7460 9618
cDepartment of Food Science, The Pennsylvania State University, University Park, PA 16802, USA
dDepartment of Food Science & Agricultural Chemistry, McGill University, 21111 Lakeshore Road, Ste-Anne-de-Bellevue, QC H9X 3V9, Canada
First published on 15th May 2023
Abstract
Partitioning and recovery of proteases from stomach extract (SE) and acidified stomach extract (ASE) of lizardfish using a three-phase partitioning (TPP) system in combination with an aqueous two-phase system (ATPS) was optimized. The highest yield and purity were obtained in the interphase of the TPP system, which consisted of a SE or ASE to t-butanol ratio of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 in the presence of 40% (w/w) (NH4)2SO4. Both TPP fractions were further subjected to ATPS. Phase compositions of ATPS including PEG molecular mass and concentrations as well as types and concentrations of salts influenced protein partitioning. The best ATPS conditions for protease partitioning into the top phase from TPP fractions of SE and ASE were 15% Na3C6H5O7–20% PEG1000 and 20% Na3C6H5O7–15% PEG1000, which increased the purity by 4-fold and 5-fold with the recovered activity of 82% and 77%, respectively. ATPS fractions of SE and ASE were subsequently mixed with several PEGs and salts for back extraction (BE). BE using 25% PEG8000–5% Na3C6H5O7 gave the highest PF and yield for both ATPS fractions. SDS-PAGE investigation revealed that the decrease in contaminating protein bands was observed after the combined partitioning systems. BE fractions of SE and ASE were quite stable at −20 and 0 °C up to 14 days. Therefore, the combination of TPP, ATPS and BE could be effectively applied to recover and purify proteases from the stomach of lizardfish.
0.5 in the presence of 40% (w/w) (NH4)2SO4. Both TPP fractions were further subjected to ATPS. Phase compositions of ATPS including PEG molecular mass and concentrations as well as types and concentrations of salts influenced protein partitioning. The best ATPS conditions for protease partitioning into the top phase from TPP fractions of SE and ASE were 15% Na3C6H5O7–20% PEG1000 and 20% Na3C6H5O7–15% PEG1000, which increased the purity by 4-fold and 5-fold with the recovered activity of 82% and 77%, respectively. ATPS fractions of SE and ASE were subsequently mixed with several PEGs and salts for back extraction (BE). BE using 25% PEG8000–5% Na3C6H5O7 gave the highest PF and yield for both ATPS fractions. SDS-PAGE investigation revealed that the decrease in contaminating protein bands was observed after the combined partitioning systems. BE fractions of SE and ASE were quite stable at −20 and 0 °C up to 14 days. Therefore, the combination of TPP, ATPS and BE could be effectively applied to recover and purify proteases from the stomach of lizardfish.
1. Introduction
Surimi, concentrated myofibrillar proteins obtained by successive washing of minced fish, is a useful ingredient for producing various processed foods with unique textural properties. Surimi-based products such as crab legs, chikuwa, fish balls, sausages, etc. have gained increasing attention because of the preferred textural properties and high nutritional value.1 Thailand is one of the most important surimi producing countries in Southeast Asia. Lizardfish (Saurida spp.) are tropical fish that have been commonly used as a potential raw material for surimi production in Thailand due to their availability, white color, good flavor, and good gel forming ability.2 During surimi processing, high amounts of processing by-products, especially viscera are discarded. Fish viscera is a potential natural source for recovering enzymes such as proteases that may have some unique properties for industrial applications e.g. in detergent, food, pharmaceutical, leather and silk industries.3 Stomach protease, especially pepsin can be recovered and applied mainly for hydrolysis purposes. Pepsin can be extracted and recovered from fish by-products, especially fish stomach, which constitute 5% of fish weight.4Nowadays, liquid–liquid extraction like a three-phase partitioning (TPP) and aqueous two-phase system (ATPS) are promising alternatives for separating biomolecules, especially proteins and enzymes.5,6 Both TPP and ATPS are relative simple and inexpensive, are easily operated and scaled-up.6 TPP is a simple bioseparation and purification technique in which a salt (e.g. ammonium sulphate) and water miscible aliphatic alcohol (e.g. t-butanol) are added to an aqueous solution containing proteins. Under optimized conditions, three phases are formed within an hour.6 Biomolecules are recovered in a purified form at the interphase, while the contaminants such as lipids, phenolics and some detergents mostly partition in t-butanol (top phase) or aqueous phase (bottom phase).7 This method is scalable and can be applied directly with crude suspensions. For ATPS, it has been an attractive technique for recovery of biological materials over other methods (precipitation, column chromatography and electrophoresis) since it constitutes gentle environmental conditions containing high water content in both liquid phases up to 70–90%.5 ATPS is generally formed by mixing aqueous conditions of two or more incompatible polymers or of polymer and salt above critical concentration.5,8 ATPS can remove contaminants, such as nucleic acids and undesirable proteins. The application of TPP or ATPS in downstream processing has been focused on partitioning, separation and recovery of various enzymes including pepsin,3 lipase,6 trypsin,8 etc. In general, the single system has been implemented for enzyme partitioning and recovery. The use of combined partitioning systems, e.g. TPP and ATPS could be an advantageous separation method to increase the purity of the target enzyme as well as to increase the recovery yield.6,9 However, there is no information regarding the use of combined TPP and ATPS for partitioning and recovery of proteases from lizardfish stomach. Therefore, the aim of this study was to optimize the separation process of proteases from the lizardfish stomach by using TPP in combination with ATPS. The storage stability of the partitioned enzymes with different temperatures and times was also investigated.
2. Materials and methods
2.1 Chemicals
Acetone, polyethylene glycol (PEG) 1000, PEG2000, PEG4000, PEG8000, tert-butanol (t-butanol) and tris(hydroxymethyl)aminomethane were obtained from Merck (Darmstadt, Germany). Hemoglobin from bovine blood, bovine serum albumin (BSA), wide range molecular weight markers and Coomassie Brilliant Blue G-250 were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Sodium dodecyl sulphate (SDS) was obtained from Fluka (Buchs, Switzerland). N,N,N′,N′-Tetramethyl ethylene diamine (TEMED) was purchased from Bio-Rad Laboratories (Hercules, CA, USA). The salts and other chemicals with the analytical grade were procured from Merck (Darmstadt, Germany).2.2 Preparation of crude extract
Lizardfish (Saurida micropectoralis), off-loaded approximately 24–36 h after capture from the Andaman Sea of Thailand were transported to Thaveelarp Fisheries Ltd, Part. Trang, Thailand. Upon arrival, lizardfish stomach was separated, collected and transported to the Department of Food Science and Technology, Thaksin University, Phatthalung, within 2 h. The samples were placed in ice using a sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ice ratio of 1
ice ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (w/w). The stomach was defatted with acetone and used for protease extraction according to the method Kuepethkaew et al.6 The dry powder referred to as “defatted stomach powder” was stored at −20 °C until used.
2 (w/w). The stomach was defatted with acetone and used for protease extraction according to the method Kuepethkaew et al.6 The dry powder referred to as “defatted stomach powder” was stored at −20 °C until used.
To prepare the crude extract, the defatted stomach powder was suspended in 50 mM Tris–HCl, pH 7.0 containing 0.2% (v/v) Tween 20 at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (w/v) and stirred continuously at 4 °C for 30 min.10 The suspension was centrifuged for 30 min at 4 °C at 5000 × g to remove the tissue debris. The supernatant was collected and referred to as “stomach extract, SE”. To activate stomach protease, SE was adjusted to pH 2.0 with 1.0 M HCl and the mixture was allowed to stand at 4 °C for 30 min.3 The suspension was centrifuged at 4 °C for 30 min at 10
9 (w/v) and stirred continuously at 4 °C for 30 min.10 The suspension was centrifuged for 30 min at 4 °C at 5000 × g to remove the tissue debris. The supernatant was collected and referred to as “stomach extract, SE”. To activate stomach protease, SE was adjusted to pH 2.0 with 1.0 M HCl and the mixture was allowed to stand at 4 °C for 30 min.3 The suspension was centrifuged at 4 °C for 30 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g using a refrigerated centrifuge (Sorvall Contifuge Stratos, Thermo Fisher Scientific, Waltham, MA, USA). The collected supernatant was referred to as “acidified stomach extract, ASE”.
000 × g using a refrigerated centrifuge (Sorvall Contifuge Stratos, Thermo Fisher Scientific, Waltham, MA, USA). The collected supernatant was referred to as “acidified stomach extract, ASE”.
2.3 Enzyme assay and protein determination
Enzyme activity was examined using hemoglobin as the substrate. The activity was tested at pH 2.0 and 40 °C for 15 min as described by Klomklao et al.11 and Kuepethkaew et al.10 The oligopeptide content in the supernatant was determined by the Lowry method12 using tyrosine as a standard. One unit of activity was defined as that releasing 1 μmol of tyrosine per min (μmol Tyr per min). A blank was run in the same manner, except that the enzyme was added into the reaction mixture after the addition of 50% (w/v) trichloroacetic acid (TCA).Protein concentration was determined by the method of Bradford using bovine serum albumin (BSA) as a protein standard.13
2.4 Three-phase partitioning (TPP) system
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (v/v). The mixtures were mixed continuously for 3 min using a vortex mixer (G506E, Scientific Industries, USA) and then allowed to stand for 60 min before subjecting to centrifuge at 7500 × g for 20 min at 4 °C to facilitate the separation of phases. Each phase was carefully separated using a Pasteur pipette. The lower aqueous layer and the interfacial phase were collected and dialyzed against 50 volumes of distilled water using a dialysis bag (MW cut-off: 14
0.5 (v/v). The mixtures were mixed continuously for 3 min using a vortex mixer (G506E, Scientific Industries, USA) and then allowed to stand for 60 min before subjecting to centrifuge at 7500 × g for 20 min at 4 °C to facilitate the separation of phases. Each phase was carefully separated using a Pasteur pipette. The lower aqueous layer and the interfacial phase were collected and dialyzed against 50 volumes of distilled water using a dialysis bag (MW cut-off: 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; EIDIA Co., Ltd, Tokyo, Japan) overnight at 4 °C with 4 changes of distilled water. After dialysis, the samples were measured for volume, protease activity and total protein content. TPP partitioning parameters including yield, specific activity (SA) and purification factor (PF) were calculated as follows:
000 Da; EIDIA Co., Ltd, Tokyo, Japan) overnight at 4 °C with 4 changes of distilled water. After dialysis, the samples were measured for volume, protease activity and total protein content. TPP partitioning parameters including yield, specific activity (SA) and purification factor (PF) were calculated as follows:where At is total protease activity in the protease rich phase and Ai is the initial protease activity of SE or ASE or fraction before being partitioned.
where SAt is the SA of the protease rich phase and SAi is the initial SA of SE or ASE or fraction before being partitioned. The system providing the highest protease recovery and purity was chosen for further study.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1.0
0.5, 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 and 1.0
1.0 and 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, w/w). Thereafter, ammonium sulphate at 40% saturation was added into the mixture. Partitioning was performed as described previously. The SE or ASE/organic solvent ratio yielding the highest enzyme recovery and purity was selected. The selected TPP fractions were used for ATPS.
1.5, w/w). Thereafter, ammonium sulphate at 40% saturation was added into the mixture. Partitioning was performed as described previously. The SE or ASE/organic solvent ratio yielding the highest enzyme recovery and purity was selected. The selected TPP fractions were used for ATPS.2.5 Aqueous two-phase system (ATPS)
ATPS was prepared in 10 mL centrifuge tubes by adding the different amounts of PEG and salts together with the selected TPP fraction of SE or ASE according to the method of Kuepethkaew et al.6 and Klomklao et al.8where VT and VB are the volume of top phase and bottom phase, respectively.
where CT and CB are concentrations of protein in top phase and bottom phase, respectively.
where ET and EB are concentrations of enzyme in top phase and bottom phase, respectively.
Based on purity and yield, the appropriate salt in ATPS rendering the most effective partitioning was selected for further study.
2.6 Back extraction (BE)
BE was used to partition the protease in PEG rich phase to aqueous salt rich phase following the method of Malpiedi et al.15 and Senphan and Benjakul9 with a slight modification. System containing 25% PEG4000 or 25% PEG8000 in the present of Na3C6H5O7 at different final concentrations (5, 10 and 15%, w/w) were used. One gram of the selected ATPS fraction of SE or ASE was added into the prepared systems. Distilled water was used to adjust the system to obtain the final weight of 5 g. The mixtures were mixed continuously for 3 min using a vortex mixer. Phase separation was achieved by centrifugation for 10 min at 5000 × g. The lower aqueous layer was collected and dialyzed against 50 volumes of distilled water using a dialysis bag (MW cut-off: 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; EIDIA Co., Ltd, Tokyo, Japan) overnight at 4 °C with 4 changes of distilled water. After dialysis, the samples were measured for enzyme activity and total protein content. Yield, SA, PF and ATPS partition parameters were calculated. BE fraction of SE or ASE with the highest purity and yield was used for further study.
000 Da; EIDIA Co., Ltd, Tokyo, Japan) overnight at 4 °C with 4 changes of distilled water. After dialysis, the samples were measured for enzyme activity and total protein content. Yield, SA, PF and ATPS partition parameters were calculated. BE fraction of SE or ASE with the highest purity and yield was used for further study.
2.7 Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE of SE, ASE and their fractionated proteases was performed using Mini-Protein II Cell apparatus (Bio-Rad Laboratories, Richmond, CA, USA) as described by Laemmli.16 In this study, acrylamide gel was made from 4.0% stacking and 12.0% separating gels. The SDS-PAGE procedure has been described previously.6,8,92.8 Storage stability of stomach protease
Protease activity of SE and ASE and their selected BE fractions (BE-SE and BE-ASE, respectively) containing 0.1% (w/v) sodium azide was monitored daily for the first 8 days and thereafter every 2 days up to 2 weeks. The storage temperature studied included −20 °C, 0 °C, 4 °C and room temperature (26–28 °C).2.9 Statistical analysis
Experiments were run in triplicate using three different lots of samples. A complete randomized design was applied in this study. Analysis of variance (ANOVA) was used to analyze the data. Values of means were compared by Duncan's multiple range tests.17 Statistical analysis was carried out by the Statistical Package for Social Science software (SPSS 24.0, SPSS Inc., Chicago, IL, USA).3. Results and discussion
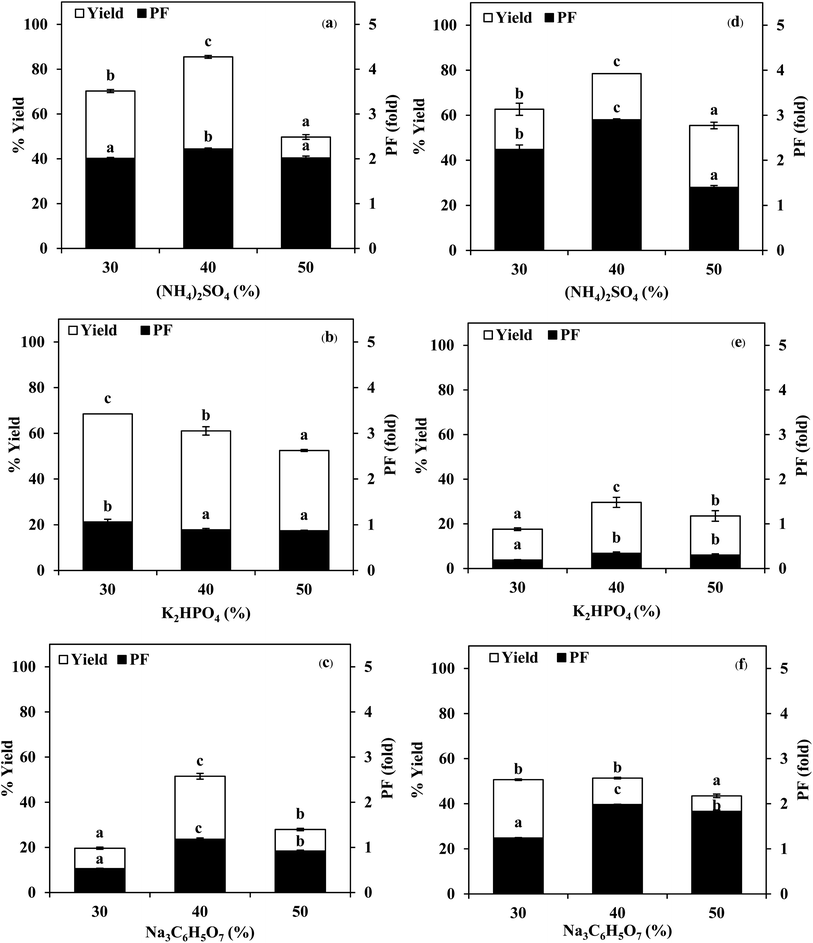
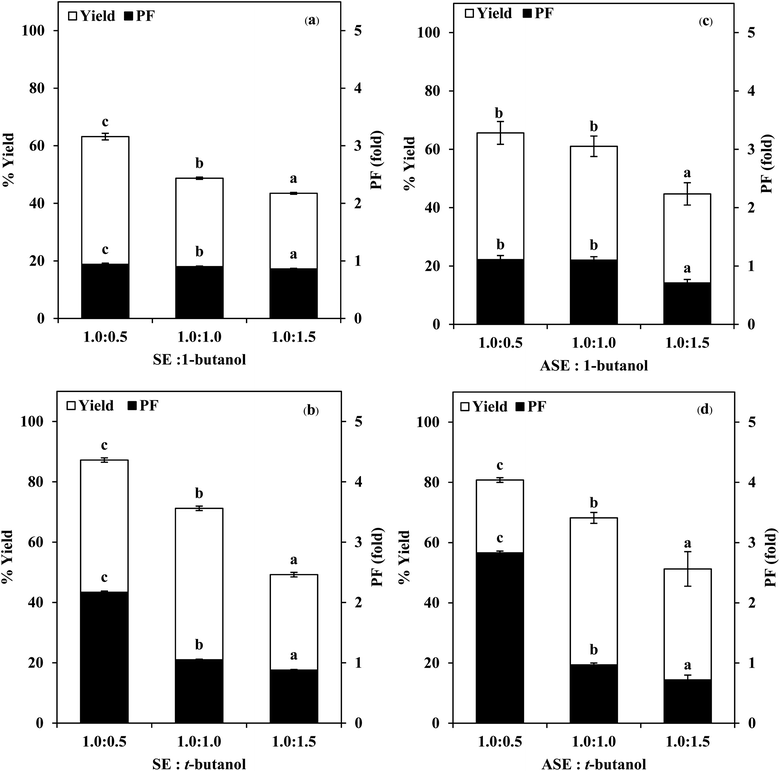
3.1 Use of TPP for stomach proteases partitioning
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1.0
0.5, 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 and 1.0
1.0 and 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 (w/w). From Fig. 2, it was observed that the yield and purity of both SE and ASE were highest at the 1.0
1.5 (w/w). From Fig. 2, it was observed that the yield and purity of both SE and ASE were highest at the 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 ratio and an increase in crude volume to organic solvents ratio has shown a significant decrease in the activity recovery and purification fold values. These results may be attributed to the synergistic effects of the increase in the concentration of organic solvents and the decrease in the saturation of ammonium sulphate.7 High organic solvent content may cause denaturation of the protein and hinders protein precipitation. Therefore, a ratio of 1.0
0.5 ratio and an increase in crude volume to organic solvents ratio has shown a significant decrease in the activity recovery and purification fold values. These results may be attributed to the synergistic effects of the increase in the concentration of organic solvents and the decrease in the saturation of ammonium sulphate.7 High organic solvent content may cause denaturation of the protein and hinders protein precipitation. Therefore, a ratio of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 of crude volume to t-butanol was considered as optimum with resulting protease recovery for SE and ASE of 87.22 and 80.77%, respectively. The crude extract to t-butanol ratio of 1.0
0.5 of crude volume to t-butanol was considered as optimum with resulting protease recovery for SE and ASE of 87.22 and 80.77%, respectively. The crude extract to t-butanol ratio of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 in the present of 50% (NH4)2SO4 was the optimal condition for the proteases partitioning from the viscera of farmed giant catfish.7 Senphan and Benjakul9 found that the optimum TPP condition of protease from hepatopancreas of Pacific white shrimp was 30% (NH4)2SO4 along with 1.0
0.5 in the present of 50% (NH4)2SO4 was the optimal condition for the proteases partitioning from the viscera of farmed giant catfish.7 Senphan and Benjakul9 found that the optimum TPP condition of protease from hepatopancreas of Pacific white shrimp was 30% (NH4)2SO4 along with 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 ratio of t-butanol to crude protease extract provided the enzyme recovery 76.0%.
1.0 ratio of t-butanol to crude protease extract provided the enzyme recovery 76.0%.
3.2 Use of ATPS for stomach proteases partitioning
| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| SEb | TPPc | SEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of SE.c Values were expressed, relative to those of TPP fraction. VR: volume ratio (top phase/bottom phase); KP: partition coefficient of protein, KE: partition coefficient of enzyme; SA: specific activity (units per mg protein); PF: purification factor; ns: no phase separation. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 20% PEG1000–15% (NH4)2SO4 | 0.71 ± 0.03d | 7.26 ± 0.10e | 30.95 ± 2.48e | 496.03 ± 39.87b | 4.22 ± 0.34b | 1.86 ± 0.15b | 62.74 ± 5.04c | 71.87 ± 5.77c |
| 20% PEG1000–20% (NH4)2SO4 | 0.50 ± 0.02b | 5.92 ± 0.05c | 6.79 ± 0.22b | 460.07 ± 15.02b | 3.91 ± 0.12b | 1.72 ± 0.06b | 50.48 ± 1.65b | 57.83 ± 1.90b |
| 20% PEG1000–25% (NH4)2SO4 | 0.40 ± 0.01a | 4.60 ± 0.04a | 0.95 ± 0.04a | 356.83 ± 14.34a | 3.04 ± 0.12a | 1.34 ± 0.06a | 32.41 ± 1.30a | 37.12 ± 1.49a |
| 20% PEG1000–15% MgSO4 | ns | ns | ns | ns | ns | ns | ns | ns |
| 20% PEG1000–20% MgSO4 | 0.89 ± 0.01e | 9.69 ± 0.04g | 26.65 ± 3.18d | 400.32 ± 47.74a | 3.41 ± 0.40a | 1.50 ± 0.18a | 57.61 ± 6.87c | 66.01 ± 7.87c |
| 20% PEG1000–25% MgSO4 | 0.65 ± 0.02c | 6.32 ± 0.02d | 20.25 ± 1.12c | 353.31 ± 19.52a | 3.01 ± 0.17a | 1.32 ± 0.07a | 35.57 ± 1.97a | 40.75 ± 2.25a |
| 20% PEG1000–15% Na3C6H5O7 | 1.02 ± 0.02f | 5.66 ± 0.02b | 56.49 ± 4.32g | 510.11 ± 39.00b | 4.34 ± 0.46b | 1.91 ± 0.14b | 83.96 ± 6.42d | 96.18 ± 7.35d |
| 20% PEG1000–20% Na3C6H5O7 | 0.64 ± 0.02c | 7.21 ± 0.14e | 45.80 ± 1.54f | 458.04 ± 15.36b | 3.89 ± 0.13b | 1.72 ± 0.06b | 80.47 ± 2.70d | 92.17 ± 3.09d |
| 20% PEG1000–25% Na3C6H5O7 | 0.47 ± 0.02b | 7.98 ± 0.05f | 63.39 ± 1.31h | 372.42 ± 7.70a | 3.17 ± 0.06a | 1.39 ± 0.03a | 77.10 ± 1.59d | 88.32 ± 1.82d |
| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| ASEb | TPPc | ASEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of ASE.c Values were expressed, relative to those of TPP fraction. Abbreviation: see legend of Table 1. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 20% PEG1000–15% (NH4)2SO4 | 0.74 ± 0.02d | 8.84 ± 0.03f | 51.18 ± 1.19e | 364.91 ± 8.52b | 4.34 ± 0.10b | 1.53 ± 0.04b | 77.54 ± 1.81c | 96.11 ± 2.24c |
| 20% PEG1000–20% (NH4)2SO4 | 0.49 ± 0.03b | 6.44 ± 0.01d | 10.72 ± 0.58a | 356.32 ± 19.26b | 4.24 ± 0.23b | 1.50 ± 0.08b | 62.96 ± 3.40b | 78.04 ± 4.22b |
| 20% PEG1000–25% (NH4)2SO4 | 0.40 ± 0.01a | 5.13 ± 0.04b | 20.21 ± 1.78b | 347.11 ± 30.56b | 4.13 ± 0.37b | 1.46 ± 0.13b | 53.58 ± 4.72a | 66.41 ± 5.85a |
| 20% PEG1000–15% MgSO4 | ns | ns | ns | ns | ns | ns | ns | ns |
| 20% PEG1000–20% MgSO4 | 0.87 ± 0.01e | 13.20 ± 0.24g | 11.47 ± 1.09a | 277.27 ± 26.34a | 3.30 ± 0.31a | 1.17 ± 0.11a | 65.16 ± 6.19b | 80.76 ± 7.67b |
| 20% PEG1000–25% MgSO4 | 0.64 ± 0.03c | 7.60 ± 0.05e | 24.53 ± 2.57c | 340.34 ± 35.63b | 4.05 ± 0.42b | 1.43 ± 0.15b | 60.40 ± 6.32ab | 74.86 ± 7.84ab |
| 20% PEG1000–15% Na3C6H5O7 | ns | ns | ns | ns | ns | ns | ns | ns |
| 20% PEG1000–20% Na3C6H5O7 | 0.62 ± 0.02c | 5.99 ± 0.10c | 66.32 ± 2.41f | 383.73 ± 13.91b | 4.56 ± 0.16b | 1.62 ± 0.06b | 75.37 ± 2.73c | 93.41 ± 3.39c |
| 20% PEG1000–25% Na3C6H5O7 | 0.49 ± 0.01b | 4.86 ± 0.13a | 45.04 ± 2.86d | 348.80 ± 22.17b | 4.15 ± 0.26b | 1.47 ± 0.09b | 59.72 ± 3.80ab | 74.02 ± 4.70ab |
From the result, the VR of all ATPS investigated of both SE and ASE was decreased when salt concentration increased. Enhancement of salt quantity provided a higher proportion of salt-rich bottom phase leading to practically reduced VR. The distribution of the protein and proteases in ATPS are reported by KP and KE, respectively. The distribution of protein in ATPS of SE and ASE likely depended on the differences in surface charge of protease and others protein contaminants, which were governed by different salts.3,8 High in KP values indicating most of proteins were more partitioning to the top phase, while the high in KE implying only the target enzyme was partitioned to the top phase. Generally, ATPS with 20% and 25% Na3C6H5O7 showed higher KE. In general, increasing salts concentration for both SE and ASE partitioning resulted in lower KP. Salts might increase the migration of the proteins to the lower phase by electrostatic repulsion effects. Tables 1 and 2 also exhibited that a less protease activity recovery was observed with increasing salt concentration. It can be found that increase in salt quantity provided the salting-out effect.8 Ketnawa et al.21 reported that the highest purification factor and yield in partitioning of alkaline protease from farmed giant catfish (Pangasianodon gigas) viscera was obtained by using 15% PEG2000–15% sodium citrate.
| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| SEb | TPPc | SEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of SE.c Values were expressed, relative to those of TPP fraction. Abbreviation: see legend of Table 1. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 15% PEG1000–15% Na3C6H5O7 | ns | ns | ns | ns | ns | ns | ns | ns |
| 20% PEG1000–15% Na3C6H5O7 | 1.01 ± 0.01f | 6.74 ± 0.21f | 51.16 ± 0.91f | 526.94 ± 9.38c | 4.48 ± 0.08c | 1.97 ± 0.04c | 82.97 ± 1.48e | 95.04 ± 1.70e |
| 25% PEG1000–15% Na3C6H5O7 | 1.07 ± 0.01g | 6.91 ± 0.23f | 30.23 ± 2.89e | 499.79 ± 47.79ab | 4.25 ± 0.41bc | 1.87 ± 0.18bc | 65.38 ± 6.25d | 74.89 ± 7.06d |
| 15% PEG2000–15% Na3C6H5O7 | 0.67 ± 0.01c | 3.14 ± 0.01b | 6.49 ± 0.51bc | 300.09 ± 23.64a | 2.55 ± 0.20a | 1.12 ± 0.09a | 27.21 ± 2.15b | 31.17 ± 2.45b |
| 20% PEG2000–15% Na3C6H5O7 | 0.82 ± 0.01d | 4.52 ± 0.04d | 11.20 ± 0.85d | 501.78 ± 37.90ab | 4.27 ± 0.32bc | 1.88 ± 0.14bc | 51.47 ± 3.89c | 58.96 ± 4.45c |
| 25% PEG2000–15% Na3C6H5O7 | 0.94 ± 0.02e | 5.18 ± 0.04e | 7.43 ± 0.71c | 439.73 ± 42.18b | 3.74 ± 0.36b | 1.65 ± 0.16b | 49.20 ± 4.72c | 56.36 ± 5.41c |
| 15% PEG4000–15% Na3C6H5O7 | 0.47 ± 0.02a | 2.58 ± 0.01a | 2.83 ± 0.51a | 249.74 ± 44.74a | 2.12 ± 0.38a | 0.94 ± 0.17a | 17.95 ± 3.21a | 20.57 ± 3.68a |
| 20% PEG4000–15% Na3C6H5O7 | 0.54 ± 0.01b | 3.27 ± 0.03b | 4.91 ± 0.31b | 509.94 ± 31.64c | 4.34 ± 0.27c | 1.91 ± 0.12c | 44.45 ± 2.76c | 50.92 ± 3.16c |
| 25% PEG4000–15% Na3C6H5O7 | 0.69 ± 0.01c | 4.27 ± 0.01c | 7.92 ± 0.60c | 435.41 ± 33.26b | 3.70 ± 0.28b | 1.63 ± 0.12b | 46.01 ± 3.51c | 52.71 ± 4.03c |
| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| ASEb | TPPc | ASEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of ASE.c Values were expressed, relative to those of TPP fraction. Abbreviation: see legend of Table 1. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 15% PEG1000–20% Na3C6H5O7 | 0.50 ± 0.03b | 5.18 ± 0.01d | 29.24 ± 0.99g | 431.33 ± 14.63d | 5.13 ± 0.17d | 1.81 ± 0.06d | 77.52 ± 2.63f | 96.09 ± 3.26f |
| 20% PEG1000–20% Na3C6H5O7 | 0.70 ± 0.01d | 6.06 ± 0.02f | 65.81 ± 0.97h | 390.19 ± 5.75c | 4.64 ± 0.07c | 1.64 ± 0.03c | 74.79 ± 1.10ef | 92.69 ± 1.37ef |
| 25% PEG1000–20% Na3C6H5O7 | 0.80 ± 0.01e | 4.40 ± 0.03g | 22.53 ± 0.90f | 386.97 ± 15.48c | 4.60 ± 0.18c | 1.63 ± 0.07c | 59.73 ± 2.39c | 74.03 ± 2.96c |
| 15% PEG2000–20% Na3C6H5O7 | 0.42 ± 0.01a | 4.03 ± 0.02b | 9.52 ± 0.16cd | 410.61 ± 6.77d | 4.88 ± 0.08d | 1.73 ± 0.03d | 59.49 ± 1.01c | 73.74 ± 1.21c |
| 20% PEG2000–20% Na3C6H5O7 | 0.63 ± 0.02c | 5.22 ± 0.07d | 16.04 ± 0.24e | 416.63 ± 6.10d | 4.96 ± 0.08d | 1.75 ± 0.03d | 72.91 ± 1.07e | 90.37 ± 1.32e |
| 25% PEG2000–20% Na3C6H5O7 | 0.68 ± 0.01cd | 5.44 ± 0.02e | 9.96 ± 0.25d | 312.17 ± 7.86b | 3.71 ± 0.09b | 1.31 ± 0.03b | 56.58 ± 1.43bc | 70.13 ± 1.76bc |
| 15% PEG4000–20% Na3C6H5O7 | 0.36 ± 0.04a | 3.12 ± 0.02a | 4.77 ± 0.14a | 428.20 ± 12.23d | 5.09 ± 0.15d | 1.80 ± 0.05d | 53.37 ± 1.52b | 66.14 ± 1.89b |
| 20% PEG4000–20% Na3C6H5O7 | 0.49 ± 0.04b | 4.15 ± 0.12c | 8.83 ± 0.21c | 425.52 ± 9.98d | 5.06 ± 0.12d | 1.79 ± 0.04d | 65.20 ± 1.53d | 80.81 ± 1.90d |
| 25% PEG4000–20% Na3C6H5O7 | 0.62 ± 0.05c | 5.14 ± 0.10d | 6.81 ± 0.57b | 218.21 ± 18.28a | 2.60 ± 0.22a | 0.92 ± 0.08a | 38.68 ± 3.24a | 47.94 ± 4.01a |
3.3 Use of BE for stomach proteases partitioning
To separate the protease in the PEG-rich phase after ATPS separation, different methods can be applied.3 The target protease can be separated from PEG by back-extraction, in which a fresh salt phase is added to the collected PEG-rich phase containing the target enzyme. By changing the system conditions, target protease can be directed to the salt phase. Thereafter, salt can be removed by dialysis or membrane filtration.9,15 Therefore, protease separation from PEG of SE and ASE was investigated. In the present investigation, ATPS containing 25% PEG4000 and 8000 in the presence of Na3C6H5O7 at varying concentrations were used for back extraction as displayed in Tables 5 and 6. For both SE and ASE, BE using ATPS containing 25% PEG8000 and 5% Na3C6H5O7 produced the highest SA and PF (p < 0.05). Under the optimal conditions of the BE system, the highest purity by 7.85-fold with a recovered activity of 54.35% and 8.41-fold with a recovered activity of 64.81% was obtained for SE and ASE, respectively, compared with SE and ASE. In the presence of 5, 10 or 15% Na3C6H5O7, PEG8000 exhibited higher SA and PF than PEG4000. PEG of the highest MW (PEG8000) excludes the protein from the top phase, driven by an entropically unfavourable term.6,9 BE for proteases from Pacific white shrimp hepatopancreas was achieved using high MW PEG (25% PEG8000).9 From the results, ATPS containing 25% PEG8000 and 5% Na3C6H5O7 was shown to be the effective BE to transfer proteases of SE and ASE to the salt phase. Those phase systems exhibiting the highest PF with appropriate yield for SE or ASE were selected for further investigation and those fractions were referred to “BE-SE” and “BE-ASE”, respectively.| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| SEb | TPPc | SEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of SE.c Values were expressed, relative to those of TPP fraction. Abbreviation: see legend of Table 1. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 25% PEG4000–5% Na3C6H5O7 | 5.93 ± 0.11d | 0.32 ± 0.01e | 1.56 ± 0.01e | 583.01 ± 6.74b | 4.96 ± 0.06b | 2.18 ± 0.03b | 24.82 ± 0.29a | 28.43 ± 0.33a |
| 25% PEG4000–10% Na3C6H5O7 | 1.86 ± 0.08b | 0.14 ± 0.01b | 0.64 ± 0.03c | 559.90 ± 2.68ab | 4.76 ± 0.02ab | 2.10 ± 0.01ab | 56.37 ± 0.27c | 64.57 ± 0.31c |
| 25% PEG4000–15% Na3C6H5O7 | 1.21 ± 0.03a | 0.09 ± 0.01a | 0.36 ± 0.07a | 546.50 ± 31.65a | 4.65 ± 0.27a | 2.05 ± 0.12a | 72.01 ± 4.17d | 82.48 ± 4.78d |
| 25% PEG8000–5% Na3C6H5O7 | 6.24 ± 0.07e | 0.28 ± 0.01d | 0.73 ± 0.02d | 922.43 ± 16.73d | 7.85 ± 0.14d | 3.46 ± 0.06d | 54.35 ± 0.98bc | 62.26 ± 1.13bc |
| 25% PEG8000–10% Na3C6H5O7 | 2.34 ± 0.01c | 0.15 ± 0.01c | 0.58 ± 0.02c | 624.27 ± 27.26c | 5.31 ± 0.24c | 2.34 ± 0.10c | 52.73 ± 2.30bc | 60.40 ± 2.64bc |
| 25% PEG8000–15% Na3C6H5O7 | 2.36 ± 0.02c | 0.14 ± 0.01b | 0.46 ± 0.08b | 591.69 ± 10.78bc | 5.03 ± 0.09bc | 2.22 ± 0.04bc | 51.49 ± 0.94b | 58.99 ± 1.08b |
| Phase composition (%, w/w) | VR | KP | KE | SA | PF | Yield (%) | ||
|---|---|---|---|---|---|---|---|---|
| ASEb | TPPc | ASEb | TPPc | |||||
| a Means ± SD from the triplicate determinations.b Values were expressed, relative to those of ASE.c Values were expressed, relative to those of TPP fraction. Abbreviation: see legend of Table 1. The different superscripts in the same column denote the significant difference (p < 0.05). | ||||||||
| 25% PEG4000–5% Na3C6H5O7 | 6.92 ± 0.16e | 0.79 ± 0.02f | 3.58 ± 0.05e | 485.52 ± 30.71bc | 5.77 ± 0.36bc | 2.04 ± 0.13bc | 20.69 ± 1.31a | 25.64 ± 1.62a |
| 25% PEG4000–10% Na3C6H5O7 | 2.59 ± 0.21c | 0.30 ± 0.01c | 1.67 ± 0.05c | 390.83 ± 33.17a | 4.65 ± 0.40a | 1.64 ± 0.14a | 36.71 ± 3.11c | 45.50 ± 3.86bc |
| 25% PEG4000–15% Na3C6H5O7 | 1.43 ± 0.03a | 0.16 ± 0.01a | 0.83 ± 0.03a | 445.94 ± 16.27ab | 5.30 ± 0.20ab | 1.88 ± 0.07ab | 61.48 ± 2.24d | 67.21 ± 12.94d |
| 25% PEG8000–5% Na3C6H5O7 | 1.74 ± 0.06ab | 0.27 ± 0.01b | 0.83 ± 0.02a | 707.48 ± 30.33d | 8.41 ± 0.36d | 2.98 ± 0.13d | 64.18 ± 2.75d | 79.55 ± 3.41e |
| 25% PEG8000–10% Na3C6H5O7 | 2.02 ± 0.03b | 0.33 ± 0.01d | 1.32 ± 0.02b | 529.57 ± 77.74c | 6.30 ± 0.92c | 2.22 ± 0.33c | 41.53 ± 6.10c | 51.46 ± 7.55c |
| 25% PEG8000–15% Na3C6H5O7 | 6.16 ± 0.26d | 0.60 ± 0.01e | 2.34 ± 0.04d | 524.55 ± 35.31c | 6.24 ± 0.42c | 2.21 ± 0.15c | 28.91 ± 1.95b | 35.83 ± 2.41ab |
3.4 Protein pattern of stomach proteases partitioned with combined partitioning system
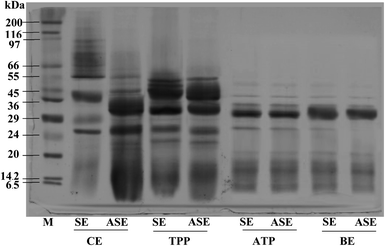
The purity of protease from the lizardfish stomach after the combined partitioning system process was determined using SDS-PAGE. As can be observed in Fig. 3, both SE and ASE contained a variety of proteins with different MW. SE contained proteins with MW of 39, 29 and 26 kDa as the major proteins. After the acidification process, some protein contaminants were removed and the major band with MW of 32 and 26 kDa was found. Also, a large number of contaminating proteins were removed after partitioning with TPP, ATPS and BE, particularly proteins with higher and lower MW. As a result, a higher purity of proteases was obtained. A greater intensity of the major protein band (32 kDa) was obtained in both SE and ASE after BE process, in comparison with that observed in SE and ASE. The migration of major protein band of SE to lower molecular weight positions after acidification or ATPS partitioning might be because of the autocatalytic activation of pepsinogen to pepsin under acidic conditions and with ATPS systems, respectively. Based on our previous investigation, the aspartic protease, most likely pepsin was the major protease found in lizardfish stomachs.10 Generally, pepsins from marine animals were reported to have molecular weights ranging from 27 to 42 kDa.24 Compared with pepsin, pepsinogen contains an additional 44 amino acids, but when exposed to the hydrochloric acid (HCl) present in gastric juice (pH of 1.5–2.0), the 44 amino acids are proteolytically removed in an autocatalytic way to activate it to pepsin.4 From these results, the major protein band with the MW of 32 kDa was found after BE process of both SE and ASE. Therefore, the electrophoretic results clearly showed that this TPP system in combination with ATPS and BE substantially primarily purified the protease from the lizardfish stomach.3.5 Storage stability of stomach proteases partitioned with combined partitioning system
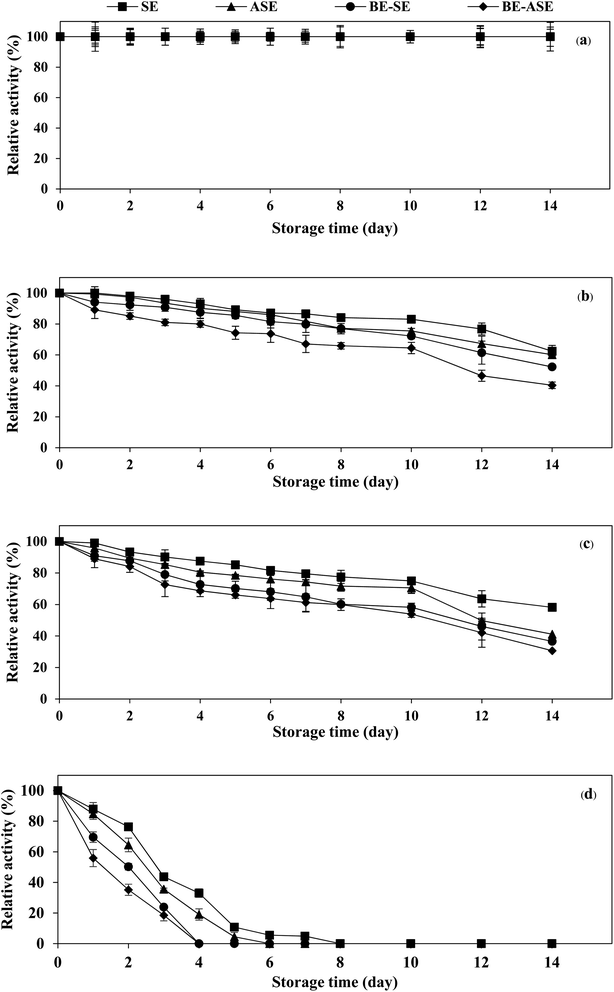
The stability of the enzyme is of great importance for the economy of their industrial application. Temperature is an important limiting factor for the storage of enzymes. Fig. 4 displays the changes in protease activity of SE and ASE as well as their BE fractions during storage at different temperatures (−20, 0, 4 °C and room temperature) up to 2 weeks. All samples exhibited greater stability at −20 °C. Freezing at −20 or −80 °C is the more common form of frozen protein storage. SE and ASE were quite stable for 14 days of storage at 0 and 4 °C. Nevertheless, at 0 and 4 °C, the stability of BE-SE and BE-ASE was lower as evidenced by a continuous decline in protease activity with increasing time of storage. The highest decline in protease activity was found when kept at ambient temperature. At room temperature, SE showed good stability compared to the other samples. At day 4 of storage at room temperature, for ASE, BE-SE and BE-ASE, more than 80% of the original activity was lost. The decline in protease activity noticed for all samples at room temperature may be due to increased degradation of enzyme active site with increased temperature. Storage at room temperature often leads to protein degradation and/or inactivity. The decrease in activity was higher when stored for a longer duration at higher temperatures. The least activity was observed when kept for 8 days at room temperature. A similar result was observed in the case of protease from albacore tuna (Thunnus alalunga) stored at room temperature.3 In general, enzymes are proteins and undergo essentially irreversible denaturation at temperatures above those to which they are ordinarily exposed in the environment. The results suggest that the stability of protease from lizardfish stomachs most likely depended on the form of enzyme and storage temperature. Also, low temperature, especially −20 °C was the ideal storage condition for the preservation of proteases from the lizardfish stomach.4. Conclusion
The combined partitioning systems including TPP, ATPS, and BE effectively recovered and partitioned the proteases from the lizardfish stomach. TPP with SE or ASE to t-butanol ratio of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 and 40% (NH4)2SO4 was firstly implemented. Both TPP fractions were further subjected to ATPS. For ATPS studied, the optimum condition for partitioning of SE and ASE was 20% PEG1000–15% Na3C6H5O7 and 15% PEG1000–20% Na3C6H5O7, respectively, and BE including 25% PEG8000–5% Na3C6H5O7 was applied in last step for both samples. The highest PF of 7.85-fold and 8.41-fold, with the yield of 54.35% and 64.18% was obtained for SE and ASE, respectively. Based on SDS-PAGE, higher purity was obtained after combined partitioning systems. Protease in SE and ASE as well as their BE fractions were stable at −20 °C throughout 14 days of storage. The instability of protease was more pronounced in higher storage temperatures.
0.5 and 40% (NH4)2SO4 was firstly implemented. Both TPP fractions were further subjected to ATPS. For ATPS studied, the optimum condition for partitioning of SE and ASE was 20% PEG1000–15% Na3C6H5O7 and 15% PEG1000–20% Na3C6H5O7, respectively, and BE including 25% PEG8000–5% Na3C6H5O7 was applied in last step for both samples. The highest PF of 7.85-fold and 8.41-fold, with the yield of 54.35% and 64.18% was obtained for SE and ASE, respectively. Based on SDS-PAGE, higher purity was obtained after combined partitioning systems. Protease in SE and ASE as well as their BE fractions were stable at −20 °C throughout 14 days of storage. The instability of protease was more pronounced in higher storage temperatures.
Author contributions
Sakonwat Kuepethkaew: investigation, methodology, writing – original draft. Sappasith Klomklao: conceptualization, investigation, writing – review & editing. Yi Zhang: writing – review & editing. Benjamin K. Simpson: conceptualization, supervision.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the National Research Council of Thailand as of fiscal year 2021 and the Thailand Science Research and Innovation (TSRI) under the Research and Researchers for Industries (RRI) to Sakonwat Kuepethkaew (PHD60I0088). The National Research Council of Thailand and Thaksin University for Project No. NRCI5-RSA63005 are also acknowledged.References
- T. Petcharat and S. Benjakul, Food Hydrocolloids, 2018, 77, 746–753 CrossRef CAS.
- S. Chanarat and S. Benjakul, Food Hydrocolloids, 2013, 30, 704–711 CrossRef CAS.
- S. Nalinanon, S. Benjakul, W. Visessanguan and H. Kishimura, Process Biochem., 2009, 44, 471–476 CrossRef CAS.
- L. Zhao, S. M. Budge, A. E. Ghaly, M. S. Brooks and D. Dave, J. Food Process. Technol., 2011, 2, 1–14 Search PubMed.
- K. Ratanapongleka, IJCEA, 2010, 1, 191–198 CrossRef CAS.
- S. Kuepethkaew, K. Sangkharak, S. Benjakul and S. Klomklao, J. Food Sci. Technol., 2017, 54, 3880–3891 CrossRef CAS PubMed.
- S. Rawdkuen, A. Vanabun and S. Benjakul, Process Biochem., 2012, 47, 2566–2569 CrossRef CAS.
- S. Klomklao, S. Benjakul, W. Visessanguan, B. K. Simpson and H. Kishimura, Process Biochem., 2005, 40, 3061–3067 CrossRef CAS.
- T. Senphan and S. Benjakul, Sep. Purif. Technol., 2014, 129, 57–63 CrossRef CAS.
- S. Kuepethkaew, S. Damodaran and S. Klomklao, Chiang Mai J. Sci., 2021, 48, 1322–1337 CAS.
- S. Klomklao, H. Kishimura, M. Yabe and S. Benjakul, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2007, 147, 682–689 CrossRef.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS PubMed.
- M. M. Bradford, Biochemistry, 1976, 72, 248–254 CAS.
- I. Roy and M. N. Gupta, Anal. Biochem., 2002, 300, 11–14 CrossRef CAS PubMed.
- L. P. Malpiedi, G. A. Picó and B. B. Nerli, Sep. Purif. Technol., 2011, 78, 91–96 CrossRef CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- R. G. D. Steel and J. H. Torrie, Principles and Procedures of Statistics, 1980 Search PubMed.
- M. Gagaoua and H. Kahina, Biosens. J., 2016, 135, 2–4 Search PubMed.
- G. A. Gomes, A. M. Azevedo, M. R. Aires-Barros and D. M. F. Prazeres, Sep. Purif. Technol., 2009, 65, 22–30 CrossRef CAS.
- R. K. Pérez, D. B. Loureiro, B. B. Neril and G. Tubio, Protein Expression Purif., 2015, 106, 66–71 CrossRef PubMed.
- S. Ketnawa, S. Benjakul, T. C. Ling, O. Martínez-Alvarez and S. Rawdkuen, Chem. Cent. J., 2013, 7, 1–9 CrossRef PubMed.
- K. Bhavsar, V. R. Kumar and J. M. Khire, Process Biochem., 2012, 47, 1066–1072 CrossRef CAS.
- B. R. Babu, N. K. Rastogi and K. S. M. S. Raghavarao, Chem. Eng. Process., 2008, 47, 83–89 CrossRef CAS.
- B. K. Simpson, in Seafood enzymes: utilization and influence on postharvest seafood quality, ed. N. F. Haard and B. K. Simpson, Marcel Dekker, New York, 2000, ch. 8, pp. 531–540 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |