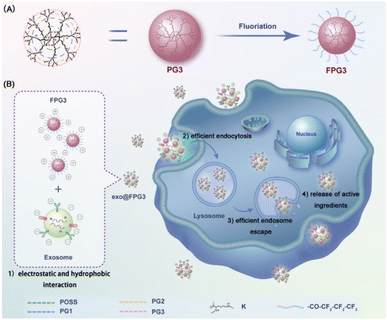
Open Access Article
Open Access ArticleImproved intracellular delivery of exosomes by surface modification with fluorinated peptide dendrimers for promoting angiogenesis and migration of HUVECs†
Shengnan Ma a,
Lei Songa,
Yueyue Baib,
Shihao Wanga,
Jiao Wanga,
Haohao Zhanga,
Fazhan Wang
a,
Lei Songa,
Yueyue Baib,
Shihao Wanga,
Jiao Wanga,
Haohao Zhanga,
Fazhan Wang *b,
Yiyan He*c,
Chuntao Tiand and
Guijun Qin*a
*b,
Yiyan He*c,
Chuntao Tiand and
Guijun Qin*a
aDepartment of Endocrinology and Metabolism, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou 450052, Henan, China. E-mail: hyqingj@zzu.edu.cn
bMedical Research Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou 450052, Henan, China. E-mail: fazhanwang_20@zzu.edu.cn
cCollege of Materials Science and Engineering, Nanjing Tech University, Nanjing 211816, Jiangsu, China. E-mail: yiyanhe@njtech.edu.cn
dDepartment of Oncology, Sanmenxia Central Hospital, Sanmenxia 472000, Henan, China
First published on 11th April 2023
Abstract
Exosomes exhibit great potential as novel therapeutics for tissue regeneration, including cell migration and angiogenesis. However, the limited intracellular delivery efficiency of exosomes might reduce their biological effects. Here, exosomes secreted by adipose-derived mesenchymal stem cells were recombined with fluorinated peptide dendrimers (FPG3) to form the fluorine-engineered exosomes (exo@FPG3), which was intended to promote the cytosolic release and the biological function of exosomes. The mass ratio of FPG3 to exosomes at 5 was used to investigate its cellular uptake efficiency and bioactivity in HUVECs, as the charge of exo@FPG3 tended to be stable even more FPG3 was applied. It was found that exo@FPG3 could enter HUVECs through a variety of pathways, in which the clathrin-mediated endocytosis played an important role. Compared with exosomes modified with peptide dendrimers (exo@PG3) and exosomes alone, the cellular uptake efficiency of exo@FPG3 was significantly increased. Moreover, exo@FPG3 significantly enhanced the angiogenesis and migration of HUVECs in vitro as compared to exo@PG3 and exosomes. It is concluded that surface fluorine modification of exosomes with FPG3 is conducive to the cellular uptake and bioactivity of the exosome, which provides a novel strategy for engineered exosomes to enhance the biological effects of exosome-based drug delivery.
1. Introduction
In order to better engage exosomes in the diagnosis and treatment of diseases, engineered exosome strategies have been proposed to improve their bioavailability, targeting and presentation.1,2 Exosomes secreted by mesenchymal stem cells have the ability to promote tissue regeneration including cell migration and angiogenesis.3,4 Meanwhile, the programmable engineered exosomes may be used to optimize wound healing and neovascularization therapy.5 The multifunctional hydrogels maximize exosomes' therapeutic efficacy by extending the local exosomes' availability during the process of tissue regeneration.6–10 In addition, the bioengineering technologies involved in the design of engineered exosomes could solve their limitations before becoming an effective therapeutic tool for various diseases, such as myocardial infarction,11,12 cancer13–15 or neurodegenerative disorders.16,17The cell membrane is one of the biological barriers for exogenous substances to enter cells. The intracellular delivery of exosomes appears to be influenced by the interaction between exosomes and cell membranes.18 The internalization mechanism of exosomes, such as micropinocytosis, micropinocytosis or receptor-mediated endocytosis, depends on the cell type,19,20 which can be intervened to regulate uptake efficiency. Yang et al. constructed the engineered exosomes modified with caveolae associated protein 2 through gene editing technology to improve the endocytosis activity of nucleus pulposus cells, resulting in increasing the uptake rate of exosomes and retarding the progression of intervertebral disc degeneration.21 Another, exogenous lipids in engineered exosomes could regulate the interactions between exosomes and cells. Alkyl chains could be inserted into exosomal membranes through hydrophobic interactions. For example, PEG-lipid conjugates were anchored into the lipid bilayer of exosomes to increase long circulation in vivo.22 Meanwhile, the fusion efficiency of non-PEGylated liposomes with exosomes was higher than that of PEGylated liposomes, and more non-PEGylated exosomes was in macrophages.23 Moreover, the phosphatidylcholine-engineered exosomes designed by Kang et al. significantly enhanced tumor cell internalization compared to native exosomes, which as carrier loading drug or RNA improved in vitro anti-tumor efficiency.24 In addition to the above techniques, the reversal of exosome surface potential also contributes to intracellular delivery and accumulation in cartilage matrix. Wang et al. proposed reversing the surface charge of small extracellular vesicles by cationic amphiphilic macromolecule to improve the intra-articular bioavailability of extracellular vesicles.25
Nevertheless, another crucial issue is the fate of exosomes within recipient cells. A large proportion of the internalized exosomes are processed in the endolysosome pathway and eventually digested in the lysosome.19 In reality the colocalization efficiency of internalized exosomes with lysosomes reached approximately 60% after 48 h of incubation with the recipient cells.26 Engineered exosome strategies have potential to promote the endosomal escape. pH-sensitive fusogenic peptide that was combined with exosomes could induce the membrane disruption and enhance the cytosolic release of exosomal cargo after the exosomes internalization.27 The arginine residues number in cell-penetrating peptides modifying the on exosomal membranes could affect the cytosolic release efficacy of exosomes and biological activity.28–30
Herein, we innovatively propose the fluorine-engineered exosomes (exo@FPG3) generated from surface engineering of exosomes with the fluorinated peptide dendrimers (FPG3) to enhance the intracellular delivery and biological activity of exosomes. In recent years, fluorinated molecules have been used to regulate cell membrane permeability and drug pharmacokinetics, greatly promoting the cytosolic delivery of biological macromolecules and chemotherapy drugs.31–33 Cheng et al. have gradually developed various fluoropolymers to achieve the efficient cytosolic delivery of gene, protein and peptides delivery, which could be attributed to the effects of fluoridation on serum tolerance, cellular uptake and endosomal escape.34–36 Considering the advantage of fluorine in medical applications, the fluorine-engineered exosomes are expected to achieve good performance in exosome uptake efficacy and cytoplasmic release of contents. As illustrated in Scheme 1, the generation 3 poly(L-lysine) dendrons (PG3) with abundant amino groups on the surface were partially fluorinated, obtaining the FPG3. Exosomes secreted by adipose-derived mesenchymal stem cells were modified with FPG3 through electrostatic and hydrophobic interaction, altering the surface potential and lipid bilayers of exosomes. It is very interesting to note that the fluorinated peptide dendrimers are beneficial for the cytosolic delivery of exosomes, which are greatly superior to free exosomes and peptide dendrimers modified exosomes. In addition, the biological functions of exosomes in angiogenesis and cell migration were further assessed by tube formation and scratch wound.
2. Results and discussion
2.1. Synthesis and characterization of fluorinated peptide dendrimer
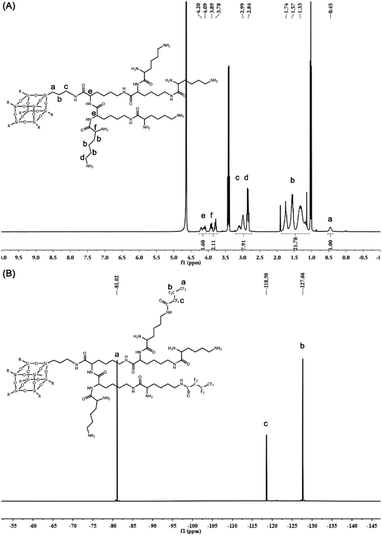
The peptide dendrimers not only have highly branched 3D structures with abundant surface groups that can be used for covalent modification but also show their capabilities in biodegradability and biocompatibility.37 To prepare the generation 3 poly(L-lysine) dendrimer (PG3), polyhedral oligomeric silsesquioxane (POSS) as the core for poly(L-lysine) dendrimers was first synthesized in one step. L-lysine as monomer was successively concatenated by condensation reaction to obtain the PG3 with 64 peripheral amino groups (Fig. 1A). The synthesis process and the characterization of the generation 1 poly(L-lysine) dendrimer (PG1), generation 2 poly(L-lysine) dendrimer (PG2) and PG3 are presented in the ESI (Fig. S1–S9†).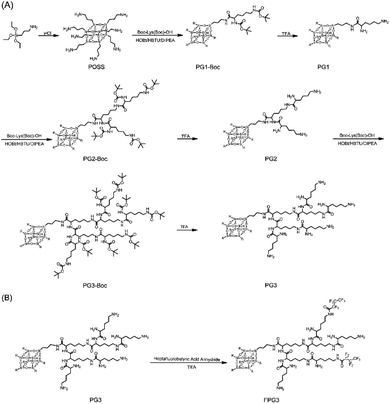 | ||
| Fig. 1 Synthetic routes of POSS core-based peptide poly(L-lysine) dendrimers (A) and fluorinated peptide dendrimer (B). | ||
All the compounds were confirmed by mass spectrometry (MS) or nuclear magnetic resonance (NMR). The most abundant peak at m/z = 8082.38 corresponded to the [M + Na]+ signal of PG3 (M = 8059) (Fig. S10†), indicating that the Boc groups on the PG3-Boc were fully removed. The structure of PG3 was further analyzed by 1H NMR (Fig. 2A). The integral area at δ 0.45 ppm for the side chain of silsesquioxane relative to the peak of δ 4.20–4.09 ppm, δ 3.89–3.78 ppm for the lysine was 2/7, proving the successful synthesis of PG3.
Then the heptafluorobutyric anhydride was used to partially functionalize the peripheral amino groups of PG3 (Fig. 1B). The ninhydrin assay was conducted to determine the fluorination degree of PG3. The calibration curve of primary amine was linear within the range of 0.05 to 0.3 mM and the linear relation formula was y = 4.6931x + 0.0145 (r2 = 0.999) (Fig. S11A†). As calculated from the equation (Fig. S11B†), the average number of fluoride groups modified on each PG3 is about 16.42, and the fluoride degree of FPG3 was 25.6%. Meanwhile, the peaks at δ 81.02 ppm, 118.50 ppm and 127.66 ppm in the spectrum of 19F NMR for FPG3 (Fig. 2B) were from the fluorocarbon chain modified on the PG3. The characterization analysis confirmed the preparation for FPG3.
2.2. Identification of ADSCs and exosomes
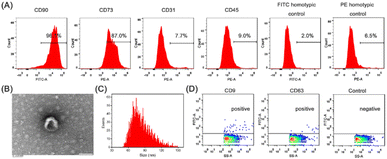
It has been reported that exosomes secreted from mesenchymal stem cells could promote the wound-healing process.38 Herein, adipose-derived mesenchymal stem cells (ADSCs) isolated from rats were stained with cell surface markers and detected with flow cytometry. The previous reports have shown that ADSCs could express CD29, CD73, CD90 and CD105, but not CD19, CD31, CD34 and CD45.39–41 In this section, the CD90, CD73, CD31 and CD45 markers were used to identify ADSCs. The fraction positive of CD90 was 96.1% and CD73 was 87.9%, whereas the expression of CD31 and CD45 were negligible with respect to the PE homotypic control (Fig. 3A). It can be proved that ADSCs were successfully isolated. Further, the exosomes secreted by ADSCs were collected by differential centrifugation. TEM image of exosomes showed saucer-like structures (Fig. 3B) that is the characteristic of general exosomes.42 The average size of exosomes measured by nanoparticle flow cytometry was about 83.68 ± 17.63 nm (Fig. 3C). Moreover, CD9 and CD63, known as typical surface markers of exosomes,43 were confirmed by flow cytometric analysis (Fig. 3D). Then, the prepared exosomes were used in the following study.2.3. Exo@FPG3 hybrid preparation and analysis
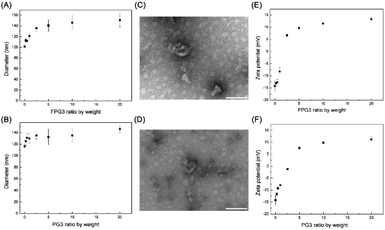
Then, the FPG3 and PG3 was applied to modify the exosomes at various weight ratios. The sizes of exo@FPG3 and exo@PG3 were measured by dynamic light scattering (DLS) (Fig. 4A and B). Compared to exosome alone, the sizes of exo@FPG3 and exo@PG3 increase by about 10–40 nm with increasing mass ratio. The morphologies of exo@FPG3 and exo@PG3 with ratio of 5 were observed by TEM, indicating that the surface of exosome was aggregated by FPG3 or PG3 (Fig. 4C and D). The interaction of exosomes with FPG3 was further evaluated by measuring the zeta potential of exo@FPG3 (Fig. 4E). Before the addition of FPG3, the average zeta potential of original exosomes was −14.2 ± 2.1 mV. The zeta potential of exo@FPG3 increased with the ascending weight ratio FPG3 to exosome, while no significant difference was observed at the ratios of 5, 10, or 20. The zeta potential was 9.6 ± 0.6 mV at the ratio of 5, which was similar to the surface charge of exosomes reversed by cationic amphiphilic macromolecule in a report.25 Meanwhile, the trend of exosome surface charge for exo@PG3 in Fig. 4F was similar to that of exo@FPG3. Then, the exo@FPG3 and exo@PG3 were prepared at a weight ratio of 5 for the next experiment.2.4. In vitro cytotoxicity
Before investigating the uptake and function of engineered exosomes, the biocompatibility of biological materials should be considered. The cytotoxicity of FPG3 or PG3 was evaluated after 24 h of treatment in human umbilical vein endothelial cells (HUVECs), respectively. The cell viability of HUVECs was presented in Fig. 5. Both FPG3 and PG3 did not influence the viability of the cells in the concentration range of 5 μg mL−1 to 150 μg mL−1. All the cell viability of the samples remained close to 90% even when the concentration of FPG3 or PG3 was as high as 150 μg mL−1, which was much higher than the concentration used to modify exosomes. The results showed that FPD3 or PD3 for the intracellular delivery of exosomes had good biocompatibility.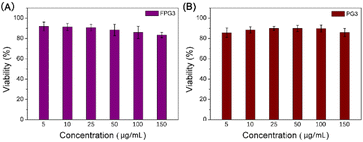 | ||
| Fig. 5 (A) Cytotoxicity of the FPG3 against HUVECs after 24 h incubation. (B) Cytotoxicity of the PG3 against HUVECs after 24 h incubation. | ||
2.5. Flow cytometer analysis and inhibition of specific endocytic pathways
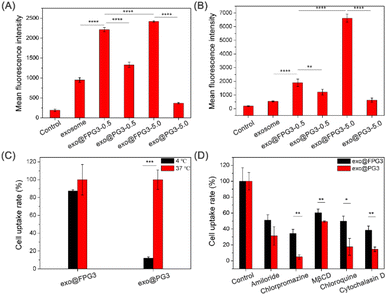
The biological activity of exosomes for therapy depends on the successful delivery of their cargos to target cells.44 Therefore, we then investigated the cellular uptake of exosomes modified with FPG3 or PG3 by HUVECs. After 2 h of incubation in the medium containing 10% FBS, the fluorescence intensity of exo@FPG3 (w/w, 0.5 and 5) group was much higher than that of exosomes and exo@PG3 groups (Fig. 6A), illustrating the ability of FPG3 for promoting the intracellular delivery of exosome. Meanwhile, in the serum-free medium, the fluorescence intensity for exo@FPG3 (w/w, 5.0) group was increased 3.4-fold compared with exo@FPG3 (w/w, 0.5) group that was incubated with cells. Moreover, the fluorescence intensity for exo@FPG3 (w/w, 5.0) group was 12.4-fold and 10.6-fold enhanced compared with exosomes and exo@PG3 (w/w, 5.0) groups (Fig. 6B), respectively. Thus, the efficiency of cellular uptake of exosomes depended on the concentrations of FPG3. In addition, FPG3 used for surface modification of exosomes have the advantage of the intracellular delivery of exosomes over PG3. Piffoux et al. reported that PEGylated liposomes were less efficient at fusion with exosomes than non-PEGylated liposomes.23 Cellular uptake studies also indicated that exogenous lipid components could regulate the interaction between exosomes and cells.45 Given the excellent performance of FPG3 prepared at a low ratio, the efficient cellular uptake of exo@FPG3 would not only attribute to the charge reversal of exosomes but also primarily own to the interaction of the highly hydrophobic fluorinated chains with the lipid bilayer of the exosome.46Encouraged by the above results, further research in endocytic pathways for exo@FPG3 or exo@PG3 was performed. HUVECs were treated with the engineered exosomes at a low temperature of 4 °C to analyze whether the intracellular uptake was energy-dependent. The quantitative results of both exo@FPG3 and exo@PG3 analyzed by flow cytometry demonstrated that the uptake efficiency of engineered exosomes was decreased under a low temperature (4 °C) (Fig. 6C), indicating that the main cellular uptake mechanism of engineered exosomes was endocytosis. It has been reported that cells take up exosomes through various endocytic pathways, such as clathrin-dependent endocytosis, macropinocytosis, caveolin-mediated uptake, phagocytosis, and lipid raft-mediated internalization.19 Then, amiloride (a macropinocytosis inhibitor in the epithelial sodium channel), chlorpromazine (an endocytic inhibitor in the clathrin-mediated pathway), MβCD (a lipid raft inhibitor blocking the caveolin-mediated endocytosis), chloroquine (a lysosome acidification inhibitor) and cytochalasin D (an actin polymerization inhibitor) were used to analyze the uptake mechanism and efficiency between exo@FPG3 and exo@PG3. The cells were pretreated with several endocytic inhibitors and supplemented with the exo@FPG3 or exo@PG3, followed by an assessment of the fluorescence intensity using flow cytometry after 2 h. Cells treated with engineered exosomes but without any inhibitors were set as controls. The intracellular uptake of exo@FPG3 and exo@PG3 was significantly repressed by any inhibitor, implying that the hybrids were internalized by multiple pathways (Fig. 6D). By comparison, the chlorpromazine and cytochalasin D significantly reduced the intracellular uptake efficiency of exo@FPG3 and exo@PG3, indicating that the internalization of hybrids was more associated with clathrin-mediated endocytosis.47
2.6. Intracellular internalization process observed by CLSM
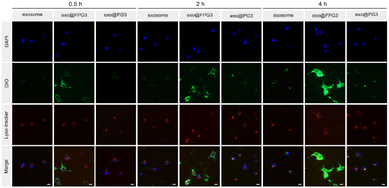
Furthermore, confocal laser scanning microscopy (CLSM) was utilized to investigate the intracellular internalization process of exo@FPG3 in HUVECs. As shown in Fig. 7, the green fluorescent signals in exosomes and exo@PG3 groups were negligible, whereas the strong green fluorescence from the exo@FPG3 group was clearly visualized in the cells after incubation 0.5 h. As the duration extended to 4 h, the green fluorescence for exosomes and exo@PG3 groups slightly enhanced over time, which was mainly co-located with the red fluorescence of endosome marker. In addition, the fluorescence for exo@FPG3 group filling in the whole cytoplasm was significantly increased over the incubation durations. These results revealed that the intracellular uptake of exosomes and exo@FPG3 by HUVECs was time dependent. The fluorescent images of cellular uptake demonstrated that exo@FPG3 had advantages over exo@PG3 and exosomes, which was in accordance with the results analyzed using flow cytometry. Meanwhile, the co-localization ratio of lyso-tracker and DiO-labeled exo@FPG3 was 0.39 after 4 h incubation (Fig. S12†), which was lower than that of exosome (0.62) and exo@PG3 (0.68). The difference in person correlation coefficient more intuitively demonstrated better lysosome escape by exo@FPG3. Probably due to the lysosomal escape ability of the fluorocarbon chains,48 most exo@FPG3 did not colocalize with intracellular lysosomes, indicating that the contents of exo@FPG3 would be efficiently released in the cytosol. The intracellular efficiency of exosomes promoted by exo@FPG3 might contribute to improve the biological effects of exosomes.2.7. Angiogenic effects of exo@FPG3 on HUVECs
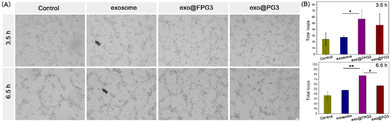
According to the above experiments, exo@FPG3 were efficiently swallowed by HUVECs. The effects of exo@FPG3 on cell function were sequentially examined. As angiogenesis plays an important role in skin wound healing, we then assessed the impact of exo@FPG3 on the angiogenic tube formation of HUVECs. After seeding on Matrigel, HUVECs were co-cultured with exosomes, exo@FPG3, exo@PG3 or PBS for 3.5 h and 6.5 h to investigate the formation of tube networks (Fig. 8A). As the activity of exosomes in tissue engineering, exosome or exo@PG3 could enhance the tube-forming ability of HUVECs compared with control group. Whereas, HUVECs treated with exo@FPG3 showed the best tube formation performance among all intervention groups after 6.5 h. Quantitative analysis of the total loops further confirmed that exo@FPG3 played a positive role in tube formation (Fig. 8B). These results revealed that the promoting effect of exo@FPG3 on angiogenesis can be attributable to the improved intracellular delivery of exosomes assisted by FPG3.2.8. Migration effects of exo@FPG3 on HUVECs
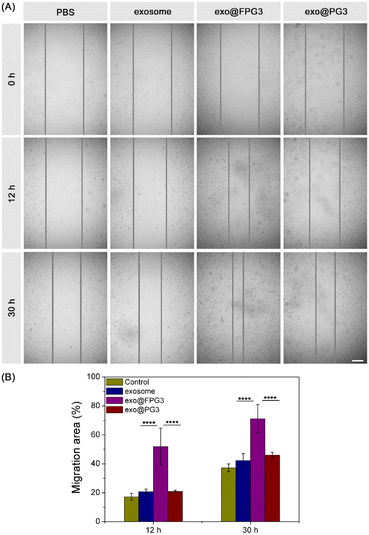
The scratch wound assay was further conducted to evaluate the motility of HUVECs. As shown in Fig. 9A, the scratched area of cell was decreased to different degrees with the extension of culture time. Compared with PBS-treated HUVECs, the exosome group and exo@PG3 group showed little improvement in the number of migrating cells. However, the scratch area of HUVECs was significantly reduced after treatment with exo@FPG3. Quantitative analysis of the scratch wound assay, the percent of migration area for exo@FPG3 group reached 71.2%, which was much higher than the other groups (Fig. 9B). The result demonstrated that the mobility of HUVECs was remarkably accelerated in the presence of exo@FPG3, which may be related to the higher bioavailability of exosomes.3. Experimental
3.1. Materials
(3-Aminopropyl) triethoxysilane, heptafluorobutyric anhydride and triethylamine (TEA), ninhydrin, hydrindantin dihydrate, chlorpromazine hydrochloride, amiloride hydrochloride dihydrate, MβCD and cytochalasin D were provided from Aladdin® Ltd (Shanghai, China). Chloroquine was ordered from Yuanye Biotechnology Co. Ltd (Shanghai, China). Boc-L-Lys (Boc)-OH, N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole hydrate (HOBT), N,N-diisopropylethylamine (DIPEA) and trifluoroacetic acid (TFA) were ordered from GL Biochem. Ltd (Shanghai, China). Biotechnology-grade cellulose ester (CE) membranes (MWCO = 100–500 Da) was obtained from Spectrum/Por. 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO), Lyso-tracker Red and 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) were purchased from Beyotime Biotechnology Co. Ltd (Shanghai, China). Exosome spin columns were produced by Rengen Biosciences Biotechnology Co. Ltd (Liaoning, China). Penicillin-streptomycin, trypsin and sterile PBS were from Solarbio® Life Sciences (Beijing, China). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were ordered from Life Technologies Corporation (Gibco®, USA). The BCA protein assay kit was purchased from Seven Biotech. (Beijing, China). CD90 (561![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 973), CD45 (554
973), CD45 (554![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 878), CD73 (551
878), CD73 (551![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 123), CD31 (555
123), CD31 (555![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027), IgG (550
027), IgG (550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 616, 550
616, 550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617) and stain buffer were produced by BD Pharmingen (USA). Cell counter kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). Matrigel was commercially available from Corning (USA). Sprague–Dawley rats were provided by Henan Experimental Animal Center (Zhengzhou, China). The study involving animals was conducted in accordance with the Principles of Laboratory Animal Care (GB/T 35
617) and stain buffer were produced by BD Pharmingen (USA). Cell counter kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). Matrigel was commercially available from Corning (USA). Sprague–Dawley rats were provided by Henan Experimental Animal Center (Zhengzhou, China). The study involving animals was conducted in accordance with the Principles of Laboratory Animal Care (GB/T 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892–2018), and was approved by the Institutional Animal Ethics Committee of the First Affiliated Hospital of Zhengzhou University (approval number: 2021-KY-0885). The informed consent was obtained for any experimentation with animal subjects. All other chemicals were of analytical grade.
892–2018), and was approved by the Institutional Animal Ethics Committee of the First Affiliated Hospital of Zhengzhou University (approval number: 2021-KY-0885). The informed consent was obtained for any experimentation with animal subjects. All other chemicals were of analytical grade.
3.2. Synthesis and characterization of fluorinated peptide dendrimer
The POSS core-based peptide poly(L-lysine) dendrimers were synthesized according to the previously reported method.49 In detail, the synthesis and characterization of POSS core, generation 1, 2 and 3 dendrimers (PG1, PG2, PG3) were presented in the ESI.† Then, PG3 (1.0 g, 0.12 mmol) was dissolved in anhydrous methanol (15 mL). The heptafluorobutyric anhydride (1.07 mL) and triethylamine (0.78 mL) were respectively added into the reaction bulb. The reaction solution was stirred in an ice bath for 30 min. The reaction sequentially proceeded for 3 days at 30 °C. After the removal of the excess solvent by rotational evaporation, the concentrated solution was added to anhydrous diethyl ether for further precipitation. A CE membrane with a molecular weight of 100–500 Da was used to purify the crude product. After dialysis for 3 days and lyophilization, the fluorinated peptide dendrimer (FPG3) was acquired and confirmed by mass spectrometry.The ninhydrin assay was used to test the primary amine groups, which was further to predict the number of fluorinated groups on PG3 peptide dendrimer. First, 85 mg of ninhydrin and 15 mg of hydrindantin dihydrate were dissolved in 10 mL of ethylene glycol-methyl ether, which was mixed with sodium acetate buffer (0.2 M, pH 5.4) at the same volume ratio. The mixture solution (400 μL) was added with different amounts of PG3 or FPG3 and the final volume of solution was adjusted to 600 μL using deionized water. All samples were incubated in boiling water for 10 minutes. The solution was cooled to room temperature, followed by addition with 600 μL of mixture of ethanol and water (v/v = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). UV spectrophotometer was used to test the absorbance of samples at 570 nm. Further, the fluoride degree of FPG3 was figured out according to the calibration curve given by the absorbance and the peptide dendrimer concentration.
4). UV spectrophotometer was used to test the absorbance of samples at 570 nm. Further, the fluoride degree of FPG3 was figured out according to the calibration curve given by the absorbance and the peptide dendrimer concentration.
3.3. Isolation and characterization of exosome secreted from ADSCs
Firstly, ADSCs sourced from adult female Sprague–Dawley rats were incubated as follows. In detail, rats were euthanized and soaked in ethyl alcohol for 15 min. The adipose tissue around the kidney and groin was carefully excised, avoiding the blood vessels. Washed with the sterile PBS containing 1% penicillin-streptomycin, the tissue in the cell culture dish was cut into pieces using sterile surgical scissors. Then, the homogenized adipose tissue was transferred to a 50 mL tube and digested by trypsin. The tube with contents was incubated with continuous shaking for 1 h at 37 °C water bath. An equal volume of DMEM with 10% FBS and 1% penicillin-streptomycin was added into the tube to terminate the digestion, followed by centrifugation at 1000 rpm for 5 minutes to remove the fat layer. The cells redispersed by DMEM medium were transferred to a 15 mL centrifuge tube and centrifuged again at 1000 rpm for 5 minutes. After removing the supernatant liquid, the cell was adhered for 5 days in the cell culture flask at 37 °C with 5% CO2. During the period, the culture medium was replaced with the fresh medium. The cells grown to passage 3 were harvested and assigned into a tube, which was stained by 1 μL FITC or PE-conjugated antibodies of CD90, CD73, CD31 and CD45 at room temperature for 15 min in the dark. The FITC or PE-labeled IgG antibody was used as an isotype control. Then, the flow cytometric analysis was performed to identify the characteristics of cells.Then, ADSCs were grown in DMEM containing 10% exosome-free FBS for reaching complete confluency. The cell medium was collected and prepared to isolate exosomes secreted by ADSCs using ultracentrifugation. All the following procedures were performed at 4 °C. In brief, the culture medium of ADSCs was centrifuged at 2000 g for 30 min to get rid of dead cells and debris. The supernatants were carefully transferred into a new tube for further centrifuging treatment at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 45 min to separate the large vesicle. After filtration through a 0.45 μm filter, the resulting supernatants were ultra-centrifuged at 100
000 g for 45 min to separate the large vesicle. After filtration through a 0.45 μm filter, the resulting supernatants were ultra-centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 70 min to obtain exosomes. For further purification, the supernatants were discarded and the remaining was resuspended with 10 mL precooled PBS and centrifuged at 100
000 g for 70 min to obtain exosomes. For further purification, the supernatants were discarded and the remaining was resuspended with 10 mL precooled PBS and centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for another 70 min. Finally, the supernatants were discarded again and exosomes were resuspended in cold 500 μL PBS. The morphology and size characterization of exosomes were observed and detected respectively by transmission electron microscopy (TEM) and NanoFCM (N30E). Binding by anti-CD9 antibody and anti-CD63 antibody, the specific proteins of exosome were also detected. Meanwhile, the partially isolated exosomes were treated with RIPA lysate and the protein concentration of exosomes was measured by BCA Assay Kit according to the operation instructions. The exosome left was stored at −80 °C until further use.
000 g for another 70 min. Finally, the supernatants were discarded again and exosomes were resuspended in cold 500 μL PBS. The morphology and size characterization of exosomes were observed and detected respectively by transmission electron microscopy (TEM) and NanoFCM (N30E). Binding by anti-CD9 antibody and anti-CD63 antibody, the specific proteins of exosome were also detected. Meanwhile, the partially isolated exosomes were treated with RIPA lysate and the protein concentration of exosomes was measured by BCA Assay Kit according to the operation instructions. The exosome left was stored at −80 °C until further use.
3.4. Exo@FPG3 hybrid preparation and analysis
Different dilutions of the FPG3 or PG3 were respectively mixed with exosomes (5 μg protein) at mass ratios of 0.25, 0.5, 1, 2.5, 5, 10 and 20 for 20 min at room temperature to form the exo@FPG3 and exo@PG3. Then, the suspensions were ultrafiltered with 300 K Nanosep. PBS was added to the hybrid solution to 1 mL, followed by the detection of size and zeta potential using Malvern spray analyzer. Meanwhile, the morphologies of exo@FPG3 and exo@PG3 with ratio of 5 were conducted by TEM.3.5. In vitro cytotoxicity assay
The cytotoxicity of fluorinated peptide dendrimer or peptide dendrimer was detected with CCK-8 viability assay against HUVECs according to the manufacturer's instructions. Briefly, HUVECs was grown in DMEM containing 10% FBS and 1% penicillin-streptomycin, which were cultured at 37 °C in an incubator supplied with 5% CO2. Then, 100 μL of HUVECs suspension (5 × 103 cells per well) were seeded into 96-well plates for overnight culture. The cell medium was replaced with complete medium containing FPG3 or PG3 at different concentrations from 5 to 150 μg mL−1. The cells were continued in the incubator for 24 h. 100 μL of serum-free medium containing 10% (v/v) was added to 96-well plates and incubated with cells for 1.5 h at 37 °C. The absorbance value at 450 nm was quantificationally measured by microplate reader.3.6. Cellular uptake and mechanism analyzed using flow cytometry
The effect of exosome surface modification on intracellular uptake was investigated by flow cytometry. Firstly, exosomes (400 μg as protein) were tagged with 20 μg mL−1 DiO for 1 h at room temperature in 200 μL of PBS. Then, exosome spin columns (MW 4000 Da) were used to remove the unincorporated DiO from labeled exosome preparations according to the operating instruction. The labeled exosomes were mixed with FPG3 or PG3 at weight ratios of 0.5 and 5 for 30 min respectively. HUVECs (1 × 105 cells per well) were seeded in 12-well plates and incubated for 24 h at 37 °C in 5% CO2. Then, the original medium in the plates was removed. HUVECs were incubated with exosome, exo@FPG3 and exo@PG3 (5 μg protein per well) in serum-free medium or medium with 10% FBS for 2 h at 37 °C in 5% CO2. After the removal of the culture medium with exosomes, the cells were washed with PBS, treated with trypsin and dispersed into DMEM medium. After centrifugation and rinsing, the cell was resuspended in PBS, and the fluorescence was subsequently measured by flow cytometry.Based on the above results, the mass ratio of FPG3 or PG3 to exosome of 5 was chosen for the following experiments. To investigate the energy-dependent endocytosis, HUVECs were pretreated at 4 °C for 1 h, and the serum-free medium supplemented with exo@FPG3, exo@PG3 or exosome was exposed to cells at 4 °C for 2 h. The controls were cells incubated with exosome hybrid or exosome at the same amounts of protein at 37 °C in 5% CO2. Meanwhile, the mechanism of cellular uptake was further studied, and several inhibitors of cellular pathways were put to use. HUVECs were pretreated with the following inhibitors diluted with medium serum-free: chlorpromazine hydrochloride (10 μg mL−1), amiloride hydrochloride dihydrate (0.3 mg mL−1), chloroquine (15 μg mL−1), MβCD (4.0 mg mL−1) or cytochalasin D (5 μM) for 1 h at 37 °C in 5% CO2. Subsequently, the cells were incubated with medium containing exo@FPG3, exo@PG3 or exosome for 2 h in presence of inhibitors. Then, the cell samples were performed as above for fluorescence detection using flow cytometry. The percent of mean fluorescence for all groups relative to the controls was calculated.
3.7. Intracellular localization observed by CLSM
The DiO-labeled exosome hybrids were prepared as above operation to further investigate the intracellular location using confocal laser scanning microscopy. HUVECs (5 × 104 cells per dish) were seeded in a 35 × 12 mm glass-bottomed dish and cultivated overnight to allow cell adherence, which was incubated with exo@FPG3, exo@PG3 or exosome (5 μg protein per group) for a scheduled time respectively. After the removal of the DiO-labeled exosome, the cells were washed with PBS and stained with Lyso-tracker Red (0.2 μM) solution for 30 min. Rinsed again with PBS, the cells were fixed with 4% paraformaldehyde and further stained with DAPI for 15 min. Then, the cells immersed in PBS were observed using CLSM. Further, the person correlation coefficient of Lyso-tracker and DiO-labeled exosome, exo@FPG3 or exo@PG3 was calculated by ImageJ software.3.8. Tube formation assay
Angiogenesis was tested by performing tube formation assays. 50 μL of Matrigel was added into each well of a 96-well plate and solidified at 37 °C for 30 min. Then, HUVECs (2 × 104 cells per well) were seeded onto the above 96-well plate and cultured in serum-free medium supplemented with exosomes (25 μg mL−1 of protein) from different groups. The control group was the cell with the addition of an equal volume of PBS. After 3.5 h and 6.5 h incubation at 37 °C, tube formation was observed under an optical microscope. The total loops were analyzed by using ImageJ software to evaluate the abilities of tube formation.3.9. Scratch wound assay
For the scratch wound assay, HUVECs (1 × 105 cells per well) were seeded in 96-well plate and grown in an incubator until reaching 100% confluency. A sterile 200 μL pipette tip was used to scratch the cell monolayer uprightly across each well. The cells were washed twice with PBS to remove the exfoliated cells and respectively replenished with 100 μL basal medium containing 25 μg mL−1 exosome protein for exo@FPG3, exo@PG3 or exosome group. The images of scratch wounds were captured at 0 h, 12 h and 30 h and analyzed by ImageJ software. The cell migration ability was evaluated by the ratio of closure area to initial wound.3.10. Statistical analysis
All dates were presented as the mean value ± standard deviation. A two-tailed Student's t-test or one-way ANOVA test was utilized to compare the statistical differences between groups. GraphPad Prism version 9.0.1. was used for all testing. P values < 0.05 was considered statistically significant.4. Conclusion
In this study, we proposed an original fluorine-engineered exosomes (exo@FPG3) to overcome the biological barriers for intracellular delivery and enhance biological function of exosomes in cell migration and angiogenesis. It was revealed that the intracellular uptake of exo@FPG3 was energy-dependent, and the clathrin-mediated endocytosis was an important pathway for exo@FPG3 to enter HUVECs. Compared to the surface potential reversal of exo@FPG3, fluorous effect is more important in promoting cellular uptake efficiency and endosomal escape of exosomes. Additionally, the efficient cytosolic delivery of exo@FPG3 would further improve the ability of cell migration and angiogenesis in vitro. In summary, the first design of fluorine-engineered exosomes and their excellent performance in regulating cell uptake and function lay foundation for the development of exosome-based therapeutic systems for various diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded by National Natural Science Foundation of China (grant number: 32000940, 31871000), Health Commission of Henan Province (grant number: SBGJ202103047), the Henan Province Youth Talent Promoting Project (2022HYTP047) and the China Postdoctoral Science Foundation (grant number: 2019M662538).Notes and references
- M. Richter, P. Vader and G. Fuhrmann, Adv. Drug Delivery Rev., 2021, 173, 416–426 CrossRef CAS PubMed.
- J. Mondal, S. Pillarisetti, V. Junnuthula, M. Saha, S. R. Hwang, I. K. Park and Y. K. Lee, J. Controlled Release, 2022, 353, 1127–1149 CrossRef PubMed.
- A. Casado-Diaz, J. M. Quesada-Gomez and G. Dorado, Front. Bioeng. Biotechnol., 2020, 8, 146 CrossRef PubMed.
- J. Wang, H. Wu, Y. Peng, Y. Zhao, Y. Qin, Y. Zhang and Z. Xiao, J. Nanobiotechnol., 2021, 19, 202 CrossRef CAS PubMed.
- O. Staufer, F. Dietrich, R. Rimal, M. Schröter, S. Fabritz, H. Boehm, S. Singh, M. Möller, I. Platzman and J. P. Spatz, Sci. Adv., 2021, 7, eabg6666 CrossRef CAS PubMed.
- Y. Xiong, L. Chen, P. Liu, T. Yu, C. Lin, C. Yan, Y. Hu, W. Zhou, Y. Sun, A. C. Panayi, F. Cao, H. Xue, L. Hu, Z. Lin, X. Xie, X. Xiao, Q. Feng, B. Mi and G. Liu, Small, 2022, 18, e2104229 CrossRef PubMed.
- Q. Tang, B. Lu, J. He, X. Chen, Q. Fu, H. Han, C. Luo, H. Yin, Z. Qin, D. Lyu, L. Zhang, M. Zhou and K. Yao, Biomaterials, 2022, 280, 121320 CrossRef CAS PubMed.
- G. Kwak, J. Cheng, H. Kim, S. Song, S. J. Lee, Y. Yang, J. H. Jeong, J. E. Lee, P. B. Messersmith and S. H. Kim, Small, 2022, 18, e2200060 CrossRef PubMed.
- M. Wang, C. Wang, M. Chen, Y. Xi, W. Cheng, C. Mao, T. Xu, X. Zhang, C. Lin, W. Gao, Y. Guo and B. Lei, ACS Nano, 2019, 13, 10279–10293 CrossRef CAS PubMed.
- H. Henriques-Antunes, R. M. S. Cardoso, A. Zonari, J. Correia, E. C. Leal, A. Jiménez-Balsa, M. M. Lino, A. Barradas, I. Kostic, C. Gomes, J. M. Karp, E. Carvalho and L. Ferreira, ACS Nano, 2019, 13, 8694–8707 CrossRef CAS PubMed.
- R. C. de Abreu, H. Fernandes, P. A. da Costa Martins, S. Sahoo, C. Emanueli and L. Ferreira, Nat. Rev. Cardiol., 2020, 17, 685–697 CrossRef PubMed.
- Q. Wang, L. Zhang, Z. Sun, B. Chi, A. Zou, L. Mao, X. Xiong, J. Jiang, L. Sun, W. Zhu and Y. Ji, Mater. Today Bio, 2021, 12, 100171 CrossRef CAS PubMed.
- J. Zhang, C. Ji, H. Zhang, H. Shi, F. Mao, H. Qian, W. Xu, D. Wang, J. Pan, X. Fang, H. A. Santos and X. Zhang, Sci. Adv., 2022, 8, eabj8207 CrossRef CAS PubMed.
- L. Cheng, X. Zhang, J. Tang, Q. Lv and J. Liu, Biomaterials, 2021, 275, 120964 CrossRef CAS PubMed.
- T. Wu, Y. Liu, Y. Cao and Z. Liu, Adv. Mater., 2022, 34, e2110364 CrossRef PubMed.
- M. Xu, T. Feng, B. Liu, F. Qiu, Y. Xu, Y. Zhao and Y. Zheng, Theranostics, 2021, 11, 8926–8944 CrossRef CAS PubMed.
- Q. Wang, T. Li, J. Yang, Z. Zhao, K. Tan, S. Tang, M. Wan and C. Mao, Adv. Mater., 2022, 34, e2201406 CrossRef PubMed.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Théry, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- L. A. Mulcahy, R. C. Pink and D. R. Carter, J. Extracell. Vesicles, 2014, 3, 24641 CrossRef PubMed.
- A. Gonda, J. Kabagwira, G. N. Senthil and N. R. Wall, Mol. Cancer Res., 2019, 17, 337–347 CrossRef CAS PubMed.
- Z. Liao, H. Liu, L. Ma, J. Lei, B. Tong, G. Li, W. Ke, K. Wang, X. Feng, W. Hua, S. Li and C. Yang, ACS Nano, 2021, 15, 14709–14724 CrossRef CAS PubMed.
- S. A. A. Kooijmans, L. A. L. Fliervoet, R. van der Meel, M. H. A. M. Fens, H. F. G. Heijnen, P. M. P. van Bergen En Henegouwen, P. Vader and R. M. Schiffelers, J. Controlled Release, 2016, 224, 77–85 CrossRef CAS PubMed.
- M. Piffoux, A. K. A. Silva, C. Wilhelm, F. Gazeau and D. Tareste, ACS Nano, 2018, 12, 6830–6842 CrossRef CAS PubMed.
- Q. Zhan, K. Yi, X. Li, X. Cui, E. Yang, N. Chen, X. Yuan, J. Zhao, X. Hou and C. Kang, Macromol. Biosci., 2021, 21, e2100042 CrossRef PubMed.
- K. Feng, X. Xie, J. Yuan, L. Gong, Z. Zhu, J. Zhang, H. Li, Y. Yang and Y. Wang, J. Extracell. Vesicles, 2021, 10, e12160 CAS.
- W. Heusermann, J. Hean, D. Trojer, E. Steib, S. von Bueren, A. Graff-Meyer, C. Genoud, K. Martin, N. Pizzato, J. Voshol, D. V. Morrissey, S. E. Andaloussi, M. J. Wood and N. C. Meisner-Kober, J. Cell Biol., 2016, 213, 173–184 CrossRef CAS PubMed.
- I. Nakase and S. Futaki, Sci. Rep., 2015, 5, 10112 CrossRef PubMed.
- I. Nakase, Processes, 2021, 9, 224 CrossRef CAS.
- S. Futaki and I. Nakase, Acc. Chem. Res., 2017, 50, 2449–2456 CrossRef CAS PubMed.
- I. Nakase, M. Niwa, T. Takeuchi, K. Sonomura, N. Kawabata, Y. Koike, M. Takehashi, S. Tanaka, K. Ueda, J. C. Simpson, A. T. Jones, Y. Sugiura and S. Futaki, Mol. Ther., 2004, 10, 1011–1022 CrossRef CAS PubMed.
- J. Lv and Y. Cheng, Chem. Soc. Rev., 2021, 50, 5435–5467 RSC.
- C. Zhang, K. Yan, C. Fu, H. Peng, C. J. Hawker and A. K. Whittaker, Chem. Rev., 2022, 122, 167–208 CrossRef CAS PubMed.
- J. Lv, H. Wang, G. Rong and Y. Cheng, Acc. Chem. Res., 2022, 55, 722–733 CrossRef CAS PubMed.
- Z. Zhang, W. Shen, J. Ling, Y. Yan, J. Hu and Y. Cheng, Nat. Commun., 2018, 9, 1377 CrossRef PubMed.
- G. Rong, C. Wang, L. Chen, Y. Yan and Y. Cheng, Sci. Adv., 2020, 6, eaaz1774 CrossRef CAS PubMed.
- M. Wang, H. Liu, L. Li and Y. Cheng, Nat. Commun., 2014, 5, 3053 CrossRef PubMed.
- F. S. Tabatabaei Mirakabad, M. S. Khoramgah, K. Keshavarz F, M. Tabarzad and J. Ranjbari, Life Sci., 2019, 233, 116754 CrossRef CAS PubMed.
- Y. Hu, R. Tao, L. Chen, Y. Xiong, H. Xue, L. Hu, C. Yan, X. Xie, Z. Lin, A. C. Panayi, B. Mi and G. Liu, J. Nanobiotechnol., 2021, 19, 150 CrossRef CAS PubMed.
- Y.-T. Chen, C.-C. Yang, Y.-Y. Zhen, C. G. Wallace, J.-L. Yang, C.-K. Sun, T.-H. Tsai, J.-J. Sheu, S. Chua, C.-L. Chang, C.-L. Cho, S. Leu and H.-K. Yip, Stem Cell Res. Ther., 2013, 4, 62 CrossRef CAS PubMed.
- S. Pak, S. W. Hwang, I. K. Shim, S. M. Bae, Y.-M. Ryu, H.-B. Kim, E.-J. Do, H.-N. Son, E.-J. Choi, S.-H. Park, S.-Y. Kim, S. H. Park, B. D. Ye, S.-K. Yang, N. Kanai, M. Maeda, T. Okano, D.-H. Yang, J.-S. Byeon and S.-J. Myung, Sci. Rep., 2018, 8, 11314 CrossRef PubMed.
- B. Li, S. Luan, J. Chen, Y. Zhou, T. Wang, Z. Li, Y. Fu, A. Zhai and C. Bi, Mol. Ther.–Nucleic Acids, 2020, 19, 814–826 CrossRef CAS PubMed.
- C. Théry, L. Zitvogel and S. Amigorena, Nat. Rev. Immunol., 2002, 2, 569–579 CrossRef PubMed.
- L. Zhu, H.-T. Sun, S. Wang, S.-L. Huang, Y. Zheng, C.-Q. Wang, B.-Y. Hu, W. Qin, T.-T. Zou, Y. Fu, X.-T. Shen, W.-W. Zhu, Y. Geng, L. Lu, H.-L. Jia, L.-X. Qin and Q.-Z. Dong, J. Hematol. Oncol., 2020, 13, 152 CrossRef PubMed.
- Y. Liang, L. Duan, J. Lu and J. Xia, Theranostics, 2021, 11, 3183–3195 CrossRef CAS PubMed.
- Y. T. Sato, K. Umezaki, S. Sawada, S.-a. Mukai, Y. Sasaki, N. Harada, H. Shiku and K. Akiyoshi, Sci. Rep., 2016, 6, 21933 CrossRef CAS PubMed.
- M. C. Z. Kasuya, S. Nakano, R. Katayama and K. Hatanaka, J. Fluorine Chem., 2011, 132, 202–206 CrossRef CAS.
- H. Xu, C. Liao, S. Liang and B.-C. Ye, ACS Appl. Mater. Interfaces, 2021, 13, 10760–10767 CrossRef CAS PubMed.
- T. Zhu, L. Shi, C. Ma, L. Xu, J. Yang, G. Zhou, X. Zhu and L. Shen, Biomater. Sci., 2021, 9, 658–662 RSC.
- X. Cai, R. Jin, J. Wang, D. Yue, Q. Jiang, Y. Wu and Z. Gu, ACS Appl. Mater. Interfaces, 2016, 8, 5821–5832 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra00300k |
| This journal is © The Royal Society of Chemistry 2023 |