DOI:
10.1039/D2RA07338B
(Paper)
RSC Adv., 2023,
13, 19725-19737
Optimization, fractional characterization, and antioxidant potential of exopolysaccharides from Levilactobacillus brevis NCCP 963 isolated from “kanji”†
Received
18th November 2022
, Accepted 15th June 2023
First published on 30th June 2023
Abstract
A novel exopolysaccharide (EPS) was obtained from Levilactobacillus brevis NCCP 963 isolated from a black carrot drink named “kanji”. The culture conditions for maximum EPS yield were explored by the Plackett–Burman (PB) design and response surface methodology (RSM) along with the fractional characterization and antioxidant potential of EPSs. The PB design screened out five significant factors, namely, glucose, sucrose, tryptone, CaCl2, and di-potassium phosphate out of eleven independent factors. The RSM indicated glucose and CaCl2 as significant factors in EPS production and a maximum EPS production of 968.89 mg L−1 was obtained at optimized levels of 10.56% glucose, 9.23% sucrose, 0.75% tryptone, 0.446% CaCl2, and 0.385% K2HPO4. A R2 value above 93% indicates higher variability, depicting the validity of the model. The obtained EPS has a molecular weight of 5.48 × 104 Da and is a homopolysaccharide in nature with glucose monosaccharides. FT-IR analysis showed significant band stretching of C–H, O–H, C–O and C–C and indicated the β-glucan nature of EPSs. The comprehensive antioxidant investigation showed significant in vitro DPPH, ABTS, hydroxyl, and superoxide scavenging capacity with EC50 values of 1.56, 0.31, 2.1, and 6.7 mg mL−1 respectively. Curd formation from the resulting strain prevented syneresis.
1. Introduction
With the advance in technology, natural polymers have gained great interest for various health, food, chemical, pharmaceutical and biotechnological applications,1 leading to increased demand for microbial exopolysaccharides (EPSs).2 Several microorganisms such as lactic acid bacteria (LAB), algae, and fungi amalgamate polysaccharides in the extracellular space and discharge them outside the cell in the form of either insoluble or soluble polymers. However, in the last decade, LAB was under extensive study because of its potential to synthesize EPSs of high molecular weight,3 serve as a probiotic with other nutritional attributes, offer protection against bowel disorders, diarrhea, and allergies as well as boost the immune system.4 Many beneficial effects including antioxidant, antimicrobial, anti-inflammatory, antiviral, immunomodulatory, antitumor, and cholesterol adsorption activities of LAB-derived EPSs have been reported.3,5,6 Furthermore, LAB-derived EPSs are used as syneresis-lowering agents, texturizers, viscosifiers, and emulsifiers, and also exhibited film-making capabilities and pseudoplastic rheological behavior in the food industry.7
Literature indicates that there are a number of factors such as carbon and nitrogen sources, pH of the culture media, growth medium, incubation temperature, enzyme production, and process conditions that affect the EPS production from LAB. Previously, single-factor optimization, i.e. change of one factor at a time keeping the other factors constant, has been used for bioprocess optimization. However, this method could not assess the affect between the interactions of the factors. Therefore, this has been done through appropriate statistical designs such as central composite rotatory design (CCRD) and response surface methodology (RSM) in recent years to optimize the conditions of a targeted response.8
Lactic acid food fermentation is an old process, whereby microbes convert fermentable sugars into lactic acid in the food substrate and other limited products via enzymatic activities. “Kanji” is one of the fermentation products made from black carrots (Daucus carota), which is commonly consumed in the Indo-Pak subcontinent. Black carrots are rich in anthocyanin pigment with high antioxidant activity. Kanji made from these carrots is a spontaneous lactic acid fermentation product with the addition of mustard, salt and/or red chilli powder. This drink is popular as a remedy for indigestion, anorexia and liver disorders. It is consumed extensively in the early summer season as an appetizer.9
Despite the obvious beneficial effects of black carrots and their drink, there are not many studies available in the literature on their processing. Although LAB has been explored in the kanji, the production of EPSs under optimized conditions has not been reported yet. Therefore, keeping in view the economic production of EPSs through LAB from fermentation products, this present study aimed to isolate LAB from kanji, explore the EPS-producing LAB and optimize the media composition for maximum EPS yield by the Plackett–Burman design and response surface methodology. The fractional characterization and antioxidant potential of the selected lactic acid strain have also been studied.
2. Materials and methods
2.1 Sample collection
The conventional kanji product available in the local market was collected, brought to the laboratory in sterilized bottles and kept at 4 °C until use.
2.2 Screening of the isolates for EPS production
For the isolation of LAB, MRS/M-17 (Oxoid-UK) media supplemented with 2% sucrose were used. Serially diluted samples were inoculated on MRS/M-17 agar plates and anaerobically incubated at 30 and 37 °C for 48–72 h. LAB were initially screened out keeping in view the ropiness, stickiness, and mucoid behavior of colonies on an agar medium. Screened out strains were further inoculated in their respective broth media (50 mL MRS/M-17 supplemented with 2% sucrose) under the above-mentioned incubation conditions.10
2.3 Preliminary identification of strains by biochemical attributes
The colonies isolated from the cultured media on MRS/M17 agar were recognized/identified via microscopic examination, gram staining, biochemical tests, and carbohydrate fermentation methods.9 Colonies on the MRS/M17 agar with sliminess, mucoid or ropy behavior were first separated. Then, the isolated strains that showed purple colored cells, either coccus or rod, via gram staining were selected for further identification by a catalase test. An individual colony was picked via an inoculation loop and smeared on the cleaned surface of a glass slide. Few drops of hydrogen peroxide (Merck, Germany, H2O2 – 3%) were dropped onto the colonies and oxygen bubble formation was observed. LAB isolates were further characterized by sugar fermentation assays using API 50 CH strips (BioMérieux, France).11
2.4 Identification of main EPS-producing strains by using 16S rRNA
The high yielding EPS strain NCCP 963 (strain deposit no from NCCP) was selected for further species identification. Concisely, the 16S sRNA gene was amplified using primers ZU1F-AGAGTTTGATCCTGGCTCAG and ZU1R-GGTTACCTTGTTACGACTT and Premix Taq™ DNA polymerase (Ex Taq™ Version 2.0, Takara Cat# RR003A). All the PCRs were performed in a 15 μL volume mixture comprising 2 μL DNA, 7.5 μL master mix, 3.5 μL PCR water and 1 μL of each primer [10 pmol L−1]. The resulting intensifications were initiated with a DNA thermal cycler made by Veriti® California, USA. The PCR product was purified and sequenced using 16S rRNA gene sequencing primers (27F, 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R, 5′-TACGGYTACCTTGTTACGACTT-3′) at Macrogen, Korea (https://dna.macrogen.com/). Finally, the resulting sequences of both primers were quality checked and assembled using BioEdit. Identification was done using the EzBioCloud server (https://www.ezbiocloud.net/).
2.5 Phylogenetic tree assessment
After sequencing the DNA data, the phylogenetic tree was constructed using MEGA SOFTWARE 11.0 by the neighbor joining (NJ) method for the alignment of sequences.12
2.6 Acid and bile tolerance
The main EPS-producing strain was assessed by its growth at higher concentrations of bile salts and low pH.13
2.7 Isolation and purification of EPSs
The EPS was produced by culturing the isolated strain Levilactobacillus brevis NCCP 963 on MRS broth at 37 °C for 3–4 days under anaerobic conditions with slight alterations.14 Briefly, the obtained turbid culture was heated at 80 °C for 10 min to denature the enzymes capable of polymer degradation. Then, centrifugation was performed at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C and the supernatant was precipitated by the addition of chilled ethanol in a double volume and kept overnight at 4 °C. It was followed by centrifugation at 11
000 rpm for 15 min at 4 °C and the supernatant was precipitated by the addition of chilled ethanol in a double volume and kept overnight at 4 °C. It was followed by centrifugation at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C. Proteins were removed as per centrifugation conditions described earlier. The pH of the resulting supernatant was increased to 8.5 by adding 6 M NaOH drop by drop and again centrifuged at 11
000 rpm for 15 min at 4 °C. Proteins were removed as per centrifugation conditions described earlier. The pH of the resulting supernatant was increased to 8.5 by adding 6 M NaOH drop by drop and again centrifuged at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The pH of the resulting supernatant was lowered to neutral by adding 1 mL of 10% TCA, and purified exopolysaccharide was gathered by adding chilled ethanol in an equal volume. Furthermore, the resulting pellet was liquefied in deionized H2O, dialyzed (molecular weight cutoff of 12
000 rpm for 15 min. The pH of the resulting supernatant was lowered to neutral by adding 1 mL of 10% TCA, and purified exopolysaccharide was gathered by adding chilled ethanol in an equal volume. Furthermore, the resulting pellet was liquefied in deionized H2O, dialyzed (molecular weight cutoff of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 14
000 to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kDa) for 48 h against distilled water with four times replacement of water per day and finally lyophilized. The total sugar content of EPSs was determined by the phenol–sulfuric method, and protein fractions were estimated by the Bradford method.8
000 kDa) for 48 h against distilled water with four times replacement of water per day and finally lyophilized. The total sugar content of EPSs was determined by the phenol–sulfuric method, and protein fractions were estimated by the Bradford method.8
2.8 Initial screening of media components
The Plackett–Burman (PB) design was used for screening the effect of most effective factors (glucose, fructose, lactose, sucrose, yeast extract, beef extract, tryptone, MgSO4, CaCl2, K2HPO4, and NaH2PO4) on bacterial growth using design expert software version 11.1.2.0.8 Each of the eleven (11) factors were given a low (−) and a high (+) value (Table 1). Subsequently, the EPS yield as a response is expressed in mg L−1 (Table S1†). The experimental order was fully randomized to have protection against the effects of prowling variables.
Table 1 Levels of independent variables used for the Plackett–Burman (PB) screening design
| Factor |
Name (%) |
Low |
High |
| A |
Glucose |
1.0 |
5.0 |
| B |
Fructose |
1.0 |
5.0 |
| C |
Lactose |
1.0 |
5.0 |
| D |
Sucrose |
1.0 |
5.0 |
| E |
Yeast extract |
0.01 |
0.05 |
| F |
Beef extract |
0.05 |
0.15 |
| G |
Tryptone |
0.25 |
1.50 |
| H |
MgSO4 |
0.15 |
0.25 |
| J |
CaCl2 |
0.005 |
0.015 |
| K |
K2HPO4 |
0.02 |
0.07 |
| L |
NaH2PO4 |
0.15 |
0.25 |
2.9 Medium optimization by central composite rotatory design (CCRD)
Five significant physiological factors, namely, glucose, sucrose, tryptone, CaCl2 and K2HPO4 were screened out by the PB design. However, further statistical optimization was done for maximum EPS production by RSM using the Design-Expert® 11 software (Stat Ease Inc., Minneapolis, USA),15 and the central composite rotatable design was adopted. The design comprised 45 runs in total (3 central points, 10 axial and 32 points factorial). The levels of five factors were coded as −α, −1, 0, +1 and +α (Table 2), and the design matrix is presented in Table 3.
Table 2 Levels of coded values for the CCRD design
| Variable (%) |
Levels |
| −α |
−1 |
0 |
+1 |
+α |
| Glucose |
1.55 |
5 |
7.5 |
10 |
13.45 |
| Sucrose |
1.55 |
5 |
7.5 |
10 |
13.45 |
| Tryptone |
0.15 |
0.7 |
1.1 |
1.5 |
2.05 |
| Calcium chloride (CaCl2) |
0.16 |
0.3 |
0.4 |
0.5 |
0.64 |
| Di-potassium hydrogen phosphate (K2HPO4) |
0.09 |
0.3 |
0.45 |
0.6 |
0.81 |
2.10 Fourier transform-infrared (FT-IR) spectroscopy
A FT-IR spectrophotometer (PerkinElmer, USA) was used to analyze the functional groups existing in the EPS in the spectrum range of 4000 to 600 cm−1. The sample pellets were prepared by mixing 1 mg of purified EPS with 100 mg of potassium bromide KBr and scanned in the above-mentioned spectrum range.16
2.11 Monosaccharide composition of EPSs
For the monosaccharide composition, the isolated exopolysaccharide was hydrolyzed by heating 2 M TFA at 120 °C for 2 h.17 The free monomers were adapted to their alditol acetates and further examined by GC. The standard alditol acetates for the monosaccharide mixture were generated under the same experimental conditions used to prepare the exopolysaccharide sample. Analysis was performed using a Varian GC/MS 4000 instrument (USA) equipped with a VF-5ms 30 m × 0.25 mm × 0.10 μm column under the following conditions—injector temperature: 320 °C; split ratio: 10; column flow: 1 mL min; carrier gas: He 99.999%; column oven temperature: 150 °C (hold time 2 min) to 300 °C (hold time 2 min) reached via a rising gradient of 10 °C min−1; ionization: EI scan type full.
2.12 Molecular weight (Mw) determination
For the molecular weight (Mw), the refined freeze-dried exopolysaccharide produced from Levilactobacillus brevis NCCP 963 was analyzed using an Agilent 1100 series HPLC system (Agilent technologie Palo AHO, CA, USA).17 The system was linked with a RI-refractive index detector (TOSOH TSK-G4000 PWxl) column 7.8 mm × 30 cm × 10 μm, (TOSOH Corp., Tokyo, Japan). First, 20 μL of sample was injected at a flow rate of 0.5 mL min−1 and a column temperature of 35 °C. Separation was carried out using 0.71% Na2SO4 as a mobile phase. Dextran (D2000) with a Mw of 2 × 106 Da, D8 with a mol. wt. of 1.338 × 105 Da, D7 with a mol. wt of 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 Da, D5 with a mol. wt of 21
100 Da, D5 with a mol. wt of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 Da, and D4 with a mol. wt of 10
400 Da, and D4 with a mol. wt of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da were used. D0 glucose with a Mw of 180 Da was used as the reference compound. These reference standards were added into the mobile phase at a rate of 10 mg per 1 mL solution.10
000 Da were used. D0 glucose with a Mw of 180 Da was used as the reference compound. These reference standards were added into the mobile phase at a rate of 10 mg per 1 mL solution.10
2.13 In vitro antioxidant analysis
2.13.1 DPPH radical scavenging activity. In vitro, the DPPH free radical scavenging activity was studied to assess the antioxidant potential of the exopolysaccharide.8 First, 0.5 mL of the sample with different concentrations of EPSs, i.e., 2, 4, 6, 8 and 10 mg mL−1 was mixed with 0.5 mL of DPPH solution (0.1 mmol L−1) in ethanol (95%) and kept in the darkness for 30 min. The absorbance level was checked at 517 nm against the blank solution. Methanol was used as a blank and ascorbic acid was used as a positive control. The scavenging activity of the DPPH radical was measured as follows:| | |
DPPH radical scavenging activity (%) = (A0 − A1/A0) × 100
| (1) |
where A1 is the absorbance of the sample and A0 is the blank solution absorbance. The EC50 value was considered as the concentration of EPS solutions that scavenged the 50% DPPH free radicals, used for the evaluation of this radical scavenging activity.
2.13.2 ABTS˙+ radical scavenging activity. The ABTS+ activity was performed by the reaction of 7 nM ABTS in distilled water containing 2.45 mM K2S2O8, and the reaction mixture was placed in the darkness at room temperature for 16 hours.18 Then, 2 mL of EPS solution with different concentrations of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7 and 0.8 mg mL−1 were added to the ABTS˙+ (4 mL) solution. After this, the mixture was incubated for 5 min in the darkness at ambient temperature, checked against the absorbance level of 734 nm along with control (ascorbic acid) and measured as follows:| | |
ABTS˙+ radical scavenging activity (%) = (1 − A1/A0) × 100
| (2) |
where A1 is the absorbance of the sample and A0 is the absorbance of the control.
2.13.3 Hydroxyl radical scavenging activity. The hydroxyl radical scavenging activity was also done with slight modifications.19,20 Briefly, 1 mL of EPS solution with different concentrations of 1, 2, 4, 6, 8 and 10 mg mL−1 was mixed with 1.8 mM salicylic acid, 1.8 mM FeSO4 and 0.3% H2O2. After placing in an incubator at 37 °C for 30 min and centrifugation at 3000 rpm for 5 min, the supernatant's absorbance was checked at 520 nm. Ascorbic acid was considered as the positive control and the calculation method was similar to that of DPPH radical activity. The absorbance of the control sample was checked by the replacement of salicylic acid with distilled water.| | |
Hydroxyl radical scavenging activity (%) = (A1 − A0)/(A − A0) × 100
| (3) |
where A1 represented the absorbance of the sample, A0 reflected the absorbance of the control in the absence of sample and A represented the absorbance without the sample.
2.13.4 Superoxide anion scavenging activity. This activity was assessed by a pyrogallol autoxidation system under alkalescent conditions.21 Briefly, 1 mL of different concentrations of EPSs, i.e. 1, 2, 4, 6, 8 and 10 mg mL−1, was added to 3 mL of 0.05 M Tris-HCl at a hydrogen concentration of 8.2, and kept for incubation at 25 °C for 10 min. Afterwards, 200 μL of preheated (25 °C) 30 mM pyrogallol was mixed and keep at the same temperature for 4 min, and the process was crossed by the addition of concentrated HCl (0.5 mL) and the absorbance of the resulting sample was measured at 329 nm along with ascorbic acid as the control. The superoxide anion scavenging activity was calculated as that of the DPPH activity with replacement of pyrogallol with distilled water for the absorbance of the positive control.| | |
Superoxide anion scavenging activity on (%) = (A0 − A1/A0) × 100
| (4) |
where A0 is the absorbance of blank and A1 is the absorbance of the sample.
2.14 Milk coagulation test 5% (v/v)
Levilactobacillus brevis pre-cultured in a MRS broth was inoculated in 10% (w/v) sterile skim milk supplemented with 3% (w/v) sucrose and milk coagulation was observed after an incubation time period of 48 h at 30 °C.
3. Results and discussion
3.1 Screening of EPS-producing isolates
In total, twelve EPS-producing strains were selected based on the morphological attributes presented in Tables S2 and S3.† Only one strain (Fig. 1), laterally identified as Levilactobacillus brevis having opaque small colonies on MRS media, was selected depending upon the ability of high EPS production (initial stage: 196.66 mg L−1). The maximum yielding strain was later deposited at National Culture Collection of Pakistan (NCCP), National Agriculture Research Center (NARC), Pakistan and was assigned number NCCP 963. Morphologically, Levilactobacillus brevis NCCP 963 was rod shaped with catalase and oxidase negative outcomes (Table S2†).
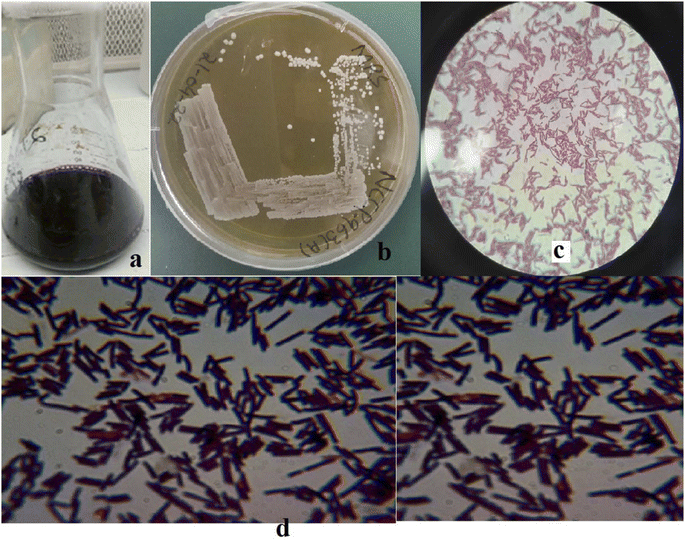 |
| | Fig. 1 (a) Kanji drink, (b) Levilactobacillus brevis NCCP 963 growth on MRS agar. (c) and (d) Morphological view of NCCP 963 under a microscope. | |
3.2 Levilactobacillus brevis 16S rRNA analysis
The genomic DNA of the isolated strain was extracted and amplified for 16S rRNA gene sequencing using ZUIF and ZUIR primers. The 16S rRNA sequences (Fig. S2 and S3†) of isolated strain were submitted to the NCBI database attributed to accession number LC712923, revealing 99.65% of identity with strains of Levilactobacillus brevis available on database.
3.3 Acid and bile salt tolerance
NCCP 963 presented a better existence under the pH conditions of 2, 3, 4 and 7 and bile concentrations of 0.2% and 0.4%. The results were in accordance with those reported in the literature where several L. brevis strains showed acid and bile tolerance behavior. Several L. brevis strains with probiotic nature have been reported previously.4,23,24 L. brevis B13-2 isolated from Chinese Kimchi revealed tolerance against bile salts and acids in an artificial gastric environment and adhered to HT-29 cells.22 Moreover, probiotic attributes, especially resistance to bile, were linked with production of EPSs from L. brevis strain25 and indicated the probiotic behavior of the isolated strain.
3.4 Initial screening of media composition
For the determination of key factors significantly affecting the EPS production/yield, a total of eleven factors, namely, glucose, fructose, lactose, sucrose, yeast extract, beef extract, tryptone, MgSO4, CaCl2, K2HPO4, & NaH2PO4 at different concentrations were optimized. Based on the PB design (Table S1†), a fitted first-order model, presenting the correlation among the 11 variables and EPS production could be described as follows:| |
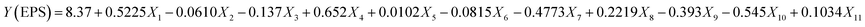 | (5) |
where Y was denoted as the yield of EPSs and Xi are the coded independent variable values and the coefficient of each variable representing the extent of the effect on EPS yield. The linear regression coefficient R2 was found to be 0.9957, and adjusted coefficient determination was adj-R2 = 0.9724, indicating that the variability of the data in the response yield could be well presented by the model. In this case, the p-values for the five variables (glucose, sucrose, tryptone, CaCl2 and K2HPO4) were <0.05, indicating that these independent variables were the important factors that affect the EPS production (Table 4). The significance of factors decreased in the following order: sucrose > glucose > K2HPO4 > tryptone > CaCl2 > MgSO4 > lactose > fructose > beef extract > yeast extract, accounting for 29.7%, 20.9%, 19.6%, 15.4%, 9.1% 2.7%, 0.5%, 0.36%, 0.31% and 0.0018% respectively. It is evident that sucrose, yeast extract and peptone enhanced the yield of EPSs synthesized by Leuconostoc dextranicum NRRL B-1146 strain to increase the EPS yield. Similarly, in another study, tryptone and sucrose had a great impact on the yield of EPSs from Leuconostoc mesenteroides DRP105.12 Ref. 26 reported that Levilactobacillus sakei predominantly utilized glucose as a major carbon source for EPS production. It was earlier reported that glucose appeared as a better substrate than lactose for more EPS yields by Lactobacillus delbrueckii subsp. bulgaricus.27 In a recent study, it has been concluded that addition of CaCl2 increases the EPS yield without altering the growth of Lactobacillus plantarum K25 strain.28 Furthermore, by the addition of K2HPO4, a significant increase could be seen in the EPS yield from 438 mg L−1 to 1673 mg L−1 by L. rhamnosus.29
3.5 Optimization of significant factors using CCRD
The experimental runs of CCRD along with the response yield are presented in Table 3. The relationships between the EPS yield and tested variables described in terms of the equation are described below. Y is denoted as predicted EPS and XA, XD, XC, XD and XE are the coded values of glucose, sucrose, tryptone, CaCl2, and K2HPO4 concentrations respectively.| |
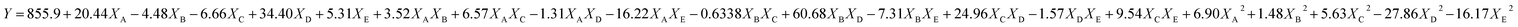 | (6) |
Table 3 Actual and predicted values for EPS yields
| Run |
A-glucose (%) |
B-sucrose (%) |
C-tryptone (%) |
D-CaCl2 (%) |
E-K2HPO4 (%) |
Actual EPS yield (mg L−1) |
Predicted value |
| 1 |
7.5 |
7.5 |
0.148 |
0.4 |
0.45 |
889.44 |
903.56 |
| 2 |
7.5 |
7.5 |
1.1 |
0.4 |
0.806 |
771.67 |
777.06 |
| 3 |
13.446 |
7.5 |
1.1 |
0.4 |
0.45 |
936.44 |
943.54 |
| 4 |
1.553 |
7.5 |
1.1 |
0.4 |
0.45 |
832.02 |
846.29 |
| 5 |
10 |
10 |
1.5 |
0.3 |
0.3 |
724.95 |
755.23 |
| 6 |
10 |
5 |
1.5 |
0.5 |
0.3 |
852.67 |
840.81 |
| 7 |
5 |
5 |
0.7 |
0.5 |
0.6 |
837.44 |
816.04 |
| 8 |
10 |
5 |
0.7 |
0.5 |
0.3 |
820.66 |
786.65 |
| 9 |
7.5 |
7.5 |
2.051 |
0.4 |
0.45 |
864.66 |
871.90 |
| 10 |
10 |
10 |
0.7 |
0.5 |
0.6 |
902.14 |
907.78 |
| 11 |
7.5 |
7.5 |
1.1 |
0.4 |
0.45 |
865.89 |
855.9 |
| 12 |
7.5 |
7.5 |
1.1 |
0.4 |
0.45 |
860.89 |
855.9 |
| 13 |
5 |
5 |
1.5 |
0.3 |
0.6 |
802.81 |
818.58 |
| 14 |
5 |
5 |
0.7 |
0.5 |
0.3 |
734.44 |
736.13 |
| 15 |
10 |
5 |
0.7 |
0.5 |
0.6 |
793.66 |
801.68 |
| 16 |
5 |
10 |
1.5 |
0.3 |
0.3 |
684.51 |
659.10 |
| 17 |
10 |
10 |
1.5 |
0.5 |
0.3 |
954.89 |
973.62 |
| 18 |
10 |
10 |
1.5 |
0.3 |
0.6 |
685.51 |
696.57 |
| 19 |
5 |
10 |
0.7 |
0.5 |
0.6 |
905.05 |
908.04 |
| 20 |
10 |
10 |
0.7 |
0.3 |
0.6 |
789.41 |
751.09 |
| 21 |
5 |
10 |
0.7 |
0.5 |
0.3 |
867.05 |
857.38 |
| 22 |
10 |
5 |
1.5 |
0.5 |
0.6 |
858.67 |
849.55 |
| 23 |
5 |
5 |
0.7 |
0.3 |
0.3 |
860.19 |
855.07 |
| 24 |
7.5 |
7.5 |
1.1 |
0.4 |
0.45 |
826.89 |
855.9 |
| 25 |
10 |
10 |
0.7 |
0.3 |
0.3 |
850.04 |
803.45 |
| 26 |
5 |
10 |
1.5 |
0.3 |
0.6 |
665.78 |
665.32 |
| 27 |
7.5 |
7.5 |
1.1 |
0.637 |
0.45 |
773.14 |
780.10 |
| 28 |
5 |
10 |
1.5 |
0.5 |
0.3 |
923.78 |
882.74 |
| 29 |
5 |
10 |
0.7 |
0.3 |
0.6 |
731.65 |
746.1 |
| 30 |
7.5 |
7.5 |
1.1 |
0.162 |
0.45 |
602.05 |
616.46 |
| 31 |
10 |
5 |
1.5 |
0.3 |
0.3 |
878.89 |
865.16 |
| 32 |
7.5 |
13.446 |
1.1 |
0.4 |
0.45 |
849.51 |
853.61 |
| 33 |
10 |
10 |
0.7 |
0.5 |
0.3 |
905.14 |
921.99 |
| 34 |
10 |
5 |
1.5 |
0.3 |
0.6 |
867.19 |
835.74 |
| 35 |
10 |
10 |
1.5 |
0.5 |
0.6 |
968.89 |
953.11 |
| 36 |
10 |
5 |
0.7 |
0.3 |
0.6 |
837.44 |
887.72 |
| 37 |
7.5 |
1.553 |
1.1 |
0.4 |
0.45 |
857.66 |
874.93 |
| 38 |
5 |
5 |
1.5 |
0.5 |
0.3 |
736.81 |
764.03 |
| 39 |
5 |
5 |
1.5 |
0.5 |
0.6 |
831.81 |
837.64 |
| 40 |
5 |
10 |
0.7 |
0.3 |
0.3 |
705.43 |
733.59 |
| 41 |
7.5 |
7.5 |
1.1 |
0.4 |
0.093 |
735.81 |
751.79 |
| 42 |
5 |
5 |
1.5 |
0.3 |
0.3 |
799.74 |
783.12 |
| 43 |
5 |
10 |
1.5 |
0.5 |
0.6 |
922.79 |
927.11 |
| 44 |
5 |
5 |
0.7 |
0.3 |
0.6 |
946.51 |
896.82 |
| 45 |
10 |
5 |
0.7 |
0.3 |
0.3 |
902.78 |
910.85 |
The results (Table 4) of the quadratic regression model suggested that the model was highly suitable and significant for EPS yield prediction, as evidenced by the model F-value (16.64), which was statistically significant (p < 0.0001). The regression coefficient values were found to be R2 = 0.9327 and non-significant lack of fit value (p = 0.3848). The present study indicated a wide range of EPS yield extending from 602.05 to 968.89 mg L−1, whereas the ratio of maximum to the minimum value was attained to be 1.61. It is important to mention here that as per the general accepted notion whenever the ratio is bigger than ten (10), it indicates that transformation is required; however, if the ratio is <3%, the power transform has little effects. Therefore, in this background, the obtained results of experiment were adequate. The model equation was validated by performing one experiment under the conditions existing within the experimental range. The optimized value of tested independent factors was 10.56% glucose, 9.23% sucrose, 0.75% tryptone, 0.446% CaCl2 and 0.385% K2HPO4. Under such conditions, the predicted value for EPS yield was 973.615 mg L−1 and the actual experimental value was 968.89 mg L−1. The best value of EPS yield was at high concentrations of glucose and K2HPO4 and slightly low concentrations of sucrose, tryptone and CaCl2. It is concluded from the present study findings that the selected solution is best for the optimization of responses defined at the start of the study.
Table 4 ANOVA for the selected factorial model (PB and CCRD designs)
| Initial screening of media composition ANOVA for selected factorial model (PB) |
Optimization of significant factors ANOVA for selected quadratic model (CCRD) |
| Source |
Sum of squares |
df |
Mean square |
F-value |
p-value |
Source |
Sum of squares |
df |
Mean square |
F-value |
p-value |
| Model |
17.61 |
11 |
1.60 |
42.77 |
0.0231* |
Model |
2.890 × 105 |
20 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450.18 450.18 |
16.64 |
<0.0001 |
| A-glucose |
3.28 |
1 |
3.28 |
87.54 |
0.0112* |
A-glucose |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.81 102.81 |
1 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.81 102.81 |
20.84 |
0.0001** |
| B-fructose |
0.0447 |
1 |
0.0447 |
1.19 |
0.3887 |
B-sucrose |
869.67 |
1 |
869.67 |
1.00 |
0.3270 |
| C-lactose |
0.2251 |
1 |
0.2251 |
6.01 |
0.1337 |
C-tryptone |
1918.65 |
1 |
1918.65 |
2.21 |
0.1502 |
| D-sucrose |
5.10 |
1 |
5.10 |
136.29 |
0.0073** |
D-CaCl2 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255.11 255.11 |
1 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255.11 255.11 |
59.01 |
<0.0001** |
| E-yeast extract |
0.0013 |
1 |
0.0013 |
0.0335 |
0.8717 |
E-K2HPO4 |
1222.07 |
1 |
1222.07 |
1.41 |
0.2472 |
| F-beef extract |
0.0797 |
1 |
0.0797 |
2.13 |
0.2818 |
AB |
397.06 |
1 |
397.06 |
0.457 |
0.5054 |
| G-tryptone |
2.73 |
1 |
2.73 |
73.03 |
0.0134 |
AC |
1379.70 |
1 |
1379.70 |
1.59 |
0.2197 |
| H-MgSO4 |
0.5909 |
1 |
0.5909 |
15.79 |
0.0579 |
AD |
55.23 |
1 |
55.23 |
0.063 |
0.8031 |
| J-CaCl2 |
1.86 |
1 |
1.86 |
49.61 |
0.0196* |
AE |
8417.53 |
1 |
8417.53 |
9.69 |
0.0047* |
| K-K2HPO4 |
3.57 |
1 |
3.57 |
95.43 |
0.0103** |
BC |
12.85 |
1 |
12.85 |
0.014 |
0.9042 |
| L-NaH2PO4 |
0.1282 |
1 |
0.1282 |
3.43 |
0.2054 |
BD |
1.178 × 105 |
1 |
1.178 × 105 |
135.6 |
<0.0001** |
| BE |
1709.96 |
1 |
1709.96 |
1.97 |
0.1734 |
| CD |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940.04 940.04 |
1 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940.04 940.04 |
22.96 |
<0.0001** |
| CE |
79.25 |
1 |
79.25 |
0.091 |
0.7652 |
| DE |
2911.61 |
1 |
2911.61 |
3.35 |
0.0796* |
| A2 |
1875.14 |
1 |
1875.14 |
2.16 |
0.1547 |
| B2 |
86.26 |
1 |
86.26 |
0.099 |
0.7554 |
| C2 |
1248.44 |
1 |
1248.44 |
1.44 |
0.2423 |
| D2 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 610.10 610.10 |
1 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 610.10 610.10 |
35.24 |
<0.0001** |
| E2 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309.96 309.96 |
1 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309.96 309.96 |
11.87 |
0.0021* |
| Residual |
0.0749 |
2 |
0.0374 |
|
|
|
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845.26 845.26 |
24 |
868.55 |
|
|
| Lack of fit |
0.0264 |
1 |
0.0264 |
0.5461 |
0.5948 |
|
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944.60 944.60 |
22 |
906.57 |
2.01 |
0.3848 |
| Pure error |
0.0484 |
1 |
0.0484 |
|
|
|
900.67 |
2 |
450.33 |
|
|
| Cor total |
17.68 |
13 |
|
|
|
|
3.098 × 105 |
44 |
|
|
|
The results exhibited that linear coefficients (XA, and XD), quadratic coefficients (D2 and E2) and the interaction coefficients (XAXB, XAXD, XAXE, XBXD, XCXD, XCXE, and XDXE) proved significant with p-values less than 0.05 and 0.01. Depending upon the obtained model, generally, the 3-D had been found adequate for explaining the results regarding maximum EPS production, as previously done for L. delbrueckii and L. acidophilus.8,30 Variables such as glucose and CaCl2 showed significant effects (p ≤ 0.0001) independently on the EPS yield (Fig. 2). However, both also showed significance with other variables such as glucose vs. K2HPO4 (p = 0.0047), sucrose vs. CaCl2 (p ≤ 0.0001), tryptone vs. CaCl2 (p ≤ 0.0001) and quadratic of CaCl2 (p ≤ 0.0001) and K2HPO4 (p = 0.0021), indicating that the mean EPS yield close to the predicted value resulted in the successful validated model. In case of glucose, it became evident that the consistent and optimum range affecting the response was lying between 4.526 and 10.473% and the maximum value for response went above 950 mg L−1, whereas the main effect of sucrose was almost similar to glucose. Tryptone was concentrated almost within the region of 0.624 and 1.575% and CaCl2 was prominent in the range of 0.281 to 0.518%. K2HPO4 concentrated in the region of 0.271 to 0.628% and most of the values of response lied above the moderate range.
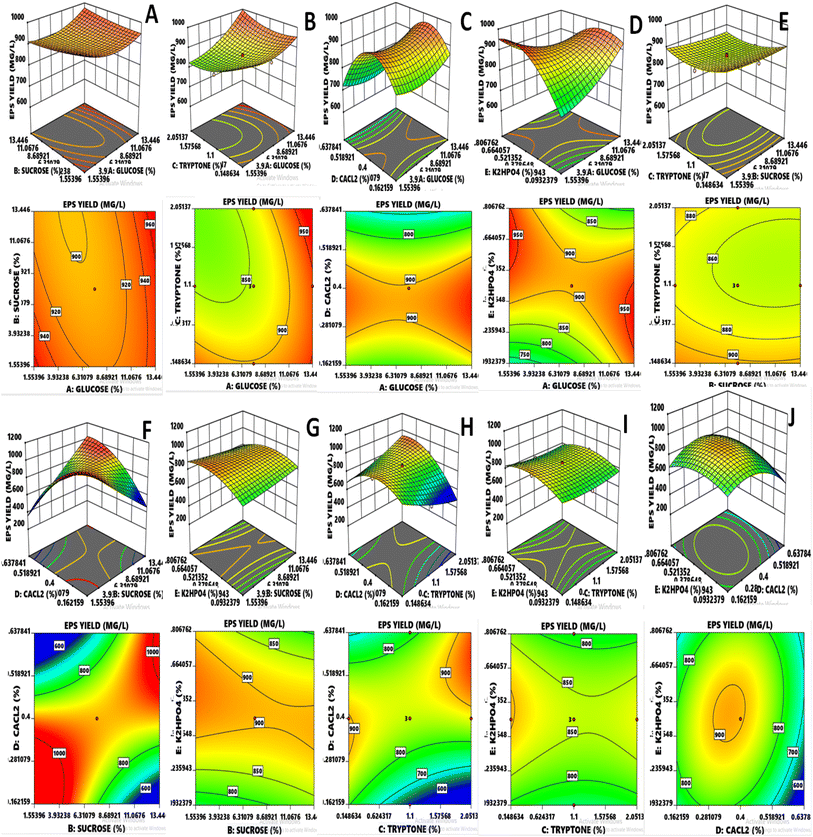 |
| | Fig. 2 (A–J) RSM three-dimensional and contour plots indicate the effect of two independent variables on response EPS yield. | |
3.6 Isolation and quantification of EPSs
In the present study, the final EPS yield of Levilactobacillus brevis NCCP 963 was 968.89 mg L−1 that was very much higher than that of the EPS produced (160 mg L−1) from Lactobacillus brevis isolated from Chinese koumiss.31 Similarly, Lactobacillus rhamnosus was able to produce 495 mg L−1 yield,32 whereas Lactobacillus plantarum, an isolate from fermented milk, resulted in an even higher yield of 956 mg L−1.26 Heating helped to get rid from the EPS-degrading enzymes, as reported earlier. Moreover, after heating, the cell-bound polysaccharide was also liberated in the medium and increased the overall yield. Similarly, further purification steps such as acidic/basic treatment and dialysis also played a significant role in the purification of extracted EPSs. The total sugar content was found to be 96.45 ± 0.06 g. 100−1 g while the absence of protein or nucleic acid resulted in a response to the Bradford test in the UV spectrum of 280 nm and 260 nm. Similarly, the total carbohydrate content in EPSs produced by Lactobacillus plantarum YW11 was reported to be 92.35%,10 and that in EPS produced by Lactobacillus helviticus MB2-1 was 95.45%.13
3.7 FT-IR analysis of EPSs
The spectrum estimation of EPSs was conceded out for the determination of functional groups in the EPS by FTIR spectroscopy. Hydroxyl group stretching from glycosides was depicted at a wave number of 3275.48 cm−1, as reported previously.33 The peak at 2919.62 cm−1 was related to the C–H stretching vibration. The indication of absorption bands of C–C group stretching was also visible at 1644.73 cm−1. The band ranging from 1320 to 1000(s) wave numbers specifically represented carboxylic acids, alcohols, ether and ester groups (C–O), showing the typical attributes for polysaccharide. Therefore, in the present study, a sharp peak at 1010.61 cm−1 indicated the C–O stretching vibration (Fig. 3). It may also be deduced that the appearance of stretching absorption at 1147 cm−1 assured the presence of β-1-4 glycosidic linkage. This absorption frequency at 1147 cm−1 is due to characteristic antisymmetric stretching of the β C1–O–C4 bond. The absence of peak in the range of 850 cm−1 depicted the absence of α linkage in the exopolysaccharide.34 Thus, it was shown that the produced EPS is an intricate polysaccharide having various functional groups, as reported earlier in various studies.14
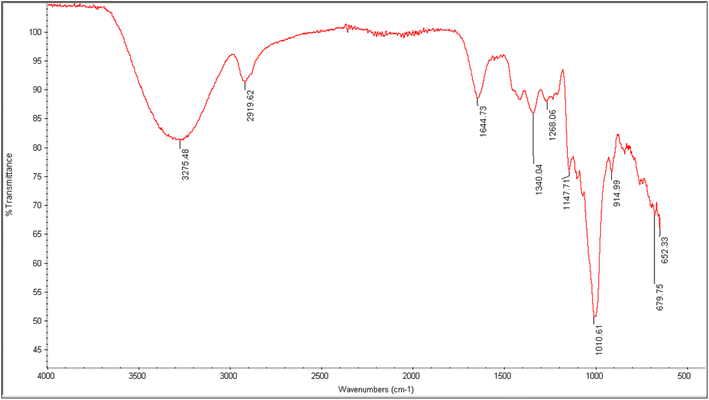 |
| | Fig. 3 FTIR analysis of NCCP 963 exopolysaccharide. | |
3.8 Monosaccharide composition and molecular weight (Mw)
The monosaccharide composition analysis depicted that NCCP 963 EPS was homopolysaccharide in nature (Fig. 4a), consisting of only glucose monomers. Lactobacillus brevis isolated from Chinese koumiss,35 Lactobacillus brevis TMW 1.2112 from beer36 and Lactobacillus brevis E25 isolated from sourdough37 have been reported to produce the EPS which was glucan in nature. Similarly, other LAB such as Lactobacillus plantarum CIDCA 8327 also produced α-glucan polysaccharide.38
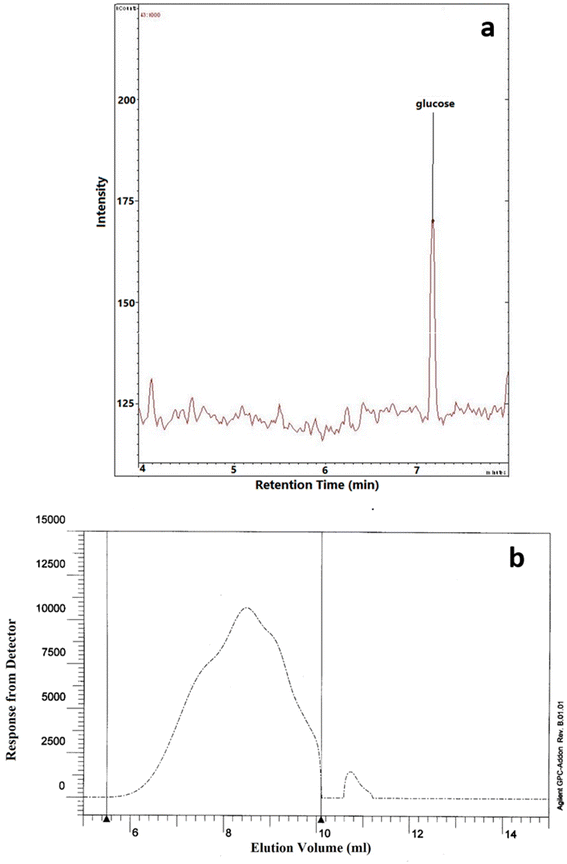 |
| | Fig. 4 (a) Monomer composition and (b) molecular weight of NCCP 963 exopolysaccharide. | |
The average Mw of exopolysaccharide produced from Levilactobacillus brevis NCCP 963 was 5.48 × 104 Da (Fig. 4b). Lactobacillus brevis TMW 1.2112 produced exopolysaccharide with a Mw in the range of 104–105.36 However several lactobacillus have been reported to produce the exopolysaccharide with a high Mw, i.e. in the range of 106–107.39 Lactobacillus plantarum DM5 produced the glucan having an average Mw of 1.11 × 106 Da.40
3.9 In vitro antioxidant activity
Four different assays, namely, DPPH, ABTS˙+, hydroxyl, and superoxide were done to get the comprehensive estimate of antioxidant activity with ascorbic acid as the control. The results are depicted in Fig. 5.
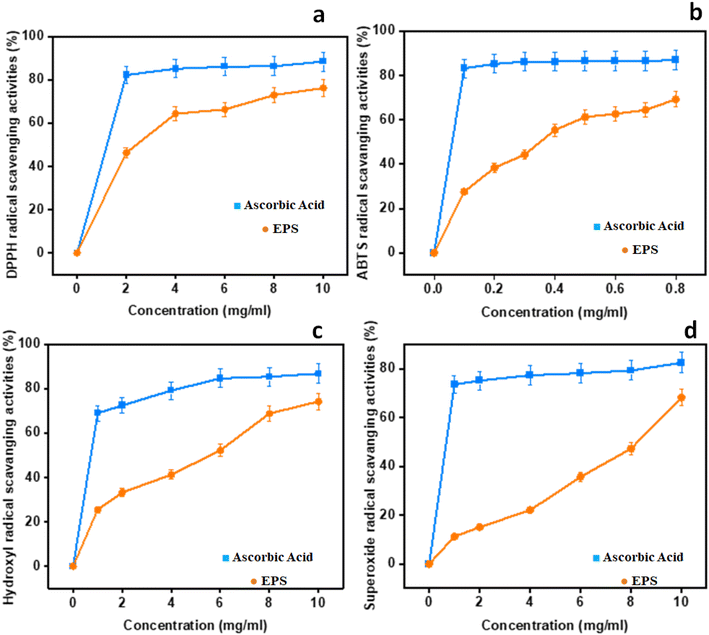 |
| | Fig. 5 In vitro antioxidant potential of EPSs: (a) DPPH radical scavenging activities; (b) ABTS; (c) hydroxyl radical scavenging activities; (d) superoxide radical scavenging activities. | |
3.9.1 DPPH radical scavenging activity. As for the DPPH radical scavenging activity of NCCP 963 was concerned, an increasing trend was observed with the increase in concentration ranging from 0 to 10 mg mL−1, as shown in Fig. 5a. Maximum activity was found to be 88.45% at the concentration level of 10 mg mL−1 with an EC50 value of 1.56 mg mL−1. This is due to the existence of hydroxyl and other functional groups in the EPS-producing strain that may give electrons for the reduction of free radicals into a more stable form.12 In the present study, this activity of the EPS was found to be 6% higher than the EPS formed from L. plantarum LP6 (ref. 41) and almost similar, 88.60%, to the EPS isolated form L. plantarum C88.42
3.9.2 ABTS˙+ radical scavenging activity. ABTS radical scavenging activity is a convenient model to assess the antioxidant activity of organic compounds such as exopolysaccharide from Lactobacillus strains.41 In the present study, all of the levels displayed unpredictable levels of antioxidant activity in a dose-dependent manner, as shown in Fig. 5b. The ABTS rate was found to be closer to that of the reference control at 0.8 mg mL−1 and an EC50 value of 0.31 mg mL−1, which showed significant ABTS radical activity even at a very low concentration in comparison to other kinds of EPSs reported earlier.43,44 When it was compared with the DPPH activity, it was found that the EPS employed stronger ABTS scavenge radical ability, due to the diversified properties of the radical model. Naturally isolated EPSs possessed abundant hydroxyl groups essential for the antioxidant activity, which provides electrons for the reduction of radicals into a more stable form or may interact with free radicals to break the chain reaction of radicals.45 Moreover, it is also reported that high levels of protein content and a low molecular weight also had the same effect on the antioxidant activities8,42 radical scavenging activities; (c) hydroxyl radical scavenging activities; and (d) superoxide radical scavenging activities.
3.9.3 Hydroxyl radical scavenging activity. The hydroxyl antioxidant activity of isolated EPSs and ascorbic acid (used as standard) is depicted in Fig. 5c. The maximum activity was found at the level of 86.78% at 10 mg mL−1 with an EC50 value of 2.1 mg mL−1 and findings were more or less similar, as previously reported in the literature.41 The reaction mechanism of the EPS in hunting hydroxyl radicals seemed to be different from the ABTS and DPPH radicals. The hydroxyl antioxidant capability of the EPS is attributed to inhibited to the free production of OH groups by chelating metallic ions such as Cu+2 and Fe+2.46 As per the findings of the present study, the behaviors of hydroxyl groups and the DPPH activity of exopolysaccharide were almost similar. However, the ABTS radical scavenging activity was found to be different. This varied behavior of the EPS may be attributed to multiple mechanisms such as binding to transition metal ion catalysts, prevention of chain initiation, and breakdown of peroxides.2
3.9.4 Superoxide anion scavenging activity. Biological macromolecules and tissue damage could also be caused by the presence of superoxide radicals with the generation of other ROS species such as hydroxyl radicals, singlet oxygen and H2O2 that further lead to severe oxidative damage to living cells.41 The findings indicating the superoxide radical scavenging activity with different concentrations of EPSs are presented in Fig. 5d. A similar trend of dose dependence was found as other antioxidant activity models, with the highest rate of 82.45% at 10 mg mL−1 and an EC50 value of 6.7 mg mL−1. The in vitro antioxidant activity of L. brevis on albino mice found that this strain displayed marked improvement in scavenging the superoxide radicals in all tissues.47
3.10 Solidification/fermentation effects of Levilactobacillus brevis NCCP 963
Syneresis is a significant issue in yoghurt industry. EPS-producing bacteria that have strong water holding capability could be used as the adjunct culture along with the traditional culture.12 Levilactobacillus brevis NCCP 963 strain formed intense curd and did not show any syneresis after 5 days in the refrigerator (Fig. S3†). Hence, they could be used in yogurt and dahi industry as an adjunct culture. Generally, solidification of 10% skim milk is difficult because of the low protein content and lactic acid formed by fermenting bacteria.48 Addition of 3% sucrose in skim milk is also attributed to interactions between proteins and EPSs, present in milk.21,31
4. Conclusion
Levilactobacillus brevis isolated from kanji showed maximum EPS production of 968.89 mg L−1 at optimized levels of 10.56% glucose, 9.23% sucrose, 0.75% tryptone, 0.446% CaCl2 and 0.385% di-potassium phosphate. The obtained EPS was homopolysaccharide in nature with a high molecular weight. The FT-IR spectrum indicates the presence of sugar fractions. The significant antioxidant potential with a high free radical scavenging capacity of DPPH, ABTS, hydroxyl, and superoxide alluded towards high potential usage in the pharmaceutical industry. Levilactobacillus brevis NCCP 963 has a high molecular weight of 5.48 × 104 Da. Based on the characterization data, the extracted EPS have a great potential to be used in dairy, yoghurt or similar industries, in which the hydrocolloid nature of product is required.
Abbreviations
| EPS | Exopolysaccharide |
| PB | Plackett–Burman |
| RSM | Response surface methodology |
| CaCl2 | Calcium chloride |
| K2HPO4 | Di-potassium phosphate |
| LAB | Lactic acid bacteria |
| CCRD | Central composite rotatory design |
| pH | Hydrogen ion concentration |
| MRS agar | De Man, Rogosa and sharpe agar |
| API | Analytical profile index |
| NJ | Neighbor joining |
| TCA | Trichloroacetic acid |
| NaOH | Sodium hydroxide |
| NCCP | National collection culture point |
| KDa | 1000 Daltons |
| MgSO4 | Magnesium sulfate |
| NaH2PO4 | Sodium dihydrogen phosphate |
| FT-IR | Fourier transform-infrared spectroscopy |
| TFA | Trifluoroacetic acid |
| GC/MS | Gas chromatography mass spectrometer |
| HPLC | High-performance liquid chromatography |
| Mw | Molecular weight |
| Na2SO4 | Sodium sulfate |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| EC50 | Half maximal effective concentration |
| K2S2O8 | Potassium persulfate |
| ABTS | 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) |
| FeSO4 | Ferrous sulfate |
| H2O2 | Hydrogen peroxide |
| Tris–HCl | Tris(hydroxymethyl)aminomethane (THAM) hydrochloride |
| NARC | National Agriculture Research Center |
| ROS | Reactive oxygen species |
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
Study was supported from Higher Education Commission of Pakistan Project No. NRPU-8709.
References
- S. Bhattacharyya, Heliyon, 2022, 8, e09250, DOI:10.1016/j.heliyon.2022.e09250.
- A. Kumar, T. Joishy, S. Das, M. C. Kalita, A. K. Mukherjee and M. R. Khan, Antioxidants, 2022, 11, 268, DOI:10.3390/antiox11020268.
- E. Korcz and L. Varga, Trends Food Sci. Technol., 2021 DOI:10.1016/j.tifs.2021.02.014.
- K. M. G. M. M. Kariyawasam, S. J. Yang, N.-K. Lee and H.-D. Paik, Food Sci. Anim. Resour., 2020, 40, 297, DOI:10.5851/kosfa.2020.e15.
- M. Ayyash, B. Abu-Jdayil, A. Olaimat, G. Esposito, P. Itsaranuwat, T. Osaili, R. Obaid, J. Kizhakkayil and S.-Q. Liu, Carbohydr. Polym., 2020, 229, 115462, DOI:10.1016/j.carbpol.2019.115462.
- J. Yu, Y. Hang, W. Sun, G. Wang, Z. Xiong, L. Ai and Y. Xia, Biology, 2022, 11, 359, DOI:10.3390/biology11030359.
- Y. Wang, Z. Ahmed, W. Feng, C. Li and S. Song, Int. J. Biol. Macromol., 2008, 43, 283–288, DOI:10.1016/j.ijbiomac.2008.06.011.
- B. Wang, Q. Song, F. Zhao, Y. Han and Z. Zhou, Int. J. Biol. Macromol., 2019, 141, 21–28, DOI:10.1016/j.ijbiomac.2008.06.011.
- J. Lamba, S. Goomer and S. Saxena, Int. J. Gastron Food Sci., 2019, 16, 100143, DOI:10.1016/j.ijgfs.2019.100143.
- J. Wang, X. Zhao, Z. Tian, Y. Yang and Z. Yang, Carbohydr. Polym., 2015, 125, 16–25, DOI:10.1016/j.carbpol.2015.03.003.
- F. M. Kilau, N. I. Mohamad, N. Saari and N. A. Sani, in AIP Conference Proceedings, AIP Publishing LLC, 2019, vol. 2111(1), p. 040006, DOI:10.1063/1.5111245.
- L. Wang, Y. Gu, X. Zheng, Y. Zhang, K. Deng, T. Wu and H. Cheng, Lebensm. Wiss. Technol., 2021, 146, 111449, DOI:10.1063/1.5111245.
- W. Li, X. Xia, W. Tang, J. Ji, X. Rui, X. Chen, M. Jiang, J. Zhou, Q. Zhang and M. Dong, J. Agric. Food Chem., 2015, 63, 3454–3463, DOI:10.1021/acs.jafc.5b01086.
- S. Tiwari, D. Kavitake, P. B. Devi and P. S. Halady, Int. J. Biol. Macromol., 2021, 183, 1585–1595, DOI:10.1016/j.ijbiomac.2021.05.14.
- D. Zhao, J. Jiang, L. Liu, S. Wang, W. Ping and J. Ge, Int. J. Biol. Macromol., 2021, 178, 306–315, DOI:10.1016/j.ijbiomac.2021.02.
- S. V. Dilna, H. Surya, R. G. Aswathy, K. K. Varsha, D. N. Sakthikumar, A. Pandey and K. M. Nampoothiri, LWT – Food Sci. Technol., 2015, 64, 1179–1186, DOI:10.1016/j.lwt.2015.07.040.
- Y. Li, Y. Liu, C. Cao, X. Zhu, C. Wang, R. Wu and J. Wu, Int. J. Biol. Macromol., 2020, 157, 36–44, DOI:10.1016/j.ijbiomac.2020.04.150.
- A. Braca, N. De Tommasi, L. Di Bari, C. Pizza, M. Politi and I. Morelli, J. Nat. Prod., 2001, 64, 892–895, DOI:10.12691/jfnr-7-4-10.
- X. Wang, L. Zhang, J. Wu, W. Xu, X. Wang and X. Lü, Food Chem., 2017, 220, 198–207, DOI:10.1016/j.foodchem.2016.10.008.
- Y. Yang, F. Feng, Q. Zhou, F. Zhao, R. Du, Z. Zhou and Y. Han, Trans. Tianjin Univ., 2019, 25, 161–168, DOI:10.1007/s12209-018-0143-9.
- B. Wang, Q. Song, F. Zhao, L. Zhang, Y. Han and Z. Zhou, Carbohydr. Polym., 2019, 223, 115111, DOI:10.1016/j.ijbiomac.2008.06.011.
- H. J. Jang, N.-K. Lee and H.-D. Paik, Food Sci. Biotechnol., 2019, 28, 1521–1528, DOI:10.1007/2Fs10068-019-00576-x.
- N. Noohi, M. Papizadeh, M. Rohani, M. Talebi and M. R. Pourshafie, Anim. Nutr., 2021, 7, 119–126, DOI:10.1016/j.aninu.2020.07.005.
- K. M. G. M. M. Kariyawasam, N.-K. Lee and H.-D. Paik, Food Biosci., 2021, 39, 100835, DOI:10.5851/kosfa.2020.e15.
- M. Fukao, T. Zendo, T. Inoue, J. Nakayama, S. Suzuki, T. Fukaya, N. Yajima and K. Sonomoto, J. Biosci. Bioeng., 2019, 128, 391–397, DOI:10.1016/j.jbiosc.2019.04.008.
- M. Y. M. Imran, N. Reehana, K. A. Jayaraj, A. A. P. Ahamed, D. Dhanasekaran, N. Thajuddin, N. S. Alharbi and G. Muralitharan, Int. J. Biol. Macromol., 2016, 93, 731–745, DOI:10.1016/j.ijbiomac.2016.09.007.
- M. M. I. N. R. Ka and A. Jayaraj, Process Biochem., 2005, 40, 229–246, DOI:10.1016/j.ab.2004.12.001.
- Y. Jiang, M. Zhang, Y. Zhang, J. Zulewska and Z. Yang, J. Dairy Sci., 2021, 104, 2693–2708, DOI:10.3168/jds.2020-19237.
- H. M. Sørensen, K. D. Rochfort, S. Maye, G. MacLeod, D. Brabazon, C. Loscher and B. Freeland, Nutrients, 2022, 14, 2938, DOI:10.3390/nu14142938.
- V. Deepak, S. Ram Kumar Pandian, S. D. Sivasubramaniam, H. Nellaiah and K. Sundar, Prep. Biochem. Biotechnol., 2016, 46, 288–297, DOI:10.1080/10826068.2015.1031386.
- M. S. R. Rajoka, Y. Wu, H. M. Mehwish, M. Bansal and L. Zhao, Trends Food Sci. Technol., 2020, 103, 36–48, DOI:10.1016/j.tifs.2020.06.003.
- P. L. Pham, I. Dupont, D. Roy, G. Lapointe and J. Cerning, Appl. Environ. Microbiol., 2000, 66, 2302–2310, DOI:10.1128/aem.66.6.2302-2310.2000.
- D. Kavitake, F. Z. Marchawala, C. Delattre, P. H. Shetty, H. Pathak and P. Andhare, Bioact. Carbohydr. Diet. Fibre, 2019, 20, 100202, DOI:10.1016/j.bcdf.2019.100202.
- F. Carfì Pavia, V. La Carrubba, V. Brucato, F. S. Palumbo and G. Giammona, Mater. Sci. Eng. C Mater. Biol. Appl., 2014, 41, 301–308, DOI:10.1016/j.msec.2014.04.045.
- M. S. R. Rajoka, H. M. Mehwish, H. Zhang, M. Ashraf, H. Fang, X. Zeng, Y. Wu, M. Khurshid, L. Zhao and Z. He, Colloids Surf., B, 2020, 186, 110734, DOI:10.1016/j.colsurfb.2019.110734.
- M. E. Fraunhofer, A. J. Geissler, D. Wefers, M. Bunzel, F. Jakob and R. F. Vogel, Int. J. Biol. Macromol., 2018, 107, 874–881, DOI:10.1016/j.ijbiomac.2017.09.063.
- E. Dertli, I. J. Colquhoun, G. L. Côté, G. Le Gall and A. Narbad, Food Chem., 2018, 242, 45–52, DOI:10.1016/j.foodchem.2017.09.017.
- M. V. Gangoiti, A. Puertas, M. F. Hamet, P. J. Peruzzo, M. Llamas, M. Medrano, A. Prieto, M. T. Dueñas and A. G. Abraham, Carbohydr. Polym., 2017, 170, 52–59, DOI:10.1016/j.carbpol.2017.04.053.
- Z. Chen, D. Ni, W. Zhang, T. Stressler and W. Mu, Biotechnol. Adv., 2021, 47, 107708, DOI:10.1016/j.biotechadv.2021.107708.
- D. Das and A. Goyal, J. Sci. Food Agric., 2014, 94, 683–690, DOI:10.1002/jsfa.6305.
- X. Wang, C. Shao, L. Liu, X. Guo, Y. Xu and X. Lü, Int. J. Biol. Macromol., 2017, 103, 1173–1184, DOI:10.1016/j.foodchem.2016.10.008.
- L. Zhang, C. Liu, D. Li, Y. Zhao, X. Zhang, X. Zeng, Z. Yang and S. Li, Int. J. Biol. Macromol., 2013, 54, 270–275, DOI:10.1016/j.ijbiomac.2012.12.037.
- X. Wang and X. Lü, Carbohydr. Polym., 2014, 102, 174–184, DOI:10.1016/j.carbpol.2013.11.012.
- X. Ding, Y.-l. Hou and W.-r. Hou, Int. J. Biol. Macromol., 2012, 50, 613–618, DOI:10.1016/j.ijbiomac.2012.01.021.
- J.-W. Shen, C.-W. Shi and C.-P. Xu, Food Technol. Biotechnol., 2013, 51, 520–527 CAS , https://www.proquest.com/scholarly-journals/exopolysaccharides-pleurotus-pulmonarius/docview/1503667045/se-2.
- I. Trabelsi, N. Ktari, M. Triki, I. Bkhairia, S. B. Slima, S. S. Aydi, S. Aydi, A. Abdeslam and R. B. Salah, Int. J. Biol. Macromol., 2018, 111, 11–18, DOI:10.1016/j.ijbiomac.2017.12.127.
- S. Noureen, A. Riaz, M. Arshad and N. Arshad, J. Appl. Microbiol., 2019, 126, 1221–1232, DOI:10.1111/jam.1418.
- W. Bejar, V. Gabriel, M. Amari, S. Morel, M. Mezghani, E. Maguin, C. Fontagné-Faucher, S. Bejar and H. Chouayekh, Int. J. Biol. Macromol., 2013, 52, 125–132, DOI:10.1016/j.ijbiomac.2012.09.014.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a and
Nauman Khalidbc
*a and
Nauman Khalidbc
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C and the supernatant was precipitated by the addition of chilled ethanol in a double volume and kept overnight at 4 °C. It was followed by centrifugation at 11
000 rpm for 15 min at 4 °C and the supernatant was precipitated by the addition of chilled ethanol in a double volume and kept overnight at 4 °C. It was followed by centrifugation at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C. Proteins were removed as per centrifugation conditions described earlier. The pH of the resulting supernatant was increased to 8.5 by adding 6 M NaOH drop by drop and again centrifuged at 11
000 rpm for 15 min at 4 °C. Proteins were removed as per centrifugation conditions described earlier. The pH of the resulting supernatant was increased to 8.5 by adding 6 M NaOH drop by drop and again centrifuged at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The pH of the resulting supernatant was lowered to neutral by adding 1 mL of 10% TCA, and purified exopolysaccharide was gathered by adding chilled ethanol in an equal volume. Furthermore, the resulting pellet was liquefied in deionized H2O, dialyzed (molecular weight cutoff of 12
000 rpm for 15 min. The pH of the resulting supernatant was lowered to neutral by adding 1 mL of 10% TCA, and purified exopolysaccharide was gathered by adding chilled ethanol in an equal volume. Furthermore, the resulting pellet was liquefied in deionized H2O, dialyzed (molecular weight cutoff of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 14
000 to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kDa) for 48 h against distilled water with four times replacement of water per day and finally lyophilized. The total sugar content of EPSs was determined by the phenol–sulfuric method, and protein fractions were estimated by the Bradford method.8
000 kDa) for 48 h against distilled water with four times replacement of water per day and finally lyophilized. The total sugar content of EPSs was determined by the phenol–sulfuric method, and protein fractions were estimated by the Bradford method.8
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 Da, D5 with a mol. wt of 21
100 Da, D5 with a mol. wt of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 Da, and D4 with a mol. wt of 10
400 Da, and D4 with a mol. wt of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da were used. D0 glucose with a Mw of 180 Da was used as the reference compound. These reference standards were added into the mobile phase at a rate of 10 mg per 1 mL solution.10
000 Da were used. D0 glucose with a Mw of 180 Da was used as the reference compound. These reference standards were added into the mobile phase at a rate of 10 mg per 1 mL solution.10

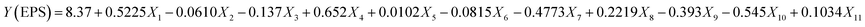
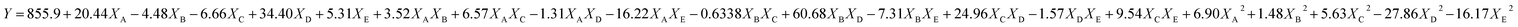
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450.18
450.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.81
102.81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.81
102.81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255.11
255.11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255.11
255.11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940.04
940.04![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940.04
940.04![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 610.10
610.10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 610.10
610.10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309.96
309.96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309.96
309.96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845.26
845.26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944.60
944.60



